Abstract
Engineering cell factories for producing biofuels and pharmaceuticals has spurred great interests to develop rapid and efficient synthetic biology tools customized for modular pathway engineering. Along the way, combinatorial gene expression control through modification of regulatory element offered tremendous opportunity for fine-tuning gene expression and generating digital-like genetic circuits. In this report, we present an efficient evolutionary approach to build a range of regulatory control elements. The reported method allows for rapid construction of promoter, 5′UTR, terminator and trans-activating RNA libraries. Synthetic overlapping oligos with high portion of degenerate nucleotides flanking the regulatory element could be efficiently assembled to a vector expressing fluorescence reporter. This approach combines high mutation rate of the synthetic DNA with the high assembly efficiency of Gibson Mix. Our constructed library demonstrates broad range of transcriptional or translational gene expression dynamics. Specifically, both the promoter library and 5′UTR library exhibits gene expression dynamics spanning across three order of magnitude. The terminator library and trans-activating RNA library displays relatively narrowed gene expression pattern. The reported study provides a versatile toolbox for rapidly constructing a large family of prokaryotic regulatory elements. These libraries also facilitate the implementation of combinatorial pathway engineering principles and the engineering of more efficient microbial cell factory for various biomanufacturing applications.
Keywords: Promoter, 5′UTR, Terminator, Riboregulatory, Trans-activating RNA, Library, Synthetic biology
1. Introduction
The plummeting cost of gene synthesis and facile gene assembly platforms allow metabolic engineers to rapidly reprogram a cell's genetic blueprint at a speed and scale never seen before [1]. Metabolic engineering has become an enabling technology to construct efficient microbial cell factories and a major driver for next-generation bio-economy [2]. By engineering heterologous pathways or endogenous metabolism, metabolic engineers have now been able to produce a large portfolio of commodity chemicals [3], [4], novel materials [5], sustainable fuels [6], [7] and pharmaceuticals [8], [9] from renewable feedstocks. This is often achieved through sophisticated metabolic engineering strategies including overexpression of rate-limiting steps [10], deletion of competing pathways [11], managing ATP [12], [13] and recycling NADPH and other cofactors [13]. While these approaches have been shown to effectively improve cellular productivity and yield, the attempt to engineer a specific pathway often requires proper balancing of precursor pathway and fine-tuning of gene expression.
Engineering microbial overproduction phenotypes remains a daunting task as it usually involves the manipulation of a handful of precursor or rate-rating pathways that are subject to tight cellular regulation. For example, precursor flux improvement by overexpression of heterologous pathways may not be accommodated by downstream pathways; intermediate accumulation or depletion may compromise cell viability and pathway productivity [14]; and overexpressed heterologous protein may penalize the cell with additional energy cost and elicit cellular stress response [15]. As an attempt to address these issues, the practice of metabolic balancing has shifted towards redistributing metabolic flux through synthetic biology approaches, including modification of plasmid copy number [16], promoter strength [17], gene codon usage [18] and RBS strength [15], [19].
Along this line, combinatorial transcriptional engineering coupled with efficient gene assembly tools has offered tremendous opportunities for customized optimization of multi-gene pathways. Excellent examples include construction of yeast xylose pathways by promoter shuffling [20], heterologous production of anti-cancer taxol precursors in E. coli [21] and rapid assembly and screening multi-gene mutant pathway libraries in E. coli and yeast [22], [23], and combinatorial optimization of fatty acids pathway to produce advanced biofuels [24], [25], multiplexed regulatory RNA [26] and global transcriptional machinery engineering [27] to produce l-tyrosine. Design of experiment (DoE) statistical procedures have been recently applied to rapidly locate the overproduction genotype out of a large number of pathway candidates. Without testing all the possible gene expression combinations, factorial design and empirical equations allow metabolic engineers to correlate gene expression level with metabolites titer; and facilitate the extrapolation of the optimal gene expression patterns leading to the desired overproduction phenotype [28], [29].
