An analysis of exonic and intronic signatures revealed that there was post-transcriptional regulation of cytosolic transcripts during C4 leaf ontogeny.
Keywords: C4, Cleomaceae, EISA, leaf ontogeny, photosynthesis, post-transcription
Abstract
C4 photosynthesis allows highly efficient carbon fixation that originates from tightly regulated anatomical and biochemical modifications of leaf architecture. Recent studies showed that leaf transcriptome modifications during leaf ontogeny of closely related C3 (Tarenaya hassleriana) and C4 (Gynandropsis gynandra) species within the Cleomaceae family existed but they did not identify any dedicated transcriptional networks or factors specifically driving C4 leaf ontogeny. RNAseq analysis provides a steady-state quantification of whole-cell mRNAs but does not allow any discrimination between transcriptional and post-transcriptional processes that may occur simultaneously during leaf ontogeny. Here we use exon–intron split analysis (EISA) to determine the extent to which transcriptional and post-transcriptional processes are involved in the regulation of gene expression between young and expanded leaves in both species. C4-specific changes in post-transcriptional regulation were observed for genes involved in the Calvin–Benson cycle and some photosystem components but not for C4 core-cycle genes. Overall, this study provides an unbiased genome-wide insight into the post-transcriptional mechanisms that regulate gene expression through the control of mRNA levels and could be central to the onset of C4 photosynthesis. This mechanism is cytosolic which implies cell-specific modifications of mRNA stability. Understanding this mechanism may be crucial when aiming to transform C3 crops into C4 crops.
Introduction
C4 photosynthesis is one of the most efficient carbon-fixing reactions on Earth (Hatch and Slack, 1966). It is widespread in many ecosystems and has existed since the late Miocene (Ehleringer et al., 1997). Based on ancestral C3 photosynthesis, it is a fantastic example of convergent evolution in response to variable atmospheric CO2 in angiosperms (Aubry et al., 2011; Williams et al., 2012). Interestingly, the independent evolution of at least 18 C4 plant families seem to be constrained to a recurrent sequence of small evolutionary steps that progressively increased the carbon concentration in the vicinity of the leaf vasculature (Heckmann et al., 2013; Williams et al., 2013). Briefly, in order to favour the ribulose 1,5-bisphosphatase carboxylase/oxygenase (RuBisCO) carboxylation reaction over its undesired oxygenation reaction, C4 plants concentrate CO2 around the enzyme and thereby drastically limit photorespiration (and therefore increase photosynthetic activity). In order to allow carbon concentration, the carbon cycle is segregated into two compartments, each containing one of the two carboxylases involved: the phosphoenolpyruvate carboxylase (PPC) in mesophyll (M) cells and RuBisCO in bundle-sheath (BS) cells. After diffusing into the leaf through stomata that have acquired the ability specifically to regulate their opening in otherwise unfavourable CO2 and humidity conditions (Aubry et al., 2016), CO2 is primarily fixed by the O2-insensitive PPC. It is subsequently exported as a four carbon compound to the BS cells where a decarboxylase charges the Calvin–Benson cycle (CBC) to feed the final carboxylation by RuBisCO. The biochemistry of various subtypes of C4 photosynthesis is classified by one of the three different decarboxylases recruited (Furbank, 2011).
Several complementary studies using high-throughput transcriptomics approaches performed multiple comparisons of steady-state transcript levels between C3 and C4 leaves as well as during their ontogeny (Brautigam et al., 2010; Gowik et al., 2011; Aubry et al., 2014a; Külahoglu et al., 2014). These ‘omics’ approaches are aimed at identifying new elements of transcriptional networks that regulate the C4 pathway (reviewed in Burgess and Hibberd, 2015). However, they also confirmed the assumption that multiple levels of regulation—transcriptional (Brown et al., 2011), post-transcriptional (Patel and Berry, 2008), and post-translational (Pick et al., 2011)—contribute to the onset and maintenance of the C4 metabolic system (Hibberd and Covshoff, 2010).
Modifications in gene expression are thought to be associated with many of the C4-specific anatomical or biochemical features, such as increased vein density, changes in stomata regulation, chloroplast dimorphism, chloroplast positioning, BS suberization, and endoreduplication (Li et al., 2010; Majeran et al., 2010; Gowik et al., 2011; Pick et al., 2011; Aubry et al., 2014a, 2016; Külahoglu et al., 2014). Further studies focused on differences in the transcriptome between specialized leaf cell types—bundle-sheath (BS) and mesophyll (M) cells—and seem to confirm the function of transcriptional regulation for gene expression tuning in C4 cycle regulation but, so far, without identifying any dedicated transcriptional networks (Li et al., 2010; Chang et al., 2012; John et al., 2014; Aubry et al., 2014a; Burgess and Hibberd, 2015). It is noteworthy that, in C4 leaves, cell-specificity is not limited to the core C4 cycle (i.e. from the initial fixation until the RuBisCO carboxylation step) but applies to other parts of the photosynthetic apparatus. In Flaveria and Cleome, transcriptional signatures show a clear segregation of photosystem I (PSI) and photosystem II (PSII) elements between BS and M cells (Gowik et al., 2011; Aubry et al., 2014a). In C4 species, from the NADP malic enzyme (NADPME)-subtype (like Flaveria and maize) BS chloroplasts are mostly agranal and there is a strong transcriptional investment in ATP production by cyclic electron flow that includes genes encoding proteins of PSI, cytochrome b6/f and NDH (NADH dehydrogenase) complexes (Nakamura et al., 2013; Aubry et al., 2014a). RuBisCO transcript levels (and most other CBC transcripts) are largely reduced in C4 compared with C3 leaves (Ku et al., 1979; Bräutigam et al., 2010). This reduction in RuBisCO is thought to be one of the major advantages of C4 nitrogen use efficiency (Oaks, 1994; Ghanoum et al., 2005). Mechanisms that restrain RuBisCO from accumulating in BS are complex and involve a mixture of transcriptional and post-transcriptional features (Berry et al., 2016). In both monocots and dicots, BS-specific expression of RuBisCO is under robust post-transcriptional control. Thus, during leaf development, expression of RbcS and RbcL have been shown to be uncoupled from translation (Patel and Berry, 2008) and RbcS untranslated regions (UTR) are suggested to play a role in RNA stability during the later stages of leaf development. Post-transcriptional regulation of gene expression may not be restricted to RuBisCO but may also be a more widely used strategy for other (photosynthesis-related) gene regulation, especially during C4 leaf development.
