Abstract
Background
Carbapenem resistance is a critical healthcare challenge worldwide. Particularly concerning is the widespread dissemination of Klebsiella pneumoniae carbapenemase (KPC). Klebsiella pneumoniae harboring blaKPC (KPC-Kpn) is endemic in many areas including the United States, where the epidemic was primarily mediated by the clonal dissemination of Kpn ST258. We postulated that the spread of blaKPC in other regions occurs by different and more complex mechanisms. To test this, we investigated the evolution and dynamics of spread of KPC-Kpn in Colombia, where KPC became rapidly endemic after emerging in 2005.
Methods
We sequenced the genomes of 133 clinical isolates recovered from 24 tertiary care hospitals located in 10 cities throughout Colombia, between 2002 (before the emergence of KPC-Kpn) and 2014. Phylogenetic reconstructions and evolutionary mapping were performed to determine temporal and genetic associations between the isolates.
Results
Our results indicate that the start of the epidemic was driven by horizontal dissemination of mobile genetic elements carrying blaKPC-2, followed by the introduction and subsequent spread of clonal group 258 (CG258) isolates containing blaKPC-3.
Conclusions
The combination of 2 evolutionary mechanisms of KPC-Kpn within a challenged health system of a developing country created the “perfect storm” for sustained endemicity of these multidrug-resistant organisms in Colombia.
Keywords: Colombia, Klebsiella pneumoniae, KPC
The spread of Gram-negative bacteria resistant to carbapenems is an urgent public health threat and a critical priority according to the World Health Organization [1]. The speed of dissemination of these pathogens, and the lack of new antibiotics active against them, threaten the medical care of hospitalized patients, especially those who are critically ill [2, 3]. Several classes of carbapenemases have emerged in members of the Enterobacteriaceae family, including metallo-β-lactamases (eg, New Delhi Metallo-β-lactmase - NDM), class D carbapenemases (eg, OXA-48), and class A carbapenemase Klebsiella pneumoniae carbapenemase (KPC) [4]. KPC was initially identified in 1996 in a K. pneumoniae (Kpn) isolate from North Carolina [5]. Since then, 24 variants have been described, and KPC-producing Gram-negative organisms with endemic levels have been reported in several countries, including the United States, Greece, Italy, Israel, and Colombia [6, 7].
KPC is commonly associated with transposable elements (eg. Tn4401a-g) and plasmids [7, 8]. The rapid dissemination of K. pneumoniae carrying blaKPC has been primarily associated with the spread of a single clonal group (CG) designated CG258, a large cluster containing 43 different sequence types (STs), with ST258 and ST512 being the two predominant STs. For instance, KPC-Kpn ST258 is responsible for 90% of all infections caused by K. pneumoniae in Israel; and causes 80% of all the KPC-Kpn outbreaks in the United States [7, 9–11]. Colombia was the first country in South America where KPC was reported; the blaKPC gene was initially identified in K. pneumoniae isolates in 2005 [12]. Since then, it has been discovered in other species of Enterobacteriaceae and other Gram-negative bacteria (including Pseudomonas aeruginosa), becoming endemic in Colombia [6]. Preliminary molecular studies suggested that clonal spread of CG258 is not the primary mechanism of dissemination of blaKPC in Colombia, as it is in other parts of the world [13].
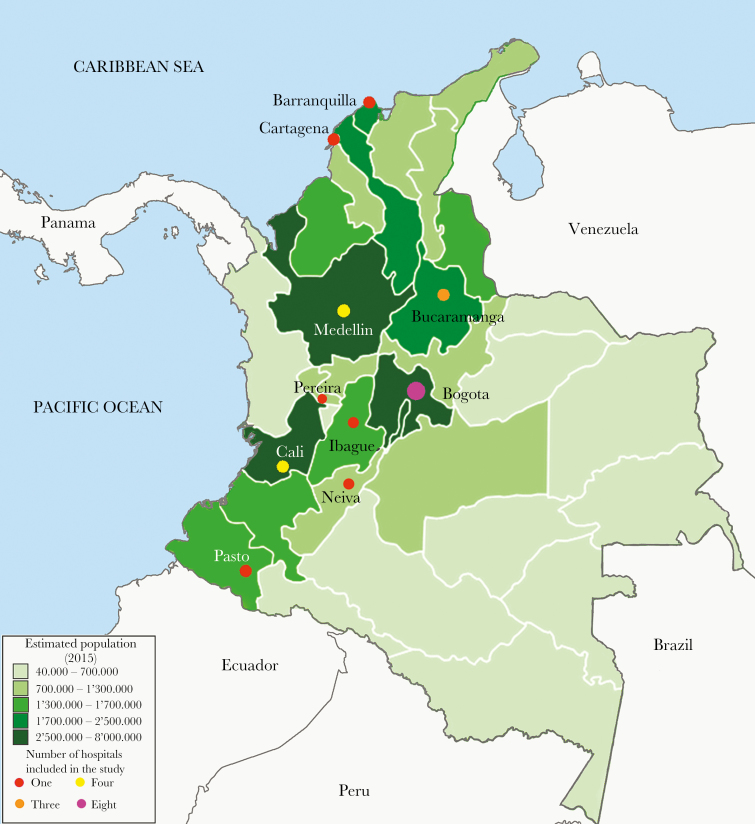
We postulated that the mechanisms of dissemination of blaKPC are more complex within Colombia. To gain insights into the evolutionary events related to the epidemic occurrence of KPC-Kpn in Colombia, we performed detailed genomic analyses of 133 KPC-Kpn isolates collected before and after the identification of the first KPC-Kpn in 2005 [12]. Isolates included were recovered from 2002 to 2014, in 24 hospitals from 10 of the most densely populated Colombian cities (Figure 1).
