Meta-analysis of auxin-responsive transcriptome data identified both known and novel cis-regulatory elements associated with specific response modes.
Keywords: Auxin, AuxRE, ARF, bZIP, bHLH, transcriptional regulation, chromatin states, bioinformatics
Abstract
The phytohormone auxin regulates virtually every developmental process in land plants. This regulation is mediated via de-repression of DNA-binding auxin response factors (ARFs). ARFs bind TGTC-containing auxin response cis-elements (AuxREs), but there is growing evidence that additional cis-elements occur in auxin-responsive regulatory regions. The repertoire of auxin-related cis-elements and their involvement in different modes of auxin response are not yet known. Here we analyze the enrichment of nucleotide hexamers in upstream regions of auxin-responsive genes associated with auxin up- or down-regulation, with early or late response, ARF-binding domains, and with different chromatin states. Intriguingly, hexamers potentially bound by basic helix–loop–helix (bHLH) and basic leucine zipper (bZIP) factors as well as a family of A/T-rich hexamers are more highly enriched in auxin-responsive regions than canonical TGTC-containing AuxREs. We classify and annotate the whole spectrum of enriched hexamers and discuss their patterns of enrichment related to different modes of auxin response.
Introduction
Auxin, a key plant hormone, regulates many processes via modulation of gene expression at the transcriptional level (reviewed in Paque and Weijers, 2016; Strader and Zhao, 2016; Weijers and Wagner, 2016). The signals regulating transcription are integrated at gene promoters, where transcription factors bind specific cis-regulatory elements, generally through direct interaction with a short DNA-binding site 6–12 bp in length (reviewed in Franco-Zorrilla and Solano, 2017).
A family of related AUXIN RESPONSE FACTORS (ARFs) mediates the primary transcriptional response to auxin (reviewed in Guilfoyle and Hagen, 2007). Different ARFs bind TGTC-containing cis-elements called AuxREs (Auxin Response Elements) presumably regardless of the cellular auxin level (Ulmasov et al., 1997, 1999). When auxin levels are low, ARFs form heterodimers with their repressors, the Aux/IAA proteins (reviewed in Paque and Weijers, 2016; Strader and Zhao, 2016). Aux/IAAs inhibit ARF function by preventing their contact with the transcription initiation complex (Ito et al., 2016) and/or through ensuring a repressive chromatin state mediated by their binding to TPL/TPR (TOPLESS and related) proteins that are hypothesized to recruit histone deacetylases (Long et al., 2006; Szemenyei et al., 2008). When auxin levels are high, Aux/IAA proteins are bound by TIR1/AFB auxin receptors and subsequently polyubiquitinated and degraded. Derepressed ARFs trigger the transcription changes, a process that may involve recruitment of SWI/SNF (SWITCH/SUCROSE NONFERMENTING) chromatin-remodeling ATPases (Wu et al., 2015). The latter make the chromatin region more accessible for other transcription factors.
The complexity and diversity of auxin transcriptional response is provided by an abundance of family members in auxin receptors, Aux/IAAs, ARFs, and their cofactors (reviewed in Weijers and Wagner, 2016). ARFs can homodimerize on DNA (Ulmasov et al., 1997; Guilfoyle et al., 1998; Vernoux et al., 2011; Boer et al., 2014) and they also are able to heterodimerize with other transcription factors. Interactions were shown between ARF7 and MYB77 (Shin et al., 2007); ARF8 and the basic helix–loop–helix (bHLH) factor BPEp (Varaud et al., 2011); ARF6 with the bHLH (PIF4) and BZR1/BES1 factors (Oh et al., 2014); ARF6/8 and MADS factor FUL (Ripoll et al., 2015); and ARF3 with representatives of G2-like (KAN1 and KAN4) (Kelley et al., 2012), bHLH (IND), Homeobox (RPL, KNAT1, and KNAT3), AP2 (BBM and PLT5), and TCP (TCP4 and TCP18) families (Simonini et al., 2016). For some cofactors, the binding sites were found in close vicinity to the ARF-binding site, forming a composite AuxRE. In the case of ARF3, whose interaction with other transcription factors is directly influenced by auxin, auxin-dependent gene regulation may occur via the DNA-binding site of the partner proteins, and thus not require a core AuxRE (Simonini et al., 2016).
Among coupling elements, the ABRE (abscisic acid response element) ACGTG(G/T)C (Choi et al., 2000) was first described as a part of the composite auxin response element in the soybean GH3 promoter, and was shown to bind a bZIP (basic leucine zipper) transcription factor (Ulmasov et al., 1995; Liu et al., 1997). Direct interaction between bZIP and ARF transcription factors has not been shown, but Arabidopsis bZIP11-related transcription factors mediate auxin response via interaction with chromatin modulator ADA2b, a subunit of a histone acetylation complex (Weiste and Dröge-Laser, 2014). Whereas bZIP-binding sites are not sufficient to mediate auxin response themselves, they couple to AuxREs and enhance auxin-mediated transcription of a GH3 gene in an auxin concentration-dependent manner (Ulmasov et al., 1995; Weiste and Dröge-Laser, 2014).
Along with ABRE, plant bZIP transcription factors bind other ACGT-containing sites; among them, A-box (TACGTA), C-box (GACGTC), and G-box (CACGTG) sequences are bound more preferentially (Izawa et al., 1993; reviewed in Foster et al., 1994; Jakoby et al., 2002). The G-box is highly enriched in ARF6-binding regions (Oh et al., 2014), but it should be noted that the G-box is not restricted as a binding site for bZIPs. PIFs and MYCs of the bHLH family (Martínez-García et al., 2000; Dombrecht et al., 2007; Fernandez-Calvo et al., 2011; Oh et al., 2012; Kim et al., 2016), AP2/ERF ABI4 (Zhang et al., 2013), and BZR1/BES1 (Yu et al., 2011; Oh et al., 2012) transcription factors can all also bind this core. Transcription factors that interact with a common cis-element may compete (Zhang et al., 2013) or co-operatively regulate (Oh et al., 2012) the target gene.
Together with bZIP-binding sites, bHLH and the BZR1/BES1-binding HUD (Hormone Up at Dawn) motif CACATG (Walcher and Nemhauser, 2012; Oh et al., 2014), MYB factor-binding site MRE (AACC) and MYB core (CNGTTR) (Shin et al., 2007), and MADS-binding CArG box (CC[A/T]6GG) (Ripoll et al., 2015) were shown to reside close to functional AuxREs. Thus, studying the footprints of transcription factor DNA binding might be an efficient way to indicate those factors involved in auxin response.