As an emerging discipline, synthetic biology is becoming increasingly important to design, construct and optimize metabolic pathways leading to desired overproduction phenotype in genetically tractable organisms. One of the major challenges for heterologous expression of multi-gene pathways is to balance the expression level of each of the enzymes among the selected pathways and achieve optimal catalytic efficiency [30]. Thus, delicately designed molecular control elements has been integrated into cell chassis to enable the host strain to precisely respond to environmental stimuli or cellular intermediates and drive carbon flux toward a target pathway. For example, engineering promoter architecture has achieved tunable gene expression in both E. coli [31] and yeast [32] at transcriptional levels; engineered metabolite-responsive riboswitches [33] and synthetic ribosome binding sites [19] can be used to precisely control protein expression at the translational level; metabolic flux channeling by spatial recruitment of desired metabolic enzymes at stoichiometric ratio on a synthetic protein scaffold can efficiently prevent the loss of intermediates due to diffusion [34]. In a word, advances in synthetic biology have accelerated our ability to design and construct cell factories for metabolic engineering applications.
Regardless of this development, it is necessary to further expand synthetic biology toolbox and diversify the number of regulatory elements that could be used for pathway fine-tuning. In this report, we present an evolutionary approach to efficiently construct regulatory element libraries that cover a broad range of gene expression dynamics. Our library construction approach has been successfully applied to construct the T7 promoter library, lactose repressor binding site (lacO) library, prokaryotic ribosome binding site library, T7 terminator library and the trans-activating RNA library. The constructed regulatory element libraries will find wide applications in transcriptional and translational fine-tuning, as well as sampling the multidimensional gene expression space to rapidly locate the desired phenotype. We envision this evolutionary approach will provide an efficient solution to balance multiple gene pathway and accelerate strain engineering for cost-competitive manufacturing of bioproducts.
2. Results and discussion
2.1. An efficient evolutionary approach to construct regulatory element library
Traditional regulatory element library construction are primarily relied on error-prone PCR [35], which typically suffers from biased mutational spectrum with transition mutation occurring more frequent than transversion mutation. Here we adopted a novel library construction approach [36] that capitalizes on partially overlapped 60-mer synthetic oligos from DNA vendors like IDT and Sigma. Customized oligos with point mutations or degenerate bases flanking the regulatory element region can be easily fused to a plasmid backbone via highly efficient Gibson assembly. The vector backbone contains the replication origin, antibiotic maker and a reporter gene, allowing for easy transformation, plasmid propagation and reporter activity screening.
Gibson assembly mix contains polymerase, ligase and T5 exonuclease activity. T5 exonuclease chews back the 5′ overhang and introduces a 3′ overhang which complements the primers. Then the polymerase extends the single strand DNA and fills up the gaps. Taq ligase joins the 5′-phosphate and 3′-hyroxyl group to form a phosphodiester bond. Because of the activity of the T5 exonuclease, a special consideration for designing the synthetic overlapping oligos is to leave the overhang at the 3′ end to avoid the digestion by T5 exonuclease (Fig. 1). Another important aspect is to keep the completeness of the library. One cannot pick a single colony to retrieve the library. Instead, a bacteria “lawn” from the agar plates were directly retrieved for plasmid mini-preparation. The plasmid library was later retransformed to the proper chassis strains (in our case, BL21(DE3)star) for proper protein expression and reporter activity quantification (Fig. 1). This approach combines the high mutation frequency of the degenerate synthetic oligos with the highly efficient gene assembly platform (specifically Gibson Assembly), offers us a rapid evolutionary strategy to construct regulatory element libraries.
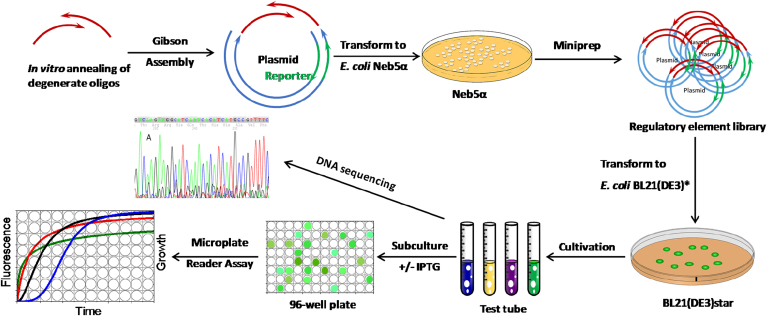
Fig. 1.