In order to assess the involvement of post-transcriptional regulation of gene expression, we took advantage of comparative experiments on closely related species during leaf ontogeny in both C3 and C4 leaves (Külahoglu et al., 2014) using a computational approach called exon–intron split analysis (EISA). EISA takes advantage of the presence of both mature and intron-containing pre-mRNAs in RNAseq samples to discriminate transcriptional from post-transcriptional effects on gene expression (Gaidatzis et al., 2015). While the abundance of transcripts may vary between two experimental conditions (such as here during leaf development), a gene that is exclusively subjected to transcriptional regulation should show tightly correlated proportions of reads mapping to introns and exons. By contrast, a gene that is subjected to post-transcriptional regulation is expected to exhibit a decreased amount of reads mapping to exons compared with reads mapping to introns, as only mature intron-free cytosolic mRNA should be impacted. This has been shown to be a possible proxy to predict gene regulation status (Gaidatzis et al., 2015). For example, a significant increase in intronic RNA expression without an increase of corresponding exonic RNA has been observed in some animal systems after antioxidant treatment, even if the physiological significance of such mechanisms remains unclear (Menon et al., 2016). We applied EISA to leaf development transcriptome series from Tarenaya hassleriana (C3) and Gynandropsis gynandra (C4), two closely related species within the Cleomaceae (Külahoglu et al., 2014). Despite some anatomical and biochemical modifications, gene expression profiles of the two species appeared relatively well conserved during leaf ontogeny (Aubry et al., 2014a; Külahoglu et al., 2014). Nevertheless genes encoding photosynthetic components and CBC enzymes appear to be subject to post-transcriptional regulation at the level of mRNA accumulation. This process is likely to be a key driver of C4 leaf ontogeny.
Materials and methods
Data access
RNAseq data were obtained from NCBI (accessions: SRP036637 and SRP036837, for G. gynandra and T. hassleriana, respectively).
The reference genome for Arabidopsis was accessed from Phytozome (http://www.phytozome.net) with the most up-to-date gene model. The genome sequences from G. gynandra and T. hassleriana were accessed as described previously (Aubry et al., 2016; Cheng et al., 2013).
Processing of the reads and quantification of exonic and intronic expression levels
The EISA method was implemented to obtain expression values for reads mapping to introns or exons as published (Gaidatzis et al., 2015) with the following criteria used. Only transcripts that map to a unique position in the genome were considered. Only reads that map inside the body of the gene (no UTRs) were taken into account and only introns enclosed by exons were considered. Exon/intron counts were quantified using qCount from QuasR (Lun et al., 2016). A threshold requiring at least two reads for every exon and intron was applied. Normalization based on the total number of reads for each library was performed separately for exons and introns. Overlapping genes were not considered. ΔExon and ΔIntron were defined as the difference in log2 of exonic or intronic expression levels between respective compartments. The model for statistical significance of differential post-transcriptional regulation was implemented in edgeR (Lun et al., 2016). Generally, the lack of reliable genome annotation, especially a definition of exon–intron boundaries, can limit the interpretation of our data (Weber, 2015). We validated our approach using publically available transcriptome data from the ago1-27 mutant (accession number: GSE77211, see Supplementary Table S1 at JXB online). This analysis showed that suppression of ARGONAUTE1 (AGO1) protein, a core component of the gene-silencing machinery (Morel et al., 2002; Vaucheret et al., 2004) modifies the extent of post-transcriptional regulation of 216 genes, especially RNA-metabolic genes (GO term enriched: RNA splicing, RNA processing, and RNA metabolism, Supplementary Table S1). It should be noted that the number of biological replicates and the coverage of the sequencing are crucial when intending to use EISA for a comprehensive evaluation of post-transcriptional regulation of gene expression.
Analysis of differential gene expression and gene ontology enrichment
Reads from RNAseq libraries of young and expanded leaves were aligned to protein-coding transcripts from the respective genomes of G. gynandra and T. hassleriana (Cheng et al., 2013; Aubry et al., 2016). Transcript expression was quantified using RSEM version 1.2.30 (Li and Dewey, 2011). Posterior probability of differential expression (PPDE) between the two conditions was estimated using the empirical Bayesian approach implemented in EBSeq version 1.1.6 (Leng et al., 2013). Gene ontology enrichment was performed using a corrected Benjamini–Hochberg enrichment score implemented in Pageman (Usadel et al., 2006).