However, several analyses of auxin-responsive upstream regions have indicated the enrichment of additional non-TGTC-containing motifs (Pufky et al., 2003; Doi et al., 2008; Berendzen et al., 2012; Mironova et al., 2014). Cis-elements associated with early transcriptional activation attracted more attention than those for auxin inhibition and late response. To unravel the repertoire of auxin response elements and their association with up- or down-regulation, early or late response, we develop a bioinformatics approach for the systematic identification of hexamers enriched in auxin-responsive upstream regions. We apply that approach to a wide variety of publicly available transcriptome data sets on auxin response studies.
Materials and methods
Data sets
We collected all available data sets on exogenous auxin treatment from the GEO database. RNA-Seq was normalized with the TMM method from the ‘edgeR’ package (Robinson et al., 2010; McCarthy et al., 2012) and transformed with ‘voom’ (Law et al., 2014) from the ‘limma’ package (Ritchie et al., 2015; Phipson et al., 2016). We processed all data from ATH1 microarrays by the ‘limma’ package or took pre-processed data when they were publicly available. For all RNA-Seq and microarray data, we applied the Benjamini–Yekutieli method (Benjamini and Yekutieli, 2001) to control the false discovery rate (FDR), and we report for each data set the subset of genes corresponding to the FDR of 0.05 and a fold change >3/2 or <2/3, which we call differentially expressed genes (DEGs). We applied this procedure to all available data sets and obtained 21 data sets containing at least 10 DEGs (Supplementary Table S1 at JXB online) (Armstrong et al., 2004; Nemhauser et al., 2004; Redman et al., 2004; Okushima et al., 2005; Delker et al., 2010; De Rybel et al., 2012; Bargmann et al., 2013; Lewis et al., 2013; Chaiwanon and Wang, 2015; Xuan et al., 2015).
For annotating putative AuxREs in these 21 data sets, we compiled control and positive gene sets as follows: the control gene set contains 11 223 genes, not differentially expressed in any of the experiments from Supplementary Table S1. The positive gene set contains 2451 genes, differentially expressed in at least three data sets.
We obtained Arabidopsis genome sequence and annotation from TAIR10 and retrieved [–1500; +1] upstream regions relative to the transcription start site of 21 098 genes with a unique probe in the ATH1 microarray.
Association of hexamers with auxin response
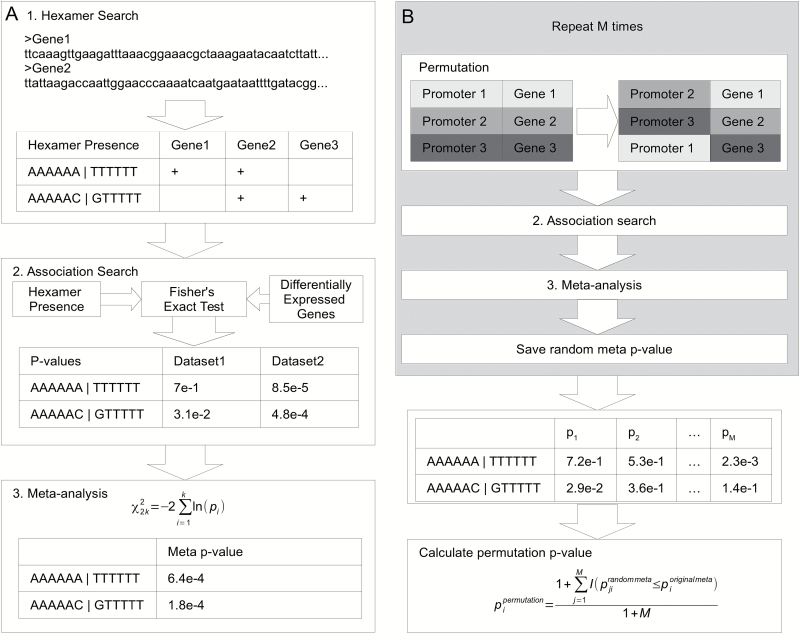
The bioinformatics approach for predicting putative AuxREs is the further elaboration of our transcriptome meta-analysis method (Mironova et al., 2014; Zemlyanskaya et al., 2016). The approach consists of the following three-step procedure (Fig. 1).
Fig. 1.
The pipeline for genome-wide association analysis for putative cis-elements associated with auxin response. (A) Three steps of the pipeline: (1) exhaustive hexamer search in the upstream regions; (2) analysis of association between the hexamer’s presence in the upstream region and auxin up- (down-) regulation of the gene; and (3) meta-analysis across all data sets. (B) Permutation test scheme, part of step (3).
In the first step, we searched for all possible hexamers in each of the upstream regions. Assuming the equivalence of hexamers on both DNA strands, we reduced the set of 4096 possible hexamers to 2080 non-redundant hexamers comprising 2016 complementary pairs and 64 palindromes. After this step, we obtained a list of hexamers per gene that occurred in its [–1500; +1] upstream region at least once.
In the second step, we analyzed for each hexamer and each data set the association of the presence of this hexamer in the gene upstream region and the gene status of being up- or down-regulated by auxin (Fig. 1A). We assess the significance of this association by the one-sided version of Fisher’s exact test (Table 1) for each data set, each hexamer, and each status. We combined for each hexamer the P-values for different data sets and different statuses using Fisher’s method:
Table 1.
The contingency table for the analysis of associations between the presence of a hexamer in the upstream region of a gene ([–1500; +1] to transcription start site) and its auxin responsiveness
| Genes | Auxin up- (down-) regulation | Totals | |
|---|---|---|---|
| Yes | No | ||
| Hexamer (+) | A | B | A+B |
| Hexamer (–) | C | D | C+D |
| Totals | A+C | B+D | A+B+C+D |
The data for every of 2080 hexamers in each whole-genome data set (Supplementary Table S1) were analyzed in the 2 × 2 contingency table using the Fisher’s exact test. The sum A+B+C+D is the number of all genes that have a unique probe on the ATH1 microarray platform.
| (1) |
where Pi is a P-value from a data set i, k is the number of data sets, and is a chi-squared statistic with 2k degrees of freedom. The resulting meta P-value is the probability of obtaining a more extreme chi-squared statistic than the calculated one under the assumption that the Pi values are statistically independent. The hexamers with Bonferroni-corrected meta P-values <0.005 were selected for the next step.
