Abstract
Melanosome biogenesis and function were studied after purification of early stage melanosomes and characterization of specific proteins sorted to that organelle. Melanosomes were isolated from highly pigmented human MNT1 melanoma cells after disruption and initial separation by sucrose density gradient centrifugation. Low-density sucrose fractions were found by electron microscopy to be enriched in stage I and stage II melanosomes, and these fractions were further separated and purified by free flow electrophoresis. Tyrosinase and dopachrome tautomerase (DCT) activities were found exclusively in stage II melanosomes, even though DCT (and to some extent tyrosinase) proteins were sorted to stage I melanosomes. Western immunoblotting revealed that these catalytic proteins, as well as TYRP1, MART1, and GP100, were cleaved and inactivated in stage I melanosomes. Proteolytic cleavage was critical for the refolding of GP100 within the melanosomal milieu, and subsequent reorganization of amorphous stage I melanosomes into fibrillar, ovoid, and highly organized stage II melanosomes appears to stabilize the catalytic functions of melanosomal enzymes and allows melanin biosynthesis to begin. These results provide a better understanding of the structural features seen during melanosome biogenesis, and they yield further clues as to the physiological regulation of pigmentation.
Keywords: pigment, melanin, tyrosinase, melanoma
More than 95 distinct genes that play direct or indirect roles in mammalian pigmentation have been identified. Many of these genes encode proteins that are localized in melanosomes, specialized pigment organelles produced only by melanocytes. These gene products alter the quality or quantity of melanin produced and/or the processing and distribution of melanosomes. The known melanosomal proteins are involved in melanogenesis as catalytic and/or structural components. These include tyrosinase (TYR), the tyrosinase-related proteins-1 and -2 (TYRP1/TRP1 and DCT/TRP2, respectively; refs. 1–3), GP100/Pmel17/silver (4–6), and Pink (7–9). In addition, another protein, termed MART1 (melanoma antigen recognized by T lymphocytes), has been cloned that localizes in melanosomes (10, 11), but its function there remains unclear.
The advent of electron microscopy (EM) and the development of methods for subcellular fractionation allowed the site of melanin synthesis and deposition to be identified many years ago as the membrane-bound granule, termed a melanosome (12, 13). Based in part on those early studies, several stages in the biogenesis and maturation of melanosomes have since been delineated: stage I, originally termed a “premelanosome,” is a relatively spherical organelle with an amorphous matrix; in stage II, the organelle is ovoid and contains a fibrillar internal matrix; in stage III, the deposition of melanin on the melanosomal matrix becomes evident; and in stage IV, the organelle is completely filled with melanin (reviewed in ref. 14). However, the sequence in which melanosomal proteins are sorted to the organelle and the role(s) they play in its maturation remain largely unknown. Melanosomes are related to lysosomes, and both types of organelles evolve initially via the same pathway (15–20). Raposo et al. (21) recently reported that the intracellular processing of GP100 follows a unique route to form stage I melanosomes that diverges from conventional lysosomes just past the early endosome stage.
This study reports the initial phases of a long-term project aimed at mapping the melanosome, i.e., identifying and characterizing all of its components. Our goal is to purify and characterize early stage melanosomes, which have thus far resisted all efforts at purification because they cosediment on sucrose gradients with other membrane-bound subcellular organelles. To isolate these early stage melanosomes, we have used free-flow electrophoresis (FFE), a method used to separate lysosomes, endosomes, and other membrane-bound vesicles (22, 23). This FFE method allows the recovery of organelles and their constituent proteins in a form suitable for studying enzyme activity, morphology, cytochemistry, etc. (23). In addition, proteins in organelles separated by FFE can be analyzed by high-resolution PAGE, mass spectrometric microsequencing, and related techniques to identify novel components. This approach has provided important clues to melanosome biogenesis and function.
Materials and Methods
Cell Culture and Preparation of Homogenates.
MNT1 cells are highly pigmented human melanoma cells grown at 37°C in an atmosphere of 95% air/5% CO2 in 150-cm2 culture dishes in MEM (all culture reagents from Life Technologies, Grand Island, NY). The medium is supplemented to a final concentration of 20% heat-inactivated FBS (Atlanta Biologicals, Norcross, GA), 10% AIM-V medium, 20 mM Hepes, 1% antibiotic–antimycotic solution, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 2 mM l-glutamine, 30 ng/ml gentamycin, and 3.7 μg/ml sodium bicarbonate. Confluent monolayers of MNT1 cells were harvested with 0.05% trypsin/0.53 mM EDTA, and washed once in 0.25 M sucrose by centrifugation at 1,000 × g for 5 min at 4°C. They were then homogenized on ice by using 20 strokes of a glass/glass tissue grinder and centrifuged at 1,000 × g for 10 min at 4°C.
Antibodies.
