Abstract
Species with a large geographic distributions present a challenge for phylogeographic studies due to logistic difficulties of obtaining adequate sampling. For instance, in most species with a Holarctic distribution, the majority of studies has concentrated on the European or North American part of the distribution, with the Eastern Palearctic region being notably understudied. Here, we study the phylogeography of the freshwater cladoceran Daphnia magna Straus, 1820 (Crustacea: Cladocera), based on partial mitochondrial COI sequences and using specimens from populations spread longitudinally from westernmost Europe to easternmost Asia, with many samples from previously strongly understudied regions in Siberia and Eastern Asia. The results confirm the previously suspected deep split between Eastern and Western mitochondrial haplotype super-clades. We find a narrow contact zone between these two super-clades in the eastern part of Western Siberia, with proven co-occurrence in a single lake in the Novosibirsk region. However, at present there is no evidence suggesting that the two mitochondrial super-clades represent cryptic species. Rather, they may be explained by secondary contact after expansion from different refugia. Interestingly, Central Siberia has previously been found to be an important contact zone also in other cladoceran species, and may thus be a crucial area for understanding the Eurasian phylogeography of freshwater invertebrates. Together, our study provides an unprecedented complete, while still not global, picture of the phylogeography of this important model species.
Introduction
Planktonic water fleas (Crustacea: Cladocera) are attractive models for phylogeographic investigations [1–4]. Indeed, planktonic samples usually contain many (tens, hundreds or even thousands) of specimen of several cladoceran taxa. Moreover, freshwater animals are highly suitable for phylogeographic studies because they have well-defined populations in water bodies separated by unsuitable habitat [5].
Many cladocerans are thought to be distributed across the entire Holarctic region, with populations being common throughout the range, which would potentially make them excellent models for Pan-Holarctic phylogeographic studies. However, our conclusions on their large distribution ranges may be too preliminary, because each taxon may in fact consist of several “cryptic” species with more local distribution ranges [6–12]. One major difficulty for phylogeographic studies of potentially Pan-Holarctic species is the need to cover large territories by adequate sampling. For instance, clustered sampling of species with uniform isolation by distance may lead to erroneous identification of genetic clusters, which may falsely be interpreted as cryptic species [13].
Most previous phylogeographic studies of cladocerans concentrated on North America [1] and Europe [14–16]. More recently, several studies on the Asian regions were published [17–20]. However, North-Eastern Eurasia is still strongly underrepresented, even in studies that otherwise cover a large geographic region [21–28]. This situation is likely explained by the logistic problems with access to different parts of Siberia, a huge territory with almost no human infrastructure. During the last decade, our team has conducted a special sampling program in this region (e.g., [29,30]). The genetic analysis of these samples has revealed phylogeographic patterns specific to Siberia and the northern half of Asia [31–33]. For two taxa (Moina and Chydorus), we demonstrated the existence of two faunistic super-complexes: (1) a European-Western Siberian super-complex and (2) a Eastern Siberian-Far Eastern super-complex, with a transition zone in the Yenisey basin, approximately near the boundary between Western and Eastern Siberia [31]. We also identified a special role of a Beringian region (including the Chukot Area of Russia, Kamchatka Peninsula, Alaska and surrounding territories) as a center of dispersion of cladocerans in Eurasia [32]. The generality of these patterns needs further verification, based on analyses of other taxa, as the identification of general patterns has the potential to strongly improve our understanding of the history of the whole freshwater fauna of Northern Eurasia.
Among numerous taxa of the cladocerans, one particular species, Daphnia (Ctenodaphnia) magna Straus, 1820, is the object of hundreds of scientific publications per year. Besides being used for evolutionary, ecological, and physiological studies [34,35], it is also a popular model for toxicological studies [36,37], and is used in aquaculture, mainly as food for aquarium fish and for juveniles of commercial fish species. Earlier studies on the phylogeography of D. magna have concentrated on Europe [14,38,39] and Turkey [20]. But D. magna is a common species also in the Middle East, Central Asia, several regions of Western and Eastern Siberia, China, and Japan (though the latter could be a dubious record, D. Ebert, personal communication), and is also found in North Africa, South Africa, and North America [40,41,9]. Several strongly divergent COI sequences have been reported from some of these regions [42,43], though it is unknown whether the strong divergence is explained by the existence of two or more highly divergent groups within the taxon or due to limited sampling of localities with intermediate haplotypes.
The aim of our study is to address these taxonomic, biogeographic, and phylogenetic knowledge gaps for Daphnia magna in the Palaearctic, with special reference to the Eastern Palearctic region. Through concentrating on this region we also aimed at increasing our knowledge of potentially general phylogeographic patterns among cladocerans (and perhaps other freshwater-inhabiting taxa) in the Palaearctic. We use partial COI sequences of a large number of specimens from many populations spread across the Northern part of the region, with fewer specimens also from other parts of the region (North Africa, Middle East, Caucasus) and from North America. These samples are complemented with already published data on specimens of known origin.
Material and methods
Field collection
We collected a total of 174 specimens from 67 populations of Daphnia magna, three specimens from a single population of D. similis, and 17 specimens from 14 populations of D. sinensis (S1 Table). The latter two species also belong to the subgenus Ctenodaphnia and are used as outgroup species. Specimens were collected with plankton nets (diameter: 20–40 cm, mesh size: 30–50 μm) or rectangular dip nets (handle length: 0.5–2 m, width: 0.2–0.3 m, mesh size: 30–50 μm) in different types of freshwater habitats and preserved in 90–96% ethanol. Before DNA extraction, each specimen was preliminarily identified to species level (D. magna or other species of the subgenus Ctenodaphnia), based on its morphology according to existing identification keys [9,33,41].
DNA sequencing
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions, and fragments were amplified with primers listed in Table 1. Note that, in addition to COI, we amplified fragments of an additional mitochondrial and three nuclear genes (Table 1) in a subset of samples. However, the results on these additional fragments were inconclusive due to insufficient resolution and sample size. We therefore present them only in S1 Text.
Table 1. Primers used to amplify mitochondrial and nuclear fragments used in this study.
| Gene fragment | Primers |
|---|---|
| 5’ region of the mitochondrial cytochrome c oxidase subunit I (COI)gene | jgLCO1490+jgHCO1490 [44], Dm_FL+RL [45], ZooplF+R [42], COI-F+R [43] |
| 5’ region of the mitochondrial rDNA 16S gene | 16Sin-F + 16Sin-R [17] |
| 5’ region of the nuclear rDNA 18S gene | 18a1 +700R [46] |
| 5’ region of the nuclear heat shock protein 90 kDa gene | HSP-90_F+R [47] |
| Region of the nuclear histone H-3 gene | H3F+R [48] |
Polymerase chain reactions (PCR) were carried out in a total volume of 25 μl, consisting of 2 μl of genomic DNA, 8.5 μl of bi-distilled water, 1 μl of each primer (10 mM) and 5 μl PCR 5x Taq ScreenMix-HS (Evrogen, Moscow, Russia). The PCR conditions for the amplification followed those described in the papers mentioned in Table 1. The PCR products were electrophoresed together with a 0.1–3 kb DNA ladder (SibEnzyme, Novosibirsk, Russia) on a 1.5% agarose gel stained with ethidium bromide and visualized under UV light. The obtained PCR products were reprecipitated at room temperature with addition of ethanol (final concentration 70%) and ammonium acetate (final concentration 125 mM). The DNA precipitate was washed with 70% ethanol, dried, and dissolved in bi-distilled water. About 0.3 pmol of the PCR product and 3.2 pmol of the relevant primer were used for Sanger sequencing. Each PCR product was sequenced bi-directionally on an ABI 3730 DNA Analyzer with the ABI PRISM BigDye Terminator v. 3.1 sequencing kit (Applied Biosystems, USA). A single consensus sequence was assembled based on the forward and reverse sequences using CodonCode Aligner v. 6.0.2 (CodonCode Corp, USA). DNA sequences were submitted to the NCBI GenBank database (S1 Table).
Phylogenetic analyses
The authenticity of the sequences was verified by BLAST comparisons with published D. magna sequences [49]. COI Sequence JF821194 from Turkey was excluded from the analysis because a preliminary tree suggested that this sequence belongs to a separate clade, which, however, could be an artifact and needs further confirmation. Sequences were edited and assembled in uGene v.1.26 [50]. The original sequences of the present study (S1 Table) and sequences from previous publications [51,52,53,54,55,56,57,58,59,60] deposited to the GenBank (S2 Table) were used for multi-sequence alignments. The sequences were first automatically aligned using the T-Coffee algorithm [61] with default options of the uGene package. Due to the strong variability of the COI flanking regions in Crustacea, no universal primers for barcoding exist, in contrast, for example, to fishes [62]. As a consequence, sequences from GenBank had been amplified with varying primers, explaining the variation in fragment lengths. Hence, all COI sequences were cropped to the minimal overlapping length of 563 bp.