In the third step, we determined the statistical significance of selected hexamers by a permutation test. In each permutation, we mixed promoters between genes so that each promoter was used exactly once (Fig. 1B). Then we performed the second and the third steps recording the meta P-values from Equation 1 for each permutation. After performing M permutations (M=1e+6) for each hexamer, we computed the permutation P-value by P=(m+1)/(M+1), where m is a number of recorded P-values not greater than the meta P-value.
We considered the association between the presence of the hexamer and the auxin responsiveness as significant with the permutation Bonferroni-corrected P-value <0.005.
Comparison of identified hexamers with known cis-elements
We compared the detected hexamers with known cis-regulatory elements using TOMTOM (Gupta et al., 2007) via DAP-seq (O’Malley et al., 2016), PBM (Franco-Zorrilla et al., 2014), and CIS-BP DNA databases (Arabidopsis thaliana). We considered the matches with an E-value <0.05 as significant. For hexamers without significant matches, we additionally screened the literature.
Hexamer enrichment within ARF-binding regions
To study if the obtained hexamers are enriched within ARF-binding regions, we used available whole-genome data on ARF6 ChIP-Seq (Oh et al., 2014; GSM1252254), ARF2, and ARF5 DAP-Seq (O’Malley et al., 2016; GSM1925138, GSM1925826, and GSM1925827). For each hexamer, we compared two proportions via one-tailed Fisher test: (i) the number of positions a hexamer occupies across all 21 098 promoters [–1500; +1] relative to the total number of all possible positions; and (ii) the same across all peaks in a particular peak set. We adjusted P-values of enrichment with Bonferroni multiple testing correction. We considered over-representation as significant at a family-wise error rate (FWER) below 0.05.
Hexamer enrichment in promoters without simple repeats
We applied RepeatMasker (Smit et al., 2013–2015) with -noint -s parameters to all upstream regions for eliminating simple and tandem repeats from the initial set of 21 098 promoters. We searched for the hexamers within masked upstream regions and used the resulting lists through the pipeline (Fig. 1).
Hexamer enrichment in promoters with different chromatin states
We used the data (Sequeira-Mendes et al., 2014) on the distribution of nine chromatin states in the Arabidopsis genome to characterize [–1500; +1] upstream regions of auxin-responsive genes and putative AuxREs. First, we estimated if the chromatin states are uniformly distributed within upstream regions of auxin-responsive genes. For this, we performed the second and the third step of the association analysis (Fig. 1) with the chromatin state domains instead of hexamers.
Secondly, we tested if the hexamers were enriched in promoter segments associated with a specific chromatin state. For each hexamer, we compared two proportions via one-tailed Fisher’s exact test: (i) the number of positions which this hexamer occupies across all upstream regions of 21 098 genes relative to the total number of all possible positions; and (ii) the similar one across all promoter segments associated with a specific chromatin state.
Thirdly, we tested if the hexamers were enriched in the upstream regions located within specific chromatin states of auxin-responsive genes (positive set) relative to the same promoter segments of non-regulated genes (control set). For each hexamer, we compared two proportions via one-tailed Fisher’s exact test: (i) the number of positions this hexamer occupies in the promoter segments of a specific chromatin state within a positive gene set relative to the cumulative number of all possible positions in these segments; and (ii) the same proportion for the control gene set.
In each step, we adjusted P-values of enrichment with Bonferroni multiple testing correction, considering the association as significant at FWER <0.05.
Results and Discussion
Identification of auxin response cis-elements
To expand our knowledge on the scope of transcriptional regulation in auxin response, we aimed to detect putative AuxREs from meta-analysis of auxin-responsive transcriptome data sets without prior assumptions on the transcription factors binding these.
Dozens of auxin-related transcriptome data sets are publicly available in A. thaliana (Supplementary Table S1). Although the experiments were not designed to test the same hypothesis (they differ in dosage of applied auxin, duration of treatment, and tissue samples), systematic association of the same hexamer with auxin response in different experiments will diminish the probability that association of a hexamer is a random result. We developed a bioinformatics approach to search for putative AuxREs using many transcriptome inputs (see the Materials and methods; Fig. 1).
The procedure generated a list of hexamers (147 in total), which were substantially enriched in upstream regions of auxin-responsive genes (Supplementary Table S2). We considered these hexamers as putative novel AuxREs.
A census of AuxREs
We found the canonical AuxRE core TGTCTC and its analog TGTCCC enriched in upstream regions of auxin-up-regulated genes, thus confirming previous findings (Ulmasov et al., 1995; Xu et al., 1997; Berendzen et al., 2012). bZIP-binding ACGT-containing elements and the bHLH-binding HUD motif (CACATG), shown earlier as mediating auxin response (Ulmasov et al., 1995; Liu et al., 1997; Walcher and Nemhauser, 2012; Oh et al., 2014; Weiste and Dröge-Laser, 2014), were also among those significantly associated with auxin response (Supplementary Table S2). These matches may be considered as an indication of the adequacy of the developed method.
Intriguingly, beyond the expected and known motifs, we found A/T-rich hexamers (not more than one G/C) to be the most abundant and the most significant in our search (Tables 2, 3). Two-thirds of the enriched hexamers were A/T rich, and they were not simple repeats, as we detected them even after filtering out these repeats from the upstream regions (see the Materials and methods). The relevance of TATA-box-like sequences to early auxin response was shown previously (Trenner et al., 2017); however, only a part of the A/T-rich hexamers resembled TATA-box sequences (according to Yamamoto et al., 2009) and peaked at the transcription start site (Supplementary Fig. S2). In Arabidopsis, many transcription factors bind cis-elements with more than five A/Ts in a row (O’Malley et al., 2016); however, such an abundance might also be a sign of their involvement in epigenetic regulation (Roy et al., 2016).
Table 2.
Summary statistics on the number of detected auxin-associated cis-regulatory elements
| Early (≤2 h) | Late (>2 h) | |
|---|---|---|
| Up | 24 a | 78 |
| Down | 3 | 121 |
| (i) Without A/T-rich hexamers | ||
| Up | 8a | 16 |
| Down | 3 | 26 |
| (ii) Specific in time of response and regulation | ||
| Up | 6a | 18 |
| Down | 0 | 59 |
| (iii) Enriched in ARF-binding regions | ||
| Up | 11a | 25 |
| Down | 3 | 37 |
a Including TGTCTC.
