The two IDA-LIKE signalling peptides, IDL6 and IDL7, suppress expression of a number of stress-related transcription factors; functional studies suggest a role in modulation of stress-induced ROS signalling.
Keywords: Abiotic stress, Arabidopsis, IDA-LIKE, peptide ligand, ROS, transcriptome, ZAT12
Abstract
Small signalling peptides have emerged as important cell to cell messengers in plant development and stress responses. However, only a few of the predicted peptides have been functionally characterized. Here, we present functional characterization of two members of the IDA-LIKE (IDL) peptide family in Arabidopsis thaliana, IDL6 and IDL7. Localization studies suggest that the peptides require a signal peptide and C-terminal processing to be correctly transported out of the cell. Both IDL6 and IDL7 appear to be unstable transcripts under post-transcriptional regulation. Treatment of plants with synthetic IDL6 and IDL7 peptides resulted in down-regulation of a broad range of stress-responsive genes, including early stress-responsive transcripts, dominated by a large group of ZINC FINGER PROTEIN (ZFP) genes, WRKY genes, and genes encoding calcium-dependent proteins. IDL7 expression was rapidly induced by hydrogen peroxide, and idl7 and idl6 idl7 double mutants displayed reduced cell death upon exposure to extracellular reactive oxygen species (ROS). Co-treatment of the bacterial elicitor flg22 with IDL7 peptide attenuated the rapid ROS burst induced by treatment with flg22 alone. Taken together, our results suggest that IDL7, and possibly IDL6, act as negative modulators of stress-induced ROS signalling in Arabidopsis.
Introduction
Plants cannot escape their environment, and are thus exposed to herbivore grazing, pathogen attacks, and other environmental perturbations such as drought, temperature changes, and high salinity. Plants have evolved sophisticated mechanisms in order to meet these challenges. At the cellular level, environmental cues are perceived either directly or indirectly and transduced through a complex signalling network, resulting in an appropriate response by changing the chemical environment of the cells. This transduction typically involves alternations of secondary messengers and signalling molecules such as calcium and phytohormones, but also reactive oxygen species (ROS).
Most types of environmental stresses such as wounding, drought, salinity, heat, cold, and pathogen attack lead to early and rapid accumulation of ROS (Miller et al., 2008; Torres, 2010; Sierla et al., 2013; Gilroy et al., 2014), known as the oxidative burst. Depending on the type of stress and the host involved, the oxidative burst relies on different types of enzymes, including cell wall peroxidases (Bindschedler et al., 2006) and plasma membrane NADPH oxidases (Torres et al., 2002; Miller et al., 2009; Torres, 2010). NADPH oxidases, often referred to as respiratory burst oxidase homologues (Rbohs), are transmembrane proteins responsible for the production of extracellular superoxide (O2·−) upon pathogen attack (Torres et al., 2002) or abiotic stresses (Kwak et al., 2003; Miller et al., 2009). Both abiotic and biotic stresses trigger a systemic autopropagating wave of ROS, mediated by the NADPH oxidase RBOHD, that travels rapidly in the apoplast from the affected tissue to the entire plant and activates a systemic response to the stress (Miller et al., 2009; Dubiella et al., 2013; Gilroy et al., 2014).
External stress triggers and activates intercellular signalling, essential for cell to cell communication. Since the discovery of systemin in tomato (Pearce et al., 1991), small post-translationally modified peptides have been recognized as important mediators of cell to cell communication (Butenko et al., 2009; Matsubayashi, 2014). They are characterized as small proteins with an N-terminal signal peptide directing the protein to the secretory pathway, a variable region, and a conserved C-terminal part containing the active peptide that after translation undergoes a series of modifications (Matsubayashi, 2014). The translated propeptide is proteolytically processed into the shorter active peptide, which may be further modified (Murphy et al., 2012) before the mature peptide can bind its receptor(s). One well-studied peptide is CLAVATA3 (CLV3) (Clark et al., 1995; Fletcher et al., 1999). Mature CLV3 is modified with two hydroxyprolines; one of these is further modified with three arabinose molecules, enhancing the binding affinity of the peptide for its receptor (Ohyama et al., 2009).
Although several peptides have been linked to growth and development, there are a few studies in Arabidopsis linking signalling peptides to plant stress responses. In Arabidopsis, the first defence-related peptide identified was the plant elicitor peptide (Pep) AtPep1, derived from the precursor peptide PROPEP1 (Huffaker et al., 2006). AtPep1 binds the two leucine repeat-rich receptor-like kinases (LRR-RLKs) PEPR1 and PEPR2 (Yamaguchi et al., 2006, 2010), and acts as a damage-associated molecular pattern (DAMP) promoting the expression of pathogen defence genes such as PLANT DEFENSIN 1.2 (PDF1.2) and PATHOGEN-RELATED 1 (PR1), thus amplifying the defence response upon pathogen attack (Yamaguchi et al., 2006, 2010). The growth-promoting phytosulfokine (PSK) peptide has also been linked to plant defence. It has been shown that PSK through its receptor PSK RECEPTOR 1 (PSKR1) attenuates pattern-trigged immunity in Arabidopsis, suggesting that PSK/PSKR1 is part of a mechanism that controls the allocation of resources between growth and immunity (Igarashi et al., 2012). Knockout plants of the C-TERMINALLY ENCODED PEPTIDE family member CEP3 show increased root and shoot growth compared with the wild type under several abiotic stress conditions, suggesting a role for CEP3 as a negative regulator of growth in response to changing environmental conditions (Delay et al., 2013). Overexpression of oxidative stress-induced peptide OSIP108 resulted in increased tolerance to the ROS inducer paraquat (De Coninck et al., 2013), and proteolytic processing of apoplastic GRIM REAPER by METACASPASE-9 produced an 11 amino acid ligand for its receptor PRK5 that is active in RbohD-dependent cell death (Wrzaczek et al., 2015). The PAMP-induced secreted peptide 1 (PIP1) and 2 (PIP2) have been shown to be induced by a variety of pathogens and elicitors, and PIP1 was found to amplify the immune response in a PEP1-like fashion through the LRR-RLK RLK7 (Hou et al., 2014). Finally, the cysteine-rich peptide AtCAPE1 confers salt sensitivity, in line with a negative regulatory role in salt stress tolerance (Chien et al., 2015).
We have recently performed a genome-wide screen for genes encoding small signalling peptides with similarity to INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) in Arabidopsis thaliana (Vie et al., 2015). IDA belongs to a family of nine genes, where IDA-LIKE 1 (IDL1) to IDL5 have been postulated to play roles during plant development (Butenko et al., 2003; Stenvik et al., 2008). Two newly discovered members of this subfamily, IDL6 and IDL7, are closely related, and expression analysis revealed that these two genes are induced by various stress treatments (Vie et al., 2015). In this study, we aimed to characterize the putative ligands IDL6 and IDL7 and their roles in modulating stress responses.
Materials and methods
Plant material
Seeds of the Arabidopsis thaliana ecotype Col-0 (N1092), idl6-1 (SALK_074245), idl6-2 (SALK_126026) (Alonso et al., 2003), and idl7 (WDL293-296; Woody et al., 2007) mutants were obtained from the European Arabidopsis Stock Centre (NASC, Nottingham, UK), and T-DNA insertions were confirmed by PCR using gene- and T-DNA-specific primers (see Supplementary Table S1 at JXB online). Verified homozygous lines were back-crossed to Col-0 wild type to ensure single knockout lines. Double knockout lines were obtained by crossing idl7 (pollen) to idl6-1 (mother plant) and idl6-2 (pollen) to idl7 (mother plant). Double homozygous lines were verified by PCR using gene- and T-DNA-specific primers (Supplementary Table S1).
IDL6 and IDL7 promoter:β-glucuronidase (GUS) fusions were generated using Gateway technology (Invitrogen). The intergenic regions upstream of IDL6 (869 bp) and IDL7 (1306 bp) were amplified from genomic DNA fromthe Col-0 ecotype using the primers pIDL6attB1 and pIDL6attB2 for the IDL6 promoter region, and pIDL7attB1 and pIDL7attB2 for the IDL7 promoter region (Supplementary Table S1). The fragments were cloned upstream of the GUS gene in the destination vector pMDC163 (Curtis and Grossniklaus, 2003) via the pDONR/ZEO vector (Invitrogen). Complementation lines were generated by amplifying the region surrounding IDL6 (860 bp upstream and 142 bp downstream coding sequence) and IDL7 (1306 bp upstream and 336 bp downstream coding sequence) from genomic DNA from the Col-0 ecotype using the primers pIDL6attB1 and IDL6comp attB2 for IDL6 and pIDL7atttB1 and IDL7comp attB2 for IDL7 (Supplementary Table S1). The fragments were cloned into the destination vector pMDC99 (Curtis and Grossniklaus, 2003). The constructs were introduced into Agrobacterium tumefaciens strain C58C1 pGV2260 and transformed into Arabidopsis Col-0 ecotype using the ‘floral dip’ method (Clough and Bent, 1998). Positive transformants were selected on half-strength solid Murashige and Skoog (MS) medium containing the T-DNA-specific selection marker hygromycin (20 µg ml–1).
Subcellular localization
Full-length (IDL7FL) or mutated [IDL7∆SP, IDL7 lacking the predicted signal peptide (SP); IDL7∆SP∆C, IDL7 lacking both the SP and the four last C-terminal amino acids; IDL7∆C, full-length IDL7 lacking the four last C-terminal amino acids] coding sequences of IDL7 were amplified from Col-0 ecotype genomic DNA using the primers SPIDL7attB1, IDL7DSPattB1, IDL7USattB2, and ILD7EPIPattB2 (Supplementary Table S1), and cloned into the destination vector pEG103 (Earley et al., 2006) by Gateway technology (Invitrogen). The vectors were introduced into A. tumefaciens strain C58C1 pGV2260, and transformed cultures [optical density at 600 nm (OD600)=0.05, 10 mM MES, 10 mM MgCl2, and 100 µM acetosyringone] were used for infiltration of leaves of 3- to 4-week-old Nicotiana benthamiana plants grown on soil under a 16 h photoperiod (70 µmol m−2 s−1) at 22 °C as described (Sparkes et al., 2006). Infiltrated leaves were investigated 2–4 d post-infection using a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) with a ×50 water immersion objective. Green fluorescent protein (GFP) was excited using an argon laser at 488 nm, and emission was detected at 495–550 nm.
