Abstract
CRISPR-Cas are nucleic acid-based prokaryotic immune systems. CRISPR arrays accumulate spacers from foreign DNA and provide resistance to mobile genetic elements containing identical or similar sequences. Thus, the set of spacers present in a given bacterium can be regarded as a record of encounters of its ancestors with genetic invaders. Such records should be specific for different lineages and change with time, as earlier acquired spacers get obsolete and are lost. Here, we studied type I-E CRISPR spacers of Escherichia coli from extinct pachyderm. We find that many spacers recovered from intestines of a 42 000-year-old mammoth match spacers of present-day E. coli. Present-day CRISPR arrays can be reconstructed from palaeo sequences, indicating that the order of spacers has also been preserved. The results suggest that E. coli CRISPR arrays were not subject to intensive change through adaptive acquisition during this time.
Keywords: Escherichia coli, CRISPR spacers, CRISPR arrays, palaeo DNA
Introduction
Prokaryotic CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR-associated proteins) systems comprise noncoding CRISPR DNA arrays containing variable spacers separated by identical or almost identical repeats and cas genes (Makarova et al. 2015). Upon CRISPR array transcription and processing, individual CRISPR RNAs containing a single spacer and flanking repeat fragments are bound by Cas proteins. Resulting ribonucleoprotein complexes recognize nucleic acids with sequences matching CRISPR RNA spacer and subsequently degrade them (Barrangou et al. 2007; Brouns et al. 2008; Marraffini & Sontheimer 2008). New spacers are acquired into one end of CRISPR arrays during a Cas protein-catalysed process referred to as ‘CRISPR adaptation’ (van der Oost et al. 2009). Bioinformatics analysis revealed that some CRISPR spacers are derived from viral and plasmid sequences (Bolotin et al. 2005; Mojica et al. 2005; Pourcel et al. 2005) and it is now commonly accepted that CRISPR-Cas systems control the spread of mobile genetic elements such as plasmids and phages by providing prokaryotes with immunity, which is both adaptive and heritable. Mobile genetic elements can escape the CRISPR-Cas defence by altering sequences recognized by CRISPR RNAs through random mutations or recombination, rendering CRISPR defence inefficient (Andersson & Banfield 2008; Deveau et al. 2008; Paez-espino et al. 2015) and necessitating acquisition of additional spacers. In several cases, studies of temporal dynamics of bacterial–bacteriophage populations in nature indeed revealed a continuous evolutionary arms race between phages and their hosts driven by cycles of new spacer acquisition followed by accumulation of phage mutants (Andersson & Banfield 2008; Sun et al. 2016). Similar dynamics was observed during long-term laboratory cultivation experiments with Streptococcus thermophilus (Paez-Espino et al. 2013).
The type I-E CRISPR-Cas system of model bacterium Escherichia coli is repressed at laboratory conditions (Pougach et al. 2010; Pul et al. 2010). However, when induced by means of genetic engineering, it efficiently prevents transformation with plasmids and/or infection by phages harbouring sequences matching spacers (Brouns et al. 2008; Pougach et al. 2010) and is also capable of highly efficient spacer acquisition (Datsenko et al. 2012; Yosef et al. 2012). The spacer content of natural isolates of E. coli is highly variable with overall diversity being higher at CRISPR arrays ends where new spacers are acquired (Diez-Villasenor et al. 2010; Touchon et al. 2011; Sheludchenko et al. 2015), suggesting that the CRISPR-Cas system is active in natural E. coli populations. However, compared to some other bacteria, very few E. coli spacers match known bacteriophages and plasmids, a surprising result considering the number of known E. coli mobile genetic elements (Diez-Villasenor et al. 2010; Touchon et al. 2011).
Analysis of palaeo DNA offers an unprecedented ability to analyse sequences from distant past and compare them to modern sequences (Hofreiter et al. 2015). CRISPR spacers are particularly attractive for such comparative analysis for their small size favours their preservation despite the fragmentation and deterioration of ancient DNA (Dabney et al. 2013), while the adaptive nature of CRISPR immunity implies significant turnover of spacers over time. Here, we studied spacers associated with type I-E E. coli CRISPR repeats from an extinct pachyderm, a baby mammoth Lyuba that died about 42 000 years ago (Fisher et al. 2009), and compared them with annotated contemporary CRISPR spacers available in public databases. To our surprise, we found no evidence of E. coli CRISPR spacer turnover. Multiple cases of palaeo CRISPR arrays preservation over the course of 42 000 years have been revealed, implying overall stability of the locus.