Figure 1.
Map of Colombia showing the locations where isolates were collected. The 10 cities included in the study are located in the most densely populated areas of the country (as indicated by darker hues). The number of hospitals per city is indicated by colored circles.
METHODS
Strain Selection
Strains were collected between 2002 and 2014 from Colombian hospitals belonging to a bacterial resistance surveillance network coordinated by Centro Internacional de Entrenamiento e Investigaciones Medicas (CIDEIM), Cali-Colombia. The network is composed of tertiary care hospitals located in 10 cities throughout the country (Figure 1). Once isolates were collected at each hospital, they were sent to CIDEIM and kept in a repository. From this collection, a set of isolates was selected based on (1) year of isolation, focusing before and after the identification of the first KPC-carrying K. pneumoniae isolate in Colombia (2005), to achieve the best temporary spread; (2) geographical diversity, encompassing the most densely populated cities across Colombia (Figure 1); (3) isolates recovered from a fluid that likely represented an infection in an attempt to avoid colonizing isolates; and (4) representative isolates from previously characterized outbreaks ([14]) (Supplementary Table 1). The identity of each isolate was confirmed using a MALDI Biotyper (RAB Lab; Bruker Daltonics, Bremen, Germany) and prepared for sequencing.
Deoxyribonucleic Acid Preparation, Library Construction, Sequencing, Assembly, and Annotation
Deoxyribonucleic acid (DNA) was isolated with the MasterPure Gram-positive DNA purification kit (Epicenter Biosciences). Illumina sequencing libraries were prepared using the TruSeq kit with Illumina indexed-encoded adapters. Libraries were pooled for whole-genome sequencing on Illumina MiSeq, NextSeq, or HiSeq 2500, and paired-end sequence reads were obtained representing at least 100-fold genome coverage. Using CLC Bio Workbench (CLC Bio QIAGEN), reads were trimmed for quality (score limit = 0.03, maximum 2 ambiguous nucleotides, 45 minimum nucleotides in reads) after removing adapters. Trimmed reads were de novo assembled with automatic bubble and word size, 1000 base pairs as the minimum contig size, auto-detecting paired distances, and mapping reads back to contigs. Genes were annotated in each genome assembly using the RAST server (http://rast.nmpdr.org). The sequence data for the isolates included in this study have been submitted to GenBank under Bioproject number PRJNA378654.
In Silico Multilocus Sequence Typing Analysis, Identification of Antimicrobial Resistance Determinants, Virulence Factors, Plasmid Typing, and Capsular Typing
We determined the STs of all isolates in silico using the MLST (Multilocus Sequence Typing) 1.8 server (https://cge.cbs.dtu.dk/services/MLST/). Using assemblies as input, resistance and plasmid replicon genes were identified using Resfinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/) and Plasmidfinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/), respectively, using a coverage cutoff of 90%. Virulence genes and capsular typing (wzi sequencing) were identified using the Institut Pasteur’s Klebsiella BigsDB site (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). The presence of Tn4401 was confirmed via BLASTn, and its isoform was determined as described previously [15].
Phylogenetic Analyses
To assess phylogenetic relationship among the 133 Colombian isolates, a core genome was generated for each isolate by excluding all regions annotated by RAST as horizontally transferred elements in the reference strain K. pneumoniae 30660/NJST258_1 (accession number NZ_CP006923) (Supplementary Table 3), in addition to the recombination region as previously defined [7]. In addition, 34 other publicly available genome sequences from National Center for Biotechnology Information (NCBI) were included (Supplementary Table 2), as geographical references. The nucleotide sequences of each one of the orthogroups defined in the core genome were aligned and concatenated to obtain a phylogenetic matrix. The matrix was used to reconstruct the phylogeny of these strains with RAxML [16], using a General Time Reversible (GTR) evolution model and a GAMMA model of rate heterogeneity selecting the best from 20 different runs and 1000 bootstrap resampling. For the CG258 phylogenetic reconstruction, 33 isolates sequenced in this study and 298 publicly available genome sequences from NCBI were included (Supplementary Table 1). Single nucleotide polymorphisms matrices were generated using pairwise whole genome alignment to the NZ_CP006923 reference with Mummer [17], after masking the recombination region as defined by Chen et al [7]. Maximum likelihood reconstructions were generated using RAxML with a GTR evolution model, a GAMMA model of rate heterogeneity, Lewis ascertainment bias, and a 100-bootstrap resampling. The trees were edited and plotted using iTOL version 3.2.4 [18].