General procedures to construct regulatory element library from synthetic overlapping degenerate oligos. These oligos were annealed and later fused to a plasmid backbone containing the reporter gene via highly efficient Gibson Assembly. Plasmid library was directly prepared from agar plates (instead of liquid culture). Activity of the regulatory element library was later screened by fluorescence readout and sequenced by Genewiz.
2.2. A hybrid T7 promoter and lactose repressor-binding site library
Promoter is the DNA sequence that caries transcriptional start signal. Promoters consists of specific response elements that provide the binding site for RNA polymerase and for transcription factors to regulate transcription. These transcription factors could be activators or repressors that are responsible for turning on or shutting down gene expression. Prokaryotic promoter generally contains two conserved sequence elements: the -10 (TATAAT box) and -35 (TTGACA sequence) region. Recruited by sigma factors, RNA polymerase will specifically interact with the -10/-35 region and form a RNAP-promoter complex to initiate transcription.
We first test if we can modify the bacteriophage T7 promoter as it is widely used in various metabolic engineering applications. The strength and efficiency of T7 RNA polymerase (RNAP) are determined by two factors: the recruitment of T7 RNAP and sigma factors on the promoter core region (-35 and -10) and the proceeding of the RNAP across the repressor binding region (lacO) [37]. To generate the T7 promoter library, we designed the synthetic overlapping oligos in such a way that we can mutate both the sigma factor binding region (-35 and -10) and the lacI repressor binding region (lacO) (Fig. 2A). IPTG induces the dissociation of lacI repressor from the lacO site and thus allows T7 RNAP to read across the DNA template and give rise to fluorescence signals. With green fluorescence protein as readout, we quantified the T7 promoter transcriptional activity of the constructed promoter candidates. Reporter output results indicate that the constructed library covers a broad range of transcriptional dynamics spanning across three orders of magnitude (Fig. 2B). For instance, the highest transcriptional activity obtained (21,340 FU/OD with promoter No. 2) was about 7-fold higher than the original T7 promoter (3942 FU/OD with promoter No. 48); the lowest transcriptional activity (22.3 FU/OD with promoter No. 46) achieved was about three order of magnitude lower than the highest promoter (No. 2).
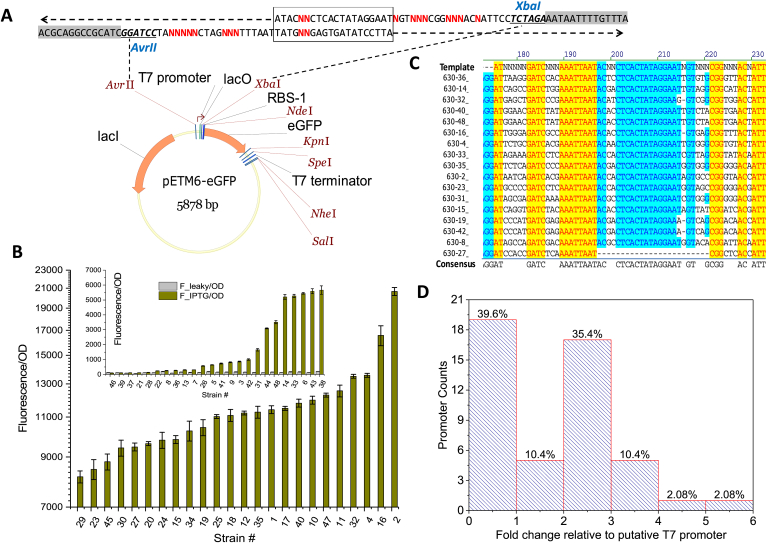
Fig. 2.
T7 promoter library. (A) Synthetic overlapping oligos was annealed together and assembled into the AvrII and XbaI digested pETM6-eGFP vector backbone. Box region shows the overlapping region and sequence flanked by AvrII and XbaI is the core of the degenerate T7 promoter and lactose-repressor binding site. Strategically designed unique restriction enzyme sites are flanking the promoter, 5′UTR and terminator region to facilitate the insertion of regulatory element libraries. (B) Promoter strength screened by green fluorescence protein. (C) DAN sequence of representative promoters. (D) Statistical distribution of promoter strength across the engineered promoter library.
