Abstract
Humans have been fascinated by sleep for millennia. After almost a century of scientific interrogation, significant progress has been made in understanding the neuronal regulation and functions of sleep. The application of new methods in neuroscience that enable the analysis of genetically defined neuronal circuits with unprecedented specificity and precision has been paramount in this endeavor. In this review, we first discuss electrophysiological and behavioral features of sleep/wake states and the principal neuronal populations involved in their regulation. Next, we describe the main modulatory drives of sleep and wakefulness, including homeostatic, circadian, and motivational processes. Finally, we describe a revised integrative model for sleep/wake regulation.
Introduction
Sleep is a complex biological state characterized by behavioral, physiological, and electrophysiological parameters. All animals studied thus far show characteristic signs of sleep, including mammals, birds, fish, insects, roundworms, and jellyfish (Cirelli and Tononi, 2008; Eban-Rothschild and Bloch, 2008; Hendricks et al, 2000; Kaiser, 1988; Nath et al, 2017; Prober et al, 2006; Raizen et al, 2008; Ramon et al, 2004; Shaw et al, 2000; Shein-Idelson et al, 2016; Vorster et al, 2014; Yokogawa et al, 2007). The transition between wakefulness and sleep involves profound changes in motor control, cognition, brain activity, and consciousness (McGinley et al, 2015). In mammals and birds, sleep and wake states are typically determined using electroencephalogram (EEG) and electromyogram (EMG) recordings, which measure global cortical and muscular activity, respectively. The awake state is heterogeneous, characterized by desynchronized EEG oscillations of low amplitude and mixed frequencies, and a variable amount of muscle activity. Active or motivated wakefulness is rich in theta (4–9 Hz) and gamma (40–300 Hz) EEG frequency ranges, whereas quiet wakefulness is characterized by slower EEG frequencies, including alpha (7–15 Hz) and beta (8–30 Hz). Using EEG and EMG recordings it is possible to recognize two distinct sleep states: non-rapid eye movement (NREM), and rapid eye movement (REM) sleep—that alternate cyclically across sleep. NREM sleep is characterized by high amplitude low-frequency delta oscillations (0.5–4 Hz) and spindles (bursts of 7–15 Hz oscillations) in the EEG, and low postural muscle tone. The EEG during REM sleep is dominated by theta and gamma oscillations, with a complete loss of muscle tone in axial posture muscles (REM muscle atonia). The sleep/wake states also differ in various physiological parameters, such as thermoregulation, cardiac activity, metabolism, and respiration (Brown et al, 2012; Saper et al, 2010). The characteristic EEG patterns are biomarkers of the different arousal states, and although most non-mammalian species lack the brain structures producing these EEG oscillations, they nonetheless show behavioral and physiological criteria for sleep (Allada and Siegel, 2008; Cirelli and Tononi, 2008; Zimmerman et al, 2008). Behaviorally, sleep is defined as a period of quiescence, with a species-specific body posture and/or sleeping site, and an elevated arousal threshold (Campbell and Tobler, 1984). Sleep is also defined by its homeostatic regulation, which is manifested by an increase in sleep need following extended wakefulness. Non-mammalian model organisms, such as the fruit fly (Drosophila melanogaster), the nematode worm (Caenorhabditis elegans), and zebrafish (Danio rerio) are extremely useful for screening novel sleep mutants and investigations on the organizational principles and genetic control of sleep (Chakravarti et al, 2017; Levitas-Djerbi and Appelbaum, 2017; Trojanowski and Raizen, 2016). In addition, various non-model organisms, such as the pectoral sandpiper (Calidris melanotos), the northern fur seal (Callorhinus ursinus), the cavefish (Astyanax mexicanus), and the honey bee (Apis mellifera), can serve as excellent systems to study the adaptive values of sleep, plasticity during sleep, and the effects of social and environmental cues on sleep (Aulsebrook et al, 2016; Duboue et al, 2011; Eban-Rothschild and Bloch, 2012, 2015; Lesku et al, 2012; Lyamin et al, 2008a; Lyamin et al, 2017).
Dissecting the neuronal mechanisms that regulate sleep/wake states
von Economo, Ranson, and Moruzzi and Magoun (von Economo, 1930; Moruzzi and Magoun, 1949; Ranson, 1939) were among the first to examine a neuronal mechanism for sleep/wake regulation in mammals and many subsequent studies have contributed to the characterizing of the principal regions and neuronal modulators of sleep/wake states. Over several decades, researchers have used various experimental techniques including lesions, pharmacological and genetic manipulations, electrical stimulations, and electrophysiological recordings, to establish a solid base for our current understanding of sleep/wake circuitry. Nonetheless, brain nuclei regulating sleep/wake states are typically heterogeneous and contain intermingled neuronal populations that frequently support different arousal states. Moreover, traditional techniques typically lack temporal resolution and/or can involve compensatory mechanisms in the sleep/wake circuitry. With the application of novel methods to manipulate specific cell types in restricted brain regions in vivo with high temporal precision (Kim et al, 2017; Lin and Schnitzer, 2016; Roth, 2016), it is now possible to reveal the activity patterns and functionally interrogate the roles of genetically defined neuronal populations involved in sleep/wake regulation. A better understanding of brain mechanisms controlling sleep/wake states would allow us to gain new insights on the functions of sleep and improve the capacity to treat sleep disorders.
Mammalian Neuronal Circuitry of Sleep/Wake States
How does the mammalian brain control sleep and wakefulness? It is currently understood that complex interactions between subcortical neuromodulatory neurons in the brainstem, midbrain, hypothalamus, and basal forebrain (BF), the thalamus, and the cortex drive behavioral, physiological, and electrocortical sleep/wake states. Wake-promoting populations project to various structures through a dorsal and a ventral pathway. The dorsal pathway innervates the thalamus—which facilitates transmission of sensory information to the cortex. The ventral pathway innervates the hypothalamus, BF, and other forebrain structures—which together excite the cortex. It is thought that wakefulness is achieved when both the dorsal and the ventral pathways are activated. In this review, we will mainly focus on wake-promoting neuronal circuits, as excellent recent reviews have covered the neuronal mechanisms underlying NREM and REM sleep (Arrigoni et al, 2016; Luppi et al, 2017; Peever and Fuller, 2017; Scammell et al, 2017; Weber and Dan, 2016).
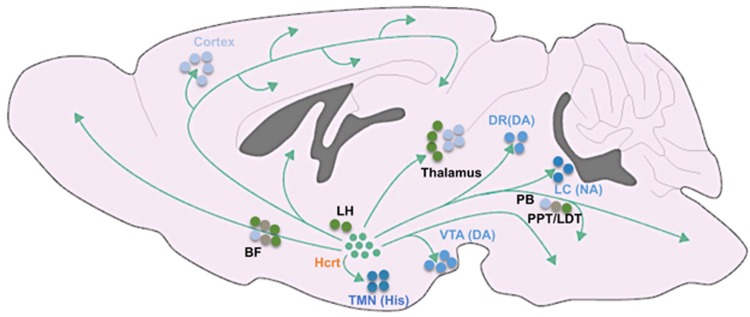
Historically, monoaminergic and cholinergic producing neurons were assigned as key players in the regulation of arousal states (Jouvet, 1972), yet recent findings reveal important roles for fast gamma-Aminobutyric acid (GABA)and glutamate neurotransmission in sleep/wake regulation (Saper and Fuller, 2017). Among the wake-promoting monoaminergic and cholinergic populations are the noradrenergic locus coeruleus (LC), dopaminergic ventral tegmental area (VTA), dopaminergic and serotonergic (5HT) dorsal raphe nucleus (DRN), histaminergic tuberomammillary nucleus (TMN), and cholinergic pedunculopontine and laterodorsal tegmental nuclei (PPT/LDT) and BF neurons (Jones, 2003; Scammell et al, 2017) (Figure 1). Importantly, monoaminergic and cholinergic neurons are essential for learning, motivation, attention, reward, mood, and locomotor behaviors in addition to sleep/wake regulation (Lorincz and Adamantidis, 2017).
Figure 1.
Schematic of neuronal wake-promoting populations. BF, basal forebrain; DA, dopamine; DR, dorsal raphe; Hcrt, hypocretin; His, histamine; LC, locus coeruleus; LH, lateral hypothalamus; NA, noradrenaline; PB, parabrachial area; PPT/LDT, pedunculopontine and laterodorsal tegmental nuclei; TMN, tuberomammillary nucleus; VTA, ventral tegmental area.
Hypocretin Neurons
Two decades ago, hypocretin/orexin (Hcrt) neurons of the lateral hypothalamus (LH) emerged as essential orchestrators of sleep and wakefulness. The hypocretins are two alternatively spliced neuropeptides, Hcrt-1 and Hcrt-2, produced from a single-precursor gene (de Lecea et al, 1998; Sakurai et al, 1998). Hcrt neurons show high discharge rates during active waking and goal-oriented behaviors, decrease their activity during quiet wakefulness, and are silent during sleep (Hassani et al, 2009b; Lee et al, 2005b; Mileykovskiy et al, 2005; Takahashi et al, 2008). Hcrt neurons strongly project to various wake-promoting populations in the brain, including cholinergic neurons of the BF and brainstem, noradrenergic neurons of the LC, and dopaminergic neurons of the VTA. The first in vivo application of optogenetics was performed by the de Lecea group to reveal that photostimulation of Hcrt neurons increases the probability of sleep-to-wake transitions and promotes wakefulness (Adamantidis et al, 2007). Loss of Hcrt-producing neurons or their receptors in humans, dogs and rodents causes narcolepsy with cataplexy—a neurological disorder characterized by an inability to control the boundaries between sleep–wake states (Chemelli et al, 1999; Lin et al, 1999; Peyron et al, 2000; Thannickal et al, 2000). In narcoleptics with cataplexy, periods of wakefulness are interrupted by unexpected sleep episodes, and REM-like episodes co-exist with conscious wakefulness (de Lecea, 2015). Nonetheless, Hcrt neurons are not essential for sleep or wake generation—as narcoleptic patients and transgenic mice lacking Hcrt neurons or Hcrt receptor-2 display normal daily amounts of sleep (Branch et al, 2016; Chemelli et al, 1999; de Lecea and Sutcliffe, 2005). Rather, the coordination between sleep/wake states and the stability of arousal appears to be localized in Hcrt neurons.
The Locus Coeruleus
The LC is the main source of noradrenergic transmission to the cortex. LC neurons fire tonically at 1–3 Hz during wakefulness, reduce their activity during NREM sleep and are nearly silent during REM sleep (Aston-Jones and Bloom, 1981a; Hobson et al, 1975). In response to salient stimuli, LC neurons transition to phasic burst firing at 8–10 Hz (Aston-Jones and Bloom, 1981b). The release of noradrenaline in the cortex increases firing rates of pyramidal neurons and electrocortical arousal, while blockade of noradrenergic pathways induces synaptic quiescence during wakefulness (Constantinople and Bruno, 2011). Administration of noradrenaline directly to the ventricles or forebrain promotes wakefulness (Flicker and Geyer, 1982; Segal and Mandell, 1970), while pharmacological inhibition of noradrenergic neurons increases NREM sleep (Berridge and Espana, 2000; De Sarro et al, 1987). Nonetheless, neuronal lesions of the LC fail to produce a consistent effect on cortical EEG (Blanco-Centurion et al, 2007; Lidbrink, 1974; Tononi et al, 1991). Optogenetic interrogations revealed that activity in noradrenergic tyrosine hydroxylase (Th) positive LC neurons is sufficient to induce wakefulness at physiologically relevant frequencies (Carter et al, 2010). Moreover, LC activity seems to be essential for Hcrt-induced awakenings, as optogenetic inhibition of Th+ LC neurons during photostimulation of Hcrt neurons prevents Hcrt-mediated sleep-to-wake transitions (Carter et al, 2012). Reciprocally, increasing the excitability of Th+ LC neurons dramatically reduces the latency of Hcrt-mediated awakenings (Carter et al, 2012). These results strongly support the premise that LC is a major output effector of Hcrt neurons in regard to arousal regulation (Carter et al, 2013).