RESULTS
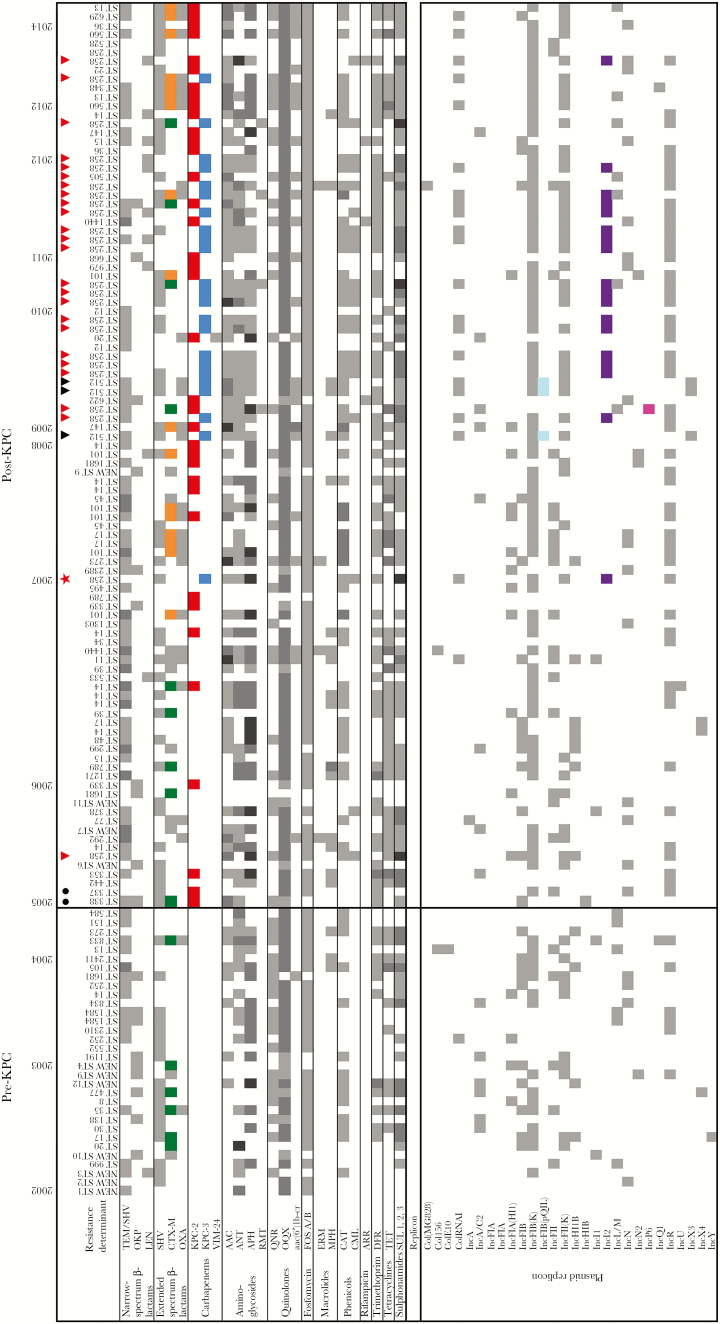
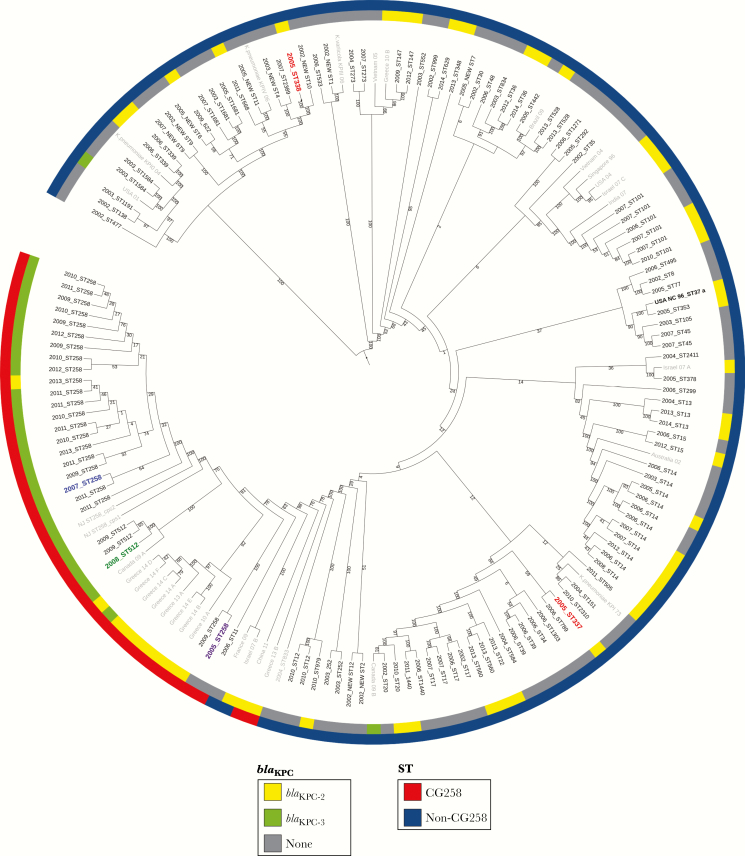
In the period before the initial detection and emergence of blaKPC in Colombia (“pre-KPC” period, 2002–2005), a variety of K. pneumoniae strains of different STs (n = 41; including 9 newly described STs) were circulating in Colombia. Most isolates carried genes conferring resistance to several antibiotics including aminoglycosides, quinolones, fosfomycin, trimetroprim, tetracyclines, sulfonamides, and β-lactams (mostly genes encoding SHV-like and CTX-M extended spectrum β-lactamases [ESBLs]). In addition, isolates were found to carry a variety of plasmid types (Figure 2). In 2005, the first KPC-Kpn isolates were collected from 2 different hospitals in Medellin, the second largest city in Colombia (Figure 1) [12]. These isolates belonged to ST338 and ST337 (Figure 4, indicated in red), not related to CG258. In addition to carrying different resistance determinants and plasmids, typing of wzi (a conserved gene from the capsular polysaccharide synthesis -cps- gene cluster) demonstrated that these isolates had different capsular types (wzi 442 and 108, respectively) (Figures 2 and 3). Both isolates carried blaKPC-2 within a Tn3-like Tn4401b structure located on an IncFIB(K) plasmid [11]. Phylogenetic analyses demonstrated that these 2 isolates were distantly related to the original North Carolina isolate from 1996 (ST37) (Figure 4). Of note, none of the blaKPC-harboring isolates to that date belonged to CG258; however, there was one isolate, also from 2005, that belonged to ST258 but did not carry blaKPC, an occurrence previously reported in Israel [19] (Figure 2).