We then sequenced 18 promoters and the sequencing results indicated all desired mutations occurred at the designed genetic loci (-35/-10 and lacI repressor binding regions) (Fig. 2C). It is generally believed that T7 promoter activity is stringently controlled by the amount of lacI repressor protein inside the cell. Mutation in the lacI repressor binding region (lacO) probably will alter the lacI repressor and lacO binding affinity and lead to protein leaky expression. Interestingly, promoter libraries with mutant lacI repressor binding region exhibit relatively small leaky expression discrepancy (eGFP expression in the absence of IPTG induction) ranging roughly from 107 FU/OD to 311 FU/OD (Fig. 2B), indicating our designed lacO region may effectively block the proceedings of the T7 RNAP. The 1000-fold transcriptional dynamics of the constructed promoter library is primarily ascribed to the synergy interaction between the promoter core region (-35 and -10) and the repressor binding region (lacO).
Statistical analysis indicates that only 4% of the promoter library spans above 4-fold gene expression change. About 40% of the promoters demonstrate transcriptional activity lower than the original promoter (Fig. 2D). Taken together, simultaneous mutation of both the core promoter region (-35/-10) and the repressor binding (lacO) region, we constructed artificial T7 promoters with transcriptional activity spanning across three orders of magnitude and relatively low leaky expression, a feature that would not be easily attainable by conventional promoter engineering approaches.
2.3. 5′-Untranslated region (5′UTR) library
Transcribed RNA typically consists of a 5′ UTR (untranslated region) to control and regulate protein synthesis. Part of the UTR region is the ribosome-binding site (RBS), which contains string of DNA upstream of the start codon (AUG) and is responsible for recruiting ribosome to promote efficient and accurate translation of mRNA [38]. The RBS in E. coli is a purine-rich sequence, which complements with the core sequence of the 3′-end of 16S rRNA of the 30S small ribosomal subunit. Base pairing of the RBS with the 16S rRNA promotes translation initiation. Activity of a RBS is determined by the length and nucleotide sequence of the spacer separating the RBS and the start codon AUG.
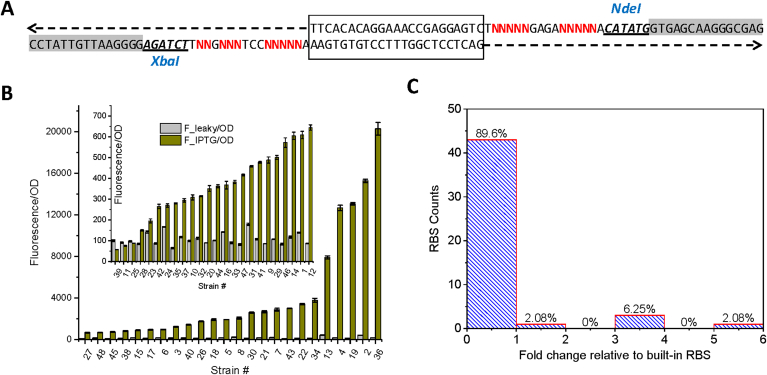
Due to its direct involvement in protein translation, RBS has been identified as the rate-limiting step for gene expression. It is found that protein expression is more dependent on translational activity (RBS strength) than transcriptional activity (promoter strength). To expand genetic parts for translational fine-tuning, we then set about to mutate the 5′UTR region upstream of the reporter gene in the pETM6 vector backbone [39]. In the same way, we designed the synthetic overlapping oligos (Fig. 3A) and annealed them together in the duplex form with 3′-overhangs to avoid T5 exonuclease digestion. The annealed synthetic oligos contain degenerate nucleotides flaking the overlapping region and later was Gibson assembled into the XbaI and NdeI digested pETM6-eGFP vector. Representative sequencing results for 5′UTR library constructed can be found in Supplementary Figure S1. Fluorescence screening of the 5′UTR library indicates about 90% of the mutant UTR are less active than the original 5′UTR (Fig. 3B and C). We believe the original UTR has been an optimized sequence, it is hard to further improve the translational activity of the built-in 5′UTR. Interestingly, there is only 2% artificial UTRs demonstrating 5–6 fold higher translational activity compared to the original 5′UTR.
Fig. 3.
5′-Untranslated region (5′UTR) library. (A) Synthetic overlapping oligos was annealed together and assembled into the XbaI and NdeI digested pETM6-eGFP vector backbone. Box region shows the overlapping region and sequence flanked by XbaI and NdeI is the core of the degenerate 5′UTR region. (B) Translational strength of 5′UTR screened by green fluorescence protein. (C) Statistical distribution of 5′UTR strength across the engineered 5′UTR library.
