Abstract
Eukaryotic genomes are packaged into chromatin, a highly organized structure consisting of DNA and histone proteins. All nuclear processes take place in the context of chromatin. Modifications of either DNA or histone proteins have fundamental effects on chromatin structure and function, and thus influence processes such as transcription, replication or recombination. In this review we highlight histone modifications specifically associated with gene transcription by RNA polymerase II and summarize their genomic distributions. Finally, we discuss how (mis-)regulation of these histone modifications perturbs chromatin organization over coding regions and results in the appearance of aberrant, intragenic transcription.
1. Introduction
Chromatin is a nucleoprotein complex built from nucleosomal repeat units. Nucleosomes themselves consist of 147 bp of DNA wrapped 1.7 times around a histone octamer core particle (two copies of histones H2A, H2B, H3 and H4 each) [1]. Chromatin not only allows for the compaction of DNA within the nucleus, it also ensures that a large fraction of genomic DNA is not readily accessible and thus has drastic consequences for the regulation of gene expression. Transcription, as well as other cellular processes, require a veritable arsenal of factors in the form of activators and repressors that enable correct temporal and spatial access to specific DNA sequences. Nucleosome dynamics, histone modifications and chromatin remodeling are three aspects of chromatin structure that are closely interlinked, and perturbation in any one part can have severe consequences for a number of cellular processes.
2. The basics of RNA polymerase II transcription
2.1 Transcription of chromatin
Polynucleosomes are extremely stable and represent the first order of packaging, often referred to as “beads-on-a-string” or 11 nm fiber [2]. While further compaction of chromatin into higher order structures does take place, most chromatin is transcribed in this configuration. Nucleosomes represent a major barrier for Pol II transcription in vivo and in vitro. Unlike phage SP6 RNA polymerase or yeast Pol III, Pol II cannot transcribe efficiently through an intact nucleosome by itself but requires additional factors that enable it to overcome the barrier represented by nucleosomes [3, 4].
2.2 Pol II transcription cycle
Transcription by Pol II (reviewed in [5]) is initiated by activator proteins binding upstream of the core promoter and signals for the subsequent recruitment of coactivators such as mediator or the SAGA histone acetyltransferase (HAT), as well as chromatin remodelers whose function it is to alter chromatin architecture for assembly of the general transcription machinery. A series of protein-protein interactions results in the recruitment of Pol II and general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH and formation of the pre-initiation complex (PIC). Following PIC assembly, the DNA at the transcription start site (TSS) is unwound by Rad25 (human XBP of TFIIH), thus allowing the single-stranded DNA template to be positioned in the Pol II active site. Concomitantly, the carboxy-terminal heptad repeat sequences (C-terminal domain; CTD) of Rpb1, the largest Pol II subunit, are phosphorylated by Kin28 (CDK7 in human TFIIH) specifically on Ser5. Pol II then loses contact with some GTFs and escapes into early elongation.
Efficient elongation requires a further phosphorylation step by Ctk1 (human P-TEFb) on the Ser2 position of the Pol II CTD that helps to recruit factors important for transcription elongation, termination and mRNA processing as well as histone modifiers and remodelers. Throughout the transcriptional cycle the Pol II CTD – whether phosphorylated or not – functions as a recruitment platform for a large number of stage-specific factors required for efficient transcription.
3. Influencing nucleosome dynamics
3.1 Chromatin remodeling and histone dynamics
One way of changing chromatin structure is by using chromatin remodelers. This class of enzymes uses the energy of ATP hydrolysis to break existing DNA-histone contacts in order to slide or evict histones/nucleosomes from the DNA (reviewed in [6]). Nucleosomes are turned over at different rates depending on their genomic location as well as on their modification status [7]. Several studies have shown that H2A/H2B dimers are rapidly exchanged in and out of existing nucleosomes over transcribed regions [8, 9]. In contrast, histone exchange of H3/H4 tetramers occurs at high rates over the promoters of actively transcribed genes, but is limited to highly transcribed genes over ORFs [8, 10, 11]. This is linked to the observation that in vitro Pol II can transcribe through hexasomal nucleosomes following the eviction of a single H2A/H2B dimer, while the H3/H4 tetramer is retained on the DNA [12, 13]. Only in highly transcribed genes that contain multiple elongating Pol II molecules is there evidence to suggest complete dissociation of histone octamers from the DNA over coding regions, which are subsequently reassembled in the wake of Pol II passage. Both eviction and reassembly of nucleosomes depend on histone chaperones, such as Asf1, Nap1, Spt6 or FACT which often work in conjunction with remodeling complexes [14-20].
3.2 Histone variant incorporation
Apart from the canonical versions of histone proteins, there are several variant forms that perform specialized functions. Variants can differ in histone tails, histone fold domains or amino acid sequence. In higher eukaryotes histone H3.3 is incorporated preferentially over transcribed regions independent of DNA replication [21]. Interestingly, the single version of histone H3 present in yeast most closely resembles the H3.3 variant rather than the replication-dependent H3.1 [22].
Histone H2A.Z (Htz1 in yeast) is another important histone variant involved in a variety of different and sometimes opposing processes. Initially identified in preventing the spread of heterochromatin to euchromatic regions [23, 24], it is involved in gene activation, gene silencing, nucleosome turnover, chromosome segregation and differentiation (reviewed in [25]). H2A.Z is highly conserved from yeast to humans. It is not essential in yeast, although deletion of HTZ1 results in transcriptional defects [26, 27]. However, loss of H2A.Z is lethal in higher eukaryotes [25]. Genome-wide studies have found H2A.Z associated with promoters at practically all +1 nucleosomes (relative to the transcription start site) and also at a large proportion of −1 nucleosomes [28-33]. A similar pattern is also found in human cells, although enhancers and insulators are also marked by H2A.Z [34]. Interestingly, in Drosophila H2A.Z associates only with the +1 nucleosome [33].
