Abstract
Background.
Kaposi’s sarcoma–associated herpesvirus (KSHV) is the causative agent of Kaposi sarcoma (KS), one of the leading cancers in human immunodeficiency virus (HIV)–infected patients in Zambia. KSHV was detected in the human central nervous system (CNS) by polymerase chain reaction (PCR) analysis, but tissue location and cell tropism for KSHV infection has not been established. Given the neurotropism exhibited by other herpesviruses and the frequent coinfection of HIV-positive individuals by KSHV, we sought to determine whether the central nervous system (CNS) can be infected by KSHV in HIV-positive Zambian individuals.
Methods.
Postmortem brain tissue specimens were collected from individuals coinfected with KSHV and HIV. PCR and Southern blots were performed on DNA extracted from the brain tissue specimens to verify KSHV infection. Immunohistochemical analysis and immunofluorescent microscopy were used to localize and identify KSHV-infected cells. Tropism was further established by in vitro infection of primary human neurons with rKSHV.219.
Results.
KSHV DNA was detected in the CNS from 4 of 11 HIV-positive individuals. Immunohistochemical analysis and immunofluorescent microscopy demonstrated that KSHV infected neurons and oligodendrocytes in parenchymal brain tissues. KSHV infection of neurons was confirmed by in vitro infection of primary human neurons with rKSHV.219.
Conclusion.
Our study showed that KSHV infects human CNS-resident cells, primarily neurons, in HIV-positive Zambian individuals.
Keywords: Kaposi’s sarcoma-associated herpesvirus, HIV, central nervous system, neurons.
Kaposi sarcoma (KS) remains one of the most prevalent malignancies associated with human immunodeficiency virus (HIV)–infected individuals in sub-Saharan African countries, such as Zambia. KS is the leading cancer in both incidence and mortality among Zambian men and the third leading cancer among Zambian women, behind cervical and breast cancers [1].
Immune suppression, resulting from age, anti–transplant rejection therapy, and T-cell depletion, is associated with classical, iatrogenic, and AIDS-associated forms of KS, respectively [2–5]. The etiological agent of KS is now known to be KS-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8). KSHV is the most recently discovered human herpesvirus and is a member of the gamma-herpesvirus or lymphocryptovirus subgroup, along with Epstein-Barr virus (EBV). KSHV was first identified in AIDS-associated KS lesions, by representational difference analysis [6]. It was also found to be associated with 2 other lymphoproliferative disorders, primary effusion lymphoma and multicentric Castleman disease, an inflammatory disease associated with HIV-infected individual [7, 8]. Like other herpesviruses, KSHV exhibits 2 distinct phases of infection: lytic replication and latency. During latency, the gene expression pattern is thought to promote immune evasion, prevent programmed cell death, and enable the virus to establish a persistent infection [9–13]. Lytic reactivation leads to virus production, which enables the spread of viruses from infected tissue compartments into the periphery. This, in turn, leads to viremia and to increased potentials for HSHV transmission and KS development [14, 15].
The association of KS development with various forms of immune suppression suggests that immune surveillance is an essential component in limiting KSHV-induced pathogenesis. The precise nature of the immune response necessary to prevent KS remains unclear, but it is evident that the virus can establish persistent infection in different cell types and encodes a number of immunomodulatory proteins that support KSHV persistence, replication, and transmission [11, 16–22]. One incompletely defined component of KSHV infection is an understanding of tissue locations where the virus has the potential to persist. The central nervous system (CNS) is one such location and where a number of other human herpesviruses have been shown to persist [23].
The known cellular tropism for KSHV includes endothelial, epithelial, and B cells and, perhaps, macrophages [7, 17, 20, 23–27], but since most herpesviruses demonstrate some degree of neurotropism, we sought to determine whether KSHV can establish infection in parenchymal cells of the CNS. Previous reports suggested the detection of viral nucleic acids in CNS samples, but these findings could be confounded by virus in immune cells circulating through brain vasculature [28–33]. We now report that KSHV can be detected in various brain tissues from individuals for whom KSHV infection was confirmed on the basis of KSHV DNA detection by PCR or KSHV seropositivity was confirmed on the basis of immunofluorescence assay (IFA) of serum or cerebrospinal fluid (CSF) samples. These individuals were also HIV coinfected. KSHV CNS infection was found to be latent, as indicated by expression of latency-associated nuclear antigen (LANA), and the majority of infected cells were identified as neurons. This result was recapitulated in infection of mixed primary human neuron cultures in vitro. Our results indicate that KSHV can gain entry into the CNS and infect neurons in HIV-positive patients. To our knowledge, this is the first study to demonstrate in vivo infection of human neurons by KSHV.
METHODS
Patient Samples
Autopsy samples were collected at the University Teaching Hospital in Lusaka, Zambia, from HIV-positive patients and HIV-negative patients. Consent was obtained from families of the deceased patients prior to specimen collection. Sample sets included frozen and formalin-fixed paraffin-embedded tissue from the frontal lobe, parietal lobe, temporal lobe, occipital lobe, hippocampus, cerebellum, and basal ganglia. CSF and plasma specimens were collected where possible.
HIV Serological Testing
Blood was screened in Zambia for HIV-specific antibodies, using HIV rapid serologic tests (Abbott-Determine and Unigold) according to the manufacturer’s instructions.
DNA Extraction From Postmortem Tissue Specimens
Fresh-frozen tissue specimens (weight, 100 mg) were pulverized by cryo-cracking with liquid nitrogen. Cells were lysed using QIAzol lysis reagent (Qiagen, Hilden, Germany) according to manufacturer’s protocol, with the following modifications. The upper aqueous phase was discarded after chloroform extraction. To precipitate the DNA, 100% ethanol was added to the lower organic phase. The DNA pellet was washed twice with 0.1 M sodium citrate, followed by another wash with 75% ethanol, and was air dried. The final extracted DNA was dissolved in Tris–ethylenediaminetetraacetic acid (TE) buffer and incubated at 65°C for 1 hour, followed by incubation overnight at room temperature.