The total number of detected hexamers (top) and their classification by three characteristics: (i) belonging to non-A/T-rich elements; (ii) hexamers which were associated specifically with one of four gene groups (up/down and early/late); and (iii) hexamers significantly enriched in at least one peak set: ARF2-, ARF5- (O’Malley et al., 2016), or ARF6-binding regions (Oh et al., 2014).
For the source data see Supplementary Table S2.
Table 3.
Overview on the statistical analysis results for predicted cis-regulatory elements associated with early auxin response
| Hexamer | Transcriptome analysis, time of response | ChIP(DAP)-Seq data analysis | Description | |||||
|---|---|---|---|---|---|---|---|---|
| Early (≤2 h) | Late (>2 h) |
Enrichment in ARF-binding regions | ||||||
| Up | Down | Up | Down | ARF2 | ARF5 | ARF6 | ||
| Known and putative ARF-binding sites | ||||||||
| TGTCTC | ** | *** | *** | *** | Classical AuxRE, ARF-binding core (Ulmasov et al., 1997). | |||
| TGTCCC | *** | *** | *** | *** | AuxRE (Xu et al., 1997; Weiste and Dröge- Laser, 2014). | |||
| GTCCCC | ** | *** | *** | *** | Putative AuxRE or TCP-binding core (Supplementary Fig. S1). | |||
| TGTGGG | *** | *** | ||||||
| bHLH- and BZR1/BES1-binding site | ||||||||
| CACATG | *** | *** | *** | *** | HUD motif, enriched in ARF6-binding regions (Oh et al., 2014). | |||
| Putative MYB-binding site | ||||||||
| GATAAG | *** | ** | *** | MYB-binding core, I-box (Rose et al., 1999) (Supplementary Fig. S1). | ||||
| Known and putative bZIP-binding sites | ||||||||
| TACGTA | ** | *** | *** | *** | A-box, bound by bZIP factors (Izawa et al., 1993). | |||
| ACGTAT | *** | *** | *** | *** | A-box-related | |||
| ACGTAG | ** | *** | ||||||
| ACGTGT | *** | *** | *** | ** | *** | G-box-related, ABRE, the binding sites for AREB/ABF factors (Yamaguchi-Shinozaki and Shinozaki, 2005). | ||
| ACGTGG | *** | *** | *** | |||||
| TATA-box-like, putative TBP-binding | ||||||||
| TATAAA | *** | *** | *** | Classical TATA-box (Heard et al., 1993) | ||||
| TATATA, ATATAT, ATATAC, ATATAG | *** | *** | *** | TATA-like (Yamamoto et al., 2009). Enriched near transcription start site (Supplementary Fig. S2). | ||||
| ATATAA | *** | *** | *** | ** | ||||
| Non-TATA-box A/T-rich | ||||||||
| AACATT | ** | *** | *** | Unknown A/T-rich, depleted near transcription start site (Supplementary Fig. S2). | ||||
| CATAAT, GATTAA | *** | *** | *** | |||||
| ACTATA, TATTAA |
** | *** | *** | |||||
| ATTAGA, AAATAC |
** | *** | ||||||
| CATATT | ** | *** | * | |||||
| CATTAT | ** | *** | *** |
|
||||
| TAATTA | ** | *** | *** | Putative ATHB-binding site (Supplementary Fig. S1). | ||||
In the meta-analysis we did not distinguish between the hexamer and its reverse complement. The data presented are only for the hexamers detected for the early responsive data sets (Supplementary Table S1); the complete data are given in Supplementary Table S2.
ChIP-Seq data for ARF6 were taken from (Oh et al., 2014).
DAP-Seq data for ARF2 and ARF5 were taken from O’Malley et al., 2016 (see the Materials and Methods).
*FWER <0.05; **FWER <0.01. ***FWER <0.001.
Analyzing the A/T-rich hexamers against the data on Arabidopsis transcription factor-binding sites generated by Franco-Zorrilla et al. (2014) and O’Malley et al. (2016) within the TOMTOM tool (Gupta et al., 2007), we found 28 significant matches (E-value <0.05). Presumable A/T-rich binding sites for MYB-related (LCL1, LHY1, RVE1, EPR1, and others), G2-like (KAN4), AT-Hook (AHL20), B3 (VRN1, REM), and Homeobox (HAT1, 2, 5, 22; ATHB6, 13, 15, 18, 20, 23–24, 53; LMI1; PHV) transcription factors were detected here to be associated with the auxin response (Supplementary Table S3; Supplementary Fig. S1).
Among non-A/T-rich hexamers, TOMTOM found significant matches with MYB-binding sites for GATAAG and AGGGTT, a FUS3-binding site for CATGCA, TCP-binding sites for TGGGCC and GTCCCC, and a number of ACGT-containing sequences, which resemble the binding sites for several transcription factors families (bHLH, bZIP, NAC, and BZR1/BES) (Supplementary Fig. S1). A closer look at the ACGT-containing sequences and their auxin response pattern allowed identification of two major groups, G-box-related (CACGTG[G/T]) and A-box-related TACGTA[A/T][A/T] (Table 3).
What could over-representation of these hexamers in auxin-responsive regulatory regions mean? The identified hexamers could be the core sequences for transcription factor-binding sites mediating primary or secondary response. They could be the coupling hexamers for TGTC-containing AuxRE, constituting with it the composite element and bound by ARF partner transcription factors (Ulmasov et al., 1995). Finally, some of the identified hexamers could influence formation of specific DNA conformations important for binding chromatin factors and thereby auxin response. In the next sections, we classify the identified hexamers to these groups.
Putative AuxREs in early and late responses
Auxin-induced transcription occurs in temporal waves regulated by ARFs and their targets. The time of transcriptional response to auxin differs for various genes; for example, even among the early responding Aux/IAA gene family in Arabidopsis some genes respond within minutes while others only respond after 2 h (Abel et al., 1995). To distinguish putative AuxREs mediating early and late responses, we performed a meta-analysis separately for the data sets with auxin treatment during <2 h (10 data sets) and the remainder (11 data sets). As a result, we identified 27 (24 up; 3 down) and 140 (78 up; 121 down) hexamers associated with early and late response, respectively (Table 2; Supplementary Table S2).
Early response
Notably, for early activation and inhibition, we detected non-overlapping sets of hexamers (Fig. 2; Table 3).