Results and discussion
Application of exon–intron split analysis to plant datasets
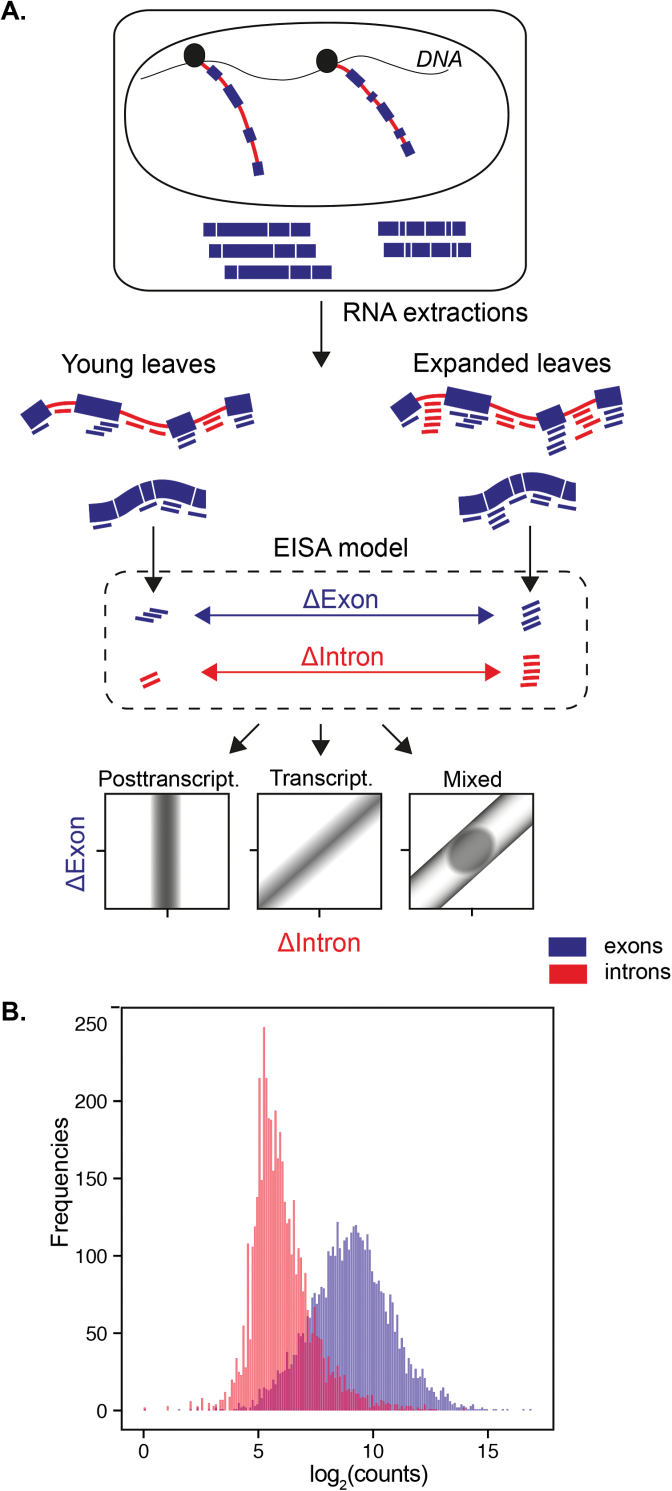
During standard RNAseq library preparation, the total extraction of mRNAs results in a mixture of mature cytosolic RNA and nuclear nascent or unspliced RNAs (Fig. 1A). Even if the vast majority of the reads analysed originate from exons, a fraction of the reads maps as introns (Fig. 1B). These might originate from genomic contamination, wrong gene models (mis-annotated exons) or from premature RNAs. Levels of intron-specific reads have been proposed to correlate with transcriptional activity and to be used as a proxy for analysing transcription (Zeisel et al., 2011; Hendriks et al., 2014). Intronic read levels often, but not always, correlate with exonic reads when compared across experimental conditions. In the case of post-transcriptionally regulated genes that are for example under miRNA control, the relative level of mature transcripts declines as the silencing machinery exclusively acts on cytosolic mature RNAs. It is of note that there are fewer intronic reads in the preparation when mRNA is selected by poly-A purification compared with total RNA extractions by ribosomal RNA depletion (Trapnell et al., 2010). However, data that are derived from both methods are still suitable for EISA, given a sufficient coverage of the transcriptome (Gaidatzis et al., 2015).
Fig. 1.
The exon–intron split analysis (EISA) detects the extent of gene expression regulation under post-transcriptional control between two experimental conditions. EISA was applied during leaf ontogeny between young and expanded leaves of G. gynandra and T. hassleriana. (A) Pre-mRNA comprising introns (red) and exons (blue) are transcribed in the nucleus, then spliced and translocated to the cytosol where the mature mRNA will be translated. EISA counts the differences in intronic (ΔIntron) and exonic (ΔExons) levels between the experimental conditions and compares them as a measure of post-transcriptional controls. The bottom part shows plots expressing ΔExon as a function of ΔIntron to show the possible patterns that are representative of the predominant mechanisms regulating gene expression (mostly transcriptional, post-transcriptional or a mixture of the two). The schematic representation was adapted from Gaidatzis et al. (2015). (B) RNA extraction results in a majority of reads mapping to exons but also reads originating from intronic segments.
The EISA method, by quantifying reads mapping exclusively to either introns or exons (see Materials and methods for details on filtering), aims to identify transcripts for which the stoichiometry between intron and exon is lost when comparing two experimental conditions (Fig. 1A). In the present study, it was possible successfully to apply EISA to recent congeneric Cleomaceae whole leaf gradient data (Külahoglu et al., 2014) but cell-specific data sets were not sequenced deep-enough to provide a statistically robust result (data not shown; Li et al., 2010; Chang et al., 2012; Aubry et al., 2014a; John et al., 2014).
Genome-scale prediction of transcriptional and post-transcriptional regulation in Cleomaceae leaf gradients
Leaf ontogeny variations between C3 and C4 leaves observed from comparisons between congeneric species have been well documented for Cleome, Flaveria, and Alloteropsis species (Gowik et al., 2011; Christin et al., 2012; Külahoglu et al., 2014). Here we used publically available transcriptomic data from congeneric Cleomaceae leaf gradients (Külahoglu et al., 2014). For each species, we selected the extreme stages of the leaf gradient (stage 0: ‘Young’ and stage 5: ‘Expanded’) analysed in the study (Külahoglu et al., 2014). 8 855 (37% of all genes detected) and 6 672 genes (28%) were differentially expressed between these two conditions in G. gynandra and T. hassleriana, respectively. EBSeq was applied using a posterior probability of being differentially expressed ≥0.95 and a minimal fold change of 2 between the two conditions. The results are summarized in Table 1 and Supplementary Table S2).
Table 1.
Genes that were detected and that have been filtered (minimal coverage of two reads per exon or intron) by the EISA pipeline and the number of genes differentially transcribed between young and expanded leaves (FDR ≤0.05) in the two Cleomaceae species The proportion of genes DT compared with the total number of genes can be found within the brackets. Differential expression of genes was calculated between young and expanded leaves using EBSeq. Overlaps between EISA and EBSeq output are shown in Fig. 2.