PCR for Detection of β-actin, KSHV Open Reading Frame 26 (ORF26), and LANA
To confirm the quality of extracted DNA, genomic DNA from all tissues was screened by β-actin PCR (primers 5′-TTCTACAATGAGCTGCGTGT-3′ [forward] and 5′-GCCAGACAGCACTGTGTTGG-3′ [reverse]). The presence of the KSHV genome in brain tissues was detected by PCR for the KSHV ORF26 gene (primers 5′-AGCCGAAAGATTCCACCAT-3′ [KS-1] and 5′-TCCGTGTTGTCTACGTCCAG-3′ [KS-2]) and nested-PCR for the LANA gene (primers 5′-AAGGAATGGGAGCCACC-3′ [NLana_1F], 5′-GCGCCCTTAACGAGAG-3′ [NLana_1R], 5′-AGGACTAGACACAAATGCTGG-3′ [NLana_2F], and 5′-AAACGAAACAGGTCTCCGG-3′ [NLana_2R]). Thermal cycling parameters for the amplicons, using Takara Ex Taq DNA polymerase, were as follows: 95°C for 5 minutes; 35 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds; 72°C for 5 minutes; and final hold at 4°C.
Southern Blot for Detection of KSHV ORF26 and LANA
To confirm PCR detection of KSHV ORF26 and LANA in brain tissue specimens, Southern blots with digoxigenin (DIG)–labeled ORF26 and LANA DNA probes were performed. The probes were prepared by amplifying BC-3 genomic DNA with primers 5′- CGAATCCAACGGATTTGACCTC-3′ (KS-4) and 5′-CCCATAAATGACACATTGGTGGTA-3′ (KS-5) for ORF26 and 5′-AGGACTAGACACAAATGCTGG-3′ (NLana_2F) and 5′-AAACGAAACAGGTCTCCGG-3′ (NLana_2R) for LANA, using the PCR DIG Probe Synthesis Kit (Roche, Basel, Switzerland ) according to the manufacturer’s protocol. Ten microliters of first-round ORF26 PCR and 10 μL of second-round LANA PCR amplification products were analyzed by standard Southern blot with overnight hybridization at 42°C. Two stages of stringent washes were performed after hybridization to remove unbound probe and incomplete or off-target hybridizations. Hybridized probes were detected with alkaline phosphatase–conjugated anti-DIG antibody and BCIP/NBT chromagen deposition reagent (DIG DNA Labeling and Detection Kit; Roche).
IFA
An IFA using KSHV-infected BC3 cells was used to detect KSHV seropositivity in all sampled individuals as previously described [37]. Briefly, 1 × 104 BC3 cells per well were spotted onto 12-well slides. Plasma or CSF samples obtained from autopsied subjects or HIV/KSHV-negative controls were diluted at 1:40, added to the affixed BC3 cells, and incubated at 37°C for 30 minutes. Slides were washed and incubated with murine α-human immunoglobulin G (IgG) antibody at 37°C for 30 minutes. Slides were washed and incubated with donkey α-mouse IgG antibody conjugated to DyLight 488 at 37°C for 30 minutes. Slides were washed and counterstained with 0.004% Evans blue for 5 minutes, and each slide was read independently by 2 laboratory workers. A sample was considered KSHV seropositive if concordant results were reported from both readers. Owing to postmortem blood clotting, plasma samples were not collectable for all subjects (Table 1).
Table 1.
Demographic and Clinical Information and Human Immunodeficiency Virus (HIV)/Kaposi’s Sarcoma–Associated Herpesvirus (KSHV) Status Among Study Patients
| Subject | Age, y | Sex | Cause of Deatha | Received ART | Serostatusb | KSHV-Specific PCR/Southern Blot Result, by Brain Regionb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV | KSHV | FL | PL | TL | OL | Hip | Cer | BG | |||||
| 92 | 32 | Male | Tuberculosis | No | + | + | − | − | − | + | + | + | − |
| 111 | 19 | Male | RVD and anemia | Yes | + | + | − | − | − | − | − | − | − |
| 116 | 44 | Male | Meningitis, R/0 tuberculosis | No | + | + | + | + | + | + | + | + | + |
| 221 | 26 | Female | CNS OI, malaria, metabolic derangements | Yes | + | + | − | − | − | − | − | − | − |
| 228 | 34 | Female | RVD, tuberculosis-associated meningitis | No | + | + | − | − | − | − | − | − | − |
| 230 | 30 | Male | Hemiplegia, RVD, possible toxoplasmosis | No | + | + | − | − | + | − | − | − | − |
| 283 | 34 | Female | RVD, tuberculosis | Yes | + | + | − | − | − | − | − | − | − |
| 315 | 24 | Male | RVD, meningitis | No | + | + | − | − | − | − | − | − | − |
| 320 | 40 | Female | Disseminated tuberculosis | Unknown | + | + | − | − | − | − | − | − | − |
| 321 | 35 | Female | Peripheral neuropathy and disseminated tuberculosis | Unknown | + | + | − | − | − | − | + | − | − |
| 231 | 34 | Male | RVD, toxic epidermal necrolysis | Yes | + | − | − | − | − | − | − | − | − |
| 001 | 47 | Male | Homicide/murder | No | − | − | − | − | − | − | − | − | − |
Abbreviations: ART, antiretroviral therapy; CNS, central nervous system; BG, basal ganglia; Cer, cerebellum; FL, frontal lobe; Hip, hippocampus; OI, opportunistic infection; OL, occipital lobe; PCR, polymerase chain reaction; PL, parietal lobe; RVD, retroviral disease; TL, temporal lobe.
aBased on report from University Teaching Hospital (Lusaka).
bA plus sign indicates seropositivity for HIV/KSHV antibody or a positive result of KSHV-specific PCR and Southern blot in various brain tissue specimens, and a minus sign indicates seronegativity for HIV/KSHV antibody or a negative result of KSHV-specific PCR and Southern blot in various brain tissue specimens.
