Abstract
Background
Although metabolomic profiling offers promise for prediction of coronary heart disease (CHD), and metabolic risk factors are more strongly associated with CHD in women than men, limited data are available for women.
Methods
We applied a liquid chromatography-tandem mass spectrometry metabolomics platform to measure 371 metabolites in a discovery set of postmenopausal women (472 incident CHD cases, 472 controls) with validation in an independent set of postmenopausal women (312 incident CHD cases, 315 controls).
Results
Eight metabolites, primarily oxidized lipids, were significantly dysregulated in cases after adjustment for matching and CHD risk factors in both the discovery and validation datasets. One oxidized phospholipid, C34:2 hydroxy-phosphatidylcholine (C34:2 hydroxy-PC), remained associated with CHD after further adjustment for other validated metabolites. Subjects with C34:2 hydroxy-PC levels in the highest quartile had a 4.7-fold increase in CHD odds compared to the lowest quartile; C34:2 hydroxy-PC also significantly improved the AUC (p<0.01) for CHD. The C34:2 hydroxy-PC findings replicated in a third replication dataset of 980 men and women (230 cardiovascular events) with a stronger association observed in women.
Conclusions
These data replicate known metabolite predictors, identify novel markers, and support the relationship between lipid oxidation and subsequent CHD.
Keywords: metabolomics, women, coronary heart disease risk, oxidized lipids
Introduction/Background
Coronary heart disease remains the leading cause of death in women, as in men. However, the clinical manifestations of CHD differs, with a greater preponderance of non-obstructive atherosclerosis and microvascular disease1 and higher case fatality rates in women as compared to men. Metabolic risk factors, especially diabetes, are particularly strong risk factors for CHD in women. Women with type 2 diabetes have over 40% higher risk of CHD than men with diabetes2, 3. However, data on the relationship of metabolites and CHD using metabolomic profiling have been very limited in women.
Examinations of the relationship of CHD with circulating blood metabolite levels measured using metabolomics have illuminated different potential pathways to CHD development. Incident cardiovascular disease and longevity have been associated with metabolites in the citric acid cycle as well as gut microbe-associated trimethylamine N-oxide (TMAO), choline, and multiple lipid-related metabolites4–11. Glutamate and glutamine have also been associated with diabetes12 as well as with cardiovascular disease in previous studies4, 6, 9, as have branched chain and aromatic amino acids12, 13. However, despite evidence of sexual dimorphism in the metabolome14, 15, these associations have not been examined in detail in women. Further, metabolite profiles may also change during menopause16, a time coinciding with increased CHD risk. Therefore, the examination of metabolomics profiles in women may help to identify new metabolic associations and mechanistic pathways underlying CHD in women, which may extend also to men.
Here, we used metabolomics to identify novel metabolic signatures for incident CHD in female participants of the Women’s Health Initiative17, with independent discovery and validation sets derived from the Observational Study (WHI-OS)18 and the placebo arms of the Hormone Trials (WHI-HT)19, respectively. We also performed external replication using an independent mixed sex study from the Prevención con Dieta Mediterránea (PREDIMED) Trial20, 21 to assess generalizability of significant metabolites to cardiovascular disease and both sexes.
Methods
This study was approved by Partners Human Research Committee, which is the IRB of Brigham and Women’s Hospital. Metabolite data from the discovery and validation datasets used in this study have been deposited in and are available from the dbGaP database under dbGaP accession phs001334.v1.p3.22 All other data will not be made available by the authors, nor will analytic methods or study materials.
Human Studies
WHI-OS (Discovery)
The discovery set was drawn from the Women’s Health Initiative-Observational Study (WHI-OS). The full WHI-OS consisted of 93,676 postmenopausal women ineligible or unwilling to participate in the related hormone trials, enrolled between 1994 and 1998 in the US. All participants provided written informed consent. For this study, 472 participants who developed CHD after the baseline examination (cases) were selected, including all non-white cases with an eligible control and a sample of the white cases, and 472 participants who did not develop CHD frequency matched on 5-year age, race/ethnicity, hysterectomy status, and 2-year enrollment groups. Only 21 hormone therapy users were included in the discovery set and use was not adjusted for, but a sensitivity analysis removing the hormone therapy users did not change the effect estimates.
WHI-HT (Validation)
The validation set was drawn from the placebo arms of the Women Health Initiative Hormone Therapy Trials. One of the trials randomized 16,608 postmenopausal women to estrogen plus progesterone or placebo, while the other randomized 10,739 women with prior hysterectomy to estrogen or placebo. All participants provided written informed consent. We selected 312 cases and 315 frequency-matched controls from the two placebo arms with matching on 5-year age, race/ethnicity, hysterectomy status, and 2-year enrollment window.
PREDIMED (Replication)
The replication set was drawn from the Prevención con Dieta Mediterránea (PREDIMED) trial which also utilized the Broad Institute metabolomics platform. The full PREDIMED trial randomized men and women at high risk of developing cardiovascular disease to one of 3 interventions: a Mediterranean dietary intervention supplemented with virgin olive oil, a Mediterranean dietary intervention supplemented with nuts, or advice about a low-fat diet. All participants provided written informed consent. The primary endpoint for this trial was a composite of MI, stroke and CVD death. For this study, a case-cohort design as outlined by Prentice23 was used, which included all 230 cases (74 MI, 115 stroke, and 31 CVD death) and a random 10% sample of participants at baseline (787 participants). Of the 787 sub-cohort participants, 37 had a primary endpoint during follow-up (and thus were also included in the 230 cases) while 750 did not, resulting in 980 unique participants for analysis.
Metabolomics
Plasma samples were collected using EDTA tubes in the WHI and PREDIMED cohorts and processed immediately. All WHI specimens were stored in a −70° freezer within 2 hours of collection or stored at −20° for up to 2 days and shipped on dry ice and stored at −70° until processing. The majority of the WHI samples had been thawed once prior to our study, with 7 samples (6 cases and 1 control) having been thawed twice. PREDIMED samples were shipped at −4°, and then stored at −70° until analysis. Metabolomic measurements were made using four complimentary LC-MS methods resulting in 371 metabolites for the WHI samples. Only the HILIC-positive and C8-positive methods were available in the PREDIMED samples. For each method, pooled plasma reference samples were included every 20 samples and results were standardized using the ratio of the value of the sample to the value of the nearest pooled reference multiplied by the median of all reference values for the metabolite.
HILIC analyses of water soluble metabolites in the positive ionization mode (HILIC-pos) were conducted using an LC-MS system comprised of a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.; Marlborough, MA) coupled to a Q Exactive hybrid quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Plasma samples (10 µL) were prepared via protein precipitation with the addition of nine volumes of 74.9:24.9:0.2 v/v/v acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (valine-d8, Sigma-Aldrich; St. Louis, MO; and phenylalanine-d8, Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 9,000 × g, 4°C), and the supernatants were injected directly onto a 150 × 2 mm, 3 µm Atlantis HILIC column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 250 µL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 0.5 minute followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 minutes. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over 70–800 m/z at 70,000 resolution and 3 Hz data acquisition rate. Other MS settings were: sheath gas 40, sweep gas 2, spray voltage 3.5 kV, capillary temperature 350°C, S-lens RF 40, heater temperature 300°C, microscans 1, automatic gain control target 1e6, and maximum ion time 250 ms.