Histochemical GUS assays
Histochemical GUS assay was performed as described by Butenko et al. (2003).
RNA decay analysis
RNA decay assays were carried out as described (Zhang et al., 2010). Four-day-old seedlings were pre-treated in incubation buffer (1 mM PIPES, pH 6.25, 1 mM sodium citrate, 1 mM KCI, 15 mM sucrose), and then supplied with cordycepin (Sigma-Aldrich, St Louis, MO, USA) to a final concentration of 1 mM. Samples were collected before (0 min) and 30 min and 60 min after cordycepin addition. Tissues for each time point were flash-frozen in liquid nitrogen and stored at –80 °C.
Peptide treatments
Synthetic peptides (Biomatik, Canada) used in this study are listed in Table 1. Seeds of the Col-0 ecotype were surface-sterilized and sown out on half-strength MS plates at a density of 20 seeds per Petri dish (14 cm diameter), and stratified for 3 d at 4 °C. Plates were grown under a 16 h photoperiod (70 µmol m−2 s−1) at 22 °C for 2 weeks. Seedlings were sprayed with an aqueous peptide solution (100 nM) supplemented with 0.02% Silwet L-77 (Lehle Seeds, Round Rock, TX, USA) and vacuum infiltrated at 20 inches Hg for 1 min. Whole rosettes were collected 1, 2, and 3 h after treatment, snap-frozen in liquid nitrogen, and stored at –80 °C.
Table 1.
Synthetic peptides used in this study
| Name | Position | Sequence |
|---|---|---|
| IDL6-EPIP | 75–98 | FGSLVLNALP10KGSVPASGPS20KRIN |
| IDL7-EPIP | 70–93 | FGSLVLNALP10KGSRPGSGPS20KKTN |
| IDL7-PIP | 82–93 | SRPGSGPSKK10TN |
| IDL7-PIPo | 82–93 | SRPGSGhypSKK10TNa |
| MOCKIDL7 | 75–98 | LSPGKNLSAP10GRVGSNPFTK20LRGS |
ahyp, hydroxyproline
Expression analyses
RNA isolation, cDNA synthesis, real-time quantitative reverse transcription–PCR (qRT–PCR), microarray experiments, statistical analysis, and Gene Ontology (GO) analysis were performed as described in Vie et al. (2015). In brief, RNA was extracted from 100 mg of frozen plant tissue each from four biological replicas using the Spectrum Plant Total RNA kit (Sigma-Aldrich). cDNA synthesis was performed on 1 µg of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany), following the supplier’s instructions. qRT–PCR was performed on a LightCycler 480 using the LightCycler 480 SYBR Green I Master kit (Roche Applied Science, Mannheim, Germany), with PCR parameters as recommended by the supplier. PCR efficiencies and Ct values were calculated by linear regression using the LinRegPCR software (Ramakers et al., 2003; Ruijter et al., 2009), and mean PCR efficiency was calculated for each pair of primers. Ct values and PCR efficiencies were then imported into the REST 2008 software (Pfaffl et al., 2002) to calculate the statistical significance of differences in expression levels upon various treatments. Primers used are listed in Supplementary Table S1. Genome-wide expression analysis was performed using the Arabidopsis (V4) Gene Expression Microarray 4 × 44K (Agilent Technology, USA) as described in the supplier’s manual. The data were analysed using the limma package (Smyth, 2004) and the R statistical data analysis program package (R 2.10.1). The Benjamini and Hochberg method to control the false discovery rate was used to identify differentially regulated genes (Benjamini and Hochberg, 1995). Genes with an adjusted P-value <0.05 were regarded as significantly differentially expressed. The microarray study is MIAME compliant. Raw data have been deposited in GEO (accession GSE77467).
Luciferase assay
Three-week-old transgenic plants containing the ZAT12::luc reporter system (Miller et al., 2009) were grown under a 16 h photoperiod regime and sprayed with 1 mM luciferin the evening prior to the experiment. The next day, plants were uniformly sprayed with luciferin supplemented with each of the three peptides (100 nM). At 65 min after treatment, the plants were injured by pricking one leaf. In total three plants per peptide were treated in each live imaging experiment. Luminescence imaging and measuring was conducted using the NightOWL in vivo imaging system (Berthold Technologies, Germany).
flg22 and H2O2 treatment
Seeds of the Col-0 ecotype were surface-sterilized and sown out on half-strength MS plates at a density of 20 seeds per Petri dish (14 cm diameter), and stratified for 3 d at 4 °C. Plates were then grown under a 16 h photoperiod (70 µmol m−2 s−1) at 22 °C for 2 weeks. Seedlings were sprayed with either flg22 (100 nM) or H2O2 (20 mM) in deionized (DI) water containing 0.02% Silwet L-77 and vacuum infiltrated at 20 inches Hg for 1 min. As control, plants were treated with DI water supplemented with 0.02% Silwet-L77 and vacuum infiltrated at 20 inches Hg for 1 min. Whole rosettes were harvested from four biological replicas at 5, 10, 15, 30, and 60 min after treatment, snap-frozen in liquid nitrogen, and stored at −80 °C.
Seedling pathogen assay
The protocol was modified from Lee et al. (2011): surface-sterilized seeds were sown in 24-well plates containing 2 ml of liquid half-strength MS, with a density of three seedlings per well, and stratified for 3 d at 4 °C. Plants were grown in a growth chamber (VB1514, Vötsch Industrietechnik, Balingen, Germany) under a 16 h photoperiod (70 µmol m−2 s−1) at 22 °C. Pseudomonas syringae pv. tomato DC3000 and P. syringae pv. tomato AvrRPM1 cultures were grown overnight in liquid King’s B medium supplemented with the appropriate antibiotics (50 µg ml−1 rifampicin for the DC3000 strain, 50 µg ml−1 rifampicin, and 50 µg ml−1 tetracycline for the AvrRPM1 strain) in a shaker at 28 °C. The cultures were washed twice with 10 mM MgCl2 and diluted to OD600=0.002 (~2.5 × 106 CFU ml−1). Seven-day-old seedlings were transferred to new 24-well plates containing fresh half-strength MS without sucrose, and 100 µl of diluted bacterial culture was added. For measurements of bacterial growth, co-cultivated seedlings were washed with 70% ethanol for 10 s and rinsed in water. Three seedlings were put into eppendorf tubes containing 100 µl of 10 mM MgCl2 and ground with a pestle. A 10 µl aliquot of serial dilutions was plated out on LA medium containing the appropriate antibiotics and incubated for 2 d at 28 °C. All pathogen assays were repeated at least three times with similar results.
Stress assays
Seeds of wild-type Col-0 and loss-of-function mutants of IDL6 and IDL7 were surface-sterilized, placed in rows on square half-strength MS plates (control) and half-strength MS with added NaCl (100 mM), mannitol (300 mM), or flg22 (100 nM), and stratified for 3 d at 4 °C. Plates were then placed vertically in a growth chamber (VB1514, Vötsch Industrietechnik) under a 16 h photopheriod (70 µmol m−2 s−1) at 22 °C. Root lengths were scored 1 week after seed plating for NaCl and mannitol treatments, and at 10 d for flg22 treatments. The X/XO assay was performed as described (Overmyer et al., 2000). All stress assays were repeated at least three times with similar results.
ROS assay
ROS measurements on Arabidopsis leaf disks were performed as described by Bisceglia et al. (2015). Briefly, rosette leaves from 4-week-old plants grown at 22 °C, with a 12 h light/12 h dark photoperiod were detached and rinsed in DI water. Leaf disks were made using a puncher, incubated for 2 h in DI water, and transferred to a 96-well plate containing DI water. The plate with leaf disks was incubated in darkness overnight. DI water was then replaced with luminol solution (30 μg luminol ml–1, 20 μg of horseradish peroxidase ml–1). Water or 100 nM peptide was added and ROS production was detected as luminescence was measured in a plate reader (Synergy H1; BioTek, Winooski, VT, USA) for 40 min.