KSHV LANA and K8.1 Immunohistochemical (IHC) Analysis
Formalin-fixed, paraffin-embedded brain tissue specimens from HIV-positive Zambian individuals were divided into 5-μm sections. Slides were deparaffinized and rehydrated by means of standard xylene and ethanol washes. Endogenous peroxidase activity was blocked using a hydrogen peroxide–methanol solution, and antigen retrieval was performed in sodium citrate solution at 98°C for 15 minutes. Slides were blocked with 10% normal goat serum in 1X PBS and incubated overnight at 4°C with murine anti-LANA monoclonal antibody (kindly provided by Dr Bala Chandran, Rosalind Franklin University of Medicine and Science, Chicago, IL) at a dilution of 1:750 for KSHV LANA protein detection or with murine anti-K8.1 monoclonal antibody (Advanced Biotechnologies, Eldersburg, MD) at the manufacturer’s recommended dilution for KSHV K8.1 protein detection. Slides were washed and incubated with horseradish peroxidase (HRP)–labeled polymeric anti-mouse secondary antibody (Dako, Santa Clara, CA). Diaminobenzidine (Dako) and hematoxylin were used for chromogen deposition and counterstaining, respectively.
Cell Lineage and KSHV Immunofluorescence-Based Detection
Dual-staining immunofluorescence (IF)–based detection was performed using a primary antibody mixture of mouse anti-KSHV LANA (as described above in the section titled “IFA”) and various anti-human cell lineage–specific antisera. The lineage-specific antisera included polyclonal rabbit antisera against feminizing locus on X-3, Fox-3/ hexaribonucleotide binding protein-3 (NeuN [Abcam, Cambridge, MA]; diluted 1:100), glial fibrillary acidic protein (GFAP [Dako]; diluted 1:500), oligodendrocyte transcription factor (Olig2 [Abcam]; diluted 1:100), ionized calcium binding adaptor molecule 1 (Iba-1 [Wako, Richmond, VA]; diluted 1:500), CD20 (Dako; diluted 1:200), von Willebrand factor (VWF [Dako]; diluted 1:2000), and cluster of differentiation 45 (CD45) (Dako, 1:500). These reagent identified neurons, astrocytes, oligodendrocytes, microglia/macrophages, B cells, endothelial cells, and lymphocytes, respectively. Slides were incubated overnight with primary antibodies at 4°C in Tris-NaCl blocking buffer (TNB). After 3 washes with TNB, the slides were stained with donkey anti-rabbit AlexaFluor 488 and anti-mouse AlexaFluor 647 secondary antibodies (Invitrogen, Waltham, MA; both diluted 1:500) and with DAPI (4′,6-diamidino-2-phenylindole 1 µg/mL; diluted 1:750) for 2 hours at room temperature in TNB with 5% milk. Slides were then counterstained for 10 minutes with Sudan Black B (Sigma-Aldrich, St. Louis, MO) in 70% ethanol to reduce autofluorescence due to lipofuscin and mounted with Fluoro-gel (Electron Microscopy Sciences, Hatfield, PA).
Recombinant KSHV Production and Primary Human Neuron Infection
Recombinant KSHV (rKSHV.219) that expresses green fluorescent protein (GFP) under the cellular EF-1α promoter and red fluorescent protein (RFP) under the viral lytic PAN promoter was produced as previously described [34]. Briefly, Vero.219 cells, which encode the recombinant KSHV, were infected with a KSHV RTA–expressing baculovirus for 4 hours, followed by overnight stimulation with 1.25 mM sodium butyrate. Cells were then washed and incubated with fresh growth medium for 72 hours before the supernatant was filtered (pore size, 0.45 µm) and collected. The rKSHV.219 virus titer was then determined using 293-T cells according to the standard Reed Muench method [35].
Neurons were obtained from the Comprehensive NeuroAIDS Center at Temple University (Philadelphia, PA). The single-cell suspension was plated at a density of 1.8 × 106 cells/60-mm poly-D lysine–coated dish and cultured in neurobasal medium with B27 supplement, horse serum, and gentamicin (NM5). After 2 hours, medium was changed, and unattached cells were discarded. Twenty-four hours later, cultures were refed with a complete change of NM5 lacking horse serum (NM0). Four days later, 25% of the medium was removed and replaced with NM0 supplemented with fluoro-deoxyuridine and uridine (dilution 1:4000) for 4 days. Cells were then cultured in NM0, with 25% of the medium changed every third day, and maintained for 2–3 weeks until infection with virus. Purity of neuronal cultures was assessed by immunolabeling with anti-MAP2 and neurofilament for detection of neurons. Neural cultures were infected with rKSHV.219 at multiplicity of infection of 0.5 for 72 hours prior to evaluation by microscopy or processing for IF-based detection of antigens.