| Differential transcription (EISA) | G. gynandra | T. hassleriana |
|---|---|---|
| Total no. of genes | 23 340 | 22 227 |
| Genes selected for EISA | 4 769 | 5 720 |
| Post-transcriptionally regulated (FDR ≤0.05) | 678 (15%) | 263 (5%) |
| Differential expression (EBSeq) | ||
| No of genes DE (FC ≥2, PPDE ≥0.95) | 8 855 | 6 672 |
| Up in young leaves | 3 949 | 2,756 |
| Up in expanded leaves | 4 906 | 3,916 |
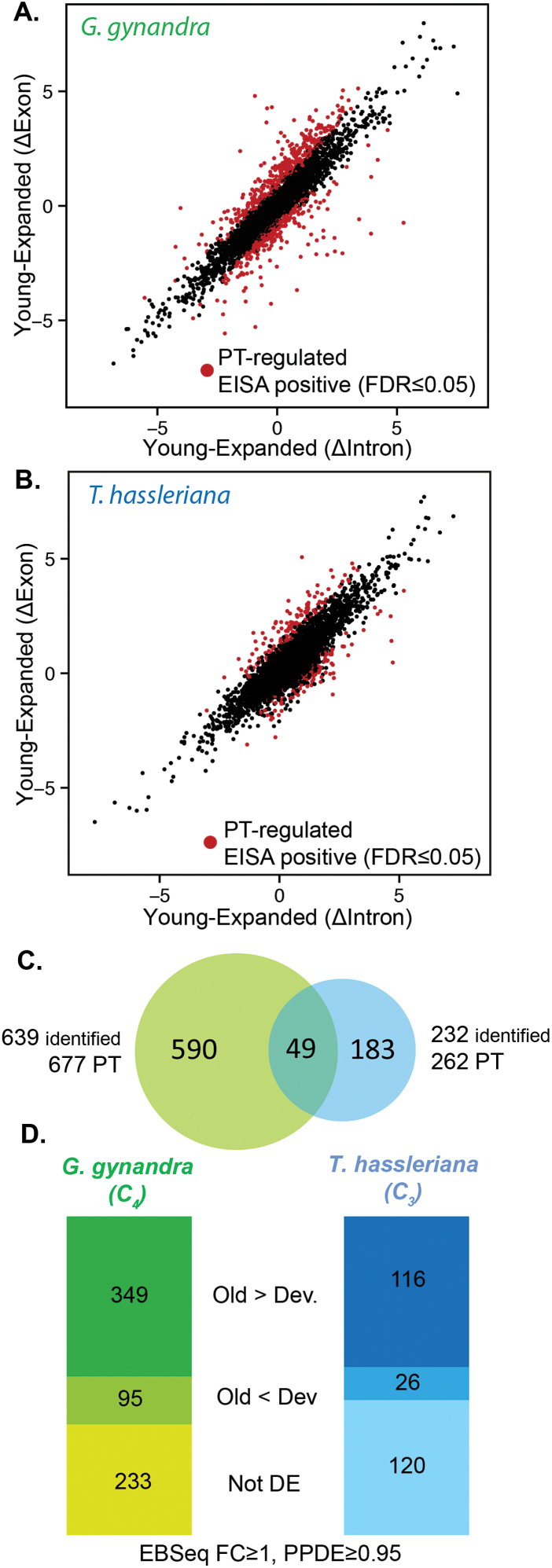
The EISA method was used on all expressed genes to identify post-transcriptional events occurring between young and expanded leaves in both C3T. hassleriana and C4G. gynandra (Fig. 2A, B). This method requires complete coverage in exons as well as introns. In order to limit false positives, a stringent minimal read cut-off was applied (see Materials and methods for details). This filtering left a quarter of the genes for both species: 4 769 and 5 720 in G. gynandra and T. hassleriana, respectively(Table 1). Of these genes, 678 in G. gynandra and 263 genes in T. hassleriana were most likely post-transcriptionally regulated (EISA positive) between young and expanded leaves with a false discovery rate FDR ≤0.05 (Table 1). Finally, 49 post-transcriptionally regulated genes were common to both species, including, for example, the gene encoding phosphoribulokinase (PRK) (Fig. 2C; Supplementary Table S3). This is consistent with a global rewiring of gene expression at the post-transcriptional level specific to the C4 leaf. A majority of the predicted post-transcriptionally regulated genes: 349 (51%) for G. gynandra and 116 (44%) for T. hassleriana were also significantly over-expressed during leaf ontogeny (Fig. 2D). Gene Ontology (GO) terms associated with photosystems and the CBC were significantly over-represented in genes that were post-transcriptionally regulated between young and expanded leaves from G. gynandra.
Fig. 2.
Changes in intronic and exonic levels during leaf development for (A) G. gynandra and (B) T. hassleriana. Significantly up- or down-regulated genes after EISA analysis (FDR ≤0.05) are shown in red. (C) Overlap between the genes that were post-transcriptionally regulated and identified as potential orthologues by BLAST analysis across the two species. (D) Among post-transcriptionally regulated genes, a majority of genes were differentially expressed between young and expanded leaves in each species (differential analysis using EBSeq with a PPDE ≥0.95 and a minimal fold change of 2). PT, post-transcriptionally regulated.
This approach was further corroborated by looking at genes that were previously shown to be under post-transcriptional regulation, for example, maize glutathione reductase 1 (GR1) is under post-transcriptional regulation in bundle-sheath cells (Pastori et al., 2000). In G. gynandra, GR1 is under post-transcriptional regulation according to the EISA prediction. A significant drop in ΔExon during leaf development (ΔExon–ΔIntron=–2.66; Supplementary Table S4) was detected. BS-specific post-transcriptional regulation of GR1 might result from convergent evolution or is ancestral to the monocot/dicot divergence as previously suggested for some C4 genes (Aubry et al., 2014a).
C4 cycle genes are mostly transcriptionally regulated during G. gynandra leaf ontogeny
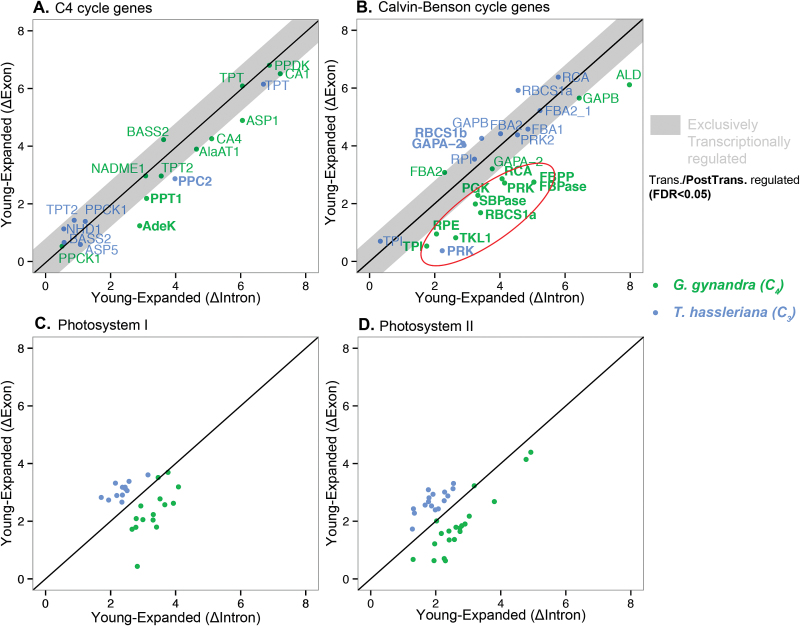
Expression of genes encoding proteins of the C4 cycle is up-regulated during leaf development in both species, but to a much larger extent in the C4 species (see Supplementary Fig. S1A at JXB online). Increased expression of C4 cycle genes in the C3 species during leaf development has previously been shown in many other dicotyledonous systems. This suggests that C4 gene regulatory networks pre-existed in C3 species and were associated with their ancestral anaplerotic function (Aubry et al., 2011; Wang et al., 2013). The EISA model tries to identify genes that show expression changes at the exon level that are not accounted for at the intron level. Genes encoding C4 core-cycle proteins are not post-transcriptionally regulated: variations of the number of reads that map introns and exons are correlated for most of these genes except for two, GgPPT1 (phosphoenolpyruvate/phosphate translocator) and ThPPC2 (phosphoenolpyruvate carboxylase 2) (Fig. 3A). This shows that, in both C3 and C4 species, C4 cycle genes are subjected to an increase in amplitude as the leaf matures but the magnitude of this amplitude is much higher in C4 leaves.