HILIC analyses of water soluble metabolites in the negative ionization mode (HILIC-neg) were conducted using an LC-MS system comprised of an AQUITY UPLC system (Waters; Milford, MA and a 5500 QTRAP mass spectrometer (SCIEX; Framingham, MA). Plasma samples (30 µL) were prepared via protein precipitation with the addition of four volumes of 80% methanol containing inosine-15N4, thymine-d4 and glycocholate-d4 internal standards (Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 9,000 × g, 4°C), and the supernatants were injected directly onto a 150 × 2.0 mm Luna NH2 column (Phenomenex; Torrance, CA). The column was eluted at a flow rate of 400 µL/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide in water) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 v/v acetonitrile/methanol) followed by a 10 min linear gradient to 100% mobile phase A. MS analyses were carried out using electrospray ionization and selective multiple reaction monitoring scans in the negative ion mode. To create the method, declustering potentials and collision energies were optimized for each metabolite by infusion of reference standards. The ion spray voltage was −4.5 kV and the source temperature was 500°C.
Positive ion mode analyses of polar and non-polar plasma lipids (C8-pos) were conducted using an LC-MS system comprised of a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.; Marlborough, MA) coupled to a Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Plasma samples (10 µL) were extracted for lipid analyses using 190 µL of isopropanol containing 1,2-didodecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids; Alabaster, AL). After centrifugation, supernatants were injected directly onto a 100 × 2.1 mm, 1.7 µm ACQUITY BEH C8 column (Waters; Milford, MA). The column was eluted isocratically with 80% mobile phase A (95:5:0.1 vol/vol/vol 10mM ammonium acetate/methanol/formic acid) for 1 minute followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/formic acid) over 2 minutes, a linear gradient to 100% mobile phase B over 7 minutes, then 3 minutes at 100% mobile-phase B. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over 200–1000 m/z at 70,000 resolution and 3 Hz data acquisition rate. Other MS settings were: sheath gas 50, in source CID 5 eV, sweep gas 5, spray voltage 3 kV, capillary temperature 300°C, S-lens RF 60, heater temperature 300°C, microscans 1, automatic gain control target 1e6, and maximum ion time 100 ms. Lipid identities were determined based on comparison to reference plasma extracts and were denoted by total number of carbons in the lipid acyl chain(s) and total number of double bonds in the lipid acyl chain(s).
Negative ion mode analyses of free fatty acids and bile acids (C18-neg) were conducted using an LC-MS system comprised of a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.; Marlborough, MA) coupled to a Q Exactive hybrid quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Plasma samples (30 µL) were extracted using 90 uL of methanol containing PGE2-d4 (Cayman Chemical Co.; Ann Arbor, MI) and centrifuged (10 min, 9,000 × g, 4°C). The samples were injected onto a 150 × 2 mm ACQUITY T3 column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 400 µL/min with 60% mobile phase A (0.1% formic acid in water) for 4 minutes followed by a linear gradient to 100% mobile phase B (acetonitrile with 0.1% formic acid) over 8 minutes. MS analyses were carried out in the negative ion mode using electrospray ionization, full scan MS acquisition over 200–550 m/z, and a resolution setting of 70,000. Metabolite identities were confirmed using authentic reference standards. Other MS settings were: sheath gas 45, sweep gas 5, spray voltage −3.5 kV, capillary temperature 320°C, S-lens RF 60, heater temperature 300°C, microscans 1, automatic gain control target 1e6, and maximum ion time 250 ms.
Raw data from Q Exactive/Exactive Plus instruments were processed using TraceFinder software (Thermo Fisher Scientific; Waltham, MA) and Progenesis QI (Nonlinear Dynamics; Newcastle upon Tyne, UK) while MultiQuant (SCIEX; Framingham, MA) was used to process 5500 QTRAP data. For each method, metabolite identities were confirmed using authentic reference standards or reference samples. CVs were calculated using pooled plasma samples from the first 800 WHI-OS participants.
Statistical Analysis
Two sets of covariates were used in WHI analyses. The first set included the frequency matching factors (age, race/ethnicity, hysterectomy status and enrollment window). The second set included the matching factors plus traditional CHD risk factors (smoking, systolic blood pressure, diabetes, total and HDL cholesterol) and CHD medication use (aspirin, statins, anti-hyperglycemics and anti-hypertensives). All metabolites were scaled and log-normalized and missing values below the limit of detection were assigned to half the lowest observed value. R was used for all analyses with packages noted below.
Discovery methods
For the discovery analysis in the WHI-OS, each metabolite was examined individually in logistic models including both sets of covariates. Statistically significant associations satisfied FDR adjusted p<0.05 using the two-stage step-up procedure24 – this procedure allows for dependencies between the metabolites and is implemented using the corresponding “TSBH” option of the mt.rawp2adjp function in the multtest package with a nominal alpha of 0.05. Unconditional logistic regression was chosen over conditional to improve precision of estimates while still controlling for the frequency matching factors25.
Validation methods
Metabolite associations with CHD discovered in WHI-OS were validated in the independent WHI-HT data set. Metabolites were examined individually using the same covariate sets used for discovery – thus, a metabolite that was selected in the discovery process when adjusted for matching factors but not when adjusted for additional risk factors would only be validated with adjustment for matching factors. Metabolite associations were considered to have validated if the FDR-adjusted p value (same procedure as discovery) for the odds ratio after adjustment was less than 0.05 and the nominal p value was less than 0.025.
AUC Calculation
By combining data from the parent WHI cohort with that generated in the matched case control study, we estimated the prediction score for each subject using methods described by Qian and colleagues26. To estimate the Receiver Operating Characteristics (ROC) curve and associated AUC, we reweighted our sample to account for the frequency matched selection of cases and controls from the source populations in the WHI. We used 1000 bootstrap samples that preserved the frequency matching to construct the confidence interval for the difference in the AUC comparing models with and without C34:2 hydroxy-PC in addition to matching and other risk factors.
Derived networks
A correlation network was generated using the full WHI dataset in which nodes represent individual metabolites and edges between node pairs indicate a partial correlation (adjusting for case status) that exceeded 0.7 in magnitude. This procedure resulted in a network of 371 nodes and 1256 edges. The connected components of the partial correlation network were identified using the clusters function in the R igraph package. Fishers exact tests were then used to test whether each connected set of metabolites included more validated metabolites associated with CHD than expected by chance.
Enantomeric composition
Since the reversed phase separation used in the LC-MS method used to profile the WHI dataset does not distinguish stereoisomers, enantiomeric purity was investigated using chiral chromatography following the method of Oh et al27. LC-MS tracings for representative samples from the WHI dataset were compared to enantiomerically pure standards as well as a racemic mixture. An Agilent 4695 mass spectrometer was used to measure the characteristic negative ion mode 319.2/115.1 transition at a collision energy setting of 13.
Replication methods
Four metabolites from the two available metabolomics platforms in PREDIMED were examined individually in weighted Cox proportional hazards models, adjusting for age, sex, intervention group, systolic blood pressure, total cholesterol, HDL cholesterol, diabetes, smoking, and statin use. The Barlow weighting method28 was used to account for the random, non-stratified sampling design along with the exact case-cohort pseudolikelihood estimators with robust variance estimators, as proposed by Langholz and Jiao 29 for the Cox proportional hazards models. Metabolite associations were considered to have replicated if the p value for the hazard ratio was less than 0.05. Interaction p-values were determined using the Wald test for the interaction coefficient.