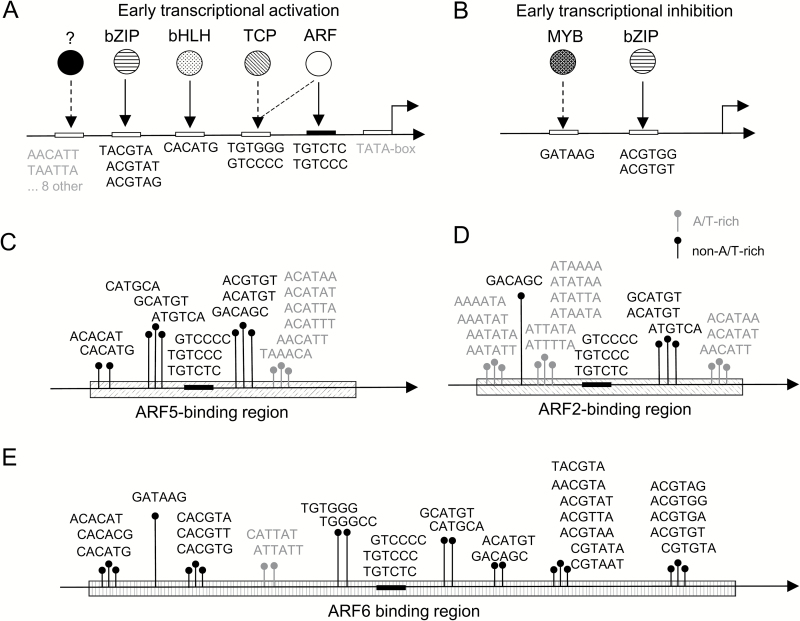
Fig. 2.
Scheme of the auxin response network reconstructed on the basis of predicted AuxREs. (A and B) Cis-regulatory elements conferring early auxin response (Table 3). (A) Activation of transcription. (B) Inhibition of transcription. (C–E) Potential coupling hexamers in composite AuxREs out of the whole list of auxin-associated cis-regulatory elements (Supplementary Table S2). The hexamers found significantly enriched within ARF5- (C); ARF2- (D), and ARF6-binding regions (E). The experimental data were taken from DAP-Seq analysis for ARF2 and ARF5 (O’Malley et al., 2016) and ChIP-Seq analysis for ARF6 (Oh et al., 2014). Pins were placed in random positions, as in this analysis we did not study the influence of orientation and relative position of the hexamers towards the TGTC-containing core.
The canonical AuxRE TGTCTC and its analog TGTCCC were specifically associated with activation of gene expression in early response to auxin (Tables 2, 23). The same association was found for TGTGGG and GTCCCC, which might be atypical ARF-binding AuxREs, or part of a TCP-binding site (Kosugi and Ohashi, 2002), as predicted by the TOMTOM tool for the latter (Supplementary Fig. S1). TATA-like hexamers (TATAAA, TATATA, ATATAA, ATATAT, ATATAC, and ATATAG), 10 non-TATA-like A/T-rich hexamers (depleted at the transcription start site; Supplementary Fig. S2), A-box-related putative bZIP-binding (TACGTA, ACGTAT, and ACGTAG), and bHLH-binding (CACATG) hexamers were also associated with early auxin-responsive transcriptional activation (Fig. 2A; Table 3).
No TGTC-containing elements were found to be associated with auxin-dependent down-regulation, which is consistent with findings published earlier (Mironova et al., 2014; Zemlyanskaya et al., 2016). However, we found G-box-related bZIP-binding (ACGTG[T/G]) and MYB-binding I-box (GATAAG; Supplementary Fig. S1) significantly associated with both early and late transcriptional repression (Fig. 2A).
Late response
There were a number of hexamers specifically associated with late up-regulation: BZR1-binding CACACG (He et al., 2005), putative TCP-binding GGCCCA, and putative MYB-binding AACCCT (Supplementary Fig. S1; Supplementary Table S3) as well as a number of A/T-rich hexamers (Supplementary Table S2). However, the lists of hexamers associated with up- and down-regulation in late response significantly overlapped (Table 2); for instance, most of the A-box-related hexamers (including early response-related TACGTA and ACGTAT) and two G-box-related (TACGTG and ACGTGT) were found for both up-/down-regulation in late response (Supplementary Table S2). The remaining G-box-related hexamers, including the classical G-box hexamer CACGTG, were specifically associated with late repression. The abundance of potential bZIP-binding hexamers among detected putative AuxREs and their segregation between up- and down-regulation, early and late responses support the findings of bZIP factors as important modulators of auxin response (Weiste and Dröge-Laser, 2014; Ulmasov et al., 1995).
The list of hexamers associated with late auxin down-regulation is almost twice longer than that for late up-regulation (Table 2). Besides ACGT-containing hexamers, EIN3-binding core ATGTA[T/C] (Kosugi and Ohashi, 2000) and a suite of A/T-rich elements are specific for late inhibition (Supplementary Table S2). The role of auxin in modulating EIN3 protein nuclear accumulation was shown earlier (He et al., 2011); our data suggest that the EIN3-mediated secondary response to auxin also occurs systematically. A great abundance of A/T-rich hexamers within upstream regions of down-regulated genes suggests that they may function in converting chromatin into a packed inactive state (discussed below). However, among A/T-rich motifs specifically associated with the late inhibitory response, TOMTOM predicts the binding sites for Homeobox, MYB, and GATA factors (Supplementary Fig. S1).
Do identified hexamers represent coupling elements to canonical AuxREs?
ARFs are known to heterodimerize with other transcription factors (Shin et al., 2007; Varaud et al., 2011; Oh et al., 2014). We thus asked if the identified hexamers (Supplementary Table S2) are coupling hexamers to the canonical AuxRE. To test this hypothesis, we explored the available data on whole-genome ARF-binding sites mapping by ChIP-Seq (ARF6; Oh et al., 2014) or DAP-Seq (ARF2 and ARF5; O’Malley et al., 2016) methods. We estimated whether ARF-binding regions were enriched with the identified hexamers (Supplementary Table S2; see the Materials and methods). Early responsive TGTCTC and TGTCCC were found prominently within the binding regions of all three ARFs, supporting the adequacy of the applied method (Table 3; Supplementary Table S2). The non-A/T-rich hexamers associated with early auxin response were significantly enriched within ARF6-binding regions; in addition, bHLH-binding CACATG and bZIP-binding ACGTGT were linked to ARF5-binding regions (Fig. 2B; Table 3).