αPEP1h, αPEP7h, αPEP8h, and αPEP13 antibodies were generated in rabbits against synthetic peptides corresponding to the carboxyl termini of human TYRP1, tyrosinase, DCT, and GP100, respectively, as described (2, 5, 24, 25). Other antibodies to melanosomal proteins used in this study included Clone M2–9E3, a mouse mAb against human MART1 (11) (Neomarkers, Fremont, CA), and HMB45, a mAb reactive against human GP100 (26) (Dako). We used other antibodies to check for subcellular organelles as follows: CD107a, a mouse mAb that reacts with the human lysosomal associated membrane protein (LAMP1), and Clone 113–1, a mouse anti-human mitochondrial antibody (Research Diagnosis, Fladres, NJ); and Bip/GRP78, GM130, and EEA1, which are monoclonal antibodies reactive with endoplasmic reticulum, Golgi apparatus, and early endosomes (Transduction Laboratories, Lexington, KY), respectively.
Preparation of Sucrose Density Gradient-Purified Melanosomes.
Melanosomes were purified by ultracentrifugation, as described (5, 27). Briefly, the cellular homogenate was layered on a discontinuous gradient of 1.0, 1.2, 1.4, 1.5, 1.6, 1.8, and 2.0 M sucrose (in 10 mM Hepes, pH 7.0) and centrifuged at 100,000 × g in a Beckman SW28 swinging-bucket rotor for 1 h at 4°C. Melanosomes that localized at various layers of the gradient were recovered by pipette, and were further purified by FFE, or were analyzed by EM and by Western immunoblotting as detailed below.
FFE.
Early melanosomes were recovered from the 1.0- to 1.2-M sucrose interface and were injected into the right inlet of an Octopus-PZE FFE apparatus (Weber, Kirchheim, Germany) at 2.0 ml/hr. FFE was performed at 1000–1100 V and ≈110–125 mA by using 0.25 M sucrose in triethanolamine, pH 7.4, as the chamber buffer and an elution flow rate of 3–4 ml/min. The temperature of the sample was kept at 4°C, and the chamber was maintained at 10°C. Fractions were collected and analyzed for enzyme activity, ultrastructure, immunoreactivity, and/or protein content, as detailed below.
Melanogenic Assays.
One milliliter of each FFE fraction was recovered and centrifuged at 14,000 × g for 30 min. The pellets were resuspended in 30 μl of extraction buffer [1% Nonidet P-40/0.01% SDS/0.1 M Tris⋅HCl, pH 7.2, and a protein inhibitor mixture (Roche Molecular Biochemicals)], vortexed, and kept at 4°C for 1 h. TYR and DCT activities were then measured as described (2, 28, 29).
EM.
Fractions to be examined were fixed overnight at 4°C in 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.3. The samples were then stored in PBS at 4°C until they were embedded in epoxy resin. Thin sections were cut, stained with uranyl acetate and lead citrate, and then examined with a Zeiss EM 912 electron microscope.
For immuno-EM, fractions were reacted for 1 h at 23°C with the primary antibody (at 1:100 dilution), washed twice in PBS, then incubated with the appropriate 10-nm gold-labeled goat anti-rabbit or anti-mouse IgG (at 1:25 dilution) for 1 h at 4°C. Controls included samples with no primary antibody. The fractions were washed again twice in PBS, then fixed and processed for EM as described above.
Western Immunoblotting Analysis.
Proteins (5 μg of protein per well) from sucrose density gradient fractions and from FFE fractions were separated on 8% polyacrylamide or 14% SDS gels, and were transferred electrophoretically to poly(vinylidene difluoride) membranes (Immobilon-P; Millipore). The blots were incubated with primary antibodies (at 1/500 or 1/1000 dilution) for 1 h at 23°C and washed several times; subsequent visualization of antibody binding was carried out with enhanced chemiluminescence (ECL; Amersham Pharmacia), according to the manufacturer's instructions.
Confocal Microscopy.
MNT1 cells were plated in 2-well Lab-Tek chamber slides (Nalge Nunc, Naperville, IL) 24 or 48 h before each experiment, and were then fixed in 4% paraformaldehyde for 15 min at 4°C. After washing in PBS, the cells were incubated in PBS containing 10% normal goat serum and 10% normal horse serum for 1 h at 23°C as a blocking step. The cells were then incubated in PBS containing 5% normal goat and horse serum and a mixture of monoclonal and polyclonal antibodies overnight at 4°C. This was followed by incubation with the corresponding secondary antibodies, i.e., goat anti-rabbit IgG labeled with Texas red (Vector, dilution, 1:100) and horse anti-mouse labeled with fluorescein isothiocyanate (Vector; dilution, 1:100). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. All preparations were examined with a confocal microscope (model TCS4D/DMIRBE; Leica, Heidelberg, Germany) equipped with argon and argon–krypton laser sources. Controls included sections stained as detailed above, but omitting the first antibody.
Miscellaneous.
Protein concentrations were usually determined with the bicinchoninic acid (BCA) assay kit (Pierce), using BSA as a standard, except for FFE fractions analyzed for TYR assay that were determined by the Bio-Rad protein assay (Bio-Rad).
Results
Purification of Early Melanosomes.