The Basal Forebrain
The BF contains three major cell groups: cholinergic, GABAergic, and glutamatergic neurons. This brain structure has been traditionally regarded as a wake-promoting region, as its activation induce wakefulness and its lesion results in slow oscillations in the EEG and a coma-like state (Buzsaki et al, 1988; Fuller et al, 2011). Nonetheless, the BF is functionally and anatomically heterogeneous and contains intermingled neurons maximally active during different arousal states (Jones, 2017). Cholinergic neurons of the BF are strongly activated during wakefulness and REM sleep (Boucetta et al, 2014; Lee et al, 2005a; Xu et al, 2015), and their optogenetic stimulation induces cortical activation and transitions from NREM sleep to wakefulness (Han et al, 2014; Irmak and de Lecea, 2014; Xu et al, 2015). Nonetheless, chemogenetic activation of BF cholinergic neurons mainly reduces delta activity during NREM sleep, rather than substantially increasing wakefulness (Anaclet et al, 2015; Chen et al, 2016a). The activity pattern of GABAergic neurons of the BF is highly heterogeneous—a subset of neurons are maximally active during wakefulness and REM sleep, another subset during NREM sleep, and a minority are maximally active during REM sleep (Duque et al, 2000; Hassani et al, 2009a; Jones, 2017). Chemogenetic activation of the entire BF GABAergic population strongly promotes wakefulness and high-frequency cortical EEG, whereas their inhibition increases NREM sleep and slow-frequency oscillations (Anaclet et al, 2015). BF parvalbumin (PV) positive GABAergic neurons are wake-active and their optogenetic stimulation induces wakefulness, whereas somatostatin-positive BF GABAergic neurons are functionally heterogeneous and may facilitate NREM sleep (Xu et al, 2015). Cortically projecting PV+ BF neurons were also demonstrated to promote cortical arousal, specifically in the gamma band (Kim et al, 2015). The neurochemical identity of BF GABAergic neurons maximally active during REM sleep remains to be elucidated. The activity of BF glutamatergic neurons is also heterogeneous (Jones, 2017), and while their optogenetic stimulation induces sleep-to-wake transitions (Xu et al, 2015), their chemogenetic activation does not modify sleep/wake architecture (Anaclet et al, 2015). Future studies using projection-specific and/or additional neurochemical markers for subpopulations of BF neurons would provide important insights onto the complex roles of this structure in sleep/wake regulation.
The Ventral Tegmental Area
The VTA is a key structure in the regulation of motivational processes, and contains dopaminergic, GABAergic, and glutamatergic neurons (Morales and Margolis, 2017). Although the roles of GABAergic and glutamatergic VTA neurons in sleep/wake regulation remain to be elucidated, much progress has been made in recent years in delineating the roles of the dopaminergic population. It has long been known that stimulants that enhance dopaminergic tone are potent wake-promoting substances (Boutrel and Koob, 2004), and their arousing effects are abolished in mice deficient in dopamine signaling (Qu et al, 2008; Wisor et al, 2001). Nonetheless, dopaminergic neurons have been considered for several decades as the only monoamine group not involved in sleep/wake regulation (Espana and Scammell, 2011; Saper et al, 2010), mainly since early electrophysiological findings suggested that VTA and substantia nigra pars compacta dopaminergic neurons do not change their mean firing rates across sleep–wake states (Miller et al, 1983; Steinfels et al, 1983; Trulson and Preussler, 1984; Trulson et al, 1981). Moreover, lesions to the VTA and substantia nigra pars compacta were not found to modify the time spent in electrocortical wakefulness (Jones et al, 1973). Recently, this view has been changed as we and others have provided compelling evidence that VTA dopaminergic neurons are key modulators of sleep and wakefulness (Dahan et al, 2007; Eban-Rothschild et al, 2016; Oishi et al, 2017a; Taylor et al, 2016).
During wakefulness, VTA dopaminergic neurons are strongly activated by various salient stimuli, including sensory cues, social cues, and rewards (Brischoux et al, 2009; Cohen et al, 2012; Gunaydin et al, 2014; Lammel et al, 2012). Using unit recording in head restrained rats, Dahan et al. revealed a robust increase in bursting activity of VTA dopaminergic neurons during REM sleep, compared to both quiet wakefulness and NREM sleep (Dahan et al, 2007). The activity pattern of VTA dopaminergic neurons during REM sleep was found to be similar to that measured during the consumption of palatable food (Dahan et al, 2007). Recently using simultaneous calcium-dependent fiber-photometry and EEG/EMG recordings we further demonstrated that VTA dopaminergic neurons reduce their activity during NREM sleep compared to both wakefulness and REM sleep (Eban-Rothschild et al, 2016). Using optogenetic and chemogenetic interrogations a causal role for VTA dopaminergic neurons in sleep/wake regulation has been demonstrated. Activation of VTA dopaminergic neurons was found to induce and maintain wakefulness during a period of high sleep pressure (Eban-Rothschild et al, 2016; Oishi et al, 2017a) and even during 0.8–0.9% isoflurane anesthesia (Taylor et al, 2016). We also demonstrated that chemogenetic inhibition of VTA dopaminergic neurons during the dark/active phase promotes sleep—strongly suggesting that activity in these neurons is necessary for wakefulness (Eban-Rothschild et al, 2016). Inhibition of VTA dopaminergic neurons promoted sleep even in the presence of various salient stimuli, including a potential mate, predator odor, and palatable food—suggesting a prominent role in salience-induced arousal (Eban-Rothschild et al, 2016). We further characterized the contribution of different output pathways of VTA dopaminergic neurons in the regulation of arousal. By employing projection-specific optogenetic manipulations, we demonstrated that stimulation of nucleus accumbens (NAc), central amygdala (CeA), and dorsolateral striatum (DLS), but not medial prefrontal cortex (mPFC) projections promoted a transition from NREM sleep to wakefulness. In addition, stimulation of mPFC and CeA projections promoted a transition from REM sleep to wakefulness (Eban-Rothschild et al, 2016). Nonetheless, only the NAc projection was found to have a role in wake maintenance and its stimulation suppressed sleep (Eban-Rothschild et al, 2016)—further supporting a role for the NAc in sleep/wake regulation (Lazarus et al, 2013; Oishi et al, 2017b; Qiu et al, 2012). These findings reveal a complex circuitry for VTA dopaminergic regulation of arousal, and highlight the importance of using cell type and projection-specific in vivo tools for functional interrogations of circuits participating in sleep/wake regulation.
Most monoaminergic neurons involved in sleep/wake regulation share similar firing patterns—with high rates of firing during wakefulness, slow firing during NREM sleep, and a virtual cessation of firing during REM sleep (Jones, 2003; Scammell et al, 2017). In contrast, VTA dopaminergic neurons show strong activation during REM sleep (Dahan et al, 2007; Eban-Rothschild et al, 2016), as do cholinergic BF neurons (Boucetta et al, 2014; Lee et al, 2005a; Xu et al, 2015). Activation of cholinergic BF neurons has been implicated in cortical arousal (Goard and Dan, 2009; Pinto et al, 2013), yet the precise role of dopaminergic activity during REM sleep remains to be elucidated.
The Dorsal Raphe Nucleus/Ventral Periaqueductal Gray
The DRN is a heterogeneous brainstem nucleus that innervates many brain regions implicated in sleep/wake regulation. DRN neurons express various neurotransmitters, including serotonin and dopamine. DRN serotonin neurons were initially hypothesized to play an important role in the initiation and maintenance of NREM sleep, as lesions to the DRN in rats and cats resulted in insomnia (Jouvet, 1972). Nonetheless, further examinations of the activity pattern of serotonergic DRN neurons across arousal states revealed that they are wake-active and predominantly silent during NREM and REM sleep (McGinty and Harper, 1976; Trulson and Jacobs, 1979). Later work suggested that serotonergic DRN neurons facilitate quiet wakefulness and inhibit REM sleep (Jacobs and Fornal, 1993). Further cell type, receptor- and projection-specific functional interrogations of serotonergic DRN circuitry are required to reveal the casual role(s) of this population in sleep/wake regulation.
The DRN or ventral periaqueductal gray (vPAG) contains an additional neuronal population implicated in sleep/wake regulation: the dopaminergic neurons. vPAG dopaminergic neurons show c-Fos immunoreactivity during wakefulness and not during sleep, and their lesion reduces wakefulness and increases sleep (Lu et al, 2006). Recently, arousal-state-dependent alterations in population activity of vPAG/DR dopaminergic neurons have been demonstrated and a causal role in sleep/wake regulation (Cho et al, 2017). DRN dopaminergic neurons where shown to be wake-active and their optogenetic activation induces wakefulness from sleep (Cho et al, 2017). In contrast, chemogenetic inhibition of DRN dopaminergic neurons promotes NREM sleep, even in the presence of various salient stimuli (Cho et al, 2017). Interestingly, optogenetic inhibition of DRN dopaminergic neurons during NREM sleep decreases the probability of auditory cues to induce wakefulness (Cho et al, 2017)—suggesting a role for these neurons in the arousal response to sensory stimuli.
It is important to note that not all dopaminergic populations promote wakefulness, as SNc dopaminergic neurons projecting to the dorsal striatum have been suggested to promote NREM sleep (Qiu et al, 2016b). It would be of interest for future studies to determine the functional role of additional dopaminergic populations, including the caudal hypothalamic A11, arcuate nucleus A12 and the zona incerta A13 groups. In addition, it would be useful to determine whether dopaminergic neurons act in concert or independently to generate, regulate, and/or maintain sleep and wake states.
The Tuberomammillary Nucleus
The TMN of the hypothalamus is the sole source of histamine in the brain (Haas et al, 2008). Histaminergic TMN neurons send widespread, yet diffused, projections to various sleep/wake regulatory regions, including to the ventrolateral preoptic nucleus and the basal forebrain (Haas et al, 2008). Histaminergic TMN neurons show arousal-state depended alterations in activity. They fire most rapidly during attentive wakefulness, reduce their activity during quiet wakefulness and are silent during NREM and REM sleep (Steininger et al, 1999; Takahashi et al, 2006). Histamine has long been implicated in wake maintenance, as antihistamines (histamine H1 receptor antagonists) promote sleep and slow-wave activity (Haas et al, 2008; Ikeda-Sagara et al, 2012). Mice lacking histamine H1 receptor show reduced wakefulness in the beginning of the active/dark phase, and a reduced arousal response in a novel environment (ie, shorter latency to enter NREM sleep) (Parmentier et al, 2016). Nonetheless, ablation of histaminergic TMN neurons produce only a modest effect on sleep/wake patterns (Anaclet et al, 2009; Parmentier et al, 2002). Recently, activity in TMN histaminergic neurons has been casually demonstrated to be necessary for wakefulness—as their optogenetic inhibition promotes NREM sleep (Fujita et al, 2017). Interestingly, mice lacking GABA receptors only in histaminergic neurons show an increased arousal response in a novel environment (ie, longer latency to enter NREM sleep) (Zecharia et al, 2012). Histaminergic TMN neurons express GABA, GAD67, and VGAT (Airaksinen et al, 1992; Takeda et al, 1984). These neurons may also co-release GABA which has been suggested to synergistically promote cortical activation and wakefulness (Yu et al, 2015)—yet additional evidence is needed to further support this finding. Future studies should further elucidate the importance of neurotransmitter co-transmission for sleep/wake regulation, in the TMN and additional sleep/wake regulatory populations. Moreover, it remains to be resolved which aspects of arousal histaminergic TMN neurons are contributing.
Glutamatergic Neurons
Glutamate is the principle excitatory neurotransmitter in the brain, and global modulation of glutamatergic signaling pathways dose-dependently modulates sleep/wake states (Maclver et al, 1996; Yamamura et al, 1990). Glutamate levels in the posterior hypothalamus and cortex rise progressively during wakefulness, decline during NREM sleep and increase rapidly at the onset of REM sleep (John et al, 2008; Naylor et al, 2012). There are several glutamatergic populations implicated in promoting arousal, including the parabrachial (PB), PPT, and supramammillary nuclei neurons (Pedersen et al, 2017; Saper and Fuller, 2017). For example, PB glutamatergic neurons send strong projections to the BF, LH, and cortex (Bernard et al, 1991; Fuller et al, 2011; Fulwiler and Saper, 1984; Hur and Zaborszky, 2005; Saper and Loewy, 1980). Partial or complete lesions to the PB cause either a reduction in wakefulness or a coma-like state with permanent slow EEG oscillations, respectively (Fuller et al, 2011). Genetically driven disruption of glutamate transmission in PB neurons also results in a significant reduction in total wakefulness and an increase in delta oscillations in the waking EEG (Kaur et al, 2013). Chemogenetic activation of PB neurons (both glutamatergic and non-glutamatergic) in rats increases wakefulness and decreases both NREM and REM sleep during the light/inactive phase (Qiu et al, 2016a). Furthermore, the LH and preoptic-BF area, but not the thalamus, where shown to be the principle output pathways in PB modulation of wakefulness (Qiu et al, 2016a). Recently, chemogenetic activation of glutamatergic PPT (and possibly also PB neurons) has been demonstrated to induce prolonged cortical activation and behavioral wakefulness, whereas their chemogenetic inhibition reduces wakefulness and increases NREM sleep (Kroeger et al, 2017). As different neuromodulatory populations also release glutamate (and/or GABA), future studies should determine the precise role neuromodulators and fast neurotransmitters play in sleep/wake regulation independently.