Most of the auxin late response non-A/T-rich hexamers were also found enriched within ARF-binding regions (Fig. 2B), with the exception of EIN3-binding ATGTA[T/C] (Kosugi and Ohashi, 2000), the putative MYB-binding site AACCCT, and seven other as yet unknown hexamers (Supplementary Table S2). These results support previously published data on enrichment of bZIP-, MYB-, and bHLH-binding core sequences in close proximity to TGTC-containing AuxREs (Shin et al., 2007; Berendzen et al., 2012; Walcher and Nemhauser, 2012; Oh et al., 2014). However, we found a great variety of potential bZIP- and MYB-related hexamers which might be the binding sites for different homologs (Supplementary Table S3). We also predict FUS3- and TCP-binding sites to be the coupling elements in composite AuxREs.
A/T-rich hexamers showed a distinct pattern: most were scant in ARF-binding regions, except for two groups of hexamers (Fig. 2C, D). Hexamers of the first group are enriched within ARF2/5-binding regions; when aligned they gave an extended motif TAAACAT[A/T][A/T] (Fig. 2B), which significantly matched the YAB5-binding site in TOMTOM (E-value <0.05; Supplementary Fig. S1). A representative hexamer AACATT was specifically associated with early auxin up-regulation (Table 3).
The group of poly(A/T) hexamers is significantly enriched within ARF2-binding regions (Fig. 2B), which predicts that ARF2 has a partner with an A/T-rich transcription factor-binding site.
Available whole-genome maps (Oh et al., 2014; O’Malley et al., 2016) do not provide a complete picture for ARF-binding regions, as they were generated for only three transcription factors of the wide ARF family. Thus, we cannot exclude that the remaining hexamers adjoin binding sites for other ARFs, or operate in other conditions. The single hexamers also could be the binding sites for ARF interaction partners, that anchor ARFs on the DNA without requiring a canonical AuxRE.
Association of AuxREs with different chromatin states
The abundance of non-TATA-box A/T-rich hexamers in the list of putative AuxREs (Supplementary Table S2) raised the question of whether these motifs function in building a specific chromatin landscape rather than in binding transcription factors. To test this hypothesis, we used the chromatin map generated by Sequeira-Mendes et al. (2014), where nine chromatin states were determined, each with distinctive properties in DNA sequence, CG methylation, nucleosome density, histone variants, and modifications. Upstream regions of genes are mainly composed of blocks of chromatin states 1 (core promoter), 2 (proximal promoter), 4 (distal promoter), and 5 (Polycomb-regulated repressed chromatin type). Chromatin states 3, 6, and 7 are more associated with intragenic regions, and states 8 and 9 correspond to heterochromatin.
First we tested whether the promoters of auxin-responsive genes have a bias in location within any of the chromatin states (see the Materials and methods). Upstream regions of both auxin up- and down-regulated genes appeared to be enriched with the chromatin in state 4 (FWER <0.001). Up-regulated genes additionally possess significantly higher portions of chromatin state 1 (FWER <0.001) and state 3 (FWER <0.05) in the upstream regions comparing non-responsive with auxin-responsive genes. Down-regulated genes were additionally enriched with chromatin states 2 (FWER <0.01) and 5 (FWER <0.001).
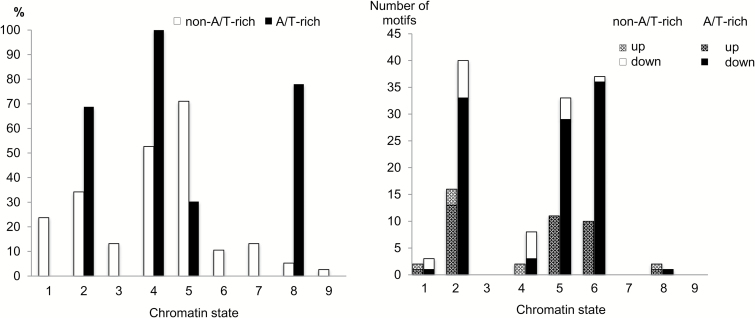
Analyzing the distribution of putative AuxREs (Supplementary Table S4) over different chromatin states (see the Materials and methods), we found that all A/T-rich hexamers were significantly enriched in chromatin state 4, and large portions of putative AuxREs were also enriched in chromatin states 8 (72%), 2 (60 %), and 5 (26 %) (Fig. 3). Thus, A/T-rich hexamers might appear in our search results because of their abundance within specific chromatin states, which make up a notable part of auxin-responsive upstream regions. It was proposed that readers of short A/T-rich hexamers might restrain gene expression, for example by recruitment of DNA methylation or repressive histone marks (Quante and Bird, 2016).
Fig. 3.
Putative AuxREs within chromatin context. (A) The portion of putative AuxREs that were found significantly enriched in the upstream regions associated with nine chromatin states (Sequeira-Mendes et al., 2014). Significance was estimated via one-tailed Fisher’s exact test (see the Materials and methods). One hexamer can be enriched in more than one state. (B) The number of putative AuxREs specifically enriched in the chromatin state islands within the upstream regions of auxin-responsive genes relative to not auxin-responsive genes. Hexamers enriched in both up- and down-regulation are counted twice. A/T-rich hexamers are shown in gray.
We next tested if putative AuxREs (Supplementary Table S4) are specifically distributed within certain chromatin domains of auxin-responsive upstream regions compared with non-regulated ones (see the Materials and methods). While we did not find any specific AuxRE association with chromatin state 3, it was the case for other chromatin states enriched in auxin-responsive genes (Figs 3B, 4).
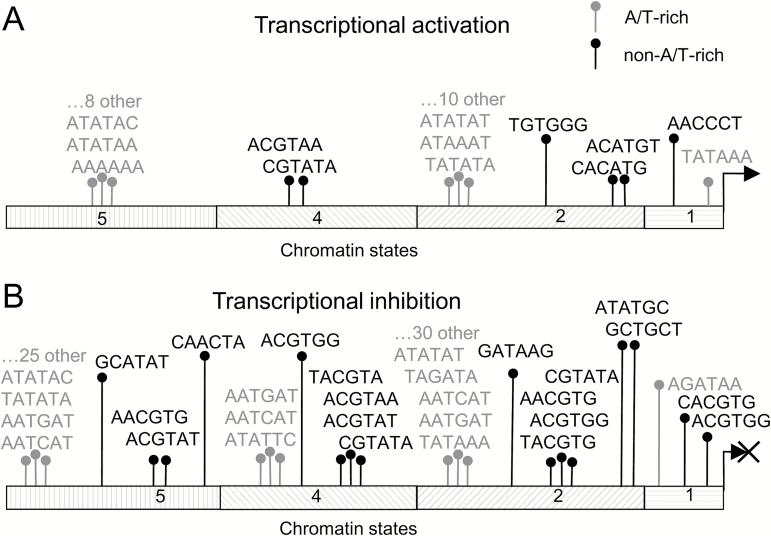
Fig. 4.