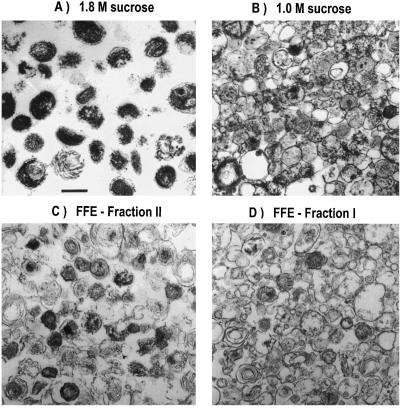
Sucrose density-gradient ultracentrifugation has been known for some time as a successful method to prepare highly purified, melanin-laden melanosomes (5, 12). However, because melanin might impair the solubilization of melanosomal components, we were interested in purifying early stage melanosomes to maximize the opportunity to detect novel proteins by using high-sensitivity mass spectrometric analysis. As we previously reported (5), the dense sucrose fractions contained essentially only stage III and stage IV melanosomes (Fig. 1A). Ultrastructural analysis of bands higher in the sucrose gradient revealed that progressively fewer melanized melanosomes were seen in less dense fractions. The 1.6 and 1.4 M sucrose fractions contained essentially stage III melanosomes, with some stage II and stage IV melanosomes and other vesicular contaminants (data not shown). The 1.0 and 1.2 M sucrose fractions contained the majority of stage I and stage II melanosomes, along with many other membrane-bound organelles, including mitochondria (Fig. 1B).
Figure 1.
Ultrastructure of sucrose density gradient and FFE purified fractions. (A) The 1.8 M sucrose fraction. (B) The 1.0 M sucrose fraction. (C) Fraction II, purified from the 1.0 M sucrose fraction by FFE. (D) Fraction I, protein-rich fraction purified from the 1.0 M sucrose fraction by FFE. (Bar = 0.5 μm.)
We used Western blotting to examine the distribution of melanosomal proteins over the entire sucrose density gradient (summarized in the upper portion of Table 1). Such analysis revealed, as might be expected, that TYR was significantly more abundant in the denser part of the gradient, where the more pigmented melanosomes were found. Surprisingly, the distribution of other melanosomal proteins differed from TYR. DCT, MART1, and GP100 (as detected by HMB45) were evenly distributed throughout the gradient, whereas TYRP1 and GP100 (as detected by αPEP13) were actually more abundant in fractions containing earlier-stage melanosomes, and were detected at minor or negligible levels in stage III and IV melanosomes.
Table 1.
Distribution of melanosomal proteins by Western immunoblotting
| Fraction | Stage | Tyrosinase (αPEP7h) | TYRP1 (αPEP1h) | DCT (αPEP8h) | GP100 (αPEP13) | MART1 (M2-9E3) | GP100 (HMB45) | Lysosome (CD107a) | ER (Bip) | Mitochondria (113-1) | Golgi (GM130) | Endosome (EEA1) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homogenate | I–IV | ++ | + | + | + | + | + | + | + | + | ND | ND | ||
| 1.0 M sucrose | I, II | + | + | + | + | + | + | + | + | + | − | − | ||
| 1.2 M sucrose | I, II | + | ++ | ± | ++ | ± | − | ± | + | +++ | ND | ND | ||
| 1.4 M sucrose | II, III | ++ | + | + | + | ± | + | + | + | ++ | ND | ND | ||
| 1.6 M sucrose | III, IV | +++ | ± | + | − | ± | + | + | + | ± | ND | ND | ||
| 1.8 M sucrose | III, IV | +++ | ± | + | − | ± | ± | + | + | ± | ND | ND | ||
| 1.0 M sucrose | I, II | + | + | + | + | + | + | + | + | + | − | − | ||
| FFE Fx 29–31 | II | ++ | − | + | − | ++ | ++ | ++ | ++ | − | − | − | ||
| FFE Fx 32–34 | II | ++ | − | ++ | − | ++ | ++ | ++ | + | − | − | − | ||
| FFE Fx 35–37 | II | ++ | − | ++ | − | +++ | + | ++ | ND | ± | ND | ND | ||
| FFE Fx 38–40 | II | ++ | − | + | + | +++ | + | ++ | ++ | + | − | − | ||
| FFE Fx 41–43 | I | + | +* | +* | ++ | ++ | − | + | +++ | +++ | − | − | ||
| FFE Fx 44–46 | I | − | +* | +* | +++ | − | − | + | ND | +++ | ND | ND | ||
| FFE Fx 47–49 | I | − | ++* | ±* | +++ | − | − | + | +++ | +++ | − | − | ||
| FFE Fx 50–52 | I | − | ++* | ±* | +++ | − | − | + | +++ | +++ | − | − |
Antibodies are given in parentheses. nd = not done; Fx, fraction.
Cleaved low molecular weight bands recognized.