To analyze the genetic variation among samples, we estimated the following parameters for each gene fragment and for each species separately: number of haplotypes (Nh), number of variable (polymorphic) sites (Nv), number of parsimony informative sites (Np), haplotype diversity (Hd), nucleotide diversity (Pi), and average number of nucleotide differences (k) [63,64]. We also carried out neutrality tests and assessed mismatch distributions. All analyses were performed in DnaSP v.5.1 [65] and MEGA v.7 [66].
The best-fitting model of nucleotide substitution was selected using the ModelFinder web application [67], based on likelihood scores for 154 different models and the Bayesian information criterion [68]. Within- and among-clade distances were calculated in MEGA using p-distance [69]. Phylogenetic trees were constructed using D. similis and D. sinensis as outgroup species. A maximum likelihood (ML) phylogenetic reconstruction was performed using the IQ-TREE web server [70] and ultrafast bootstrap [71] resampled 10000 times. Maximum parsimony (MP) analyses were performed in PAUP*4.0a152 [72]. A heuristic MP searches was done using equal weighting, 10 random sequence addition replicates and TBR branch swapping. Non-parametric bootstrapping was performed to assess the nodal support, using 1000 pseudoreplicates for MP. Bayesian analyses (BI) were performed in BEAST v2.4.6 [73]. Six independent Markov chain Monte Carlo (MCMC) analyses were run simultaneously for 50 million generations and sampled every 1000 generations. The first 50% of the generations were discarded as burn-in. A 50% majority rule consensus tree was generated from the remaining trees, and the posterior probability of each node was estimated as the percentage of trees recovering any particular node. MP and BI analyses were performed on the computer cluster CIPRES Science Gateway v.3.3 [74].
To establish whether there was any evidence for cryptic species, we performed a Bayesian generalized mixed Yule coalescent (bGMYC) analysis [75] with a threshold of 0.5, using the last 100 trees of the BEAST MCMC file and the GMYC-web server [76]. Furthermore, we used our haplotypes to construct a network in the program PopART [77] to show relationships among the individuals sampled from different locations. The TCS algorithm was selected for this network, based on the implemented statistical parsimony [78]. Second, the Network package (version 5, http://www.fluxus-technology.com) was used to construct a median-joining haplotype network with Steiner maximum parsimony post-processing [79], putting equal weight on each variable nucleotide site. Before constructing this network, data were processed using the star contraction algorithm [80] to reduce of number of similar haplotypes and hence to reduce the complexity of the network. As an additional test for presence of distinct clades, we used the program ABGD [81] to assess p-distances [69] with 100 replicate steps in this reduced-complexity network.
The reduced-complexity network was also used for a nested clade analysis, which identifies significant phylogroups, and to test for historical demographic processes within the major phylogroups. The nested clade phylogeographic analysis (NCPA) was performed in the program ANeCA v.1.2 [82] with integrated module GeoDis v.2.6 for calculation based on a new approach to the interpretation of biological processes according to [83]. Subsequently, to test for neutrality within individual clades, Tajima’s D tests [84] and Fu’s FS tests [85] were performed in Arlequin v.3.5 [86] with 1000 permutations. We also performed R2-tests [87] in DnaSP, again with 1000 permutations. Mismatch distributions were investigated for some groups to evaluate a model of exponential population growth [88]. For the latter, a goodness of fit test was performed, using a parametric bootstrap approach based on the sum of squared deviations (SSD) between the observed and simulated mismatch distributions [89]. The demographic parameter Tau was estimated using a generalized nonlinear least square approach, and the confidence interval of this parameter was computed using parametric bootstrap with 1000 replicates in Arlequin.
Divergence times were estimated using a relaxed molecular clock approach with uncorrelated lognormal distributions of branch rates in the program BEST v.1.6.1, following methods and calibrations of [90]. A second variant of this analysis was done using the additional calibration point of [91] for the Dapnia/Ctenodaphnia split (145 MYA), which suggests that the speed of nucleotide substitutions is significantly lower than suggested by [90].
Results
Genetic diversity
We obtained 174 original COI sequences of three Daphnia species (D. magna, D. similis, D. sinensis) from different regions of the Palaearctic (S1 Table, Fig 1). They were analyzed together with 609 COI sequences obtained from Genbank (S2 Table). As we found that these haplotypes belong to two strongly divergent clades (“super- clades” A and B, see below), we analyzed genetic diversity in the whole sample as well as within each of the two super-clades (S3 Table) and carried out neutrality tests and assessed mismatch distributions also for each super-clade separately (S4 Table).
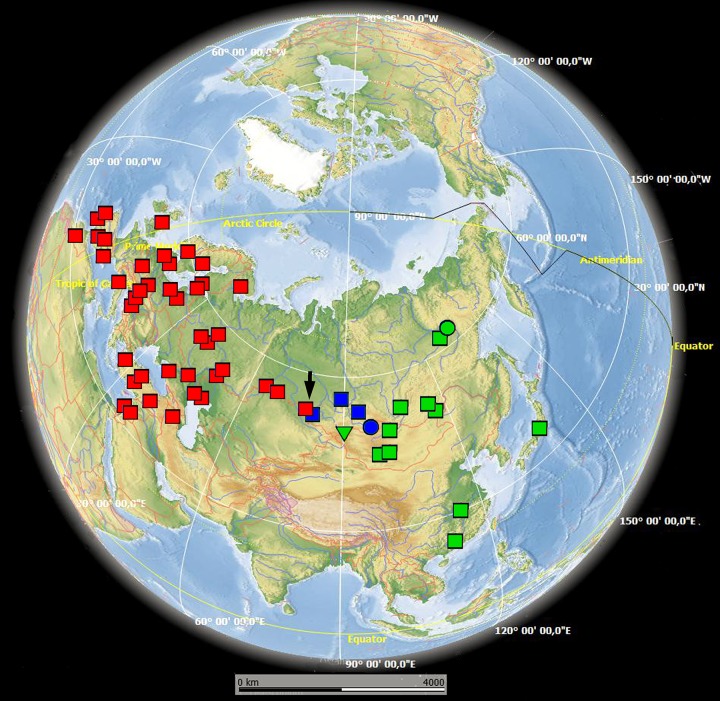
Fig 1. Sampling sites and distribution of major COI haplotype clades for Eurasian accessions of Daphnia magna (both original and sequences retrieved from GenBank).
Colors and symbols correspond to those used in subsequent figures. The base map was obtained from the open domain plain map available at https://marble.kde.org/.
The genetic diversity at COI was high in the entire sample as well as in super-clades A and B. Haplotypic diversity was higher in super-clade A (mainly European populations) than in super-clade B (mainly Asian populations), possibly explained by the wider geographic distribution of super-clade A. However, nucleotide diversity and the number of polymorphic sites were almost identical between the two clades (S3 and S4 Tables). Within- and among-clade distances are represented in S5 Table. The samples apparently originated from the laboratory cultures are listed in S6 Table.