Figure 2.
Resistance determinants by antibiotic class and plasmid replicons identified in the 133 Klebsiella pneumoniae isolates included in the study. Isolates are chronologically organized by year of isolation (2002 to 2014). Klebsiella pneumoniae isolates belonging to ST258 are identified by a red triangle; isolates belonging to ST512 are identified by a black triangle; first blaKPC-2 harboring isolates recovered in 2005 are denoted by black circles. For all classes of resistance determinants, white means absence and any color means presence. Gray scale indicates the number of copies of the same gene, regardless of the variant found (light, 1 copy; medium, 2 copies; dark, 3 copies). β-lactamases of special interest are highlighted in color: blaCTX-M-12 (green), blaCTX-M-15 (orange), blaKPC-2 (red), and blaKPC-3 (blue). Resistance determinants detected in the isolates include: narrow spectrum β-lactamases (TEM-1, SHV-1 SHV-11, OKP-1, LEN-1/12/16); extended spectrum β-lactamases (SHV-5/11/12/25/27/31/33/101/108/129, CTX-M 2/12/15/96, OXA-1/2/9/47), carbapenemases (KPC-2/3, VIM-24); aminoglycoside-modifying enzymes [aminoglycoside N-acetyltransferases (AAC), aminoglycoside O-nucleotidyltranferases (ANT), aminoglycoside O-phosphotransferases (APH)]; 16S rRNA-modifying enzymes [methyltransferases (RMT)]; quinolone conferring resistance enzymes [plasmid-mediated quinolone-resistance (QNR), OqxA-B efflux pump, and N-acetyltransferase AAC(6′)-Ib-cr]; fosfomycin resistance proteins (FosA, FosB); macrolides, lincosamides, and streptogramins (MLS) resistance determinants [ErmB rRNA methylase (ERM) and macrolide phosphorylases (MphA/B)]; chloramphenicol resistance determinants [chloramphenicol acetyltransferase (CatA1) and chloramphenicol resistance efflux protein, CmlA]; rifampin ADP-ribosyltransferase (Arr); trimethoprim dihydrofolate reductase (DfrA); tetracycline efflux pump (TetA-C); sul1, sul2, and sul3 genes encoding dihydropteroate synthetase (DHPS). For plasmid replicons, an 80% similarity cutoff in PlasmidFinder was used; gray indicates presence, and color indicates plasmid types of interest: IncFIB(pQIL), cyan; IncI2, purple; IncP6, magenta.
Figure 4.
Circular representation of the transformed phylogenetic tree (ignoring branch lengths) generated by RAxML and drawn using iTOL version 3.2.4, showing the genetic relationships among 133 Colombian Klebsiella pneumoniae isolates. Isolates in gray correspond to K. pneumoniae isolates from different parts of the world added as reference. Isolates in red correspond to the first 2 Colombian K. pneumoniae isolates described, carrying blaKPC-2. Isolate in purple corresponds to the oldest ST258 isolate in our collection. Isolate in blue corresponds to the oldest ST258 KPC-Kpn isolate in our collection. Isolate in green corresponds to the index isolate from the “Israeli outbreak” in Medellin. aFirst KPC-Kpn reported in the world (North Carolina).
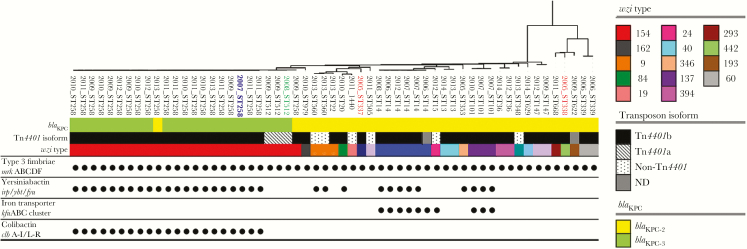
Figure 3.
Transposable element carrying blaKPC, virulence factors and capsular diversity as revealed by wzi typing in the blaKPC harboring K. pneumoniae isolates, identified by year of isolation and ST. Indicated in red are isolates corresponding to the first blaKPC-harboring isolates reported in Colombia; in blue is the oldest blaKPC-harboring CG258 isolate found in our collection; in green is the index isolate from the “Israeli clone”. Genes encoding for the main virulence factors described in K. pneumoniae are grouped (type 3 fimbriae, yerisiniabactin, iron transporter, colibactin); a dot indicates presence of at least one gene from the group, blank indicates absence. Each wzi type is indicated by a different color. For the transposable element, ND indicates not determined.
Between 2005 and 2007, blaKPC-2 was identified in several cities among K. pneumoniae isolates with heterogeneous genetic backgrounds, as evidenced by 8 different STs: 338, 339, 353, 337, 789, 101 and 14 (ST14 being the most frequent ST, representing 30% of the isolates). These blaKPC-2-carrying isolates contained a variety of capsular types, virulence-associated genes, and plasmid replicons. However, only Tn4401b was identified in all of them (Figures 2 and 3).
