Summary
We assessed differential risk for hospital-onset Staphylococcus aureus bacteremias and used genome sequencing to discern pathways of transmission. African-American race was associated with genetic clustering of both community- and healthcare-onset infection isolates.
Keywords: MRSA, Whole genome sequencing, disparities, bacteremia.
Abstract
Background.
We examined whether disparities existed in hospital-onset (HO) Staphylococcus aureus bloodstream infections (BSIs) and used whole-genome sequencing (WGS) to identify factors associated with USA300 transmission networks.
Methods.
We evaluated HO methicillin-susceptible S. aureus (MSSA) and HO methicillin-resistant S. aureus (MRSA) BSIs for 2009–2013 at 2 hospitals and used an adjusted incidence for modeling. WGS and phylogenetic analyses were performed on a sample of USA300 BSI isolates. Epidemiologic data were analyzed in the context of phylogenetic reconstructions.
Results.
On multivariate analysis, male sex, African-American race, and non-Hispanic white race/ethnicity were significantly associated with HO-MRSA BSIs whereas Hispanic ethnicity was negatively associated (rate ratio, 0.41; P = .002). Intermixing of community-onset and HO-USA300 strains on the phylogenetic tree indicates that these strains derive from a common pool. African-American race was the only factor associated with genomic clustering of isolates.
Conclusions.
In a multicenter assessment of HO–S. aureus BSIs, African-American race was significantly associated with HO-MRSA but not MSSA BSIs. There appears to be a nexus of USA300 community and hospital transmission networks, with a community factor being the primary driver. Our data suggest that HO-USA300 BSIs likely are due to colonizing strains acquired in the community before hospitalization. Therefore, prevention efforts may need to extend to the community for maximal benefit.
(See the editorial commentary by Ray on pages 1631–3.)
The emergence of community-associated (CA) methicillin-resistant Staphylococcus aureus (MRSA) has transformed the epidemiology of MRSA over the last 15 years [1, 2]. While originally observed only in healthcare facilities, MRSA infections are now seen extensively in community settings [3]. By pulsed-field gel electrophoresis (PFGE), USA300 is the most common CA-MRSA strain in the United States [4]. USA300 MRSA has had a significant impact on community settings and now has entered healthcare facilities as an important cause of hospital-onset (HO) MRSA bloodstream infections (BSIs) [5, 6]. We previously reported that from 2007 to 2013, >50% of HO-MRSA BSIs at the major public hospital in Chicago, Illinois, were due to USA300 [6], suggesting that USA300 strains were displacing traditional hospital MRSA strains.
Community factors associated with increased risk for CA-MRSA colonization and infection include close person-to-person contact, fewer opportunities for (or less attention to issues of) infection control, and increased opportunity for skin abrasions [7, 8]. Greater risk for CA-MRSA colonization and infection has been associated with African-American race, whereas Hispanic ethnicity appears to be protective [9, 10]. It has been suggested that certain community exposures such as correctional facilities [10–12], illicit drug use [6, 13], and social networks [14, 15] may enable spread of MRSA in a community and contribute to these observed disparities in risk. While national data supports a decline over the last 10 years in hospital- and healthcare-associated invasive infections due to MRSA, the rate of CA-MRSA invasive infections has not shown similar declines [16–18], suggesting that certain community exposures continue to promote risk that needs to be further evaluated.
PFGE has limited discriminatory power for USA300 strains [19]. Whole-genome sequencing (WGS) can better characterize MRSA strains, providing an epidemiologic tool to improve our understanding of transmission dynamics that have made CA-MRSA [15] endemic in certain communities and healthcare facilities [20, 21]. Using WGS, we previously described community transmission networks of USA300 MRSA strains found to colonize individuals seeking care at the major public hospital in Chicago [15]. While it is well-recognized that USA300 strains have now entered healthcare facilities and can cause HO infections, the relative roles of community and healthcare transmission networks for these strains are unknown. Furthermore, given the complexity of colonization and transmission dynamics of USA300, it is unclear whether epidemiologic classifications such as community-associated and hospital-associated [1] remain meaningful to describe MRSA acquisition. The objective of this study was to determine whether sex and racial disparities exist for S. aureus bacteremia and to use WGS with epidemiologic data to characterize possible transmission pathways associated with USA300 BSIs in the community and the hospital.