GABAergic Neurons
GABA has been traditionally viewed as a sleep-promoting neurotransmitter. Most sleep-promoting populations are GABAergic, including those in the preoptic area (Sherin et al, 1996) and parafacial zone (Anaclet et al, 2014). Sleep-promoting medications typically target and enhance GABAergic signaling (Luppi et al, 2017; Schulz and Macdonald, 1981). Nonetheless, recent studies have identified several GABAergic populations in the LH (Herrera et al, 2016; Venner et al, 2016) and BF (Anaclet et al, 2015; Xu et al, 2015) that promote wakefulness and cortical activation. LH GABAergic neurons projecting to the thalamic reticular nucleus (TRN) where shown to increase wakefulness by inhibiting TRN GABAergic neurons (Herrera et al, 2016). Optogenetic stimulation of either LH GABAergic cell bodies or their projections to the TRN induces transitions to wakefulness from NREM sleep, while their optogenetic inhibition increases NREM sleep and delta oscillations in the EEG (Herrera et al, 2016). In addition, recent chemogenetic interrogations revealed that GABAergic neurons of the ventral LH promote wakefulness, and their inhibition increases NREM sleep, possibly via projections to the ventrolateral preoptic nucleus (Venner et al, 2016).
In summary, much progress has been made in recent years in identifying the roles of distinct neuronal populations in sleep/wake regulation. Nonetheless, it is still unclear by which pre- and post-synaptic actions sleep/wake regulatory neuromodulators regulate arousal and what role co-transmission plays in sleep/wake regulation. In addition, a major question that remains to be resolved is how information from the diverse sleep/wake regulatory populations is integrated to result in a coherent arousal state.
Neuronal Mechanisms Regulating Sleep/Wake States Across Species
All animals studied thus far, across six different phyla, sleep. The neuronal, cellular, and molecular pathways associated with sleep are highly conserved across species—suggesting an early and common origin for sleep (Allada and Siegel, 2008; Joiner, 2016; Nall and Sehgal, 2014). In zebrafish, the neurotransmitters and neuromodulators controlling sleep/wake states are located in parallel structures to mammals (Levitas-Djerbi and Appelbaum, 2017; Oikonomou and Prober, 2017; Sundvik and Panula, 2015). For example, in both mammals and zebrafish, Hcrt and histamine neurons are located in the hypothalamus (Appelbaum et al, 2009; Sundvik et al, 2011) and noradrenaline in the LC (Singh et al, 2015). Genetic and pharmacological manipulations of GABAergic, glutamatergic, monoaminergic, and neuropeptidergic neurons in zebrafish result in similar modifications to sleep and wakefulness as in mammals (Barlow and Rihel, 2017; Levitas-Djerbi and Appelbaum, 2017; Rihel et al, 2010). In zebrafish, as in mammals, Hcrt neurons send widespread projections throughout the brain and consolidate wakefulness (Elbaz et al, 2017; Faraco et al, 2006; Prober et al, 2006; Yelin-Bekerman et al, 2015; Yokogawa et al, 2007), whereas ablation of Hcrt neurons fragments sleep/wake states (Chen et al, 2016b; Elbaz et al, 2012). Mammals and insects also share similar functions in the neurotransmitters and neuromodulators controlling sleep/wake states. For example, as in mammals, dopamine is a major regulator of sleep/wake states in Drosophila, and enhancement of dopaminergic neurotransmission dramatically reduces sleep and arousal threshold, and increases locomotor activity (Andretic et al, 2005; Hendricks et al, 2003; Kume et al, 2005; Pimentel et al, 2016; Wu et al, 2008). In addition, noradrenergic and histaminergic activity promotes wakefulness in Drosophila, whereas GABAergic and serotonergic neurotransmission promotes sleep (Michel and Lyons, 2014; Nall and Sehgal, 2014). These similarities, as well as the ease of completing large unbiased screens in simple model systems and the numerous forward and reverse genetic approaches available, have placed these species as prominent models in sleep research. Their study provides important insights into the regulatory processes and functions of sleep.
PROCESSES MODULATING THE PROPENSITY TO STAY AWAKE OR FALL ASLEEP
Most behaviors, such as foraging, courting and predator defense, are incompatible with sleep, and animals need to carefully coordinate the timing of sleep and wakefulness to maximize their survival. The probability of an individual entering a particular sleep/wake state depends on multiple factors, including previous sleep history, circadian time, environmental cues, and internal needs (Figure 2). Here, we will focus on homeostatic, circadian, and motivational processes modulating sleep/wake states. A better understanding of these circuits will not only reveal how brain states are regulated, but also how the brain balances conflicting needs. It is important to note that there are additional factors that can modulate sleep/wake behaviors, such as breathing (Morrell and Twigg, 2006; Yackle et al, 2017) and immune state (Krueger et al, 2011), yet they are beyond the scope of this review.
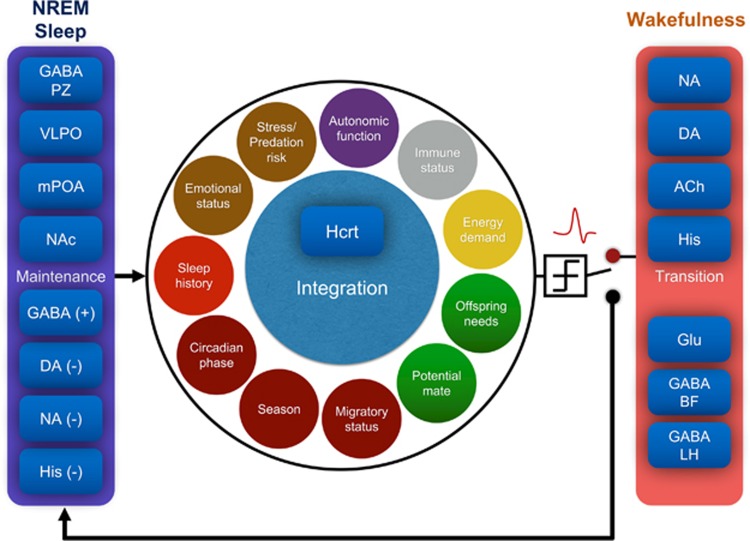
Figure 2.
Schematic of the integrative model of sleep/wake regulation. An ‘integrator’ Hcrt neuron continuously integrates information from multiple and often conflicting variables, and makes the decision whether to fire and wake up the animal, or stay silent and facilitate sleep. The probability of a sleep-to-wake transition depends on (i) the functional connectivity between different sleep/wake regulatory populations, including the GABAergic neurons of the PZ, mPOA, and VLPO and dopaminergic neurons of the VTA, and (ii) internal and external factors, including sleep history, circadian phase, and predation risk. ACh, acetylcholine; BF, basal forebrain; DA, dopamine; GABA, gamma-aminobutyric acid; Glu, glutamate; Hcrt, hypocretin; His, histamine; LH, lateral hypothalamus; mPOA, medial preoptic area; NAc, nucleus accumbens; NA, noradrenaline; PZ, parafacial zone; VLPO, ventral lateral preoptic area.
Homeostatic Regulation of Sleep/Wake States
When wakefulness is extended, sleep pressure accumulates and only dissipates during subsequent sleep. This homeostatic process was initially modeled three decades ago by Borbely in the two-process model of sleep regulation (Borbely, 1982). According to this model, sleep pressure (Process S) increases during wakefulness and declines during sleep in a cycle superimposed over the circadian (~24-h long) cycle of activity (Process C) (Borbely and Achermann, 1999; Borbely et al, 2016). While this model has been highly influential, the neuronal substrate(s) of Process S still remain elusive. More than a century ago, researchers revealed that the cerebrospinal fluid of sleep deprived animals contained substances that can promote sleep in other animals (Ishimori, 1909; Legendre and Pieron, 1913). This finding led to the premise that wake-dependent homeostatic substances accumulate with wakefulness, and when sensed by a homeostatic system promote sleep. Among the homeostatic substances and mechanisms suggested are adenosine and its receptors A1 and A2A, cytokines such as interleukin-1 and tumor necrosis factor-α, prostaglandin D2, and Nitric oxide (Benington and Heller, 1995; Brown et al, 2012; Mang and Franken, 2015; Morairty et al, 2013). Nonetheless, it is still unclear how the brain senses and responds to sleep need, where in the brain this process takes place, and whether lack of sleep is sensed by a master control center (ie, a sleep ‘homeostat’) or by various regulatory neuronal networks. These remain central questions in sleep neurobiology (Allada et al, 2017; Mang and Franken, 2015).
Delta oscillations in the EEG are homeostatically regulated and are thought to reflect the accumulation of sleep pressure during wakefulness, and its discharge during sleep (Mang and Franken, 2015; Vyazovskiy et al, 2011a). Slow oscillations increase with wakefulness, decline with sleep and reach their peak following sleep deprivation (Dijk et al, 1987; Tobler and Borbely, 1986). Interestingly, it has been proposed that sleep intensity, as reflected by delta oscillations during NREM sleep, depends on previous wake time spent in active-motivated wakefulness (theta-dominated wakefulness) rather than only on previous wake duration (Vassalli and Franken, 2017).
Recent studies in Drosophila provide important insights regarding the mechanisms and circuits that may mediate the homeostatic drive for sleep (Allada et al, 2017; Donlea et al, 2017). Sleep in flies can be induced by the activation of sleep-promoting neurons that innervate the dorsal Fan-shaped body (dFB) (Donlea et al, 2011). Sleep need was found to be reflected in dFB neuron excitability—it is elevated to promote sleep after prolonged waking and reduced to permit waking once flies are rested (Donlea et al, 2014). Furthermore, the changes in excitability of dFB neurons were shown to occur through the antagonistic action of dopamine on two potassium channels, Shaker and Sandman (Pimentel et al, 2016). Another study demonstrated that the activation of a subset of Ellipsoid Body neurons elicits a need for sleep that was only apparent following the termination of neuronal activation and not during the activation itself—suggesting a prominent role in sleep drive (Liu et al, 2016). Intriguingly, the activation of certain arousal circuits, rather that wakefulness per se, has been suggested to drive sleep homeostasis in flies (Seidner et al, 2015). Seidner et al. induced wakefulness by activating cholinergic, dopaminergic, and octopaminergic neurons, and demonstrated that only wakefulness induced by the activation of cholinergic neurons results in a sleep rebound while octopaminergic and dopaminergic neurons did not (Seidner et al, 2015). Moreover, activity in octopaminergic neurons seems to directly inhibit a homeostatic sleep response (Seidner et al, 2015). Future studies should further explore these intriguing findings and determine to what extent they can be generalized to mammals.
Recently a forward-genetic approach has been applied to reveal sleep mutants in mammals. By screening sleep patterns in more than 8000 strains of randomly mutagenized mice, researchers revealed two sleep mutants, sleepy and dreamless, that show altered sleep need (Funato et al, 2016). Sleepy mutants, that have a gain-of-function mutation in Sik3 serine-threonine protein kinase, show increased NREM sleep (Funato et al, 2016). Dreamless mutants, that have a mutation in the sodium leak channel NALCN, show a decrease in REM sleep amount and episode duration (Funato et al, 2016). Although the precise mechanisms by which these mutations modulate sleep need are still unclear, the use of unbiased forward-genetic screens in mammals will likely lead to major insights into the pathways and mechanisms of sleep regulation, as it did for circadian biology (Allada et al, 2001; Susaki et al, 2017; Vitaterna et al, 1994).
Circadian Regulation of Sleep/Wake States
Circadian rhythms in activity are apparent in the daily organization of sleep and wakefulness into a specific phase of the day-processes C in the two-process model of sleep regulation (Borbely, 1982; Borbely et al, 2016). Diurnal species such as humans, zebrafish and flies primarily sleep at night while nocturnal species, such as mice, primarily sleep during the day. This pattern of sleep/wake organization follows the light–dark cycle, but also persists in the absence of environmental time cues—demonstrating the existence of an intrinsic regulatory mechanism. This internal rhythm is generated by the circadian clock which regulates numerous physiological and behavioral processes. The circadian clock can be synchronized to the environment by different cues or Zeitgebers (Albrecht, 2012; Saper et al, 2010), such as light (Gooley et al, 2003; Whitmore et al, 2000), temperature (Buhr et al, 2010), stress and exercise (Tahara et al, 2017), food availability (Stephan, 2002), and social interactions (Bloch et al, 2013; Fuchikawa et al, 2016). Internal clocks are advantageous to animals, enabling them to anticipate, rather than react to, daily recurring events and align their physiology and behavior to the changing environment (Albrecht, 2012; Sharma, 2003).