2.4. T7 terminator library
Terminator is a nucleic acid sequence that contains the transcription termination signal. Termination process involves the interaction of mRNA secondary structure with the termination factors and RNAP complex. Prokaryotic terminator generally forms a stem-loop hairpin structure to restrict the RNAP read through the downstream template DNA and leads to the release of mRNA from the transcriptional complex. Most of the prokaryotic terminator requires the involvement of a rho-factor (ρ), which is a termination factor that help to stall the proceeding RNA polymerase. Previous research has characterized a large panel of natural and synthetic E. coli terminators with varying strength [40], paving the way for forward design of synthetic transcriptional unit.
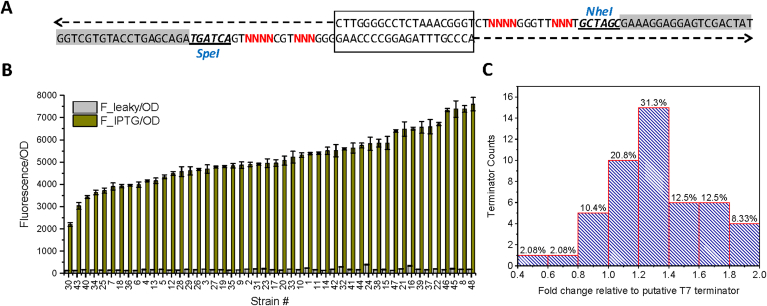
We designed the overlapping synthetic oligos and the degenerate terminator sequence is flanked with the SpeI and NheI site (Fig. 4A). The annealed terminator library could be easily assembled to the SpeI and NheI digested pETM6-eGFP vector via Gibson assembly. Representative sequencing results for T7 terminator library can be found in Supplementary Figure S2 and Figure S3. Transcriptional output was analyzed with green fluorescence protein as reporter. It was found that 92% of the terminators exhibit activity ranging from 0% to 160% of the putative T7 terminator (Fig. 4B and C). And only 8% of the terminator candidate display transcriptional strength above 180% of the putative T7 terminator. These data indicate that the recalcitrance or the rigidity of the original terminator: it might be a result of natural selection that restricted the artificial evolution approach to further optimize the sequence space of this T7 terminator.
Fig. 4.
T7 terminator library. (A) Synthetic overlapping oligos was annealed together and assembled into the SpeI and NheI digested pETM6-eGFP vector backbone. Box region shows the overlapping region and sequence flanked by SpeI and NheI is the core of the degenerate T7 terminator library. (B) Translational strength of terminator library screened by green fluorescence protein. (C) Statistical distribution of terminator strength across the engineered terminator library.
2.5. Trans-activating RNA library
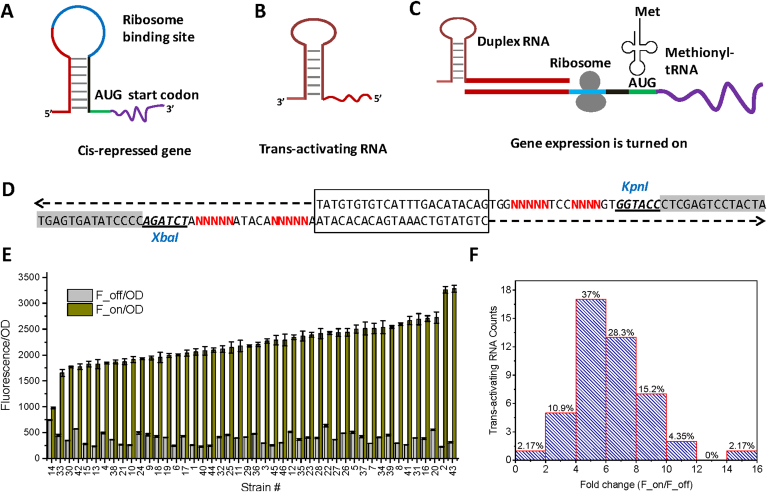
Promoters, terminators, and ribosome-binding sites have facilitated the tunable control of gene expression to coordinate carbon flux and increase the production of important metabolites. Apart from transcription and translation initiation control, non-coding small RNAs have emerged as promising alternative to tune gene expression [26], [41], [42]. Synthetic riboregulator is normally designed to carry one pair of non-coding RNAs (Fig. 5). A cis-repressing RNA is built upstream of the ribosome binding site in front of the AUG start codon to regulate protein translation initiation (Fig. 5A). Sequence complementarity allows the formation of a stable stem-loop structure that sequestrate the ribosome binding site (RBS), thus occluding the entry of the ribosomes and blocking translation (Fig. 5A). On the other hand, a trans-activating RNA (Fig. 5B) is designed to partially base-pair with the 5′ end of the cis-repressing RNA, which forms a more table RNA duplex structure to relieve the Cis-repressing RNA and RBS stem-loop structure. In this way, RBS region of the coding gene will be exposed and allows for the recruitment of ribosome to initiate protein translation (Fig. 5C). This translational on/off control offers a unique approach to regulate protein expression. Previous research has mapped the sequence feature of the cis-repressing RNA and trans-activating RNA and engineered trans-activating RNAs that span across about 200-fold gene expression activation [42], [43].