Materials and Methods
Sampling
An intact mammoth calf named Lyuba was found at Yamal Peninsula (western Siberia, Russia) in 2007 (Fisher et al. 2009) and brought to St. Petersburg without thawing. The carcass was processed in a sterilized laboratory room at −20 °C. The abdominal wall was opened from the left side. All internal organs were in a good shape. The stomach and intestines appeared full. Several grams of intestinal or stomach content were recovered and stored in sterilized packages at −20 °C until further analysis.
DNA extraction
All manipulations with ancient samples, including PCR amplification, were performed in a separate building in laboratory rooms where no prior molecular biology research was conducted. All samples were sterile as judged by the absence of colony formation after aliquots of intestinal or stomach content suspensions used for DNA purification were plated on LB agar plates. DNA was extracted by the following procedure: approximately 0.5 g of material was combined with 600 µL of preheated lysis buffer (10 mm Tris-HCl, pH 7.8, 50 mm EDTA, 150 mm NaCl, 2.5% N-lauroyl sarcosine, 500 mm β-mercaptoethanol, 400 µg/mL proteinase K and 2.5 mm N-phenacylthiazolium bromide (Poinar 1998)), and samples were incubated at 65 °C for at least 4 h with vigorous agitation and extracted with an equal volume of phenol–chloroform (1:1) mixture, followed by chloroform–octanol (24:1) mixture extraction. DNA from aqueous phase was precipitated with isopropyl alcohol (0.6 volume) and 0.1 volume of 3 m sodium acetate. Precipitated DNA was dissolved in 50–100 µL of milli-Q water. A mock control was performed by following the procedure described above with 0.5 ml of distilled water instead of palaeo material. DNA from Escherichia coli K12 cells was extracted in standard molecular biology laboratory with genomic DNA purification kit (Thermo Scientific) according to the manufacturer’s instruction. Genomic DNA prepared from E. coli K12 was shared by sonication on Vibra-Cell VCX130 machine (Sonics) at 100% power for 5 min yielding DNA fragments with a mean ~200 bp length to reproduce the state of degradation of ancient DNA extracted from the mammoth sample.
PCR and sequencing
The method used for spacer amplification is similar to those previously applied for other CRISPR-Cas systems (Sun et al. 2016; Lopatina et al. 2016). To minimize biases due to variations in individual repeat sequences, primers used for amplification were designed based on a repeat Logo determined with WebLogo 3.0 (Crooks et al. 2004) from repeats in all known type I-E E. coli CRISPR arrays. PCR amplification was performed using a forward primer Rep1-3 (CGCTGGCGCGGGGAACWC) and reverse primers Rep 2-1 (GCGCCAGCGGGGATAAACCG) and Rep 2-2 (GCGCCAGCGGGGATAAACCN). The molar ratio of Rep2-1/Rep2-2 was 3/1; the overall concentration of reverse primers was the same as that of the forward primer. 50 µL PCR reactions contained 67 mm Tris-HCl, pH 8.3, 17 mm (NH4)2SO4, 0.001% Tween 20, 2.5 mm MgCl2, 10 ng of DNA template, 25 pmol of forward primer or reverse primer mix, and 1.25 units of Encyclo Taq polymerase (Evrogen). For each DNA sample analysed, five to ten individual PCR reactions were set up. After amplification, individual reactions were pooled and processed jointly.
Amplicons corresponding to E. coli K12 and ‘mammoth’ samples were used to obtain libraries with TruSeq DNA sample preparation kit according to the manufacturer’s instructions. Paired-end sequencing was performed on Illumina MiSeq platform with MiSeq reagent kit v.2 (Illumina), in 250-bp cycles. For palaeo samples, 462, 332 and 402 thousands of pair reads were obtained for first, second and third biological replica, correspondingly. A total of 160 thousands reads were obtained for the K12 sample.