How does the circadian system regulate sleep and wakefulness? In mammals, overt rhythmicity is coordinated by the central pacemaker located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus (Saper, 2013). Lesions to the SCN abolish the daily rhythm in sleep/wake states and lead to sleep fragmentation, including in non-human primates (Edgar et al, 1993). It has been hypothesized that the major output region of the SCN, in regard to sleep/wake regulation, is the dorsomedial nucleus of the hypothalamus (via the subparaventricular zone), which heavily innervates sleep- and wake-promoting nuclei (Deurveilher and Semba, 2005; Saper, 2013). Nonetheless, the precise pathways and roles of the SCN in sleep/wake state synchronization, sleep structure, and sleep quality, remain to be elucidated.
The central circadian clock is comprised of cell-autonomous pacemaker neurons (Allada et al, 2001; Takahashi, 2017). The pacemaker neurons contain a molecular transcription–translation-based autoregulatory feedback loop—in which circadian clock genes rhythmically regulate their own expression (Takahashi, 2017), as well as the expression of thousands of other genes (Zhang et al, 2014). The core clock genes, in mammals, include the Period genes, the Cryptochrome genes, Clock, and Bmal1 (Takahashi, 2017). Besides the core molecular loop, there are additional accessory loops that contribute to the overall precision and robustness of the clock. The clock genes are not only expressed in the central pacemaker, instead, their expression is widespread throughout the body and brain (Dibner et al, 2010; Guilding and Piggins, 2007; Mohawk et al, 2012). Interestingly, recent findings suggest that clock genes outside the SCN can control sleep/wake state, as well as sleep homeostasis (Ehlen et al, 2017; Franken, 2013; Mahoney et al, 2013; Mang et al, 2016; Yu et al, 2014). For example, daily oscillations in the expression of the enzyme histidine decarboxylase in histaminergic TMN neurons depends on Bmal1 (Yu et al, 2014). Deleting Bmal1 specifically from histaminergic TMN neurons resulted in various alterations to sleep/wake architecture including sleep fragmentation, as well as reduced recovery sleep and delta power following sleep deprivation (Yu et al, 2014). A recent study also suggests that Bmal1 expression in skeletal muscle can regulate sleep (Ehlen et al, 2017).
The molecular and neuronal pathways regulating the circadian clock and the sleep/wake cycle in Drosophila have been extensively studied over the last decades (Artiushin and Sehgal, 2017). Since the identification of period (Konopka and Benzer, 1971)—a Drosophila mutant with altered circadian timekeeping—the fly has served as a prominent model for studies of the molecular and neuronal machinery of the circadian system and sleep regulation (Artiushin and Sehgal, 2017; Bargiello et al, 1984; Greenspan et al, 2001; Konopka and Benzer, 1971; Liu et al, 1992). Intriguingly, reverse genetic analysis in Drosophila has also proved a powerful model to study human sleep disorders (Chakravarti et al, 2017; Donelson and Sanyal, 2015).
The circadian pacemaker of Drosophila contains around 75 pairs of neurons that are organized in several groups among which are the pigment-dispersing factor (PDF)-positive small and large ventral lateral neurons (sLNVs and lLNVs), the dorsal neurons (DN1s, DN2s, and DN3s), and the dorsal lateral neurons (Allada and Chung, 2010). Multiple studies have demonstrated that activity in different clock neurons can modulate sleep and wakefulness (Chung et al, 2009; Donlea et al, 2009; Parisky et al, 2008; Shang et al, 2008; Shang et al, 2011; Sheeba et al, 2008). For example, a Drosophila screen identified a cycling gene wide awake (wake), which links circadian clock output to sleep onset in flies and rodents (Liu et al, 2014). WAKE expressed in clock neurons peaks in expression at dusk, and acts downstream of circadian genes to upregulate GABAA receptors, driving sleep (Funato et al, 2016). In addition, PDF secreted from sLNVs neurons was shown to act on DN1s neurons to suppress sleep in anticipation of dawn. Recently, glutamatergic DN1s clock neurons were shown to promote sleep by negative feedback onto key clock neurons that typically promote wakefulness (Guo et al, 2016). These essential studies provide mechanistic insight into the regulation of sleep by neurons that control circadian rhythms.
It is important to note that the sleep/wake state itself can also strongly influence the circadian clock, as well as various physiological rhythms, including hormonal secretion (Archer and Oster, 2015; Czeisler and Buxton, 2017). Moreover, the expression of most rhythms at the behavioral, physiologic, and biochemical levels are thought to be regulated by inputs from both the circadian oscillator and sleep/wake states. Misalignment of circadian rhythms and sleep can have adverse health outcomes, including increased risk for all-cause mortality, obesity, type-2 diabetes, and cardiovascular disease, as well as increased risk for various sleep and neuropsychiatric disorders including major depressive disorder (Archer and Oster, 2015; McClung, 2007; Morris et al, 2016; Scheer et al, 2009; Wulff et al, 2010). A better understanding of the neuronal machinery underlying the association between circadian rhythms and sleep regulation will not only reveal one of the most important regulatory processes of arousal states, but would be important for the development of improved therapeutics.
Motivational Regulation of Sleep/Wake States
Motivational processes can powerfully modulate the propensity of animals to stay awake or go to sleep. When motivated, humans can stay awake and engage in various cognitive and physical activities, such as reading a book, working on a grant, or attending a party, far beyond their physiological bedtime while ignoring homeostatic and circadian sleep drives. In the wild, foraging and mating opportunities and the presence of predators drive motivational responses and modulate arousal. Animals can stay awake for extended periods, sleep longer, sleep lighter, or only with half of the brain in response to different internal and external conditions. A better understanding of the processes modulating the propensity to stay awake or go to sleep will not only improve our understanding of brain circuits regulating arousal states but also provide valuable insights into the general problem of balancing conflicting needs.
For studies on the link between motivational processes and sleep/wake regulation, non-model organisms in natural or semi-natural conditions are of particular interest (Aulsebrook et al, 2016; Rattenborg et al, 2017), as they are typically exposed to an heterogeneous environment in which animals face various competing needs, ie, the need to forage, reproduce, evade predators, and sleep. Laboratory settings rarely provide similar conflicting needs. Moreover, arousal induced by heterogeneous sensory cues could be associated with the activation of neuronal circuits that are not necessarily recruited under standard laboratory conditions.
The energy status of individuals can strongly modulate sleep/wake states. Hunger can drive arousal, locomotor activity, and suppress sleep—probably to favor foraging behavior (Borbely, 1977; Danguir and Nicolaidis, 1979; Dewasmes et al, 1989; Jacobs and McGinty, 1971; Rattenborg et al, 2016). In contrast, satiety can promote sleep (postprandial sleep) (Jenkins et al, 2006; Murphy et al, 2016; Varin et al, 2015). The presence of predators or predator cues can modulate sleep/wake states (Lima et al, 2005), reduce sleep (Lesku et al, 2008), or induce frequent arousals (Dominguez, 2003; Gauthier-Clerc et al, 1998; Lendrem, 1984). Even the motivation to mate and reproduce can powerfully affect sleep and wake states (Lesku et al, 2012). In addition to sleep suppression over long migratory and foraging flights (Rattenborg et al, 2004; Rattenborg et al, 2016), birds and marine mammals can show unihemispheric sleep (Lyamin et al, 2008b; Rattenborg et al, 2000)—a state that allows simultaneous engagement in two otherwise mutually exclusive tasks. For example, marine mammals must continuously rise to the water's surface to breath air and take continuous care of their young during critical early life stages—thus unihemispheric sleep enables them to sleep while engaging in these essentail behaviors (Lyamin et al, 2008b; Rattenborg et al, 2000).
Which neuronal mechanisms modulate the arousal response in face of different environmental and homeostatic needs? Although various wake-promoting neuronal populations have been demonstrated to induce and promote arousal, they vary in their capacity to maintain wakefulness (Eban-Rothschild and de Lecea, 2017a; Luppi et al, 2017; Weber and Dan, 2016). Increasing evidence also suggests that different wake-promoting neuronal populations have distinct roles in supporting arousal under specific environmental and motivational conditions (Eban-Rothschild et al, 2017b; Joiner, 2016; Jones, 2003; Scammell et al, 2017).
In laboratory settings, LH Hcrt neurons have been implicated in mediating the increase in wakefulness in response to stressful conditions (Bonnavion et al, 2015; Mahler et al, 2014; Winsky-Sommerer et al, 2005), including reduced food availability (Yamanaka et al, 2003). Hcrt neurons are inhibited by an increase in extracellular glucose concentrations and circulating leptin, while they are excited by elevations in the hunger hormone ghrelin or hypoglycemia (Burdakov and Alexopoulos, 2005; Burdakov et al, 2006; Sakurai, 2007; Williams et al, 2008). Moreover, the sensitivity of Hcrt neurons to triglycerides, carbon dioxide, and pH further support a role for Hcrt neurons in sensing metabolic factors (Williams et al, 2007). Optogenetic stimulation of leptin receptor-expressing (LepRb) GABAergic neurons in the LH produces postsynaptic currents in Hcrt cells (Bonnavion et al, 2015), while other GABAergic neurons in the LH, labeled with neurotensin, interact with Hcrt cells to regulate arousal (Grossberg et al, 2011; Jennings et al, 2015). Remarkably, transgenic mice in which Hcrt neurons are ablated (Hcrt ataxin3 mice) do not show an increase in wakefulness or locomotion following fasting (Yamanaka et al, 2003). Together these findings suggest that Hcrt neurons are an important link between metabolism and arousal regulation (Bonnavion and de Lecea, 2010).
VTA dopaminergic neurons are principal regulators of motivational processes (Berridge and Robinson, 1998; Salamone et al, 2016; Schultz, 2007), and are well suited to promote arousal for the support of various motivated behaviors, including mate-seeking and predator defense. VTA dopaminergic neurons are strongly activated by salient stimuli in primates and rodents (Salamone et al, 2016; Schultz, 2007; Wise, 2004), and their chemogenetic inhibition in mice prevents the maintenance of wakefulness in the presence of various salient stimuli, including a potential mate and predator scent (Eban-Rothschild et al, 2016). Intriguingly, chemogenetic inhibition of VTA dopaminergic neurons does not suppress the motivation to engage in sleep-related and goal-directed behaviors, such as nest-building (Eban-Rothschild et al, 2016). This finding suggests that the motivation to stay awake, and engage in wake-related behaviors, is regulated by different neuronal circuits than the motivation to sleep or engage in sleep-related behaviors.
Cholinergic neurons have been associated with unihemispheric cortical activation during unihemispheric sleep (Lapierre et al, 2007). Northern fur seals (Callorhinus ursinus) show unihemispheric sleep preferentially while sleeping in the water (Lyamin et al, 2017). Acetylcholine release was found to be lateralized and tightly linked to the hemisphere that was awake, while histamine, serotonin, and noradrenaline were found to be synchronously released from both the awake and asleep hemispheres (Lapierre et al, 2013; Lapierre et al, 2007; Lyamin et al, 2016). These findings suggest that acetylcholine is responsible for unihemispheric EEG activation, and future studies modulating the activity of cholinergic neurons would provide causal evidence.
In the fly, octopaminergic neurons (the Drosophila counterpart of noradrenaline), have been shown to mediate sleep suppression by male sexual arousal (Machado et al, 2017). Male flies suppress their sleep in the presence of females, and female-induced arousal is attenuated by sexual satiety or elevated sleep drive (Machado et al, 2017). Recently, a small number of octopaminergic neurons, designated as male-specific 1 (MS1) neurons, were found to regulate male sleep in a sexual context. The activation of MS1 neurons in isolated males reduces sleep, while their inhibition in the presence of a female attenuate mating-induced arousal (Machado et al, 2017).
Taken together, the work reviewed here suggests that different arousal systems support wakefulness under distinct environmental and internal needs. A distinct role for each system in support of a distinct behavioral state could account for the apparent redundancy in wake-promoting circuits (Eban-Rothschild et al, 2017a; Jones, 2003).