Fig. 5.
Trans-activating RNA library. (A) Cis-repressing RNA is designed to partially complement itself and sequestrate the ribosome binding site. (B) Structure of trans-activating RNA. (C) Interaction of trans-activating RNA with cis-repressing RNA relieves the sequestration and allows the recruitment of ribosome and translation initiation. (D) Synthetic overlapping oligos was annealed together and assembled into the XbaI and KpnI digested pCDM4 vector backbone. Box region shows the overlapping region and sequence flanked by XbaI and KpnI is the core of the degenerate trans-activating RNA. (E) Translational strength of trans-activating RNA library screened by green fluorescence protein. (F) Statistical distribution of gene expression activation across the engineered trans-activating RNA library.
We first inserted a cis-repressing RNA to the 5′UTR region of the pETM6-eGFP vector. This cis-repressing RNA effectively blocked eGFP translation and the reporter output only led to 16% of the fluorescence intensity of the original 5′UTR without cis-repressing RNA. This repressed state (or OFF state) could be reversed by the addition of a second vector expressing the trans-activating RNA library. We designed overlapping synthetic oligos and the degenerate trans-activating RNA sequence is flanked with the XbaI and KpnI site (Fig. 5D). The annealed trans-activating RNA library was assembled to the XbaI and KpnI digested pCDM4 vector via Gibson assembly. Representative sequencing results for trans-activating RNA library can be found in Supplementary Figure S4, Figure S5, Figure S6 and Figure S7. Reporter output was quantified to evaluate to which extent the transformed trans-activating RNA could restore gene expression from the “repressed” states. Fluorescence intensity for both repressed state (OFF state) and activated state (ON state) were measured (Fig. 5E). Fold change (FON/FOFF) of gene expression at the ON state relative to the OFF state was calculated and statistically analyzed (Fig. 5F). It is interesting to find that about 90% of the trans-activating RNA results in a translational activity change spanning from 2-fold to 12-fold (Fig. 5E). Around 2% of the trans-activating RNA leads to a 16-fold gene expression change. This result clearly demonstrates that trans-activating RNAs are effective regulatory control elements for gene expression fine-tuning.
3. Conclusion
Combinatorial gene expression control through modification of regulatory element has offered tremendous opportunities for fine-tuning gene expression and generating digital-like genetic circuits. Our evolutionary approach allows for rapid construction of promoter, 5′UTR, terminator and trans-activating RNA libraries. The synthetic overlapping oligos has the feature of high mutation rate, easy assembly to the vector, representing a powerful tool for generating regulatory control elements. In addition, our constructed library demonstrates broad range of transcriptional or translational gene expression activities. Both the promoter library and 5′UTR library exhibits gene expression dynamics spanning across three order of magnitude. The terminator library and trans-activating RNA library displays relatively narrowed gene expression pattern. Rapid construction of regulatory element library would be critical to expand our ability in applying combinatorial engineering principles to optimize microbial cell factories for various bio-manufacturing applications.