Results
IDL7 is processed C-terminally prior to export from the cell
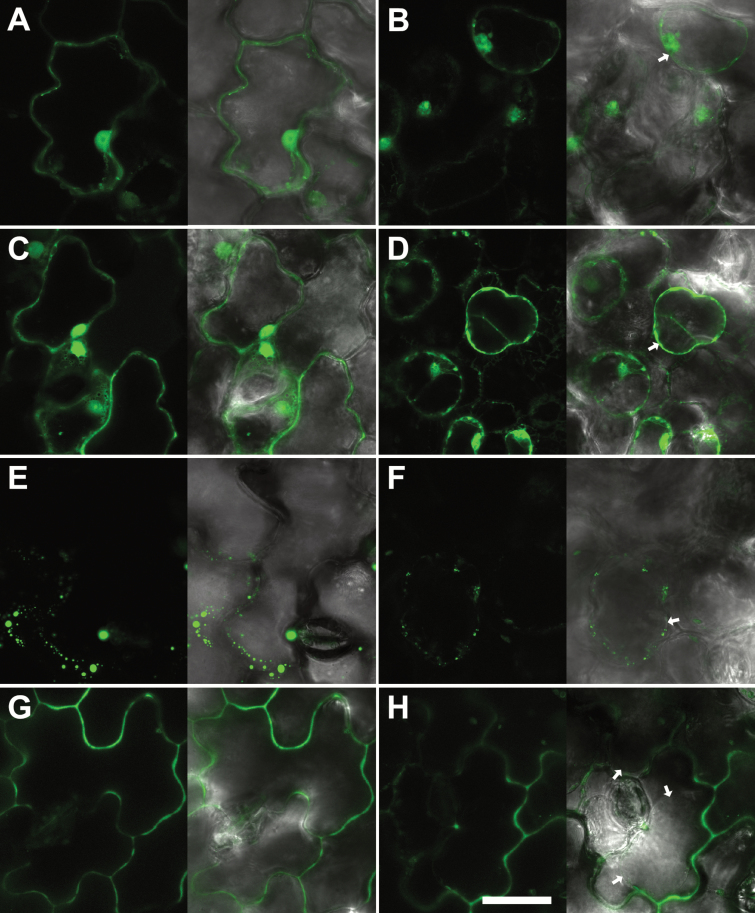
One of the hallmarks of the IDA family proteins is the presence of an N-terminal SP for the secretory pathway. IDA has previously been shown to be localized to the apoplast (Butenko et al., 2003). To investigate whether IDL7 is secreted out of the cell, we created constructs containing translational fusions between GFP and full-length IDL7 cDNA (IDL7FL, corresponding to amino acids 1–97) or IDL7 without the predicted N-terminal SP (IDL7∆SP, corresponding to amino acids 22–97), as shown in Supplementary Fig. S1. The protein fusions were transiently expressed in N. benthamiana leaves, and the subcellular localization was examined by confocal microscopy. All constructs produced a strong fluorescence signal; the GFP control was located in the cytosol and nucleus (Fig. 1A). IDL7∆SP::GFP was, as expected, located in the cytosol, and could also be found in the nucleus (Fig. 1C), resembling the GFP control. This localization was confirmed by plasmolysis with 1 M NaCl (GFP control, Fig. 1B; and IDL7∆SP::GFP, Fig. 1D). IDL7FL::GFP localization appeared to be intracellular (Fig. 1E), with the formation of fluorescent aggregates, in addition to a weak extracellular localization. Plasmolysis verified the intracellular localization (Fig. 1F).
Fig. 1.
IDL7 is localized to the apoplast. C-terminal GFP tags were cloned in-frame with IDL7, expressed in N. benthamiana epidermal cells under control of the 35S promoter, and analyzed using confocal laser scanning microscopy. The GFP control was localized in the cytosol and the nucleus (A), confirmed by plasmolysis (1 M NaCl, 30 min) (B). (C) IDL7∆SP–GFP was localized in the cytosol, confirmed by plasmolysis (1 M NaCl, 30 min) (D). (E) IDL7FL–GFP was found clogged in vesicular compartments, confirmed by plasmolysis (1 M NaCl, 30 min) (F). (G) IDL7∆C–GFP was localized in the apoplastic space, confirmed by plasmolysis (1 M NaCl, 30 min) (H). Arrows indicate plasmolysed plasma membrane. Scale bar=20 ìm.
It has been suggested that small signalling peptides might be processed C-terminally as well as N-terminally (Matsubayashi, 2014). The fluorescent aggregates seen in IDL7FL::GFP-transformed cells might thus result from proteolytic cleavage of the IDL7 C-terminal end fused with GFP. We therefore generated a new IDL7–GFP fusion without the four C-terminal amino acids of IDL7, corresponding to the C-terminal end of the IDA EPIP (extended PIP) peptide (Stenvik et al., 2008; Supplementary Fig. S1). In cells expressing IDL7∆C::GFP, however, the fluorescent signal was detected at the surface of the cell (Fig. 1G). In plasmolysed cells expressing IDL7∆C::GFP, fluorescence was observed in the extracellular space, between the detached plasma membrane and the cell wall (Fig. 1H). Altogether, these results show that IDL7 is indeed exported from the cell, and that IDL7 is processed C-terminally as well as N-terminally during maturation.
Rapid turnover of IDL6 and IDL7 mRNA
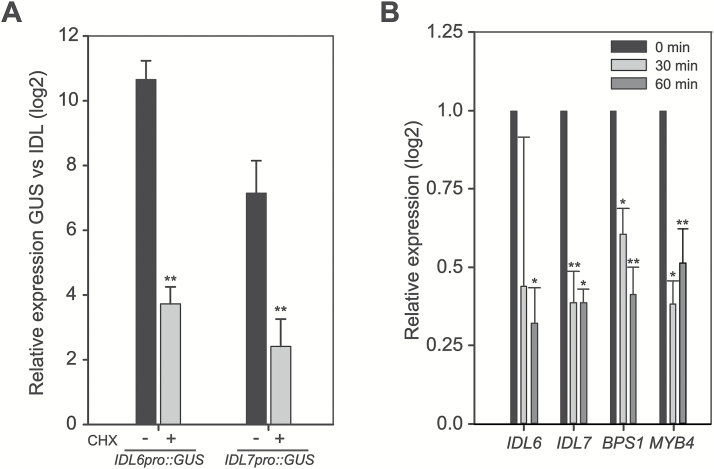
IDL6 pro :GUS and IDL7pro:GUS lines were generated to investigate the expression pattern of these genes. However, the expression patterns indicated by GUS staining did not match the expression pattern found by qRT–PCR (Vie et al., 2015); instead, GUS appeared to be constitutively expressed at high levels in most tissues (Supplementary Fig. S2). This, together with the elevated expression level of IDL6 and IDL7 observed upon cycloheximide (CHX) treatment (Vie et al., 2015), led us to perform a comparison of the expression levels of IDL6/IDL7 and GUS in four independent lines each of IDL6pro:GUS and IDL7pro:GUS. GUS expression in IDL6pro:GUS and IDL7pro:GUS lines was 1000-fold and 200-fold higher than the IDL6 and IDL7 expression levels, respectively (Fig. 2A). In comparison, the GUS expression after CHX treatment in IDL6pro:GUS lines was only up-regulated 13-fold compared with IDL6, and 5-fold up-regulated in the IDL7pro:GUS lines compared with IDL7 (Fig. 2A; Supplementary Fig. S2). One consequence of the inhibitory effect of CHX on translation is that mRNA molecules become trapped on polysomes, thereby preventing mRNA degradation (Cochran et al., 1983; Edwards and Mahadevan, 1992). Therefore, we measured mRNA decay on IDL6 and IDL7 at early growth stages using the transcription inhibitor cordycepin (Johnson et al., 2000). mRNA from both IDL6 and IDL7 showed a rapid decay shortly after cordycepin treatment (Fig. 2B), confirming the GUS assay results. Taken together, the results suggest that IDL6 and IDL7 are under strong regulation, at both transcriptional and post-transcriptional level.
Fig. 2.
Rapid turnover of IDL6 and IDL7 mRNA. (A) Ten-day-old reporter lines (n=3) expressing GUS under control of the IDL6 and IDL7 promoters were analysed for GUS and IDL6/IDL7 expression through qRT–PCR with (two independent lines) or without (four independent lines) CHX treatment (10 µg ml–1, 3 h). The mean and SD of the expression ratio (log2) between GUS and IDL6/IDL7 for each construct is shown. (B) mRNA decay analysis showing the relative gene expression level of IDL6 and IDL7 at 30 min and 60 min after adding of cordycepin (1 mM) to the media. BPS1 and MYB4 were included as controls (n=4). Statistical differences (REST analysis: *P-value <0.05; **P-value <0.01) between the time point samples and control are indicated. Error bars indicate SDs.
IDL7 peptide down-regulates many stress-responsive genes
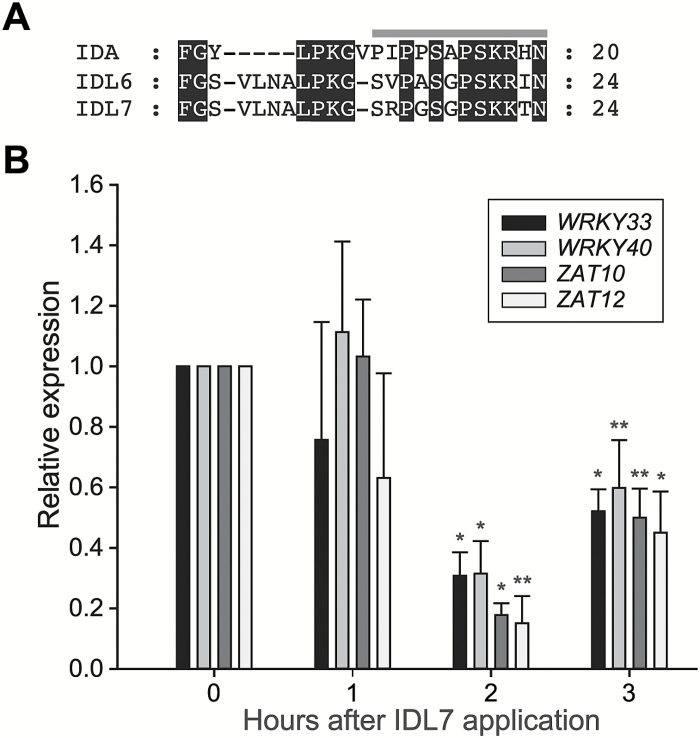
The bioactive part of the IDA peptide has been identified to be within the C-terminal part, between amino acids 50 and 69. This 20 amino acid peptide, named IDA EPIP, is sufficient to rescue the floral abscission-deficient phenotype of the ida mutant in vitro (Stenvik et al., 2008). The corresponding EPIP motifs from IDL6 and IDL7, while somewhat longer (24 amino acids), are similar to IDA in 12 positions (Fig. 3A). Peptides corresponding to the EPIP motifs of IDL6 and IDL7 were synthesized and tested for bioactivity in a pilot experiment by spraying wild-type plants with a 10 µM peptide solution. No phenotypes were observed on the plants after treatment; however, a microarray analysis of plants 3 h after treatment suggested that both peptides induced significant transcriptome changes (results not shown). Among the most down-regulated genes were the stress-related transcription factor genes WRKY33, WRKY40, ZAT10, and ZAT12. These genes were selected for a closer analysis of the timing of the response to IDL7 peptide. The peptide concentration was reduced to 100 nM, which is comparable with other studies (Yamaguchi et al., 2006; Delay et al., 2013; Chien et al., 2015; Wrzaczek et al., 2015). qRT–PCR showed that the strongest transcriptional response was observed 2 h after application of peptides (Fig. 3B). This time point was chosen for global transcription profiling.