The first reported blaKPC-3-producing Kpn isolate belonging to CG258 in Colombia was isolated in 2008; this discovery was initially described as an apparent introduction from Israel in an outbreak setting in a hospital in Medellin [14]. Despite this event being considered the principal introduction of CG258 and blaKPC-3 in Colombia, our results indicate that KPC-3-producing K. pneumoniae belonging to this genetic lineage were already circulating undetected in the country. In fact, we identified an isolate carrying blaKPC-3 recovered in 2007 (1 year before the apparent introduction of the Israeli clone) (Figures 3 and 4, indicated in blue) from a patient that was hospitalized in a different city (Ibague, 127 miles from Medellin; see Figure 1). The blaKPC-3 gene in this isolate (KPC_48) was located on a Tn4401b transposon within a ≈80-Kb plasmid (Supplementary Figure S1). Four plasmid replicons were identified for this isolate, including IncI2. Basic Local Alignment Search Tool (BLAST) analyses revealed that the plasmid contained in this strain was highly similar to the completely sequenced IncI2 plasmid pBK15692 (NC_022520.1) (Supplementary Figure S2), the most widely found in the New York/New Jersey area since 2005 [20]. In addition, KPC_48 shares the same virulence factors as the majority of ST258 isolates collected in Colombia in subsequent years, namely, type 3 fimbriae mrkABCDF, yersiniabactin irp, ybt, fyu, and colibactin clbA-I/L-R gene clusters. In contrast, ST512 strains associated with the Israeli outbreak only contain the type 3 fimbriae (mrkABCDF) gene cluster, carry blaKPC-3 within a Tn4401a structure, and share the same set of plasmid replicons, including pQIL (Figures 2 and 3).
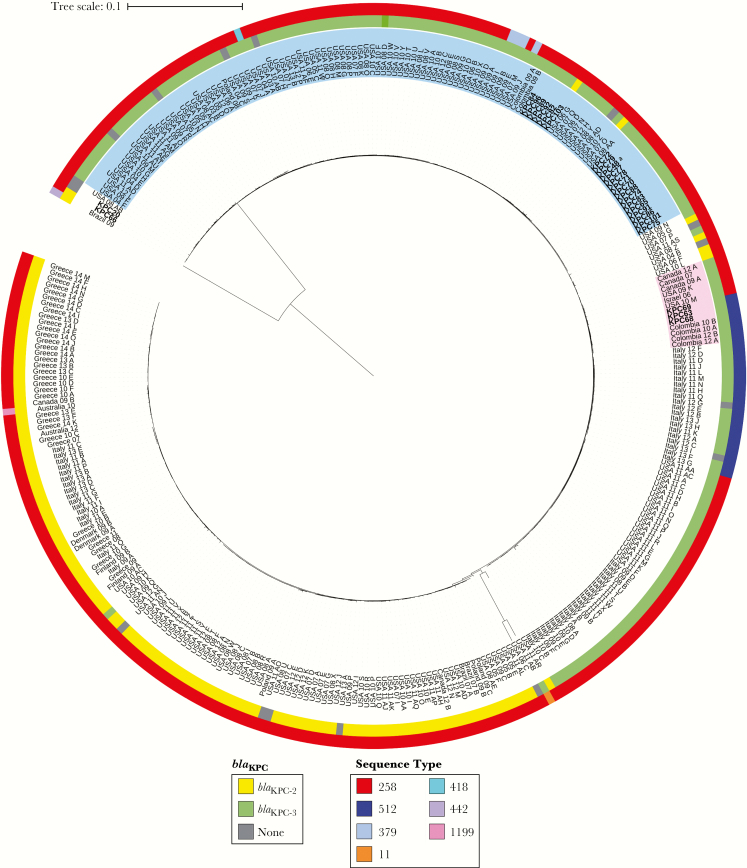
Figure 5 illustrates the comparative genomic analyses of Colombian K. pneumoniae belonging to CG258. Our results clearly indicate that the K. pneumoniae ST512 isolates harboring blaKPC-3 collected in Medellin in 2008 (identified as the “Israeli outbreak” index strain, KPC_63) and 2 other KPC-3-positive isolates from 2009 isolated in the same hospital (KPC_68 and 69), were more closely related to each other than to the rest of the ST258 blaKPC-3-harboring isolates. Moreover, these isolates clustered together with a 2006, KPC-3-producing isolate from Israel (Figure 5, highlighted in pink). In contrast, all of the remaining 21 blaKPC-3-containing K. pneumoniae, recovered in all 10 cities country-wide, are closely related to the first KPC-harboring ST258 isolate recovered in 2007 (KPC-48), and they were grouped together with isolates recovered in the New York/New Jersey area from 2006 to 2012 (Figure 5, highlighted in blue).
Figure 5.
Circular representation of the transformed phylogenetic tree generated by RAxML and drawn using iTOL version 3.2.4, showing the genetic relationships among 331 CG258 Klebsiella pneumoniae isolates. Isolates shown in bold correspond to Colombian isolates collected in this study, all other isolates were obtained from GenBank and are named with the respective country where they were collected followed by 2 digits indicating the year of isolation. Outer ring indicates the sequence type (ST); inner ring indicates the KPC variant carried by each isolate. Isolates highlighted in pink indicate the cluster including the “Israeli outbreak”-related Colombian isolates (ST512), as well as other worldwide isolates found on GenBank. Isolates highlighted in blue indicate the cluster including the oldest ST258 isolate in our collection (KPC48a) as well as most of the other ST258 Colombian isolates reported in this study.