Results
The baseline characteristics of the 944 women in the WHI-OS discovery set (472 cases and 472 controls) and the 627 women in the WHI-HT validation set (312 cases and 315 controls) are shown in Table 1. The mean age was similar in both sets. The median time to CHD event for the cases, defined as myocardial infarction or death due to CHD, was 5.8 years in the discovery set and 3.7 years in the validation set. A total of 371 metabolites of known identity were profiled using liquid chromatography tandem mass spectroscopy (LC-MS) with a median CV of 6.28%, as measured in replicate quality control pooled plasma samples included throughout the analyses.
Table 1.
Baseline Characteristics of WHI Datasets
| WHI-OS Discovery (N=944) |
WHI-HT Validation (N=627) |
|||||
|---|---|---|---|---|---|---|
|
| ||||||
| Cases N = 472 |
Controls N = 472 |
p-value | Cases N = 312 |
Controls N = 315 |
p-value | |
| Age, years | 68 (7) | 67 (7) | 0.51 | 67 (7) | 66 (7) | 0.64 |
|
| ||||||
| Race | 1.00 | 1.00 | ||||
| White | 348 (73%) | 348 (73%) | 254 (81%) | 256 (81%) | ||
| Black | 69 (15%) | 69 (15%) | 37 (12%) | 38 (12%) | ||
| Other | 55 (12%) | 55 (12%) | 21 (7%) | 21 (7%) | ||
|
| ||||||
| Systolic blood pressure, mmHg | 135 (19) | 130 (18) | <0.001 | 137 (19) | 131 (18) | <0.001 |
|
| ||||||
| Diabetes | 78 (17%) | 25 (5%) | <0.001 | 65 (21%) | 31 (10%) | <0.001 |
|
| ||||||
| BMI, m2/kg | 29 (6) | 27 (6) | 0.001 | 30 (6) | 29 (6) | 0.21 |
|
| ||||||
| Total cholesterol, mg/dL | 231 (47) | 232 (47) | 0.51 | 236 (43) | 235 (39) | 0.57 |
|
| ||||||
| HDL cholesterol, mg/dL | 51 (16) | 57 (17) | <0.001 | 47 (12) | 52 (12) | <0.001 |
|
| ||||||
| Smoking status, | 0.016 | 0.033 | ||||
| Current | 48 (10%) | 28 (6%) | 56 (18%) | 34 (11%) | ||
| Former | 209 (44%) | 243 (51%) | 145 (46%) | 166 (53%) | ||
| Never | 215 (46%) | 201 (43%) | 111 (36%) | 115 (36%) | ||
|
| ||||||
| Aspirin use | 116 (25%) | 98 (21%) | 0.16 | 93 (30%) | 78 (25%) | 0.16 |
|
| ||||||
| Statin use | 42 (9%) | 39 (8%) | 0.72 | 60 (19%) | 36 (11%) | 0.007 |
|
| ||||||
| Anti-hypertensive use | 144 (31%) | 91 (19%) | <0.001 | 103 (33%) | 83 (26%) | 0.068 |
|
| ||||||
| Anti-hyperglycemic use | 47 (10%) | 13 (3%) | <0.001 | 52 (17%) | 18 (6%) | <0.001 |
N(%) or mean (SD)
The relationship between individual metabolites and CHD was assessed using logistic regression controlling for matching variables only or matching variables plus CHD risk factors. In the discovery set, 60 of the 371 metabolites were identified as significant (two stage FDR<0.05) controlling for matching variables alone and 36 (17 also identified in the matching analysis) were identified controlling for matching plus CHD risk factors. Results of the discovery analysis are shown in Supplemental Table 1.
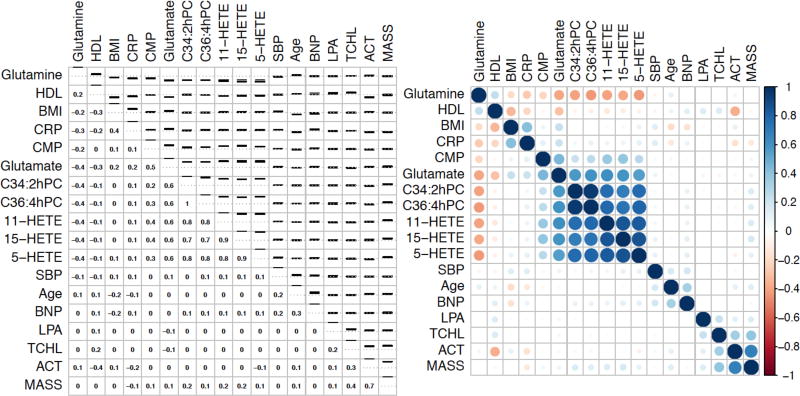
All metabolites discovered when controlling for matching variables were then tested in a separate validation set of 627 women from the placebo arms of WHI-HT (312 cases and 315 controls), and 33 were significantly associated with CHD (FDR p<0.05, nominal p<0.025). Similarly, 8 of the 36 metabolites discovered with adjustment for matching plus CHD risk factors remained significant in the independent validation dataset (nominal p<0.0014). Table 2 presents the results only for validated metabolites; the results for all discovered metabolites can be found in Supplemental Table 2. Of the 8 metabolites that were significantly associated with CHD after adjustment for traditional CHD risk factors, 7 were associated with increased risk: C34:2 hydroxy-phosphatidylcholine (hydroxy-PC), C36:4 hydroxy-PC, 5-hydroxyeicosatetraenoic acid (HETE), 15-HETE, 11-HETE, glutamate, and cytidine monophosphate (CMP). A single metabolite, glutamine, was associated with decreased risk of CHD. Associations between the 8 metabolites and other biomarkers and CHD risk factors are shown in Figure 1. Of note, none of the oxidized lipid metabolites (C34:2 hydroxy-PC, C36:4 hydroxy-PC, 5-HETE, 15-HETE, and 11-HETE) were correlated with C-reactive protein, lipoprotein(a), or Lp-PLA2 activity or mass. Coefficients of variation and percent of samples with values below the limit of detection for validated metabolites are provided in Supplemental Table 3.
Table 2.
