Vitamin B1 biosynthesis de novo is regulated by duplicated genes and a conserved riboswitch in cassava. Diversity of vitamin B1 levels in cassava leaves negatively correlates with expression of selected vitamin B1 biosynthesis genes.
Keywords: Cassava, crop, diversity, vitamers, vitamin B1, riboswitch
Abstract
Vitamin B1, which consists of the vitamers thiamin and its phosphorylated derivatives, is an essential micronutrient for all living organisms because it is required as a metabolic cofactor in several enzymatic reactions. Genetic diversity of vitamin B1 biosynthesis and accumulation has not been investigated in major crop species other than rice and potato. We analyzed cassava germplasm for accumulation of B1 vitamers. Vitamin B1 content in leaves and roots of 41 cassava accessions showed significant variation between accessions. HPLC analyses of B1 vitamers revealed distinct profiles in cassava leaves and storage roots, with nearly equal relative levels of thiamin pyrophosphate and thiamin monophosphate in leaves, but mostly thiamin pyrophosphate in storage roots. Unusually, the cassava genome has two genes encoding the 4-amino-2-methyl-5-hydroxymethylpyrimidine phosphate synthase, THIC (MeTHIC1 and MeTHIC2), both of which carry a riboswitch in the 3ʹ-UTR, as well as the adenylated thiazole synthase, THI1 (MeTHI1a and MeTHI1b). The THIC and THI1 genes are expressed at very low levels in storage roots compared with the accumulation of vitamin B1, indicating only limited biosynthesis de novo therein. In leaves, vitamin B1 content is negatively correlated with THIC and THI1 expression levels, suggesting post-transcriptional regulation of THIC by the riboswitch present in the 3ʹ-UTR of the THIC mRNA and regulation of THI1 by promoter activity or alternative post-transcriptional mechanisms.
Introduction
Vitamin B1 is essential for all living organisms. It functions as a cofactor for various enzymes involved in key metabolic pathways, including glycolysis, the citric acid cycle, branched-chain amino acid biosynthesis, and the cytosolic non-oxidative stage of the pentose phosphate pathway (Goyer, 2010; Rapala-Kozik, 2011). Mammals, including humans, lack the ability to biosynthesize vitamin B1 and therefore crop plants are one of the major dietary sources of this micronutrient. In humans, an acute lack of vitamin B1 can lead to various chronic diseases, including cardiovascular diseases known as ‘wet’ beriberi and neurological disorders termed ‘dry’ beriberi (Rapala-Kozik, 2011). Globally, nearly two billion people suffer from deficiencies in one or more essential micronutrients (Thompson and Amoroso, 2011; von Grebmer et al., 2014; Singh et al., 2016); this is especially the case in low- and middle-income countries whose populations have low dietary diversity and limited access to supplementation strategies (Muthayya et al., 2013). Although rice, maize, and wheat remain the world’s leading cultivated crops, cassava is the most widely grown orphan food crop and is consumed predominantly in developing countries (Sayre et al., 2011; Varshney et al., 2012). Raw cassava storage roots have a vitamin B1 level that only partially covers the human daily requirement (Fitzpatrick et al., 2012). Moreover, cassava leaves and storage roots are usually soaked and boiled in water for the purpose of cyanide detoxification before consumption (Padmaja, 1995; Ufuan Achidi et al., 2005; Muoki and Maziya-Dixon, 2010). This processing can affect the nutritive value through modification and losses of nutrients, including the water-soluble and/or heat-labile vitamins (Montagnac et al., 2009b; Li et al., 2015).
Vitamin B1 biosynthesis de novo in plants has been mostly characterized in the model plant Arabidopsis (Goyer, 2010; Rapala-Kozik, 2011; Fitzpatrick and Thore, 2014). Vitamin B1 is present as three predominant vitamers in the cell, namely thiamin, thiamin monophosphate (TMP), and thiamin pyrophosphate (TPP). Triphosphorylated and adenylated forms of thiamin also exist in plants and animals but are far less abundant (Bettendorff et al., 2007; Gangolf et al., 2010). TMP is generated by the fusion of 4-methyl-5-β-hydroxyethylthiazole phosphate (HET-P) and 4-amino-2-methyl-5-hydroxymethylpyrimidine pyrophosphate (HMP-PP), which are independently biosynthesized. Biosynthesis of the thiazole moiety in plants is assumed to occur via a pathway similar to that described in yeast, in which adenosine diphospho-5-(β-ethyl)-4-methylthiazole-2-carboxylic acid (ADT) synthase (THI4) catalyzes the conversion of NAD+, glycine, and a sulfur atom from the THI4 protein itself to the adenylated thiazole intermediate, ADT (Chatterjee et al., 2011; Goyer, 2017). ADT is subsequently transformed to HET-P by an uncharacterized NUDIX hydrolase (Goyer et al., 2013; Goyer, 2017). ADT synthase is a single-turnover enzyme, which after the donation of sulfur is assumed to become catalytically inactive with regard to thiamin biosynthesis (Chatterjee et al., 2011; Fitzpatrick and Thore, 2014). Based on sequence homology, orthologs of THI4 (named THI1) have been characterized in several plant species (Belanger et al., 1995; Machado et al., 1996; Wang et al., 2006). The vitamin B1 pyrimidine moiety in plants is biosynthesized via a pathway similar to the one characterized in bacteria, during which 4-amino-2-methyl-5-hydroxymethylpyrimidine phosphate (HMP-P) synthase (THIC) catalyzes a complex rearrangement of 5-aminoimidazole ribonucleotide (AIR) to HMP-P (Lawhorn et al., 2004; Raschke et al., 2007; Kong et al., 2008; Coquille et al., 2013). In plants, HMP-P is further phosphorylated to HMP-PP by a bifunctional enzyme named TH1 in Arabidopsis (Ajjawi et al., 2007b) and THI3 in maize (Rapala-Kozik et al., 2007), which subsequently catalyzes the condensation of HMP-PP and HET-P to TMP. In plants, biosynthesized TMP is first dephosphorylated to thiamin and subsequently pyrophosphorylated to TPP. Plant enzymes from the haloacid dehalogenase (HAD) phosphatase family have recently been demonstrated to have a TMP-selective phosphatase activity, with the enzyme TH2 purportedly specific for TMP (Hasnain et al., 2016; Mimura et al., 2016). The conversion of thiamin to TPP is catalyzed by thiamin pyrophosphokinase (TPK) (Rapala-Kozik et al., 2009). Most of the enzymes involved in vitamin B1 biosynthesis de novo are localized in the chloroplast (Belanger et al., 1995; Chabregas et al., 2001; Ajjawi et al., 2007b; Raschke et al., 2007; Kong et al., 2008), except for TPKs, which are localized in the cytosol (Ajjawi et al., 2007a), and TH2, which is also cytosolic as well as being potentially targeted to the mitochondria (Mimura et al., 2016).
In Arabidopsis, THIC transcript levels are regulated by light (Raschke et al., 2007), the circadian clock (Bocobza et al., 2013), and a riboswitch in the 3ʹ-UTR of THIC mRNA (Sudarsan et al., 2003; Bocobza et al., 2007, 2013; Wachter et al., 2007), which collectively contribute to the regulation of vitamin B1 biosynthesis. The current model states that the THIC riboswitch undergoes alternative splicing in the 3ʹ-UTR region as a function of TPP, leading to the formation of transcripts with different 3ʹ-UTR lengths that affect mRNA stability (Bocobza et al., 2007; Wachter et al., 2007). When the intracellular TPP concentration is high, binding of this ligand to the riboswitch changes its conformation, exposing a splice site in the THIC 3ʹ-UTR. The consequent splicing eliminates the consensus polyadenylation signal and results in unstable long 3ʹ-UTR transcripts, reducing THIC protein levels and subsequently decreasing biosynthesis de novo of TPP.
Acquiring data about micronutrient contents in staple crops is essential to understand the potential for exploiting genetic diversity for increased micronutrient content and therefore human health. Different varieties of the same species, as well as wild species, can display considerable variation in micronutrient contents (Bouis and Welch, 2010). The exploitation of natural variation further assists in the identification of markers for candidate genes that control micronutrient accumulation (Conn et al., 2012), as it has been shown for vitamin A in rice (Vallabhaneni et al., 2009; Yan et al., 2010). Diversity of vitamin B1 content has so far been analyzed only in rice and potato germplasm (Villareal and Juliano, 1989; Sotelo et al., 1990; Kennedy and Burlingame, 2003; Goyer and Haynes, 2011; Goyer and Sweek, 2011). Characterization of the diversity of vitamin B1 accumulation in the germplasm of staple crops could help in the implementation of biofortification approaches to reduce vitamin B1 deficiencies. Such deficiencies occur at high frequency in populations whose diets are either poor in sources of vitamin B1 or rich in thiaminase, a thiamine-degrading enzyme, which is abundantly present in raw and fermented fish sauce (a common Asian delicacy) as well as certain vegetables and roasted insects consumed primarily in Africa and Asia (Boros, 2000; Barennes et al., 2015).
Here, we report the natural variation of vitamin B1 content in 41 cassava accessions grown under controlled conditions, and further investigate vitamin B1 biosynthesis and regulation. We quantified the total vitamin B1 content in leaves and storage roots by HPLC, and characterized the B1 vitamer profiles in both tissues. In order to identify potential determinants of vitamin B1 accumulation, we analyzed the transcriptional regulation of genes encoding key enzymes involved in vitamin B1 biosynthesis de novo in accessions contrasting in vitamin B1 content.