COI phylogenetic tree
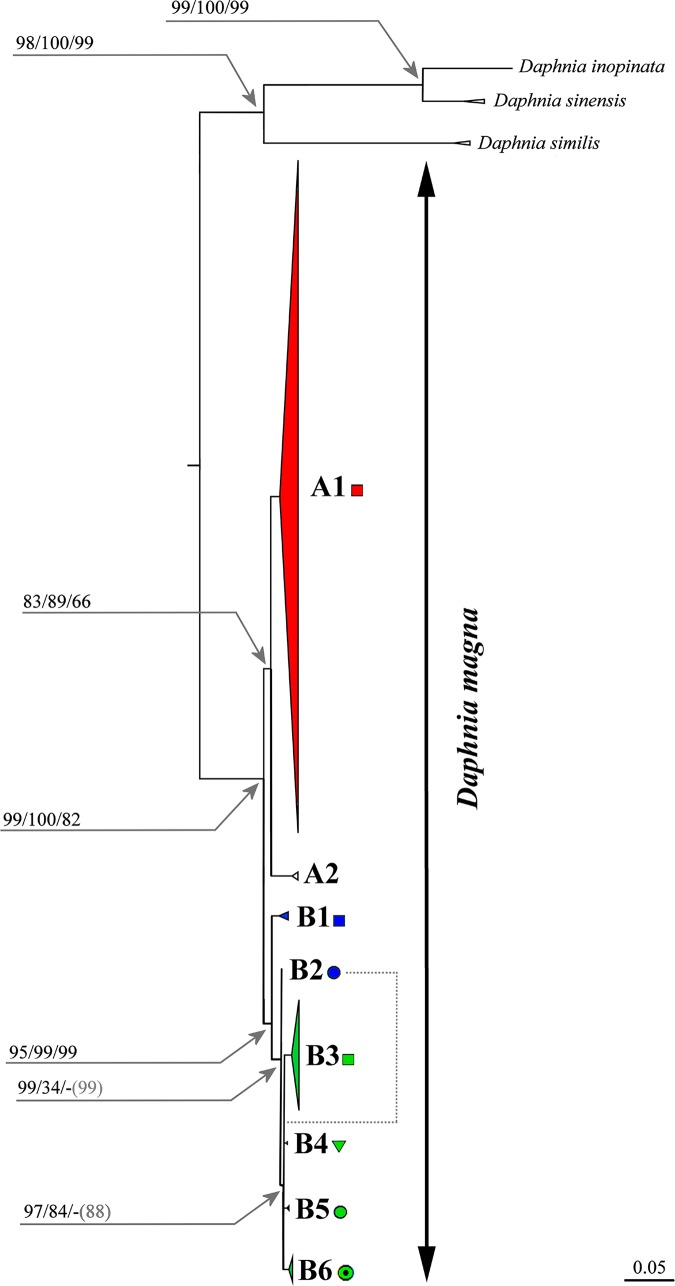
The best-fit model [92] for the whole COI data set (our sequences and GenBank sequences combined) was HKY with four categories of gamma (+G4, shape alpha 1.61) and a proportion of invariable sites (+I, 0.602) with a relative BIC weight of 0.3648. Based on the cumulative relative weight, two other models were also within the 95% confidence interval. One best ML tree was found, with an overall likelihood score of -ln L = -2775.1574. These analyses revealed the existence of two highly divergent and well-supported main super-clades within Dapnia magna (clades A and B, Fig 2). Each of these two clades was subdivided into several sub-clades Fig 2). Furthermore, the outgroup species formed three clades of the D. similis group, which were well separated from D. magna, as already found in previous studies [23,33]. Trees constructed with different methods were congruent with this finding (with a single exception, Fig 2), but the clade support differed among analyses (Fig 2). All deep branches had a strong support in ML and BI. In general, the clades that have a strong support in ML had a weaker support in BI.
Fig 2. Maximum likelihood tree based on all analyzed COI.
The support values of individual nodes are based on: Maximum likelihood (ML) / Maximum Parsimony (MP) / Bayesian Inference (BI). Dotted line indicates an incongruence between best-supported topologies using the three methods, with grey numbers indicating BI support for an alternative BI tree topology.
The super clade A includes clade A1, which was found in samples from Europe, the Mediterranean region (including North Africa), the Middle East, Turkey and Caucasus, as well as in a few populations in Western Siberia. Within this clade, there are several sequences from the Genbank labeled as originating from North America (Mexico, USA, Canada), but in reality most of these may have originated from laboratory cultures, presumable of European origin (S6 Table). Few others are very likely cases of recent human-mediated invasions (see Discussion). The American individuals with A1 haplotypes were therefore excluded from further analyses and are not represented on any of the figures, except for proven laboratory clones, which are included in S4 Table. The super clade also includes clade A2, which is present exclusively in samples from North America. Unfortunately, there is no exact information on the sampling localities of the sequences deposited to the GenBank, only stated “Manitoba, Canada” [51], and only names of sampling sites (“Round Lake” and “Sue”) with unclear exact geographic locations are provided in [14]. We hence decided to omit all North American samples from the map (Fig 1).
The super-clade B includes several Asian and North American lineages and co-occurs with clade A1 in a single lake in the Novosibirsk Area (Western Siberia) (Fig 1, black arrow). No occurrences of clade A1 were found to the East of this location, nor of super-clade B to the West of this location. Clade B1 is relatively locally distributed in the southern parts of Asian Russia, at the boundary between Western (Ob' basin) and Eastern (Enisey Basin) Siberia. Clade B2 is found in a single water body in North Mongolia. Clade B3 is widely distributed in Eastern Siberia, Japan, and East China. Clade B4 is found in a single water body in Ob' basin (easternmost Western Siberia). Clade B5 is found only in Central Yakutia (Eastern Siberia), and clade B6 is present in Arctic Canada only.
COI median haplotype network
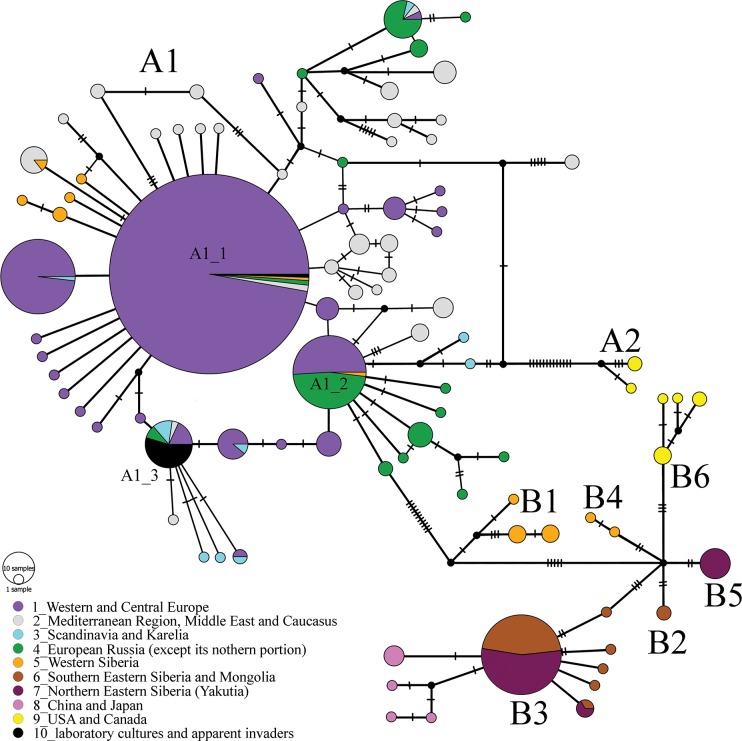
The TCS network consists of the 93 different COI haplotypes (Fig 3). Each of the clades identified in the phylogenetic trees (see above) is well-recognizable in this network.
Fig 3. Median-joining COI haplotype network.
Median vectors are indicated by small black circles.
Super-clade A is again clearly subdivided into a diverse clade A1 and a distant clade A2 (North American samples). Within the A1 clade, we found several sub-networks with a star-like topology, suggesting recent population expansion. The central haplotype of sub-network A1_1 (Fig 3) was found in samples from many European countries, the Mediterranean region, European Russia, and a single population in Western Siberia. The central haplotype of sub-network A1_2 had a similar geographic origin as the major haplotype of A1_1, but was more common in European Russia and was not found in the Mediterranean region nor the Middle East. The central haplotype of sub-network A1_3 again had a similar geographic distribution, but was particularly common among samples from the Middle East.
Among the super-clade B, the only clade B3 showed a star-like topology, with its central haplotype being found in samples from Mongolia, China, and several areas of Eastern Siberia (Yakutia, Zabaikalsky Territory, Irkutsk Area), but not penetrating to the west of Lake Baikal. The clade B1 is separated from others by five mutations. All other clades (B2, B3, B4, B5, B6) form a remarkable monophyletic group interconnected through a hypothetical (extinct or un-sampled) haplotype. Tajima's D and mismatch distribution analyses (S4 Table) were consistent with the hypothesis of recent population expansion for all these clades, except B6.
Nested clade phylogeographic analysis
The maximum parsimony network of the major COI clades of D. magna (S1 Fig) shows the two main clades of the highest rank, corresponding to super-clades A and B, to be clearly separated, with demographic processes (sensu Templeton 2004) of the long-distance colonization type (S4 Table). In the European-Western Siberian clade A1, we can see a complicated network with several equally probable topologies. Separation of the clade B6 from North America is likely explained by isolation by distance (S4 Table).
bGMYC
The results of the bMYGC analysis (S2 Fig) provide no strong support for separate species (i.e., no populations or groups of populations from geographically close localities with posterior probabilities of conspecifity of < 0.05). However, the deep branches are well-supported, especially between super-clades A and B, which have a posterior probability of conspecifity of less than 0.1 (though > 0.05). Within the super-clade A, the European-Western Siberian clade A1 and the North American clade A2 are differentiated. The internal structure of the super-clade B is more complicated: it contains at least three sub-groups: A Western Siberian group (s34, 35, 36) corresponding to clade B1 of the TCS network, an Eastern Siberian-Far Eastern group (s44, 45, 46) corresponding to clades B2-B5 of the network, and a Canadian group (s40, 41, 42) corresponding to clade B6. Therefore, this analysis confirms the complexity of the D. magna phylogeographic structure, especially in the Asian Palaearctic region. Furthermore, the high genetic diversity of D. magna is confirmed by the p-distances, which range up to >5%. At the same time, outgroup species differ from any clades within D. magna by 14–17%.