METHODS
Study Population for Assessing Disparities of Hospital-Onset S. aureus BSIs
We previously reported on HO–S. aureus BSIs from 2007 to 2013 [6] from individuals seeking care at Stroger (formerly Cook County) Hospital (CCH), a 464-bed facility and the major public hospital in Chicago, Illinois. To put these data into a community context, we electronically queried existing microbiologic surveillance data for HO–S. aureus BSIs from Rush University Medical Center (RUMC), a tertiary care 650-bed academic medical center that has a campus adjacent to CCH. We examined counts of HO methicillin-susceptible S. aureus (MSSA) and HO-MRSA BSIs for 2009–2013 at both institutions and recorded sex, race, and ethnicity for each individual with a HO–S. aureus bacteremia. For the denominator of patient-days, we stratified the yearly inpatient denominator for each institution by sex, race, and ethnicity.
A race-, sex-, and ethnicity-adjusted incidence of S. aureus bacteremia was used for modeling, with incidence used as the outcome measure and race, sex, ethnicity, and institution as covariates. Univariate analysis was performed to assess individual effects of each covariate on the incidence. Multivariate analysis was used with all covariates of interest included in the model, consistent with our primary study objective, which was to determine if sex, racial, and ethnic disparities existed in a large urban area. Yearly central line–associated BSI (CLABSI) standardized infection ratio (SIR) data were obtained for both institutions as a surrogate for infection control practice at that hospital.
Univariate and multivariate models were constructed using Poisson regression with incidence of BSI as the outcome variable. Separate models were constructed for HO-MRSA and HO-MSSA bacteremia. Analyses were conducted with SAS version 9.3 (Cary, North Carolina), using the Proc Genmod statement. Approval for the study was obtained by the CCH and RUMC institutional review boards.
Whole-Genome Sequencing
From the CCH dataset, a subset of BSIs underwent genomic analysis. Both community-onset (CO; infection presenting in an outpatient setting or within 3 days of hospitalization) and HO (infection occurring >3 days into hospitalization) MRSA BSI isolates from 2009 to 2013 had undergone PFGE analysis, as previously reported [6], with repeat isolates from an individual excluded if within 30 days of the index isolate. As our primary objective in this study was to examine HO-BSIs, all isolates identified as USA300 by PFGE that were classified as HO were included for WGS analysis. A random sample of a similar number of USA300 strains classified as CO was chosen for WGS for comparison. For all isolates undergoing WGS, 76% were from males and 64% were from African-American patients (Supplementary Table 1). A large proportion of individuals had hospitalization at CCH in the prior 3 months (41%), and 20% of individuals were current users of illicit drugs.
DNA was extracted from 80 MRSA isolates from 79 patients and prepared for sequencing on an Illumina HiSeq2500 instrument using standard library preparation approaches and sample-specific barcoding, as described in prior work [22]. Libraries from isolates were pooled together before sequencing to an average depth of 200–300 times coverage per genome. Library preparation and sequencing were performed at the DNA Services facility at the University of Illinois at Chicago. The quality of reads was assessed with Fastqc [23] and Trimmomatic [24] was used for trimming the adapter sequences and low-quality bases. Reference genomes for variant calling were chosen based on the sample’s relatedness to MRSA USA300 (GenBank accession number NC_002952) and MRSA USA100 (PATRIC Genome ID 1422125.3 S. aureus DAR3548) on a single-nucleotide polymorphism (SNP)–based maximum likelihood phylogenetic tree generated using kSNP software [25]. Sixty-three isolates were designated as USA300, 14 as USA100, and 3 as “other” due to their placement away from USA100 and USA300 clades on the kSNP phylogeny (Supplementary Figure 1). These classifications are supported by in silico multilocus sequence typing, with USA300 and USA100 genomes being predominantly ST8 and ST5, respectively (Supplementary Table 2). Sequencing reads from 2 USA300 genomes showed evidence of being a mixture of multiple strains, based on the number of positions with reads supporting multiple alleles. These 2 samples (7303 and 7268) were therefore excluded from phylogenetic analyses. One pair of isolates (8234 and 8381) were from the same person; the epidemiologic classification for both bacteremias was HO, with isolates separated in time by >3 months and occurring during different hospitalizations. These isolates clustered together on the phylogenetic tree; only one of these isolates (8381) was included in analyses aimed at understanding epidemiological factors associated with clustering.