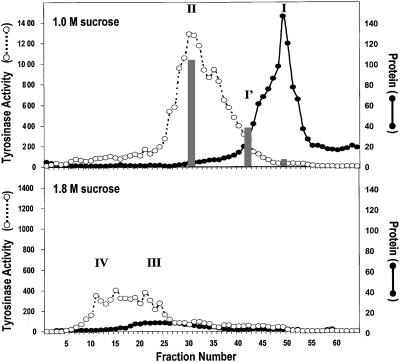
From these results it was clear that the 1.0 and 1.2 M sucrose fractions contained the early melanosomes of interest. We then further separated these fractions by FFE (taking advantage of charge differences in the organelles) and examined the distribution of proteins and melanogenic activities (Fig. 2 Upper). Results for the 1.0 M sucrose fraction are presented throughout this report, but essentially identical results were obtained with the 1.2-M sucrose fraction. The bulk of proteins eluted at very distinct and discrete parts of the gradient, which were separated from the catalytic activities. The TYR- and DCT-rich fraction II consisted almost exclusively of stage II melanosomes, and eluted more toward the anode than did the general protein fraction I (EM micrographs of those fractions are shown in Fig. 1 C and D). The protein-rich fraction I contained mitochondria, endoplasmic reticulum (ER), and other membrane-bound organelles, including stage I melanosomes. The specific activity of TYR was enriched about 80-fold in fraction II, compared with the 1.0 M sucrose starting material, with 40% recovery of enzyme activity. DCT activity showed a similar distribution pattern to TYR, most of that catalytic activity eluting in fraction II; DCT and TYR activities in the protein-rich fraction I were at background levels.
Figure 2.
Distribution curves of TYR and proteins after FFE. (Upper) Elution of the 1.0 M sucrose layer separated by FFE. The broken line with open symbols represents TYR enzyme activity. DCT activity is shown as the gray bars, and it was assessed by HPLC. The solid line with closed symbols shows protein content. Fraction I (at fraction no. 49) TYR activity was 24.8 cpm/hr per 1 μg of protein, DCT was at background, and protein was 146.0 μg/ml. Fraction II (at fraction no. 30) TYR activity was 1289.8 cpm/hr per 1 μg of protein, DCT activity was 25.2 pmol/hr per 1 μg of protein, and protein was 3.7 μg/ml. Fraction I′ was taken at the point where the TYR and protein elution profiles intersect. (Lower) Elution of FFE fractions derived from the 1.8-M sucrose layer; lines and symbols are as reported for the Upper panel.
Distribution of Melanosomal Proteins After Purification by FFE.
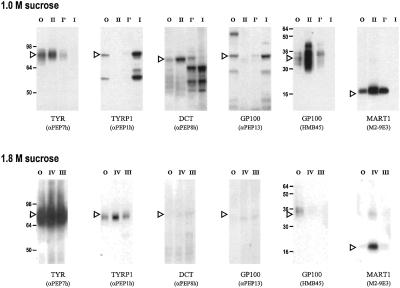
Western blot analysis of the distribution of melanosomal proteins in the 1.0-M sucrose starting material and in the FFE-purified fractions I, I′, and II are shown in Fig. 3 Upper and are summarized in the bottom half of Table 1. TYR, GP100 (as recognized by HMB45), and MART1 localization in the fractions coincided with the profile of TYR and DCT enzymatic activities, i.e., they were virtually negative in fraction I and were highly concentrated in fraction II. In contrast, GP100 (as detected by αPEP13) and TYRP1 were abundant in fraction I and were detectable at much lower or negligible levels in fraction II. DCT was actually detectable in all fractions assessed, but it was intact in fraction II, and was found as lower molecular weight and cleaved fragments in fraction I. Because the catalytic function of DCT was detectable only in fraction II whereas the bulk of its protein (present as cleaved bands) was found in fraction I, it is clear that its catalytic activity is impaired by that cleavage and is functional only after the transition from stage I to stage II melanosomes. It is of further interest to note that GP100, as detected by αPEP13 in stage I melanosomes (fraction I), was full-length (i.e., ≈80 kDa), whereas GP100, as detected by HMB45 in stage II melanosomes (fraction II), shows a smaller mass (≈33 kDa). Approximately 50% of immunoreactive TYRP1 is cleaved in the stage I melanosomes present in fraction I.
Figure 3.
Immunoblot analysis of sucrose gradient and FFE-purified fractions. Equal quantities of protein (5 μg) were subjected to SDS/PAGE and immunoblotting. (Upper) O, original starting material (1.0 M sucrose fraction); I, I′, and II as detailed for Fig. 2. Primary antibodies used were as noted in parentheses; exposure time, ≈30 sec. Numbers at the left indicate the migration of known molecular mass markers (in kDa). Arrowheads indicate the migration positions of full-length proteins. (Lower) Western blots for the 1.8 M sucrose fraction separated by FFE; exposure time, ≈2 min (about 5 times longer); O, original starting material (1.8 M sucrose fraction), and fractions III and IV were as noted for Fig. 2.