Fig. 3.
Changes in intronic and exonic levels during leaf development for genes involved in specific pathways. Genes encoding for the C4 cycle (A), the Calvin–Benson cycle (B), photosystem I (C), and photosystem II (D) show various levels of post-transcriptional regulation between young and expanded leaves in G. gynandra (green dots) and T. hassleriana (blue dots). The grey area contains genes that are transcriptionally regulated, i.e. with ΔExon and ΔIntron correlated during leaf development. The names of post-transcriptionally regulated genes are in bold and EISA-positive genes from the Calvin–Benson cycle are highlighted within a red circle. The dots in plot (C) represent subunits of photosystem I: Lhca1, Lhca2, Lhca3, Lhca4, Lhca5, Lhca6, Ohp2, PsaN, PsaL, PsaD1, Ptac8, PsaE2, PsaG, PsaH2, PsAf, PsaK, PsaO, PsaD2; and those in plot (D) represent subunits of photosystem II: Lhcb6, Cab1, Lhcb2.1, Lhcb4.3, Lhcb5, Lhcb4.1, Lhcb3, Psb27, Lpa19, Psbp-1, Pql1, Npq4, PsbY, PsbR, Pql3, PsbX, PsbW, Ppl2, PsbO2, PsbQ, Psb28, Ohp, Lpa2 (for single gene ΔExon and ΔIntron data, see Supplementary Table S4). Abbreviations: AdeK, adenylate kinase; AlaAT1, alanine aminotransferase; ALD, aldolase; ASP1/5, aspartate aminotransferase 1/5; BASS2, bile acid symporter; CA1/4, carbonic anhydrase 1/4; FBA2_1, fructose bisphosphate aldolase 2_1; FBPase, fructose-bisphosphate aldolase; FBPP, fructose 1,6-bisphosphate phosphatase; GAP-A -B, GADPDH subunit-A or –B; NADME1, NAD-dependent malic enzyme 1; NHD1, sodium:proton antiporter; PGK, phosphoglycerate kinase; PPC2, phosphoenolpyruvate carboxylase; PPCK1, phosphoenolpyruvate carboxykinase 1; PPDK, pyruvate-orthophosphate dikinase; PPT1, phosphoenolpyruvate/phosphate translocator; PRK1/2, phosphoribulokinase; SBPase, sedoheptulose-bisphosphatase; RBCS1a/b, RuBisCO small subunit 1a/b; RCA, RuBisCO activase; RPE, d-ribulose-5-phosphate-3-epimerase; RPI, ribose-5-phosphate isomerase; RuBisCO, ribulose 1,5-bisphosphatase carboxylase; TKL1, transketolase 1; TPI, triose phosphate isomerase; TPT1 & 2, triose phosphate/phosphate translocator.
Interestingly, this increase in expression does not seem to be regulated at the post-transcriptional level according to EISA. This is consistent with data previously published suggesting a strong involvement of the transcriptional machinery to the C4 cycle (Bräutigam et al., 2010; Aubry et al., 2014a). However, regulation of M specificity of C4 cycle genes like GgPPDK (pyruvate-orthophosphate dikinase) or GgCA4 (β-carbonic anhydrase 4) is regulated by post-transcriptional mechanisms involving untranslated regions (Williams et al., 2016). This suggests a complex interplay between transcriptional and post-transcriptional regulation of gene expression for these genes.
Calvin–Benson cycle and photosystem genes are post-transcriptionally regulated in the C4 leaf gradient
Genes encoding proteins involved in the CBC are up-regulated during leaf development in both C3 and C4 species (Supplementary Fig. S1). In contrast to the core C4 cycle, there is a tendency towards a lower expression of CBC genes in C4 leaves (Bräutigam et al., 2010). In G. gynandra, most genes (10 out of 13 detected by EISA) showed a significant (FDR ≤0.05) drop in ΔExon compared with ΔIntron (Fig. 3B). In most C4 species, the CBC is split between BS and M cells (Majeran et al., 2005, 2010; Aubry et al., 2014a; John et al., 2014). Interestingly, all genes involved in the CBC known to be specifically expressed in BS cells are predicted as being post-transcriptionally regulated (FDR ≤0.05, Fig. 3B). Genes that are preferentially expressed in M, like triose phosphate isomerase (TPI) or the GAPDH subunit B (GAP-B) are also post-transcriptionally regulated, suggesting this mechanism is not exclusive to BS genes. These genes, nonetheless, have their expression level increased at the tissue level, proportional to the leaf age (Supplementary Fig. S1). We postulate that the significant decrease in ΔExon during leaf development is a mixture of transcriptional and post-transcriptional regulation. None of the T. hassleriana CBC genes, except phosphoribulokinase (PRK), were predicted to be under post-transcriptional regulation during leaf development (blue dots in Fig. 3B). This suggests a post-transcriptional regulation mechanism specific to the C4 leaf. It is known that most CBC genes are expressed cell-specifically in G. gynandra leaves. Therefore, we propose a model of post-transcriptional regulation that operates between BS and M cells on a specific subset of transcripts. For example, sedoheptulose-bisphosphatase (SBPA) is mostly expressed in BS cells and the ΔExon/Δintron correlation is lost during leaf development (Fig. 3B). This can be interpreted as an increase in expression at the transcriptional level in BS cells, combined with a mesophyll-specific degradation of exon-only mature transcripts. In a similar manner, a large proportion, i.e. 18 and 23 transcripts encoding for PSI and PSII proteins, respectively, were post-transcriptionally regulated (decrease in ΔExon, Fig. 3C, D). These results indicate that regulation of C4 core genes is somehow distinct from photosynthetic genes during C4 leaf ontogeny and that this mechanism could be the basis for the actual cell-specific regulation of photosynthesis in C4 leaves.