Putative AuxREs specifically enriched within chromatin states 1, 2, 4, and 5 of auxin-responsive genes. Core promoters tends to possess transcriptionally active chromatin state 1, proximal promoters usually belong to chromatin state 2, and distal promoters to state 4 or state 5 (Sequeira-Mendes et al., 2014). (A) Association with transcriptional activation. (B) Association with transcriptional inhibition. A/T-rich hexamers are shown in gray. For details see Supplementary Table S4.
Many A/T-rich putative AuxREs are specifically over-represented in chromatin states 2 and 5 of both auxin-activated and repressed genes, with the repressed genes showing a greater variety (Fig. 4; Supplementary Table S4). Most of them are unknown; however, TOMTOM predicts (E-value <0.05) a number of transcription factor-binding sites: Homeobox-related AAT[G/C]AT and KAN4-binding core ATATTC were significantly enriched within chromatin states 2, 4, and 5 of auxin-repressed genes. The TATA-box sequence TATAAA was enriched in core promoters (chromatin state 1) of only auxin-activated genes.
Negatively and positively auxin-responsive genes differed by non-A/T-rich hexamers associated with specific chromatin states (Fig. 4; Supplementary Table S4). All chromatin regions of auxin-inhibited genes were enriched with A-box- and G-box-related ACGT-containing hexamers; in auxin-activated genes, only chromatin state 4 was enriched with A-box-related ACGTA[A/T]A hexamers. In general, the chromatin landscape of upstream regions in auxin-inhibited genes was richer in specific hexamers than those of auxin-activated genes (Fig. 4B).
Interestingly TGTC-containing cores were not enriched in any chromatin state. Recently we showed that ethylene-responsive genes possess an EIN3-binding site within specific chromatin state 4 (Zemlyanskaya et al., 2016); the present data predict a similar preference to bind DNA within specific chromatin domains for ARF partners, but not ARFs themselves.
The auxin transcriptional regulation machinery involves a number of chromatin-remodeling factors (Szemenyei et al., 2008; Weiste and Dröge-Laser, 2014; Wu et al., 2015); the variety of auxin-associated hexamers specifically enriched within a certain chromatin context suggests that there are as yet unknown players in epigenetic regulation of auxin response. While the transcription factors bound to non-A/T-rich motifs might recruit chromatin-remodeling complexes, A/T-rich motifs might facilitate binding with these complexes, or directly influence nucleosome positioning.
Conclusions
In recent years, plant biologists have generated an enormous amount of whole-genome expression profiling data. Development of high-throughput sequencing technologies makes the growth of these big data even faster. Despite providing a challenge for comprehensive analysis, accumulation of the data also provides benefits when studying the intricate features which are highlighted under systematic analysis. A search for cis-regulatory elements mediating a complex response is an example, as the binding sites for major regulators should be over-represented in the promoters of DEGs in many data sets wherein the regulator is involved. Development of a bioinformatics method which detects systematically over-represented motifs over many related transcriptome data sets (Fig. 1), helped us to identify a comprehensive set of auxin-response elements, and most of them were novel. Our results predict the key players in early and late auxin response (Fig. 2A–B; Table 3), and expand our knowledge on potential ARF partners whose binding sites are enriched within ARF-binding regions (Fig. 2C–E). Application of the meta-analysis pipeline on the data for the chromatin landscape of the A. thaliana genome (Sequeira-Mendes et al., 2014) suggested which cis-regulatory elements might be involved in epigenetic regulation of auxin response (Fig. 4). These results also highlight the benefits of employment of independent data in meta-analyses, which promise that new findings will appear from as yet understudied whole-genome data.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Transcriptome data sets used in the meta-analysis.
Table S2. Hexamers significantly associated with auxin response: meta- and permutation P-values; statistics for the hexamer over-representation in ARF-binding regions.
Table S3. Hexamers associated with auxin response: matches with the transcription factor-binding sites from DAP-seq (O’Malley et al., 2016), PBM (Franco-Zorrilla et al., 2014), and CIS-BP DNA databases (A. thaliana) found by the TOMTOM tool (Gupta et al., 2007).
Table S4. Analysis of the hexamer enrichment within specific chromatin states (Sequeira-Mendes et al., 2014) of auxin-responsive upstream regions.
Fig. S1. Significant matches (E-value <0.05) of putative AuxREs with the transcription factor-binding sites identified by TOMTOM (Gupta et al., 2007).
Fig. S2. Distribution of A/T-rich putative AuxREs along the upstream regions of auxin-responsive genes.
Supplementary Material
Acknowledgements
We thank Elena Zemlyanskaya, Daniil Wiebe, Yuri Matushkin, Sergey Lashin, and Fedor Kazantsev for fruitful discussions, Dmitry Oschepkov and Sumanth Mutte for providing pre-processed ChIP-Seq/DAP-Seq data, and the Siberian Branch of the Russian Academy of Sciences (SB RAS) Siberian Supercomputer Center for providing their supercomputer facilities. The work was partly supported by the Russian Ministry of Science and Education under the 5-100 Excellence Programme and by the project 0324-2016-0008 from the Russian State Budget. DN is supported by a PhD sandwich fellowship from Wageningen Graduate School.
Glossary
Abbreviations:
- AuxRE
auxin response element
- DEG
differentially expressed gene.