Variants were identified by mapping filtered paired-reads to the appropriate reference genomes using the Burrows-Wheeler short-read aligner, discarding polymerase chain reaction duplicates with Picard and calling variants with SAMtools and bcftools. Variants were filtered from raw results using GATK’s VariantFiltration (QUAL >100, MQ >50, >15 reads supporting variant, FQ <.025) (QUAL and FQ: http://samtools.sourceforge.net/mpileup.shtml [Accessed January 2016], MQ: http://samtools.github.io/hts-specs/VCFv4.3.pdf [Accessed January 2016]). In addition, a custom python script was used to filter out SNPs that were (1) <5 bp in proximity to indels; (2) <10 bp in proximity to each other; or (3) non-core SNPs.
Phylogenetic Analysis and Linkage of Epidemiologic Data
Maximum likelihood trees were constructed in RAxML [26] with parameters set to use a general time-reversible model with site-specific rate categories (–m GTRCAT). Bootstrap analysis was performed with the number of bootstrap replicates determined using the bootstrap convergence test and the autoMRE convergence criteria (–N autoMRE). Bootstrap support values are displayed on the best-scoring tree identified during rapid bootstrap analysis (–f a).
To elucidate putative pathways of USA300 transmission, we quantified the extent to which different epidemiological factors could explain the clustering of bloodstream infection isolates on the phylogenetic tree. For this analysis, we constructed a tree containing USA300 bloodstream infection isolates from the current study and a set of previously published USA300 isolates taken from individuals in New York City, with the latter acting as an outgroup by which regional clusters were partitioned. Regional clusters were defined as subclades on the tree that met the following criteria: (1) They contained only isolates from the current study, (2) they contained isolates from 2 or more individuals, and (3) the subclade that defined the cluster was observed in at least 90% of bootstrapped trees. Within each regional cluster, we then tallied the number of isolates belonging to subclades of size 2 or greater that were homogeneous for the considered epidemiological factor (eg, exclusively African-American or white). The significance of the number of isolates in uniform clusters was evaluated by performing 1000 random permutations of epidemiological labels and constructing an empirical distribution for the number of isolates expected to fall into homogeneous clusters given the constraints of the tree structure and the label frequencies associated with a given factor.
To test whether any epidemiological factors were associated with acquisition of USA100 strains, we performed χ2 tests comparing the 14 patients with USA100 strains to the 65 patients with non-USA100 strains.
Data Access
The genome sequence data from this study have been submitted to the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJNA345238.
RESULTS
Bacteremia Disparities Assessment
There were 156 HO-MRSA and 256 HO-MSSA BSIs during 2009–2013 for the 2 institutions, with an overall unadjusted incidence of 0.10/1000 patient-days and 0.18/1000 patient-days for MRSA and MSSA, respectively. On univariate analysis (Table 1), male sex, African-American race, and non-Hispanic white race/ethnicity were each significantly associated with risk for HO-MRSA BSI. For HO-MSSA BSIs, only male sex was a significant predictor. Hispanic ethnicity was negatively associated with HO-MRSA bacteremia (rate ratio, 0.45; 95% confidence interval [CI], .25–.8; P = .006) but not with HO-MSSA bacteremia (P = .78).
Table 1.
Univariate Analysis of Risk Factors Associated With a Race- and Sex-Adjusted Hospital-Associated Staphylococcus aureus Bacteremia Incidence
| MRSA | MSSA | |||||
|---|---|---|---|---|---|---|
| Factor | OR | (95% CI) | P Value | OR | (95% CI) | P Value |
| Sex | ||||||
| Male | 2 | (1.41–2.84) | .0001 | 2.21 | (1.7–2.87) | <.0001 |
| Female | Reference | Reference | ||||
| Race/Ethnicity | ||||||
| Black | 2.46 | (1.37–4.43) | .0026 | 0.92 | (.65–1.3) | .64 |
| Other | 0.73 | (.21–2.57) | .63 | 1.38 | (.82–2.34) | .23 |
| White, non-Hispanic | 2.15 | (1.16–3.99) | .015 | 1.17 | (.82–1.68) | .38 |
| Hispanic | Reference | Reference | ||||
| Institution | ||||||
| CCH | 1.76 | (1.26–2.47) | .001 | 1.68 | (1.31–2.15) | <.0001 |
| RUMC | Reference | Reference | ||||
Abbreviations: CCH, Stroger (formerly Cook County) Hospital; CI, confidence interval; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; OR, odds ratio; RUMC, Rush University Medical Center.