Bioinformatics analysis
Raw sequencing data were analysed using shortread and biostrings packages (Morgan et al. 2009). Illumina-sequencing reads were filtered for quality scores of ≥. Reads that contained 32-bp sequences between two CRISPR repeats were selected, and the intervening 32-bp sequences were considered as spacers.
The spacer clustering procedure is presented in detail in the Supporting Information section. Briefly, each spacer was represented as a 32 × 4 = 128 dimensional numerical vector in which information about each nucleotide is stored in four corresponding dimensions. The distance between two spacers or clusters was defined as a sum over 128 dimensions of the absolute values of the difference between their coordinates. Spacers were clustered into a three-level branching structure with each subsequent level having clusters of progressively higher similarity between its members. At the last level of segregation, clusters had radii approximately equal to 3, which reflects the maximum number of substitutions between spacers. The code was written in F# and is available upon request. To verify robustness, clustering was performed repeatedly starting with different randomly chosen initial spacer sequences. The procedure converged to same cluster sets for major (N > 10) clusters. Next, consensus sequences of each cluster were compared to each other using standard pairwise BLASTn algorithms with an e-value less than 10−9. When trivial matchings of each cluster to itself were excluded, overlapping of 0.15% or less of the clusters was detected, indicating that underclustering was minimal. As an independent verification of the clustering procedure, a data set of 30 000 spacers acquired from pG8-C1T plasmid (Shmakov et al. 2014) was clustered alone or together with one of the spacer sets analysed in this work. The average number of plasmid-derived spacer clusters corresponded to known number of plasmid protospacers (the ratio did not exceed 1.2), while clustering of combined set of plasmid-derived and palaeo spacers was found to proceed independently, as should be expected because no palaeo spacers match the pG8-C1T plasmid sequences.
The spacer diversity saturation was calculated according to Good’s formula: C = 1 − (n1/N), where n1 is the number of sequences that occurred only once and N is the sample size (Good 1953). Spacer clusters of three biological replicates were merged based on pairwise comparison with up to three mismatches tolerated using shortread and biostrings r packages (Morgan et al. 2009). Spacers from annotated CRISPR arrays of Salmonella and E. coli downloaded from GenBank were extracted and clustered in the same way. Pairwise comparison with up to three mismatches tolerated was also used to find intersections between spacer clusters from the mammoth sample and annotated arrays. Two benchmark groups of ‘recent’ and ‘ancient’ spacers were composed, correspondingly, from three leader-proximal and three leader-distant spacers from each known array. For each spacer, the frequency of its belonging to one of these groups was determined. Then, the sums of ‘recent’ and ‘ancient’ frequency values were next calculated.
To search for protospacers matching spacer sequences, cluster consensus sequences were aligned to nt (2016) databases using BLASTn algorithm adjusted for short sequences. Hits with an e-value>0.001 or matching CRISPR arrays were filtered out.
Reads containing two or three spacers were extracted and grouped with up to three mismatches tolerated in each spacer. Comparisons with fragments of E. coli CRISPR arrays present in public databases were performed using shortread and biostrings packages (Morgan et al. 2009) with up to three mismatches per each spacer allowed.
To reconstruct CRISPR allele fragments, pairs of neighbouring spacers were represented as a directed graph, where vertices were spacers and edges connecting vertices represented spacers present in one read. Each edge had its own weight reflecting the frequency of two spacers’ co-occurrence. To reconstruct most common arrays, we considered only edges with weights above 30. After decomposition of resulting subgraphs into connected components, the longest path for each component was determined. Vertices in the longest path corresponded to spacer of a reconstructed array. Described algorithms were implemented using shortread and biostrings packages (Morgan et al. 2009). Scripts are available from the authors upon request.