When the heavily pigmented 1.8 M sucrose fraction was separated by FFE (Fig. 2 Lower), a broad peak that eluted even closer to the anode was found (presumably because of the higher quantities of charged melanins present). All detectable protein was eluted from the gradient in the TYR-positive fractions III and IV, and no protein-rich fraction was eluted later in the gradient, which demonstrates the highly purified nature of melanosomes in this fraction. The distribution of melanosomal proteins in those fractions (Fig. 3 Lower) was as expected, based on analysis of the sucrose gradient shown above. The levels of TYR activity in these fractions were significantly lower than those derived from the 1.0-M sucrose fraction, and the Western blots shown in Fig. 3 Lower were exposed ≈5 times longer than those in the top half to visualize those bands. TYR was by far the most abundant melanosomal protein found in these melanized melanosomes; TYRP1 and MART1 were present at minor levels in the most pigmented fraction IV, whereas DCT and GP100 were just barely detectable. TYR, TYRP1, DCT, MART1, and GP100 in fractions III and IV were full-length proteins when detectable.
Evaluation of these FFE fractions by using antibodies specific for other organelles was also highly informative (summarized in the five right columns of Table 1). LAMP1 (detected by CD107a) is a marker for organelles of the endosomal–lysosomal lineage (30, 31). Antibodies to LAMP1 were reactive with all FFE fractions, but were especially reactive with the TYR-rich fraction II. Antibodies specific for the ER (Bip/GRP78, a chaperone in the ER lumen) also reacted with all FFE fractions, but reactivity was strongest with the general protein fraction I. Nevertheless, significant reactivity with ER markers was seen in the purified stage II melanosomes and also in the highly purified 1.8 M sucrose fraction. Staining for a mitochondrial marker was positive for fraction I and was negative for fraction II, which corroborated our EM observations on the large mitochondrial component of that fraction (Fig. 1). None of the FFE-purified fractions were recognized by antibodies against a Golgi marker (GM130, a structural element of the Golgi apparatus) or an early endosome marker (EEA1, an early endosome peripheral membrane protein).
Immuno-EM.
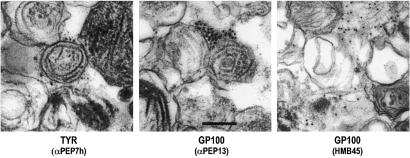
The two antibodies that recognize GP100 (i.e., αPEP13 and HMB45) show radically different reactivity patterns. The epitope for HMB45 is on an interior motif of the protein whereas αPEP13 recognizes the carboxyl terminus (5, 6, 33). Previous metabolic labeling and pulse–chase analysis combined with sucrose density gradient subcellular fractionation previously showed that processing of GP100 is very quick compared with the TYRPs, and that it is not processed through the trans-Golgi network, but follows a route distinct from the TYRPs to be sorted to melanosomes (5). Almost immediately after GP100 is sorted to the melanosome, its carboxyl terminus (and membrane-spanning motif) is cleaved, which releases the bulk of the protein into the intramelanosomal milieu. This fragment was postulated to be associated with, and perhaps even become, the fibrillar melanosomal matrix, a conclusion further supported by the recent study of Raposo et al. (21). We thus used immuno-EM to confirm the specificity of epitope recognition by these antibodies.
The immunoreactivity of αPEP7h shows that, indeed, gold particles decorate the exterior face of the melanosomal membrane (Fig. 4). The number of labeled melanosomes is particularly evident in FFE fraction II, which contains the majority of stage II melanosomes but can also be seen in a few melanosomes in fraction I. Reactivity with αPEP13 shows a similar staining pattern on the exterior surface of stage I and II melanosomal membranes in fraction I (and to some extent in fraction II, where they could be found occasionally). In sharp contrast, reactivity with HMB45 is seen only associated with the internal matrix of melanosomes (primarily in fraction II) whose limiting membrane has been disrupted. No gold particles were seen in control samples, in which the primary antibody was omitted. It is clear that αPEP13 recognizes the carboxyl tail of GP100 on the cytosolic side of stage I melanosomes, whereas HMB45 specifically recognizes the luminal fragment of GP100, but only after it has been cleaved and is incorporated or associated with the fibrillar matrix of stage II melanosomes.
Figure 4.
Immuno-EM gold staining of FFE fractions. Immunostaining of FFE fraction II. (Left) Stained with αPEP7h for TYR. (Center) stained with αPEP13 for GP100. (Right) Stained with HMB45 for GP100. (All sections, ×25,000.)
Confocal Microscopy.
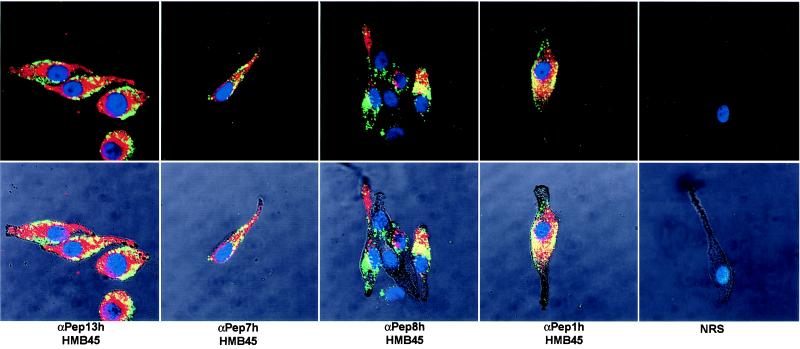
The above results demonstrate that the de novo form of GP100 (recognized by αPEP13) is associated with stage I melanosomes, and that the processed, lower molecular weight form of GP100 (recognized by HMB45) is found only in stage II melanosomes. We therefore used dual labeling and confocal microscopy to confirm this point (Fig. 5). It is clear from the merged image that particulate melanosomes in MNT1 melanoma cells stain with αPEP13 (red) or with HMB45 (green), but only rarely with both (yellow). In general, the stage I melanosomes are found in the perinuclear region, whereas the stage II melanosomes are more toward the periphery of the cells. However, αPEP13-reactive melanosomes are also found in the dendrites, suggesting that the small transmembrane fragment cleaved from GP100 remains recognizable by the αPEP13 antibody as the melanosome continues to mature. TYR, TYRP1, and DCT are all found in the perinuclear area (i.e., in stage I melanosomes) and colocalize with GP100 (recognized by HMB45) in stage II melanosomes. Interestingly, heavily pigmented melanosomes in the dendrites show little or no staining with these antibodies, demonstrating that the epitopes are lost or are masked by melanin deposition.