Materials and methods
Plant material
Cassava accessions were obtained as in vitro plantlet material from germplasm collections at ETH Zurich (Swiss Federal Institute of Technology, Switzerland), IITA (International Institute of Tropical Agriculture, Nigeria), CIAT (International Center for Tropical Agriculture, Columbia), MARI (Mikocheni Agricultural Research Institute, Tanzania), and CTCRI (Central Tuber Crops Research Institute, India) (see Supplementary Table S1 at JXB online). Each accession was vegetatively propagated in vitro on cassava basic medium [CBM: 1× Murashige and Skoog (MS) medium including vitamins (Duchefa), 2% (w/v) sucrose, 2 μM copper(II) sulfate, and 0.3% (w/v) gelrite; pH 5.8] and grown for 1 month in a climate chamber at 28 °C under a 16/8 h light/dark regime. Plantlets were then transferred to soil following a previously described procedure (Bull et al., 2009) and grown under greenhouse conditions (16 h light at 26 °C and 60% humidity, 8 h dark at 17 °C and 50% humidity). Leaves and storage roots from 5-month-old cassava plants were sampled for analysis. Three replicates for each of the 41 cassava accessions were used in the preliminary screening. The confirmation screening on the 18 selected cassava accessions included four biological replicates for each accession. The three youngest fully expanded leaves were sampled (without petioles) from the top of the plants and immediately frozen in liquid nitrogen. The storage roots were washed with water, peeled, and the starchy tissue immediately frozen in liquid nitrogen. To ensure that the moisture content of the tissues did not influence the analyses of vitamin B1 content, the dry matter content was calculated on the basis of the mass difference after drying the samples at 40 °C for 1 week.
For the time-course experiment, selected cassava accessions were first propagated in vitro and then grown under greenhouse conditions for 7 months following the above-described procedure. Plants were sampled every 4 h for 24 h as well as 1 h before the end of the sunlight period, 1 h before the end of the supplementary artificial light period, and 1 h before the end of the dark period. At each time point, a pool of leaf portions (corresponding to half to one lobe) from four fully expanded apical leaves was sampled and immediately frozen in liquid nitrogen.
For the sequencing of cassava THIC genes, cv. 60444 plantlets were vegetatively propagated in vitro on CBM without vitamins and grown for 10 days in a climate chamber at 28 °C under a 16/8 h light/dark regime. Plantlets were then transferred to CBM without vitamins, or supplemented with 10 μM of commercial thiamin hydrochloride (Sigma-Aldrich) for 24 h, prior to leaf sampling.
Thermal processing experiments were performed with approximately 30 cm-long commercial waxed cassava roots imported from Costa Rica. They were processed in two different ways for the evaluation of vitamin B1: (i) storage roots were peeled, sliced into ~70 g sections, and boiled in 2 l of tap water for 30 min; (ii) storage roots were peeled, sliced into ~70 g sections and soaked in 1 l of tap water for 90 min, then rinsed and boiled in fresh tap water (2 l) for 30 min. The dry matter in each sample was evaluated by drying samples for 5 days at 50 °C.
Vitamin B1 quantification
Yeast bioassay
Yeast bioassays for vitamin B1 content were performed according to a method previously established with the thi4 auxotrophic strain of Saccharomyces cerevisiae (Raschke et al., 2007). Vitamin B1 was extracted from 50 mg of leaves and 100 mg of storage roots. Frozen ground tissues were resuspended in 20 mM sulfuric acid (ratio: 100 mg tissue:1 ml extraction buffer) and incubated at room temperature for 30 min in the dark, and the extract was sterilized at 100 °C for 1 h. After extraction, the solution was adjusted to pH 5.7 using 3 M sodium acetate and centrifuged. The supernatant was then treated with acid phosphatase type I (Sigma) (0.2 U/10 μl in 50 μl plant extract) for 12–15 h at 37 °C to convert the phosphorylated forms of vitamin B1 to non-phosphorylated forms. Total vitamin B1 content was calculated from the linear range of a dose–response curve established with known amounts of commercial thiamin hydrochloride (Sigma-Aldrich).
HPLC measurements
Quantification of B1 vitamers was performed according to a previously established HPLC method with minor modifications (Moulin et al., 2013). B1 vitamers were extracted from 50 mg of frozen ground tissues using 100 μl of 1% (v/v) trichloroacetic acid. The mixture was vortexed at room temperature for 30 min and centrifuged at 16100 g for 10 min at room temperature. The clear supernatant was neutralized by adding 10% of the final volume of 3 M sodium acetate. Samples were oxidized by the addition of 30 mM potassium ferricyanide before separation on a Cosmosil π-NAP column (150 × 4.6 mm, 3 μm pore size). Quantification of TMP, TPP, and thiamin thiochrome derivatives was performed by integrating the corresponding fluorescent peak areas extrapolated from standard curves of similarly treated commercial B1 vitamers (TMP chloride, Fluka; TPP chloride, Sigma; thiamin hydrochloride, Fluka). Data were normalized to tissue fresh weight.
Sequencing and expression analysis of cassava genes from the vitamin B1 biosynthesis de novo pathway
RNA extraction
Total RNA from leaves and storage roots was extracted according to a method previously reported by Cazzonelli et al. (1998) with some minor modifications. Approximately 200–300 mg of frozen ground tissues were mixed with 1 ml of lysis buffer [150 mM Tris base adjusted to pH 7.5 with boric acid, containing 2% (w/v) SDS and 50 mM EDTA], vortexed for 5 min, and centrifuged at 16100 g for 3 min at room temperature. Absolute ethanol (0.25 volumes) and 5 M potassium acetate (0.11 volumes) were added to 800 μl of the supernatant. The mixture was extracted twice with 1 volume of chloroform:isoamylalcohol (24:1, pH 7.5–8.0) and 1 volume of phenol:chloroform:isoamylalcohol (25:24:1, pH 7.5–8.0). Nucleic acids from the recovered aqueous phase were precipitated in 1 ml of absolute ethanol for 30 min at –80 °C and centrifuged at 16100 g for 30 min at 4 °C. The pellet was washed with 80% ethanol and resuspended in 200 μl of diethylpyrocarbonate (DEPC)-treated water. DEPC-treated water was prepared by incubating 0.1% DEPC in distilled water overnight, and subsequently autoclaved. RNA was precipitated with lithium chloride (final concentration 2 M) overnight at 4 °C. The extract was centrifuged for 30 min at 4 °C and the RNA pellet washed with 80% and 100% ethanol, vacuum dried, and resuspended in DEPC-treated water.
In silico identification of cassava vitamin B1 biosynthetic genes
The cassava orthologs of the Arabidopsis genes encoding vitamin B1 biosynthetic enzymes were identified by BLASTing the Arabidopsis protein sequences from the TAIR10 database (Lamesch et al., 2012) against the available translated Manihot esculenta v6.1 genome in Phytozome (Prochnik et al., 2012).
Amplification and sequencing of MeTHIC1 and MeTHIC2 3ʹ-UTR splice variants
RNA was extracted from leaves using the above-described protocol. cDNA was synthesized with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific AG) according to the manufacturer’s instructions, using 1 μg total RNA. cDNA was synthesized in three different ways: (i) using oligo-(dT)18 primers, (ii) using random hexamer oligonucleotides, and (iii) by a 3ʹ-RACE (rapid amplification of cDNA ends) procedure, using oligo-(dT)18-adapter primers [GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18] taking advantage of the natural poly(A) tail in mRNA as a generic priming site for PCR. For cDNA synthesized with oligo-(dT)18 and random hexamer oligonucelotide primers, the 3ʹ-UTR splice variants were amplified by PCR using MeTHIC1 and MeTHIC2 specific primer sets. Forward primers were located at the 3ʹ end of THIC exon 4 and reverse primers at the end of the 3ʹ-UTR sequence predicted by Phytozome (Prochnik et al., 2012). For cDNA synthesized with oligo-(dT)18-adapter primers, the 3ʹ-UTR splice variants were amplified by PCR using MeTHIC1 and MeTHIC2 specific forward primers located at the end of THIC exon 4 and reverse primers in the adapter region. A first PCR was performed using 3ʹ-RACE primer as the reverse primer and a second one, with the PCR product as template, was performed using 3ʹ-RACE nested primer. PCR products were run on a 1% (w/v) agarose gel and the different bands, corresponding to THIC 3ʹ-UTR splice variants, were extracted, ligated into the pJET1.2/blunt vector with the Clone PCR cloning Kit (Thermo Scientific) according to the manufacturer’s instructions, and transformed into thermocompetent Escherichia coli cells. Two to six clones were sequenced for each 3ʹ-UTR splicing variant using the Sanger sequencing method. Primer sequences are reported in Supplementary Table S2.
Real-time quantitative PCR analysis
cDNA was synthesized using random hexamer oligonucleotide primers with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific AG) according to the manufacturer’s instructions, using 1 μg total RNA for leaves and 500 ng total RNA for storage roots. Real-time quantitative PCR (RT-qPCR) reactions were performed using the LightCycler 480 II system (Roche Diagnostics AG) and Fast SYBR® Green Master Mix (Applied Biosystems, Thermo Fisher Scientific). The reaction mixture contained 1 μl DEPC-treated water, 1 μM of each primer, 5 μl SYBR® Green Master Mix and 2 μl cDNA diluted 10-fold and 3-fold, respectively, for leaves and storage roots. PCR cycling conditions were as follows: initial denaturation at 95 °C for 2 min followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 30 s. Relative target gene expression levels were normalized to three reference genes, MePP2A, MeUBQ10, and MevATPs (Moreno et al., 2011), averaged by their geometric mean (Vandesompele et al., 2002). Efficiencies of primer pairs were measured and confirmed to be similar, allowing the use of the 2-ΔΔCT method (Livak and Schmittgen, 2001) to calculate relative gene expression levels. Primer sequences are reported in Supplementary Table S3.