Validation in the independent WHI-HT dataset of metabolites associations with future CHD discovered in the WHI-OS metabolites.
| Metabolite | WHI-HT Adjusted for matching* |
WHI-HT Adjusted for risk factors† |
||
|---|---|---|---|---|
| Odds Ratio‡ | P-value§ | Odds Ratio‡ | P-value§ | |
| C34:2 hydroxy-PC | 1.83 (1.54, 2.17) | 5.4E-13 | 1.76 (1.47, 2.10) | 2.5E-10 |
| C36:4 hydroxy-PC | 1.66 (1.39, 1.99) | 2.4E-09 | 1.56 (1.30, 1.87) | 7.0E-07 |
| 15-HETE | 1.73 (1.44, 2.07) | 2.3E-10 | 1.65 (1.36, 1.99) | 5.6E-08 |
| 5-HETE | 1.71 (1.43, 2.04) | 3.0E-10 | 1.65 (1.37, 1.99) | 2.6E-08 |
| 11-HETE | 1.68 (1.41, 2.00) | 9.0E-10 | 1.64 (1.36, 1.97) | 4.3E-08 |
| 12-HETE | 1.64 (1.38, 1.95) | 3.9E-09 | ||
| glutamate | 1.64 (1.38, 1.95) | 5.5E-09 | 1.50 (1.25, 1.80) | 8.3E-06 |
| glutamine | 0.61 (0.50, 0.73) | 1.4E-08 | 0.67 (0.55, 0.82) | 2.5E-05 |
| asparagine | 0.75 (0.64, 0.88) | 4.2E-04 | ||
| CMP | 1.38 (1.17, 1.63) | 1.1E-04 | 1.33 (1.11, 1.59) | 1.4E-03 |
| uracil | 0.80 (0.68, 0.94) | 6.5E-03 | ||
| xanthosine | 0.80 (0.68, 0.95) | 7.5E-03 | ||
| glucose | 1.42 (1.19, 1.69) | 3.4E-05 | ||
| sucrose | 1.40 (1.18, 1.65) | 7.2E-05 | ||
| sorbitol | 1.28 (1.08, 1.51) | 3.4E-03 | ||
| 2-hydroxyglutarate | 1.45 (1.22, 1.72) | 1.0E-05 | ||
| fumarate/maleate | 1.22 (1.03, 1.44) | 2.1E-02 | ||
| isocitrate | 1.21 (1.02, 1.42) | 2.4E-02 | ||
| C50:0 TAG | 1.37 (1.16, 1.61) | 1.4E-04 | ||
| C50:1 TAG | 1.34 (1.14, 1.58) | 3.6E-04 | ||
| C52:0 TAG | 1.42 (1.20, 1.67) | 2.5E-05 | ||
| C52:1 TAG | 1.38 (1.17, 1.63) | 8.1E-05 | ||
| C52:2 TAG | 1.32 (1.12, 1.55) | 7.8E-04 | ||
| C54:1 TAG | 1.34 (1.14, 1.58) | 3.3E-04 | ||
| C54:2 TAG | 1.36 (1.15, 1.60) | 2.0E-04 | ||
| C54:3 TAG | 1.25 (1.06, 1.47) | 6.7E-03 | ||
| C56:2 TAG | 1.34 (1.14, 1.59) | 3.4E-04 | ||
| C56:3 TAG | 1.23 (1.05, 1.45) | 1.2E-02 | ||
| C34:0 DAG | 1.26 (1.06, 1.49) | 6.5E-03 | ||
| C34:1 DAG | 1.40 (1.18, 1.65) | 5.9E-05 | ||
| C36:1 DAG | 1.41 (1.19, 1.67) | 3.9E-05 | ||
| C36:2 DAG | 1.31 (1.11, 1.54) | 1.3E-03 | ||
| C40:10 PC | 0.82 (0.70, 0.97) | 1.8E-02 | ||
adjusted for baseline age, race/ethnicity, hysterectomy status, and enrollment window
adjusted for baseline age, race/ethnicity, hysterectomy status, enrollment window, aspirin use, statin use, anti-hyperglycemic use, anti-hypertensive use, smoking, systolic blood pressure, diabetes, total and HDL cholesterol.
odds ratios are per 1 standard deviation of log transformed values
Nominal p-values presented, cutoff for validation was nominal p<0.025 and FDR adjusted p-value less than 0.05
Figure 1.
Correlations between validated metabolites, cardiovascular biomarkers, age, BMI, and systolic blood pressure. Left panel shows spearman correlations on bottom and confidence interval on top; right panel shows heat map. Both figures are ordered using hierarchical clustering. SBP: Systolic blood pressure, TCHL: Total cholesterol, HDL: HDL cholesterol, BMI: Body mass index, CRP: log C-reactive protein, ACT: Lp-PLA2 activity, LPA: Lipoprotein(a), MASS: Lp-PLA2 mass, BNP: N-terminal pro-BNP
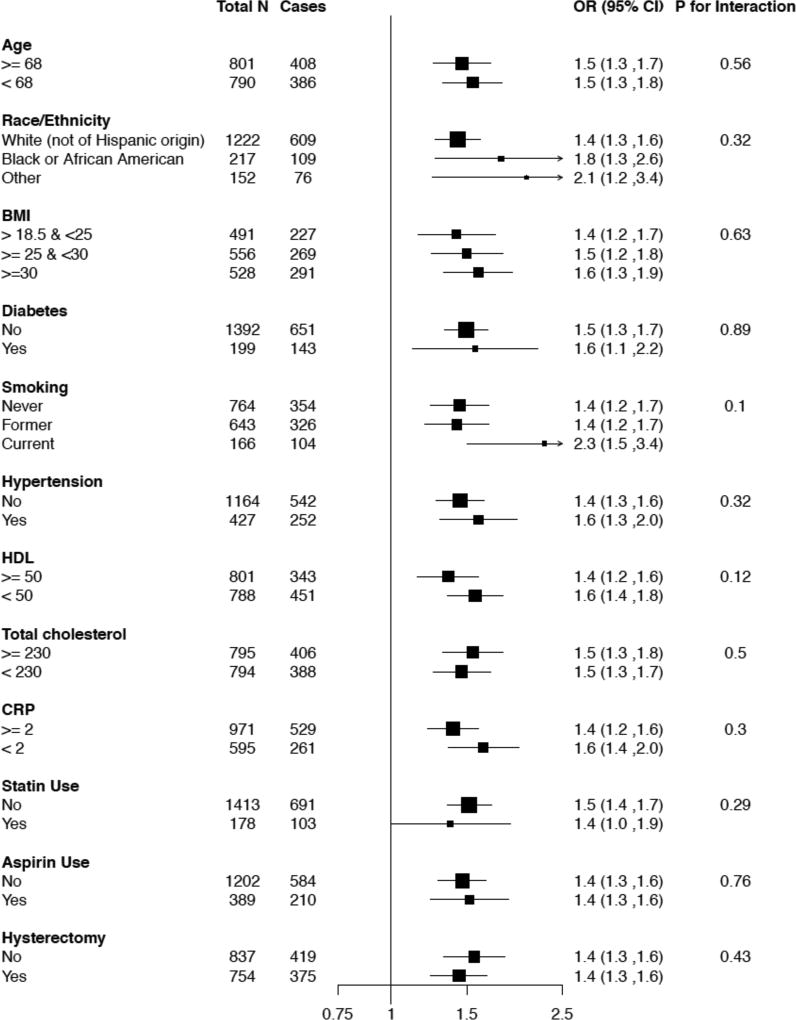
When the 8 validated metabolites were mutually adjusted for in models controlling for matching factors and CHD risk factors, only C34:2 hydroxy-PC remained significantly associated with CHD case status in the WHI. Women in the highest quartile of C34:2 hydroxy-PC had an OR of CHD of 4.69 (2.84, 7.90) compared to those in the lowest quartile. The relationship between C34:2 hydroxy-PC and CHD remained consistent across subgroups of baseline characteristics as shown in Figure 2. In order to evaluate the impact on prediction of CHD, we estimated the prediction score by integrating metabolomics data from the matched case control study to existing data on risk factors available for the full WHI cohort26. We estimated the area under the receiver operator characteristic curve (AUC) by incorporating inverse-probability weights to re-weight the distribution of risk factors in the controls to reflect the full WHI-OS and WHI-HT cohorts. When C34:2 hydroxy-PC was added to the model with traditional CHD risk factors and matching factors, the AUC improved significantly from 0.76 to 0.79 with a change of 0.03 (95% CI: 0.01, 0.05, p<0.01).