ABGD test
The ABDG test based on haplotype groups (s_numbers) according to the star contraction algorithm [80] revealed the following large groups within D. magna: Group 1 –Eastern Siberian clade B3 (s44, 45, 46), Group 2 –Eastern Siberian clades B2, B4, B5 (s37, 38, 39, 43), Group 3—Western Siberian clade B1 (s34, 35, 36), Group 4 –Canadian clade B6 (s40, 41, 42), Group 5 –North American clade A2 (s17, 18), and Group 6 –European clade A1.
Divergence time estimates
The divergence time estimates differed strongly, depending on the method used. Following the procedures and calibrations of [90], we obtained divergence time estimates between clades A and B of 2.2–2.8 MYA, 2.5 MY between clades A1 and A2, 2.6 MY between the Canadian B6 and other B-subclades, and 10–11 MY between D. magna and the outgroup species. However, using the additional calibration of the sub-genera Ctenodaphnia / Daphnia split, which was estimated at 145 MYA [91], resulted in almost ten times larger divergence time estimates, 17–20 MY between clades A and B, 15 MY between A1 and A2, 22 MY between B6 and the other B-subclades, and 100–120 MY between D. magna and other Ctenodaphnia species (S5 Table).
Results on one additional mitochondrial and three nuclear gene fragments
As mentioned above, the results on the other gene fragments were inconclusive and therefore only presented in S1 and S3 Tables, S1 Text and S3 and S4 Figs.
Discussion
Phylogeography: General patterns
Our analyses consistently and unambiguously identified a deep split between Eastern and Western mitochondrial haplotypes of D. magna in Eurasia, in particular between the European-Middle Eastern-Western Siberian = Western Eurasian clade (A1) and the Eastern Siberian-Far Eastern-Chinese-"Beringian" = East Asian ones (B1-B5). Sampling locations were distributed almost evenly from Western Europe to Easternmost Asia, and the transition between the two groups appears to be rather abrupt. The network structure suggests that this suture zone (sensu [93–95]) is a secondary contact zone between the two super-clades. The width of this zone and its North-South extension cannot currently be assessed due to a lack of available samples. However, the lake in which the two super-clades are found to co-occur is located in the Ob' River basin, in the Easternmost part of Western Siberia, somewhat west to the transition zone between two super-groups of haplotypes previously identified in other cladocerans [31,32]. Regardless, our findings provide strong additional evidence for a marked longitudinal (East-West) differentiation of the cladoceran fauna in Northern Eurasia.
Phylogeography: Local patterns of mitochondrial haplotype distributions
On a more local scale, we find little evidence for a strong geographic structure of mitochondrial haplotypes within the Western A1 clade, consistent with previous studies [14]. The main A1 subclades show a wide geographic distribution, and co-occur in different regions, though at different frequencies. Together with the evidence for relatively recent population expansion, a possible scenario is expansion from several refugia and subsequent mixing through migration. The current data do not allow us to identify the location of these refugia, though the different frequencies of the subclades in different regions may provide some indications. Haplotype diversity is especially high in the Mediterranean region and the Middle East and involves many peripheral haplotypes in the star-like network. These might be relicts of a pre-Pleistocene diversity of the clade A1, with only a subset of this diversity being involved in recent population expansion.
The individual clades within super-clade B are more distinct, though the existence of intermediate haplotypes in non-sampled populations remains a realistic possibility. Several clades (B1, B2, B4, B5) were found only in a small region or even only at a single location, either at the boundary between Eastern and Western Siberia (B1, B2, B4) or in Northeastern Siberia (B5). All aforementioned B clades may be relicts of a previous, hypothetical "pan-Beringian" population (East Asia plus North America, with no evidence for contemporary occurrence of D. magna in easternmost Siberia nor westernmost North America). Interestingly, the clade B1, which is the clade that occurs in the contact zone between A and B super-clades, has an intermediate position between the two super-clades in the haplotype network (though it clearly belongs to super-clade B, and hence has a more recent common ancestor with other B clades than with A clades). The interpretation of this observation needs further investigation, but again highlights the special interest of the boundary between Western and Eastern Siberia.
The super clade B also contains the more widespread clade B3, distributed across large parts of Eastern Siberia, Northeastern Siberia, as well as the Far East, with evidence for recent population expansion. The fact that haplotypes from China and Japan do not belong to the central group of the B3 clade, but are offshoots of it, suggests that these more southern regions may have been colonized from the north (Eastern Siberia, Mongolia), in contrast to the more common northward expansion that is assumed for most postglacial colonization events [32].
Two clades A2 and B6 were found exclusively in North America. We will discuss the phylogeography of D. magna in North America only very briefly here, as it is clear that too few samples from that continent were available for our study to reach more detailed conclusions. Furthermore, the precise sampling locations of this material are unclear (see above). Nonetheless, the observation that both super-clades seem to be present in North America, with haplotypes that are clearly distinct from the haplotype clades found in Europe and Asia, suggests the possibility that North America may historically have been colonized from both sides (for evidence of more recent colonization, see below).
We have used two common approaches to separate evolutionary significant groups, the Nested Clade Phylogeographical Analysis [96,83] and a Bayesian approach realized in the GMYC model [75]. The NCPA has been widely used in cladoceran phylogeography [14,27,97], but its use has been criticized [98] and its results must be discussed with a great care. In most cases, separation of the highest rank of clades in NCPA is unproblematic, but the adequateness of lower-rank clades is questionable, despite the reduction of phylogenetic noise with the star contraction algorithm [80]. In our case, the NCPA suggested for almost all groups of D. magna a good fit with the RGF (restricted gene flow) model, which is in agreement with the cladoceran biology (existence of isolated populations with a large number of generations per season). For the high-ranking super-clades (A and B) the finding of correspondence to the RGF model is a useful conclusion, as it suggests long-term (but potentially incomplete) isolation.
The bGMYC, on the other hand, is very sensitive to the mathematical model parameters [83], but apparently gives better results for long-term phylogenetic reconstructions [75]. The considerable COI polymorphism in D. magna allowed us to use this method here, with apparently more adequate conclusions concerning larger clades. In the literature, methods of statistical analysis of phylogeographic patterns appear to be chosen somewhat haphazardly, rather than according to the advantages and limits of each method for a given data set (see [83,98]). Moreover, complicated models are often used, which are too sensitive to their parameters [63]. We therefore think that results of sophisticated methods must be interpreted with a great care, and should be verified with independent data and/or alternative methods. Moreover, simpler methods, such as the separation of evolutionary significant units according to genetic distance [63] may be preferable in some cases, compared to more sophisticated ones [99]. Therefore, the application of algorithms of phylogeographic reconstructions should be accompanied by a critical analysis of the results, taking into account species biology.
No evidence for cryptic species
Many previous genetic studies have advocated the existence of species complexes instead of single species among different species groups of Daphnia (see [9,12]). Although we detected two highly distinct mitochondrial super-clades within Daphnia magna, which may be interpreted as separate taxa (e.g., by the ABGD test), several lines of evidence strongly suggest that they represent two divergent super-clades within a single species. First, the genetic distance between the two super clades is much smaller than between D. similis, D. sinensis, and D. inopinata, which are good, though very closely related biological species with reproductive isolation strongly supported by differences in male morphology [33]. Second, no concordant subdivision was observed in nuclear genes (S1 Text, though this may be due to a lack of sufficient variability of these genes), and, third, and most importantly, individuals from the two super-clades readily interbreed, at least in the laboratory [100]. Therefore, to date there is no evidence of the existence of cryptic species within D. magna.
Invasions
Cladocerans, and particularly some species of Daphnia, are very popular laboratory animals and are often used as food in aquaculture. Moreover, their resting stages are long-lived, which favors dispersal by human activities (e.g., the ephippia of D. magna of super-clade A were found in ballast water [56]). Many COI sequences of D. magna are marked on GenBank as belonging to "Mexican" or "Canadian" populations (S6 Table). However, closer investigation led to the conclusion that these contain many specimens that originated from laboratory cultures in those countries. The genetic variability among specimens with known laboratory origin (“cultures” in S6 Table) is very low. Possibly, all these specimens derive from commercial clones (or perhaps from a single clone) from Aquatic BioSystems Inc., Fort Collins, Colorado, USA, which ships ephippia to customers. Likely, one of these clones was introduced to the lake near Dorset Environmental Science Center, Ontario (as it has the same haplotype as the "cultures"). Therefore we hypothesize that only clades A2 and B6 represent specimens of true North American origin, whereas North American specimens belonging to A1 are likely explained by anthropogenic invasion.
In general, D. magna does not seems to be a species with a strong invasive potential–in contrast to D. lumholtzi, D. cf. pulex, and D. galeata [101–103]. But recent anthropogenic invasions are revealed here, at least of the clade A1 probably in North America. Such anthropogenic invasions could strongly affect phylogeograpic conclusions. For example, it is possible that the distributional ranges of the three central haplotypes in Europe have been strongly enlarged by human activities. Therefore invasions (both detected and undetected ones) complicate phylogeographic analyses of Daphnia and many other organisms, and require a critical interpretation of the obtained results.