Statistical analysis
The effect of the cultivars on vitamin B1 content, gene expression levels, and phenotype was evaluated by one-way analysis of variance (ANOVA) at the 0.05 significance level. When the effect was significant, the ANOVA was followed by a Tukey’s test for post-hoc pairwise comparisons (α=0.05). Normality was assessed using the Shapiro-Wilk test on residuals (α=0.01), and homoscedasticity was tested using Bartlett’s test (α=0.01). The gene expression levels in leaves and storage roots of each cultivar were compared using Student’s t-test. A bilateral test was applied and type 2 (homoscedasticity) or 3 (hetereoscedasticity) was determined by Fisher’s test of equality of variance (F-test; α=0.05). Expression values of MeTHIC1, MeTHI1a, and MeTHI1b in leaves and underground fresh weight of the confirmation screening of 18 accessions displayed moderate deviation from normality and homoscedasticity, possibly associated with the small sample size.
Results
Selection of cassava accessions
We selected 41 cassava accessions for a representative diversity of germplasm from different regions in Africa, Central and South America, and Asia. Cultivars were provided by CIAT (http://ciat.cgiar.org/what-we-do/crop-conservation-and-use/cassava-diversity/) and IITA (http://my.iita.org/accession2/), and included traditional cultivars, improved varieties, and elite breeding lines. Our selection was also based on criteria such as tolerance and susceptibility to viruses and Xanthomonas axonopodis pv. manihotis. Two additional wild relative species (Manihot pseudoglaziovii and Manihot tristis) were included in the selection to provide a wider range of genetic diversity (Supplementary Table S1). Cassava plants were grown under controlled conditions in a greenhouse. The range of phenotypic variation in stems, leaves, and storage roots was similar in two independent experiments, and the distribution of the accessions according to their phenotypic characteristics (in particular, plant height, above-ground fresh weight, and underground fresh weight) was nearly identical in two independent experiments (Supplementary Table S4).
Total vitamin B1 content in leaves was stable during the sampling period
In Arabidopsis, vitamin B1 biosynthesis de novo is regulated by the circadian clock, resulting in a reported significant oscillation of the TMP vitamer detected in a single period (Bocobza et al., 2013). AtTHIC transcript analysis also revealed induction of the gene by light (Raschke et al., 2007). To determine whether vitamin B1 biosynthesis is regulated diurnally in cassava, we measured vitamin B1 levels at 4-hour intervals during a 24-hour period in accessions ARG 13, cv. 60444, and BRA 132, using a yeast bioassay (Raschke et al., 2007). Vitamin B1 levels in leaves remained stable during the 24-hour period, except for BRA 132, which showed a small but statistically significant decrease during the dark period (Supplementary Fig. S1). Samples were therefore collected from all 41 cassava accessions between 13.30 and 17.00 h, when vitamin B1 levels remained stable in the three control accessions (Supplementary Fig. S1).
Natural variation of vitamin B1 levels in greenhouse-grown cassava accessions
For screening purposes, we first measured vitamin B1 content in the 41 accessions selected using a yeast bioassay, which is a cost-effective and high-throughput method. We found statistically significant differences for vitamin B1 accumulation in leaf and root tissues between the selected accessions (Supplementary Table S5). Accessions with a vitamin B1 content below the 25th percentile of the distribution were considered to be accessions with low vitamin B1 content, whereas those with a vitamin content above the 75th percentile were considered to be accessions with high vitamin B1 content. Eighteen cassava accessions contrasting in terms of the distribution of vitamin B1 content in leaves and storage roots were selected for additional independent measurements (Supplementary Table S6). Most accessions had a similar distribution of vitamin B1 content in leaves and storage roots in both independent experiments (Supplementary Tables S5 and S6); however, it should be noted that vitamin B1 levels and the extent of the variation differed between the two experiments. The distribution of accessions according to their vitamin B1 contents in leaves and storage roots was similar based on either per gram of fresh weight or per gram of dry weight (Supplementary Table S6). Our analysis revealed no correlation between the vitamin B1 contents in leaves and storage roots (Supplementary Fig. S2). In addition, no correlation was observed between vitamin B1 content and biomass (Supplementary Fig. S3). Combining the two independent measurements, we prioritized eight cassava accessions with low, medium, and high vitamin B1 contents in leaves and storage roots for further analysis.
We performed HPLC analyses on the eight selected contrasting accessions to provide an accurate quantification of B1 vitamers (Fig. 1). Vitamin B1 contents determined in the selected accessions were in line with those initially determined using the yeast bioassay, although some discrepancies were observed (Fig. 1C, D; Supplementary Table S6A, B). HPLC analyses confirmed the significant differences of vitamin B1 contents in leaves and storage roots, showing a 5.3 (± 3.6)- and a 2.7 (± 0.6)-fold difference between the cassava accessions, respectively (Fig. 1C, D).
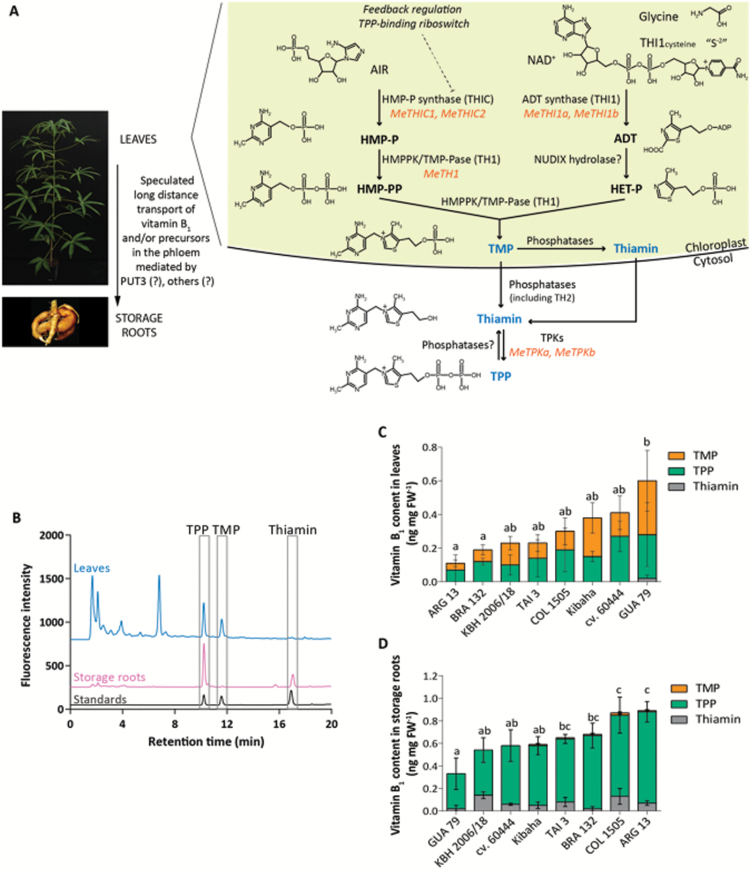
Fig. 1.
Natural variation of vitamin B1 content in selected cassava accessions. (A) Scheme of the vitamin B1 biosynthesis pathway as described in Arabidopsis and extrapolated to cassava. The three measured B1 vitamers, TMP (thiamin monophosphate), thiamin, and TPP (thiamin pyrophosphate), are shown in blue text. The pyrimidine and thiazole moieties, HMP-P (4-amino-2-methyl-5-hydroxymethylpyrimidine phosphate), HMP-PP (4-amino-2-methyl-5-hydroxymethylpyrimidine pyrophosphate), an adenylated thiazole intermediate (ADT), and HET-P (4-methyl-5-β-hydroxyethylthiazole phosphate), are shown in bold black text. The identified cassava orthologs coding for the known biosynthetic enzymes, HMP-P synthase encoded by THIC (MeTHIC1: Manes.02G121700; MeTHIC2: Manes.01G164200), ADT synthase encoded by THI1 (MeTHI1a: Manes.15G075600; MeTHI1b: Manes.03G123800), HMPPK/TMP-Pase (2-methyl-4-amino-5-hydroxymethylpyrimidine phosphate kinase/thiamin monophosphate pyrophosphorylase) encoded by TH1 (MeTH1: Manes.10G122900), and TPKs (thiamin pyrophosphokinases) encoded by TPKs (MeTPKa: Manes.05G063600; MeTPKb: Manes.01G217400), are shown in orange italic text. (B) HPLC chromatograms of leaf and storage root extracts. To facilitate visualization, the profile of storage roots was offset by 200 and the profile of leaves was offset by 750 fluorescence units, relative to the baseline. (C, D) HPLC analysis of vitamin B1 in (C) leaves and (D) storage roots. Data presented are the mean±SD of four biological replicates. Significant differences (P<0.05; Tukey’s multiple comparison test) are indicated by different letters.
The phosphorylated forms of vitamin B1 are the most abundant in cassava and B1 vitamer profiles differ in leaves and storage roots
Vitamin B1 is mainly present as phosphorylated esters and predominantly as the coenzyme vitamer, TPP, in unicellular organisms (Schweingruber et al., 1991; Moulin et al., 2013) and in leaves of higher plants, including Arabidopsis (Rapala-Kozik et al., 2012; Bocobza et al., 2013; Pourcel et al., 2013) and rice (Dong et al., 2016). Cassava fully expanded leaves also mostly accumulate the phosphorylated forms of vitamin B1, with TPP and TMP respectively accounting for 40–66% and 37–58% of total vitamin B1 levels. The thiamin vitamer was not detected in most leaf samples (Table 1A). Cassava storage roots mainly accumulated TPP (73–94% of total vitamin B1) and thiamin was detected in all accessions (6–26% of total vitamin B1), while TMP constituted less than 2% of total vitamin B1 (Table 1B).
Table 1.