Results and Discussion
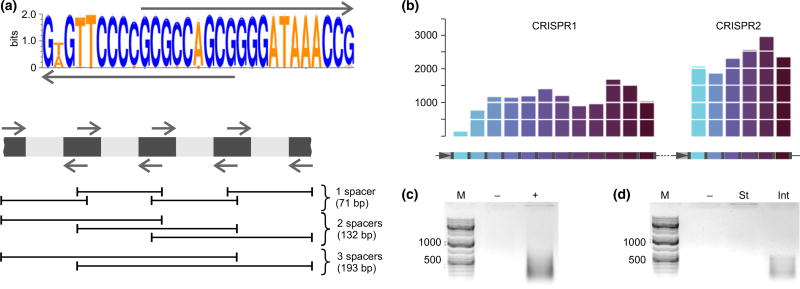
To determine the overall diversity of spacers associated with Escherichia coli type I-E CRISPR repeat in an intestinal sample, a PCR-based method amplifying short spacer-containing fragments of CRISPR arrays with partially overlapping primers complementary to CRISPR repeat was applied (Sun et al. 2016; Lopatina et al. 2016) (Fig. 1a). The procedure should allow amplification of the entire complement of spacers associated with chosen CRISPR repeat and is particularly well suited for analysis of palaeo DNA which is usually degraded to 50–400-bp fragments (Dabney et al. 2013). It should be noted that type I-E CRISPR repeat sequences of E. coli and Salmonella are identical (Touchon & Rocha 2010), so our procedure cannot distinguish spacers originating from these bacteria. To evaluate the procedure, we applied it to a laboratory E. coli strain K12, which contains two CRISPR arrays, CRISPR1 and CRISPR2 according to the classification of Sun et al. 2016; with twelve and six different spacers, correspondingly (Fig. 1b) (Diez-Villasenor et al. 2010). The K12 genomic DNA was disrupted by sonication to give a mean fragment size of ~200 bp to mimic palaeo DNA. Amplified PCR fragments (Fig. 1c) were purified and subjected to high-density Illumina sequencing. Spacers (defined as 32-nt-long sequences bracketed by CRISPR repeats) were extracted from individual reads and mapped to K12 CRISPR arrays. Reads corresponding to every K12 spacer were obtained (Fig. 1b). The frequency of reads corresponding to different spacers within each array and the mean number of spacers amplified from CRISPR1 and CRISPR2 arrays were not equal, indicating that our procedure provides a representative qualitative but not quantitative view of type I-E repeat-associated spacers. Many of the longer reads contained more than one spacer-repeat unit. When neighbouring spacers from longer reads were analysed, their order matched the order of neighbouring spacers in K12 CRISPR arrays.
Fig. 1.
Escherichia coli type I-E CRISPR-Cas system spacer retrieval from K12 strain and a palaeo DNA sample. (a) A Logo of the E. coli type I-E CRISPR repeat is shown at the top. The arrows above and below the Logo indicate primers used in PCR amplification. A scheme showing expected products of PCR amplification from an E. coli type I-E CRISPR array using repeat-specific primers is presented below. Repeats are dark grey, and spacers are light grey. Expected amplification products are shown below as black lines with their sizes indicated. (b) The procedure outlined in (a) was applied to E. coli K12 strain containing two CRISPR arrays (CRISPR1 and CRISPR2, schematically shown at the bottom, with repeats indicated in grey, and spacers are in colour). Rightward horizontal arrows indicate promoters in the leader of each array. Leader-proximal spacers are coloured with lighter shades of blue, while leader-distant spacers are shown in progressively darker colours. The number of Illumina reads corresponding to each spacer is shown on the histograms above. (c, d) Results of E. coli type I-E CRISPR spacer amplification from K12 strain (c) and mammoth intestinal (‘Int’) and stomach (‘St’) content samples (d). Lanes marked as ‘−’ show results obtained with mock-purified DNA.
Spacer content in samples from baby mammoth Lyuba (Fisher et al. 2009) was next investigated. Amplification products were obtained in reactions containing DNA purified from samples of mammoth intestinal content but not in control reactions containing mock-purified DNA or DNA purified from a sample of mammoth stomach content where no E. coli was expected (Fig. 1d).