RESULTS
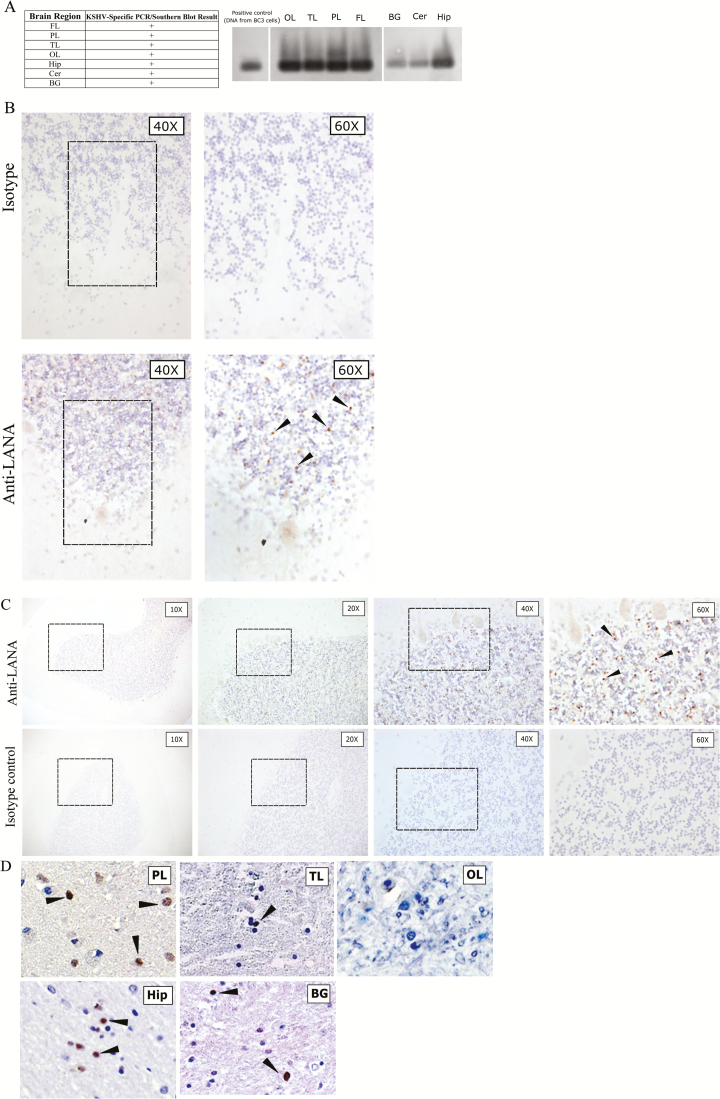
Given the adult KSHV seroprevalence in Zambia of 48% [36–38], with an even higher prevalence among HIV-infected individuals, and the demonstrated neurotropism of many other members of the Herpesviridae family, we worked closely with colleagues at the University Teaching Hospital to initiate collection of postmortem brain tissue specimens to determine whether KSHV could be directly detected in the brain parenchyma. Formalin-fixed, paraffin-embedded, and fresh-frozen tissue specimens were collected and analyzed. We first examined the brain tissue specimens from an HIV-positive individual who did not have cutaneous or visceral KS (subject 116; Table 1). To investigate whether KSHV infects the CNS and whether it exhibits regional or cellular tropism, DNA was extracted from various frozen brain tissue specimens, including the frontal, temporal, parietal, and occipital lobes; the basal ganglion; the hippocampus; and the cerebellum. The extracted DNA was amplified by primers specific for KSHV capsid protein, encoded by ORF26. We found that tissue specimens from different brain regions of subject 116 were positive for KSHV by PCR, and the results were confirmed by Southern blot analysis using a KSHV-specific ORF26 probe (Figure 1A). The presence of KSHV DNA was further confirmed for cerebellum and hippocampus tissue specimens from subject 116, using PCR specific for KSHV LANA, followed by Southern blot confirmation with a KSHV LANA–specific probe.
Figure 1.
A, Southern blot hybridization–confirmed polymerase chain reaction (PCR) results for a human immunodeficiency virus (HIV)–positive, Kaposi’s sarcoma (KS)–associated herpesvirus (KSHV)–seropositive, but KS-asymptomatic individual (subject 116), showing that most central nervous system (CNS) tissues harbored KSHV genomic DNA. Southern blot with probe against KSHV ORF26 is shown. Demographic and clinical characteristics of subject 116 are available in Table 1. B, Findings of immunohistochemical (IHC) analysis of CNS tissue specimens from subject 116, using near-adjacent cerebellum (Cer) sections for isotype and anti–latency-associated nuclear antigen (LANA) staining. The different magnifications are of the same cerebellar folium. LANA-positive cells appeared as brown punctate staining, and representative cells are designated by arrowheads. C, Progressive magnifications of anti-LANA IHC staining of Cer sections from subject 116 (top) in comparison to isotype control IHC staining of a near-adjacent section (bottom). The architecture of the folia is evident at lower magnification, and the Purkinje cells bordering the granular layer are resolved at higher magnifications. Boxes denote the field magnified in the subsequent panel. LANA-positive cells appeared as brown punctate staining, and representative cells are designated by arrowheads. D, IHC staining of addition brain regions from subject 116 reveals KSHV LANA in the parietal lobe (PL), temporal lobe (TL), hippocampus (Hip), and basal ganglia (BG) but not in the occipital lobe (OL). LANA-positive cells appeared as brown punctate staining, and representative cells are designated by arrowheads. Isotype control staining of the same tissue specimens was negative (not shown). Control staining with anti-LANA in a KSHV-seronegative subject is shown in Supplementary Figure 1. ART, antiretroviral therapy; +, positive.
Since the brain is a highly vascular organ and because KSHV is known to have tropism for B cells that comprise approximately 5%–10% of blood leukocytes, it remained possible that detection of KSHV resulted from amplification of viral sequences in vascular cells. Alternately, KSHV could be present in the brain parenchyma, either in extravasating immune cells or in CNS-resident cell types. To discriminate between these possibilities, we tested for the presence of LANA antigen in the PCR-positive brain tissue specimens, since it is the primary KSHV protein expressed during latency [9, 12, 13]. LANA localization is typically nuclear, and its staining pattern is typically punctate [39, 40], although recent reports suggest some isoforms with alternative subcellular localization [41]. Robust expression of KSHV LANA was detected in subject 116. An example of the KSHV detection by IHC analysis in the cerebellar tissue is shown in Figure 1B, together with specificity controls. Surprisingly, we detected very little KSHV antigen localized to brain vasculature (Figure 1C). Rather, labeling was evident, although sparse, in various regions of the parenchyma and highest in the gray matter, where the larger proportion of brain cell soma are localized. In the cerebral cortex, KSHV latent antigen was localized to cells in the deeper gray matter, near the white matter border. A representative panel of KSHV LANA staining by IHC analysis in the sampled brain tissue specimens for subject 116 is shown in Figure 1D. Despite staining of adjacent or near-adjacent sections, expression of the KSHV lytic antigen K8.1 was not detected in any brain tissue (data not shown).