The involvement of H2A.Z in transcription regulation has been clearly established, yet the mechanistic details remain a focus of ongoing research. Suggestions range from H2A.Z-mediated effects on nucleosome stability, nucleosome positioning and establishing contacts with the transcriptional machinery to maintaining active genes close to the nuclear periphery [27, 30, 31, 35, 36]. In yeast the presence of H2A.Z-containing nucleosomes at gene promoters is inversely proportional to their transcription rates [29-31]. However, the opposite applies to human cells and Drosophila where H2A.Z shows a high degree of colocalization with Pol II [33, 34, 36] and is required for transactivation during hormone receptor signaling [37]. Yeast H2A.Z is thought to mark promoters that have undergone Pol II transcription initiation, as untranscribed genes do not contain Htz1. Since most genes in yeast are actively transcribed, this explains the wide-spread presence of Htz1 at most promoters.
Replacement of H2A by H2A.Z at nucleosomes is catalyzed by the Swr1 complex [38-40] and its metazoan orthologs SRCAP and p400 [41, 42] and requires prior acetylation of histones H3 and H4 [32, 43, 44]. The Ino80 chromatin remodeler mediates the reverse reaction, substituting H2A.Z with H2A [45].
3.3 Histone modifications
Histones are subject to a vast number of post-translational modifications (PTMs), such as methylation of arginine (R) residues; methylation, acetylation, ubiquitination, ADP-ribosylation and sumoylation of lysine (K) residues; and phosphorylation of serine (S) and threonine (T) residues (Fig. 1) (reviewed in [46]). Modification of histones are carried out by specialized enzymes, some of which display rather broad specificities such as the Gcn5 histone acetyltransferase, while yet others are known to modify single sites only, eg. the Set1 and Set2 lysine methyltransferases (KMTs) (Table 1). Certain modifications are generally associated with actively transcribed (euchromatin) or repressed chromatin (heterochromatin) states. Most modifications exhibit distinct spatial and/or temporal distributions and are associated with enhancers, promoters, open reading frames (ORFs), differentiation states or cell-cycle stages (Fig. 2). Histone modifications by themselves can alter the charge distribution and DNA contacts with histone octamers, thus influencing chromatin structure directly. Most prominently, acetylation of histone H4K16 has been shown to directly prevent folding of chromatin into higher-order structures [47]. Furthermore, histone modifications also serve to recruit downstream effectors that influence chromatin structure. A number of domains have been identified that can interact specifically with modified histones (Table 1) (reviewed in [48, 49]).
Figure 1.
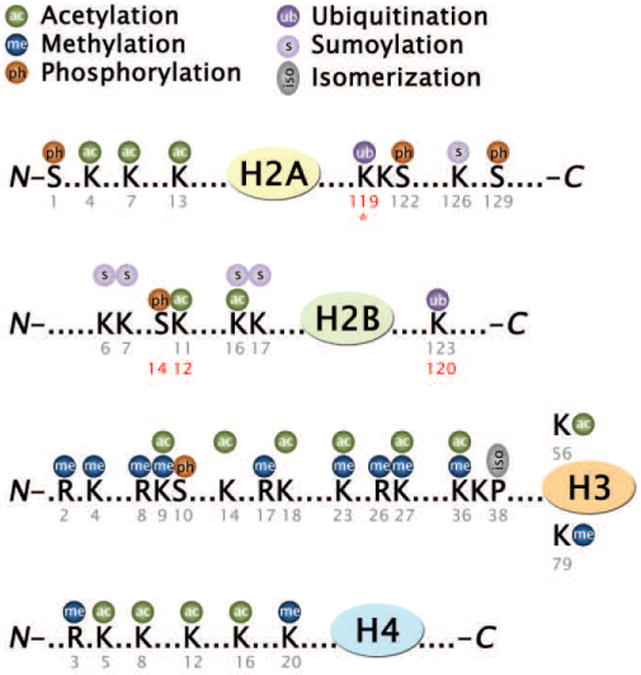
Posttranslational modifications associated with yeast histones. Alternative residue numbers that refer to mammalian histones are shown in red. Ubiquitination of histone H2A on K119 does not exist in yeast (*).
Table 1.
Common histone modifications. Alternative residue numbers that refer to mammalian histones are shown in red. Enzymes known to mediate as well as remove any given modification are shown for Saccharomyces cerevisiae (S.c.), Drosophila melanogaster (D.m.) and their mammalian counterparts. Recognition modules that bind specific modifications are also indicated. If interaction partners but no specific domain(s) were identified, then the proteins are shown in brackets.