Molecular clock and dating of divergence times
The two molecular clock scenarios used yielded strongly different results on the timing of divergence of D. magna from its sister species, as well as of the super-clades and clades within the species. Currently the issue remains unresolved, and its resolution will have to take into account Frey's [6,7] paradigm of the cladoceran biogeography, morphological stasis during the last millions of year, up to ten-fold differences in evolutionary rates between sister clades [104], as well as the possibility of decreasing mutation rates as a function of time [105,106]. In the end, it seems that the question can only be resolved with appropriate palaeontological records, which are, unfortunately, still far too rare for pre-Pleistocene cladocerans [107].
Supporting information
Numbers identify star groups of haplotypes (see S2 Table). Circle size is proportional to the frequency of the haplotypes. Small dark circles indicate unsampled or extinct haplotypes. Geographic regions for star groups are as follows: s1 –Western, Central Europe, Mediterranean region, Middle East; s2 –Western Europe; s3 –Scandinavian Peninsula and Western Europe; s4-s8 –Mediterranean region; s9 –Mediterranean region and Near East; s10 –Western Europe; s11 –European Russia; s12—Near East; s13-s14 –European Russia; s15-s16 –Mediterranean region; s17-s18 –North America; s19 –European Russia s20 –Mediterranean region; s21 –Western Europe; s22 –Western and Central Europe; s23 –Central and Eastern Europe; s24-s25 –Western Europe; s26 –Western Europe and Scandinavian Peninsula; s27 –different regions of Europe and laboratory cultures; s28 –Mediterranean region; s29 –Western and Central Europe; s30 –Scandinavian Peninsula; s31-s33 –European Russia; s34-s36 –Western Siberia; s37 –Eastern Siberia; s38-s39 –Western Siberia; s40-s42 –North America; s43-s45 –Eastern Siberia; s46 –Far East of Asia.
(TIF)
Clades highlighted in red represent the maximum likelihood limits. The colored lines correspond to a sequence-by-sequence matrix, with lines colored according to the posterior probability that the sequences are conspecific, which allows visualizing uncertainty in group limits.
(TIF)
All original samples have prefix zh, while the Genbank samples have no this prefix. GenBank accession numbers for original samples are given in S1 Table, for sequences obtained from GenBank they are: USA–AY921452, Denmark–DQ470575, Belgium–AM490278, China1 –KF993366, China2 –KM244710, China3 –KP296147, China4 –NC_026914, Novosibirsk1 –JN874603, Novosibirsk2 –JN874602, Novosibirsk3 –JN874604, Novosibirsk4 –JN874601.
(TIF)
All original samples have prefix zh, while the Genbank samples do not have this prefix. GenBank accession numbers for original samples are given in S1 Table, for sequences obtained from GenBank they are: USA—DQ845268.
(PDF)
COI clade designations as defined in Fig 2.
(XLS)
(XLS)
n—sample size, Nh—number of haplotypes, Nv—number of variable (polymorphic) sites, Np—number of parsimony informative sites, Hd—haplotype diversity, Pi—nucleotide diversity, k—average number of nucleotide differences.
(DOC)
n—sample size, Nh—number of haplotypes, Nv—number of variable (polymorphic) sites, Np—number of parsimony informative sites, Hd—haplotype diversity, Pi—nucleotide diversity, k—average number of nucleotide differences; R2 population size expansion test and results of Tajima’s D, Fu’s FS and mismatch distributions: tau-parameter, SSD (sum of squares deviation) and Harpending's Raggedness index including associated p-values. Biological processes are (Templeton, 2004): RGF–restricted gene flow; D–dispersal; LDD–long-distance dispersal; IBD–isolation by distance; AF–allopatric fragmentation; PF–past fragmentation; LDC–long-distance colonization.
(DOC)
In diagonal, bold–estimates of average evolutionary divergence over sequence pairs within groups. Below diagonal–estimates of evolutionary divergence over sequence pairs between groups. The presence of n/c in the results denotes cases in which it was not possible to estimate evolutionary distances. Above diagonal–time divergence, in MYA; first digit–“fast clock” from (Schwentner et al., 2013), second digit–“slow clock” from (Kotov & Taylor, 2011).
(DOC)
(DOC)
(DOCX)
Acknowledgments
Field collection in Russia was carried out by our team or by colleagues as part of a governmental project "Ecology and biodiversity of aquatic ecosystems and invasions of alien species" (no. 0109-2014-0008), with governmental permission to collect samples from public property. Sampling in the natural reserves of Russia was conducted with special permissions of their Directors (M.A. Kirgintsev, Belozersky Natural Reserve; Yu.P. Suschitsky, State Khankaisky Biosphere Reserve), we also thank the Administration of these nature reserves for assistance during the sampling. Verbal permission to collect in private farm ponds was obtained from local owners. Mongolian samples were collected by the Joint Russian-Mongolian Complex Biological Expedition with permission of the Ministry of Nature, Environment and Tourism of Mongolia. Some samples from Finland and Germany were provided by colleagues who collected them with permissions linked to their activity as hydrobiologists in governmental institutions of their countries. The study did not involve any endangered or protected species. We are very grateful to A. I. Klimovsky, A. A. Zharov for their assistance with sampling in different regions of Russia. Many thanks to V.A. Artamonova, D.S. Dorofeev, D. Ebert, A.V. Krylov, A.A. Makhrov, F. Marrone, A.V. Tchabovsky, L.E. Savinetskaya, E.I. Zuykova for contributing samples of D. magna and D. cf. similis.
Data Availability
All data are freely available at Genbank (see S1 Table for accession numbers).
Funding Statement
This work was supported by the European Union (Marie Curie Career Integration Grant DAMANMP for CRH) and the Russian Government Program of Competitive Growth of Kazan Federal University (for AAK), and the Russian Foundation for Basic Research (grant 15-29-02-447 Офи-м for TVN). The part concerning biological invasions was funded by the Russian Foundation for Basic Research (project № 17-05-00782-a for DPK), the part concerning nuclear loci was supported by the Russian Science Foundation (grant № 16-14-10173 for TVN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Taylor DJ, Finston TL, Hebert PDN. Biogeography of a widespread freshwater crustacean: Pseudocongruence and cryptic endemism in the North American Daphnia laevis complex. Evolution. 1998;52(6): 1648–1670. doi: 10.1111/j.1558-5646.1998.tb02245.x [DOI] [PubMed] [Google Scholar]
- 2.Weider LJ, Hobaek A, Colbourne JK, Crease TJ, Dufresne F, Hebert PDN. Holarctic phylogeography of an asexual species complex. I. Mitochondrial DNA variation in arctic Daphnia. Evolution. 1999;53(3): 777–792. doi: 10.1111/j.1558-5646.1999.tb05372.x [DOI] [PubMed] [Google Scholar]
- 3.Weider LJ, Hobaek A, Hebert PDN, Crease TJ. Holarctic phylogeography of an asexual species complex. II. Allozymic variation and clonal structure in arctic Daphnia. Molecular ecology. 1999;8(1): 1–13. doi: 10.1046/j.1365-294X.1999.00522.x [Google Scholar]
- 4.Cox AJ, Hebert PDN. Colonization, extinction, and phylogeographic patterning in a freshwater crustacean. Molecular ecology. 2001;10(2): 371–386. doi: 10.1046/j.1365-294X.2001.01188.x [DOI] [PubMed] [Google Scholar]
- 5.Incagnone G, Marrone F, Barone R, Robba L, Naselli-Flores L. How do freshwater organisms cross the “dry ocean”. A review on passive dispersal and colonization processes with a special focus on temporary ponds. Hydrobiologia. 2015;750(1): 103–123. doi: 10.1007/s10750-014-2110-3 [Google Scholar]
- 6.Frey DG. Questions concerning cosmopolitanism in Cladocera. Archiv für Hydrobiologie. 1982;93: 484–502 [Google Scholar]
- 7.Frey DG. The taxonomy and biogeography of the Cladocera. Hydrobiologia. 1987;145(1): 5–17. doi: 10.1007/BF02530260 [Google Scholar]
- 8.Smirnov NN. Cladocera: The Chydorinae and Sayciinae (Chydoridae) of the World Amsterdam: SPB Academic; 1996. 1–197 p. [Google Scholar]
- 9.Benzie JAH. The genus Daphnia (including Daphniopsis): Anomopoda: Daphniidae Ghent: Kenobi Productions; 2005. 1–376 p. [Google Scholar]