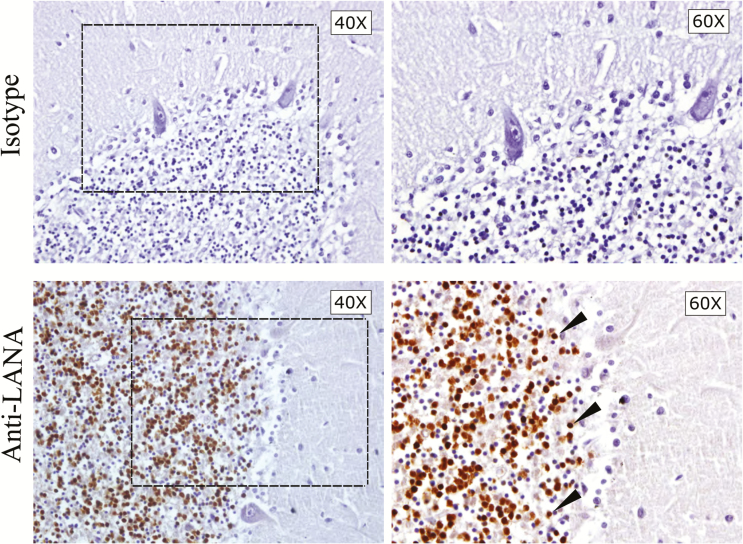
To confirm that KSHV can be found in brain tissues of other infected individuals, we further analyzed brain tissue specimens from 10 additional HIV/KSHV-coinfected individuals, a KSHV-seronegative but HIV-positive individual (subject 231), and a KSHV-seronegative, HIV-negative individual (subject 001; Table 1). Interestingly, unlike subject 116, no KSHV was detected in the CNS from a number of KSHV-seropositive subjects, whereas for others, such as subject 92, only specimens from certain tissues (ie, the occipital lobe, hippocampus, and cerebellum) were found to be PCR positive. The PCR-positive results were confirmed by IHC analysis, and an example of the IHC findings is shown in Figure 2. Similar to subject 116, there was high expression of LANA in the granular layer of the cerebellar cortex of subject 92.
Figure 2.
Immunohistochemical staining for latency-associated nuclear antigen (LANA) on cerebellum (Cer) sections from subject 92. Sections within 30 µm of one another were stained with isotype control sera or anti-LANA. The different magnifications are of the same cerebellar folium in near-adjacent sections. The box indicates the selected area of the granular layer magnified in the right panel. LANA-positive cells appeared as brown punctate staining, and representative cells are designated by arrowheads.
Based on our analysis of these subjects, we found that 4 of 10 KSHV-seropositive individuals harbor KSHV DNA in the CNS. We also found that distinct brain compartments can be infected by KSHV but that the sites of infection vary among individuals. Immunolabeling of the HIV-negative, KSHV-seronegative control (subject 001; Supplementary Figure 1) and the HIV-positive, KSHV-seronegative control (data not shown) did not reveal any evidence of anti-LANA cross-reactivity in CNS tissue specimens, demonstrating the specificity of detection.
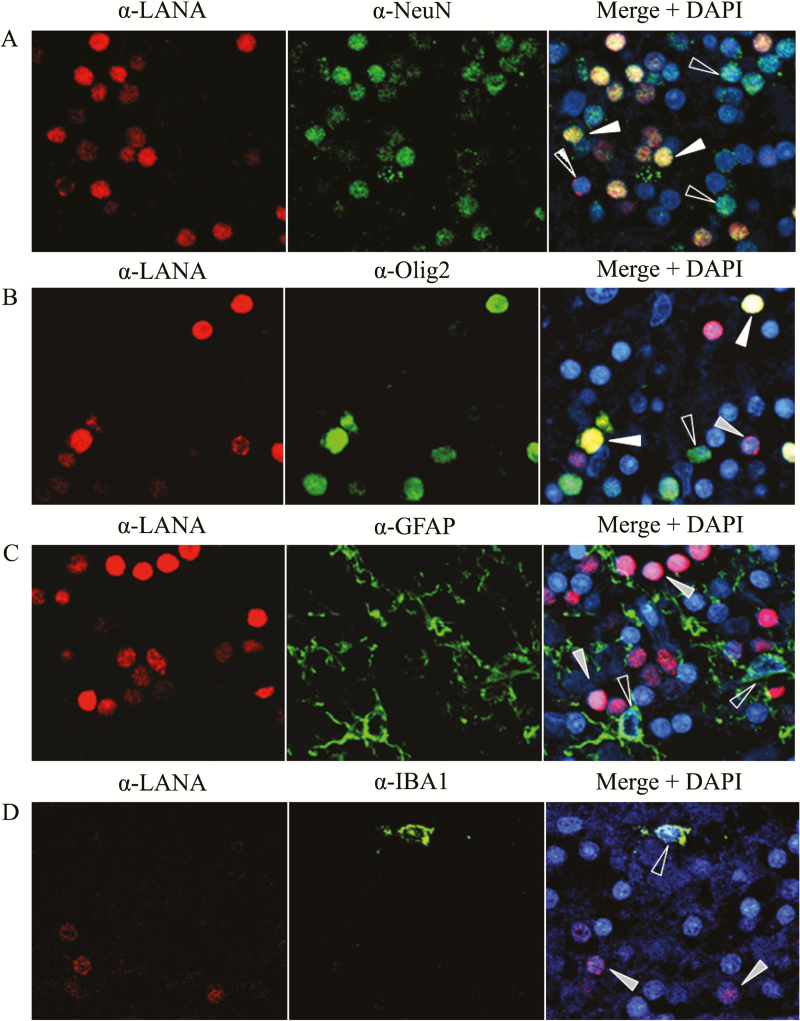
To identify the cell types harboring KSHV in the brain, we conducted dual-staining IF using CNS cell lineage-specific antibodies in combination with anti-LANA to identify the KSHV-infected cells. Antibodies against NeuN, GFAP, Olig2, Iba-1, CD20, VWF, and CD45 were used to identify neurons, astrocytes, oligodendrocytes, microglia/macrophages, B cells, endothelial cells, and leukocytes, respectively. Representative IF data from the cerebellum of subject 92 demonstrates that KSHV LANA signal colocalized with neuronal and, to a lesser extent, oligodendrocyte markers (Figure 3A and 3B) but not with specific markers for astrocyte or microglia/macrophage lineages (Figure 3C and 3D). Our findings demonstrate that KSHV can infect neurons and potentially neuronal support cells in the brain parenchyma from KSHV-seropositive human brain tissues. No colocalization of KSHV LANA signal with B cells (CD20), endothelial cells (VWF), or lymphocytes (CD45) was detected in brain parenchymal tissue specimens (Supplementary Figure 2).