| PTM | Position | Enzyme | Function | Recognized by | Reversed by | ||||
|---|---|---|---|---|---|---|---|---|---|
| S.c. | D.m. | Mammals | S.c. | D.m. | Mammals | ||||
| Lys Acetylation | H2AK4 K5 | Esa1 [172] | Tip60 [173] | TIP60, P300/CBP, HAT1 [174-176] | Transcription activation | Rpd3 [177] | |||
| H2AK7 | Esa1 [172] | ||||||||
| H2BK11 K12 | Gcn5 [178] | P300/CBP, ATF2 [175, 179] | Transcription activation | Rpd3, Hda1 [177] | |||||
| H2BK16 K15 | Gcn5, Esa1 [178] | P300/CBP, ATF2 [175, 179] | |||||||
| H3K9 | Gcn5, Rtt109 [180, 181] | dGcn5 [182] | SRC1, PCAF [175, 183] | Transcription activation | Rpd3, Set3, Hda1, Hos2, Hst1 [177, 184] | dHDAC1 [185] | SIRT1/6 [186] | ||
| H3K14 | Gcn5, Hpa2, Esa1, Elp3, Sas2, Sas3 [172, 178, 180, 187-190] | dGcn5, Taf1, dCBP [182, 191] | P300/CBP, TAF1, hGCN5, PCAF, MOZ, MORF, TIP60, SRC1, HBO1 [174, 175, 183, 191, 192] | Bromo, PHD [193, 194] | |||||
| H3K18 | Gcn5 [180] | dCBP | P300/CBP [175, 195] | ||||||
| H3K23 | Gcn5, Sas3 [178, 190] | P300/CBP [175] | |||||||
| H3K27 | Gcn5, Rtt109 [178, 196] | dCBP [197] | |||||||
| H3K36 | Gcn5 [198] | ||||||||
| H3K56 | Rtt109 [51] | dCBP [199] | P300/CBP [199] | PH, (Snf5) [200, 201] | Hst3/4 [202] | Sir2 [199] | SIRT1/2/6 [199, 203] | ||
| H4K5 | Esa1 | Hat1, dCBP | HAT1, TIP60, P300, HBO1 [174, 204] | Transcription activation | Bromo [205] | Rpd3, Set3, Hos2, Hst1 [177, 184, 206] | |||
| H4K8 | Esa1, Elp3, Gcn5 [188] | dCBP | TIP60, P300, HBO1 [174, 204] | Bromo [205] | |||||
| H4K12 | Hat1, Esa1, Hpa2 [187] | TIP60, P300, HBO1 [174, 204] | |||||||
| H4K16 | Sas2, Esa1 [172, 189] | Mof, Atac2 [207, 208] | hMOF, TIP60, ATF2 [174, 179, 209] | Bromo [210] | Sir2 [211] | Sir2 [199] | SIRT1/2 [186, 212] | ||
| Lys Methylation | H3K4 | Set1 [55] | Trx, Trr, Ash1, Set1 [213-216] | SET1, NSD2-3, SET7/9, MLL1-4, SMYD3, ASH1L [217-220] | Transcription activation | PHD, Chromo, WD40, ADD, Tudor, MBT, Zf-CW [68, 73, 74, 221-225] | Jhd2 [226] | Lid, Su(var)3-3 [227, 228] | LSD1, AOF1, JARID1A-D [229-231] |
| H3K9 | Su(var)3-9, Ash1, G9a [215, 232, 233] | SUV39H1/2, G9a, Eu-HMT1, SETDB1, RIZ1, ASH1 [232, 234-237] | Silencing | PHD, Chromo, Tudor, WD40, Ankyrin [223, 238-241] | Rph1 [242] | dKDM4B [243] | LSD1, JHDM2A/B, JMJD2A-D, KIAA1718, PHF8 [244-248] | ||
| H3K23 | Chromo [249] | ||||||||
| H3K27 | E(z) [250] | EZH1/2, NSD2-3, G9a [217, 234, 251] | Silencing | Chromo, WD40 [240, 252] | |||||
| H3K36 | Set2 [113] | Set2, Mes4, Ash1 [253-255] | SETD2-3, NSD1-3, SMYD2, ASH1L, SETMAR [254, 256-258] | Transcription elongation | Chromo, PHD, PWWP [128, 259, 260] | Jhd1, Rph1, Gis1 [242, 261, 262] | dKDM4A/B [243] | JHDM1A/B, JMJD2A-C [246, 263] | |
| H3K79 | Dot1 [92] | Grappa [109] | DOT1L [93] | Tudor [112] | |||||
| H4K20 | Pr-set7, Suv4-20, Ash1 [215, 264-266] | ASH1L, NSD1, Pr-SET7, SUV4-20H, NSD2 [257, 264, 265, 267, 268] | Transcription activation/ repression | Tudor, MBT, PWWP, WD40 [70, 224, 269] | KIAA1718, PHF8 [247, 248] | ||||
| Arg Methylation | H3R2me2a me2s | ? [270] | PRMT6 PRMT5/7 [86, 271] | Transcription repression/ activation | WD40 [86] | JMJD6 [272] | |||
| H3R8 | PRMT5 [273] | Transcription repression | PAD4 [274] | ||||||
| H3R17 | CARM1 [195] | Transcription activation | Tudor [275] | PAD4 [274] | |||||
| H3R26 | CARM1 [276] | ||||||||
| H4R3 | Rmt1 [91] | PRMT1, PRMT5 [273, 277] | Transcription activation | Tudor, ADD, (p300/PCAF) [275, 278, 279] | JMJD6, PAD4 [272, 274] | ||||
| Phos | H2BS10 S14 | Ste20 [280] | MST1 [281] | Apopotosis | |||||
| H3S10 | Snf1, Ipl1 [282, 283] | Jil1, Aurora B [284, 285] | MSK1/2, IKKα, PKB, RSK2, PIM1, Aurora B, JNK[286-291] | Transcription activation, mitosis, meiosis | (Gcn5), 14-3-3 [292, 293] | Glc7 [283] | PP2A [294] | ||
| Ub | H2AK119 | Ring1B [295] | Ring1B, 2A-HUB [296, 297] | Transcription repression | Calypso [298] | USP3, USP16, USP21, USP22, 2A-DUB [299-303] | |||
| H2BK123 K120 | Rad6, Bre1 [58, 304] | Rad6, Bre1 [305, 306] | HR6A/B, RNF20/40, UbcH6 [307, 308] | Transcription activation | (Cps35) [85] | Ubp8, Ubp10 [309, 310] | Nonstop,Scrawny,USP7[311-313] | USP3, USP22 [299, 314] | |
| Sumo | H2AK126 | Ubc9, Siz1, Siz2 [315] | Transcription repression | ||||||
| H2BK6/7 | |||||||||
| H2BK16/K17 | |||||||||
| H4K5 K8 K12 K16 K20 | Ubc9, Siz1, Siz2 [315] | UBC9 | |||||||
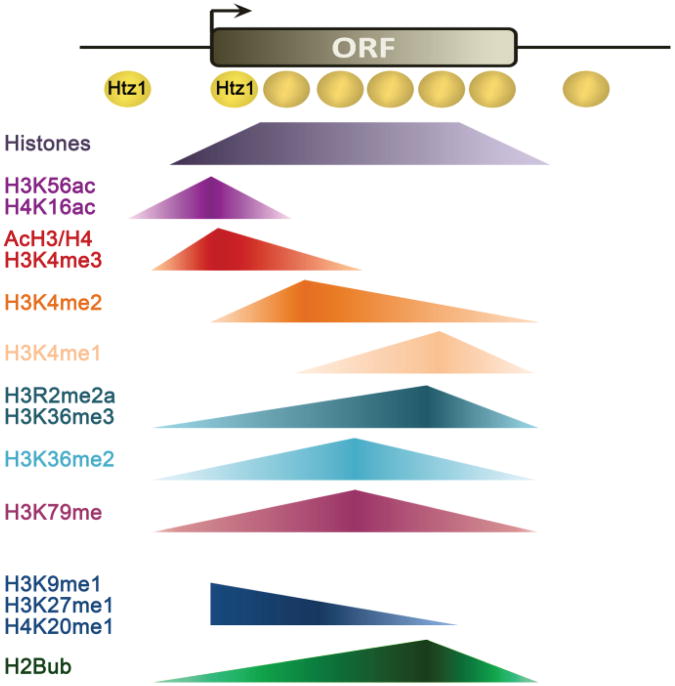
Figure 2.