- 10.Korovchinsky NM. The Cladocera (Crustacea: Branchiopoda) as a relict group. Zoological Journal of the Linnean Society. 2006;147(1): 109–124. doi: 10.1111/j.1096-3642.2006.00217.x [Google Scholar]
- 11.Van Damme K, Sinev AY, Dumont HJ. Separation of Anthalona gen.n. from Alona Baird, 1843 (Branchiopoda: Cladocera: Anomopoda): Morphology and evolution of scraping stenothermic alonines. Zootaxa. 2011;2875: 1–64 [Google Scholar]
- 12.Kotov AA. A critical review of the current taxonomy of the genus Daphnia O. F. Müller, 1785 (Anomopoda, Cladocera). Zootaxa. 2015;3911(2): 184–200. doi: 10.11646/zootaxa.3911.2.2 [DOI] [PubMed] [Google Scholar]
- 13.Tucker JM, Schwartz MK, Truex RL, Wisely SM, Allendorf FW. Sampling affects the detection of genetic subdivision and conservation implications for fisher in the Sierra Nevada. Conservation Genetics. 2014;15(1): 123–136. doi: 10.1007/s10592-013-0525-4 [Google Scholar]
- 14.DeGelas K, DeMeester L. Phylogeography of Daphnia magna in Europe. Molecular ecology. 2005;14(3): 753–764. doi: 10.1111/j.1365-294X.2004.02434.x [DOI] [PubMed] [Google Scholar]
- 15.Faustova M, Sacherovà V, Svensson J-E, Taylor DJ. Radiation of European Eubosmina (Cladocera) from Bosmina (E.) longispina–concordance of multipopulation molecular data with paleolimnology. Limnology and Oceanography. 2011;56(2): 440–450. doi: 10.4319/lo.2011.56.2.0440 [Google Scholar]
- 16.Hamrová EVA, Krajicek M, Karanovic T, Černý M, Petrusek A. Congruent patterns of lineage diversity in two species complexes of planktonic crustaceans, Daphnia longispina (Cladocera) and Eucyclops serrulatus (Copepoda), in East European mountain lakes. Zoological Journal of the Linnean Society. 2012;166(4): 754–767. doi: 10.1111/j.1096-3642.2012.00864.x [Google Scholar]
- 17.Zuykova EI, Bochkarev NA, Katokhin AV. Identification of the Daphnia species (Crustacea: Cladocera) in the lakes of the Ob and Yenisei River basins: morphological and molecular phylogenetic approaches. Hydrobiologia. 2013;715(1): 135–150. doi: 10.1007/s10750-012-1423-3 [Google Scholar]
- 18.Ma X, Petrusek A, Wolinska J, Gieβler S, Zhong Y, Zhong Y, et al. Diversity of the Daphnia longispina species complex in Chinese lakes. A DNA taxonomy approach. Journal of Plankton Research. 2015;37(1): 56–65. doi: 10.1093/plankt/fbu091 [Google Scholar]
- 19.So M, Ohtsuki H, Makino W, Ishida S, Kumagai H, Yamaki KG, et al. Invasion and molecular evolution of Daphnia pulex in Japan. Limnology and Oceanography. 2015;60(4): 1129–1138. doi: 10.1002/lno.10087 [Google Scholar]
- 20.Özdemir E, Altındağ A, Kandemir İ. Molecular diversity of some species belonging to the genus Daphnia O. F. Müller, 1785 (Crustacea: Cladocera) in Turkey. Mitochondrial DNA. Part A, DNA mapping, sequencing, and analysis. 2017;28(3): 424–433. doi: 10.3109/19401736.2015.1136303 [DOI] [PubMed] [Google Scholar]
- 21.Ishida S, Taylor DJ. Mature habitats associated with genetic divergence despite strong dispersal ability in an arthropod. BMC evolutionary biology. 2007;7: 52 doi: 10.1186/1471-2148-7-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida S, Taylor DJ. Quaternary diversification in a sexual Holarctic zooplankter, Daphnia galeata. Molecular ecology. 2007;16(3): 569–582. doi: 10.1111/j.1365-294X.2006.03160.x [DOI] [PubMed] [Google Scholar]
- 23.Adamowicz SJ, Petrusek A, Colbourne JK, Hebert PDN, Witt JDS. The scale of divergence: A phylogenetic appraisal of intercontinental allopatric speciation in a passively dispersed freshwater zooplankton genus. Molecular phylogenetics and evolution. 2009;50(3): 423–436. doi: 10.1016/j.ympev.2008.11.026 [DOI] [PubMed] [Google Scholar]
- 24.Belyaeva M, Taylor DJ. Cryptic species within the Chydorus sphaericus species complex (Crustacea: Cladocera) revealed by molecular markers and sexual stage morphology. Molecular phylogenetics and evolution. 2009;50(3): 534–546. doi: 10.1016/j.ympev.2008.11.007 [DOI] [PubMed] [Google Scholar]
- 25.Xu S, Hebert PDN, Kotov AA, Cristescu ME. The noncosmopolitanism paradigm of freshwater zooplankton. Insights from the global phylogeography of the predatory cladoceran Polyphemus pediculus (Linnaeus, 1761) (Crustacea, Onychopoda). Molecular ecology. 2009;18(24): 5161–5179. doi: 10.1111/j.1365-294X.2009.04422.x [DOI] [PubMed] [Google Scholar]
- 26.Crease TJ, Omilian AR, Costanzo KS, Taylor DJ. Transcontinental phylogeography of the Daphnia pulex species complex. PLoS ONE. 2012;7(10): e46620 doi: 10.1371/journal.pone.0046620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millette KL, Xu S, Witt JDS, Cristescu ME. Pleistocene-driven diversification in freshwater zooplankton: Genetic patterns of refugial isolation and postglacial recolonization in Leptodora kindtii (Crustacea, Cladocera). Limnology and Oceanography. 2011;56(5): 1725–1736. doi: 10.4319/lo.2011.56.5.1725 [Google Scholar]
- 28.Xu L, Han B-P, Van Damme K, Vierstraete A, Vanfleteren JR, Humont HJ. Biogeography and evolution of the Holarctic zooplankton genus Leptodora (Crustacea: Branchiopoda: Haplopoda). Journal of Biogeography. 2011;38(2): 359–370. doi: 10.1111/j.1365-2699.2010.02409.x [Google Scholar]
- 29.Bekker EI, Kotov AA, Taylor DJ. A revision of the subgenus Eurycercus (Eurycercus) Baird, 1843 emend. nov. (Cladocera: Eurycercidae) in the Holarctic with the description of a new species from Alaska. Zootaxa. 2012;3206: 1–40 [Google Scholar]
- 30.Kotov AA. Faunistic complexes of the Cladocera (Crustacea, Branchiopoda) of Eastern Siberia and the Far East of Russia. Zoologicheskii Zhurnal. 2016;95(7): 748–768. doi: 10.7868/S0044513416070059 [Google Scholar]
- 31.Bekker EI, Karabanov DP, Galimov YR, Kotov AA. DNA barcoding reveals high cryptic diversity in the North Eurasian Moina species (Crustacea: Cladocera). PLoS ONE. 2016;11(8): e0161737 doi: 10.1371/journal.pone.0161737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotov AA, Karabanov DP, Bekker EI, Neretina TV, Taylor DJ. Phylogeography of the Chydorus sphaericus group (Cladocera: Chydoridae) in the Northern Palearctic. PLoS ONE. 2016;11(12): e0168711 doi: 10.1371/journal.pone.0168711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popova EV, Petrusek A, Kořínek V, Mergeay J, Bekker EI, Karabanov DP, et al. Revision of the Old World Daphnia (Ctenodaphnia) similis group (Cladocera: Daphniidae). Zootaxa. 2016;4161(1): 1–40. doi: 10.11646/zootaxa.4161.1.1 [DOI] [PubMed] [Google Scholar]
- 34.Lampert W. Daphnia: Development of a model organism in ecology and evolution Oldendorf/Luhe: International Ecology Institute. 2011. XIX + 250 p. [Google Scholar]
- 35.Smirnov NN. Physiology of the Cladocera Amsterdam: Academic Press; 2017. 1–418 p. [Google Scholar]
- 36.Anderson BG. The toxicity of DDT to Daphnia. Science. 1945;102(2656): 539 doi: 10.1126/science.102.2656.539 [DOI] [PubMed] [Google Scholar]
- 37.Abe T, Saito H, Niikura Y, Shigeoka T, Nakano Y. Embryonic development assay with Daphnia magna: Application to toxicity of aniline derivatives. Chemosphere. 2001;45(4–5): 487–495. doi: 10.1016/S0045-6535(01)00049-2 [DOI] [PubMed] [Google Scholar]
- 38.Galimov Y, Walser B, Haag CR. Frequency and inheritance of non-male producing clones in Daphnia magna: Evolution towards sex specialization in a cyclical parthenogen. Journal of evolutionary biology. 2011;24(7): 1572–1583. doi: 10.1111/j.1420-9101.2011.02288.x [DOI] [PubMed] [Google Scholar]
- 39.Fields PD, Reisser C, Dukić M, Haag CR, Ebert D. Genes mirror geography in Daphnia magna. Molecular ecology. 2015;24(17): 4521–4536. doi: 10.1111/mec.13324 [DOI] [PubMed] [Google Scholar]
- 40.Brooks JL. The systematics of North American Daphnia New Haven, Conn: Connecticut Academy of Arts & Sciences; 1957. 1–180 p. [Google Scholar]
- 41.Flößner D, Dahl F, Dahl M, Bischoff H. Krebstiere, Crustacea (Kiemen- und Blattfüßer, Branchiopoda, Fischläuse, Branchiura) Jena: VEB Gustav Fischer Verlag; 1972. 1–501 p. [Google Scholar]
- 42.Prosser S, Martínez-Arce A, Elías-Gutiérrez M. A new set of primers for COI amplification from freshwater microcrustaceans. Molecular ecology resources. 2013;13(6): 1151–1155. doi: 10.1111/1755-0998.12132 [DOI] [PubMed] [Google Scholar]
- 43.Xu M, Zhang HJ, Deng DG, Wang WP, Zhang XL, Zha LS. Phylogenetic relationship and taxonomic status of four Daphnia species based on 16S rDNA and COI sequence. Acta Hydrobiologica Sinica. 2014;38(6): 1040–1043. doi: 10.7541/2014.153 [Google Scholar]