Three independent mammoth intestinal content DNA purifications/amplifications were performed followed by high-density Illumina sequencing. Tens of thousands of nonredundant spacer sequences were obtained in each replicate (Table 1). Clustering of such a large number of unique sequences based on direct BLAST sequence comparisons of every spacer is a computationally intensive task. Therefore, a faster k-means hierarchical clustering-based procedure was utilized (for details of algorithm, threshold values choice and verifications tests, see Materials and Methods and Supporting Information sections). The clustering procedure reduced complexity of spacer sets from each biological replicate to 1.2–1.4 thousands spacer clusters. Sequences that fell into distinct clusters differed from each other in more than three positions. The depth of sequencing allowed us to reach 80–99% coverage of spacer diversity in each replicate as estimated by the Good’s criterion (Good 1953) (Materials and Methods and Table 1).
Table 1.
Statistics of palaeo-spacer sequencing and clustering
| Replicate | CRISPR spacers, total | CRISPR spacers, nonredundant | Clusters | Good’s criterion | Cluster combined set |
|---|---|---|---|---|---|
| I | 824 536 | 47 429 | 1411 | 0.830986 | 1883 |
| II | 448 951 | 33 226 | 1220 | 0.999231 | |
| III | 709 795 | 46 489 | 1175 | 0.875101 |
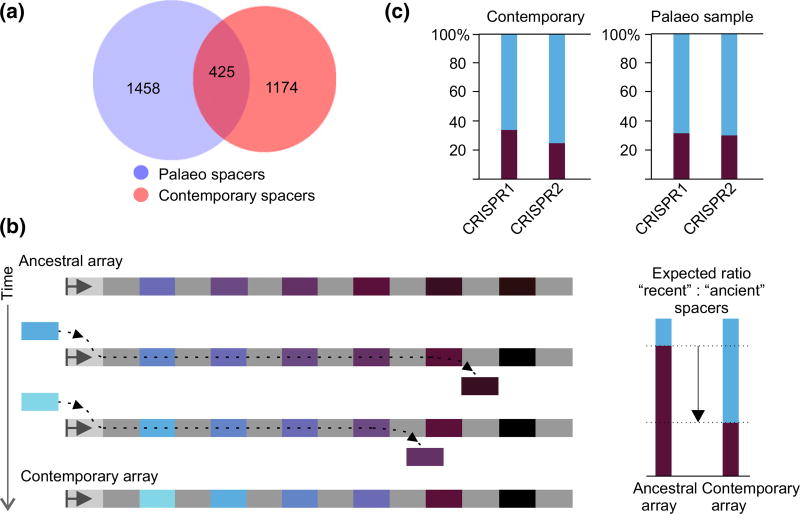
Spacer clusters present in each biological replicate were merged with up to three mismatches tolerated. In this way, a final set of 1883 unique clusters of spacers from the mammoth sample was created (Table 1). To obtain contemporary E. coli spacer set for comparison, the clustering procedure was applied to 1728 spacers from E. coli type I-E CRISPR arrays present in public databases, producing 1599 spacer clusters. Direct BLAST comparison of the mammoth and contemporary spacer cluster sets revealed 425 common clusters (Fig. 2a).
Fig. 2.
Comparison of ancient and present-day Escherichia coli type I-E CRISPR spacers. (a) Comparison of spacer cluster sets. Numbers within circles correspond to unique and overlapping spacer clusters. Blue circle represents clusters obtained from the mammoth sample; red circle represents known E. coli type I-E spacer cluster set. (b) An ancestral CRISPR array is schematically shown at the top. Repeats are light grey, and spacers are coloured. The leader (light grey rectangle with arrow) is shown on the left. With the passage of time, additional spacers (coloured with lighter shades of blue) are acquired at the leader-proximal end, while internal spacers (dark-coloured) are lost. A resulting contemporary array is shown at the bottom. Expected ratios of recently acquired (spacer-proximal) and ancient (spacer-distal) spacers in the ancestral and contemporary arrays are shown at the right. (c) The overall frequency of ‘ancient’ and ‘recent’ E. coli type I-E CRISPR spacer clusters from known CRISPR arrays present in public databases (DB) and in the mammoth sample is shown. Data for CRISPR1 and CRISPR2 arrays are shown separately.