On multivariate analysis (Table 2), male sex, African-American race, and non-Hispanic white race/ethnicity remained significantly associated with risk of HO-MRSA bacteremia, even after controlling for institution. In contrast, only male sex and non-Hispanic white race/ethnicity were associated with HO-MSSA bacteremia after adjusting for institutional effects. Even after controlling for sex and institution, Hispanic ethnicity remained negatively associated with HO-MRSA bacteremia (rate ratio, 0.41; 95% CI, .23–.72; P = .002) but not with HO-MSSA bacteremia (P = .41). The average CLABSI SIR [27] for each institution during the study period was <1, indicating better than average hospital-wide infection control efforts. The ratio of the RUMC to CCH SIR during the study period was 1.2.
Table 2.
Multivariate Analysis of Risk Factors Associated With a Race- and Sex-Adjusted Hospital-Associated Staphylococcus aureus Bacteremia Incidence
| MRSA | MSSA | |||||
|---|---|---|---|---|---|---|
| Factor | OR | (95% CI) | P Value | OR | (95% CI) | P Value |
| Sex | ||||||
| Male | 1.89 | (1.31–2.67) | .0006 | 2.04 | (1.56–2.66) | <.0001 |
| Female | reference | reference | ||||
| Race/Ethnicity | ||||||
| Black | 2.59 | (1.44–4.65) | .0015 | .97 | (.68–1.37) | .846 |
| Other | 0.72 | (.2–2.52) | .61 | 1.36 | (.8–2.3) | .253 |
| White, non-Hispanic | 2.77 | (1.47–5.22) | .0017 | 1.47 | (1.01–2.14) | .042 |
| Hispanic | reference | reference | ||||
| Institution | ||||||
| CCH | 1.85 | (1.28–2.66) | .001 | 1.72 | (1.32–2.26) | <.0001 |
| RUMC | reference | reference | ||||
Abbreviations: CCH, Stroger (formerly Cook County) Hospital; CI, confidence interval; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; OR, odds ratio; RUMC, Rush University Medical Center.
Factors That Serve as the Basis for Genomic Clustering of USA300 Strains
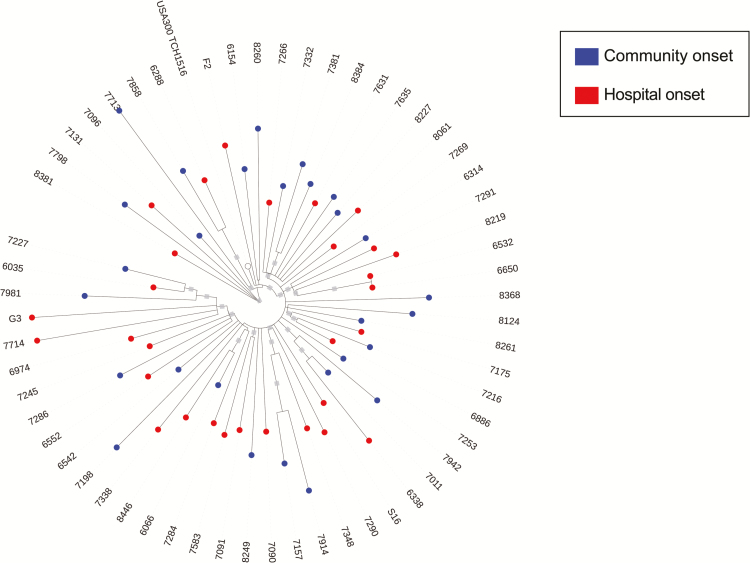
One explanation for the observed association between demographic factors and risk for HO-MRSA bacteremias is that a significant portion of HO-MRSA infections actually derive from colonizing isolates acquired prior to hospitalization. To test this hypothesis, we applied WGS and phylogenetic analyses to USA300 MRSA BSI isolates from 60 patients and evaluated both community and healthcare factors for their ability to explain genetic clustering of MRSA isolates. Isolates represented infections classified as CO-MRSA or HO-MRSA, such that we could evaluate whether the epidemiologic classifications CO and HO would lead to distinct genetic clusters (Figure 1). Although there was evidence for some transmission occurring in the hospital (isolates 6650 and 6532 were from 2 different patients hospitalized at the same time in the same intensive care unit), we observed that CO and HO USA300 strains were largely intermixed on the phylogenetic tree. This intermixing indicates that CO and HO infections derive from a common pool and hence manifest as merged USA300 transmission networks rather than separate community and hospital networks (Figure 1).