Figure 2.
Subgroup analysis for C34:2 hydroxy-PC in WHI-HT women
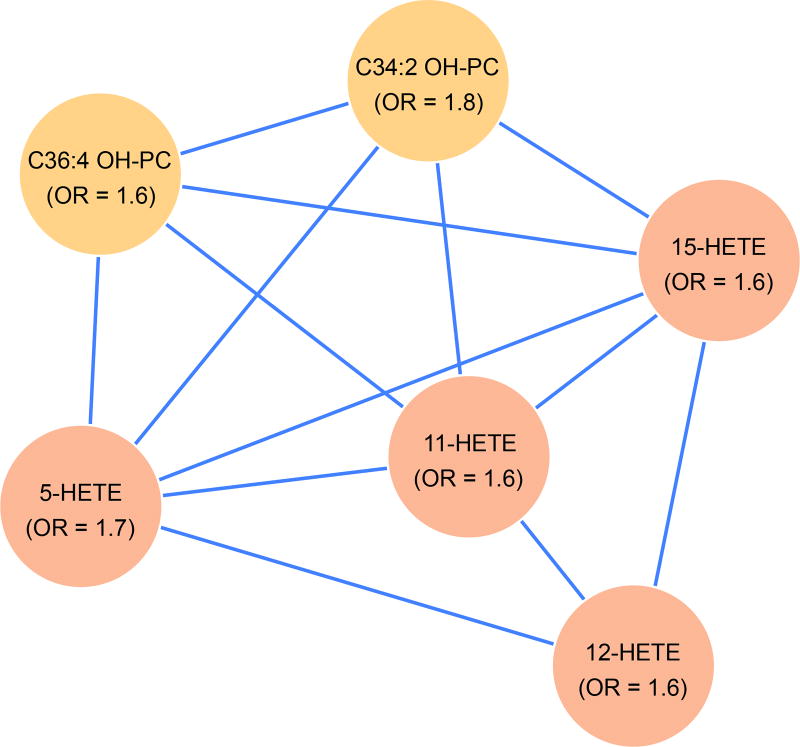
To identify connections between validated metabolites, a correlation network of the 371 metabolites was constructed using all metabolite pairs. Edges between metabolite pairs were drawn to indicate an absolute Spearman partial correlation coefficient exceeding 0.7, while adjusting for case status. An enrichment analysis was layered on the partial correlation network to discover clusters of connected (correlated) metabolites that are enriched for associations with CHD. Six clusters included validated metabolites, and of these, only 1 cluster, shown in Figure 3, contained significantly more validated metabolites than would be expected by chance (p=3.185e-07). Each node in this cluster was an oxidized lipid, including 2 hydroxy-PCs and 4 HETEs, 3 of which were validated after full adjustment for risk factors and one of which (12-HETE) was validated with adjustment for matching factors only.
Figure 3.
Cluster of 6 oxidized lipid metabolites, all 6 of which are validated markers in a network of spearman partial correlations. Enrichment p value = 3.185e-07. All markers are positively associated with increased CHD risk. All correlations (edges) are positive.
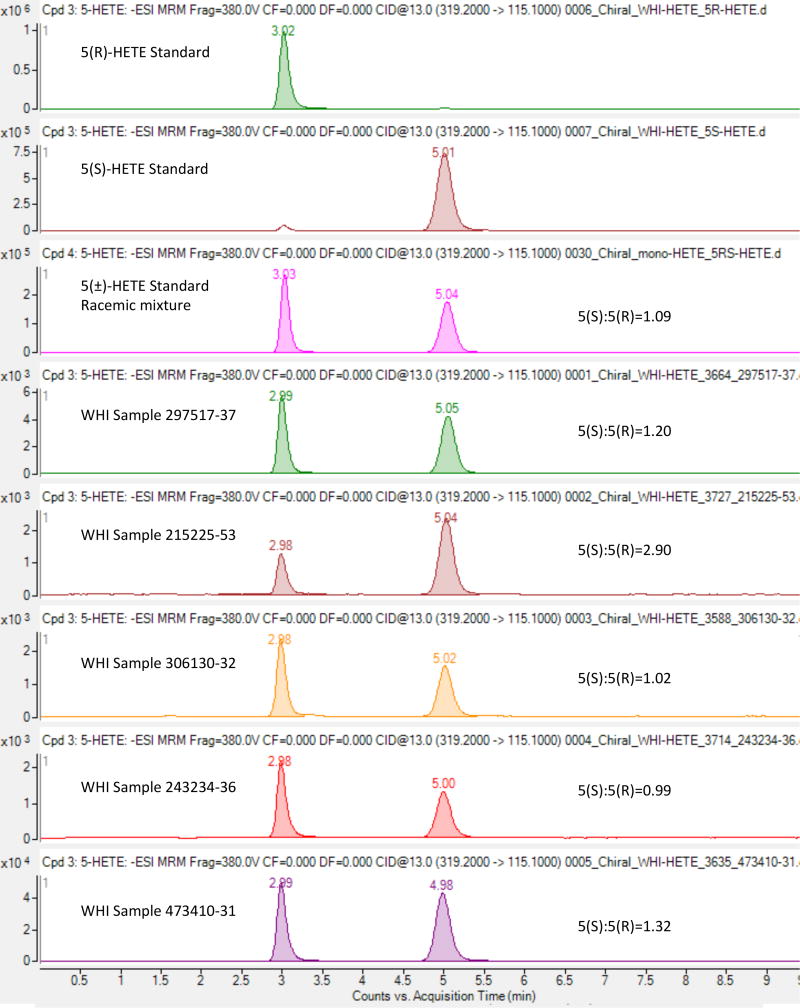
To determine whether the mono-HETEs associated with CHD were generated by enzymatic activity (single isomers) or nonenzymatic oxidation of arachidonic acid (racemic mixture), we analyzed stereoisomers of 5-HETEusing chiral chromatography. As shown in Figure 4, enantiomerically pure 5(R)-HETE and 5(S)-HETE standards were measured at nominal retention times of 3.0 and 5.0 minutes, respectively, while a racemic mixture of 5(+/−)-HETE showed nearly equivalent measured peak areas between the enantiomers. Analyses of five representative plasma samples selected from the highest quartiles of 5-HETE levels in WHI showed generally similar 5(S) and 5(R) peak areas, suggesting that the WHI plasma measured levels of 5-HETE are a mixture of R- and S-enantiomers, in similar proportions.
Figure 4.
Chiral chromatography analysis of 5-HETE. Panels (from top) are the results from agilent 4695 mass spectrometer measurements of the characteristic negative ion mode 319.2/115.1 transition at a collision energy setting of 13 for: (1) enantiomerically pure 5(R)-HETE; (2) enantiomerically pure 5(S)-HETE; (3) a racemic mixture of 5(+/−)-HETE; (4–8) five representative plasma samples selected from the highest quartiles of 5-HETE levels in WHI.