Genome-wide distribution patterns of histone modifications involved in transcription. Distributions are shown relative to an average yeast gene: the promoter, transcription start site (arrow), coding sequence (ORF) and 3′ intergenic region are shown. All data sets refer to yeast with the exception of H3K9, H3K27 and H4K20 methylation.
3.4 Histone modifications and transcription
Histone modification states associated with both silenced chromatin as well as active genes have been characterized in great detail over the last few years. In the section below we aim to focus particularly on histone modifications associated with transcriptionally active chromatin (Fig.2).
3.4.1 Histone acetylation
Newly synthesized histone H4 is acetylated at K5 and K12, while soluble histone H3 is modified at K56 [50, 51]. These marks are important for their deposition and are quickly removed following incorporation into chromatin. A second group of HATs acetylates histones at multiple sites in a chromatin-specific context (reviewed in [52]).
Histone acetylation disrupts the electrostatic interactions existing between the positively charged histones and the negatively charged DNA by neutralizing the positive charges of lysine residues [53, 54]. Thus it is not surprising that, taken as a group, acetylation correlates with transcription activation (Table 1). Acetylated lysines are generally recognized by bromodomains that are found in a large number of other factors, eg. the RSC and SWI/SNF remodeling complexes (reviewed in [49]).
3.4.2 Histone H3K4 methylation and H2B monoubiquitination
Methylation of H3K4 in yeast is carried out by a single methyltransferase, the Set1 complex (COMPASS) within a pathway highly conserved from yeast to humans [55, 56]. In comparison, Drosophila have three H3K4 methylase complexes, namely Trithorax (Trx), Trithorax-related (Trr) and dSet1. Mammals contain at least six such complexes: SET1A, SET1B, as well as MLL1- 4. SET1A and SET1B are orthologs of dSet1; MLL1 and MLL2 are related to Trx, while MLL3 and MLL4 derived from Trr (reviewed in [57]). All COMPASS-like complexes are built around the catalytic Set1 or MLL protein and core subunits Cps60/ASH2, Cps30/WDR5 and Cps50/RBBP5, in addition to several complex-specific subunits [57]. H3K4 monomethylation only requires a core complex consisting of Set1, Cps30 (Swd3) and Cps50 (Swd1). Di- and trimethylation of H3K4 by Set1/COMPASS is a highly regulated process that depends on prior monoubiquitination of H2B on Lys123 (H2Bub) by the Rad6/Bre1 E2/E3 ubiquitin ligase complex [58, 59]. H2B ubiquitination itself is the product of a complex regulatory cascade for which Pol II functions as a central recruitment platform. H2Bub requires active Pol II transcription as shown by its dependence on the activity of Kin28, a Pol II Ser5-specific CTD kinase that marks the transition from Pol II initiation to elongation [60]. Transcribing Pol II stimulates recruitment of the PAF complex through its association with phosphorylated elongation factor Spt5 [61, 62]. PAF in turn associates with the Rad6/Bre1 ubiquitin ligase. Both Spt5 and Rad6 are regulated by the Bur1/Bur2 protein kinase complex, which further links PAF binding and H2B ubiquitination. H2Bub is recognized by COMPASS component Cps35 (Swd2), which then recruits the other COMPASS subunits to enable H3K4 di- and trimethylation (reviewed in [57]). Methylation of H3K4 requires H2B ubiquitination, but not vice versa, as the levels of H2Bub are not affected in an H3K4R site mutant that cannot be methylated and therefore mimics an unmodified lysine residue.
In flies and mammals dSet1 and SET1A/B are the primary H3K4 di- and trimethylase complexes, respectively. Analogous to yeast, they also rely on the PAF complex and H2B ubiquitination (though on K120 rather than K123) for H3K4 trimethylation [57]. Interestingly, the C. elegans ortholog of dSet1 plays a role in the regulation of life span, as lower levels of H3K4 methylation have been linked to extended life spans [57]. Whether this process depends on H2B ubiquitination and/or H3K4 di- and trimethylation remains to be seen.