An integrative model for sleep/wake regulation
In recent years, various theoretical models have been proposed to explain how and where the brain integrates information and computes a decision whether to wake up or stay asleep. The flip/flop model (Saper et al, 2010), posits that mutually inhibitory interactions of wake-promoting and sleep-promoting neurons produce a state similar to a flip/flop switch in an electrical circuit. This concept has served as a helpful framework to explain interactions between wake and sleep-promoting neuronal circuits that minimize intermediate states between sleep and wakefulness. However, intermediate and dissociated arousal states do exist, for example in narcolepsy and REM behavior disorder (de Lecea, 2015). In addition, unihemispheric (Lyamin et al, 2008b; Rattenborg et al, 2000) and local sleep (Huber et al, 2004; Krueger et al, 2008; Nir et al, 2011; Vyazovskiy et al, 2011b) phenomena are difficult to explain in a simple binary decision. The flip/flop switch assumes mutual inhibition dynamics with fast synaptic transmission that cannot explain the relatively slow onset of sleep. Furthermore, spontaneous awakening is associated with sleep inertia—a state of impaired cognitive and sensory/motor performance immediately after awakening. Sleep inertia is thought to result from the insertion of sleep-like patterns of cortical neuronal activity into the awake state (Vyazovskiy et al, 2014). In view of the complexity of the aforementioned arousal circuits uncovered during the last decade, the binary states proposed by the flip/flop model need to be revised.
Integrator Circuit
As an alternative to the flip/flop binary model, we propose a theoretical framework to predict sleep-to-wake transitions based on an online classification function: depending on inputs with different ‘weights’, an ‘integrator’ neuron classifies the state as sleep or wake. We hypothesize that an ‘integrator’ neuron (eg, LH Hcrt neurons) continuously integrate information from multiple variables and make the decision whether to fire and wake up the animal, or stay silent and facilitate NREM sleep (Figure 2). Comparisons of the latencies to wakefulness following optogenetic stimulations of pairs of neuronal structures (Carter and de Lecea, 2011), has prompted us to build a ‘hierarchy’ matrix that assigns probabilities of eliciting wakefulness (Sorooshyari et al, 2015). A more sophisticated iteration of this model would consider a continuous activation function that reflects probabilities of wakefulness. These probabilities would depend on (i) the functional connectivity between different neurotransmitter systems, and (ii) physiological factors, such as those discussed above (Figure 2). Some of these inputs to the integrator could be associated with individual neuronal circuits, whereas the contribution of other functions whose output depends on sequential input from multiple systems (such as metabolism and limbic structures) may be modeled by hidden layers. Optogenetic interrogations of these circuits will allow us to establish the maximum contribution of each circuit to sleep/wake dynamics (Carter and de Lecea, 2011; Sorooshyari et al, 2015).
An essential part of this model is the time scale of integration and information flow. In other words: how long does it take for an integrator neuron to decide whether it is appropriate to fire? Short integration times would be very sensitive to environmental and physiological inputs, but would make sleep bouts short and the sleep cycle fragmented. In contrast, long integration times would result in stable sleep at the expense of disconnection from the environment. The relatively long-time scale of neuromodulators seems to fit optimally with integration times lasting several seconds. These timescales are also consistent with the minimum quantum of sleep continuity necessary for memory consolidation (Rolls et al, 2011). The neuromodulation time scale could also explain the dynamics of cortical output circuits and the observed phenomenon of local sleep. A threshold function would determine whether sufficient cortical domains are engaged in local sleep to generate synchronous oscillations associated with NREM sleep.
Conclusions
The advent of in vivo cell type-specific chemogenetic and optogenetic tools and genetically encoded calcium indicators has allowed researchers to interrogate the role of specific neuronal circuits in the control of sleep/wake states. From the first optogenetic experiment in freely moving animals investigating whether the activity of Hcrt neurons is sufficient to induce sleep-to-wake transitions, multiple studies have contributed to our current understanding of the roles of different populations in sleep/wake regulation. Now, it is time to further explore how information from the diverse sleep/wake regulatory populations is integrated to control overt arousal and how diverse external and internal inputs modulate the regulatory capacities of these circuits. These studies would contribute to the understanding of the different functions of sleep and wakefulness.
Funding and disclosure
This work was supported by the Katharine McCormick Advanced postdoctoral fellowships (AE-R), the NIH grants: R01-MH087592, R01-MH102638, and R01-AG04767 (LdL), a Brain and Behavior Research Foundation (NARSAD) Grant (LdL), a Merck Grant (LdL) and a Johnson & Johnson Grant (LdL), Legacy Heritage Biomedical Program of the Israel Science Foundation (992/14, LA), Israel Science Foundation (690/15, LA) and the US Israel Binational Science Foundation (2011335, LA and LdL). The authors declare no conflict of interest.
Acknowledgments
We thank Jeremy C. Borniger, Suszie Tyree, and the referees for helpful comments on this manuscript.
References
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L (2007). Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450: 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airaksinen MS, Alanen S, Szabat E, Visser TJ, Panula P (1992). Multiple neurotransmitters in the tuberomammillary nucleus: comparison of rat, mouse, and guinea pig. J Comp Neurol 323: 103–116. [DOI] [PubMed] [Google Scholar]
- Albrecht U (2012). Timing to perfection: the biology of central and peripheral circadian clocks. Neuron 74: 246–260. [DOI] [PubMed] [Google Scholar]
- Allada R, Chung BY (2010). Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72: 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Cirelli C, Sehgal A (2017). Molecular mechanisms of sleep homeostasis in flies and mammals. Cold Spring Harbor Perspect Biol 9 pii: a027730; doi: 10.1101/cshperspect.a027730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M (2001). Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci 24: 1091–1119. [DOI] [PubMed] [Google Scholar]
- Allada R, Siegel JM (2008). Unearthing the phylogenetic roots of sleep. Curr Biol 18: R670–R679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaclet C, Ferrari L, Arrigoni E, Bass CE, Saper CB, Lu J et al (2014). The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat Neurosci 17: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaclet C, Parmentier R, Ouk K, Guidon G, Buda C, Sastre JP et al (2009). Orexin/hypocretin and histamine: distinct roles in the control of wakefulness demonstrated using knock-out mouse models. J Neurosci 29: 14423–14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaclet C, Pedersen NP, Ferrari LL, Venner A, Bass CE, Arrigoni E et al (2015). Basal forebrain control of wakefulness and cortical rhythms. Nat Commun 6: 8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ (2005). Dopaminergic modulation of arousal in Drosophila. Curr Biol 15: 1165–1175. [DOI] [PubMed] [Google Scholar]
- Appelbaum L, Wang GX, Maro GS, Mori R, Tovin A, Marin W et al (2009). Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc Natl Acad Sci USA 106: 21942–21947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SN, Oster H (2015). How sleep and wakefulness influence circadian rhythmicity: effects of insufficient and mistimed sleep on the animal and human transcriptome. J Sleep Res 24: 476–493. [DOI] [PubMed] [Google Scholar]
- Arrigoni E, Chen MC, Fuller PM (2016). The anatomical, cellular and synaptic basis of motor atonia during rapid eye movement sleep. J Physiol 594: 5391–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G, Sehgal A (2017). The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol 44: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones G, Bloom FE (1981. a). Activity of norepinephrine-containing locus coeruleus neurons in behaving rats anticipates fluctuations in the sleep-waking cycle. J Neurosci 1: 876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones G, Bloom FE (1981. b). Norepinephrine-containing locus coeruleus neurons in behaving rats exhibit pronounced responses to non-noxious environmental stimuli. J Neurosci 1: 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulsebrook AE, Jones TM, Rattenborg NC, Roth TC 2nd, Lesku JA (2016). Sleep ecophysiology: integrating neuroscience and ecology. Trends Ecol Evol 31: 590–599. [DOI] [PubMed] [Google Scholar]
- Bargiello TA, Jackson FR, Young MW (1984). Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312: 752–754. [DOI] [PubMed] [Google Scholar]
- Barlow IL, Rihel J (2017). Zebrafish sleep: from geneZZZ to neuronZZZ. Curr Opin Neurobiol 44: 65–71. [DOI] [PubMed] [Google Scholar]
- Benington JH, Heller HC (1995). Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol 45: 347–360. [DOI] [PubMed] [Google Scholar]
- Bernard JF, Carroue J, Besson JM (1991). Efferent projections from the external parabrachial area to the forebrain: a Phaseolus vulgaris leucoagglutinin study in the rat. Neurosci Lett 122: 257–260. [DOI] [PubMed] [Google Scholar]
- Berridge CW, Espana RA (2000). Synergistic sedative effects of noradrenergic alpha(1)- and beta-receptor blockade on forebrain electroencephalographic and behavioral indices. Neuroscience 99: 495–505. [DOI] [PubMed] [Google Scholar]
- Berridge KC, Robinson TE (1998). What is the role of dopamine in reward: hedonic impact, reward learning, or incentive salience? Brain Res Brain Res Rev 28: 309–369. [DOI] [PubMed] [Google Scholar]
- Blanco-Centurion C, Gerashchenko D, Shiromani PJ (2007). Effects of saporin-induced lesions of three arousal populations on daily levels of sleep and wake. J Neurosci 27: 14041–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch G, Herzog ED, Levine JD, Schwartz WJ (2013). Socially synchronized circadian oscillators. Proc Biol Sci 280: 20130035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnavion P, de Lecea L (2010). Hypocretins in the control of sleep and wakefulness. Curr Neurol Neurosci Rep 10: 174–179. [DOI] [PubMed] [Google Scholar]
- Bonnavion P, Jackson AC, Carter ME, de Lecea L (2015). Antagonistic interplay between hypocretin and leptin in the lateral hypothalamus regulates stress responses. Nat Commun 6: 6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbely AA (1977). Sleep in the rat during food deprivation and subsequent restitution of food. Brain Res 124: 457–471. [DOI] [PubMed] [Google Scholar]
- Borbely AA (1982). A two process model of sleep regulation. Hum Neurobiol 1: 195–204. [PubMed] [Google Scholar]
- Borbely AA, Achermann P (1999). Sleep homeostasis and models of sleep regulation. J Biol Rhythm 14: 557–568. [DOI] [PubMed] [Google Scholar]
- Borbely AA, Daan S, Wirz-Justice A, Deboer T (2016). The two-process model of sleep regulation: a reappraisal. J Sleep Res 25: 131–143. [DOI] [PubMed] [Google Scholar]
- Boucetta S, Cisse Y, Mainville L, Morales M, Jones BE (2014). Discharge profiles across the sleep-waking cycle of identified cholinergic, GABAergic, and glutamatergic neurons in the pontomesencephalic tegmentum of the rat. J Neurosci 34: 4708–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrel B, Koob GF (2004). What keeps us awake: the neuropharmacology of stimulants and wakefulness-promoting medications. Sleep 27: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Branch AF, Navidi W, Tabuchi S, Terao A, Yamanaka A, Scammell TE et al (2016). Progressive loss of the orexin neurons reveals dual effects on wakefulness. Sleep 39: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brischoux F, Chakraborty S, Brierley DI, Ungless MA (2009). Phasic excitation of dopamine neurons in ventral VTA by noxious stimuli. Proc Natl Acad Sci USA 106: 4894–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW (2012). Control of sleep and wakefulness. Physiol Rev 92: 1087–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS (2010). Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdakov D, Alexopoulos H (2005). Metabolic state signalling through central hypocretin/orexin neurons. J Cell Mol Med 9: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdakov D, Jensen LT, Alexopoulos H, Williams RH, Fearon IM, O'Kelly I et al (2006). Tandem-pore K+ channels mediate inhibition of orexin neurons by glucose. Neuron 50: 711–722. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Bickford RG, Ponomareff G, Thal LJ, Mandel R, Gage FH (1988). Nucleus basalis and thalamic control of neocortical activity in the freely moving rat. J Neurosci 8: 4007–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SS, Tobler I (1984). Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev 8: 269–300. [DOI] [PubMed] [Google Scholar]