Finally, we sought to test the generalizability of our findings in a new independent cohort, PREDIMED, that included both men and women and used a combined CVD endpoint of incident MI, stroke, and CHD death (230 cases) in a case-cohort design. Two of the four LC-MS methods done in the same lab used to profile the WHI data had been applied to PREDIMED, allowing us to test 4 of the 8 validated metabolites (glutamine, glutamate, C34:2 hydroxy-PC, and C36:4 hydroxy-PC). Baseline characteristics of the PREDIMED validation cohort are shown in Supplementary Table 4. For the combined CVD outcome in men and women, 3 of the 4 metabolites (both hydroxy-PCs and glutamate) replicated while glutamine did not (Table 3). All of the relationships were stronger for women, with only glutamate remaining significant in men alone. An interaction term indicated significant effect modification by sex for both hydroxy-PCs (p for interaction <0.001). In a sensitivity analysis using CHD cases only (74 cases), the hazard ratio (HR) for C34:2 hydroxy-PC was 1.56 (1.09–2.24), P=0.02. For C36:4 hydroxy-PC the CHD only HR was 1.59 (1.10–2.30) P=0.01. In a corresponding analysis using stroke cases only (115 cases), the HR for C34:2 hydroxy-PC was 1.48 (1.13–1.92), P=0.004, and for C36:4 hydroxy-PC was 1.34 (1.02–1.77) P=0.036.
Table 3.
Validation of metabolites among men and women in PREDIMED
| All CVD* (N=980, 230 cases) |
CVD Men Only* (139 cases) |
CVD Women Only* (91 cases) |
Interaction by Sex P value |
||||
|---|---|---|---|---|---|---|---|
| Metabolite | Hazard Ratio (95%CI) |
P value | Hazard Ratio (95%CI) |
P value | Hazard Ratio (95%CI) |
P value | |
| C34:2 Hydroxy-PC | 1.40 (1.15–1.70) | 0.0008 | 1.16 (0.90, 1.51) | 0.26 | 1.79 (1.30, 2.46) | 0.0004 | 0.013 |
| C36:4 Hydroxy-PC | 1.36 (1.12–1.66) | 0.0002 | 1.14 (0.86, 1.49) | 0.366 | 1.67 (1.22, 2.29) | 0.0015 | 0.026 |
| Glutamate | 1.48 (1.22–1.80) | 0.00009 | 1.39 (1.04, 1.84) | 0.024 | 1.64 (1.19, 2.24) | 0.0022 | 0.32 |
| Glutamine | 0.84 (0.64–1.11) | 0.22 | 0.83 (0.59, 1.19) | 0.32 | 0.79 (0.50, 1.26) | 0.33 | 0.65 |
adjusted for baseline age, sex, intervention group, systolic blood pressure, total cholesterol, high density lipoprotein cholesterol, diabetes status, smoking, statin use.
Discussion
In our study of the association of metabolites with CHD in post-menopausal women, we identified and validated 8 metabolites associated with future CHD in women. These metabolites included glutamine, glutamate, cytidine monophosphate (CMP), and five oxidized lipids that had not previously been associated with incident CHD in humans. The strongest associations with CHD were observed for the oxidized lipids including two species of hydroxy-PCs (C34:2 and C36:4) and three oxidized derivatives from arachidonic acid (15-HETE, 5-HETE, and 11-HETE). The hydroxy-PCs were also externally validated for CVD, CHD and stroke in an external population (PREDIMED), with stronger associations seen in women.
Arachidonic acid derived HETEs have previously been associated with atherosclerosis, but not previously with incident CHD. A previous study by Oni-Orisan et al found an association between higher levels of 11-HETE and 15-HETE and extent of prevalent coronary artery disease.30 5-HETE, 12-HETE, and 15-HETE are well characterized products of 5-, 12-, and 15-lipoxygenase activity, respectively. 5-Lipoxgenase (ALOX5) activity is essential for leukotriene biosynthesis and is therefore linked to a number of inflammatory pathologies. However, when mono-HETEs are produced by these enzymes, the stereochemistry of the hydroxyl bound carbon is in the S-configuration. Our analyses found that the HETEs associated with CHD appeared to be produced from oxidative stress. This is the first report of an association between non-enzymatically generated mono-HETEs and incidence of CHD.
We also identified two novel oxidized phospholipids, C34:2 hydroxy-PC and C36:4 hydroxy-PC, associated with CHD. C34:2 hydroxy-PC was the metabolite most strongly associated with CHD in our cohort, with a more than 4-fold increased risk of CHD for women in the highest quartile. This association was robust across all strata tested, including whether or not aspirin and statins were used, or whether there was baseline diabetes or obesity. In PREDIMED, where the majority of incident CVD cases were in men, C34:2 hydroxy-PC was validated as significantly associated with CVD, as well as CHD and stroke. When examined by sex, women had a significant 79% increased risk of CVD per SD of C34:2 hydroxy-PC but results were nonsignificant in men, despite fewer CVD events occurring in women, and the sex by metabolite interaction term was significant. Similar associations were observed for C36:4 hydroxy-PC. Of note, no antioxidants were added to the samples to prevent ex-vivo oxidation in either of the studies. Whether these biomarkers remain more strongly associated with CVD in women in other populations, as well as the underlying mechanisms, requires further exploration.
The dysregulated oxidized lipids (HETEs and hydroxy-PCs) associated with CHD may offer new markers of oxidation and risk. They were not correlated with other biomarkers measured (ρ< 0.2) including inflammation markers, traditional lipids, BMI and systolic blood pressure, but are strongly associated with each other, as illustrated in the network analysis, and weakly associated with the other metabolites. From the stereoisomer analysis, the mono-HETE profiles are consistent with a non-enzymatic oxidative mechanism, which could also include generation of the hydroxy-PCs. While we were unable to assess replication of the HETEs in the PREDIMED due to the lack of available data on this class of metabolites, the strong replication of the hydroxy-PCs along with the strong correlation between the hydroxy-PCs and the HETEs suggests that the HETEs are likely to replicate as well. Lipid oxidation has been extensively studied in the context of model systems of overt atherosclerotic lesions31. Demonstration of the relationship between significantly dysregulated oxidized lipids and cardiovascular outcomes in longitudinal human cohort studies has been more difficult but some associations with CHD outcomes32, 33 and mortality34 have been shown. Additionally, a recent study of plasma metabolites of patients with existing CHD compared to healthy controls also identified lipid peroxidation related metabolites, including 5-HETE35. Our data support the hypothesis that early mechanisms involving lipid oxidation, for which these metabolites may be part of the cause or a consequence, may contribute to the etiology of CHD years before an event.
Our results are also not inconsistent with prior studies focusing narrowly on lipidomics. Both TAGs and DAGs were associated with CHD in our sample, though the association was eliminated after adjustment for total and HDL cholesterol. Using a lipidomics platform, Stegemann et al, found that C54.2 TAG was associated with increased risk of CHD in the Bruneck Study, with validation in Twins UK.7 Methods between their study and ours differed in the metabolites measured; additionally, ours included an increased number of cases for discovery and exclusively included women for the discovery and validation sets. We did replicate the C54:2 TAG finding, though not after adjustment for traditional risk factors, but did not find a significant association for C16:1CE, C36.5 PE, or C18:1 CE. Alshehry et al also found multiple lipids associated with future cardiovascular disease in a population with type 2 diabetes. Of these, we found similar nominal associations for C16:0 Ceramide(d18:1), C22:0 Ceramide(d18:1), C40:6 PC, and C56:6 TAG36.