Post-transcriptional regulation of RbcS and cell-specificity
RuBisCO is a hexadecameric protein, composed of eight nuclear-encoded subunits (RBCS) and eight chloroplast-encoded subunits (RBCL). In C4 dicots like Flaveria and amaranths, RbcL and RbcS expression are tightly regulated and repressed in the mesophyll (Patel and Berry, 2008; Berry et al., 2016). RbcS expression at the translational level appears to be central to the early stage of development, whereas mRNA stability mediates cell-specificity at a later stage (Patel and Berry, 2008). Consistent with this, ectopic expression of RbcS under a constitutive promoter in maize failed to accumulate transcripts in the mesophyll compartment (Wostrikoff et al., 2012).
Our data imply that, despite an overall increase in RbcS expression on the whole leaf scale (Supplementary Fig. S1), at least part of the RbcS1a expression is under post-transcriptional regulation in G. gynandra (Fig. 3B). Given the high specificity of RbcS transcripts in the BS compartment at a steady-state level in G. gynandra (Aubry et al., 2014a), the mesophyllic compartment is the most obvious place for this to happen. The cytosolic degradation of mature transcripts translates to a decrease in ΔExon during leaf development. The potential influence of cytosol-based regulation of RbcS transcript levels on chloroplast expression of RbcL remains to be shown, as gene expression of both subunits is tightly linked (Wostrikoff et al., 2012).
Models for post-transcriptional regulation of photosynthesis in C4 leaves
We have shown here that post-transcriptional control of RNA stability is likely to play a key function during C4 leaf ontogeny, but the molecular basis of the mechanism involved in C4 species remains unclear. Our results confirm a large body of data indicating that post-transcriptional regulation is crucial for photosynthesis regulation as, for example, in the case of ferredoxin-1 mRNA stability controlled by light in tobacco (Petracek et al., 1998). In plants, multiple levels of control can regulate cytosolic mRNA stability: basal RNA decay machineries, sequence- and secondary structure-specific decay (that often implies cis-elements in untranslated regions), and stimulus-dependent degradation often linked to translational repression (Gutiérrez et al., 1999). The mRNA decay processes are complex and can be subjected to multiple modes of degradation: de-adenylation of the poly-A tail and/or 5′-end decapping and 3′-end uridylation potentially followed by 5′–3′ decay or 3′–5′ degradation by the exosome (Gutiérrez et al., 1999; Siwaszek et al., 2014). All of these mechanisms require a specific array of trans-acting factors. We discuss here the likelihood of each potential control mechanism to be involved in the regulation of C4 leaf ontogeny.
miRNA
miRNA-mediated degradation that is known to repress gene expression in diverse developmental processes (phase transition, leaf shape, floral organ identity; Chen, 2009) could be a possible mechanism to explain the observed post-transcriptional regulation. Unfortunately, no cell-specific miRNA profiling of C4 leaves has been published to our knowledge.
Non-sense-mediated decay (NMD)
Cytosolic aberrant mRNAs may be degraded via the non-sense-mediated mRNA decay (NMD) pathways, considered to be one of the main post-transcriptional regulation processes in eukaryotes (Siwaszek et al., 2014; Dai et al., 2016). However, genes involved in the NMD pathway in plants such as up-frameshift protein 1 and 3 (UPF1 and 3) and exoribonucleases (XRN4, 5,6) were not differentially expressed between BS and M cells in mature G. gynandra leaves and no cell-specific polymorphism could be observed between the transcripts from the two cell types (Aubry et al., 2014a). This mechanism is therefore unlikely to be recruited for transcriptional regulation of photosynthetic genes during C4 leaf ontogeny.
Translation
Interaction with the translational machinery is another way of controlling mRNA levels by translational repression (Roy and Jacobson, 2013). The relationship between mRNA decay and translation efficiency are, however, complex and differ on a gene-to-gene basis. More work is necessary to show whether ribosomal loading of mRNA is actually different in genes predicted to be post-transcriptionally regulated using EISA modelling. Interestingly, a complex regulation of translation of RbcS transcripts observed in Arabidopsis (assessed by monitoring the association of transcripts to various polysomal fractions) may indicate that already in C3 ancestors RuBisCO expression is under a specific regime of transcriptional and post-transcriptional regulation (Piques et al., 2009). Therefore, further experiments analysing the association of photosynthesis genes with ribosomes on a cell-specific basis are required. Moreover, applying recently developed ribosomal footprints to such leaf material (Ingolia et al., 2009; Juntawong et al., 2014) could shine new light on transcription–translation interactions in a C4 context.
Splicing
Differences in mRNA splicing at the pathway level that would lead to a mis-interpretation of the variations in intronic signals are unlikely. Indeed, a large proportion of genes predicted to be post-transcriptionally regulated and involved in the same pathway also present very distinct intron/exon organization (Gaidatzis et al., 2015). The median number of exons and introns of post-transcriptionally (EISA FDR ≤0.05) or transcriptionally regulated (EISA FDR >0.05) genes were compared in each species and showed no significant differences based on Wilcoxon–ann–Whitney test. This indicates that the EISA output is not biased by the structures of the genes.
Post-transcriptional co-ordination of gene expression
In G. gynandra, M specificity of two C4 genes, PPDK and CA4, is mediated by 5′ and 3′ UTR regions (Kajala et al., 2012; Williams et al., 2012, 2016). The mechanism co-ordinating the cell-specific expression involves a cis-element (MEM2) that specifically increases translation in the M compartment (Williams et al., 2016). Other examples of gene expression co-ordination of functionally related genes have been described (Keene, 2007; Goodarzi et al., 2012). Here we postulate that mechanisms similar to those co-ordinating the expression of photosynthesis genes have been recruited during C4 leaf development. More work is necessary to identify which of the 200 predicted RNA-binding proteins from six different families in Arabidopsis might regulate mRNA stability (Fedoroff, 2002; Ambrosone et al., 2012).