- Carter ME, Brill J, Bonnavion P, Huguenard JR, Huerta R, de Lecea L (2012). Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc Natl Acad Sci USA 109: E2635–E2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, de Lecea L (2011). Optogenetic investigation of neural circuits in vivo. Trends Mol Med 17: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, de Lecea L, Adamantidis A (2013). Functional wiring of hypocretin and LC-NE neurons: implications for arousal. Front Behav Neurosci 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Yizhar O, Chikahisa S, Nguyen H, Adamantidis A, Nishino S et al (2010). Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat Neurosci 13: 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti L, Moscato EH, Kayser MS (2017). Unraveling the neurobiology of sleep and sleep disorders using drosophila. Curr Top Dev Biol 121: 253–285. [DOI] [PubMed] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C et al (1999). Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98: 437–451. [DOI] [PubMed] [Google Scholar]
- Chen L, Yin D, Wang TX, Guo W, Dong H, Xu Q et al (2016. a). Basal forebrain cholinergic neurons primarily contribute to inhibition of electroencephalogram delta activity, rather than inducing behavioral wakefulness in mice. Neuropsychopharmacology 41: 2133–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chiu CN, McArthur KL, Fetcho JR, Prober DA (2016. b). TRP channel mediated neuronal activation and ablation in freely behaving zebrafish. Nat Methods 13: 147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JR, Treweek JB, Robinson JE, Xiao C, Bremner LR, Greenbaum A et al (2017). Dorsal raphe dopamine neurons modulate arousal and promote wakefulness by salient stimuli. Neuron 94: 1205–1219.e1208. [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R (2009). The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 19: 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G (2008). Is sleep essential? PLoS Biol 6: e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JY, Haesler S, Vong L, Lowell BB, Uchida N (2012). Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature 482: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinople CM, Bruno RM (2011). Effects and mechanisms of wakefulness on local cortical networks. Neuron 69: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeisler CA, Buxton OM (2017) Human Circadian Timing System and Sleep-Wake Regulation. In: Kryger Meir, Roth T, Dement WC (eds). Principles and Practice of Sleep Medicine. Sixth Edition. Elsevier. Chapter 35, pp 362–376. [Google Scholar]
- Dahan L, Astier B, Vautrelle N, Urbain N, Kocsis B, Chouvet G (2007). Prominent burst firing of dopaminergic neurons in the ventral tegmental area during paradoxical sleep. Neuropsychopharmacology 32: 1232–1241. [DOI] [PubMed] [Google Scholar]
- Danguir J, Nicolaidis S (1979). Dependence of sleep on nutrients' availability. Physiol Behav 22: 735–740. [DOI] [PubMed] [Google Scholar]
- de Lecea L (2015). Optogenetic control of hypocretin (orexin) neurons and arousal circuits. Curr Top Behav Neurosci 25: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE et al (1998). The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci USA 95: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lecea L, Sutcliffe JG (2005). The hypocretins and sleep. FEBS J 272: 5675–5688. [DOI] [PubMed] [Google Scholar]
- De Sarro GB, Ascioti C, Froio F, Libri V, Nistico G (1987). Evidence that locus coeruleus is the site where clonidine and drugs acting at alpha 1- and alpha 2-adrenoceptors affect sleep and arousal mechanisms. Br J Pharmacol 90: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurveilher S, Semba K (2005). Indirect projections from the suprachiasmatic nucleus to major arousal-promoting cell groups in rat: implications for the circadian control of behavioural state. Neuroscience 130: 165–183. [DOI] [PubMed] [Google Scholar]
- Dewasmes G, Duchamp C, Minaire Y (1989). Sleep changes in fasting rats. Physiol Behav 46: 179–184. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U (2010). The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549. [DOI] [PubMed] [Google Scholar]
- Dijk DJ, Beersma DG, Daan S (1987). EEG power density during nap sleep: reflection of an hourglass measuring the duration of prior wakefulness. J Biol Rhythm 2: 207–219. [DOI] [PubMed] [Google Scholar]
- Dominguez J (2003). Sleeping and vigilance in Black-tailed Godwit. J Ethol 21: 57–60. [Google Scholar]
- Donelson NC, Sanyal S (2015). Use of Drosophila in the investigation of sleep disorders. Exp Neurol 274(Pt A): 72–79. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Alam MN, Szymusiak R (2017). Neuronal substrates of sleep homeostasis; lessons from flies, rats and mice. Curr Opin Neurobiol 44: 228–235. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Miesenbock G (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N, Shaw PJ (2009). Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332: 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboue ER, Keene AC, Borowsky RL (2011). Evolutionary convergence on sleep loss in cavefish populations. Curr Biol 21: 671–676. [DOI] [PubMed] [Google Scholar]
- Duque A, Balatoni B, Detari L, Zaborszky L (2000). EEG correlation of the discharge properties of identified neurons in the basal forebrain. J Neurophysiol 84: 1627–1635. [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A, Bloch G (2012). Social influences on circadian rhythms and sleep in insects. Adv Genet 77: 1–32. [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A, Bloch G (2015). The colony environment modulates sleep in honey bee workers. J Exp Biol 218(Pt 3): 404–411. [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A, de Lecea L (2017. a). Neuronal substrates for initiation, maintenance, and structural organization of sleep/wake states. F1000Res 6: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eban-Rothschild A, Giardino WJ, de Lecea L (2017. b). To sleep or not to sleep: neuronal and ecological insights. Curr Opin Neurobiol 44: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eban-Rothschild A, Rothschild G, Giardino WJ, Jones JR, de Lecea L (2016). VTA dopaminergic neurons regulate ethologically relevant sleep-wake behaviors. Nat Neurosci 19: 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eban-Rothschild AD, Bloch G (2008). Differences in the sleep architecture of forager and young honeybees (Apis mellifera. J Exp Biol 211(Pt 15): 2408–2416. [DOI] [PubMed] [Google Scholar]
- Edgar DM, Dement WC, Fuller CA (1993). Effect of SCN lesions on sleep in squirrel monkeys: evidence for opponent processes in sleep-wake regulation. J Neurosci 13: 1065–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlen JC, Brager AJ, Baggs J, Pinckney L, Gray CL, DeBruyne JP et al (2017). Bmal1 function in skeletal muscle regulates sleep. eLife 6 pii: e26557; doi: 10.7554/eLife.26557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz I, Levitas-Djerbi T, Appelbaum L (2017). The hypocretin/orexin neuronal networks in zebrafish. Curr Top Behav Neurosci 33: 75–92. [DOI] [PubMed] [Google Scholar]
- Elbaz I, Yelin-Bekerman L, Nicenboim J, Vatine G, Appelbaum L (2012). Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish. J Neurosci 32: 12961–12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espana RA, Scammell TE (2011). Sleep neurobiology from a clinical perspective. Sleep 34: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco JH, Appelbaum L, Marin W, Gaus SE, Mourrain P, Mignot E (2006). Regulation of hypocretin (orexin) expression in embryonic zebrafish. J Biol Chem 281: 29753–29761. [DOI] [PubMed] [Google Scholar]
- Flicker C, Geyer MA (1982). The hippocampus as a possible site of action for increased locomotion during intracerebral infusions of norepinephrine. Behav Neural Biol 34: 421–426. [DOI] [PubMed] [Google Scholar]
- Franken P (2013). A role for clock genes in sleep homeostasis. Curr Opin Neurobiol 23: 864–872. [DOI] [PubMed] [Google Scholar]
- Fuchikawa T, Eban-Rothschild A, Nagari M, Shemesh Y, Bloch G (2016). Potent social synchronization can override photic entrainment of circadian rhythms. Nat Commun 7: 11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A, Bonnavion P, Wilson MH, Mickelsen LE, Bloit J, de Lecea L et al (2017). Hypothalamic tuberomammillary nucleus neurons: electrophysiological diversity and essential role in arousal stability. J Neurosci 37: 9574–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller PM, Sherman D, Pedersen NP, Saper CB, Lu J (2011). Reassessment of the structural basis of the ascending arousal system. J Comp Neurol 519: 933–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulwiler CE, Saper CB (1984). Subnuclear organization of the efferent connections of the parabrachial nucleus in the rat. Brain Res 319: 229–259. [DOI] [PubMed] [Google Scholar]
- Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z et al (2016). Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539: 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Clerc M, Tamisier A, Cezilly F (1998). Sleep-vigilance trade-off in Green-winged Teals (Anas crecca crecca). Can J Zool 76: 2214–2218. [Google Scholar]
- Goard M, Dan Y (2009). Basal forebrain activation enhances cortical coding of natural scenes. Nat Neurosci 12: 1444–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley JJ, Lu J, Fischer D, Saper CB (2003). A broad role for melanopsin in nonvisual photoreception. J Neurosci 23: 7093–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ, Tononi G, Cirelli C, Shaw PJ (2001). Sleep and the fruit fly. Trends Neurosci 24: 142–145. [DOI] [PubMed] [Google Scholar]
- Grossberg AJ, Zhu X, Leinninger GM, Levasseur PR, Braun TP, Myers MG Jr. et al (2011). Inflammation-induced lethargy is mediated by suppression of orexin neuron activity. J Neurosci 31: 11376–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilding C, Piggins HD (2007). Challenging the omnipotence of the suprachiasmatic timekeeper: are circadian oscillators present throughout the mammalian brain? Eur J Neurosci 25: 3195–3216. [DOI] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A et al (2014). Natural neural projection dynamics underlying social behavior. Cell 157: 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC et al (2016). Circadian neuron feedback controls the Drosophila sleep—activity profile. Nature 536: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas HL, Sergeeva OA, Selbach O (2008). Histamine in the nervous system. Physiol Rev 88: 1183–1241. [DOI] [PubMed] [Google Scholar]
- Han Y, Shi YF, Xi W, Zhou R, Tan ZB, Wang H et al (2014). Selective activation of cholinergic basal forebrain neurons induces immediate sleep-wake transitions. Curr Biol 24: 693–698. [DOI] [PubMed] [Google Scholar]
- Hassani OK, Lee MG, Henny P, Jones BE (2009. a). Discharge profiles of identified GABAergic in comparison to cholinergic and putative glutamatergic basal forebrain neurons across the sleep-wake cycle. J Neurosci 29: 11828–11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani OK, Lee MG, Jones BE (2009. b). Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci USA 106: 2418–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A et al (2000). Rest in Drosophila is a sleep-like state. Neuron 25: 129–138. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI (2003). Modafinil maintains waking in the fruit fly drosophila melanogaster. Sleep 26: 139–146. [DOI] [PubMed] [Google Scholar]
- Herrera CG, Cadavieco MC, Jego S, Ponomarenko A, Korotkova T, Adamantidis A (2016). Hypothalamic feedforward inhibition of thalamocortical network controls arousal and consciousness. Nat Neurosci 19: 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson JA, McCarley RW, Wyzinski PW (1975). Sleep cycle oscillation: reciprocal discharge by two brainstem neuronal groups. Science 189: 55–58. [DOI] [PubMed] [Google Scholar]
- Huber R, Ghilardi MF, Massimini M, Tononi G (2004). Local sleep and learning. Nature 430: 78–81. [DOI] [PubMed] [Google Scholar]
- Hur EE, Zaborszky L (2005). Vglut2 afferents to the medial prefrontal and primary somatosensory cortices: a combined retrograde tracing in situ hybridization study [corrected]. J Comp Neurol 483: 351–373. [DOI] [PubMed] [Google Scholar]
- Ikeda-Sagara M, Ozaki T, Shahid M, Morioka E, Wada K, Honda K et al (2012). Induction of prolonged, continuous slow-wave sleep by blocking cerebral H(1) histamine receptors in rats. Br J Pharmacol 165: 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmak SO, de Lecea L (2014). Basal forebrain cholinergic modulation of sleep transitions. Sleep 37: 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimori K (1909). True cause of sleep: a hypnogenic substance as evidenced in the brain of sleep-deprived animals. Tokyo Igakkai Zasshi 23: 429–457. [Google Scholar]
- Jacobs BL, Fornal CA (1993). 5-HT and motor control: a hypothesis. Trends Neurosci 16: 346–352. [DOI] [PubMed] [Google Scholar]
- Jacobs BL, McGinty DJ (1971). Effects of food deprivation on sleep and wakefulness in the rat. Exp Neurol 30: 212–222. [DOI] [PubMed] [Google Scholar]
- Jenkins JB, Omori T, Guan Z, Vgontzas AN, Bixler EO, Fang J (2006). Sleep is increased in mice with obesity induced by high-fat food. Physiol Behav 87: 255–262. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Ung RL, Resendez SL, Stamatakis AM, Taylor JG, Huang J et al (2015). Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 160: 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Ramanathan L, Siegel JM (2008). Rapid changes in glutamate levels in the posterior hypothalamus across sleep-wake states in freely behaving rats. Am J Physiol 295: R2041–R2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ (2016). Unraveling the evolutionary determinants of sleep. Curr Biol 26: R1073–R1087.27780049 [Google Scholar]
- Jones BE (2003). Arousal systems. Front Biosci 8: s438–s451. [DOI] [PubMed] [Google Scholar]