Fig. 3.
IDL7 negatively affects the expression of stress-related genes. (A) Alignment of the EPIP peptides from IDA, IDL6, and IDL7. The grey bar above the alignment indicates the PIP peptide. (B) Effect of the IDL7-EPIP peptide on gene expression of the transcription factor genes WRKY33, WRKY40, ZAT10, and ZAT12 over time. Statistical differences (REST analysis: *P-value <0.05; **P-value <0.01) between samples treated with MOCKIDL7 and IDL7 peptide (100 nM) are indicated (n=3). Error bars indicate SDs.
Global transcriptome profiles were obtained from Arabidopsis seedlings following 2 h treatments with 100 nM IDL6-EPIP or IDL7-EPIP. Treatment with the IDL7 EPIP peptide resulted in significant changes in the transcriptome compared with mock peptide-treated plants; in total, 939 genes were found to be significantly regulated (log2>0.5 and log2< –0.5; P<0.05; Supplementary Dataset S1). Table 2 presents the most highly regulated genes. Interestingly, a majority of the genes (73%) were down-regulated. Treatment with the IDL6-EPIP peptide resulted in a much weaker response, with only 42 significantly regulated genes (log2>0.5 and log2< –0.5; P<0.05; Supplementary Dataset S2). When using less strict statistical criteria for IDL6-regulated genes (log2>0.5 and log2< –0.5; P<0.1), the number of significantly regulated genes was still lower (493 genes; 270 up-regulated, 298 down-regulated) compared with IDL7-treated plants. More than half of the genes regulated by IDL6-EPIP treatment (using the less stringent statistical criteria) were also regulated by IDL7-EPIP treatment (Supplementary Fig. S3). This suggests that despite the strong homology of IDL6 and IDL7 peptide, the effect of IDL7-EPIP on the transcriptome is much stronger at the given developmental stage and environmental conditions than IDL6-EPIP.
Table 2.
The most up- and down-regulated genes 2 h after treatment with IDL7-EPIP peptide (P<0.05)
Genes with expression ratios of log2>1.0 or log2< –1.5 are listed
| Description | Locus | Log 2 fold change | Variance | Adjusted P-value |
|---|---|---|---|---|
| ASN1 (GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1) | At3g47340 | 1.519 | 0.252 | 0.013 |
| SPla/RYanodine receptor (SPRY) domain-containing proteina | At4g06536 | 1.279 | 0.125 | 0.010 |
| ADS1 (DELTA 9 DESATURASE 1)b | At1g06080 | 1.218 | 0.218 | 0.033 |
| SAUR26 (SMALL AUXIN UP RNA 26)b | At3g03850 | 1.200 | 0.022 | 0.010 |
| ATMAGL4 (MONOACYLGLYCEROL LIPASE 4)b | At1g73480 | 1.151 | 0.231 | 0.023 |
| SAUR29 (SMALL AUXIN UP RNA 29)a,b | At3g03820 | 1.127 | 0.077 | 0.008 |
| Unknown proteina | At4g33467 | 1.111 | 0.457 | 0.015 |
| HMT3 (HOMOCYSTEINE S-METHYLTRANSFERASE 3)a | At3g22740 | 1.097 | 0.084 | 0.017 |
| A_thal_3526 family proteina,b | At3g55240 | 1.030 | 0.034 | 0.013 |
| P5CS1 (DELTA1-PYRROLINE-5-CARBOXYLATE SYNTHASE 1)a | At2g39800 | 1.023 | 0.057 | 0.013 |
| IAA29 (INDOLEACETIC ACID-INDUCED PROTEIN 29)b | At4g32280 | 1.019 | 0.067 | 0.010 |
| CYP96A12 (Cytochrome P450 96A12)a | At4g39510 | 1.011 | 0.110 | 0.016 |
| DUF946 family protein | At2g44230 | 1.009 | 0.086 | 0.020 |
| ABC transporter-related protein | At3g21080 | –1.507 | 0.341 | 0.012 |
| Protein phosphatase AP2C2a | At1g07160 | –1.523 | 0.280 | 0.021 |
| ALAAT2 (ALANINE AMINOTRANSFERASE 2) | At1g72330 | –1.532 | 0.252 | 0.023 |
| Unknown proteina | At2g15020 | –1.537 | 0.028 | 0.008 |
| ST (steroid sulphotransferase) | At2g03760 | –1.558 | 0.637 | 0.040 |
| DIC2 (Mitochondrial Dicarboxylate Carrier Protein 2)a | At4g24570 | –1.560 | 0.004 | 0.008 |
| Glycine-rich proteina | At1g07135 | –1.563 | 0.014 | 0.008 |
| Unknown protein | At5g14730 | –1.571 | 0.244 | 0.017 |
| Unknown proteina | At1g25400 | –1.582 | 0.440 | 0.012 |
| ZAT11 (ZINC FINGER OF ARABIDOPSIS THALIANA 11)a | At2g37430 | –1.584 | 0.275 | 0.012 |
| UGT73C3 (UDP-GLUCOSYL TRANSFERASE 73C3) | At2g36780 | –1.605 | 0.375 | 0.029 |
| ZAT8 (ZINC FINGER OF ARABIDOPSIS THALIANA 8) | At3g46080 | –1.616 | 0.607 | 0.025 |
| Unknown protein | At4g08555 | –1.636 | 0.930 | 0.036 |
| VQ12 VQ motif-containing protein | At2g22880 | –1.643 | 0.428 | 0.021 |
| UGT73C6 (UDP-GLUCOSYL TRANSFERASE 73C6) | At2g36790 | –1.675 | 0.535 | 0.018 |
| 2-Oxoglutarate-dependent dioxygenase like protein | At5g43450 | –1.676 | 0.313 | 0.021 |
| Bifunctional inhibitor/lipid-transfer proteina | At1g62500 | –1.683 | 0.790 | 0.018 |
| HSP23.5-M (23.5 kDa heat shock protein, mitochondrial) | At5g51440 | –1.701 | 0.323 | 0.023 |
| Unknown protein | At5g24640 | –1.715 | 0.811 | 0.039 |
| Unknown protein | At2g41730 | –1.719 | 0.592 | 0.022 |
| ZAT7 (ZINC FINGER OF ARABIDOPSIS THALIANA 7)a | At3g46090 | –1.774 | 0.263 | 0.013 |
| WRKY40 (WRKY TRANSCRIPTION FACTOR 40)a | At1g80840 | –1.798 | 0.178 | 0.012 |
| ZAT6 (ZINC FINGER OF ARABIDOPSIS THALIANA 6)a,b | At5g04340 | –1.809 | 0.114 | 0.011 |
| Chaperone DnaJ-domain superfamily proteina | At1g72416 | –1.840 | 0.226 | 0.014 |
| WRKY33 (WRKY TRANSCRIPTION FACTOR 33)a | At2g38470 | –1.842 | 0.204 | 0.008 |
| PIP1 (PAMP-INDUCED SECRETED PEPTIDE 1)a | At4g28460 | –1.844 | 0.733 | 0.040 |
| Acyl-CoA N-acetyltransferasea | At2g32020 | –1.855 | 0.867 | 0.047 |
| Ethylene-responsive transcription factor ERF105a | At5g51190 | 1.856 | 0.219 | 0.008 |
| HSP15.4 (Heat shock protein class V 15.4)a | At4g21870 | –1.913 | 0.044 | 0.008 |
| AGC2-1/OXI1 (OXIDATIVE SIGNAL-INDUCIBLE 1)a | At3g25250 | –1.930 | 0.509 | 0.028 |
| Unknown protein | At4g12735 | –1.979 | 0.720 | 0.029 |
| Eukaryotic aspartyl protease family protein | At5g48430 | –1.984 | 0.020 | 0.008 |
| CYP81D8 (Cytochrome P450 81D8) | At4g37370 | –1.989 | 0.796 | 0.026 |
| AtOM66 (Outer mitochondrial membrane protein of 66 kDa) | At3g50930 | –2.102 | 0.760 | 0.031 |
| ATPP2-A5 (PHLOEM PROTEIN 2-LIKE A5)a | At1g65390 | –2.120 | 0.127 | 0.001 |
| COG4129-domain proteina | At3g09450 | –2.136 | 0.099 | 0.008 |
| UGT74E2 (UDP-GLUCOSYL TRANSFERASE 74E2) | At1g05680 | –2.151 | 1.090 | 0.048 |
| DUF295-domain protein | At5g55150 | –2.202 | 1.096 | 0.043 |
| ZAT10/STZ (SALT-TOLERANCE ZINC FINGER)a | At1g27730 | –2.215 | 0.137 | 0.008 |
| ZAT12 /RHL41 (RESPONSIVE TO HIGH LIGHT 41) | At5g59820 | –2.359 | 0.619 | 0.013 |
| CML38 (CALMODULIN-LIKE PROTEIN 38)a | At1g76650 | –2.456 | 0.121 | 0.008 |
aSignificantly regulated 2 h after treatment with IDL6 peptide (P<0.1).
bIdentified as PIF-regulated PIF4 target gene (Oh et al., 2012).