Figure 3.
Identification of Kaposi’s sarcoma–associated herpesvirus (KSHV)–infected cell types by immunofluorescence. Formalin-fixed, paraffin-embedded blocks of cerebellum tissue from subject 92 were divided into 5.0-µm sections and subjected to dual-label immunofluorescence, using mouse monoclonal anti-LANA (α-LANA) to identify cells expressing latent KSHV antigen, and cell lineage-specific antibodies directed against antigens for neurons (α-NeuN), oligodendrocytes (α-Olig2), astrocytes (α-GFAP), or activated macrophage/microglia (α-IBA1). White arrows denote infected cells costained with the lineage marker, gray arrows indicate infected cells of alternative lineage, and open arrows indicate lineage-identified uninfected cells.
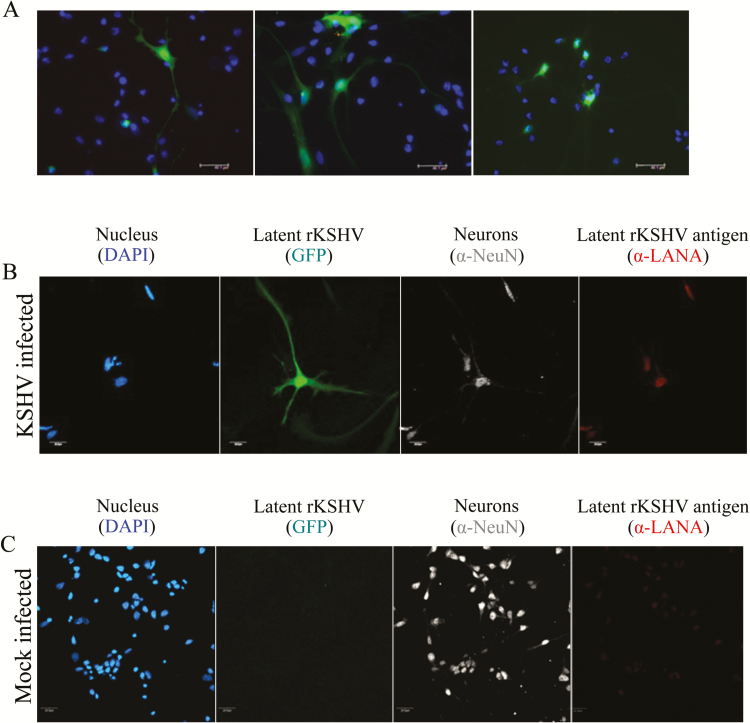
To validate the apparent tropism of KSHV for neurons in human postmortem tissue specimens, primary human neurons were infected in vitro with 0.5 MOI of a recombinant KSHV.219 that expresses GFP during latent infection and RFP during lytic infection. At 72 hours after infection, primary neurons were fixed and stained with DAPI. Confocal microscopy was used to identify cells exhibiting in situ GFP expression as a surrogate marker for KSHV latent infection (Figure 4A). The expression of GFP in cells with obvious axonal and dendritic morphology demonstrates that KSHV was capable of latent infection of neurons in vitro. Since primary neuronal cell culture contains an enriched mixture of neurons and glial support cells, dual-color IF microscopy using α-LANA and a neuronal cell marker α-NeuN, as described above, was implemented to confirm that GFP-expressing cells were indeed neurons (Figure 4B). Despite clear identification of NeuN-positive cells in the mock-infected culture, no GFP or LANA staining was evident or colocalized with NeuN expression in those cultures (Figure 4C). Consistent with the finding from human postmortem tissue specimens, KSHV lytic expression was not detected in the infected primary neuron cultures (data not shown).
Figure 4.
Kaposi’s sarcoma–associated herpesvirus (KSHV) infection of primary human neural cultures. Primary human neuron cultures were established from triturated mixed cell suspensions from neural tissue specimens. Infection with rKSHV.219, which expresses green fluorescent protein (GFP) from the EF1-a promoter, was conducted at an multiplicity of infection of 0.5. Cultures were micrographed at 488 nM excitation 72 hours after infection. A, Three representative panels show cells with apparent neuron morphology that are expressing GFP, indicative of KSHV infection. B, Similar cultures were fixed in 4% buffered paraformaldehyde and subjected to immunofluorescence-based detection of the indicated antigens or of recombinant KSHV–derived GFP in situ. C, An identically treated mock-infected control culture from the tissue donor.