- Jones BE (2017). Principal cell types of sleep-wake regulatory circuits. Curr Opin Neurobiol 44: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BE, Bobillier P, Pin C, Jouvet M (1973). The effect of lesions of catecholamine-containing neurons upon monoamine content of the brain and EEG and behavioral waking in the cat. Brain Res 58: 157–177. [DOI] [PubMed] [Google Scholar]
- Jouvet M (1972). The role of monoamines and acetylcholine-containing neurons in the regulation of the sleep-waking cycle. Ergeb Physiol 64: 166–307. [DOI] [PubMed] [Google Scholar]
- Kaiser W (1988). Busy bees need rest, too. J Comp Physiol A 163: 565–584. [Google Scholar]
- Kaur S, Pedersen NP, Yokota S, Hur EE, Fuller PM, Lazarus M et al (2013). Glutamatergic signaling from the parabrachial nucleus plays a critical role in hypercapnic arousal. J Neurosci 33: 7627–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Adhikari A, Deisseroth K (2017). Integration of optogenetics with complementary methodologies in systems neuroscience. Nat Rev Neurosci 18: 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Thankachan S, McKenna JT, McNally JM, Yang C, Choi JH et al (2015). Cortically projecting basal forebrain parvalbumin neurons regulate cortical gamma band oscillations. Proc Natl Acad Sci USA 112: 3535–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S (1971). Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger D, Ferrari LL, Petit G, Mahoney CE, Fuller PM, Arrigoni E et al (2017). Cholinergic, glutamatergic, and GABAergic neurons of the pedunculopontine tegmental nucleus have distinct effects on sleep/wake behavior in mice. J Neurosci 37: 1352–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JM, Majde JA, Rector DM (2011). Cytokines in immune function and sleep regulation. Handb Clin Neurol 98: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JM, Rector DM, Roy S, Van Dongen HP, Belenky G, Panksepp J (2008). Sleep as a fundamental property of neuronal assemblies. Nat Rev Neurosci 9: 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR (2005). Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM et al (2012). Input-specific control of reward and aversion in the ventral tegmental area. Nature 491: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre JL, Kosenko PO, Kodama T, Peever JH, Mukhametov LM, Lyamin OI et al (2013). Symmetrical serotonin release during asymmetrical slow-wave sleep: implications for the neurochemistry of sleep-waking states. J Neurosci 33: 2555–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre JL, Kosenko PO, Lyamin OI, Kodama T, Mukhametov LM, Siegel JM (2007). Cortical acetylcholine release is lateralized during asymmetrical slow-wave sleep in northern fur seals. J Neurosci 27: 11999–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M, Chen JF, Urade Y, Huang ZL (2013). Role of the basal ganglia in the control of sleep and wakefulness. Curr Opin Neurobiol 23: 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Hassani OK, Alonso A, Jones BE (2005. a). Cholinergic basal forebrain neurons burst with theta during waking and paradoxical sleep. J Neurosci 25: 4365–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Hassani OK, Jones BE (2005. b). Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. J Neurosci 25: 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre R, Pieron H (1913). Recherches sur le besoin de sommeil consecutif a une veille prolongee. Z Allgem Physiol 14: 235–262. [Google Scholar]
- Lendrem DW (1984). Sleeping and vigilance in birds, II. An experimental study of the Barbary dove (Streptopelia risoria. Animal Behaviour 32: 243–248. [Google Scholar]
- Lesku JA, Bark RJ, Martinez-Gonzalez D, Rattenborg NC, Amlaner CJ, Lima SL (2008). Predator-induced plasticity in sleep architecture in wild-caught Norway rats (Rattus norvegicus. Behav Brain Res 189: 298–305. [DOI] [PubMed] [Google Scholar]
- Lesku JA, Rattenborg NC, Valcu M, Vyssotski AL, Kuhn S, Kuemmeth F et al (2012). Adaptive sleep loss in polygynous pectoral sandpipers. Science 337: 1654–1658. [DOI] [PubMed] [Google Scholar]
- Levitas-Djerbi T, Appelbaum L (2017). Modeling sleep and neuropsychiatric disorders in zebrafish. Curr Opin Neurobiol 44: 89–93. [DOI] [PubMed] [Google Scholar]
- Lidbrink P (1974). The effect of lesions of ascending noradrenaline pathways on sleep and waking in the rat. Brain Res 74: 19–40. [DOI] [PubMed] [Google Scholar]
- Lima SL, Rattenborg NC, Lesku JA, Amlaner CJ (2005). Sleeping under the risk of predation. Anim Behav 70: 723–736. [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X et al (1999). The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98: 365–376. [DOI] [PubMed] [Google Scholar]
- Lin MZ, Schnitzer MJ (2016). Genetically encoded indicators of neuronal activity. Nat Neurosci 19: 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M et al (2014). WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, Wu MN (2016). Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165: 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zwiebel LJ, Hinton D, Benzer S, Hall JC, Rosbash M (1992). The period gene encodes a predominantly nuclear protein in adult Drosophila. J Neurosci 12: 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz ML, Adamantidis AR (2017). Monoaminergic control of brain states and sensory processing: existing knowledge and recent insights obtained with optogenetics. Prog Neurobiol 151: 237–253. [DOI] [PubMed] [Google Scholar]
- Lu J, Jhou TC, Saper CB (2006). Identification of wake-active dopaminergic neurons in the ventral periaqueductal gray matter. J Neurosci 26: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi PH, Peyron C, Fort P (2017). Not a single but multiple populations of GABAergic neurons control sleep. Sleep Med Rev 32: 85–94. [DOI] [PubMed] [Google Scholar]
- Lyamin OI, Lapierre JL, Kosenko PO, Kodama T, Bhagwandin A, Korneva SM et al (2016). Monoamine release during unihemispheric sleep and unihemispheric waking in the fur seal. Sleep 39: 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamin OI, Lapierre JL, Kosenko PO, Mukhametov LM, Siegel JM (2008. a). Electroencephalogram asymmetry and spectral power during sleep in the northern fur seal. J Sleep Res 17: 154–165. [DOI] [PubMed] [Google Scholar]
- Lyamin OI, Manger PR, Ridgway SH, Mukhametov LM, Siegel JM (2008. b). Cetacean sleep: an unusual form of mammalian sleep. Neurosci Biobehav Rev 32: 1451–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamin OI, Mukhametov LM, Siegel JM (2017). Sleep in the northern fur seal. Curr Opin Neurobiol 44: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado DR, Afonso DJ, Kenny AR, Oztu Rk-Colak A, Moscato EH, Mainwaring B et al (2017). Identification of octopaminergic neurons that modulate sleep suppression by male sex drive. eLife 6 e23130, doi: 10.7554/eLife.23130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclver MB, Mikulec AA, Amagasu SM, Monroe FA (1996). Volatile anesthetics depress glutamate transmission via presynaptic actions. Anesthesiology 85: 823–834. [DOI] [PubMed] [Google Scholar]
- Mahler SV, Moorman DE, Smith RJ, James MH, Aston-Jones G (2014). Motivational activation: a unifying hypothesis of orexin/hypocretin function. Nat Neurosci 17: 1298–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney CE, Brewer JM, Bittman EL (2013). Central control of circadian phase in arousal-promoting neurons. PloS ONE 8: e67173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang GM, Franken P (2015). Genetic dissection of sleep homeostasis. Curr Top Behav Neurosci 25: 25–63. [DOI] [PubMed] [Google Scholar]
- Mang GM, La Spada F, Emmenegger Y, Chappuis S, Ripperger JA, Albrecht U et al (2016). Altered sleep homeostasis in rev-erbalpha knockout mice. Sleep 39: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA (2007). Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther 114: 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinley MJ, Vinck M, Reimer J, Batista-Brito R, Zagha E, Cadwell CR et al (2015). Waking state: rapid variations modulate neural and behavioral responses. Neuron 87: 1143–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty DJ, Harper RM (1976). Dorsal raphe neurons: depression of firing during sleep in cats. Brain research 101: 569–575. [DOI] [PubMed] [Google Scholar]
- Michel M, Lyons LC (2014). Unraveling the complexities of circadian and sleep interactions with memory formation through invertebrate research. Front Syst Neurosci 8: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskiy BY, Kiyashchenko LI, Siegel JM (2005). Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron 46: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Farber J, Gatz P, Roffwarg H, German DC (1983). Activity of mesencephalic dopamine and non-dopamine neurons across stages of sleep and walking in the rat. Brain Res 273: 133–141. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS (2012). Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morairty SR, Dittrich L, Pasumarthi RK, Valladao D, Heiss JE, Gerashchenko D et al (2013). A role for cortical nNOS/NK1 neurons in coupling homeostatic sleep drive to EEG slow wave activity. Proc Natl Acad Sci USA 110: 20272–20277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M, Margolis EB (2017). Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat Rev Neurosci 18: 73–85. [DOI] [PubMed] [Google Scholar]
- Morrell MJ, Twigg G (2006). Neural consequences of sleep disordered breathing: the role of intermittent hypoxia. Adv Exp Med Biol 588: 75–88. [DOI] [PubMed] [Google Scholar]
- Morris CJ, Purvis TE, Hu K, Scheer FA (2016). Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci USA 113: E1402–E1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moruzzi G, Magoun HW (1949). Brain stem reticular formation and activation of the EEG. Electroencephalogr Clin Neurophysiol 1: 455–473. [PubMed] [Google Scholar]
- Murphy KR, Deshpande SA, Yurgel ME, Quinn JP, Weissbach JL, Keene AC et al (2016). Postprandial sleep mechanics in Drosophila. eLife 5 e19334; doi: 10.7554/eLife.19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall A, Sehgal A (2014). Monoamines and sleep in Drosophila. Behav Neurosci 128: 264–272. [DOI] [PubMed] [Google Scholar]
- Nath RD, Bedbrook CN, Abrams MJ, Basinger T, Bois JS, Prober DA et al (2017). The jellyfish cassiopea exhibits a sleep-like state. Curr Biol 27: e2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor E, Aillon DV, Barrett BS, Wilson GS, Johnson DA, Johnson DA et al (2012). Lactate as a biomarker for sleep. Sleep 35: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir Y, Staba RJ, Andrillon T, Vyazovskiy VV, Cirelli C, Fried I et al (2011). Regional slow waves and spindles in human sleep. Neuron 70: 153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Prober DA (2017). Attacking sleep from a new angle: contributions from zebrafish. Curr Opin Neurobiol 44: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Suzuki Y, Takahashi K, Yonezawa T, Kanda T, Takata Y et al (2017. a). Activation of ventral tegmental area dopamine neurons produces wakefulness through dopamine D2-like receptors in mice. Brain Struct Funct 222: 2907–2915. [DOI] [PubMed] [Google Scholar]
- Oishi Y, Xu Q, Wang L, Zhang BJ, Takahashi K, Takata Y et al (2017. b). Slow-wave sleep is controlled by a subset of nucleus accumbens core neurons in mice. Nat Commun 8: 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ et al (2008). PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier R, Ohtsu H, Djebbara-Hannas Z, Valatx JL, Watanabe T, Lin JS (2002). Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J Neurosci 22: 7695–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier R, Zhao Y, Perier M, Akaoka H, Lintunen M, Hou Y et al (2016). Role of histamine H1-receptor on behavioral states and wake maintenance during deficiency of a brain activating system: a study using a knockout mouse model. Neuropharmacology 106: 20–34. [DOI] [PubMed] [Google Scholar]
- Pedersen NP, Ferrari L, Venner A, Wang JL, Abbott SBG, Vujovic N et al (2017). Supramammillary glutamate neurons are a key node of the arousal system. Nat Commun 8: 1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peever J, Fuller PM (2017). The biology of REM Sleep. Curr Biol 27: R1237–R1248. [DOI] [PubMed] [Google Scholar]
- Peyron C, Faraco J, Rogers W, Ripley B, Overeem S, Charnay Y et al (2000). A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat medicine 6: 991–997. [DOI] [PubMed] [Google Scholar]
- Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, Miesenbock G (2016). Operation of a homeostatic sleep switch. Nature 536: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L, Goard MJ, Estandian D, Xu M, Kwan AC, Lee SH et al (2013). Fast modulation of visual perception by basal forebrain cholinergic neurons. Nat Neurosci 16: 1857–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF (2006). Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci 26: 13400–13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu MH, Chen MC, Fuller PM, Lu J (2016. a). Stimulation of the pontine parabrachial nucleus promotes wakefulness via extra-thalamic forebrain circuit nodes. Curr Biol 26: 2301–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu MH, Liu W, Qu WM, Urade Y, Lu J, Huang ZL (2012). The role of nucleus accumbens core/shell in sleep-wake regulation and their involvement in modafinil-induced arousal. PloS one 7: e45471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu MH, Yao QL, Vetrivelan R, Chen MC, Lu J (2016. b). Nigrostriatal dopamine acting on globus pallidus regulates sleep. Cereb Cortex 26: 1430–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu WM, Huang ZL, Xu XH, Matsumoto N, Urade Y (2008). Dopaminergic D1 and D2 receptors are essential for the arousal effect of modafinil. J Neurosci 28: 8462–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV et al (2008). Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Ramon F, Hernandez-Falcon J, Nguyen B, Bullock TH (2004). Slow wave sleep in crayfish. Proc Natl Acad Sci USA 101: 11857–11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson SW (1939). Somnolence caused by hypothalamic lesions in the monkey. Archives of Neurology & Psychiatry 41: 1–23. [Google Scholar]