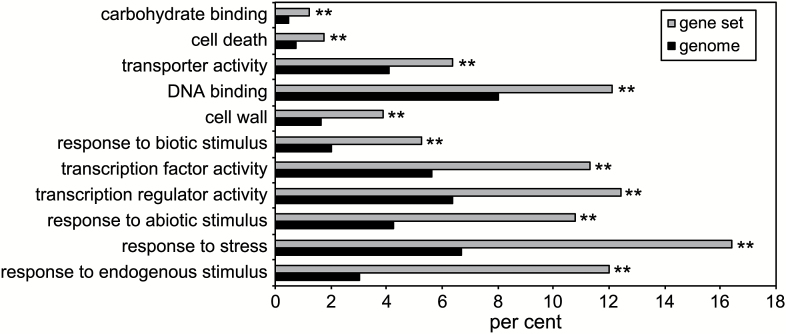
To elucidate a possible function of the IDL7-EPIP peptide, the dataset was analysed for over-representation of Gene Ontology (GO) terms (Ashburner et al., 2000) on genes with log2< –0.5; P<0.05. These results show a clear over-representation of plant stress-related classes, especially the classes for response to stress, response to endogenous stimulus, responses to abiotic stimulus, and gene regulation (Fig. 4; Supplementary Dataset S3).
Fig. 4.
GO enrichment analysis of significantly down-regulated (P-value <0.05) genes in 2-week-old seedlings 2 h after treatment with 100 nM IDL7-EPIP peptide. Control seedlings were treated with 100 nM MOCKIDL7 peptide. The 10 terms most significantly enriched in the gene set are listed from top to bottom. Bars show the frequency of each GO term in the IDL7-responsive gene set and the genome. **P-value <0.01.
The most prominent gene families in the dataset included WRKY transcription factors, ZINC FINGER PROTEINS (ZFP), MYB transcription factors, and ethylene-responsive transcription factors (Table 2; Supplementary Dataset S1). We found 11 significantly down-regulated WRKY genes after treatment with the IDL7 peptide; all of them are associated with stress responses. Nine out of 20 genes encoding ZFPs belonging to the C2H2 subfamily C1-2i, all associated with abiotic stress responses, were down-regulated by IDL7. Many other genes associated with stress adaptation or defence were down-regulated, including a large group encoding Ca2+-binding proteins, genes encoding heat shock proteins, cytochrome P450-encoding genes, and genes encoding receptor proteins (Table 2; Supplementary Dataset S1).
Genes up-regulated by IDL7-EPIP treatment included GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1 (ASN1), SMALL AUXIN UP RNA (SAUR) and INDOLEACETIC ACID-INDUCED PROTEIN 29 (IAA29) (Table 2). Since SAURs have been shown to be regulated by the basic helix–loop–helix (bHLH) transcription factor PHYTOCHROME INTERACTING FACTOR 4 (PIF4; Franklin et al., 2011), we compared the genes in Table 2 with a list of PIF4 target genes identified by ChIP sequencing (Oh et al., 2012). Six of 13 genes up-regulated by log2>1.0 by IDL7 treatment were PIF4 targets. In contrast, only one of 39 genes down-regulated by log2< –1.5 was a PIF4 target. PIF4 itself was moderately, but significantly induced by IDL7-EPIP treatment (Supplementary Dataset S1).
To gain a further overview of the genes regulated by IDL7-EPIP, a network analysis of the most down-regulated genes in the gene set (log2< –1) was performed using the STRING database (Szklarczyk et al., 2015). It is a general assumption that genes involved in the same processes are co-regulated or interact in one way or another, also known as the ‘guilt-by-association’ principle (Saito et al., 2008). As shown in Supplementary Fig. S4, a tight cluster of highly connected genes was found within our array, indicating that IDL7 is regulating this network. The cluster includes the ZFP genes ZAT6, ZAT12, ZAT10/STZ, and SZF1, the WRKY transcription factor genes WRKY33, WRKY40, and WRKY53, and CML38, ERF5, and ERF6.
IDL7 suppresses the expression of ZAT12 upon wounding
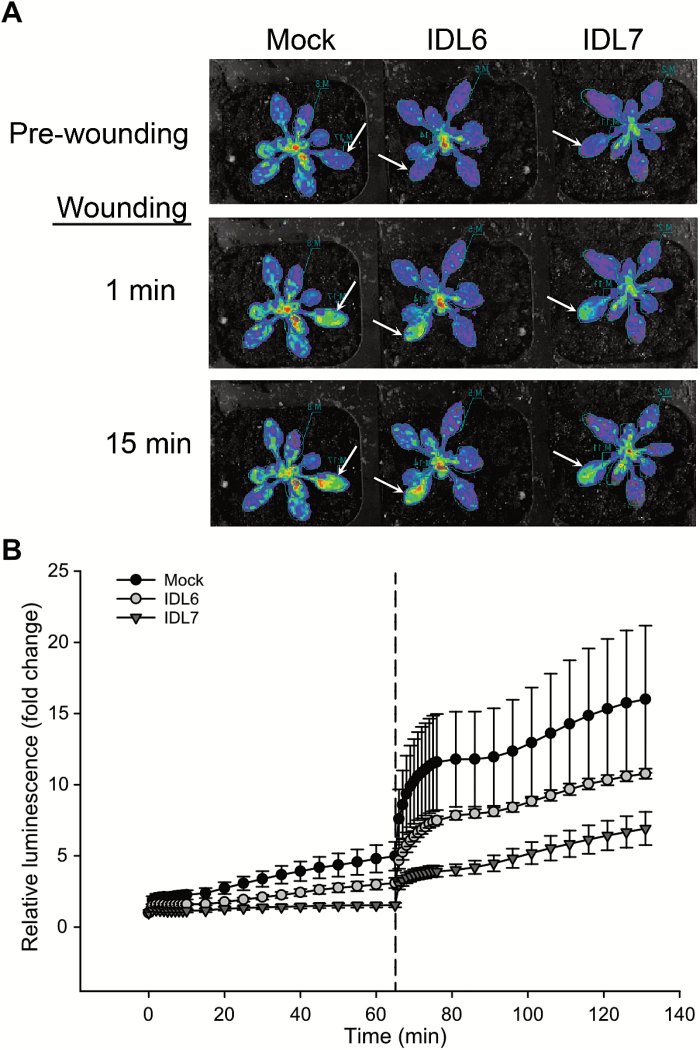
One of the most down-regulated genes in our array was the ZFP gene ZAT12. ZAT12 is highly responsive to different environmental conditions, and is rapidly induced after wounding and accumulation of ROS (Davletova et al., 2005a; Miller et al., 2009). Furthermore, in a study using the luciferase reporter gene, ZAT12 expression was shown to be systemically induced at a rate up to 8.4 cm min−1 upon wounding (Miller et al., 2009). We wanted to investigate the effect of treatment with IDL6-EPIP and IDL7-EPIP peptide prior to wounding on the expression of ZAT12 using the ZAT12::luciferase reporter plants used in the study by Miller and co-workers (Miller et al., 2009). Three plants per peptide were treated with luciferin and peptides (IDL6-EPIP, IDL7-EPIP, and MOCKIDL7), and 1 h later one rosette leaf per plant was wounded. Interestingly, as shown in Fig. 5, IDL7-EPIP was able to suppress spreading of the luciferase signal, verifying the negative effect of IDL7-EPIP on ZAT12 expression. IDL6-EPIP was also able to suppress ZAT12 expression, but the response was weaker. This trend was observed in three independent experiments.
Fig. 5.
Suppression of the rapid wound-induced signal in ZAT12::luc plants by IDL6-EPIP and IDL7-EPIP peptides. Plants pre-treated with IDL6-EPIP, IDL7-EPIP, or MOCKIDL7 (100 nM) by spraying were simultaneously wounded in one leaf (marked with arrows), and changes in luminescence were immediately measured using the NightOwl in vivo imaging system. (A) Representative pictures are shown. Injured leaves are marked with arrows. (B) Quantification of the wound-induced signals in rosette leaves over the time course before and after wounding. Error bars indicate the SD between three biological replicates. (This figure is available in color at JXB online.)
IDL6 and IDL7 are stress-responsive genes
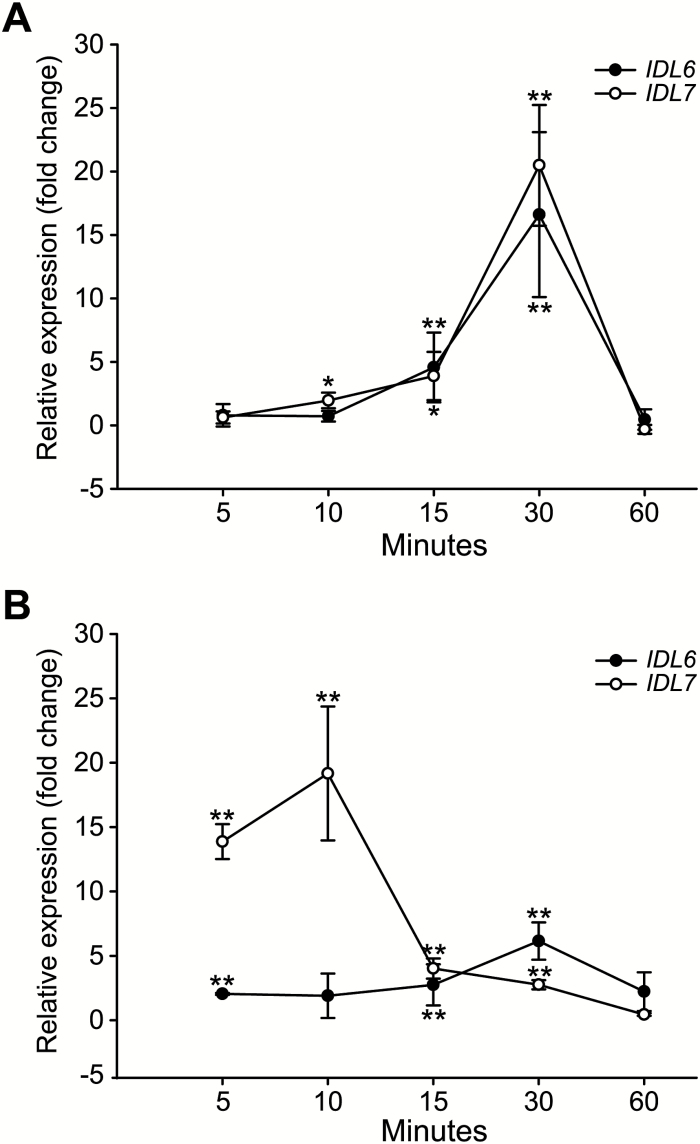
In silico data indicate that IDL6 and IDL7 are moderately expressed during development, and that both genes are rapidly induced by stresses including exposure to P. syringae and the pathogen-associated elicitor flg22, salt, UV, wounding, and ROS (Vie et al., 2015). To analyse the stress-induced expression of IDL6 and IDL7, a time course experiment with Arabidopsis seedlings treated with flg22 or H2O2 was performed. Expression analyses using qRT–PCR analysis showed that the expression of both IDL6 and IDL7 was rapidly induced 10–15 min after flg22 treatment, and reached a peak after 30 min (Fig. 6A). The transcriptional response of IDL7 to H2O2 treatment was even faster, reaching maximal levels after 10 min, whereas induction of IDL6 expression was both weaker and slower (Fig. 6B).