To further characterize the KPC-carrying K. pneumoniae Colombian isolates, we analyzed (1) the cps locus (capsule polysaccharide bio-synthesis operon) by wzi typing and (2) the plasmid replicon types. cps is one of the primary determinants of antigenicity associated with K. pneumoniae and capsule switching and is a species-specific mechanism used to escape the host immune response. In addition, DNA exchange in and around the cps region has been suggested as an important mechanism used by K. pneumoniae to rapidly diversify and evolve [21]. Interestingly, typing of the wzi locus revealed that, consistent with our genomic analyses, a remarkable degree of cps diversity is present in our collection (Figure 3). Additionally, isolates harboring blaKPC-2 exhibited 10 different cps variants (some of them not even associated with a particular K- type), whereas all isolates belonging to CG258 and harboring blaKPC-3 only belonged to wzi type 154. Moreover, the plasmids found in blaKPC-containing isolates (Figure 2 and Supplementary Figure S3) were diverse, as observed before [13]. Additionally, a variety of plasmid replicons were found in blaKPC-2-harboring isolates (including IncQ1, IncHI1B, IncP6, IncU, IncN2, IncFIA[HI1], IncA/C2, IncFII, and IncN), most of them associated with broad host range transmission. In contrast, 3 incompatibility groups were exclusively found in CG258 blaKPC-3 carrying isolates. Plasmids IncX3 and pQIL were only detected in ST512 isolates, whereas IncI2 were exclusive of the ST258 isolates.
DISCUSSION
Our detailed genomic studies reconstruct a unique and complex pattern of dissemination of KPC-Kpn in Colombia, contributing to the high endemic levels of carbapenem resistance that threaten the healthcare system in this developing country. These observations are in contrast with the data from other endemic countries including Israel, Italy, Greece, and the United States, where CG258 has been the main driver of blaKPC dissemination [6, 11, 22–24]. Our results indicate that before the emergence of KPC (“pre-KPC period,” 2002–2005), ESBLs were prevalent amongst K. pneumoniae circulating in Colombia, a finding that is consistent with previous surveillance studies conducted during this period [25]. Of note, there was a complete absence of carbapenem resistance at that time [26], suggesting that the acquisition of Tn4401b harboring blaKPC-2 by a clinical strain represented a sentinel event that influenced the subsequent spread of these organisms in the Colombian healthcare system. It is remarkable that after this sentinel event, circulation of blaKPC-2 among K. pneumoniae isolates with heterogeneous genetic backgrounds between 2005 and 2007 was rapidly documented and also coincided with the spread of blaCTX-M-15 in the country (Figure 2) [27, 28]. The high prevalence of ESBLs (up to 71% [25]) in K. pneumoniae from Colombian hospitals might have led to an increased use of carbapenems, thus providing selective pressure that favored the emergence of carbapenem resistance, mediated by the horizontal transfer and promiscuity of blaKPC-2-carrying plasmids among Gram-negative bacteria in Colombia. Indeed, during that and subsequent periods, other species carrying blaKPC-2 were identified, including Enterobacter cloacae complex, Enterobacter aerogenes, Citrobacter freundii, Serratia marcescens, Klebsiella oxytoca, Salmonella enterica sv. Typhimurium, and even Pseudomonas aeruginosa (an unusual pathogen to harbor blaKPC) [13, 29, 30]. Other factors contributing to the spread may have been related to travel within the country and sharing of patients between institutions. However, the exact driving force for this remarkable spread is unknown.
Taken together, our results suggest that the initial dissemination of blaKPC in Colombia was independent of the presence of CG258. Instead, the major factor that influenced the initial spread was high rates of horizontal plasmid transfer and/or transposition of blaKPC-2 on Tn4401b. Of note, a BLASTn analysis showed that the first blaKPC-2-carrying plasmid of P. aeruginosa (pCOL-1) was highly similar to a plasmid found in one of the K. pneumoniae isolates recovered in 2009 (Figures 2 and Supplementary Figure S4; 99% query coverage, 100% identity by BLAST), supporting interspecies transfer [31].
Upon deeper analysis, we discovered that the “Israeli clone” was not responsible for the introduction and clonal expansion of CG258. Instead, it appears that the index ST258 isolate harbors IncI2 plasmids that have been circulating in the United States (New York/New Jersey area) since 2005 [20]. It is interesting to note that the blaKPC-3-harboring plasmid carried by isolate KPC_48 is very similar to pBK15692, a 77-Kb plasmid that has a characteristic IncI2 backbone that includes genes encoding type IV pili and shufflon regions. In addition to blaKPC-3 (contained within a Tn4401b inserted into a Tn1331 element forming a nested transposon), it also carries blaOXA-9, blaTEM-1,aac(6’)-Ib-cr, and aadA1 (Supplementary Figure S2). Although the index isolate for the “Israeli clone” (obtained from a patient who traveled to Medellin to undergo a liver transplant [14]) was initially assigned to ST258, our sequencing indicates that it truly belonged to ST512 and carried blaKPC-3 within a Tn4401a structure associated with the characteristic pKpQIL plasmid (Figure 2 and Figure 3 [highlighted in green]) [32]. At the time, Israel was dealing with a countrywide outbreak of carbapenem-resistant K. pneumoniae CG258 [33]. Epidemiological studies indicated that the prevalent clone identified by pulsed-field gel electrophoresis (PFGE) and designated as “Q” grouped isolates belonging to ST258 [11]. However, in 2006, isolates belonging to ST512 (single nucleotide variant of ST258) started to appear in Israel, and in some cases, even became the prevalent ST [34]. Because isolates belonging to ST258 and ST512 have closely related PFGE patterns, it is possible that some of the isolates classified at the time as belonging to clone “Q” were actually ST512.