HPLC analysis of B1 vitamers in leaves and storage roots of selected cassava accessions
| A | |||||||
|---|---|---|---|---|---|---|---|
| Leaves | TPP | TMP | Thiamin | Total vitamin B1 (ng mg FW–1) |
|||
| ng mg FW–1 | % total vitamin B1 | ng mg FW–1 | % total vitamin B1 | ng mg FW–1 | % total vitamin B1 | ||
| ARG 13 | 0.07 ± 0.09a | 55 ± 15 | 0.04 ± 0.02a | 45 ± 15 | nd | nd | 0.11 ± 0.11a |
| BRA 132 | 0.12 ± 0.02a | 63 ± 8 | 0.07 ± 0.03a | 37 ± 8 | nd | nd | 0.19 ± 0.04a |
| KBH 2006/18 | 0.10 ± 0.06a | 44 ± 19 | 0.13 ± 0.04ab | 55 ± 19 | nd | nd | 0.21 ± 0.06ab |
| TAI 3 | 0.14 ± 0.11a | 55 ± 27 | 0.09 ± 0.05a | 45 ± 27 | nd | nd | 0.24 ± 0.07ab |
| COL 1505 | 0.19 ± 0.13a | 65 ± 6 | 0.11 ± 0.08a | 35 ± 6 | nd | nd | 0.30 ± 0.20ab |
| Kibaha | 0.15 ± 0.03a | 42 ± 13 | 0.23 ± 0.09ab | 58 ± 13 | nd | nd | 0.37 ± 0.10ab |
| cv. 60444 | 0.27 ± 0.09a | 66 ± 21 | 0.14 ± 0.10ab | 34 ± 21 | nd | nd | 0.41 ± 0.07ab |
| GUA 79 | 0.26 ± 0.19a | 40 ± 8 | 0.32 ± 0.18b | 57 ± 9 | 0.02 ± 0.02 | 3 ± 3 | 0.60 ± 0.39b |
| B | |||||||
|---|---|---|---|---|---|---|---|
| Storage roots | TPP | TMP | Thiamin | Total vitamin B1 (ng mg FW–1) |
|||
| ng mg FW–1 | % total vitamin B1 | ng mg FW–1 | % total vitamin B1 | ng mg FW–1 | % total vitamin B1 | ||
| GUA 79 | 0.31 ± 0.14a | 94 ± 5 | nd | nd | 0.02 ± 0.03a | 6 ± 5 | 0.33 ± 0.16a |
| KBH 2006/18 | 0.40 ± 0.11ab | 73 ± 9 | 0.00 ± 0.00a | 0 ± 1 | 0.14 ± 0.03c | 26 ± 8 | 0.54 ± 0.09ab |
| cv. 60444 | 0.52 ± 0.14abc | 90 ± 1 | 0.00 ± 0.00a | 0 ± 0 | 0.06 ± 0.01ab | 10 ± 1 | 0.58 ± 0.16ab |
| Kibaha | 0.53 ± 0.08abc | 91 ± 5 | 0.01 ± 0.01ab | 1 ± 1 | 0.05 ± 0.03a | 8 ± 4 | 0.59 ± 0.10ab |
| TAI 3 | 0.56 ± 0.04abcd | 86 ± 4 | 0.01 ± 0.00ab | 2 ± 1 | 0.08 ± 0.04abc | 12 ± 5 | 0.65 ± 0.06bc |
| BRA 132 | 0.65 ± 0.11bcd | 95 ± 3 | 0.01 ± 0.01ab | 2 ± 1 | 0.02 ± 0.02a | 4 ± 3 | 0.69 ± 0.12bc |
| COL 1505 | 0.72 ± 0.16cd | 83 ± 7 | 0.02 ± 0.01b | 2 ± 1 | 0.13 ± 0.07bc | 15 ± 7 | 0.87 ± 0.16c |
| ARG 13 | 0.81 ± 0.09d | 90 ± 3 | 0.01 ± 0.01ab | 2 ± 1 | 0.07 ± 0.02abc | 8 ± 3 | 0.89 ± 0.07c |
B1 vitamer distribution in (A) leaves and (B) storage roots of greenhouse-grown cassava plants. Total vitamin B1 content corresponds to the TMP, TPP, and thiamin contents for each replicate. Percentage of total vitamin B1 corresponds to the proportion of each vitamer relative to the total vitamin B1 content. Data presented are the mean±SD of four biological replicates. Significant differences (P<0.05; Tukey’s multiple comparison test) are indicated by different letters. FW, fresh weight; nd, not detected.
Vitamin B1 content in cassava storage roots is not sufficient to reach the dietary recommended daily allowance
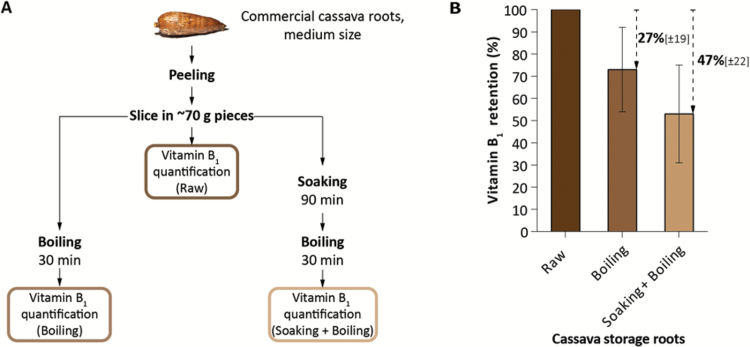
It has been reported that 100 g of raw cassava storage root contains 0.087 mg vitamin B1 (Fitzpatrick et al., 2012; USDA Nutrient Laboratory, 2016). Based on these data, the recommended daily allowance (RDA) of 1.2 mg day–1 vitamin B1 for 19- to 30-year-old female adults would be met by consuming 1.4 kg raw cassava. As vitamin B1 is known to be heat sensitive and water soluble (Fitzpatrick et al., 2012), losses are expected to occur during the processing and cooking of cassava storage roots. For example, boiling peeled potatoes for 30 minutes leads to a 12% reduction in vitamin B1 content (Augustin et al., 1978; Goyer and Haynes, 2011). Therefore, we tested whether processing and cooking alters the vitamin B1 content in cassava leaves and storage roots. We used commercial cassava roots and applied thermal processing following typical household processing methods in Northern Mozambique (Muoki and Maziya-Dixon, 2010). Cassava storage roots were peeled, sliced, and boiled in water for 30 min, or soaked for 90 min in standing water and then boiled for 30 min (Fig. 2A). Boiling cassava roots decreased vitamin B1 levels by 27 ± 19% and the additional soaking resulted in further substantial losses of up to 47 ± 22% (Fig. 2B; Supplementary Table S7). Our results highlight the importance of measuring vitamin B1 in processed and cooked cassava foodstuffs to calculate the recommended daily intake for populations relying mainly on cassava in their diet.
Fig. 2.
Thermal processing of cassava commercial storage roots and decrease in total vitamin B1 retention. (A) Cooking procedures for cassava storage roots. (B) Evaluation of vitamin B1 retention in cassava storage roots. Vitamin B1 retention was set at 100% for raw cassava; vitamin B1 losses following both processing procedures are indicated by the arrows. Vitamin B1 was measured using a yeast bioassay. Data presented are the mean±SD of four biological replicates.
Vitamin B1 biosynthesis de novo genes are duplicated in cassava and are expressed at different levels in leaves and storage roots
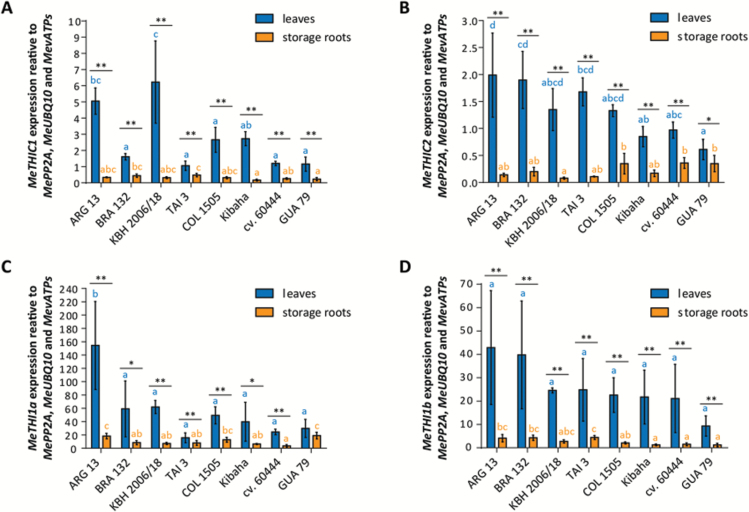
In plants, THIC and THI1 genes have been shown to regulate the production de novo and levels of vitamin B1 in leaves (Pourcel et al., 2013; Dong et al., 2015, 2016; Goyer, 2017). Using the Arabidopsis protein sequence of THIC encoded by the At2g29630 gene as a search parameter in Phytozome (Prochnik et al., 2012), two THIC genes were identified in the cassava AM560-2 reference genome v6.1. They were named MeTHIC1 (Manes.02G121700) and MeTHIC2 (Manes.01G164200), and the amino acid sequences of the corresponding encoded proteins were 95% identical. Similarly, two cassava THI1 genes were identified using the Arabidopsis THI1 protein sequence (encoded by At5g54770) as the search parameter, and were named MeTHI1a (Manes.15G075600) and MeTHI1b (Manes.03G123800). The transcript sequence of the MeTHI1b gene appeared to be incomplete in the cassava AM560-2 reference genome. However, the full MeTHI1b coding sequence could be determined on the basis of JGI Illumina sequencing data from gDNA for cassava accessions cv. 60444, TMe-3, TMe-7, SC8, and COL 22 (Bredeson et al., 2016) (Supplementary Fig. S4). The amino acid sequences of the corresponding proteins encoded by MeTHI1a and MeTHI1b appeared to be 94% identical. Despite high similarity at the coding sequence level between the cassava homologs, expression of MeTHIC1/2 and MeTHI1a/b could be discriminated by the design of sequence-specific RT-qPCR primers (Supplementary Table S3). MeTHIC and MeTHI1 genes showed differential expression in the eight selected cassava accessions in both leaves and roots, with the exception of MeTHI1b in leaf samples (Fig. 3). Expression was significantly higher in leaves than in storage roots in all of the accessions except for MeTHI1a in GUA 79. A similar pattern of expression has been found in Arabidopsis and maize, where THIC and THI1 transcription is high in green tissues and much lower in non-photosynthetic tissues (Guan et al., 2014; Colinas and Fitzpatrick, 2015). Moreover, MeTHI1 transcripts appeared to be much more abundant than MeTHIC transcripts (Fig. 3C, D). THI1 can catalyze only a single turnover and therefore its levels would be expected to be higher than those of THIC, as was also observed in Chlamydomonas (Moulin et al., 2013).