- 44.Geller J, Meyer C, Parker M, Hawk H. Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Molecular ecology resources. 2013;13(5): 851–861. doi: 10.1111/1755-0998.12138 [DOI] [PubMed] [Google Scholar]
- 45.Orsini L, Mergeay J, Vanoverbeke J, DeMeester L. The role of selection in driving landscape genomic structure of the waterflea Daphnia magna. Molecular ecology. 2013;22(3): 583–601. doi: 10.1111/mec.12117 [DOI] [PubMed] [Google Scholar]
- 46.Reumont BM von, Meusemann K, Szucsich NU, Dell'Ampio E, Gowri-Shankar V, Bartel D, et al. Can comprehensive background knowledge be incorporated into substitution models to improve phylogenetic analyses. A case study on major arthropod relationships. BMC evolutionary biology. 2009;9: 119 doi: 10.1186/1471-2148-9-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotov AA, Ishida S, Taylor DJ. A new species in the Daphnia curvirostris (Crustacea: Cladocera) complex from the eastern Palearctic with molecular phylogenetic evidence for the independent origin of neckteeth. Journal of Plankton Research. 2006;28(11): 1067–1079. doi: 10.1093/plankt/fbl041 [Google Scholar]
- 48.Colgan DJ, Ponder WF, Eggler PE. Gastropod evolutionary rates and phylogenetic relationships assessed using partial 28S rDNA and histone H3 sequences. Zoologica Scripta. 2000;29(1): 29–63. doi: 10.1046/j.1463-6409.2000.00021.x [Google Scholar]
- 49.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, et al. BLAST: A more efficient report with usability improvements. Nucleic acids research. 2013;41(Web Server issue): W29–33. doi: 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28(8): 1166–1167. doi: 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- 51.Hebert PDN, Remigio EA, Colbourne JK, Taylor DJ, Wilson CC. Accelerated molecular evolution in halophilic crustaceans. Evolution. 2002;56(5): 909–926 [DOI] [PubMed] [Google Scholar]
- 52.Adamowicz SJ, Hebert PDN, Marinone MC. Species diversity and endemism in the Daphnia of Argentina: A genetic investigation. Zoological Journal of the Linnean Society. 2004;140(2): 171–205. doi: 10.1111/j.1096-3642.2003.00089.x [Google Scholar]
- 53.Colbourne JK, Wilson CC, Hebert PDN. The systematics of Australian Daphnia and Daphniopsis (Crustacea: Cladocera): a shared phylogenetic history transformed by habitat-specific rates of evolution. Biological Journal of the Linnean Society. 2006;89(3): 469–488. doi: 10.1111/j.1095-8312.2006.00687.x [Google Scholar]
- 54.Elias-Gutierrez M, Martinez Jeronimo F, Ivanova NV, Valdez-Moreno M, Hebert PDN. DNA barcodes for Cladocera and Copepoda from Mexico and Guatemala, highlights and new discoveries. Zootaxa. 2008;1839: 1–42 [Google Scholar]
- 55.Petrusek A, Tollrian R, Schwenk K, Haas A, Laforsch C. A "crown of thorns" is an inducible defense that protects Daphnia against an ancient predator. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(7): 2248–2252. doi: 10.1073/pnas.0808075106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briski E, Cristescu ME, Bailey SA, MacIsaac HJ. Use of DNA barcoding to detect invertebrate invasive species from diapausing eggs. Biological Invasions. 2011;13(6): 1325–1340. doi: 10.1007/s10530-010-9892-7 [Google Scholar]
- 57.Jeffery NW, Elías-Gutiérrez M, Adamowicz SJ. Species diversity and phylogeographical affinities of the Branchiopoda (Crustacea) of Churchill, Manitoba, Canada. PLoS ONE. 2011;6(5): e18364 doi: 10.1371/journal.pone.0018364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang X, Shi X, Kotov AA, Gu F. Confirmation through genetic analysis of the existence of many local phyloclades of the genus Simocephalus (Crustacea, Cladocera) in China. PLoS ONE. 2014;9(11): e112808 doi: 10.1371/journal.pone.0112808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang M, Tan M, Meng G, Yang S, Su X, Liu S, et al. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic acids research. 2014;42(22): e166 doi: 10.1093/nar/gku917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma X, Wolinska J, Petrusek A, Gießler S, Hu W, Yin M. The phenotypic plasticity in Chinese populations of Daphnia similoides sinensis. Recurvate helmeted forms are associated with the presence of predators. Journal of Plankton Research. 2016;38(4): 855–864. doi: 10.1093/plankt/fbw031 [Google Scholar]
- 61.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, et al. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic acids research. 2011;39(Web Server issue): W13–7. doi: 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes. 2007;7(4): 544–548. doi: 10.1111/j.1471-8286.2007.01748.x [Google Scholar]
- 63.Nei M, Kumar S. Molecular evolution and phylogenetics New York: Oxford University Press; 2000. 1–333 p. [Google Scholar]
- 64.Avise JC. Phylogeography The history and formation of species. Cambridge, Mass., London: Harvard University Press; 2000. 1–464 p. [Google Scholar]
- 65.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11): 1451–1452. doi: 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 66.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Molecular biology and evolution. 2016;33(7): 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler A von, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature methods. 2017;14(6): 587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Posada D, Buckley TR. Model selection and model averaging in phylogenetics. Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic biology. 2004;53(5): 793–808. doi: 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- 69.Collins RA, Boykin LM, Cruickshank RH, Armstrong KF. Barcoding's next top model. An evaluation of nucleotide substitution models for specimen identification. Methods in Ecology and Evolution. 2012;3(3): 457–465. doi: 10.1111/j.2041-210X.2011.00176.x [Google Scholar]
- 70.Trifinopoulos J, Nguyen L-T, Haeseler A von, Minh BQ. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic acids research. 2016;44(W1): 232–235. doi: 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minh BQ, Nguyen MAT, Haeseler A von. Ultrafast approximation for phylogenetic bootstrap. Molecular biology and evolution. 2013;30(5): 1188–1195. doi: 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Version 4. Sunderland, MA: Sinauer Associates; 2002. 1–142 p. [Google Scholar]
- 73.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2014;10(4): e1003537 doi: 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, et al. A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evolutionary bioinformatics online. 2015;11: 43–48. doi: 10.4137/EBO.S21501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reid NM, Carstens BC. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC evolutionary biology. 2012;12: 196 doi: 10.1186/1471-2148-12-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29(22): 2869–2876. doi: 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leigh JW, Bryant D, Nakagawa S. popART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6(9): 1110–1116. doi: 10.1111/2041-210X.12410 [Google Scholar]
- 78.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Molecular ecology. 2000;9(10): 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- 79.Polzin T, Daneshmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Operations Research Letters. 2003;31(1): 12–20. doi: 10.1016/S0167-6377(02)00185-2 [Google Scholar]
- 80.Forster P, Torroni A, Renfrew C, Röhl A. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Molecular biology and evolution. 2001;18(10): 1864–1881 doi: 10.1093/oxfordjournals.molbev.a003728 [DOI] [PubMed] [Google Scholar]
- 81.Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular ecology. 2012;21(8): 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- 82.Panchal M. The automation of Nested Clade Phylogeographic Analysis. Bioinformatics. 2007;23(4): 509–510. doi: 10.1093/bioinformatics/btl614 [DOI] [PubMed] [Google Scholar]
- 83.Templeton AR. Statistical hypothesis testing in intraspecific phylogeography: Nested clade phylogeographical analysis vs. approximate Bayesian computation. Molecular ecology. 2009;18(2): 319–331. doi: 10.1111/j.1365-294X.2008.04026.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3): 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2): 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular ecology resources. 2010;10(3): 564–567. doi: 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 87.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Molecular biology and evolution. 2002;19(12): 2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034 [DOI] [PubMed] [Google Scholar]