The set of spacer clusters from public databases for Salmonella is much larger than that of E. coli (it consists of more than ~3.6 thousands clusters), but the two sets do not overlap. There was a minimal 0.04% overlap between the mammoth and the Salmonella sets, suggesting that most mammoth sample spacers correspond to E. coli type I-E CRISPR arrays spacers.
Spacers are acquired at one end of the array proximal to the leader region, and for every acquired spacer, an additional copy of CRISPR repeat is generated (Barrangou et al. 2007; Datsenko et al. 2012; Erdmann & Garrett 2012; Lopez-Sanchez et al. 2012; Swarts et al. 2012). Spacers located close to this end of the array should have been acquired more recently, while distal spacers should correspond to ancient acquisition events. As CRISPR arrays cannot grow indefinitely, the acquisition of new spacers shall be accompanied by the loss of older internal spacers (Deveau et al. 2008; Horvath et al. 2008; Lopez-Sanchez et al. 2012). As a result, a turnover in spacer composition is expected (Fig. 2b). Specifically, recently acquired spacers present in contemporary arrays should have been less frequent or even absent in ancestral arrays (Fig. 2b). For every spacer cluster from contemporary set and for overlapping spacer clusters from the mammoth set, the frequency of spacer occurrence in three leader-proximal (‘recent’) and leader-distal (‘ancient’) positions of annotated E. coli CRISPR arrays was calculated (see Materials and Methods). The overall frequency of ‘recent’ and ‘ancient’ spacer clusters was then determined by summing the values obtained for individual clusters. The spacer content in CRISPR1 and CRISPR2 arrays is unrelated (Diez-Villasenor et al. 2010; Touchon & Rocha 2010; Kupczok et al. 2015), suggesting that spacers in each array are acquired independently and there is no recombination between arrays. Therefore, ‘recent’ and ‘ancient’ spacers from CRISPR1 and CRISPR2 arrays were treated separately. In contemporary E. coli spacer set, ‘recent’ spacers constituted ~70% of the total in both arrays (Fig. 2c). Higher portion of ‘recent’ spacers arose due to higher diversity of leader-proximal spacers compared to the more homogeneous leader-distant spacers. Strikingly, the overall proportion of spacer clusters matching either ‘age’ group remained the same in the mammoth set (Fig. 2c). Thus, our analysis failed to reveal a significant turnover of spacers associated with E. coli type I-E CRISPR repeats in the course of 42 000 years that separate E. coli from mammoth and the present-day E. coli.
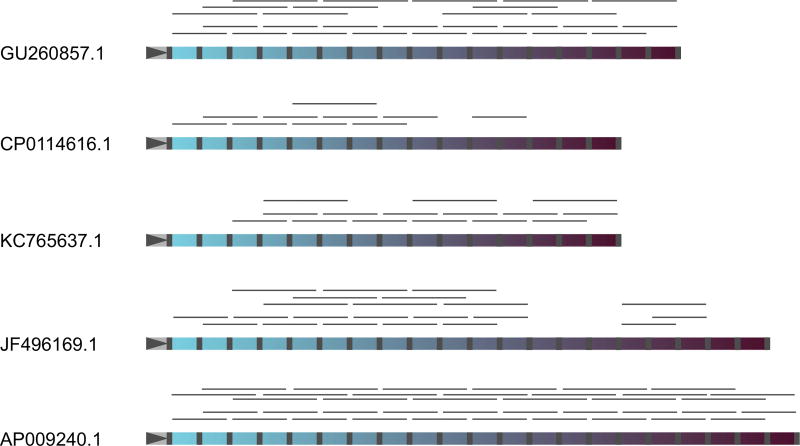
We next analysed neighbouring spacer pairs in longer high-density Illumina-sequencing reads from the mammoth sample with the hope of reconstructing CRISPR arrays. A total of 902 unique neighbouring spacer pairs were extracted from the mammoth sample and mapped to annotated E. coli CRISPR arrays, yielding 257 neighbouring spacer pairs from the mammoth sample that matched annotated CRISPR arrays. Full or almost full-length contemporary arrays could be reconstructed using these spacer pairs. Selected examples of such reconstructions are shown in Fig. 3. The same analysis was performed for triplets of spacers extracted from some of the longer reads. Of a total of 305 cases, 130 triplets corresponded to contemporary arrays, and in several cases, they could be used to reconstruct arrays identical to those reconstructed with spacer pairs (Fig. 3). Thus, some E. coli CRISPR arrays or their fragments remained unchanged for more than 40 thousand years.