Figure 5.
Confocal microscopy localization of melanosomal proteins. Antibody reactivity patterns were examined by dual labeling confocal microscopy. The following antibodies were used and detected by red fluorescence: αPEP13 (recognizing GP100, at a dilution of 1:40), αPEP7h (recognizing TYR, 1:20), αPEP8h (recognizing DCT, 1:20), αPEP1h (recognizing TYRP1, 1:20), and normal rabbit serum (NRS; 1:20) as a control. Detected by green fluorescence was HMB45 (recognizing GP100, at 1:10). (Upper) The merged images of dual staining, (yellow indicates colocalization of the signal); nuclei are stained blue with 4′,6-diamidino-2-phenylindole. Lower shows the Nomarski image and allows the cellular structure and unstained pigmented melanosomes to be seen.
Discussion
The melanosomal proteins identified and cloned to date all play important roles in the pigment function of melanosomes (as enzymes or as structural proteins), as recently reviewed in refs. 34 and 35. Genetic evidence suggests that many more remain to be discovered, and the fact that virtually all of the cloned pigment genes have been associated with human hereditary pigment diseases makes the identification and characterization of the remaining genes even more critical. Studies performed some time ago outlined the two distinct sorting pathways involved in melanosome formation, i.e., that enzymatic components were sorted to melanosomes by means of coated vesicles, whereas its structural components originated directly from the ER and/or early Golgi. This scheme has been gradually revised as the complex nature of adapter protein complexes and the sorting mechanisms they direct has evolved. It is now known that TYR (and perhaps other melanosomal proteins) are sorted by means of the AP3 system to early endosomes, then to late endosomes, and eventually to melanosomes (16, 36–39).
The results reported herein provide a better understanding of how the biogenesis of melanosomes and pigment synthesis is regulated. Each of the six melanosomal proteins has been known for some time to be detectable in mature melanosomes, but the sequence and timing of their sorting to that organelle during its formation as well as the processing that they undergo during its maturation have been a matter of conjecture. Our data demonstrate clearly that GP100 is sorted initially to stage I melanosomes, and that the other known melanosomal proteins are detectable there as well. However, these melanosomal proteins are degraded or partially cleaved, probably because of the highly proteolytic nature of the enzymes present in that milieu (15). Our results demonstrate that, for DCT and TYR, such degradation impairs catalytic function and melanin synthesis is inhibited. In the case of GP100, the proteolytic cleavage is essential to its processing from a membrane-bound form to the “free” form that then becomes part of the fibrillar component of the melanosome. Our results show clearly that the cleavage and processing of GP100 accompanies the restructuring of early melanosomes from amorphous rounded vesicles (stage I) into elongated fibrillar structures (stage II). From the absence of melanin and full-length catalytic proteins, it is clear that all melanogenic enzymes found in stage I melanosomes are effectively cleaved and inactivated. It is only after the transition to stage II melanosomes that the enzymes become resistant to proteolysis and melanin production ensues.
It is at this crucial point, when GP100 is processed to its soluble form and the melanogenic enzymes become active within the melanosome, that melanin synthesis begins. Initially, synthesis proceeds at a relatively slow rate that is sufficient to give the organelle the charge that allows it to be separated by FFE, and to be seen in fraction pellets and by EM. Other studies by our laboratory have demonstrated clearly that TYRP1 and DCT interact with TYR in a complex, and that this complex is formed early during their processing in the ER (40). MART1 is also delivered to the melanosome at this time, and its function there has so far defied all efforts at identification. However, the close relationship in the expression patterns and subcellular localization of the melanogenic enzymes and MART1 suggests that MART1 may have an important function in the melanosome.