Direct associations for glutamate and inverse associations for glutamine, initially identified by similar metabolomic profiling, have been consistently reported for both CHD and as well as diabetes4, 6, 9, 37. Cheng et al found that glutamate and glutamine were associated with markers of glucose metabolism as well as incident diabetes and showed that glutamine supplementation improved glucose tolerance in mice37. A glutamic acid related polymorphism has also been related to a higher risk of incident CHD among diabetics38. Other metabolomic studies have also found associations of glutamate and glutamine with CHD including two studies primarily in men6, 9 as well as one which replicated the association in samples with both men and women4.
Of note, TMAO, choline and branched chain and aromatic amino acids previously identified as associated with increased CHD risk were not selected using the data-driven approach of our study. TMAO and choline were not nominally significant in this cohort of women (Supplemental Table 1). Other studies have reported higher branch chain amino acid levels in men than women14 and in a pre-specified PREDIMED analysis combining men and women, branched chain amino acid levels were associated with CVD events39. We did identify a relationship between risk of CHD and isocitrate in matched analyses, but other branched chain amino acids were not associated with CHD in our cohort. Isocitrate was inversely associated with longevity and positively associated with cardiovascular disease as reported by Cheng et al, in an analysis also using the Framingham dataset11.
Our study has several strengths including a well-validated metabolomics platform, detailed covariate information, a large number of carefully adjudicated endpoints, and a robust methodology. In addition, the study provides a detailed look at metabolomics and CHD in women, which has previously been lacking. Our results did replicate in the similarly aged women of the PREDIMED study, where the mean BMI was similar. While the generalizability beyond these postmenopausal women remains to be seen, these women are representative of the current population BMI and, importantly, represent a group of women at high risk of CHD. Additional data will also be needed to identify whether these results are specific to women or menopausal status or whether there is a smaller effect in men, as seen in the PREDIMED data.
Conclusion
In two separate sets of post-menopausal women, multiple metabolites associated with CHD risk were identified and validated. These metabolites included novel metabolites, such as hydroxy-PCs and HETEs, as well as glutamate and glutamine, which have been consistently identified in other populations. The strongest marker, C34:2 hydroxy-PC, further replicated in a third set of women and men, with a suggestion of an interaction by sex. Overall, the strong association of oxidized phospholipids (hydroxy-PCs and HETEs) with CHD supports additional investigation of the mechanism and potential clinical use of oxidative risk. Further evaluation of these markers in other populations, as well as more detailed biologic inquiry, is warranted.
Supplementary Material
Clinical Perspective.
What is new?
Using a robust design, we identified and validated 33 metabolites associated with coronary heart disease (CHD) in postmenopausal women, 8 of which remained independently associated after adjustment for standard cardiovascular risk factors.
The risk association for the novel metabolite with the strongest association (C34:2 hydroxy-phosphatidylcholine) is independent of other metabolites, improves the AUC when added to standard risk factors, and replicates in an additional dataset containing both men and women as well as for both CHD and stroke events.
We have identified a cluster of novel metabolites related to oxidative stress, which are not strongly correlated with traditional biomarkers.
What are the clinical implications?
Identified metabolites may be useful as additional biomarkers for clinical risk assessment in addition to highlighting new areas of biologic exploration and potential therapeutic targets.
Metabolite precursors of CHD need to be studied in sufficient samples sizes of both men and women.
Acknowledgments
Sources of Funding
Metabolomic analysis in the WHI was funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contract HHSN268201300008C. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. A list of WHI investigators is available online at https://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf.
Footnotes
Disclosures
None
References
- 1.Shaw LJ, Bairey Merz CN, Pepine CJ, Reis SE, Bittner V, Kelsey SF, Olson M, Johnson BD, Mankad S, Sharaf BL, Rogers WJ, Wessel TR, Arant CB, Pohost GM, Lerman A, Quyyumi AA, Sopko G. Insights From the NHLBI-Sponsored Women’s Ischemia Syndrome Evaluation (WISE) Study: Part I: Gender Differences in Traditional and Novel Risk Factors, Symptom Evaluation, and Gender-Optimized Diagnostic Strategies. Journal of the American College of Cardiology. 2006;47:S4–S20. doi: 10.1016/j.jacc.2005.01.072. http://dx.doi.org/10.1016/j.jacc.2005.01.072. [DOI] [PubMed] [Google Scholar]
- 2.Regensteiner JG, Golden S, Huebschmann AG, Barrett-Connor E, Chang AY, Chyun D, Fox CS, Kim C, Mehta N, Reckelhoff JF. Sex Differences in the Cardiovascular Consequences of Diabetes Mellitus. Circulation. 2015;132:2424–2447. doi: 10.1161/CIR.0000000000000343. [DOI] [PubMed] [Google Scholar]
- 3.Peters SA, Huxley RR, Woodward M. Diabetes as a risk factor for stroke in women compared with men: a systematic review and meta-analysis of 64 cohorts, including 775 385 individuals and 12 539 strokes. The Lancet. 2014;383:1973–1980. doi: 10.1016/S0140-6736(14)60040-4. [DOI] [PubMed] [Google Scholar]
- 4.Würtz P, Havulinna AS, Soininen P, Tynkkynen T, Prieto-Merino D, Tillin T, Ghorbani A, Artati A, Wang Q, Tiainen M. Metabolite profiling and cardiovascular event risk: a prospective study of three population-based cohorts. Circulation. 2015;131:774–785. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y-M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaarhorst AA, Verhoeven A, Weller CM, Böhringer S, Göraler S, Meissner A, Deelder AM, Henneman P, Gorgels AP, van den Brandt PA. A metabolomic profile is associated with the risk of incident coronary heart disease. American heart journal. 2014;168:45–52. e7. doi: 10.1016/j.ahj.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Stegemann C, Drozdov I, Shalhoub J, Humphries J, Ladroue C, Didangelos A, Baumert M, Allen M, Davies AH, Monaco C. Comparative lipidomics profiling of human atherosclerotic plaques. Circulation: Cardiovascular Genetics. 2011;4:232–242. doi: 10.1161/CIRCGENETICS.110.959098. [DOI] [PubMed] [Google Scholar]
- 8.Shah SH, Sun J-L, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, Haynes C, Hauser ER, Kraus WE, Granger CB. Baseline metabolomic profiles predict cardiovascular events in patients at risk for coronary artery disease. American heart journal. 2012;163:844–850. e1. doi: 10.1016/j.ahj.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, Dungan J, Newby LK, Hauser ER, Ginsburg GS. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circulation: Cardiovascular Genetics. 2010 doi: 10.1161/CIRCGENETICS.109.852814. https://doi.org/10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed]