DISCUSSION
Currently, only a few PCR-based studies have suggested that the brain is susceptible to KSHV infection [28, 29, 31]. However, the detection of viral DNA by PCR alone is insufficient to demonstrate that the brain is susceptible because of the extensive vascularization of the tissue. One recent report, focused primarily on the potential neural tropism of KSHV’s nearest gamma-herpesvirus relative, Epstein-Barr virus, also showed limited in vitro evidence that KSHV can infect the SH-SY5Y neuronal cell line [42]. Using PCR-based methods, they demonstrated expression of both LANA and the lytic protein K8. We detected infection in the same cell line, using the recombinant KSHV-GFP construct, but we were unable to unambiguously detect K8.1 protein expression by using immunofluorescence (data not shown). The results of these in vitro findings were largely consistent with our demonstration of latent infection in human primary neuron cultures, as shown by GFP expression from rKSHV.219 and colocalized LANA expression, but lytic reactivation was not observed.
Our data from human samples reveal that both KSHV DNA and LANA antigen can be detected in neurons and oligodendrocyte support cells in various brain regions. However, the brain tissue specimens we analyzed did not reveal any evidence of lytic reactivation. This is perhaps not surprising since our postmortem sampling did not include any individuals with symptomatic KS. In addition, our data showed that not all KSHV/HIV-coinfected patients harbor KSHV in the brain, suggesting that KSHV CNS localization could be related to their HIV disease. A broader panel of cases with defined HIV staging and with or without KS needs to be analyzed to confirm these findings. However, such postmortem CNS tissue specimens are not currently available.
This study also raises additional questions that warrant further investigation. How frequently is the brain infected in the absence of HIV? Does KSHV demonstrate any preference for specific regions of the CNS or for specific types of neurons, as has been demonstrated for other herpesviruses? Finally, are there pathogenic or neurocognitive consequences of KSHV infection in the CNS?
The presence of KSHV in the brain suggests that the virus may potentially spread from the initial site of infection into the CNS. However, the mechanism for this dissemination remains unclear. It has been demonstrated that a number of viruses, including herpesviruses, undergo direct retrograde transport of virions through neural circuits from peripheral tissue specimens to the CNS [43]. Whether KSHV can access this type of transport route needs to be determined. In addition it is possible that the seeding of KSHV into the CNS could be mediated by peripheral blood cells. Although we did not detect KSHV infected extravasating cells in the tissue specimens analyzed, a lack of detection could be related to the timing of viral deposition or to the likelihood of detecting a rare event among a limited tissue sampling.
Given the association of HIV and KSHV infection, how HIV coinfection and immunosuppression may facilitate KSHV infection and the establishment of CNS infection needs to be analyzed in comparison to HIV-negative cases. It is plausible that HIV coinfection induces the expression of soluble mediators that alter the permeability of the blood brain barrier to enable KSHV or KSHV-infected leukocytes to achieve CNS infection. In the African setting, the role of HIV could be direct or indirect because HIV-positive individuals are often coinfected with other opportunistic infectious agents, including parasites. Indeed, it has been demonstrated that helminth infection associates with KSHV infection in Africa [44]. Whether the presence of these comorbidities influences KSHV neuroinvasion will need to be determined.
In summary, our study is the first to demonstrate that brain tissues of Zambian HIV-infected patients are capable of harboring KSHV. KSHV infection is evident in CNS-resident cells, including neurons and oligodendrocytes. There appear to be differences in KSHV distribution in the brains among asymptomatic individuals, but more postmortem samples will be needed to determine whether particular tissues are selectively targeted. Likewise, brain tissue specimens need to be collected postmortem from HIV-positive and HIV-negative individuals with symptomatic KS to determine whether KSHV infection of the brain is different in more-advanced disease states and its dependence on HIV. Our investigation revealed little or no KSHV lytic antigen expression, but a larger number of samples or alternative organ/tissue culture methods may be required to determine whether CNS-resident cells can support lytic reactivation.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank all tissue donor families for their participation in this study.
Financial support. This work was supported by the National Institutes of Health (grants NS074903, CA65903, and P30 GM103509 and Fogarty grant D43 TW01492 to C. W.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. GLOBOCAN 2012: estimated cancer incidence, mortality and prevalence worldwide in 2012. http://globocan.iarc.fr. Accessed 11 July 2016. [Google Scholar]
- 2. Alibek K, Baiken Y, Kakpenova A, et al. Implication of human herpesviruses in oncogenesis through immune evasion and suppression. Infect Agent Cancer 2014; 9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jakob L, Metzler G, Chen KM, Garbe C. Non-AIDS associated Kaposi’s sarcoma: clinical features and treatment outcome. PLoS One 2011; 6:e18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ariza-Heredia EJ, Razonable RR. Human herpes virus 8 in solid organ transplantation. Transplantation 2011; 92:837–44. [DOI] [PubMed] [Google Scholar]
- 5. Knowles DM, Cesarman E. The Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) in Kaposi’s sarcoma, malignant lymphoma, and other diseases. Ann Oncol 1997; 8(Suppl 2):123–9. [PubMed] [Google Scholar]
- 6. Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994; 266:1865–9. [DOI] [PubMed] [Google Scholar]
- 7. Staskus KA, Sun R, Miller G, et al. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. J Virol 1999; 73:4181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dittmer DP, Damania B. Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)--an update. Curr Opin Virol 2013; 3:238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. KSHV LANA--the master regulator of KSHV latency. Viruses 2014; 6:4961–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baresova P, Pitha PM, Lubyova B. Distinct roles of Kaposi’s sarcoma-associated herpesvirus-encoded viral interferon regulatory factors in inflammatory response and cancer. J Virol 2013; 87:9398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sathish N, Yuan Y. Evasion and subversion of interferon-mediated antiviral immunity by Kaposi’s sarcoma-associated herpesvirus: an overview. J Virol 2011; 85:10934–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ballestas ME, Kaye KM. The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Future Microbiol 2011; 6:1399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Komatsu T, Barbera AJ, Ballestas ME, Kaye KM. The Kaposi’ s sarcoma-associated herpesvirus latency-associated nuclear antigen. Viral Immunol 2001; 14:311–7. [DOI] [PubMed] [Google Scholar]