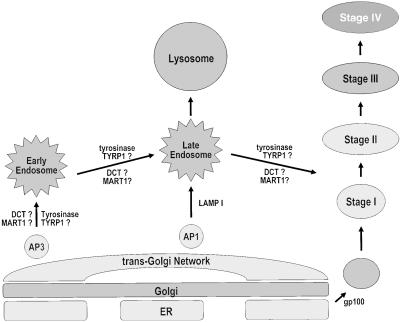
Fig. 6 illustrates our current understanding of melanosome biogenesis. Melanosomes are unique organelles by virtue of being produced in melanocytes, which express specific proteins not produced by other types of cells. The earliest form of melanosome (stage I) is structurally identical to an early endosome (15). Raposo et al. (21) have proposed that this earliest stage of melanosome is a unique component of the endosomal organelle pathway, and we would further delineate that as being derived from the ER/Golgi network. The presence of ER-specific marker proteins in all stages of melanosomes further supports this scheme, and is consistent with all other observations reported in this study. TYR (and perhaps other melanosomal proteins) is sorted to melanosomes by the AP3 system by means of early and late endosomes. The fusion of late endosomes bearing melanosomal components would give rise to their increasing size as they develop. These same early melanosomes are devoid of TYR and DCT catalytic activity, and thus no melanin can be formed. Melanosomal proteins incorporated into stage I melanosomes are cleaved (probably by endogenous proteases within), and the amino-terminal fragments are released into the internal matrix. It is at this time that several events occur rapidly: (i) the internal structure of stage I melanosomes is reorganized into the ellipsoidal, fibrillar structure characteristic of stage II melanosomes; (ii) GP100 now becomes recognizable by HMB45 (because of its conformational change into the melanosomal matrix protein); and (iii) TYR and DCT catalytic proteins become functional and melanin synthesis proceeds. Stage III and IV melanosomes are distinguished solely on the extent of melanin deposited on the fibrillar matrix.
Figure 6.
Model for melanosomal biogenesis and protein localization. Schematic of a melanocyte and the major stages of endosome/lysosome/melanosome biogenesis. The scheme is adapted from Bonifacino and Dell'Angelica (16), Raposo et al. (21), and Manga et al. (41).
Thus, in sum, the proteolytic processing of melanosomal proteins once they arrive at the de novo early melanosome (stage I) plays a crucial role in delaying catalytic function and melanin production until the melanosome has reorganized into a more mature organelle. Many cytotoxic intermediates are generated during melanin synthesis, and the melanosome must be competent to contain those derivatives to maintain viability of the cell. Delaying melanin synthesis until the melanosome reorganizes may be important to its capacity to contain those toxic intermediates. Future study should be directed to identifying other novel components of melanosomes and in characterizing their function in this unique organelle.
Abbreviations
- DCT
dopachrome tautomerase
- TYR
tyrosinase
- TYRP1
tyrosinase-related protein-1
- FFE
free-flow electrophoresis
- EM
electron microscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hearing V J, Tsukamoto K. FASEB J. 1991;5:2902–2909. [PubMed] [Google Scholar]
- 2.Tsukamoto K, Jackson I J, Urabe K, Montague P M, Hearing V J. EMBO J. 1992;11:519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon B S. J Invest Dermatol. 1993;100:134S–140S. doi: 10.1111/1523-1747.ep12465022. [DOI] [PubMed] [Google Scholar]
- 4.Kwon B S, Chintamaneni C D, Kozak C A, Copeland N G, Gilbert D J, Jenkins N A, Barton D E, Francke U, Kobayashi Y, Kim K K. Proc Natl Acad Sci USA. 1991;88:9228–9232. doi: 10.1073/pnas.88.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T, Urabe K, Orlow S J, Higashi K, Imokawa G, Kwon B S, Potterf S B, Hearing V J. J Biol Chem. 1994;269:29198–29205. [PubMed] [Google Scholar]
- 6.Zhou B K, Kobayashi T, Donatien P D, Bennett D C, Hearing V J, Orlow S J. Proc Natl Acad Sci USA. 1994;91:7076–7080. doi: 10.1073/pnas.91.15.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner J M, Nakatsu Y, Gondo Y, Lee S, Lyon M F, King R A, Brilliant M H. Science. 1992;257:1121–1124. doi: 10.1126/science.257.5073.1121. [DOI] [PubMed] [Google Scholar]
- 8.Rinchik E M, Bultman S J, Horsthemke B, Lee S T, Strunk K M, Spritz R A, Avidano K M, Jong M T C, Nicholls R D. Nature (London) 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 9.Rosemblat S, Durham-Pierre D, Gardner J M, Nakatsu Y, Brilliant M H, Orlow S J. Proc Natl Acad Sci USA. 1994;91:12071–12075. doi: 10.1073/pnas.91.25.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Rivoltini L, Topalian S L, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawakami Y, Battles J K, Kobayashi T, Wang X, Tupesis J L, Marincola F M, Robbins P F, Hearing V J, Gonda M A, Rosenberg S A. J Immunol Methods. 1997;202:13–25. doi: 10.1016/s0022-1759(96)00211-6. [DOI] [PubMed] [Google Scholar]