Figure 1.
Maximum-likelihood phylogeny of community-onset (CO) and hospital-onset (HO) methicillin-resistant Staphylococcus aureus (MRSA) genomes. A phylogeny was constructed using genome-wide variants identified in 60 MRSA bloodstream infection isolates relative to the TCH1516 USA300 reference genome. Note that 2 isolates that are more distantly related are not displayed for visualization purposes.
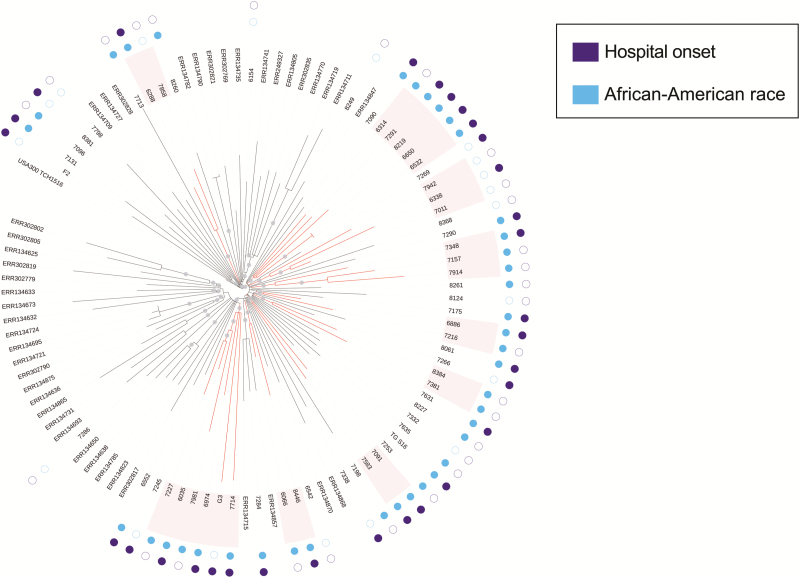
To help partition these isolates into local transmission networks, we used previously published genomes from New York [28] to act as regional out-groups (see Methods). Nine regional clusters of size 2 or greater were identified that had strong bootstrap support and were not dissected by isolates from New York (Figure 2). We next searched for epidemiological drivers of transmission by evaluating each epidemiological factor for its capacity to group subsets of patients into epidemiologically homogeneous subclusters more often than would be expected by chance (Table 3). Confirming our earlier observation of intermixing of CO and HO USA300 strains, we found that isolates failed to significantly cluster by CO/HO status. Among the set of healthcare and community factors, only African-American race (a community factor) served as the basis for clustering (P = .007). Six of the 9 regional clusters were composed solely of African-American individuals and an additional 6-person regional cluster contained a subcluster of 3 African-American individuals. Hispanic or non-Hispanic white individuals comprised one 3-person regional cluster but were otherwise largely excluded from phylogenetic clusters. Hospitalization at CCH in the prior 3 months and other healthcare-related factors were not associated with significant clustering. In addition, community factors previously identified as associated with CA-MRSA colonization and infection were not associated with genomic clustering of USA300 BSI isolates (Table 3).
Figure 2.
Maximum-likelihood phylogeny of USA300 methicillin-resistant Staphylococcus aureus (MRSA) isolates from Chicago bloodstream infections and New York household surveillance. A phylogeny was constructed using genome-wide variants for the isolates from Figure 1, and an additional set of USA300 MRSA isolates collected during a previous study of New York households. Gray circles indicate splits with >90% bootstrap support. Red branches indicate subclades with >90% bootstrap support that contain only isolates from the current study, and are therefore deemed regional clusters. The inner ring of circles adjacent to the tip labels indicates whether the isolate was taken from an African-American individual (blue) and the outer circles indicate whether an isolates was designated hospital onset (purple).
Table 3.