Fig. 3.
Reconstruction of contemporary CRISPR arrays from reads containing two or three spacers from the mammoth sample. Mapping results of neighbouring spacer pairs and triplets on five selected CRISPR arrays from contemporary Escherichia coli are shown. Repeats are grey, and spacers are coloured. The leader regions are marked by grey triangles on the left of each array. Leader-proximal spacers are coloured with lighter shades of blue, while leader-distant spacers are dark-coloured. Detected reads containing neighbouring spacer pairs or triplets are shown by thin grey lines above each array.
Most (645) neighbouring spacer pairs from the mammoth sample had no matches to contemporary E. coli arrays. They were used to reconstruct longer chains (see Materials and Methods) yielding twelve 3- to 8-spacerlong array fragments that must correspond to CRISPR arrays/array fragments that are either extinct or that have not been isolated yet in contemporary E. coli.
The collection of spacers from the ‘mammoth’ sample considerably expands the variety of unique E. coli type I-E CRISPR spacers. Only a small percentage of E. coli type I-E CRISPR spacers from the database match sequences of phages and other mobile genetic elements (Diez-Villasenor et al. 2010; Touchon & Rocha 2010). In addition to known phage-matching spacers, several novel hits of palaeo spacers to mobile genetic elements were found. However, the overall percentage of hits to genomes of known phages, plasmids and likely prophages for spacer clusters from the mammoth sample remained low (0.6%, Table 2).
Table 2.
Hits of CRISPR spacer clusters originated from the mammoth sample
| Cluster consensus sequence Hit | Hit |
|---|---|
| GCATCTCTTCCACTTAAATCTCCTTGTTACGA | Enterobacteria phage NJ0 |
| CGGGATAATTCAGCTTTCACATCACGGCAAGA | Enterobacteria phage phiEco32 |
| TGCCGGGTTCGACTGGACGCCATTTGCCATCT | Enterobacteria phage epsilon15 |
| GGTAAAAACACGGTCTGAACCGACATTCATGT* | Enterobacteria phage P7 |
| CATTTTTGCGTGGCGAGCTGCGCCGCGTTCTG* | Escherichia phage JLK-2012 |
| ACGATTGGGCAGCCAGAGTTGCCGCCGGGAAA | Escherichia coli strain T23 plasmid pEQ1 |
| CGGCCAGGCTGGATTTAAGCGGCACGGCCGCA | Uncultured bacterium plasmid pMBUI4 |
| GTCGCCTCAATAGCGCGTTTACCTTTGCTGTT | Uncultured bacterium plasmid pMBUI4 |
| GCCAGGGCAAGCGGCCCAAGGGCAAGGTCATA | Plasmid pMCBF1 |
| GGGATCTCATCGTCAAAATCGTGAGCCGGATC | Escherichia coli strain BK28960 plasmid |
| CCAGCCGTTCAGTATTGCCGGTGTCAGCAAAA* | Enterobacter cloacae strain 34983 plasmid p34983-328.905 kb |
| GCCGTCGTGCCGTGTTCACCTTTACGAACCTG* | Klebsiella pneumoniae ATCC BAA-2146 plasmid pHg |
| TAAAATGAGAGCTTTTGTTCGCTTGAGCAATA | Escherichia coli genome, fimbrial protein |
| CAAGAAGTACTGAACCGATATACTCGCCAACC | Escherichia coli genome, intergenic between two hypothetical proteins |
| AGGACAGTAAAAATGACGGAATTGTTTATCAG | Escherichia coli genome assembly FHI92, tail sheath protein |
Asterisk mark clusters found in both the mammoth and contemporary data sets.
Overall, our findings reveal that E. coli population contains a vast variety of spacers that remain stable over long periods of time. The order of spacers also appears to be preserved at least in some arrays. Most spacers have no matches to known mobile genetic elements, and their origin and sequences they target remain to be established.