- 12.Seiji M, Shimao K, Birbeck M S C, Fitzpatrick T B. Ann NY Acad Sci. 1963;100:497–533. [PubMed] [Google Scholar]
- 13.Seiji M, Fitzpatrick T B, Birbeck M S C. J Invest Dermatol. 1961;36:243–252. doi: 10.1038/jid.1961.42. [DOI] [PubMed] [Google Scholar]
- 14.Nordlund J J, Boissy R E, Hearing V J, King R A, Ortonne J P. The Pigmentary System: Physiology and Pathophysiology. New York: Oxford Univ. Press; 1998. [Google Scholar]
- 15.Diment S, Eidelman M, Rodriguez G M, Orlow S J. J Biol Chem. 1995;270:4213–4215. doi: 10.1074/jbc.270.9.4213. [DOI] [PubMed] [Google Scholar]
- 16.Bonifacino J S, Dell'Angelica E C. J Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlow S J. J Invest Dermatol. 1995;105:3–7. doi: 10.1111/1523-1747.ep12312291. [DOI] [PubMed] [Google Scholar]
- 18.Swank R T, Novak E K, McGarry M P, Zhang Y, Li W, Zhang Q, Feng L. Pigm Cell Res. 2000;13:59–67. doi: 10.1034/j.1600-0749.13.s8.12.x. [DOI] [PubMed] [Google Scholar]
- 19.Spritz R A. Trends Genet. 1999;15:337–340. doi: 10.1016/s0168-9525(99)01785-0. [DOI] [PubMed] [Google Scholar]
- 20.Dell'Angelica E C, Mullins C, Caplan S, Bonifacino J S. FASEB J. 2000;14:1265–1278. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- 21.Raposo G, Tenza D, Murphy D M, Berson J F, Marks M S. J Cell Biol. 2001;152:809–823. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh M, Schmid S, Kern H, Harms E, Male P, Mellman I, Helenius A. J Cell Biol. 1987;104:875–886. doi: 10.1083/jcb.104.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid S L, Fuchs R, Male P, Mellman I. Cell. 1988;52:73–83. doi: 10.1016/0092-8674(88)90532-6. [DOI] [PubMed] [Google Scholar]
- 24.Jiménez M, Tsukamoto K, Hearing V J. J Biol Chem. 1991;266:1147–1156. [PubMed] [Google Scholar]
- 25.Virador V, Matsunaga N, Matsunaga J, Valencia J, Oldham R J, Kameyama K, Peck G L, Abdel-Malek Z A, Hearing V J. Pigm Cell Res. 2001;14:289–297. doi: 10.1034/j.1600-0749.2001.140410.x. [DOI] [PubMed] [Google Scholar]
- 26.Gown A M, Vogel A M, Hoak D, Gough F, McNutt M A. Am J Pathol. 1984;123:195–203. [PMC free article] [PubMed] [Google Scholar]
- 27.Gahl W A, Potterf S B, Durham-Pierre D, Brilliant M H, Hearing V J. Pigm Cell Res. 1995;8:229–233. doi: 10.1111/j.1600-0749.1995.tb00668.x. [DOI] [PubMed] [Google Scholar]
- 28.Hearing V J, Ekel T M. Biochem J. 1976;157:549–557. doi: 10.1042/bj1570549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hearing V J, Ekel T M, Montague P M, Nicholson J M. Biochim Biophys Acta. 1980;611:251–268. doi: 10.1016/0005-2744(80)90061-3. [DOI] [PubMed] [Google Scholar]
- 30.Kornfeld S, Mellman I. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig T, Griffiths G, Hoflack B. J Cell Biol. 1991;115:1561–1572. doi: 10.1083/jcb.115.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlow S J, Boissy R E, Moran D J, Pifko-Hirst S. J Invest Dermatol. 1993;100:55–64. doi: 10.1111/1523-1747.ep12354138. [DOI] [PubMed] [Google Scholar]
- 33.Adema G J, de Boer A J, Vogel A M, Loenen W A M, Figdor C G. J Biol Chem. 1994;269:20126–20133. [PubMed] [Google Scholar]
- 34.Hearing V J. J Invest Dermatol Symp Proc. 1999;4:24–28. doi: 10.1038/sj.jidsp.5640176. [DOI] [PubMed] [Google Scholar]
- 35.del Marmol V, Beermann F. FEBS Lett. 1996;381:165–168. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- 36.Vijayasaradhi S, Xu Y, Bouchard B, Houghton A N. J Cell Biol. 1995;130:807–820. doi: 10.1083/jcb.130.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honing S, Sandoval I V, von Figura K. EMBO J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setaluri V. Pigm Cell Res. 2000;13:128–134. doi: 10.1034/j.1600-0749.2000.130302.x. [DOI] [PubMed] [Google Scholar]
- 39.Jimbow K, Park J S, Kato F, Hirosaki K, Toyofuku K, Hua C, Yamashita T. Pigm Cell Res. 2000;13:222–229. doi: 10.1034/j.1600-0749.2000.130403.x. [DOI] [PubMed] [Google Scholar]
- 40.Toyofuku, K., Wada, I., Valencia, J. C., Kushimoto, T., Ferrans, V. J. & Hearing, V. J. (2001) FASEB J., in press. [DOI] [PubMed]
- 41.Manga P, Samaraweera P, Rosenberg B, Orlow S J. Pigm Cell Res. 2001;14:243–248. doi: 10.1034/j.1600-0749.2001.140403.x. [DOI] [PubMed] [Google Scholar]