Two additional pieces of evidence support the possible United States-Colombia link. First, we discovered that the apparent genetic platform of dissemination (Tn4401b) is within an IncI2 plasmid (very similar to pBK15692), which is associated with the dissemination of blaKPC-3 [7, 20]. Indeed, the majority of ST258 isolates characterized in this work carried blaKPC-3 within a Tn4401b. In contrast, only three ST512 isolates (the index case, and 2 other strains isolated in Medellin from the same hospital where the Israeli outbreak occurred) carried blaKPC-3 on a Tn4401a transposon. Second, our 2007 CG258 isolate (KPC_48) shares the same virulence factors with the majority of ST258 isolates collected in Colombia after its emergence, namely, type 3 fimbriae mrkABCDF, yersiniabactin irp/ybt/fyu, and colibactin clbA-I/L-R gene clusters. In contrast, ST512 strains associated with the Israeli outbreak only contain the type 3 fimbriae (mrkABCDF) gene cluster (Figure 3).
Results from this work, and another recent study conducted in Medellin [35], suggest that since its introduction from Israel in 2008, ST512 is confined to that city, cocirculating with other K. pneumoniae carrying blaKPC-2 and blaKPC-3 from different STs. After 2008, our molecular and epidemiological data support the observation that up to 68% of nosocomial K. pneumoniae isolates harbored blaKPC, and these organisms became endemic in Colombia, and thus both KPC-2 and KPC-3 variants disseminated [36]. Our phylogenetic data shown in Figure 4 strongly suggests compartmentalization of the genes encoding these 2 variants. Indeed, blaKPC-2 continues to disseminate via horizontal gene transfer among K. pneumoniae isolates from different genetic backgrounds and capsule types, representing the most common blaKPC gene in our collection (62% of K. pneumoniae isolates). Of note, only blaKPC-2 has disseminated into other Enterobacteriaceae and P. aeruginosa isolates in Colombia [13, 29, 30]. On the other hand, a monophyletic clade of the “high risk clone” CG258 K. pneumoniae carrying blaKPC-3 is also expanding simultaneously. These results are in sharp contrast to the epidemiology described in the majority of studies of other endemic countries such as the United States, Israel, Italy, or Greece, where the main culprit for dissemination has been K. pneumoniae CG258 [7]. In 2009, the Centers for Disease Control and Prevention reported that up to 70% of the K. pneumoniae blaKPC strains in their collection belong to CG258 [11]. This number is even greater in Israel, where 90% of the strains were part of CG258 at the peak of the epidemic. It is interesting to note that the expansion of a single clone in that country may also explain why the strict infection control measures implemented were successful to combat the epidemic [22]. However, more recent observations point out that the dynamics within single institutions or smaller regions have unique characteristics [37–40] and suggest a change in the trend of dominance of CG258 for a somewhat concomitant spread of CG258 and non-CG258 in other endemic countries such as Israel, United States, Italy, Brazil, and Argentina [35, 37, 41–44].
Certain limitations of the study should be mentioned. As a nongovernmental sentinel surveillance system, CIDEIM’s antimicrobial resistance network relies on the voluntary collaboration of hospitals around the country. This limits our pool of samples and naturally, introduces a bias in the strains received per city and per year included in the study. In addition, given its retrospective nature, this work does not delineate the true incidence of Kpn-KPC infections in the population nor does it discriminate between community-acquired or nosocomial infection.
CONCLUSIONS
The combination of two evolutionary mechanisms in a challenged health system from a developing country has created the “perfect storm” for a massive epidemic of carbapenemase-producing Gram-negative bacteria. This stochastic strategy used by K. pneumoniae in Colombia may pose a significant public health threat in other developing countries with similar infrastructures and widespread use of carbapenems, where traditional infection control measures (useful in parts of the world where clonal expansion of CG258 plays a major role in KPC dissemination) would be insufficient. In these countries, the challenge of KPC-Kpn may demand novel approaches combining comprehensive molecular surveillance, innovative infection control procedures, and antibiotic stewardship strategies across different settings.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Presented in part: 55th Interscience Conference on Antimicrobial Agents and Chemotherapy, September 2015, San Diego, CA; Infectious Disease Genomics Conference, October 2015, Wellcome Genome Campus, Hinxton, Cambridge, UK.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH) or the Department of Veterans Affairs.