- 88.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular biology and evolution. 1992;9(3): 552–569 doi: 10.1093/oxfordjournals.molbev.a040727 [DOI] [PubMed] [Google Scholar]
- 89.Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics. 1999;152(3): 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schwentner M, Clavier S, Fritsch M, Olesen J, Padhye S, Timms BV, et al. Cyclestheria hislopi (Crustacea: Branchiopoda): a group of morphologically cryptic species with origins in the Cretaceous. Molecular phylogenetics and evolution. 2013;66(3): 800–810. doi: 10.1016/j.ympev.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 91.Kotov AA, Taylor DJ. Mesozoic fossils (145 Mya) suggest the antiquity of the subgenera of Daphnia and their coevolution with chaoborid predators. BMC evolutionary biology. 2011;11: 129 doi: 10.1186/1471-2148-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Systematic biology. 2001;50(4): 580–601 [PubMed] [Google Scholar]
- 93.Remington CL. Suture-zones of hybrid interaction between recently joined biotas. Evolutionary Biology. 1968;2: 321–428. doi: 10.1007/978-1-4684-8094-8_8 [Google Scholar]
- 94.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789): 907–913. doi: 10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- 95.Hewitt GM. The structure of biodiversity–insights from molecular phylogeography. Frontiers in zoology. 2004;1(1): 4 doi: 10.1186/1742-9994-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Templeton AR. Statistical phylogeography: Methods of evaluating and minimizing inference errors. Molecular ecology. 2004;13(4): 789–809 [DOI] [PubMed] [Google Scholar]
- 97.Nédli J, De Meester L, Major Á, Schwenk K, Szivák I, Forró L. Salinity and depth as structuring factors of cryptic divergence in Moina brachiata (Crustacea: Cladocera). Fundamental and Applied Limnology. 2014;184(1): 69–85. doi: 10.1127/1863-9135/2014/0462 [Google Scholar]
- 98.Beaumont MA, Nielsen R, Robert C, Hey J, Gaggiotti O, Knowles L, et al. In defence of model-based inference in phylogeography. Molecular ecology. 2010;19(3): 436–446. doi: 10.1111/j.1365-294X.2009.04515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barley AJ, Thomson RC. Assessing the performance of DNA barcoding using posterior predictive simulations. Molecular ecology. 2016;25(9): 1944–1957. doi: 10.1111/mec.13590 [DOI] [PubMed] [Google Scholar]
- 100.Reisser CMO, Fasel D, Hürlimann E, Dukic M, Haag-Liautard C, Thuillier V, et al. Transition from environmental to partial genetic sex determination in Daphnia through the evolution of a female-determining incipient W chromosome. Molecular biology and evolution. 2017;34(3): 575–588. doi: 10.1093/molbev/msw251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dzialowski AR, O'Brien WJ, Swaffar SM. Range expansion and potential dispersal mechanisms of the exotic cladoceran Daphnia lumholtzi. Journal of Plankton Research. 2000;22(12): 2205–2223. doi: 10.1093/plankt/22.12.2205 [Google Scholar]
- 102.Mergeay J, Verschuren D, De Meester L. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proceedings of the Royal Society B: Biological Sciences. 2006;273(1603): 2839–2844. doi: 10.1098/rspb.2006.3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sousa FDR, Palaoro AV, Elmoor-Loureiro LMA, Kotov AA. Predicting the invasive potential of the cladoceran Daphnia lumholtzi Sars, 1885 (Crustacea: Cladocera: Daphniidae) in the Neotropics: are generalists threatened and relicts protected by their life-history traits. Journal of Limnology. 2017;76(2): 272–280. doi: 10.4081/jlimnol.2016.1571 [Google Scholar]
- 104.Bolotov IN, Aksenova OV, Bespalaya YV, Gofarov MY, Kondakov AV, Palster IS, et al. Origin of a divergent mtDNA lineage of a freshwater snail species, Radix balthica, in Iceland: Cryptic glacial refugia or a postglacial founder event. Hydrobiologia. 2017;787(1): 73–98. doi: 10.1007/s10750-016-2946-9 [Google Scholar]
- 105.Ho SYW, Tong KJ, Foster CSP, Ritchie AM, Lo N, Crisp MD. Biogeographic calibrations for the molecular clock. Biology letters. 2015;11(9): 20150194 doi: 10.1098/rsbl.2015.0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hipsley CA, Müller J. Beyond fossil calibrations: Realities of molecular clock practices in evolutionary biology. Frontiers in genetics. 2014;5: 138 doi: 10.3389/fgene.2014.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Damme K, Kotov AA. The fossil record of the Cladocera (Crustacea: Branchiopoda): Evidence and hypotheses. Earth-Science Reviews. 2016;163: 162–189. doi: 10.1016/j.earscirev.2016.10.009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Numbers identify star groups of haplotypes (see S2 Table). Circle size is proportional to the frequency of the haplotypes. Small dark circles indicate unsampled or extinct haplotypes. Geographic regions for star groups are as follows: s1 –Western, Central Europe, Mediterranean region, Middle East; s2 –Western Europe; s3 –Scandinavian Peninsula and Western Europe; s4-s8 –Mediterranean region; s9 –Mediterranean region and Near East; s10 –Western Europe; s11 –European Russia; s12—Near East; s13-s14 –European Russia; s15-s16 –Mediterranean region; s17-s18 –North America; s19 –European Russia s20 –Mediterranean region; s21 –Western Europe; s22 –Western and Central Europe; s23 –Central and Eastern Europe; s24-s25 –Western Europe; s26 –Western Europe and Scandinavian Peninsula; s27 –different regions of Europe and laboratory cultures; s28 –Mediterranean region; s29 –Western and Central Europe; s30 –Scandinavian Peninsula; s31-s33 –European Russia; s34-s36 –Western Siberia; s37 –Eastern Siberia; s38-s39 –Western Siberia; s40-s42 –North America; s43-s45 –Eastern Siberia; s46 –Far East of Asia.
(TIF)
Clades highlighted in red represent the maximum likelihood limits. The colored lines correspond to a sequence-by-sequence matrix, with lines colored according to the posterior probability that the sequences are conspecific, which allows visualizing uncertainty in group limits.
(TIF)
All original samples have prefix zh, while the Genbank samples have no this prefix. GenBank accession numbers for original samples are given in S1 Table, for sequences obtained from GenBank they are: USA–AY921452, Denmark–DQ470575, Belgium–AM490278, China1 –KF993366, China2 –KM244710, China3 –KP296147, China4 –NC_026914, Novosibirsk1 –JN874603, Novosibirsk2 –JN874602, Novosibirsk3 –JN874604, Novosibirsk4 –JN874601.
(TIF)
All original samples have prefix zh, while the Genbank samples do not have this prefix. GenBank accession numbers for original samples are given in S1 Table, for sequences obtained from GenBank they are: USA—DQ845268.
(PDF)
COI clade designations as defined in Fig 2.
(XLS)
(XLS)
n—sample size, Nh—number of haplotypes, Nv—number of variable (polymorphic) sites, Np—number of parsimony informative sites, Hd—haplotype diversity, Pi—nucleotide diversity, k—average number of nucleotide differences.
(DOC)
n—sample size, Nh—number of haplotypes, Nv—number of variable (polymorphic) sites, Np—number of parsimony informative sites, Hd—haplotype diversity, Pi—nucleotide diversity, k—average number of nucleotide differences; R2 population size expansion test and results of Tajima’s D, Fu’s FS and mismatch distributions: tau-parameter, SSD (sum of squares deviation) and Harpending's Raggedness index including associated p-values. Biological processes are (Templeton, 2004): RGF–restricted gene flow; D–dispersal; LDD–long-distance dispersal; IBD–isolation by distance; AF–allopatric fragmentation; PF–past fragmentation; LDC–long-distance colonization.
(DOC)
In diagonal, bold–estimates of average evolutionary divergence over sequence pairs within groups. Below diagonal–estimates of evolutionary divergence over sequence pairs between groups. The presence of n/c in the results denotes cases in which it was not possible to estimate evolutionary distances. Above diagonal–time divergence, in MYA; first digit–“fast clock” from (Schwentner et al., 2013), second digit–“slow clock” from (Kotov & Taylor, 2011).
(DOC)
(DOC)
(DOCX)
Data Availability Statement
All data are freely available at Genbank (see S1 Table for accession numbers).