In light of past two decades' achievements, metabolic engineering has witnessed significant progress from stoichiometry-based pathway engineering to combinatorial pathway fine-tuning and optimization. As a result, multivariate regulatory metabolic engineering and statistical experiment design principles have emerged with the attempt to speed up the design-build-test cycle. This requirement necessitates the further development of genetic toolbox to efficiently probe the genotype-phenotype gene expression space. In this sense, our constructed promoter, 5′UTR, terminator and trans-activating RNA library will be useful to explore the optimal gene expression pattern and accelerate strain engineering.
4. Methods
4.1. Strains, media and oligos
LB broth was routinely used to cultivate E. coli NEB5α, BW27784 and BL21(DE3) star to maintain plasmid, plasmid minipreparation and promoter activity screening. Synthetic overlapping oligos were ordered from Integrated DNA Technologies (IDT). A detailed list of strains, plasmids and primers could be found in supplementary Table S1.
4.2. Regulatory element library construction and screening
Regulatory element library was constructed by Gibson assembling of synthetic overlapped oligos to replace the original regulatory elements in pETM6-eGFP. Single stranded degenerate DNA oligos (Supplementary Table S1) were annealed in 10 mM Tris-HCl and 1 mM EDTA buffer following three cycles of heating (95 °C for 2 min) and cooling (25 °C for 1.5 min) on a Biorad PCR block. Then 5 μL of 10 μM of the annealed oligos were mixed with 2.5 μL of the appropriately enzyme digested and gel purified pETM6-eGFP vector (∼25 ng plasmid DNA) and 7.5 μL of the 2 x Gibson Assembly. The mixture was kept at 50 °C for 1 h. Then 3 μL of the Gibson reaction was chemically transformed into 12 μL NEB5α high efficient competent cell. Overnight grown colonies were scraped with a razor blade and the library plasmid DNA was prepared with the Zyppy miniprep kits. 1.5 μL of the library plasmid was retransformed into 20 μL BL21(DE3) star cell using electroporation.
For regulatory element activity screening, BL21 transformants were individually inoculated into 2.5 mL LB broth supplemented with 100 μg/mL ampicillin or 50 μg/mL streptomycin and grown at 37 °C with shaking 250 rpm overnight. For each of the screening, 48 individual colonies were picked and inoculated. The next morning, 20 μL of the overnight culture was inoculated to 220 μL fresh LB (with 100 μg/mL ampicillin or 50 μg/mL streptomycin) in a Greiner Bio-One black 96-well fluorescence plate. Triplicate inoculation was performed for each of the colony. 10 μL 5 mM IPTG (two lanes of inoculation for testing inducible expression) or sterile water (one lane of inoculation for testing leaky expression) was added to each well with a multichannel pipette. The entire plate was incubated at a 30 °C vortemp plate shaker with shaking at 450 rpm. Then green fluorescence was detected with a SpectraMax microplate reader (Molecular device) with excitation at 495 nm and emission at 512 nm every 45 min for 6 h. Regulatory element activity was calculated with the rate of green fluorescence accumulation divided by the rate of cell density increase. Almost all the samples have a linear response curve with the R2 above 0.95. All experiments were performed in triplicates to ensure reproducibility.
4.3. Data analysis and model fitting
Statistical data analysis and model fitting were performed with the OriginPro 2017. All quantitative plots were made by OriginPro 2017.
Contributions
PX designed the study. EJ performed the study with input from other coauthors. PX and EJ analyzed the data and wrote the manuscript.