- 14. Purushothaman P, Uppal T, Verma SC. Molecular biology of KSHV lytic reactivation. Viruses 2015; 7:116–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lukac DM, Yuan Y. Reactivation and lytic replication of KSHV. In: Arvin A, Campadelli-Fiume G, Mocarski Eet al. eds. Human herpesviruses: biology, therapy, and immunoprophylaxis Ch 26. Cambridge, United Kingdom: Cambridge University Press, 2007. [Google Scholar]
- 16. Hu Z, Usherwood EJ. Immune escape of gamma-herpesviruses from adaptive immunity. Rev Med Virol 2014; 24:365–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dollery SJ, Santiago-Crespo RJ, Kardava L, Moir S, Berger EA. Efficient infection of a human B cell line with cell-free Kaposi’s sarcoma-associated herpesvirus. J Virol 2014; 88:1748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee HR, Brulois K, Wong L, Jung JU. Modulation of immune system by Kaposi’s sarcoma-associated herpesvirus: lessons from viral evasion strategies. Front Microbiol 2012; 3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riva G, Barozzi P, Torelli G, Luppi M. Immunological and inflammatory features of Kaposi’s sarcoma and other Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8-associated neoplasias. AIDS Rev 2010; 12:40–51. [PubMed] [Google Scholar]
- 20. Coleman CB, Nealy MS, Tibbetts SA. Immature and transitional B cells are latency reservoirs for a gammaherpesvirus. J Virol 2010; 84:13045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Means RE, Lang SM, Jung JU. Human gammaherpesvirus immune evasion strategies. In: Arvin A, Campadelli-Fiume G, Mocarski Eet al. eds. Human herpesviruses: biology, therapy, and immunoprophylaxis. Ch 31. Cambridge, United Kingdom: Cambridge University Press, 2007. [PubMed] [Google Scholar]
- 22. Touloumi G, Hatzakis A, Potouridou I, et al. The role of immunosuppression and immune-activation in classic Kaposi’s sarcoma. Int J Cancer 1999; 82:817–21. [DOI] [PubMed] [Google Scholar]
- 23. Chen T, Hudnall SD. Anatomical mapping of human herpesvirus reservoirs of infection. Mod Pathol 2006; 19:726–37. [DOI] [PubMed] [Google Scholar]
- 24. Veettil MV, Bandyopadhyay C, Dutta D, Chandran B. Interaction of KSHV with host cell surface receptors and cell entry. Viruses 2014; 6:4024–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parsons CH, Adang LA, Overdevest J, et al. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J Clin Invest 2006; 116:1963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dittmer D, Stoddart C, Renne R, et al. Experimental transmission of Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) to SCID-hu Thy/Liv mice. J Exp Med 1999; 190:1857–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kikuta H, Itakura O, Taneichi K, Kohno M. Tropism of human herpesvirus 8 for peripheral blood lymphocytes in patients with Castleman’s disease. Br J Haematol 1997; 99:790–3. [DOI] [PubMed] [Google Scholar]
- 28. Volpi A. Epstein-Barr virus and human herpesvirus type 8 infections of the central nervous system. Herpes 2004; 11Suppl 2:120A–7A. [PubMed] [Google Scholar]
- 29. Corboy JR, Garl PJ, Kleinschmidt-DeMasters BK. Human herpesvirus 8 DNA in CNS lymphomas from patients with and without AIDS. Neurology 1998; 50:335–40. [DOI] [PubMed] [Google Scholar]
- 30. Brink NS, Sharvell Y, Howard MR, Fox JD, Harrison MJ, Miller RF. Detection of Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus DNA in CSF from persons infected with HIV who had neurological disease. J Neurol Neurosurg Psychiatry 1998; 65:191–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan PK, Ng HK, Cheung JL, Cheng AF. Survey for the presence and distribution of human herpesvirus 8 in healthy brain. J Clin Microbiol 2000; 38:2772–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Broccolo F, Iuliano R, Careddu AM, et al. Detection of lymphotropic herpesvirus DNA by polymerase chain reaction in cerebrospinal fluid of AIDS patients with neurological disease. Acta Virol 2000; 44:137–43. [PubMed] [Google Scholar]
- 33. Said JW, Tasaka T, de Vos S, Koeffler HP. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus type 8 encephalitis in HIV-positive and -negative individuals. AIDS 1997; 11:1119–22. [DOI] [PubMed] [Google Scholar]
- 34. Vieira J, O’Hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 2004; 325:225–40. [DOI] [PubMed] [Google Scholar]
- 35. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg 1938; 27:493–7. [Google Scholar]
- 36. He J, Bhat G, Kankasa C, et al. Seroprevalence of human herpesvirus 8 among Zambian women of childbearing age without Kaposi’s sarcoma (KS) and mother-child pairs with KS. J Infect Dis 1998; 178:1787–90. [DOI] [PubMed] [Google Scholar]
- 37. Minhas V, Brayfield BP, Crabtree KL, Kankasa C, Mitchell CD, Wood C. Primary gamma-herpesviral infection in Zambian children. BMC Infect Dis 2010; 10:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar P, Kuwa NY, Minhas V, et al. Higher levels of neutralizing antibodies against KSHV in KS patients compared to asymptomatic individuals from Zambia. PLoS One 2013; 8:e71254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kedes DH, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Invest 1997; 100:2606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minhas V, Crosby LN, Crabtree KL, et al. Development of an immunofluorescence assay using recombinant proteins expressed in insect cells to screen and confirm presence of human herpesvirus 8-specific antibodies. Clin Vaccine Immunol 2008; 15:1259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang G, Chan B, Samarina N, et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A 2016; 113:E1034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jha HC, Mehta D, Lu J, et al. Gammaherpesvirus infection of human neuronal cells. MBio 2015; 6:e01844–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Antinone SE, Smith GA. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol 2010; 84:1504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wakeham K, Webb EL, Sebina I, et al. Parasite infection is associated with Kaposi’s sarcoma associated herpesvirus (KSHV) in Ugandan women. Infect Agent Cancer 2011; 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.