Fig. 3.
Expression levels of selected vitamin B1 biosynthesis de novo genes in leaves and storage roots of eight selected cassava accessions. (A) MeTHIC1, (B) MeTHIC2, (C) MeTHI1a, and (D) MeTHI1b expression levels in leaves and storage roots of selected cassava accessions contrasting in vitamin B1 content in these tissues. Data presented are the mean±SD of four biological replicates [except for leaves: MeTHI1a and MeTHI1b, TAI 3 (n=3) and KBH 2006/18 (n=3)]. Significant differences in gene expression levels between accessions in leaves and in storage roots (P<0.05; Tukey’s multiple comparison test) are indicated by different letters. Significant differences in gene expression levels between leaves and storage roots in each accession (Student’s t-test): *P<0.05, **P<0.01.
Leaf total vitamin B1 content is negatively correlated with expression levels of the MeTHIC and MeTHI1 homologs
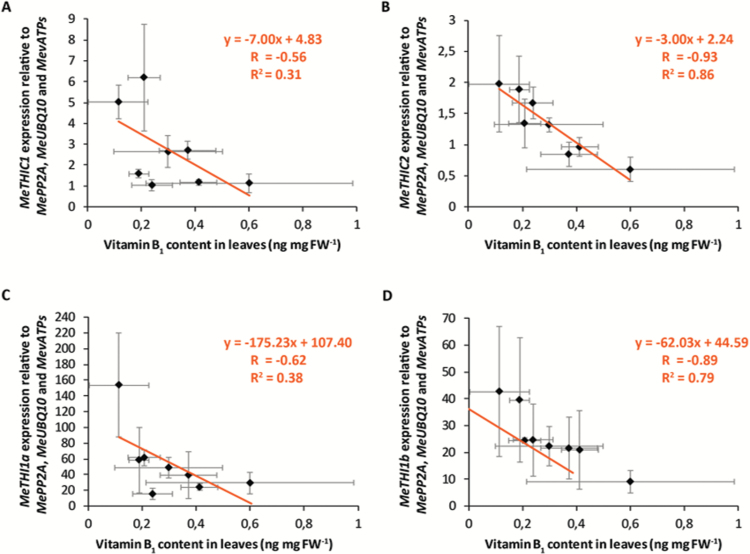
Plotting either MeTHIC1, MeTHIC2, MeTHI1a, or MeTHI1b gene expression levels in cassava leaves against the vitamin B1 content measured by HPLC for each accession revealed a negative correlation in each case (Fig. 4). The strongest negative correlation was observed for MeTHIC2 (Pearson’s correlation coefficient, R= –0.93, R2=0.86) and MeTHI1b (R= –0.89, R2=0.79) (Fig. 4B, D). Our results suggest that the negative correlation between MeTHIC transcripts and vitamin B1 content could result from the post-transcriptional regulation of THIC gene expression mediated by the 3ʹ-UTR riboswitch, as demonstrated in other plant species (Sudarsan et al., 2003; Bocobza et al., 2007, 2013; Wachter et al., 2007). The TPP riboswitch was lost from the THI1 gene family during gymnosperm evolution (Bocobza et al., 2007), and therefore regulation of MeTHI1b transcripts may occur via regulation of MeTHI1b promoter activity or alternative post-transcriptional mechanisms.
Fig. 4.
Correlation between vitamin B1 content quantified by HPLC and expression levels of selected vitamin B1 biosynthetic genes, (A) MeTHIC1, (B) MeTHIC2, (C) MeTHI1a, and (D) MeTHI1b, in cassava leaves. Data presented are the mean±SD of four biological replicates for vitamin B1 content and for gene expression [except for MeTHI1a and MeTHI1b, TAI 3 (n=3) and KBH 2006/18 (n=3)].
MeTHIC mRNAs exist as different splice variants
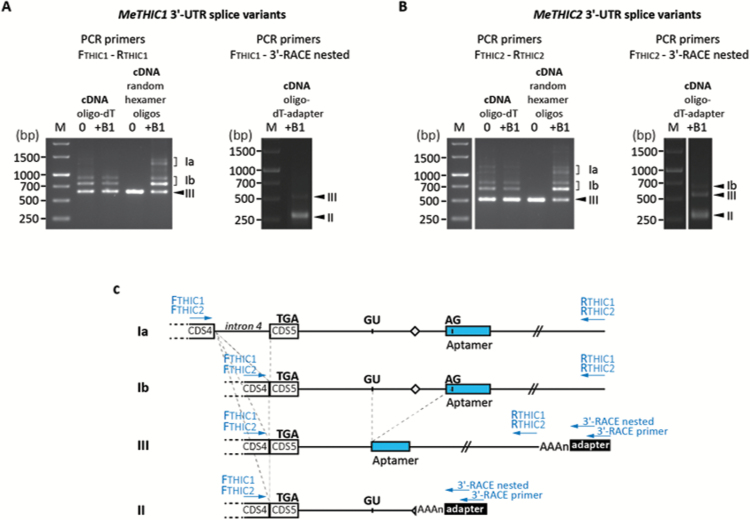
Our analysis of the MeTHIC homologs revealed that both contain a TPP riboswitch in their respective 3ʹ-UTR, based on sequence homology (Supplementary Fig. S5). In silico analysis predicted that the MeTHIC1 and MeTHIC2 3ʹ-UTRs display a structure similar to those predicted for THIC 3ʹ-UTRs in other plant species (Wachter et al., 2007). In Arabidopsis, three THIC transcript types (I–III) have been identified on the basis of their varying lengths at the 3ʹ-UTR. Type I (THIC-I) corresponds to the precursor RNA retaining the complete aptamer. THIC-I can either be processed upstream of the aptamer region to result in type II (THIC-II, intron-retained variant) with a shorter 3ʹ-UTR, or be spliced into type III (THIC-III, intron-spliced variant) with 7 bp missing at the 5ʹ end of the aptamer and a longer 3ʹ-UTR (Wachter et al., 2007). We analyzed MeTHIC1 and MeTHIC2 3ʹ-UTRs in cv. 60444, using cDNA synthesized with oligo-(dT)18 primers, random hexamer oligonucleotides, or oligo-(dT)18-aptamer primers. We were able to amplify and sequence four 3ʹ-UTR splice variants—THIC-Ia retaining intron 4, THIC-Ib spliced intron 4 upstream of the stop codon, a THIC-II variant, and a THIC-III variant—for both MeTHIC1 and MeTHIC2 using specific primer pairs (Supplementary Table S2; Fig. 5; Supplementary Fig. S5). PCR primer pairs FTHIC1-RTHIC1 (Fig. 5A, left panel) and FTHIC2-RTHIC2 (Fig. 5B, left panel) allowed the detection of type I (MeTHIC1-I, MeTHIC2-I) and the intron-spliced type III (MeTHIC1-III, MeTHIC2-III) transcripts (Fig. 5C). For both MeTHIC homologs, the smallest PCR product corresponds to MeTHIC-III, whereas the upper bands correspond to MeTHIC-Ia and MeTHIC-Ib splicing variants, as well as to products derived from these two splicing forms as previously reported in Arabidopsis (Wachter et al., 2007). The primer pairs FTHIC1-3ʹ-RACE nested (Fig. 5A, right panel) and FTHIC2-3ʹ-RACE nested (Fig. 5B, right panel) allow the detection of all three splicing variants if they are polyadenylated. Under these conditions, MeTHIC1-I could not be detected (Fig. 5A, right panel) and MeTHIC2-I was barely detectable (Fig. 5B, right panel), which suggests that only a minor fraction of the type I transcript is polyadenylated. These results support previous findings in Arabidopsis, where AtTHIC-I is likely to be the unprocessed AtTHIC pre-mRNA that is further processed and polyadenylated into type II and III mRNA variants (Wachter et al., 2007).
Fig. 5.
Organization of the cassava MeTHIC1 and MeTHIC2 3ʹ-UTR regions. PCR amplification of (A) MeTHIC1 and (B) MeTHIC2 3ʹ-UTR regions. Plants were grown in vitro in cassava basic medium without vitamin B1 (indicated by 0) or in medium supplemented with 10 μM of vitamin B1 (indicated by +B1). cDNA was synthesized using oligo-(dT)18 primers, random hexamer oligonucleotides, or oligo-(dT)18-adapter primers, and PCR was performed using two different sets of primers for both MeTHIC homologs. (C) Organization of the MeTHIC1 and MeTHIC2 3ʹ-UTR regions. CDS4 and CDS5 represent the last exons of the transcripts. An intron (intron 4) is located in front of the stop codon indicated by TGA and is spliced in transcripts Ib, II, and III. GU and AG identify the splice sites to obtain form III and the dashed lines indicate splicing events. The diamond indicates the transcript processing site. Primers used for PCR amplification of MeTHIC1 and MeTHIC2 3ʹ-UTR are indicated by arrows.