Acknowledgements
The authors would like to acknowledge the Department of Chemical, Biochemical and Environmental Engineering, College of Engineering and Information Technology, Office of the Vice President for Research (gratn number 10145-1113-021-STRT7XUP-MAIN) at University of Maryland Baltimore County for funding support.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.synbio.2017.10.003.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Xu P., Bhan N., Koffas M.A.G. Engineering plant metabolism into microbes: from systems biology to synthetic biology. Curr Opin Biotechnol. 2013;24(2):291–299. doi: 10.1016/j.copbio.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Biggs B.W. Multivariate modular metabolic engineering for pathway and strain optimization. Curr Opin Biotechnol. 2014;29:156–162. doi: 10.1016/j.copbio.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Lee J.W. Systems metabolic engineering for chemicals and materials. Trends Biotechnol. 2011;29(8):370–378. doi: 10.1016/j.tibtech.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Tai Y.S., Xiong M., Zhang K. Engineered biosynthesis of medium-chain esters in Escherichia coli. Metab Eng. 2015;27:20–28. doi: 10.1016/j.ymben.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Xiong M. Scalable production of mechanically tunable block polymers from sugar. Proc Natl Acad Sci. 2014;111(23):8357–8362. doi: 10.1073/pnas.1404596111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao K. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab Eng. 2015;29:56–65. doi: 10.1016/j.ymben.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Xu P. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U. S. A. 2014;111(31):11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y. Microbial biosynthesis of the anticoagulant precursor 4-hydroxycoumarin. Nat Commun. 2013;4:2603. doi: 10.1038/ncomms3603. [DOI] [PubMed] [Google Scholar]
- 9.Thodey K., Galanie S., Smolke C.D. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10(10):837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai M., Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15(1):1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Stephanopoulos G. Synthetic biology and metabolic engineering. ACS Synth Biol. 2012;1(11):514–525. doi: 10.1021/sb300094q. [DOI] [PubMed] [Google Scholar]
- 12.Lan E.I., Liao J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc Natl Acad Sci. 2012;109(16):6018–6023. doi: 10.1073/pnas.1200074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh A. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab Eng. 2011:76–81. doi: 10.1016/j.ymben.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Leonard E. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci U. S. A. 2010;107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelcbuch L. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013;41(9) doi: 10.1093/nar/gkt151. e98-e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juminaga D. Modular engineering of L-tyrosine production in Escherichia coli. Appl Environ Microbiol. 2012;78(1):89–98. doi: 10.1128/AEM.06017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anthony J. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2009;11(1):13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Bokinsky G. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci U. S. A. 2011;108(50):19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salis H., Mirsky E., Voigt C. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C., Jeffries T. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol. 2007;73(19):6072–6077. doi: 10.1128/AEM.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajikumar P. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du J. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012;40(18):e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramon A., Smith H.O. Single-step linker-based combinatorial assembly of promoter and gene cassettes for pathway engineering. Biotechnol Lett. 2011;33(3):549–555. doi: 10.1007/s10529-010-0455-x. [DOI] [PubMed] [Google Scholar]
- 24.Xu P. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- 25.Chen B., Lee D.-Y., Chang M.W. Combinatorial metabolic engineering of Saccharomyces cerevisiae for terminal alkene production. Metab Eng. 2015;31:53–61. doi: 10.1016/j.ymben.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Na D. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotech. 2013;31(2):170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 27.Santos C.N.S., Xiao W., Stephanopoulos G. Rational, combinatorial, and genomic approaches for engineering L-tyrosine production in Escherichia coli. Proc Natl Acad Sci. 2012;109(34):13538–13543. doi: 10.1073/pnas.1206346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu P. Improving metabolic pathway efficiency by statistical model-based multivariate regulatory metabolic engineering. ACS Synth Biol. 2016;6(1):148–158. doi: 10.1021/acssynbio.6b00187. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H. Algorithmic co-optimization of genetic constructs and growth conditions: application to 6-ACA, a potential nylon-6 precursor. Nucleic Acids Res. 2015;43(21):10560–10570. doi: 10.1093/nar/gkv1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong L. YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metab Eng Commun. 2017;5(Supplement C):68–77. doi: 10.1016/j.meteno.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox R., Surette M., Elowitz M. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007:3. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blazeck J. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol. 2011;77(22):7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michener J.K. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab Eng. 2011;14(3):212–222. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dueber J. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 35.Alper H. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U. S. A. 2005;102(36):12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coussement P. One step DNA assembly for combinatorial metabolic engineering. Metab Eng. 2014;23:70–77. doi: 10.1016/j.ymben.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Temme K. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012;40(17):8773–8781. doi: 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou S. Obtaining a panel of cascade promoter-5′-UTR complexes in Escherichia coli. ACS Synth Biol. 2017;6(6):1065–1075. doi: 10.1021/acssynbio.7b00006. [DOI] [PubMed] [Google Scholar]
- 39.Xu P. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E-coli. ACS Synth Biol. 2012;1(7):256–266. doi: 10.1021/sb300016b. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y.-J. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Meth. 2013;10(7):659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 41.Callura J.M. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci. 2010;107(36):15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green Alexander A. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159(4):925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucks J.B. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U. S. A. 2011;108(21):8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.