The MLL complexes lack the ortholog of yeast Cps35 (mammalian WDR82) and are likely recruited independently of H2Bub. Instead, they function as transcriptional coactivators, involved in processes such as activation of the developmentally important Hox genes or nuclear receptor transactivation (reviewed in [57]). Misregulation of any of these complexes can have serious consequences: chromosomal translocations of the MLL1 gene are associated with acute myeloid and lymphoid leukemia, while mutations in MLL4 have been linked to non-Hodgkin's lymphoma [57].
Trimethylated H3K4 is a hallmark of active promoters and the 5′ ends of ORFs and correlates well with increased levels of gene expression [63]. H3K4me2 is found throughout coding regions, while H3K4me1 localizes mostly towards the 3′ ends of ORFs [64].
In higher eukaryotes H3K4 monomethylation has also become a reliable indicator of gene enhancers [65, 66]. Furthermore, promoters of developmentally important genes in mammalian stem cells are marked by both H3K4me3 as well as H3K27 trimethylation (a repressive modification). Such “bivalency” is critical for proper gene regulation and stem cell commitment during differentiation (reviewed in [57]). However, we have yet to define the COMPASS-like complex(es) involved in the regulation of H3K4 methylation of either phenomenon.
H3K4 methylation does not affect either elongation rate or processivity of Pol II by itself [67]. Rather it functions as a signaling platform that is recognized by a host of other factors through recognition modules that may specifically recognize a single modification state or exhibit somewhat broader specificity. Thus, unmethylated H3K4 recruits proteins through their PHD, WD40 or ADD domains. Many more proteins are known to bind to methylated H3K4 through PHD, Chromo, Tudor, MBT and Zf-CW domains (reviewed in [48]). Proteins recruited through H3K4 methylation fulfill a number of different functions: many have been shown to be involved in chromatin remodeling and histone modification and play important roles during transcription, such as the human CHD1 and BPTF ATPases, or Sgf29 and Yng1 which are part of the yeast SAGA and NuA3 HATs, respectively [68-72]. However, other proteins involved in diverse processes such as DNA methylation (Dnmt3L) or recombination (RAG2) are also recruited by H3K4 methylation [73, 74].
3.4.3 Histone H2B monoubiquitination
Monoubiquitination of histone H2B is a modification found both at promoters and over open reading frames [60, 75-78] and has functions independent of its involvement in histone H3 lysine methylation. While incorporation of ubiquitin does not greatly affect nucleosome structure [79, 80], recent work shows that H2Bub prevents compaction of the chromatin fiber into a higher-order structure [81]. In this respect the effects of H2B ubiquitination are similar to those of H4K16 acetylation, although these two histone modifications function in parallel pathways [81]. Furthermore, H2B ubiquitination has been shown to promote reassembly of nucleosomes in the wake of elongating Pol II [76, 79, 82]. Reduced nucleosome occupancy was observed for an H2BK123A site mutant, whereby the most highly expressed genes showed the largest reductions in nucleosome occupancies [76]. Reassembly seems to involve the Chd1 remodeling enzyme, although it is currently not clear how this is achieved mechanistically [83]. Pol II elongation is further helped by increased DNA accessibility as a result of H2Bub-dependent stimulation of FACT activity [82, 84]. No bona fide ubiquitin-binding domain has been identified so far, although H2Bub is required for the binding of the COMPASS Cps35 subunit to chromatin [85].
3.4.4 Histone H3R2 methylation
Three modification states exist for arginine residues: monomethylation, symmetric dimethylation (Rme2s) and asymmetric dimethylation (Rme2a). Symmetric H3R2 methylation has only been observed in higher eukaryotes so far and is mediated by PRMT5 and PRMT7 [86]. In contrast, asymmetric H3R2 methylation exists in both yeast and metazoans, although the methyltransferase responsible, PRMT6, has only been identified in higher eukaryotes [87, 88].
The status of histone H3R2 methylation plays an important role for H3K4 methylation and thus for gene expression. Asymmetric H3R2me2 is mutually exclusive with trimethylated H3K4 and accumulates over mid- to 3′-regions of ORFs as well as over the promoters of inactive genes [87, 89]. In yeast, H3R2me2a abolishes binding of the COMPASS subunit Cps40 (Spp1) to mono- and dimethylated H3K4 through its PHD domain due to steric hindrance [89]. This interaction is however necessary for efficient H3K4 trimethylation [90]. Similarly, in humans presence of H3R2me2a inhibits binding of the MLL methyltransferase complex via the WD40 domain of its WDR5 subunit, with negative consequences for H3K4me3 [87, 88]. Vice versa, presence of the H3K4me3 mark also interferes with PRMT6-mediated methylation of H3R2 [87, 88].
Symmetric methylation of H3R2 has the opposite effects when compared to H3R2me2a. It is found at the −1 nucleosome of promoters as well as at promoter-distal sites [86]. H3R2me2s enhances binding of WDR5 which results in increased levels of H3K4me3. Conversely, depletion of H3R2me2s via knock-down of PRMT5 and PRMT7 also reduced H3K4me3 levels. Furthermore, the presence of H3R2me2s also blocked binding of RBBP7, a component of several co-repressor complexes such as the Sin3a histone deacetylase complex [86].
3.4.5 Histone H3K79 methylation
Methylation of H3K79 is mediated by Dot1 [91, 92] which preferentially methylates histone H3 in a nucleosomal context [93]. Similarly to H3K4, efficient trimethylation of H3K79 requires prior ubiquitination of histone H2B [94-96], which is thought to improve Dot1 processivity, possibly through allosteric changes [97-99]. Interestingly, H2Bub seems to stimulate Dot1-mediated H3K79 methylation both directly and indirectly: Dot1 directly binds ubiquitin [100], but it also associates indirectly with H2Bub through other proteins such as proteasomal ATPases Rpt4 and Rpt6 [101] or the Set1/COMPASS subunit Cps35 when overexpressed [85]. Currently, methylated H3K79 is the only histone methyl mark where no corresponding demethylase has been identified, although there are indications that H3K79 methylation can be reversed in vivo [102].