Probability of Genomic Clustering of 60 USA300 Isolates as Defined by Whole-Genome Sequencing According to Community- and Healthcare-Related Factors
| Factor | Probability of Clusters by Factor |
|---|---|
| Epidemiologic classification of bacteremia (CO vs HA) | 0.827 |
| Community factors | |
| Sex | 0.579 |
| Race | 0.007 |
| Ethnicity | 0.011 |
| Current illicit drug use | 0.678 |
| Current incarceration | 0.718 |
| Currently in unstable housinga | 0.636 |
| Healthcare factors | |
| Year of bloodstream infection | 0.325 |
| Hospitalized in the prior 3 months | 0.739 |
| Malignancy | 0.633 |
| Hemodialysis | 0.330 |
| Length of stay | 0.827 |
| Admission to the intensive care unit | 0.575 |
Clustering was defined as containing >1 USA300 isolate with a common epidemiologic factor.
Abbreviations: CO, community-onset; HA, hospital-onset.
aUnstable housing was defined as currently being homeless, living in a homeless shelter, living in a substance abuse center, living in subsidized housing, or staying in homes of friends. When we tested by type of unstable housing, the probability was 0.682.
To assess if the associations seen extended to other strains of MRSA, we evaluated whether a group of 14 USA100 strains was enriched with community and healthcare factors compared to the other BSI isolates that underwent WGS (Supplementary Table 3). We found no community factors associated with USA100 strains. In contrast, individuals with USA100 strains were significantly enriched for having an active malignancy (odds ratio, 1.72; 95% CI, 1.01–2.94; P = .002).
DISCUSSION
In this multicenter assessment of HO–S. aureus BSIs, we found that African-American race was significantly associated with HO-MRSA BSIs, even when adjusting for sex and institution effects. Furthermore, Hispanic ethnicity was negatively associated with HO-MRSA BSIs after adjustment for sex and institution effects. In a nested analysis of the genomic data from WGS of USA300 MRSA bloodstream isolates, African-American race was identified as the only factor associated with phylogenetic clustering of patient isolates. Whether a BSI was classified as CO or HO was not associated with genomic clustering of USA300 MRSA strains, suggesting a nexus of USA300 community and hospital transmission networks.
We specifically focused our assessment of racial and ethnic disparities on HO-BSIs because we hypothesized that nosocomial events would be the major driver of these infections and that these disparities would be less frequent. We have previously demonstrated racial and ethnic disparities in CA-MRSA colonization and infection in a population seeking care at the major public hospital in Chicago and had postulated that differences in community exposures and social networks accounted for these disparities [9, 10, 13]. However, we have now observed that these disparities exist for nosocomial MRSA BSIs and are present in 2 institutions in Chicago.
In contrast to our findings for MRSA, race and ethnicity were not associated with risk for HO-MSSA BSIs. Prior work in Chicago [9] compared risk factors for skin infections due to CA-MRSA in comparison to CA-MSSA and observed that African-American race was a significant predictor of CA-MRSA whereas Hispanic ethnicity was negatively associated with CA-MRSA. The current study further supports the unique epidemiology of MRSA, even for infections occurring during hospitalization. It has been purported that CA-MRSA emerged in addition to MSSA and not in replacement of it [7, 9]. We speculate that, because MSSA has been endemic for so long in community and healthcare settings, transmission networks are not necessary for continued spread. In contrast, as the current study supports, various epidemiologic drivers likely enhance the spread of CA-MRSA strains, allowing for distinct transmission networks.
In our prior work using genomic data to extend our epidemiologic assessments of CA-MRSA, we had observed that African-American race served as the basis of genomic transmission networks among USA300 isolates colonizing human immunodeficiency virus (HIV)–infected and HIV-uninfected individuals, with no individuals of Hispanic ethnicity belonging to an identified network [15]. Prior work has also supported the notion of uneven geographic distribution of CA-MRSA in urban areas in the United States and has suggested that there is differential risk for transmission of CA-MRSA in certain neighborhoods [7, 9, 14]. We further explore these findings in the current study by including a population of largely HIV-uninfected individuals and by analyzing BSIs. We did not find that previously recognized risk factors for USA300 MRSA [8]—illicit drug use, incarceration exposure, and unstable housing—served as the basis of identified USA300 transmission networks. In addition, recent prior hospitalization was not associated with genomic transmission networks, making differential exposure to healthcare settings less likely to account for genomic clustering. African-American race, a community factor, was the only factor associated with transmission networks, suggesting that community factors may be an important driver of USA300 BSIs, even those that are hospital-onset. A study by Chang et al [29] observed genetically similar strains between highly connected healthcare facilities but that genetic similarity was not associated with patient transfer networks between institutions. Their finding supports our hypothesis that community transmission networks may be driving a portion of HO-MRSA BSIs. Given the extensive size of the community population, specific community factors need to be identified to improve our ability to control the spread of MRSA. Further assessment with a tool such as social network analysis may be needed to understand networks that are driving spread of USA300 in the community, whether previously reported high-risk exposures such as incarceration serve to propagate spread of strains in the community, and how community networks impact infections in acute care settings.