Supplementary Material
Fig. S1 A plot showing the dependence of the number of secondary clusters N vs. the cutoff distance between clusters D.
Acknowledgments
We thank Dr. Jaroslav Ispolatov for advice. This work was supported by grants from Russian Science Foundation [14-14-00988 to KS], NIH RO1 grant GM10407 to KS and Russian Foundation for Basic Research [16-04-00767 to E.S and 17-04-02144 to IIA].
Footnotes
E.S. designed research, performed experiments, analysed data and wrote the manuscript, A.L. performed experiments, M.K. designed and implemented clustering procedure and analysed data, S.M. and S.S. analysed data, A.T. collected and provided mammoth samples, I.I.A. helped analyse data, M.L. performed high-throughput sequencing, and K.S. designed research, analysed data and wrote the manuscript.
Data accessibility
The cluster sets of palaeo spacers and annotated spacers from E. coli and Salmonella arrays used in this work as well as CRISPR array fragments reconstituted from palaeo sample can be downloaded from https://doi.org/10.6084/m9.figshare.1613398, and raw data can be download from https://doi.org/10.6084/m9.figshare.4244501.v1.
Additional supporting information may be found in the online version of this article.
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Dusko Ehrlich S. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney J, Meyer M, Paabo S. Ancient DNA damage. Cold Spring Harbor Perspectives in Biology. 2013;5:a012567. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature Communications. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. Journal of Bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Villasenor C, Almendros C, Garcia-Martinez J, Mojica FJM. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156:1351–1361. doi: 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Molecular Microbiology. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D, Rountrey A, Tikhonov A, et al. Life history of remarkably preserved woolly mammoth calf from the Yamal peninsula, northwestern Siberia. Journal of Vertebrate Paleontology. 2009;29:96A. [Google Scholar]
- Good IL. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Hofreiter M, Paijmans JL, Goodchild H, et al. The future of ancient DNA: Technical advances and conceptual shifts. Bioassay. 2015;37:284–293. doi: 10.1002/bies.201400160. [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin A-C, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. Journal of Bacteriology. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupczok A, Landan G, Dagan T. The contribution of genetic recombination to CRISPR array evolution. Genome Biology and Evolution. 2015;7:1925–1939. doi: 10.1093/gbe/evv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina A, Medvedeva S, Shmakov S, et al. Metagenomic analysis of bacterial communities of antarctic surface snow. Frontiers in Microbiology. 2016;7:398. doi: 10.3389/fmicb.2016.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez MJ, Sauvage E, Da Cunha V, et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Molecular Microbiology. 2012;85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nature Reviews Microbiology. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science (New York, N.Y.) 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Morgan M, Anders S, Lawrence M, et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics (Oxford, England) 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends in Biochemical Sciences. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, et al. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nature Communications. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- Paez-espino D, Sharon I, Morovic W, et al. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio. 2015;6:1–9. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar HN. Molecular coproscopy: dung and diet of the extinct ground sloth nothrotheriops shastensis. Science. 1998;281:402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- Pougach K, Semenova E, Bogdanova E, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Molecular Microbiology. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Pul Ü, Wurm R, Arslan Z, et al. Identification and characterization of E. coli CRISPR- cas promoters and their silencing by H-NS. Molecular Microbiology. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- Sheludchenko MS, Huygens F, Stratton H, Hargreaves M. CRISPR diversity in E. coli isolates from australian animals, humans and environmental waters. PLoS One. 2015;10:e0124090. doi: 10.1371/journal.pone.0124090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Savitskaya E, Semenova E, et al. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Research. 2014;42:5907–5916. doi: 10.1093/nar/gku226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CL, Thomas BC, Barrangou R, Banfield JF. Metagenomic reconstructions of bacterial CRISPR loci constrain population histories. The ISME Journal. 2016;10:858–870. doi: 10.1038/ismej.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MWJ, Brouns SJJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Rocha EPC. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010;5:e11126. doi: 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Charpentier S, Clermont O, et al. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. Journal of Bacteriology. 2011;193:2460–2467. doi: 10.1128/JB.01307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Research. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 A plot showing the dependence of the number of secondary clusters N vs. the cutoff distance between clusters D.