While we have a relatively good understanding of the role of H3K79 methylation in DNA damage response and cell cycle regulation, its link to transcription is less clear. In yeast, methylated H3K79 is depleted from telomeric, mating-type and ribosomal DNA, but ubiquitous everywhere else which accounts for ∼90% of the yeast genome [63, 103]. Also, H3K79 methylation restricts recruitment of Sir proteins to heterochromatic regions and thus generally coincides with euchromatin [103]. H3K79 methylation is associated with transcribed genes in flies, mice and humans [104-106].
Several DOT1L-associated complexes have been identified in mammals that also contain the Pol II Ser2-specific CTD kinase P-TEFb, thus further suggesting involvement of Dot1 in transcription elongation [107, 108]. Purification of the DOT1L-containing complex DotCom also pulled down members of the Wnt pathway. P-TEFb was not isolated with this particular complex. However, DOT1L was nevertheless required for the expression of Wingless target genes, thus further supporting its role in transcription activation [109]. Similarly, a recent paper also implicates DOT1L in the regulation of JAK-STAT-dependent genes [110]. In mice DOT1L-mediated H3K79 methylation directly regulates expression of dystrophin, leading to defects during cardiac development when mutated [111]. However, the mechanism linking H3K79 methylation to transcription activation and elongation remains unclear and requires further investigation. Only a single recognition module for methylated H3K79 has been identified: however, the Tudor domain of 53BP1 has been shown to be involved in DNA repair rather than transcription [112].
3.4.6 Histone H3K36 methylation
Methylation of H3K36 is a widespread histone modification associated with ORFs [63]. Methylation of H3K36 is mediated by Set2, the sole histone H3K36 methyltransferase in yeast [113, 114]. While mono-and dimethylation of H3K36 by the Set2 catalytic domain require no other factors, H3K36me3 is dependent on full-length Set2 and its association with Pol II [115]. In particular, phosphorylation of the Pol II CTD by Ctk1 on Ser2 specifically stimulates Set2 binding [116-119] and is thought to positively affect Set2 protein stability [120]. Ctk1 is required for proper H3K36 trimethylation [115, 117, 119], which accumulates towards the 3′ ends of ORFs [63] (Fig.2). H3K36 methylation is also affected by the proline isomerase Fpr4, which acts on H3P38 and antagonizes H3K36me levels in vivo [121].
H3K36me2 and H3K36me3 are generally associated with actively transcribed genes, yet only H3K36me3 levels correlate with transcription rates [63, 122]. H3K36 methylation is associated with transcribed genes and hence usually referred to as an activating histone mark. However, it actually exerts a repressive effect on chromatin structure as H3K36 di- and trimethylation signal for the deacetylation of histones H3 and H4 in the wake of Pol II passage through activation of the Rpd3S histone deacetylase complex [123-125]. Methylated H3K36 can be read by a number of different recognition modules. Rpd3S contains a chromo-as well as a PHD domain on its Eaf3 and Rco1 subunits. While the Eaf3 chromodomain recognizes H3K36me3 specifically, it functions in combination with the Rco1 PHD domain to bind H3K36 methylated nucleosomes [123, 126]. An increasing number of PWWP-domain proteins also bind preferentially to H3K36 trimethylated nucleosomes, such as the BRPF1 subunit of human MOZ acetyltransferase, which together with H3K36me3 is important for Hox gene expression [127, 128]. Other examples include the chromatin-associated Psip1 short (p52) isoform which plays a role in alternative splicing [129] and the Ioc4 subunit of the yeast Isw1b chromatin remodeler [130, 131] which relies on H3K36 methylation to maintain ordered chromatin over transcribed ORFs (see below) [130].
In contrast to yeast, eight different H3K36 methyltransferases have been identified in higher eukaryotes so far: NSD1-3, SETD2/3, ASH1L, MES4, SETMAR and SMYD2 (reviewed in [132]). While in vivo substrate specificities have not been determined for all enzymes as yet, SETD2 is thought to be the only human methyltransferase mediating H3K36 trimethylation in cells [133]. SETD2 is also the closest ortholog of yeast Set2 and interacts with Pol II during transcription elongation [134]. All other enzymes seem to be mono- and dimethylases. Some also act on other histone as well as non-histone targets: NSD1, for example has been reported to methylate NFκB as well as histone H4K20 [132]. NSD2 methylates H3K36 in a nucleosomal context, but prefers H4K44 when confronted with histone octamers. NSD2 is an interesting enzyme, as addition of short DNA molecules that may function as allosteric effectors, results in subsequent preferential H3K36 dimethylation of histone octamers [135]. The higher complexity of H3K36 methylases in humans also suggests more wide-spread biological involvement when compared to yeast. Indeed, in metazoans H3K36 methylation has been implicated in a number of processes, including gene activation and repression, alternative splicing, dosage compensation, as well as DNA replication, recombination and repair [132]. At the same time, dysfunctional H3K36 methylation has been linked to a large number of diseases, including for example severe developmental defects, breast, lung and prostate cancer, acute myeloid leukaemia, and neuroblastoma (reviewed in [132]). Efforts to elucidate the mechanisms governing these processes are still in the beginning stages.
4. Maintenance of genome integrity
Chromatin represents a barrier for efficient Pol II transcription, which is alleviated by the action of a number of positive transcription elongation factors. At the same time chromatin structure needs to be restored in the wake of Pol II passage. Failure to do so leads to gross perturbations in the nucleosomal organization of genes and exposure of intragenic (cryptic) promoter-like sequences that initiate aberrant transcription from inside open reading frames (Text box 1) [136].
Text box 1 What is cryptic transcription?