Fig. 6.
Time series analysis of IDL6 and IDL7 expression by qRT–PCR after flg22 treatment (A) and H2O2 treatment (B) compared with untreated tissue (n=4). Statistical differences (REST analysis: *P-value <0.05; **P-value <0.01) between the time point samples and control are indicated. Error bars indicate SDs.
T-DNA insertion lines of IDL6 and IDL7 were used for functional analysis. Loss of gene expression was verified using qRT–PCR. Two available T-DNA insertion lines were obtained for IDL6. Both idl6 [SALK_074245 (idl6-1) and SALK_126026M (idl6-2)] lines contain a T-DNA insertion 1 bp upstream of the start codon, and low IDL6 transcript levels were detected. However, IDL6 expression in these mutant lines was not inducible with flg22 (Supplementary Fig. S5A), suggesting that idl6-1 and idl6-2 are knockdown mutants. Only one T-DNA insertion line was available for IDL7. idl7 (WDL293-296) has the T-DNA inserted in the exon (after 188 bp), and is a null line with no detectable IDL7 mRNA with or without induction of flg22 (Supplementary Fig. S5B). Neither idl6 nor idl7 plants displayed any observable phenotypical differences compared with wild-type control plants (Col-0) under normal growth conditions. Therefore, crosses between idl6 and idl7 plants were performed. However, neither idl6-1 idl7 nor idl6-2 idl7 double mutants displayed any obvious phenotypes during normal growth conditions, and no additive effect of the double mutation was found (results not shown). To investigate the potential change in gene expression in the mutant lines compared with Col-0, 2-week-old seedlings were treated with flg22 (100 nM) and samples harvested after 3 h, 1 h after the peak response to IDL7-EPIP. Surprisingly, the responses of ZAT10 and WRKY40 were slightly, but not significantly, reduced in all the mutant lines compared with Col-0 wild type (Supplementary Fig. S6).
idl7 loss-of-function lines are more tolerant to salt stress
The observed induction of IDL6 and IDL7 expression upon a broad range of environmental perturbations (Vie et al., 2015) suggests that these genes are involved in modulating stress responses in plants. Seedlings of idl6 and idl7 were therefore exposed to different stresses and examined for seed germination, root growth, ion leakage, and pathogen growth. The following stresses were tested: elicitor treatment (100 nM flg22), pathogen infection (P. syringae DC3000 and P. syringae AvrRPM1), salinity stress (100 mM NaCl), and osmotic stress (300 mM mannitol).
No significant differences between wild-type plants and the mutant lines were observed after treatment with the pathogen-derived elicitor flg22 (Supplementary Fig. S7A). Similarly, no significant differences in growth of P. syringae were observed between wild-type plants and the mutant lines, neither with the virulent nor with the avirulent strain (Supplementary Fig. S7B, C). As shown in Supplementary Fig. S8A, both idl6 and idl7 lines showed significantly (P<0.01) increased tolerance to salinity stress, producing longer roots than the wild type under the same growth conditions. However, no additive effect was observed for the double loss-of-function mutant idl6-2 idl7. Complementation lines of idl6 and idl7 loss-of-function lines were used to verify the observed phenotype (Supplementary Fig. S8B). No significant differences in root growth were observed between idl6 and idl7 lines and the wild type grown in the presence of mannitol (Supplementary Fig. S8C). Hypocotyl elongation in the dark was also investigated, with no differences found between the wild type and the mutant lines (Supplementary Fig. S8D). Germination rates of idl6 and idl7 mutant lines on medium containing NaCl, mannitol, or abscisic acid (ABA) did not differ from the wild type (results not shown).
IDL7 acts as a negative modulator of stress-induced ROS signalling
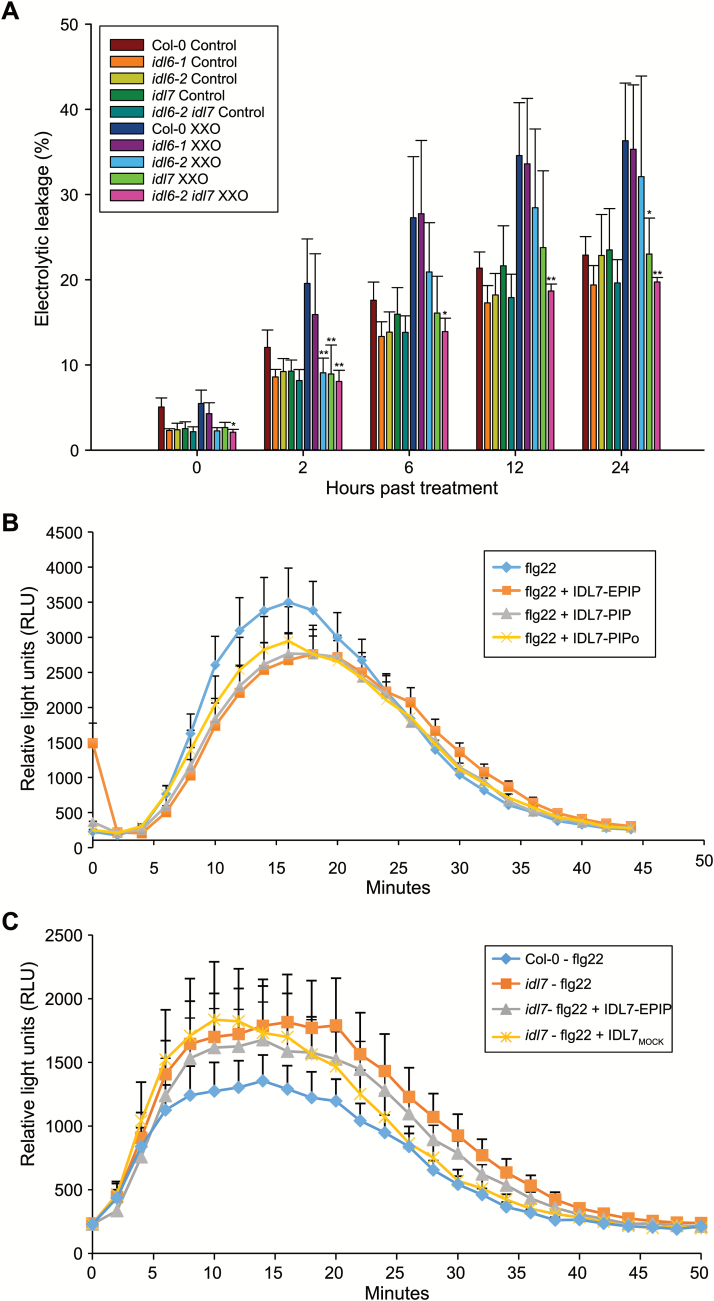
Since IDL6 and especially IDL7 expression was rapidly and strongly induced by H2O2, we investigated the response of idl6 and idl7 mutants to oxidative stress, using the xanthine/xanthine oxidase (X/XO) system. X/XO is an O2·¯-generating system that mimics and induces production of extracellular superoxide by NADPH oxidases, similar to O3. X/XO treatment results in cell death, which can be measured by electrolyte leakage (Overmyer et al., 2005). X/XO treatments of idl6-1, idl6-2, idl7, and the idl6-2 idl7 double mutant showed that idl7, but not idl6, was more tolerant to ROS, as indicated by less electrolyte leakage from idl7 compared with the wild type and idl6 over time (Fig. 7A).
Fig. 7.
IDL6 and IDL7 are negative modulators of stress-induced ROS signalling. (A) Measurements of electrolytic leakage after treatments with the ROS-generating enzyme assay xanthine/xanthine oxidase (0.05 U) 2, 6, 12, and 24 h after treatment (n=4). Statistical differences for all experiments (Student’s t-test: *P-value <0.05, **P-value <0.01, ns indicates P-value >0.05) between the wild-type Col-0 and mutants are indicated. Error bars indicate SDs. (B, C) Modulation of flg22-induced oxidative burst by IDL7 peptide. Arabidopsis Col-0 wild-type (B) or idl7 (C) leaf disks were exposed to flg22, IDL7-EPIP, IDL7-PIP, IDL7-PIPo, or MOCKIDL7 peptides (100 nM), either alone or in combination (flg22+IDL7, flg22+IDL7-PIP, flg22+IDL7-PIPo, and flg22+MOCKIDL7). Water was added as a control. ROS production measured as luminescence was monitored over time as relative light units (RLU). Error bars indicate the SE of n=12 replicates.