- 10.Ganna A, Salihovic S, Sundström J, Broeckling CD, Hedman ÅK, Magnusson PK, Pedersen NL, Larsson A, Siegbahn A, Zilmer M. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014;10:e1004801. doi: 10.1371/journal.pgen.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng S, Larson MG, McCabe EL, Murabito JM, Rhee EP, Ho JE, Jacques PF, Ghorbani A, Magnusson M, Souza AL, Deik AA, Pierce KA, Bullock K, O'Donnell CJ, Melander O, Clish CB, Vasan RS, Gerszten RE, Wang TJ. Distinct metabolomic signatures are associated with longevity in humans. Nature communications. 2015;6:6791. doi: 10.1038/ncomms7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nature medicine. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnusson M, Lewis GD, Ericson U, Orho-Melander M, Hedblad B, Engström G, Östling G, Clish C, Wang TJ, Gerszten RE. A diabetes-predictive amino acid score and future cardiovascular disease. European heart journal. 2013;34:1982–1989. doi: 10.1093/eurheartj/ehs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumsiek J, Mittelstrass K, Do KT, Stückler F, Ried J, Adamski J, Peters A, Illig T, Kronenberg F, Friedrich N, Nauck M, Pietzner M, Mook-Kanamori DO, Suhre K, Gieger C, Grallert H, Theis FJ, Kastenmüller G. Gender-specific pathway differences in the human serum metabolome. Metabolomics. 2015;11:1815–1833. doi: 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C, Roemisch-Margl W, Polonikov A, Peters A, Theis FJ. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011;7:e1002215. doi: 10.1371/journal.pgen.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auro K, Joensuu A, Fischer K, Kettunen J, Salo P, Mattsson H, Niironen M, Kaprio J, Eriksson JG, Lehtimäki T. A metabolic view on menopause and ageing. Nature communications. 2014;5:4708. doi: 10.1038/ncomms5708. [DOI] [PubMed] [Google Scholar]
- 17.The Women’s Health Initiative Study Writing Group. Design of the women’s health initiative clinical trial and observational study. Controlled clinical trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 18.Pradhan AD, Manson JE, Rossouw JE, Siscovick DS, Mouton CP, Rifai N, Wallace RB, Jackson RD, Pettinger MB, Ridker PM. Inflammatory biomarkers, hormone replacement therapy, and incident coronary heart disease: prospective analysis from the Women's Health Initiative observational study. Jama. 2002;288:980–987. doi: 10.1001/jama.288.8.980. [DOI] [PubMed] [Google Scholar]
- 19.Writing Group for the Women's Health Initiative Investigators. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women's Health Initiative randomized controlled trial. Jama. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 20.Estruch R, Ros E, Martinez-Gonzalez M. Mediterranean diet for primary prevention of cardiovascular disease. N Engl J Med. 2013;369:676–677. doi: 10.1056/NEJMc1306659. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-González MÁ, Corella D, Salas-Salvadó J, Ros E, Covas MI, Fiol M, Wärnberg J, Arós F, Ruíz-Gutiérrez V, Lamuela-Raventós RM. Cohort profile: design and methods of the PREDIMED study. International journal of epidemiology. 2012;41:377–385. doi: 10.1093/ije/dyq250. [DOI] [PubMed] [Google Scholar]
- 22.Rexrode K. DbGap. National Library of Medicine; Metabolomics of Coronary Heart Disease (CHD) in the WHI. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001334.v1.p3. [Google Scholar]
- 23.Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 24.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. [Google Scholar]
- 25.Pearce N. Analysis of matched case-control studies. BMJ. 2016;352:i969. doi: 10.1136/bmj.i969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian J, Payabvash S, Kemmling A, Lev MH, Schwamm LH, Betensky RA. Variable selection and prediction using a nested, matched case-control study: Application to hospital acquired pneumonia in stroke patients. Biometrics. 2014;70:153–163. doi: 10.1111/biom.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh SF, Vickery TW, Serhan CN. Chiral lipidomics of E-series resolvins: aspirin and the biosynthesis of novel mediators. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2011;1811:737–747. doi: 10.1016/j.bbalip.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. Journal of clinical epidemiology. 1999;52:1165–1172. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 29.Langholz B, Jiao J. Computational methods for case-cohort studies. Computational Statistics & Data Analysis. 2007;51:3737–3748. [Google Scholar]
- 30.Oni-Orisan A, Edin ML, Lee JA, Wells MA, Christensen ES, Vendrov KC, Lih FB, Tomer KB, Bai X, Taylor JM. Cytochrome P450-derived epoxyeicosatrienoic acids and coronary artery disease in humans: a targeted metabolomics study. Journal of lipid research. 2016;57:109–119. doi: 10.1194/jlr.M061697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Birukov KG, Romanoski CE, Springstead JR, Lusis AJ, Berliner JA. Role of phospholipid oxidation products in atherosclerosis. Circulation research. 2012;111:778–799. doi: 10.1161/CIRCRESAHA.111.256859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gómez M, Vila J, Elosua R, Molina L, Bruguera J, Sala J, Masià R, Covas MI, Marrugat J, Fitó M. Relationship of lipid oxidation with subclinical atherosclerosis and 10-year coronary events in general population. Atherosclerosis. 2014;232:134–140. doi: 10.1016/j.atherosclerosis.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Koenig W, Karakas M, Zierer A, Herder C, Baumert J, Meisinger C, Thorand B. Oxidized LDL and the risk of coronary heart disease: results from the MONICA/KORA Augsburg Study. Clinical Chemistry. 2011;57:1196–1200. doi: 10.1373/clinchem.2011.165134. [DOI] [PubMed] [Google Scholar]
- 34.Linna M, Ahotupa M, Löppönen MK, Irjala K, Vasankari T. Circulating oxidised LDL lipids, when proportioned to HDL-c, emerged as a risk factor of all-cause mortality in a population-based survival study. Age and ageing. 2013;42:110–113. doi: 10.1093/ageing/afs074. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Chen B, Chen T, Guo S, Xue X, Chen Q, Zhao M, Xia L, Zhu Z, Zheng L, Yin H. Comprehensive metabolomics identified lipid peroxidation as a prominent feature in human plasma of patients with coronary heart diseases. Redox Biology. 2017;12:899–907. doi: 10.1016/j.redox.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alshehry ZH, Mundra PA, Barlow CK, Mellett NA, Wong G, McConville MJ, Simes J, Tonkin AM, Sullivan DR, Barnes EH. Plasma lipidomic profiles improve upon traditional risk factors for the prediction of cardiovascular events in type 2 diabetes. Circulation. 2016 doi: 10.1161/CIRCULATIONAHA.116.023233. https://doi.org/10.1161/CIRCULATIONAHA.116.023233. [DOI] [PubMed]
- 37.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, Deik AA, Magnusson M, Fox CS, O'Donnell CJ, Vasan RS, Melander O, Clish CB, Gerszten RE, Wang TJ. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125:2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi L, Qi Q, Prudente S, Mendonca C, Andreozzi F, di Pietro N, Sturma M, Novelli V, Mannino GC, Formoso G. Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA. 2013;310:821–828. doi: 10.1001/jama.2013.276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz-Canela M, Toledo E, Clish CB, Hruby A, Liang L, Salas-Salvadó J, Razquin C, Corella D, Estruch R, Ros E. Plasma branched-chain amino acids and incident cardiovascular disease in the PREDIMED trial. Clinical chemistry. 2016;62:582–592. doi: 10.1373/clinchem.2015.251710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.