The term “cryptic” transcription is used to refer to two different, yet overlapping processes. It was first used to describe the presence of transcripts initiated from intragenic promoters that are usually inaccessible for assembly of the transcription machinery [136]. Transcription from these promoters may occur in either the sense or antisense direction, and sometimes in both [137, 138]. Furthermore, intragenic promoters can be found anywhere over ORFs and are not necessarily associated with the nucleosome-free regions of genes (see below). RNAs produced from cryptic promoters are stable and often, but not always polyadenylated, as cryptic transcripts have been identified from mRNA as well as total RNA fractions. Also, the Winston lab has shown that at least some of these intragenic transcripts are translated into proteins [137], although whether these protein products have any functional roles remains to be determined.
More recently a lot of attention has been focused on the wide-spread, “pervasive” transcription of non-coding RNAs (ncRNA), that are also often referred to as cryptic transcripts (reviewed in [139, 140]). For the purposes of this review we will refer to the intragenic transcripts described above as “cryptic” and to all others as “pervasive”. Generally these pervasive ncRNAs are further categorized as cryptic unstable transcripts (CUTs) or stable unannotated transcripts (SUTs). SUTs can be identified in wildtype yeast [141, 142], whereas CUTs are observed in yeast strains with impaired RNA degradation pathways, such as the exosome mutant rrp6Δ [143]. More recently, a third group of ncRNAs has been identified that relies on the Xrn1 5′-3′ RNA exonuclease for degradation and is therefore named Xrn1-sensitive unstable transcripts (XUTs) [144]. There is a certain amount of overlap between these groups as ∼ 30% of SUTs and ∼ 10% of CUTs are also XUTs [144]. Presumably, this also applies to the intragenic cryptic transcripts, as we have no information as to the regulation of their degradation.
CUTs, SUTs and XUTs are polyadenylated transcripts. They are generally initiated from nucleosome-free regions at either the promoter or the 3′ intergenic region and transcription occurs mostly from the antisense strand [141, 144]. In the case of divergent CUTs transcribed in both the sense and antisense direction, expression of sense CUTs correlates negatively with gene transcription rates, whereas the reverse is true for antisense CUTs, suggesting a mechanism of transcriptional interference with normal gene expression [141, 142]. A recent study found, however, that in yeast transcription is highly directional and favors the sense orientation. This preference is dependent on functional Rpd3S histone deacetylase [145].
Determining the functional significance of these ncRNAs is an ongoing focus of research, but a regulatory role in gene expression has been identified for a number of ncRNAs. For example, PHO84 antisense transcripts inhibit sense transcription of the PHO84 gene both in cis and in trans [146, 147]. In both instances silencing of PHO84 requires production of the antisense transcript. However, cis inhibition relies on the Hda1/2/3 histone deacetylase which leads to promoter deacetylation [146]. In contrast, trans silencing requires functional Set1 as increased H3K4 trimethylation promotes production of the antisense transcript [147]. Pervasive transcription has also been identified in higher eukaryotes (reviewed in [139, 140]), eg. the human HOTAIR transcript that is involved in the silencing of the HOXD locus in trans through interactions with PRC2 [148]. While most known examples of ncRNAs are linked to repression, several ncRNAs promote gene expression. The Drosophila roX RNAs mark the male X chromosome and cooperate with the MSL complex to enhance transcription elongation over genes involved in male dosage compensation [149]. Another example is HOTTIP which directly binds to the WDR5 subunit of MLL H3K4 methylases to activate transcription of HOXA in cis and results in increased levels of H3K4me3[150].
4.1 Properties of cryptic promoters
Cryptic promoters are diverse and do not fall into a single group. Some, such as the promoter present within the FLO8 gene contain a TATA-box while others, eg. at the STE11 locus do not [136, 151]. Recent work has shown that cryptic promoters are regulated independently from their respective canonical promoter [151]. Furthermore, transcription initiation from cryptic promoters relies on the same components of the transcriptional machinery as their canonical counterparts [151]. It may be expected that transcription from cryptic promoters interferes with the production of the corresponding full-length transcripts. However, several gene expression microarray experiments of mutants with known cryptic transcript phenotypes did not exhibit large-scale changes in overall gene expression and therefore argue against this idea [130, 137, 138, 152, 153]. This may be explained by the observation that cryptic transcript genes tend to be infrequently transcribed [137, 138].
4.2 Histone chaperones
The Winston laboratory used a genetic screen to systematically identify genes required for the repression of cryptic transcription. In agreement with the data published from a number of laboratories, the strongest phenotypes were associated with mutations in histones, regulators of histone genes, transcription elongation factors, histone chaperones, chromatin remodelers and histone modifiers [137, 154]. These observations argue that any mutation that results in reduced nucleosome occupancy and increased DNA accessibility results in the production of cryptic transcripts (Fig. 3). Indeed, aberrant intragenic transcription was first identified in mutants of the Spt6 and Spt16 (FACT) histone chaperones, known to play a role in nucleosome reassembly during transcription elongation [136, 155, 156]. Similar findings were also obtained for a Rtt106-deficient strain: Rtt106 is a histone chaperone associated with coding sequences and involved in histone H3 deposition over ORFs [154, 157]. Furthermore, the cryptic transcript phenotype of a rtt106Δ strain was exacerbated by the additional mutation of SPT6 [157].
Figure 3.