In order to investigate further the role of the IDL7 peptide in ROS responses, we employed a luminol-based assay to measure ROS production in Arabidopsis leaf disks. In addition to IDL7-EPIP, we included two shorter synthetic peptides in this analysis; IDL7-PIP (see Fig. 3A) and the hydroxyprolinated IDL7-PIPo, as a recent publication on IDA has shown that a hydroxyprolinated 12 amino acid IDA-PIP peptide has higher biological activity than the IDA-EPIP peptide (Butenko et al., 2014). As expected, treatment with flg22 peptide resulted in a rapid ROS burst, whereas no ROS production was observed in the control (Fig. 7B). Addition of IDL7 or MOCKIDL7 peptide alone did not lead to any ROS production (Supplementary Fig. S9). Interestingly, when IDL7-EPIP peptide was added together with flg22 peptide, the ROS burst was significantly (Student’s t-test, P<0.001) lower compared with flg22 peptide (Fig. 7B). Co-treatment with MOCKIDL7 peptide did not change the level of the ROS burst (Student’s t-test, P>0.05, Supplementary Fig. S9). However, treatments with the shorter IDL7-PIP peptide and the hydroxyprolinated IDL7-PIPo did not lead to a decrease in the ROS burst compared with IDL7-EPIP treatment (Fig. 7B). Flg22 treatment of idl7 plants resulted in increased ROS production compared with the Col-0 wild type (Student’s t-test, P<0.001); this effect was partially suppressed by co-treatment with IDL7-EPIP (Student’s t-test, P<0.001, Fig. 7C). Thus, IDL7 peptide has an attenuating effect on the flg22-induced ROS burst.
Discussion
In this study we describe two new putative peptide ligands belonging to the extended family of IDA and IDL peptides (Vie et al., 2015) and show evidence suggesting that they act as negative modulators of ROS responses.
Small transcriptionally modified peptides are characterized by an N-terminal SP that directs the peptide out of the cell. Analyses of subcellular localization of IDA and CLE peptides indicate that they are localized to the extracellular space (Butenko et al., 2003; Sharma et al., 2003). Similar to the CLE peptides and IDA, IDL6 and IDL7 contain an N-terminal SP (Vie et al., 2015). However, in addition to localization in the apoplast, a full-length fusion of IDL7::GFP was accumulated in a vesicular compartment (Fig. 1E, F). An IDL7∆SP::GFP fusion displayed cytosolic localization resembling GFP (Fig. 1A–D). Deletion of the four C-terminal amino acids of IDL7 resulted in a GFP fusion protein with apoplastic localization (Fig. 1G), which was confirmed through plasmolysis experiments (Fig. 1H). These results suggest that IDL7 is processed at both the N- and the C-terminus before or during the transport out of the cell. The EPIP/SGPS motif of all IDL subfamily members contains a C-terminal asparagine followed by between 4 and 13 poorly conserved residues (Vie et al., 2015). Removal of Asn69 abolishes IDA activity in a ROS burst assay (Butenko et al., 2014). Furthermore, IDA in complex with the ectodomain of its receptor HAESA (HAE) suggests that Asn69 constitutes the C-terminal residue of the mature IDA peptide in planta (Santiago et al., 2016). Thus, it seems likely that C-terminal processing is a general and important feature of the maturation process of these peptides. Some of the C-terminal residues may confer specificity to the mature peptides. The N-terminus of the mature IDL6 and IDL7 peptides remains unresolved.
Our analyses indicate that the IDL6 and IDL7 promoters show high basal activity (Fig. 2); a similar observation was recently made by Wang et al. (2017) using an IDL6 promoter:GUS construct. This is compensated by a high turnover rate for the transcripts. Thus, expression of a stable reporter gene transcript under control of the IDL6 or IDL7 promoter will lead to artificially high reporter levels (Supplementary Fig. S2). Alternatively, the binding site for a strong transcriptional repressor or another important regulatory unit, e.g. the 3'-untranslated region, could be missing in the promoter regions used in the IDL6pro:GUS and IDL7pro:GUS plants. A peptide ligand involved in stress signalling should be rapidly induced in order to ensure a quick response to biotic or abiotic challenges to the plant. However, the signal should also be quickly attenuated to ensure specificity and avoid runaway responses. Both IDL6 and IDL7 are transiently expressed upon H2O2 or flg22 treatment (Fig. 6; Wang et al., 2017), in line with a role in early responses to these stresses.
Transcriptome analyses of seedlings treated with either IDL6-EPIP or IDL7-EPIP indicated that these peptides act as negative regulators of genes associated with early responses to stress in Arabidopsis seedlings (Figs 3, 4; Table 2; Supplementary Dataset S1, S2). Treatments with the synthetic IDL7-EPIP resulted in a striking transcriptome response, in which 75% of the significantly regulated genes were down-regulated. Several studies have proposed a ‘universal stress response transcriptome’ cluster in plants, which includes IDL7, ZAT10, ZAT12, WRKY11, WRKY18, WRKY33, WRKY40, and CML38 (Kilian et al., 2007; Ma and Bohnert, 2007; Walley et al., 2007, Hahn et al., 2013). In addition, network analysis of the genes most down-regulated by IDL7-EPIP (Supplementary Fig. S4) illustrates that these genes are highly co-expressed and probably involved in the same biological processes. Expression of ZFP and WRKY genes is highly responsive to a great variety of stresses; these transcription factor families have emerged as key regulators and integrators of ROS signalling in plants (Miller et al., 2008). ZAT12 has been described as a hyper-responsive gene to ROS accumulation, and is expressed both locally and systemically within minutes after increased ROS production (Miller et al., 2009). ROS-induced expression of ZAT12 was strongly reduced in response to treatment with IDL7-EPIP. Both the transcriptional changes trigged by IDL7-EPIP and the increased tolerance to ROS and salinity stress in idl6 and idl7 lines (Fig. 7A; Supplementary Fig. S7) suggest that IDL6 and IDL7 are negative modulators of stress responses in Arabidopsis.
Pathogen attack or environmental perturbations such as increased salinity induce a large network of different pathways containing both positive and negative regulatory components, where negative regulators may be important factors preventing excessive and unrestricted activation of defence mechanisms. Activation of the plant defence system has been shown to have a major impact on plant growth and fitness. In line with thi0s, overexpression of ZAT7 leads to severe growth defects (Ciftci-Yilmaz et al., 2007). Mutants for the MAP kinase MPK4 display severe growth retardation, possibly through overaccumulation of the defence-related phytohormone salicylic acid (SA) and H2O2 (Petersen et al., 2000; Ichimura et al., 2006; Suarez-Rodriguez et al., 2007), and overexpression of DREB2A, a positive regulator of stress tolerance, showed reduced growth and dwarfism similar to mpk4 mutants (Sakuma et al., 2006).
Expression analyses of IDL6 and IDL7 show that these genes are responsive to a wide variety of biotic and abiotic stresses (Fig. 6; Vie et al., 2015). However, both the single and double loss-of-function mutants displayed minor phenotypes in only a subset of the stress assays performed during this study. There are several possible explanations for the lack of phenotypes. One possibility is that IDL6 and IDL7 are part of a complex signalling network with redundant pathways, and that altered expression of IDL6 and IDL7 is not enough to disrupt this network noticeably. The gene expression of ZAT10 and WRKY40 was not different in the mutant lines compared with Col-0 after flg22 (Supplementary Fig. S6), possibly reflecting the lack of phenotypes observed in our stress assays. It is possible that IDL6 and IDL7 have multiple roles in tolerance to different stresses, and that our functional characterizations have not included all of the specific stresses linked to IDL6 and IDL7. In a recent study, IDL6-overexpressing plants were found to exhibit reduced resistance to P. syringae pv. tomato DC3000, whereas IDL6 dsRNA-silenced plants showed increased resistance (Wang et al., 2017). There are major differences between the two studies, with regard both to the growth stage of the plants (1 versus 5 weeks) and to the nature of the knockdown mutants (T-DNA versus dsRNA), which could explain the different outcomes.
The fact that treatments with X/XO and NaCl (Fig. 7; Supplementary Fig. S7) led to a phenotype in idl7 lines is supported by the fact that a large fraction of the genes down-regulated by IDL7-EPIP treatment are associated with these stresses. WRKYs and ZFPs have been linked to oxidative and salinity stress (Rizhsky et al., 2004; Davletova et al., 2005a, b; Ciftci-Yilmaz et al., 2007; Jiang and Deyholos, 2009; Chen et al., 2010). Overexpression of ZAT7 and ZAT12 has been reported to increase tolerance to saline and oxidative stress (Rizhsky et al., 2004; Davletova et al., 2005b; Ciftci-Yilmaz et al., 2007). The increased tolerance to salinity stress in the idl6 and idl7 loss-of-function lines are in line with these data.
In spite of its similarity to IDL7, IDL6-EPIP displayed a far weaker effect on transcriptional changes compared with IDL7-EPIP, indicating that IDL6 has lower biological activity than IDL7. Alternatively, IDL6 and IDL7 might have different roles in stress regulation, mediated through differences in expression patterns, receptor availability, and/or peptide modifications, or that the peptide is active at different growth stages from those investigated in this study. It is worth noting that IDL6-EPIP differs in four amino acids in comparison with IDL7-EPIP, where IDL7-EPIP contains one charged amino acid (arginine versus valine) more than IDL6-EPIP in front of the SGPS motif (Fig. 3A). It is also likely that many of the transcriptional targets of the two peptides are expressed at their basal, non-induced levels during normal growth conditions, explaining the few regulated genes for IDL6 and the moderate response for IDL7.
Several lines of evidence point toward a role for IDL7, and possibly IDL6, as a modulator of ROS signalling. The transcriptional response of IDL7 to H2O2 treatment is rapid (Fig. 6B). The temporal difference in IDL7 induction by H2O2 and flg22 could correspond to the time needed for flg22 to induce a ROS burst (Gómez-Gómez et al., 1999; Fig. 7B). Application of exogenous IDL7-EPIP (and to a minor extent IDL6-EPIP) peptide blocked the rapid induction of ZAT12 expression after wounding (Fig. 5), which has been shown to be mediated by ROS (Miller et al., 2009). idl7 and idl6-2 idl7 double mutants displayed decreased sensitivity to ROS produced by X/XO (Fig. 7A). Importantly, the IDL7 peptides attenuated the flg22-induced ROS burst and partially rescued the enhanced ROS production in the idl7 background (Fig. 7B, C). The fact that the synthetic IDL7 peptide fails to rescue the idl7 ROS phenotype completely suggests that it is not identical to the mature IDL7 peptide with regard to length and/or post-translational modification. However, counterintuitively, both the shorter IDL7-PIP and the hydroxyprolinated IDL7-PIPo resulted in a similar response to the longer IDL7-EPIP peptide (Fig. 7B). At this point, we cannot draw any conclusions regarding the length of the biological peptide. It is possible that the bioactive IDL7 peptide is longer than IDA, but still contains modifications, or that the sensitivity of our assays is not high enough to detect the possible stronger effects of the more active peptide.