- Rattenborg NC, Amlaner CJ, Lima SL (2000). Behavioral, neurophysiological and evolutionary perspectives on unihemispheric sleep. Neurosci Biobehav Rev 24: 817–842. [DOI] [PubMed] [Google Scholar]
- Rattenborg NC, de la Iglesia HO, Kempenaers B, Lesku JA, Meerlo P, Scriba MF (2017). Sleep research goes wild: new methods and approaches to investigate the ecology, evolution and functions of sleep. Philos Transact R Soc Lond Ser 372 pii: 20160251; doi: 10.1098/rstb.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenborg NC, Mandt BH, Obermeyer WH, Winsauer PJ, Huber R, Wikelski M et al (2004). Migratory sleeplessness in the white-crowned sparrow (Zonotrichia leucophrys gambelii). PLoS Biol 2: E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenborg NC, Voirin B, Cruz SM, Tisdale R, Dell'Omo G, Lipp HP et al (2016). Evidence that birds sleep in mid-flight. Nat Commun 7: 12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S et al (2010). Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327: 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls A, Colas D, Adamantidis A, Carter M, Lanre-Amos T, Heller HC et al (2011). Optogenetic disruption of sleep continuity impairs memory consolidation. Proc Natl Acad Sci USA 108: 13305–13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL (2016). DREADDs for Neuroscientists. Neuron 89: 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T (2007). The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci 8: 171–181. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H et al (1998). Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92: 573–585. [DOI] [PubMed] [Google Scholar]
- Salamone JD, Pardo M, Yohn SE, Lopez-Cruz L, SanMiguel N, Correa M (2016). Mesolimbic dopamine and the regulation of motivated behavior. Curr Top Behav Neurosci 27: 231–257. [DOI] [PubMed] [Google Scholar]
- Saper CB (2013). The central circadian timing system. Curr Opin Neurobiol 23: 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Fuller PM (2017). Wake-sleep circuitry: an overview. Curr Opin Neurobiol 44: 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE (2010). Sleep state switching. Neuron 68: 1023–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper CB, Loewy AD (1980). Efferent connections of the parabrachial nucleus in the rat. Brain research 197: 291–317. [DOI] [PubMed] [Google Scholar]
- Scammell TE, Arrigoni E, Lipton JO (2017). Neural circuitry of wakefulness and sleep. Neuron 93: 747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA (2009). Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA 106: 4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W (2007). Multiple dopamine functions at different time courses. Annu Rev Neurosci 30: 259–288. [DOI] [PubMed] [Google Scholar]
- Schulz DW, Macdonald RL (1981). Barbiturate enhancement of GABA-mediated inhibition and activation of chloride ion conductance: correlation with anticonvulsant and anesthetic actions. Brain research 209: 177–188. [DOI] [PubMed] [Google Scholar]
- Segal DS, Mandell AJ (1970). Behavioral activation of rats during intraventricular infusion of norepinephrine. Proc Natl Acad Sci USA 66: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidner G, Robinson JE, Wu M, Worden K, Masek P, Roberts SW et al (2015). Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol 25: 2928–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M (2008). Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA 105: 19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Haynes P, Pirez N, Harrington KI, Guo F, Pollack J et al (2011). Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK (2003). Adaptive significance of circadian clocks. Chronobiol Int 20: 901–919. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287: 1834–1837. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK et al (2008). Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shein-Idelson M, Ondracek JM, Liaw HP, Reiter S, Laurent G (2016). Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science (New York, NY) 352: 590–595. [DOI] [PubMed] [Google Scholar]
- Sherin JE, Shiromani PJ, McCarley RW, Saper CB (1996). Activation of ventrolateral preoptic neurons during sleep. Science 271: 216–219. [DOI] [PubMed] [Google Scholar]
- Singh C, Oikonomou G, Prober DA (2015). Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. eLife 4: e07000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorooshyari S, Huerta R, de Lecea L (2015). A framework for quantitative modeling of neural circuits involved in sleep-to-wake transition. Front Neurol 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfels GF, Heym J, Strecker RE, Jacobs BL (1983). Behavioral correlates of dopaminergic unit activity in freely moving cats. Brain Res 258: 217–228. [DOI] [PubMed] [Google Scholar]
- Steininger TL, Alam MN, Gong H, Szymusiak R, McGinty D (1999). Sleep-waking discharge of neurons in the posterior lateral hypothalamus of the albino rat. Brain Res 840: 138–147. [DOI] [PubMed] [Google Scholar]
- Stephan FK (2002). The "other" circadian system: food as a Zeitgeber. J Biol Rhythms 17: 284–292. [DOI] [PubMed] [Google Scholar]
- Sundvik M, Kudo H, Toivonen P, Rozov S, Chen YC, Panula P (2011). The histaminergic system regulates wakefulness and orexin/hypocretin neuron development via histamine receptor H1 in zebrafish. Faseb j 25: 4338–4347. [DOI] [PubMed] [Google Scholar]
- Sundvik M, Panula P (2015). Interactions of the orexin/hypocretin neurones and the histaminergic system. Acta Physiol 213: 321–333. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Ukai H, Ueda HR (2017). Next-generation mammalian genetics toward organism-level systems biology. NPJ Syst Biol Appl 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y, Aoyama S, Shibata S (2017). The mammalian circadian clock and its entrainment by stress and exercise. J Physiolog Sci 67: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017). Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18: 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Lin JS, Sakai K (2006). Neuronal activity of histaminergic tuberomammillary neurons during wake-sleep states in the mouse. J Neurosci 26: 10292–10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Lin JS, Sakai K (2008). Neuronal activity of orexin and non-orexin waking-active neurons during wake-sleep states in the mouse. Neuroscience 153: 860–870. [DOI] [PubMed] [Google Scholar]
- Takeda N, Inagaki S, Shiosaka S, Taguchi Y, Oertel WH, Tohyama M et al (1984). Immunohistochemical evidence for the coexistence of histidine decarboxylase-like and glutamate decarboxylase-like immunoreactivities in nerve cells of the magnocellular nucleus of the posterior hypothalamus of rats. Proc Natl Acad Sci USA 81: 7647–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NE, Van Dort CJ, Kenny JD, Pei J, Guidera JA, Vlasov KY et al (2016). Optogenetic activation of dopamine neurons in the ventral tegmental area induces reanimation from general anesthesia. Proc Natl Acad Sci USA 113: 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M et al (2000). Reduced number of hypocretin neurons in human narcolepsy. Neuron 27: 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I, Borbely AA (1986). Sleep EEG in the rat as a function of prior waking. Electroencephalogr Clin Neurophysiol. 64: 74–76. [DOI] [PubMed] [Google Scholar]
- Tononi G, Pompeiano M, Cirelli C (1991). Effects of local pontine injection of noradrenergic agents on desynchronized sleep of the cat. Prog Brain Res 88: 545–553. [DOI] [PubMed] [Google Scholar]
- Trojanowski NF, Raizen DM (2016). Call it worm sleep. Trends Neurosci 39: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trulson ME, Jacobs BL (1979). Raphe unit activity in freely moving cats: correlation with level of behavioral arousal. Brain Res 163: 135–150. [DOI] [PubMed] [Google Scholar]
- Trulson ME, Preussler DW (1984). Dopamine-containing ventral tegmental area neurons in freely moving cats: activity during the sleep-waking cycle and effects of stress. Exp Neurol 83: 367–377. [DOI] [PubMed] [Google Scholar]
- Trulson ME, Preussler DW, Howell GA (1981). Activity of substantia nigra units across the sleep-waking cycle in freely moving cats. Neurosci Lett 26: 183–188. [DOI] [PubMed] [Google Scholar]
- Varin C, Rancillac A, Geoffroy H, Arthaud S, Fort P, Gallopin T (2015). Glucose induces slow-wave sleep by exciting the sleep-promoting neurons in the ventrolateral preoptic nucleus: a new link between sleep and metabolism. J Neurosci 35: 9900–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Franken P (2017). Hypocretin (orexin) is critical in sustaining theta/gamma-rich waking behaviors that drive sleep need. Proc Natl Acad Sci USA 114: E5464–E5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner A, Anaclet C, Broadhurst RY, Saper CB, Fuller PM (2016). A novel population of wake-promoting GABAergic neurons in the ventral lateral hypothalamus. Curr Biol 26: 2137–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD et al (1994). Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Economo CV (1930). Sleep as a problem of localization. J Nerv Mental Dis 71: 249–259. [Google Scholar]
- Vorster AP, Krishnan HC, Cirelli C, Lyons LC (2014). Characterization of sleep in Aplysia californica. Sleep 37: 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Cirelli C, Tononi G (2011. a). Electrophysiological correlates of sleep homeostasis in freely behaving rats. Prog Brain Res 193: 17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Cui N, Rodriguez AV, Funk C, Cirelli C, Tononi G (2014). The dynamics of cortical neuronal activity in the first minutes after spontaneous awakening in rats and mice. Sleep 37: 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Olcese U, Hanlon EC, Nir Y, Cirelli C, Tononi G (2011. b). Local sleep in awake rats. Nature 472: 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Dan Y (2016). Circuit-based interrogation of sleep control. Nature 538: 51–59. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Cermakian N, Crosio C, Foulkes NS, Pando MP, Travnickova Z et al (2000). A clockwork organ. Biol Chem 381: 793–800. [DOI] [PubMed] [Google Scholar]
- Williams RH, Alexopoulos H, Jensen LT, Fugger L, Burdakov D (2008). Adaptive sugar sensors in hypothalamic feeding circuits. Proc Natl Acad Sci USA 105: 11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RH, Jensen LT, Verkhratsky A, Fugger L, Burdakov D (2007). Control of hypothalamic orexin neurons by acid and CO2. Proc Natl Acad Sci USA 104: 10685–10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsky-Sommerer R, Boutrel B, de Lecea L (2005). Stress and arousal: the corticotrophin-releasing factor/hypocretin circuitry. Mol Neurobiol 32: 285–294. [DOI] [PubMed] [Google Scholar]
- Wise RA (2004). Dopamine, learning and motivation. Nat Rev Neurosci 5: 483–494. [DOI] [PubMed] [Google Scholar]
- Wisor JP, Nishino S, Sora I, Uhl GH, Mignot E, Edgar DM (2001). Dopaminergic role in stimulant-induced wakefulness. J Neurosci 21: 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Koh K, Yue Z, Joiner WJ, Sehgal A (2008). A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep 31: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Gatti S, Wettstein JG, Foster RG (2010). Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci 11: 589–599. [DOI] [PubMed] [Google Scholar]
- Xu M, Chung S, Zhang S, Zhong P, Ma C, Chang WC et al (2015). Basal forebrain circuit for sleep-wake control. Nat Neurosci 18: 1641–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yackle K, Schwarz LA, Kam K, Sorokin JM, Huguenard JR, Feldman JL et al (2017). Breathing control center neurons that promote arousal in mice. Science 355: 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura T, Harada K, Okamura A, Kemmotsu O (1990). Is the site of action of ketamine anesthesia the N-methyl-D-aspartate receptor? Anesthesiology 72: 704–710. [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Beuckmann CT, Willie JT, Hara J, Tsujino N, Mieda M et al (2003). Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron 38: 701–713. [DOI] [PubMed] [Google Scholar]
- Yelin-Bekerman L, Elbaz I, Diber A, Dahary D, Gibbs-Bar L, Alon S et al (2015). Hypocretin neuron-specific transcriptome profiling identifies the sleep modulator Kcnh4a. eLife 4: e08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T, Marin W, Faraco J, Pezeron G, Appelbaum L, Zhang J et al (2007). Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol 5: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Ye Z, Houston CM, Zecharia AY, Ma Y, Zhang Z et al (2015). Wakefulness is governed by GABA and histamine cotransmission. Neuron 87: 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zecharia A, Zhang Z, Yang Q, Yustos R, Jager P et al (2014). Circadian factor BMAL1 in histaminergic neurons regulates sleep architecture. Curr Biol 24: 2838–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecharia AY, Yu X, Gotz T, Ye Z, Carr DR, Wulff P et al (2012). GABAergic inhibition of histaminergic neurons regulates active waking but not the sleep-wake switch or propofol-induced loss of consciousness. J Neurosci 32: 13062–13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA 111: 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Naidoo N, Raizen DM, Pack AI (2008). Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci 31: 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]