Our analysis revealed that intron 4, which is spliced in MeTHIC transcripts Ib, II, and III, is located proximal to the stop codon, similar to what has been reported for LeTHIC in tomato (Wachter et al., 2007). This is different from most other analyzed plant species, in which the stop codon is immediately followed by an intron (Wachter et al., 2007). The intron 4 (length in MeTHIC1: 493 bp, length in MeTHIC2: 366 bp) present in mRNA variant Ia and spliced in Ib, II, and III (Fig. 5C) may represent regular splicing events in cassava because no alternative splice variants II and III retaining this intron could be amplified. However, analysis of MeTHIC1-Ia and MeTHIC2-Ia transcripts using the ExPASy translate tool (Artimo et al., 2012) revealed an immediate stop codon in intron 4, leading to the translation of THIC1 and THIC2 proteins with two amino acid residue truncations at the C-terminus (Supplementary Fig. S6). It remains to be demonstrated whether this truncation affects THIC functionality.
Discussion
In this study, we have shown that cassava germplasm has significant variation in vitamin B1 accumulation between accessions. Previous studies have shown that vitamin B1 content in rice germplasm varies up to 2.8-fold in white rice and up to 2.0-fold in brown rice (Villareal and Juliano, 1989; Sotelo et al., 1990). Similarly, potato germplasm studies, including primitive cultivated wild species, have shown that vitamin B1 content varies up to 2.6-fold in mature tubers (Goyer and Sweek, 2011). Our analysis shows a comparable range of variation (i.e. 2.7-fold in storage roots) in the assessed cassava germplasm. Although the 41 cassava accessions we have analyzed represent only a small fraction of the overall genetic diversity in cassava, they are representative of cassava varieties cultivated for subsistence and commercial production (Supplementary Table S1). Our measurements of vitamin B1 content in peeled cassava storage roots provide concentration values that are in the range of previous reports (Woot-Tsuen, 1968; Favier, 1977; Montagnac et al., 2009a; USDA Nutrient Laboratory, 2016). This suggests that standard greenhouse conditions and a relatively short growth cycle (i.e. 6 months) can be used to estimate vitamin B1 in cassava storage roots. However, the average vitamin B1 content we measured in cassava leaves was 5- to 5.5-fold below previously reported measurements (Wobeto et al., 2006; Rapala-Kozik et al., 2008, 2012; Shewry et al., 2011). Growing conditions, plant age, and vitamin B1 extraction method may explain at least in part these discrepancies.
The comparison of vitamin B1 levels in 100 g portions of five major crops shows that raw cassava contains intermediate levels of vitamin B1 (Fitzpatrick et al., 2012). The present study has shown that typical processing of cassava roots by soaking and boiling further decreases the amount of vitamin B1. Moreover, polyphenolic compounds that accumulate at high levels in cassava (Montagnac et al., 2009b) might reduce the bioavailability of vitamin B1 by reacting with it to yield non-absorbable vitamin B1 disulfide (Hilker and Somogyi, 1982; Hotz and Gibson, 2007; Rapala-Kozik, 2011). Several studies have reported vitamin B1 deficiencies in populations whose diet is mostly based on cassava (Adamolekun, 2011). Our HPLC data suggest that genetic variation of vitamin B1 content in cassava leaves and storage roots, which was 5.4- and 2.7-fold, respectively (Table 1), could be exploited by trait introgression to address the issue of vitamin B1 deficiencies.
We found that total vitamin B1 content in cassava leaves has no or only limited variation during a 24-hour diurnal period (Supplementary Fig. S1). Vitamin B1 biosynthesis de novo in Arabidopsis is regulated by light (Raschke et al., 2007), as well as in a circadian manner via the THIC gene promoter (Bocobza et al., 2013). The transcript levels of AtTHIC and AtCCA1, which encodes the circadian clock regulator CIRCADIAN CLOCK ASSOCIATED 1, oscillate in opposite patterns in Arabidopsis (Bocobza et al., 2013). CCA1 is a transcriptional repressor that can bind to the evening element (AAAATATCT) located 128 bp upstream of the AtTHIC 5ʹ-UTR. Our analysis here indicates the absence of CCA1 binding sites in the 1 kb region located upstream of the MeTHIC1 and MeTHIC2 5ʹ-UTRs (Supplementary Fig. S7), suggesting that both cassava THIC homologs have not retained CCA1-based circadian control of gene expression. However, the absence of circadian regulation remains to be demonstrated for the cassava THIC genes.
A direct comparison of the genes involved in the vitamin B1 biosynthesis de novo pathway in Arabidopsis indicates that the pathway is conserved in cassava, with the notable exception of the duplication of the THIC and THI1 genes. One cassava ortholog (MeTH1: Manes.10G122900) of AtTH1 (At1g22940) and two TPK genes (MeTPKa: Manes.05G063600 and MeTPKb: Manes.01G217400) corresponding to AtTPK1 (At1g02880) and AtTPK2 (At2g44750) are present in the cassava genome. The duplication of THIC genes is not specific to cassava. Ten of the 64 plant species for which genomes are available in the Phytozome database (Goodstein et al., 2012) have two or more THIC genes (Supplementary Fig. S8). In the case of THI1, 18 plant species have more than one ortholog (Supplementary Fig. S8). We focused our analysis on THIC and THI1 genes because both were previously shown to regulate the production and level of vitamin B1, using either precursor supplementation in Arabidopsis (Pourcel et al., 2013) or transgenic overexpression approaches in Arabidopsis and rice (Dong et al., 2015, 2016; Goyer, 2017). Based on biofortification studies in Arabidopsis and rice, further bottlenecks impeding higher accumulation of thiamin have been hypothesized (Dong et al., 2015; Goyer, 2017).
Sequence analysis of MeTHIC1 and MeTHIC2 indicates that both have retained a TPP riboswitch sequence in their 3ʹ-UTRs, with small differences in the P3 stem of the aptamer sequence, which is not directly involved in TPP binding (Wachter et al., 2007) (Supplementary Fig. S5). Expression profiles of MeTHIC genes in leaves and roots of selected accessions are similar, although the transcript levels of individual genes can vary (Fig. 3). An alignment of THIC protein sequences shows that the central domain containing the (β/α)8 TIM barrel, which likely includes the active site (Coquille et al., 2013), is conserved in both cassava THIC homologs (Supplementary Fig. S9). Moreover, the main residues responsible for binding the AIR substrate, which are located in the iron sulfur cluster binding loop, and the metal ion binding site (Coquille et al., 2013), are conserved in both MeTHIC1 and MeTHIC2 (Supplementary Fig. S9). Collectively, these results suggest that MeTHIC1 and MeTHIC2 have retained the same functions. Both MeTHI1a and MeTHI1b share a high similarity with AtTHI1 (Supplementary Fig. S4B) and invariant residues of ADT synthases are conserved in the MeTHI1 proteins (Godoi et al., 2006). MeTHI1 proteins are predicted by TargetP (Emanuelsson et al., 2007) to be targeted to the chloroplast.
Overall, MeTHIC and MeTHI1 transcript levels in the selected accessions were negatively correlated with vitamin B1 content in leaves (Fig. 4). The negative feedback regulation of THIC mRNAs by the TPP-binding riboswitch has been previously characterized in plants (Bocobza et al., 2007, 2013; Wachter et al., 2007). The strong negative correlation between MeTHIC2 transcript levels (combined expression of forms Ib, II, and III; Fig. 5) and vitamin B1 content (Fig. 4) suggests that a THIC riboswitch-based control also regulates vitamin B1 content in cassava. The levels of vitamin B1 might also be partially feedback-controlled by the regulation of MeTHI1b, because transcripts of MeTHI1b also have a strong negative correlation with vitamin B1 content in cassava leaves (Fig. 4). However, the mechanism regulating MeTHI1b transcript levels remains unknown.
The lower accumulation of TMP and the relatively low expression of MeTHIC and MeTHI1 genes in cassava storage roots, compared with leaves, indicate a limited capacity of storage roots to biosynthesize the pyrimidine and thiazole moieties that are the precursors of TMP. However, our results show that cassava storage roots do indeed accumulate vitamin B1, which is essential for metabolic activity, including the pentose phosphate pathway, acetyl-CoA biosynthesis, and the tricarboxylic acid cycle (Frank et al., 2007; Rapala-Kozik, 2011). In plants, vitamin B1 biosynthesis de novo predominantly occurs in green tissues (Guan et al., 2014; Colinas and Fitzpatrick, 2015; Martinis et al., 2016) because most of the biosynthetic enzymes are localized in the chloroplast (Belanger et al., 1995; Chabregas et al., 2001; Ajjawi et al., 2007b; Raschke et al., 2007; Kong et al., 2008). In Arabidopsis and maize, vitamin B1 biosynthetic genes are expressed strongly in green tissues and to a much lower extent in non-photosynthetic organs, supporting the recently proposed concept of a ‘division of labor’ between photosynthetic and non-photosynthetic tissues, which serve, respectively, as a source and a sink of vitamin B1 (Guan et al., 2014; Colinas and Fitzpatrick, 2015). High vitamin B1 biosynthesis in leaves and long-distance transport of vitamin B1 and/or vitamin B1 precursors to sink tissues could compensate for reduced vitamin B1 biosynthesis in storage roots to meet the requirements for metabolic activity. A vitamin B1 long-distance transporter has recently been identified in Arabidopsis (Martinis et al., 2016). Moreover, thiamin, TMP, and TPP are detectable in phloem sap, implying that these three vitamers could be transported from source leaves to sink organs such as roots (Martinis et al., 2016). The lack of a strong positive correlation between vitamin B1 accumulation in cassava leaves and in storage roots (Supplementary Fig. S2) suggests the existence of a complex control of vitamin B1 transport and homeostasis. Future studies should focus on the contribution of vitamin B1 biosynthesis, salvage, and transport to the accumulation of vitamin B1 in cassava organs. This knowledge will help in the implementation of strategies for the development of cassava varieties with enhanced vitamin B1 levels in organs/tissues consumed by populations deficient in this micronutrient.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Evaluation of vitamin B1 content in leaves over a 24 h period.