References
- Abel S, Nguyen MD, Theologis A. 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. Journal of Molecular Biology 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A. 2004. Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 14978–14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Vanneste S, Krouk G et al. 2013. A map of cell type-specific auxin responses. Molecular Systems Biology 9, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29, 1165–1188. [Google Scholar]
- Berendzen KW, Weiste C, Wanke D, Kilian J, Harter K, Dröge-Laser W. 2012. Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription. BMC Plant Biology 12, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY. 2015. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Current Biology 25, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. 2000. ABFs, a family of ABA-responsive element binding factors. Journal of Biological Chemistry 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Delker C, Pöschl Y, Raschke A, Ullrich K, Ettingshausen S, Hauptmann V, Grosse I, Marcel Q. 2010. Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. The Plant Cell 22, 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Xuan W et al. 2012. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nature Chemical Biology 8, 798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ et al. 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Hosaka A, Nagata T, Satoh K, Suzuki K, Mauleon R, Mendoza MJ, Bruskiewich R, Kikuchi S. 2008. Development of a novel data mining tool to find cis-elements in rice gene promoter regions. BMC Plant Biology 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua N. 1994. Plant bZIP proteins gather at ACGT elements. FASEB Journal 8, 192–200. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. 2014. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proceedings of the National Academy of Sciences, USA 111, 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Solano R. 2017. Identification of plant transcription factor target sequences. Biochimica et Biophysica Acta 1860, 21–30. [DOI] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biology 8, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Current Opinion in Plant Biology 10, 453–460. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Ulmasov T, Hagen G. 1998. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cellular and Molecular Life Sciences 54, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. 2005. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Brumos J, Li H et al. 2011. A small-molecule screen identifiesl-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. The Plant Cell 23, 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard DJ, Kiss T, Filipowicz W. 1993. Both Arabidopsis TATA binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: evidence for gene-specific changes in DNA binding specificity of TBP. EMBO Journal 12, 3519–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. 2016. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proceedings of the National Academy of Sciences, USA 113, 6562–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. 1993. Plant bZIP protein DNA binding specificity. Journal of Molecular Biology 230, 1131–1144. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. 2012. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139, 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kang H, Park J, Kim W, Yoo J, Lee N, Kim J, Yoon TY, Choi G. 2016. PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. The Plant Cell 28, 1388–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. 2000. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Research 28, 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. 2002. DNA binding and dimerization specificity and potential targets for the TCP protein family. The Plant Journal 30, 337–348. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. 2013. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. The Plant Cell 25, 3329–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Hagen G, Guilfoyle TJ. 1997. A G-box-binding protein from soybean binds to the E1 auxin-response element in the soybean GH3 promoter and contains a proline-rich repression domain. Plant Physiology 115, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. Topless regulates apical embryonic fate in arabidopsis. Science 312, 1520–1523. [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova VV, Omelyanchuk NA, Wiebe DS, Levitsky VG. 2014. Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genomics 15(Suppl 12), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biology 2, E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. 2014. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3, e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology 14, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K et al. 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Weijers D. 2016. Auxin: the plant molecule that influences almost anything. BMC Biology 14, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. 2016. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Annals of Applied Statistics 10, 946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM. 2003. The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Functional and Integrative Genomics 3, 135–143. [DOI] [PubMed] [Google Scholar]
- Quante T, Bird A. 2016. Do short, frequent DNA sequence motifs mould the epigenome?Nature Reviews. Molecular Cell Biology 17, 257–262. [DOI] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD. 2004. Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. The Plant Journal 38, 545–561. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll JJ, Bailey LJ, Mai QA, Wu SL, Hon CT, Chapman EJ, Ditta GS, Estelle M, Yanofsky MF. 2015. microRNA regulation of fruit growth. Nature Plants 1, 15036. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I, Wienand U. 1999. The tomato I-box binding factor LeMYBI is a member of a novel class of Myb-like proteins. The Plant Journal 20, 641–652. [DOI] [PubMed] [Google Scholar]
- Roy A, Dutta A, Roy D, Ganguly P, Ghosh R, Kar RK, Bhunia A, Mukhopadhyay J, Chaudhuri S. 2016. Deciphering the role of the AT-rich interaction domain and the HMG-box domain of ARID-HMG proteins of Arabidopsis thaliana. Plant Molecular Biology 92, 371–388. [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Aragüez I, Peiró R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C. 2014. The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. The Plant Cell 26, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. The Plant Cell 19, 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, Freire-Rios A, Sorefan K, Weijers D, Friml J, Østergaard L. 2016. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes and Development 30, 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0 http://www.repeatmasker.org.
- Strader LC, Zhao Y. 2016. Auxin perception and downstream events. Current Opinion in Plant Biology 33, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during arabidopsis embryogenesis. Science 319, 1384–1386. [DOI] [PubMed] [Google Scholar]
- Trenner J, Poeschl Y, Grau J, Gogol-Döring A, Quint M, Delker C. 2017. Auxin-induced expression divergence between Arabidopsis species may originate within the TIR1/AFB–AUX/IAA–ARF module. Journal of Experimental Botany 68, 539–552. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1997. ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. 1995. Composite structure of auxin response elements. The Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. The Plant Cell 23, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology 7, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher CL, Nemhauser JL. 2012. Bipartite promoter element required for auxin response. Plant Physiology 158, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annual Review of Plant Biology 67, 539–574. [DOI] [PubMed] [Google Scholar]
- Weiste C, Dröge-Laser W. 2014. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nature Communications 5, 3883. [DOI] [PubMed] [Google Scholar]
- Wu Q, Madany P, Akech J et al. 2015. The SWI/SNF ATPases are required for triple negative breast cancer cell proliferation. Journal of Cellular Physiology 230, 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Hagen G, Guilfoyle T. 1997. Multiple auxin response modules in the soybean SAUR 15A promoter. Plant Science 126, 193–201. [Google Scholar]
- Xuan W, Audenaert D, Parizot B et al. 2015. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Current Biology 25, 1381–1388. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2005. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science 10, 88–94. [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Yoshitsugu T, Sakurai T, Seki M, Shinozaki K, Obokata J. 2009. Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. The Plant Journal 60, 350–362. [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J et al. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. The Plant Journal 65, 634–646. [DOI] [PubMed] [Google Scholar]
- Zemlyanskaya EV, Wiebe DS, Omelyanchuk NA, Levitsky VG, Mironova VV. 2016. Meta-analysis of transcriptome data identified TGTCNN motif variants associated with the response to plant hormone auxin in Arabidopsis thaliana L. Journal of Bioinformatics and Computational Biology 14, 1641009. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Feng LY, Cheng J et al. 2013. The roles of two transcription factors, ABI4 and CBFA, in ABA and plastid signalling and stress responses. Plant Molecular Biology 83, 445–458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.