We also observed that epidemiologic classification of USA300 MRSA BSIs (CO vs HO) did not serve as the basis for genomic clustering, suggesting that there are not distinct community and hospital transmission networks for USA300 strains. As further support for the unique colonization and transmission dynamics of USA300, USA100 strains had significant enrichment on genomic analysis with a healthcare factor, active malignancy, a finding that likely reflects that recent or frequent healthcare exposures may still play a role in USA100 epidemiology [6]. For USA300 strains causing BSIs, epidemiologic definitions may be less valuable for predicting where strain acquisition occurs; terms now may merely describe site of infection presentation. The higher prevalence of patients already colonized with MRSA being admitted to CCH [10, 13] may explain why the rate of MRSA BSIs was higher there despite the favorable comparison of SIRs between the 2 institutions.
Our study has limitations. It was a retrospective study; however, data at both institutions were from electronic surveillance and were collected prospectively, making full data capture more likely; in addition, community and healthcare factor data were confirmed with medical record review. Second, there are likely community and healthcare factors that were not assessed for genomic clustering and warrant further analysis. Third, surveillance cultures were not collected in this study so we do not know if individuals developing HO infections were colonized at admission to the hospital. However, an objective of the study was to use genomic analysis as an epidemiologic tool to identify if there are community and hospital USA300 MRSA transmission networks; our results suggest potential sites of strain acquisition that need to be further evaluated. Fourth, because our study was limited to BSI isolates, our sample size is small; a larger sample size with MRSA isolates from other types of clinical infections would allow better resolution in identifying regional transmission networks and assigning epidemiologic and healthcare drivers. Finally, our study population includes a large proportion of individuals residing in urban areas, which may limit the generalizability of our findings; however, 80% of the US population resides in urban areas [30], and the inclusion of 2 acute care hospitals—one a public institution and the other a private university medical center—strengthens our results.
In summary, we demonstrate that there are sex, racial, and ethnic disparities for HO-MRSA BSIs across 2 medical centers. Using WGS on a sample of USA300 MRSA BSI isolates in conjunction with epidemiologic classification of infections, we found that there is an intermixing of USA300 transmission networks between the community and hospital. Only African-American race served as the basis for genomic clustering of USA300 MRSA BSIs, suggesting that community factors may be an important driver of HO-MRSA BSIs due to USA300 strains. Future infection prevention interventions for USA300 MRSA may need to extend to the community for maximal benefit.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank Yoona Rhee, MD, ScM, for her assistance with data collection. The authors thank the Collaborative Research Unit and Alex Patino (data query developer) at Stroger Hospital of Cook County for their assistance with data collection.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases (NIAID) or the National Institutes of Health.