Financial support. Funds and facilities from the Cleveland Department of Veterans Affairs, VISN 10 Geriatric Research Education and Clinical Center, and the Veterans Affairs Office Research and Development (Award Number 1I01BX001974 to RAB) supported this work. Research in this publication was also supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R21AI114508, R01AI100560, R01AI063517, and R01AI072219 (to R.A.B); K24-AI114818, R01-AI093749, R21-AI114961, and R21/R33 AI121519 (to C.A.A).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics.Geneva, Switzerland: World Health Organization; 2017. [Google Scholar]
- 2. Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 2014; 20:831–8. [DOI] [PubMed] [Google Scholar]
- 3. Logan LK. Carbapenem-resistant enterobacteriaceae: an emerging problem in children. Clin Infect Dis 2012; 55:852–9. [DOI] [PubMed] [Google Scholar]
- 4. van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017; 8:460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yigit H, Queenan AM, Anderson GJ et al. . Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 2001; 45:1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Munoz-Price LS, Poirel L, Bonomo RA et al. . Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 2014; 22:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheppard AE, Stoesser N, Wilson DJ et al. . Nested russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 2016; 60:3767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wright MS, Perez F, Brinkac L et al. . Population structure of KPC-producing Klebsiella pneumoniae isolates from midwestern U.S. hospitals. Antimicrob Agents Chemother 2014; 58:4961–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009; 9:228–36. [DOI] [PubMed] [Google Scholar]
- 11. Kitchel B, Rasheed JK, Patel JB et al. . Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 2009; 53:3365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Villegas MV, Lolans K, Correa A et al. . First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother 2006; 50:2880–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mojica MF, Correa A, Vargas DA et al. . Molecular correlates of the spread of KPC-producing Enterobacteriaceae in Colombia. Int J Antimicrob Agents 2012; 40:277–9. [DOI] [PubMed] [Google Scholar]
- 14. Lopez JA, Correa A, Navon-Venezia S et al. . Intercontinental spread from Israel to Colombia of a KPC-3-producing Klebsiella pneumoniae strain. Clin Microbiol Infect 2011; 17:52–6. [DOI] [PubMed] [Google Scholar]
- 15. Naas T, Cuzon G, Truong HV, Nordmann P. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 2012; 56:4753–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 2011; 27:334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007; 23:127–8. [DOI] [PubMed] [Google Scholar]
- 19. Adler A, Paikin S, Sterlin Y et al. . A swordless knight: epidemiology and molecular characteristics of the blaKPC-negative sequence type 258 Klebsiella pneumoniae clone. J Clin Microbiol 2012; 50:3180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L, Chavda KD, Al Laham N et al. . Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 2013; 57:5019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wyres KL, Gorrie C, Edwards DJ et al. . Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol Evol 2015; 7:1267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwaber MJ, Lev B, Israeli A et al. . Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin Infect Dis 2011; 52:848–55. [DOI] [PubMed] [Google Scholar]
- 23. Karampatakis T, Antachopoulos C, Iosifidis E, Tsakris A, Roilides E. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece. Future Microbiol 2016; 11:809–23. [DOI] [PubMed] [Google Scholar]
- 24. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017; 41:252–75. [DOI] [PubMed] [Google Scholar]
- 25. Villegas MV, Correa A, Perez F, Miranda MC, Zuluaga T, Quinn JP. Prevalence and characterization of extended-spectrum beta-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. Diagn Microbiol Infect Dis 2004; 49:217–22. [DOI] [PubMed] [Google Scholar]
- 26. Miranda MC, Pérez F, Zuluaga T et al. . Antimicrobial resistance in gram negative bacteria isolated from intensive care units of Colombian hospitals, WHONET 2003, 2004 and 2005. Biomédica 2006; 26:424–33. [PubMed] [Google Scholar]
- 27. Valenzuela de Silva EM, Mantilla Anaya JR, Reguero Reza MT et al. . Detection of CTX-M-1, CTX-M-15, and CTX-M-2 in clinical isolates of Enterobacteriaceae in Bogota, Colombia. J Clin Microbiol 2006; 44:1919–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Villegas MV, Correa A, Perez F et al. . CTX-M-12 beta-lactamase in a Klebsiella pneumoniae clinical isolate in Colombia. Antimicrob Agents Chemother 2004; 48:629–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodríguez E, Bautista A, Barrero L. First report of a Salmonella enterica serovar typhimurium isolate with carbapenemase (KPC-2) in Colombia. Antimicrob Agents Chemother 2014; 58:1263–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother 2007; 51:1553–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother 2013; 68:1757–62. [DOI] [PubMed] [Google Scholar]
- 32. Chen L, Chavda KD, Melano RG et al. . Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 2014; 58:2871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Samra Z, Ofir O, Lishtzinsky Y, Madar-Shapiro L, Bishara J. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int J Antimicrob Agents 2007; 30:525–9. [DOI] [PubMed] [Google Scholar]
- 34. Warburg G, Hidalgo-Grass C, Partridge SR et al. . A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6’)-Ib. J Antimicrob Chemother 2012; 67:898–901. [DOI] [PubMed] [Google Scholar]
- 35. Ocampo AM, Chen L, Cienfuegos AV et al. . High frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics: a two-year surveillance in five Colombian tertiary care hospitals. Antimicrob Agents Chemother 2015; DOI: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hernández-Gómez C, Blanco VM, Motoa G et al. [Evolución de la resistencia antimicrobiana en bacilos Gram negativos en unidades de cuidados intensivos en Colombia]. Biomédica 2014; 34:91–100. [DOI] [PubMed] [Google Scholar]
- 37. Mathers AJ, Stoesser N, Sheppard AE et al. . Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 2015; 59:1656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cerqueira GC, Earl AM, Ernst CM et al. . Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A 2017; 114:1135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snitkin ES, Zelazny AM, Thomas PJ et al. . Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012; 4:148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marsh JW, Krauland MG, Nelson JS et al. . Genomic epidemiology of an endoscope-associated outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae. PLoS One 2015; 10:e0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Del Franco M, Paone L, Novati R et al. . Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d’Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789). BMC Microbiol 2015; 15:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adler A, Khabra E, Paikin S, Carmeli Y. Dissemination of the blaKPC gene by clonal spread and horizontal gene transfer: comparative study of incidence and molecular mechanisms. J Antimicrob Chemother 2016; 71:2143–6. [DOI] [PubMed] [Google Scholar]
- 43. Sampaio JL, Gales AC. Antimicrobial resistance in Enterobacteriaceae in Brazil: focus on β-lactams and polymyxins. Braz J Microbiol 2016; 47Suppl 1:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gomez SA, Pasteran FG, Faccone D et al. . Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect 2011; 17:1520–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.