Fig. S2. Correlation between vitamin B1 content in leaves and storage roots.
Fig. S3. Correlation between plant phenotype and vitamin B1 content in leaves or storage roots.
Fig. S4. Determination of the MeTHI1b full coding sequence and alignment of AtTHI1, MeTHI1a, and MeTHI1b protein sequences.
Fig. S5. Sequenced MeTHIC1 and MeTHIC2 3ʹ-UTR splice variants in the cassava accession cv. 60444.
Fig. S6. Translation of MeTHIC1 and MeTHIC2 transcript forms Ia (including intron 4) and Ib (excluding intron 4).
Fig. S7. Analysis for a CCA1 binding motif upstream of the 5ʹ-UTR of Arabidopsis and cassava THIC genes.
Fig. S8. Number of THIC and THI1 homologs in the genomes available in the Phytozome database.
Fig. S9. Alignment of THIC protein sequences from Arabidopsis and cassava.
Table S1. Description of the 41 cassava accessions selected for quantification of vitamin B1 content.
Table S2. Primers used for PCR amplification of MeTHIC1 and MeTHIC2 3ʹ-UTR splice variants.
Table S3. Primers used for RT-qPCR analysis.
Table S4. Phenotypic characterization of greenhouse-grown cassava accessions.
Table S5. Preliminary quantification of vitamin B1 in 41 cassava accessions using a yeast bioassay.
Table S6. Repeated independent quantification of vitamin B1 in 18 selected cassava accessions using a yeast bioassay.
Table S7. Thermal processing of cassava commercial storage roots.
Supplementary Material
Acknowledgements
Financial support is gratefully acknowledged from the Swiss National Science Foundation (grants 31003A-141117/1 and 31003A-162555 to TBF; grant 31003A-140911 to WG, HV, and TBF), the VELUX Foundation, and the University of Geneva and the ETH Zurich. The authors thank Irene Zurkirchen (ETH Zurich) for support in the greenhouse and Michael Moulin (University of Geneva) for advice regarding the HPLC measurements and data interpretation.
References
- Adamolekun B. 2011. Neurological disorders associated with cassava diet: a review of putative etiological mechanisms. Metabolic Brain Disease 26, 79–85. [DOI] [PubMed] [Google Scholar]
- Ajjawi I, Rodriguez Milla MA, Cushman J, Shintani DK. 2007a. Thiamin pyrophosphokinase is required for thiamin cofactor activation in Arabidopsis. Plant Molecular Biology 65, 151–162. [DOI] [PubMed] [Google Scholar]
- Ajjawi I, Tsegaye Y, Shintani D. 2007b. Determination of the genetic, molecular, and biochemical basis of the Arabidopsis thaliana thiamin auxotroph th1. Archives of Biochemistry and Biophysics 459, 107–114. [DOI] [PubMed] [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K et al. . 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research 40, W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J, Johnson SR, Teitzel C, True RH, Hogan JM, Toma RB, Shaw RL, Deutsch RM. 1978. Changes in the nutrient composition of potatoes during home preparation: II. Vitamins. American Potato Journal 55, 653–662. [Google Scholar]
- Barennes H, Sengkhamyong K, René JP, Phimmasane M. 2015. Beriberi (thiamine deficiency) and high infant mortality in northern Laos. PLoS Neglected Tropical Diseases 9, e0003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger FC, Leustek T, Chu B, Kriz AL. 1995. Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Molecular Biology 29, 809–821. [DOI] [PubMed] [Google Scholar]
- Bettendorff L, Wirtzfeld B, Makarchikov AF et al. . 2007. Discovery of a natural thiamine adenine nucleotide. Nature Chemical Biology 3, 211–212. [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. 2007. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes & Development 21, 2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocobza SE, Malitsky S, Araújo WL, Nunes-Nesi A, Meir S, Shapira M, Fernie AR, Aharoni A. 2013. Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis. The Plant Cell 25, 288–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros LG. 2000. Population thiamine status and varying cancer rates between western, Asian and African countries. Anticancer Research 20, 2245–2248. [PubMed] [Google Scholar]
- Bouis HE, Welch RM. 2010. Biofortification—a sustainable agricultural strategy for reducing micronutrient malnutrition in the global South. Crop Science 50, S20–S32. [Google Scholar]
- Bredeson JV, Lyons JB, Prochnik SE et al. . 2016. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotechnology 34, 562–570. [DOI] [PubMed] [Google Scholar]
- Bull SE, Owiti JA, Niklaus M, Beeching JR, Gruissem W, Vanderschuren H. 2009. Agrobacterium-mediated transformation of friable embryogenic calli and regeneration of transgenic cassava. Nature Protocols 4, 1845–1854. [DOI] [PubMed] [Google Scholar]
- Cazzonelli C, Cavallaro A, Botella J. 1998. Cloning and characterisation of ripening-induced ethylene biosynthetic genes from non-climacteric pineapple (Ananas comosus) fruits. Functional Plant Biology 25, 513–518. [Google Scholar]
- Chabregas SM, Luche DD, Farias LP, Ribeiro AF, van Sluys MA, Menck CF, Silva-Filho MC. 2001. Dual targeting properties of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Molecular Biology 46, 639–650. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. 2011. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. 2007. Biosynthesis of thiamin thiazole in eukaryotes: conversion of NAD to an advanced intermediate. Journal of the American Chemical Society 129, 2914–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Schroeder FC, Jurgenson CT, Ealick SE, Begley TP. 2008. Biosynthesis of the thiamin-thiazole in eukaryotes: identification of a thiazole tautomer intermediate. Journal of the American Chemical Society 130, 11394–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinas M, Fitzpatrick TB. 2015. Natures balancing act: examining biosynthesis de novo, recycling and processing damaged vitamin B metabolites. Current Opinion in Plant Biology 25, 98–106. [DOI] [PubMed] [Google Scholar]
- Conn SJ, Berninger P, Broadley MR, Gilliham M. 2012. Exploiting natural variation to uncover candidate genes that control element accumulation in Arabidopsis thaliana. New Phytologist 193, 859–866. [DOI] [PubMed] [Google Scholar]
- Coquille S, Roux C, Mehta A, Begley TP, Fitzpatrick TB, Thore S. 2013. High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana. Journal of Structural Biology 184, 438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Stockwell VO, Goyer A. 2015. Enhancement of thiamin content in Arabidopsis thaliana by metabolic engineering. Plant & Cell Physiology 56, 2285–2296. [DOI] [PubMed] [Google Scholar]
- Dong W, Thomas N, Ronald PC, Goyer A. 2016. Overexpression of thiamin biosynthesis genes in rice increases leaf and unpolished grain thiamin content but not resistance to Xanthomonas oryzae pv. oryzae. Frontiers in Plant Science 7, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Favier J. 1977. Valeur alimentaire de deux aliments de base africains: le manioc et le sorgho. Paris: ORSTOM (Office de la Recherche Scientifique et Technique Outre-mer). [Google Scholar]
- Fitzpatrick TB, Basset GJ, Borel P et al. . 2012. Vitamin deficiencies in humans: can plant science help?The Plant Cell 24, 395–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Thore S. 2014. Complex behavior: from cannibalism to suicide in the vitamin B1 biosynthesis world. Current Opinion in Structural Biology 29, 34–43. [DOI] [PubMed] [Google Scholar]
- Frank RA, Leeper FJ, Luisi BF. 2007. Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cellular and Molecular Life Sciences 64, 892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangolf M, Wins P, Thiry M, El Moualij B, Bettendorff L. 2010. Thiamine triphosphate synthesis in rat brain occurs in mitochondria and is coupled to the respiratory chain. Journal of Biological Chemistry 285, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoi PH, Galhardo RS, Luche DD, Van Sluys MA, Menck CF, Oliva G. 2006. Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana. Journal of Biological Chemistry 281, 30957–30966. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R et al. . 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer A. 2010. Thiamine in plants: aspects of its metabolism and functions. Phytochemistry 71, 1615–1624. [DOI] [PubMed] [Google Scholar]
- Goyer A. 2017. Thiamin biofortification of crops. Current Opinion in Biotechnology 44, 1–7. [DOI] [PubMed] [Google Scholar]
- Goyer A, Hasnain G, Frelin O, Ralat MA, Gregory JF 3rd, Hanson AD. 2013. A cross-kingdom Nudix enzyme that pre-empts damage in thiamin metabolism. The Biochemical Journal 454, 533–542. [DOI] [PubMed] [Google Scholar]
- Goyer A, Haynes KG. 2011. Vitamin B1 content in potato: effect of genotype, tuber enlargement, and storage, and estimation of stability and broad-sense heritability. American Journal of Potato Research 88, 374–385. [Google Scholar]
- Goyer A, Sweek K. 2011. Genetic diversity of thiamin and folate in primitive cultivated and wild potato (Solanum) species. Journal of Agricultural and Food Chemistry 59, 13072–13080. [DOI] [PubMed] [Google Scholar]
- Guan JC, Hasnain G, Garrett TJ, Chase CD, Gregory J, Hanson AD, McCarty DR. 2014. Divisions of labor in the thiamin biosynthetic pathway among organs of maize. Frontiers in Plant Science 5, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain G, Roje S, Sa N, Zallot R, Ziemak MJ, de Crécy-Lagard V, Gregory JF 3rd, Hanson AD. 2016. Bacterial and plant HAD enzymes catalyse a missing phosphatase step in thiamin diphosphate biosynthesis. The Biochemical Journal 473, 157–166. [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouzé P, Brunak S. 1996. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Research 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker DM, Somogyi JC. 1982. Antithiamins of plant origin: their chemical nature and mode of action. Annals of the New York Academy of Sciences 378, 137–145. [DOI] [PubMed] [Google Scholar]