Financial support. The project described was supported by the NIAID (grant number R01AI114688; principal investigator [PI]: K. J. P.) and the Centers for Disease Control and Prevention Epicenter Grant (cooperative agreement number 1U54CK000161; PI: R. A. W.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fridkin SK, Hageman JC, Morrison M, et al. ; Active Bacterial Core Surveillance Program of the Emerging Infections Program Network Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med 2005; 352:1436–44. [DOI] [PubMed] [Google Scholar]
- 2. Moran GJ, Krishnadasan A, Gorwitz RJ, et al. ; EMERGEncy ID Net Study Group Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 2006; 355:666–74. [DOI] [PubMed] [Google Scholar]
- 3. Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 1998; 279:593–8. [DOI] [PubMed] [Google Scholar]
- 4. Tenover FC, McDougal LK, Goering RV, et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 2006; 44:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis 2008; 46:787–94. [DOI] [PubMed] [Google Scholar]
- 6. Rhee Y, Aroutcheva A, Hota B, Weinstein RA, Popovich KJ. Evolving epidemiology of Staphylococcus aureus bacteremia. Infect Control Hosp Epidemiol 2015; 36:1417–22. [DOI] [PubMed] [Google Scholar]
- 7. Popovich KJ, Weinstein RA, Aroutcheva A, Rice T, Hota B. Community-associated methicillin-resistant Staphylococcus aureus and HIV: intersecting epidemics. Clin Infect Dis 2010; 50:979–87. [DOI] [PubMed] [Google Scholar]
- 8. Miller LG, Diep BA. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 2008; 46:752–60. [DOI] [PubMed] [Google Scholar]
- 9. Hota B, Ellenbogen C, Hayden MK, Aroutcheva A, Rice TW, Weinstein RA. Community-associated methicillin-resistant Staphylococcus aureus skin and soft tissue infections at a public hospital: do public housing and incarceration amplify transmission? Arch Intern Med 2007; 167:1026–33. [DOI] [PubMed] [Google Scholar]
- 10. Popovich KJ, Smith KY, Khawcharoenporn T, et al. Community-associated methicillin-resistant Staphylococcus aureus colonization in high-risk groups of HIV-infected patients. Clin Infect Dis 2012; 54:1296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okano JT, Blower S. Are correctional facilities amplifying the epidemic of community-acquired methicillin-resistant Staphylococcus aureus? Nat Rev Microbiol 2010; 8:83. [DOI] [PubMed] [Google Scholar]
- 12. Popovich KJ, Zawitz C, Weinstein RA, Grasso AE, Hotton AL, Hota B. The intersecting epidemics of human immunodeficiency virus, community-associated methicillin-resistant Staphylococcus aureus, and incarceration. Open Forum Infect Dis 2015; 2:ofv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Popovich KJ, Hota B, Aroutcheva A, et al. Community-associated methicillin-resistant Staphylococcus aureus colonization burden in HIV-infected patients. Clin Infect Dis 2013; 56:1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diep BA, Chambers HF, Graber CJ, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med 2008; 148:249–57. [DOI] [PubMed] [Google Scholar]
- 15. Popovich KJ, Snitkin E, Green SJ, et al. Genomic epidemiology of USA300 methicillin-resistant Staphylococcus aureus in an urban community. Clin Infect Dis 2016; 62:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwamoto M, Mu Y, Lynfield R, et al. Trends in invasive methicillin-resistant Staphylococcus aureus infections. Pediatrics 2013; 132:e817–24. [DOI] [PubMed] [Google Scholar]
- 17. Dantes R, Mu Y, Belflower R, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 2013; 173:1970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen DB, Lessa FC, Belflower R, et al. Invasive methicillin-resistant Staphylococcus aureus infections among patients on chronic dialysis in the United States, 2005–2011. Clin Infect Dis 2013; 57:1393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. David MZ, Taylor A, Lynfield R, et al. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J Clin Microbiol 2013; 51:814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy CR, Hudson LO, Spratt BG, et al. Predictors of hospitals with endemic community-associated methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 2013; 34:581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dukic VM, Lauderdale DS, Wilder J, Daum RS, David MZ. Epidemics of community-associated methicillin-resistant Staphylococcus aureus in the United States: a meta-analysis. PLoS One 2013; 8:e52722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Himsworth CG, Miller RR, Montoya V, et al. Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLoS One 2014; 9:e87983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. S. A. FastQC: a quality control tool for high throughput sequence data. http://wwwbioinformaticsbabrahamacuk/projects/fastqc/. Accessed January 2016. [Google Scholar]
- 24. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gardner SN, Hall BG. When whole-genome alignments just won’t work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 2013; 8:e81760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Centers for Disease Control and Prevention First state-specific healthcare-associated infections summary data report. http://www.cdc.gov/hai/pdfs/stateplans/SIR_05_25_2010.pdf Accessed 21 September 2016.
- 28. Uhlemann AC, Dordel J, Knox JR, et al. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 2014; 111:6738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang HH, Dordel J, Donker T, et al. Identifying the effect of patient sharing on between-hospital genetic differentiation of methicillin-resistant Staphylococcus aureus. Genome Med 2016; 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. United States Census Bureau Growth in urban population outpaces rest of nation, census bureau reports. https://www.census.gov/newsroom/releases/archives/2010_census/cb12-50.html Accessed 15 August 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.