Finally, a model involving RNA binding proteins that regulate the mRNA stability of a given set of photosynthesis-related genes during leaf ontogeny of C4 might be the simplest scenario that explains our observations (Williams et al., 2012). Distinct families of RNA binding proteins could be involved (Silverman et al., 2013), such as PUF proteins that often recognize 3′ UTRs and are dependent on secondary RNA structure (Francischini and Quaggio, 2009; Lu et al., 2009) or sequence-specific PENTATRICOPEPTIDE REPEAT (PPR) that are mostly but not exclusively localized in mitochondria and chloroplasts (Colcombet et al., 2013). More work is required to understand the extent of its complexity (e.g. timing of the regulation in both cells) and diversity (e.g. how that regulation accommodates multiple types of Kranz anatomy) across other C4 species. Interestingly, this model would provide two advantages at the pathway level, (i) a reasonable control over transcripts originating from M cells in M cells themselves, and, (ii) would avoid the potential accumulation of transcripts that could have leaked from BS cells (or other cells) into M cells and is, therefore an efficient way to control translation tightly in each respective compartment.
EISA limitations and potentials for the C4 photosynthesis field
Evidence of the presence of unspliced pre-mature RNAs in total RNA samples has already been reported when comparing transcriptome and polyribosomal translatome fractions from Arabidopsis leaves (Aubry et al., 2014b; Zhang et al., 2015). When the fraction of RNA that binds ribosomes (referred to as the ‘translatome’) was immunopurified, most of the intron-containing pre-mRNAs were eliminated. This also implies that the EISA method cannot be applied to data deriving from TRAPseq experiments recently used in one of the few cell-specific expression-profiling studies of C3 BS cells from Arabidopsis (Aubry et al., 2014b).
Using data from entire leaf tissue might not be sufficient to obtain a precise cell-specific understanding of the post-transcriptional mechanisms, but does provide a genome-wide insight of their involvement in the pathway regulation. Further experiments using cell-specific fractions with deep sequencing coverage, and taking into account the untranslated regions of the genes for the EISA, are needed to refine the EISA analysis.
Conclusions
Relatively few genes are known to be post-transcriptionally regulated in C4 leaves and the extent to which this level of regulation is involved in cellular fate remains unclear. Some level of post-transcriptional regulation has been suggested to occur in a single gene-based manner, such as for CAs, PPDK, and RbcS that are specific to C4 leaves of multiple genes encoding photosynthetic machinery components. The model proposed here is reminiscent of the theory that conceptualizes high-order regulation of functionally related mRNAs (Keene, 2007; Tenenbaum et al., 2011). More work is needed to clarify the exact significance of this post-transcriptional signature in each cell type, in particular, the mesophyll compartment of C4 leaves and how this mechanism interacts with predicted transcriptional modules and translation (Williams et al., 2012; Aubry et al., 2014a; Külahoglu et al., 2014). Perhaps not surprisingly, the stability of chloroplast-encoded transcripts is also very variable among the developmental stages (Klaff and Gruissem, 1991). The fact that a large majority of the nuclear-encoded proteins in the C4 leaf under post-transcriptional regulation are eventually targeted to the chloroplast might suggest an involvement of some plastid-to-nucleus retrograde signalling in this process. Unfortunately, all high-throughput C4 cell-specific data available (Li et al., 2010; Chang et al., 2012; Aubry et al., 2014a; John et al., 2014) do not have enough coverage to use the EISA method robustly (data not shown). Therefore, in the context of C4 engineering, more research is required to identify relevant RNA binding, RNA processing (helicases) proteins, and non-coding regulatory RNA. Together with a careful monitoring of mRNA half-lives, this could set the stage for the engineering of an efficient C4 cycle in a C3 species such as rice. Our unbiased genome-wide approach shows a massive rewiring of C4 leaves at the post-transcriptional level compared with C3 leaves. It may be crucial to understand this mechanism before trying to transform C3 into C4 crops.
Supplementary data
Supplementary data can be found at JXB online.
Figure S1. Gene expression profiles in leaf gradient of T. hassleriana and G. gynandra for (A) C4 cycle genes and (B) the Calvin–Benson cycle.
Table S1. EISA analysis on a control dataset from the WT and an ago1-27 Arabidopsis mutant.
Table S2. List of transcripts that are differentially expressed between young and expanded leaves in G. gynandra and T. hassleriana.
Table S3. List of transcripts that are common to both species after EISA analysis during leaf development (described by their AGI reference number).
Table S4. List of transcripts that are predicted to be post-transcriptionally regulated by EISA between young and expanded leaves in G. gynandra and T. hassleriana.
Supplementary Material
Acknowledgements
SA is supported by the University of Zurich and the Plant Fellows programme of the Zürich–Basel Plant Science Centre. NF is supported by the Clinical Trial Unit of the University of Bern. We would like to thank Stefan Hörtensteiner and Enrico Martinoia for material support, and Julian Hibberd, Ivan Reyna-Llorens and Ben Williams for helpful discussion.