- Hotz C, Gibson RS. 2007. Traditional food-processing and preparation practices to enhance the bioavailability of micronutrients in plant-based diets. Journal of Nutrition 137, 1097–1100. [DOI] [PubMed] [Google Scholar]
- Kennedy G, Burlingame B. 2003. Analysis of food composition data on rice from a plant genetic resources perspective. Food Chemistry 80, 589–596. [Google Scholar]
- Kong D, Zhu Y, Wu H, Cheng X, Liang H, Ling HQ. 2008. AtTHIC, a gene involved in thiamine biosynthesis in Arabidopsis thaliana. Cell Research 18, 566–576. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D et al. . 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40, D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhorn BG, Mehl RA, Begley TP. 2004. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Organic & Biomolecular Chemistry 2, 2538–2546. [DOI] [PubMed] [Google Scholar]
- Li KT, Moulin M, Mangel N et al. . 2015. Increased bioavailable vitamin B6 in field-grown transgenic cassava for dietary sufficiency. Nature Biotechnology 33, 1029–1032. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Machado CR, de Oliveira RL, Boiteux S, Praekelt UM, Meacock PA, Menck CF. 1996. THI1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Molecular Biology 31, 585–593. [DOI] [PubMed] [Google Scholar]
- Martinis J, Gas-Pascual E, Szydlowski N, Crèvecoeur M, Gisler A, Bürkle L, Fitzpatrick TB. 2016. Long-distance transport of thiamine (vitamin B1) is concomitant with that of polyamines. Plant Physiology 171, 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura M, Zallot R, Niehaus TD, Hasnain G, Gidda SK, Nguyen TN, Anderson EM, Mullen RT. 2016. Arabidopsis TH2 encodes the orphan enzyme thiamin monophosphate phosphatase. The Plant Cell 28, 2683–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac JA, Davis CR, Tanumihardjo SA. 2009a. Nutritional value of cassava for use as a staple food and recent advances for improvement. Comprehensive Reviews in Food Science and Food Safety 8, 181–194. [DOI] [PubMed] [Google Scholar]
- Montagnac JA, Davis CR, Tanumihardjo SA. 2009b. Processing techniques to reduce toxicity and antinutrients of cassava for use as a staple food. Comprehensive Reviews in Food Science and Food Safety 8, 17–27. [DOI] [PubMed] [Google Scholar]
- Moreno I, Gruissem W, Vanderschuren H. 2011. Reference genes for reliable potyvirus quantitation in cassava and analysis of Cassava brown streak virus load in host varieties. Journal of Virological Methods 177, 49–54. [DOI] [PubMed] [Google Scholar]
- Moulin M, Nguyen GT, Scaife MA, Smith AG, Fitzpatrick TB. 2013. Analysis of Chlamydomonas thiamin metabolism in vivo reveals riboswitch plasticity. Proceedings of the National Academy of Sciences of the United States of America 110, 14622–14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoki PN, Maziya-Dixon B. 2010. Household utilization of Manioc (Manihot esculenta Crantz) in Northern Mozambique. Ecology of Food and Nutrition 49, 337–356. [DOI] [PubMed] [Google Scholar]
- Muthayya S, Rah JH, Sugimoto JD, Roos FF, Kraemer K, Black RE. 2013. The global hidden hunger indices and maps: an advocacy tool for action. PloS One 8, e67860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmaja G. 1995. Cyanide detoxification in cassava for food and feed uses. Critical Reviews in Food Science and Nutrition 35, 299–339. [DOI] [PubMed] [Google Scholar]
- Pourcel L, Moulin M, Fitzpatrick TB. 2013. Examining strategies to facilitate vitamin B1 biofortification of plants by genetic engineering. Frontiers in Plant Science 4, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik S, Marri PR, Desany B et al. . 2012. The cassava genome: current progress, future directions. Tropical Plant Biology 5, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapala-Kozik M. 2011. Vitamin B1 (thiamine): a cofactor for enzymes involved in the main metabolic pathways and an environmental stress protectant. Advances in Botanical Research 58, 37–90. [Google Scholar]
- Rapala-Kozik M, Gołda A, Kujda M. 2009. Enzymes that control the thiamine diphosphate pool in plant tissues. Properties of thiamine pyrophosphokinase and thiamine-(di)phosphate phosphatase purified from Zea mays seedlings. Plant Physiology and Biochemistry 47, 237–242. [DOI] [PubMed] [Google Scholar]
- Rapala-Kozik M, Kowalska E, Ostrowska K. 2008. Modulation of thiamine metabolism in Zea mays seedlings under conditions of abiotic stress. Journal of Experimental Botany 59, 4133–4143. [DOI] [PubMed] [Google Scholar]
- Rapala-Kozik M, Olczak M, Ostrowska K, Starosta A, Kozik A. 2007. Molecular characterization of the thi3 gene involved in thiamine biosynthesis in Zea mays: cDNA sequence and enzymatic and structural properties of the recombinant bifunctional protein with 4-amino-5-hydroxymethyl-2-methylpyrimidine (phosphate) kinase and thiamine monophosphate synthase activities. The Biochemical Journal 408, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapala-Kozik M, Wolak N, Kujda M, Banas AK. 2012. The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biology 12, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke M, Burkle L, Muller N, Nunes-Nesi A, Fernie AR, Arigoni D, Amrhein N, Fitzpatrick TB. 2007. Vitamin B1 biosynthesis in plants requires the essential iron sulfur cluster protein, THIC. Proceedings of the National Academy of Sciences of the United States of America 104, 19637–19642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre R, Beeching JR, Cahoon EB et al. . 2011. The BioCassava plus program: biofortification of cassava for sub-Saharan Africa. Annual Review of Plant Biology 62, 251–272. [DOI] [PubMed] [Google Scholar]
- Schweingruber AM, Dlugonski J, Edenharter E, Schweingruber ME. 1991. Thiamine in Schizosaccharomyces pombe: dephosphorylation, intracellular pool, biosynthesis and transport. Current Genetics 19, 249–254. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Van Schaik F, Ravel C, Charmet G, Rakszegi M, Bedo Z, Ward JL. 2011. Genotype and environment effects on the contents of vitamins B1, B2, B3, and B6 in wheat grain. Journal of Agricultural and Food Chemistry 59, 10564–10571. [DOI] [PubMed] [Google Scholar]
- Singh U, Praharaj CS, Singh SS, Singh NP. 2016. Biofortification of food crops. New Delhi: Springer India. [Google Scholar]
- Sotelo A, Sousa V, Montalvo I, Hernandez M, Hernandez-Aragon L. 1990. Chemical composition of different fractions of 12 Mexican varieties of rice obtained during milling. Cereal Chemistry 67, 209–212. [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B, Amoroso L. 2011. Combating micronutrient deficiencies: food-based approaches. Wallingford: CAB International; Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Ufuan Achidi A, Ajayi OA, Bokanga M, Maziya-Dixon B. 2005. The use of cassava leaves as food in Africa. Ecology of Food and Nutrition 44, 423–435. [Google Scholar]
- USDA Nutrient Laboratory 2016. National nutrient database for standard reference, release 28, basic report: 11134, Cassava, raw. Washington, DC: United States Department of Agriculture, Agricultural Research Service. [Google Scholar]
- Vallabhaneni R, Gallagher CE, Licciardello N, Cuttriss AJ, Quinlan RF, Wurtzel ET. 2009. Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiology 151, 1635–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Ribaut JM, Buckler ES, Tuberosa R, Rafalski JA, Langridge P. 2012. Can genomics boost productivity of orphan crops?Nature Biotechnology 30, 1172–1176. [DOI] [PubMed] [Google Scholar]
- Villareal CP, Juliano BO. 1989. Variability in contents of thiamine and riboflavin in brown rice, crude oil in brown rice and bran-polish, and silicon in hull of IR rices. Plant Foods for Human Nutrition 39, 287–297. [DOI] [PubMed] [Google Scholar]
- von Grebmer K, Saltzman A, Birol E et al. . 2014. 2014 Global hunger index: the challenge of hidden hunger. Bonn: Welthungerhilfe; Washington D.C: International Food Policy Research Institute; Dublin: Concern Worldwide. [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. 2007. Riboswitch control of gene expression in plants by splicing and alternative 3ʹ end processing of mRNAs. The Plant Cell 19, 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ding X, Yuan M, Qiu D, Li X, Xu C, Wang S. 2006. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation. Plant Molecular Biology 60, 437–449. [DOI] [PubMed] [Google Scholar]
- Wobeto C, Corrêa AD, de Abreu CMP, dos Santos CD, de Abreu JR. 2006. Nutrients in the cassava (Manihot esculenta Crantz) leaf meal at three ages of the plant. Food Science and Technology (Campinas) 26, 865–869. [Google Scholar]
- Woot-Tsuen WL, Félix B, Jardin C. 1968. Food composition table for use in Africa. Bethesda: U.S. Department of Health, Education, and Welfare; Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Yan J, Kandianis CB, Harjes CE et al. . 2010. Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nature Genetics 42, 322–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.