The receptor(s) for the IDL7 peptide are unknown; however, IDL7 perception is likely to involve LRR-RLKs that phosphorylate downstream targets. Binding of IDA to its receptors HAE or HAESA-LIKE2 (HSL2) activates a mitogen-activated protein (MAP) kinase cascade that results in the phosphorylation of transcription factors (Cho et al., 2008; Patharkar and Walker, 2015). Furthermore, the effect of IDL6 on resistance to P. syringae and pectin degradation was found to be dependent on HAE and HSL2 (Wang et al., 2017). A pathway possibly involving HAE/HSL2 could act downstream of IDL7 to down-regulate genes of the ‘universal stress response transcriptome’, through either activation of a transcriptional repressor, or inactivation of a transcriptional activator. However, this scenario does not explain the attenuating effect of the IDL7 peptides on ROS production when added together with flg22. Alternatively, downstream targets of IDL7 signalling could be involved in regulation of ROS levels. An attractive group of target candidates would be NADPH oxidases, especially RBOHD. In addition to calcium binding, RBOH activity is regulated by phosphorylation by several classes of protein kinases (Baxter et al., 2014; Kimura et al., 2017). IDL7 signalling could regulate RBOH phosphorylation status, either by inactivating one or more of these kinases, or by activating a phosphatase with specificity toward RBOHs. Instead of inhibiting ROS production, IDL7 signalling could potentially activate ROS scavenging enzymes.
What could be the role of IDL6/IDL7-regulated ROS attenuation? IDL7 (and possibly IDL6) may act in a negative feedback loop terminating the fast phase of the biphasic ROS burst. Mild salt treatment induces a biphasic RBOHD-dependent ROS burst similar to the one induced by flg22 (Xie et al., 2011), and the rbohd rbohf double mutant displays increased salt sensitivity (Ma et al., 2012; Ben Rejeb et al., 2015). The strong negative effect of IDL6/IDL7 treatment on wound-induced ZAT12 expression also suggests that these peptides might be involved in attenuation of the autopropagating ‘ROS wave’ leading to systemic acquired acclimation (Baxter et al., 2014; Gilroy et al., 2014). Such attenuation could be important for specificity of the signal, as well as avoiding runaway responses.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Overview of the constructs used for localization studies.
Fig. S2. Distribution of GUS mRNA directed by the IDL6 and IDL7 promoters in 10-day-old seedlings.
Fig. S3. Venn diagram of IDL6- and IDL7-responsive genes.
Fig. S4. Network analysis of genes down-regulated by IDL7-EPIP treatment.
Fig. S5. Verification of T-DNA insertion lines.
Fig. S6. Expression of IDL7-responsive genes in idl6 and idl7 mutant backgrounds.
Fig. S7. Growth arrest phenotype and susceptibility of idl6 and idl7 mutants to the phytopathogen Pseudomonas syringae.
Fig. S8. Growth arrest phenotype of idl6 and idl7 mutants to abiotic stress.
Fig. S9. Modulation of flg22-induced ROS burst by IDL7-EPIP and MOCKIDL7.
Table S1. List of PCR primers used in this study.
Dataset S1. Genes significantly regulated (P<0.05) 2 h after treatment with IDL7-EPIP.
Dataset S2. Genes significantly regulated (P<0.05) 2 h after treatment with IDL6-EPIP.
Dataset S3. Raw data from GO analysis of significantly down-regulated (P<0.05) genes by IDL7-EPIP treatments.
Supplementary Material
Acknowledgements
We appreciate the excellent technical assistance of Torfinn Sparstad. We also would like to thank the two reviewers for helpful suggestions. AKV and AMB were funded by the Research Council of Norway by grant 175238/S10. GM and EC were supported by grant 938/11 from the Israel Science Foundation, by the Marie Curie Actions-International Career Integration Grants FP7-PEOPLE-2011-CIG, and by the IS-4652 US–Israel Bi-National Agriculture Research and Development Grant. MW is funded by the University of Helsinki (Postdoc grant and 3-year-fund allocation) and the Academy of Finland (decisions #275632 and #283139).
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Ben Rejeb K, Benzarti M, Debez A, Bailly C, Savouré A, Abdelly C. 2015. NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in Arabidopsis thaliana. Journal of Plant Physiology 174, 5–15. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57, 289–300. [Google Scholar]
- Bindschedler LV, Dewdney J, Blee KA, et al. 2006. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. The Plant Journal 47, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisceglia NG, Gravino M, Savatin DG. 2015. Luminol-based assay for detection of immunity elicitor-induced hydrogen peroxide production in Arabidopsis thaliana leaves. Bio-Protocol 5, e1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB. 2003. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. The Plant Cell 15, 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. 2009. Plant peptides in signalling: looking for new partners. Trends in Plant Science 14, 255–263. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G. 2014. Tools and strategies to match peptide–ligand receptor pairs. The Plant Cell 26, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. 2010. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien PS, Nam HG, Chen YR. 2015. A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. Journal of Experimental Botany 66, 5301–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. 2008. Regulation of floral organ abscission in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105, 15629–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. 2007. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. Journal of Biological Chemistry 282, 9260–9268. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cochran BH, Reffel AC, Stiles CD. 1983. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell 33, 939–947. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. 2005a Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. The Plant Cell 17, 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. 2005b The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology 139, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coninck B, Carron D, Tavormina P, Willem L, Craik DJ, Vos C, Thevissen K, Mathys J, Cammue BP. 2013. Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. Journal of Experimental Botany 64, 5297–5307. [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. Journal of Experimental Botany 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. 2013. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proceedings of the National Academy of Sciences, USA 110, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Edwards DR, Mahadevan LC. 1992. Protein synthesis inhibitors differentially superinduce c-fos and c-jun by three distinct mechanisms: lack of evidence for labile repressors. EMBO Journal 11, 2415–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, et al. 2011. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences, USA 108, 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR, Mittler R. 2014. A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends in Plant Science 19, 623–630. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. 1999. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. The Plant Journal 18, 277–284. [DOI] [PubMed] [Google Scholar]
- Hahn A, Kilian J, Mohrholz A, Ladwig F, Peschke F, Dautel R, Harter K, Berendzen KW, Wanke D. 2013. Plant core environmental stress response genes are systemically coordinated during abiotic stresses. International Journal of Molecular Sciences 14, 7617–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W. 2014. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathogens 10, e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. 2006. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences, USA 103, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. 2006. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. Journal of Biological Chemistry 281, 36969–36976. [DOI] [PubMed] [Google Scholar]
- Igarashi D, Tsuda K, Katagiri F. 2012. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. The Plant Journal 71, 194–204. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. 2009. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology 69, 91–105. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Perez-Amador MA, Lidder P, Green PJ. 2000. Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proceedings of the National Academy of Sciences, USA 97, 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kimura S, Waszczak C, Hunter K, Wrzaczek M. 2017. Bound by fate: the role of reactive oxygen species in receptor-like kinase signaling. The Plant Cell 29, 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO Journal 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chah OK, Sheen J. 2011. Stem-cell-triggered immunity through CLV3p–FLS2 signalling. Nature 473, 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ. 2007. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biology 8, R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F. 2012. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. Journal of Experimental Botany 63, 305–317. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants. Annual Review of Plant Biology 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. 2009. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Science Signaling 2, ra45. [DOI] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. 2008. Reactive oxygen signaling and abiotic stress. Physiologia Plantarum 133, 481–489. [DOI] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. 2012. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. The Plant Cell 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology 14, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chemical Biology 5, 578–580. [DOI] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinänen M, Saarma M, Scheel D, Kangasjärvi J. 2005. Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiology 137, 1092–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Jr, Kangasjärvi J. 2000. Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. The Plant Cell 12, 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Walker JC. 2015. Floral organ abscission is regulated by a positive feedback loop. Proceedings of the National Academy of Sciences, USA 112, 2906–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. 1991. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–897. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, et al. 2000. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. 2004. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. Journal of Biological Chemistry 279, 11736–11743. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. 2008. Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends in Plant Science 13, 36–43. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell 18, 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M. 2016. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. Elife 5, e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Ramirez J, Fletcher JC. 2003. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Molecular Biology 51, 415–425. [DOI] [PubMed] [Google Scholar]
- Sierla M, Rahikainen M, Salojärvi J, Kangasjärvi J, Kangasjärvi S. 2013. Apoplastic and chloroplastic redox signaling networks in plant stress responses. Antioxidants and Redox Signaling 18, 2220–2239. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3, Article3. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. 2008. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. The Plant Cell 20, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. 2007. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiology 143, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. 2010. ROS in biotic interactions. Physiologia Plantarum 138, 414–429. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proceedings of the National Academy of Sciences, USA 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vie AK, Najafi J, Liu B, Winge P, Butenko MA, Hornslien KS, Kumpf R, Aalen RB, Bones AM, Brembu T. 2015. The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. Journal of Experimental Botany 66, 5351–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K. 2007. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genetics 3, 1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hou S, Wu Q, Lin M, Acharya BR, Wu D, Zhang W. 2017. IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. The Plant Journal 89, 250–263. [DOI] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. 2007. The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. Journal of Plant Research 120, 157–165. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Vainonen JP, Stael S, et al. 2015. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO Journal 34, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB. 2011. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. The Plant Journal 66, 280–292. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. 2010. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. The Plant Cell 22, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. 2006. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences, USA 103, 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Murphy C, Sieburth LE. 2010. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proceedings of the National Academy of Sciences, USA 107, 15981–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.