References
- Ambrosone A, Costa A, Leone A, Grillo S. 2012. Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Science 182, 12–18. [DOI] [PubMed] [Google Scholar]
- Aubry S, Aresheva O, Reyna-Llorens I, Smith-Unna RD, Hibberd JM, Genty B. 2016. A specific transcriptome signature for guard cells from the C4 plant Gynandropsis gynandra. Plant Physiology 170, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. Journal of Experimental Botany 62, 3049–3059. [DOI] [PubMed] [Google Scholar]
- Aubry S, Kelly S, Kümpers BMC, Smith-Unna RD, Hibberd JM. 2014a Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genetics 10, e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM. 2014b Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. The Plant Journal 78, 659–673. [DOI] [PubMed] [Google Scholar]
- Berry JO, Mure CM, Yerramsetty P. 2016. Regulation of Rubisco gene expression in C4 plants. Current Opinion in Plant Biology 31, 23–28. [DOI] [PubMed] [Google Scholar]
- Brautigam A, Kajala K, Wullenweber J, et al. 2010. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439. [DOI] [PubMed] [Google Scholar]
- Burgess SJ, Hibberd JM. 2015. Insights into C4 metabolism from comparative deep sequencing. Current Opinion in Plant Biology 25, 138–144. [DOI] [PubMed] [Google Scholar]
- Chang YM, Liu WY, Shih AC, et al. 2012. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiology 160, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology 25, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, van den Bergh E, Zeng P, et al. 2013. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of Crucifers. The Plant Cell 25, 2813–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, Gregory R, Kellogg EA, Hartwell J, Osborne CP. 2012. Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Current Biology 22, 445–449. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Lopez-Obando M, Heurtevin L, Bernard C, Martin K, Berthomé R, Lurin C. 2013. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biology 10, 1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Li W, An L. 2016. NMD mechanism and the functions of Upf proteins in plant. Plant Cell Reports 35, 5–15. [DOI] [PubMed] [Google Scholar]
- Ehleringer JR, Cerling TE, Helliker BR. 1997. C-4 photosynthesis, atmospheric CO2, and climate. Oecologia 112, 285–299. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. 2002. RNA-binding proteins in plants: the tip of an iceberg?Current Opinion in Plant Biology 5, 452–459. [DOI] [PubMed] [Google Scholar]
- Francischini CW, Quaggio RB. 2009. Molecular characterization of Arabidopsis thaliana PUF proteins—binding specificity and target candidates. The FEBS Journal 276, 5456–5470. [DOI] [PubMed] [Google Scholar]
- Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types?Journal of Experimental Botany 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Gaidatzis D, Burger L, Florescu M, Stadler MB. 2015. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nature Biotechnology 33, 722–729. [DOI] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, Chow WS, Andrews TJ, Conroy JP, von Caemmerer S. 2005. Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiology 137, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Najafabadi HS, Oikonomou P, Greco TM, Fish L, Salavati R, Cristea IM, Tavazoie S. 2012. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature 485, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Brautigam A, Weber KL, Weber AP, Westhoff P. 2011. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4?The Plant Cell 23, 2087–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, MacIntosh GC, Green PJ. 1999. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends in Plant Science 4, 429–438. [DOI] [PubMed] [Google Scholar]
- Hatch MD, Slack CR. 1966. Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochemical Journal 101, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber AP, Lercher MJ. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. [DOI] [PubMed] [Google Scholar]
- Hendriks G-J, Gaidatzis D, Aeschimann F, Großhans H. 2014. Extensive oscillatory gene expression during C. elegans larval development. Molecular Cell 53, 380–392. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM. 2014. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiology 165, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. 2014. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, Hibberd JM. 2012. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. The Plant Journal 69, 47–56. [DOI] [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nature Reviews Genetics 8, 533–543. [DOI] [PubMed] [Google Scholar]
- Klaff P, Gruissem W. 1991. Changes in chloroplast mRNA stability during leaf development. The Plant Cell 3, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku MSB, Schmitt MR, Edwards GE. 1979. Quantitative determination of RuBP carboxylase–oxygenase protein in leaves of several C3 and C4 plants. Journal of Experimental Botany 30, 89–98. [Google Scholar]
- Külahoglu C, Denton AK, Sommer M, et al. 2014. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. The Plant Cell 26, 3243–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNASeq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Lu G, Dolgner SJ, Hall TMT. 2009. Understanding and engineering RNA sequence specificity of PUF proteins. Current Opinion in Structural Biology 19, 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Chen Y, Smyth GK. 2016. It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods in Molecular Biology 1418, 391–416. [DOI] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. 2005. Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. The Plant Cell 17, 3111–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, et al. 2010. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. The Plant Cell 22, 3509–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Lu C, Menon R, Schwartz J, Guan Y. 2016. Effects of antioxidants in human cancers: differential effects on non-coding Intronic RNA expression. Antioxidants 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. The Plant Cell 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Iwano M, Havaux M, Yokota A, Munekage YN. 2013. Promotion of cyclic electron transport around photosystem I during the evolution of NADP–malic enzyme-type C4 photosynthesis in the genus Flaveria. The New Phytologist 199, 832–842. [DOI] [PubMed] [Google Scholar]
- Oaks A. 1994. Efficiency of nitrogen utilization in C3 and C4 cereals. Plant Physiology 106, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori GM, Mullineaux PM, Foyer CH. 2000. Post-transcriptional regulation prevents accumulation of glutathione reductase protein and activity in the bundle sheath cells of maize. Plant Physiology 122, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Berry JO. 2008. Rubisco gene expression in C4 plants. Journal of Experimental Botany 59, 1625–1634. [DOI] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M. 2009. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Molecular Systems Biology 5, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF. 1998. Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proceedings of the National Academy of Sciences, USA 95, 9009–9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Brautigam A, Schluter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant Cell 23, 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Jacobson A. 2013. The intimate relationships of mRNA decay and translation. Trends in Genetics 29, 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman IM, Li F, Gregory BD. 2013. Genomic era analyses of RNA secondary structure and RNA-binding proteins reveal their significance to post-transcriptional regulation in plants. Plant Science 205–206, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwaszek A, Ukleja M, Dziembowski A. 2014. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biology 11, 1122–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Christiansen J, Nielsen H. 2011. The post-transcriptional operon. Methods in Molecular Biology 703, 237–245. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, et al. 2006. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Development 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA. 2013. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. The Plant Journal 75, 656–670. [DOI] [PubMed] [Google Scholar]
- Weber APM. 2015. Discovering new biology through sequencing of RNA. Plant Physiology 169, 1524–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Aubry S, Hibberd JM. 2012. Molecular evolution of genes recruited into C4 photosynthesis. Trends in Plant Science 17, 213–220. [DOI] [PubMed] [Google Scholar]
- Williams BP, Burgess SJ, Reyna-Llorens I, Knerova J, Aubry S, Stanley S, Hibberd JM. 2016. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. The Plant Cell 28, 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Johnston IG, Covshoff S, Hibberd JM. 2013. Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. Elife 2, e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Clark A, Sato S, Clemente T, Stern D. 2012. Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type-specific accumulation. Plant Physiology 160, 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Köstler WJ, Molotski N, et al. 2011. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Molecular Systems Biology 7, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rosen BD, Tang H, Krishnakumar V, Town CD. 2015. Polyribosomal RNA-Seq reveals the decreased complexity and diversity of the Arabidopsis translatome. PloS One 10, e0117699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.