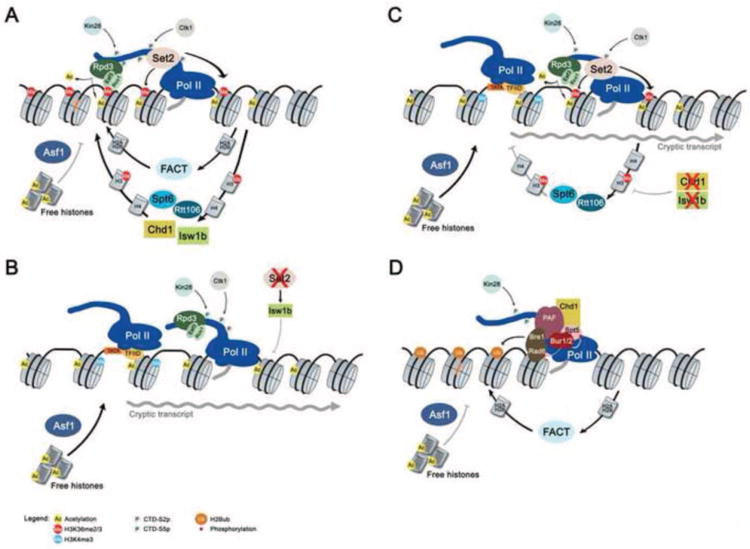
Perturbation of chromatin structure leads to aberrant intragenic transcription. (A) Histone chaperones like FACT, Spt6 and Rtt106 aid transcription elongation by disassembly of nucleosomes in front of Pol II and subsequent reassembly in its wake. Kin28 and Ctk1 specifically phosphorylate Ser5 and Ser2 within the CTD heptad repeats, respectively. Set2 is recruited to the elongating polymerase through the Ser2 phosphorylated form of the CTD and methylates Lys36 on histone H3. Methylated H3K36 recruits chromatin remodelers such as Isw1b and ensures the retention of these nucleosomes over ORFs . Maintaining H3K36 methylated nucleosomes disfavours incorporation of free, acetylated histones in their stead. Any remaining histone acetylation is removed by the Rpd3S histone deacetylase. Rpd3S associates with the Ser5 phosphorylated form of the Pol II CTD and recognizes methylated H3K36 through the chromo- and PHD-domains of its Eaf3 and Rco1 subunits, respectively. H3K36 di- and trimethylation stimulates Rpd3S activity and hence the removal of acetyl marks from transcribed chromatin [171]. (B) Mutations in key proteins involved in this pathway lead to perturbations in chromatin structure. Deletion of SET2 completely abolishes H3K36 methylation and simultaneously allows for the incorporation of free, acetylated histones. Rpd3S is still recruited to ORFs through its association with Pol II. However, in the absence of H3K36 methylation it can no longer catalyze the removal of acetyl marks from histones H3 and H4. Alterations in the chromatin architecture and increased exposure of internal promoter-like sites lead to PIC formation and initiation of cryptic transcription from inside of open reading frames. (C) Absence of chromatin remodelers Isw1b and/or Chd1impairs the retention of H3K36-methylated nucleosomes in cis and leads to increased incorporation of free, acetylated histones over ORFs despite continued Set2 and Rpd3S activities. The resulting alterations in chromatin structure lead to PIC formation and cryptic transcription. (D) H2Bub exerts a stabilizing influence on nucleosomes. Establishment requires a complex cascade of factors: H2Bub requires active Pol II transcription as shown by its dependence on Ser5 CTD phosphorylation by Kin28. Transcribing Pol II further stimulates recruitment of the PAF complex through its association with phosphorylated Spt5 [61, 62]. PAF in turn associates with the Rad6/Bre1 ubiquitin ligase. Both Spt5 and Rad6 are also regulated by the Bur1/Bur2 protein kinase complex, further linking PAF binding and H2B ubiquitination. Chd1 may also be involved in this pathway since it is known to interact both with PAF and Spt5 [164]. Disruption of this cascade is envisaged to result in lower levels of H2Bub over ORFs and consequently reduced nucleosome occupancy which is thought to expose cryptic promoters.
4.3 Set2-Rpd3S pathway
Deletion of components involved in the Set2-Rpd3S pathway such as Set2 or Rco1 exhibit the same phenotype [123, 124]. Either mutation results in the generation of hyperacetylated chromatin over open reading frames which is thought to adopt a less compact structure and thus allow for intragenic transcription initiation.Similarly, mutation of key residues in histones H4 (K44), H2A (L116, L117) and H3 thought to interfere with nucleosome binding by Set2 result in lower levels of H3K36 di- and trimethylation, increased levels of histone H4 acetylation and the appearance of cryptic transcripts [158-160] (Fig. 3B). Deletion of Ctk1, a kinase that specifically phosphorylates Ser2 of the Pol II CTD and vital for recruitment of Set2 to elongating Pol II also causes a severe cryptic transcript phenotype due to a complete lack of di- and trimethylated H3K36 in this mutant [137].
Recent work from our laboratory has shown that H3K36 methylation plays a more fundamental role in preserving chromatin architecture than previously thought. We found that H3K36 methylation affects histone dynamics, as it prevents the incorporation of new, acetylated histones over transcribed ORFs and thereby promotes the retention of H3K36 methylated nucleosomes in cis (Fig. 3A) [153]. Furthermore, increased histone exchange over ORFs seems to be responsible for the bulk of histone hyperacetylation in a SET2 mutant, suggesting that higher histone turnover contributes significantly to the increased acessibility of cryptic promoters (Fig. 3B) [153].
Retention of H3K36-methylated nucleosomes over ORFs relies on chromatin remodelers Isw1b and Chd1. Deletion of these remodelers results in increased histone turnover, histone acetylation and wide-spread cryptic transcription, while having no or little effect on H3K36 methylation levels (Fig.3C) [130, 161]. Both remodelers act within the Set2 pathway as deletion of either in a set2Δ background shows similar levels of cryptic transcription and histone acetylation when compared to a set2Δ single deletion [130]. Isw1b is recruited directly by H3K36 methylated nucleosomes in vivo and in vitro [130, 131]. Chd1 has not been shown to interact preferentially with H3K36 methylated nucleosomes [162, 163], but it is known to bind to several transcription elongation factors such as the PAF complex and Spt5 [164] and thus may be brought in by Pol II itself.
Suppression of cryptic initiation by H3K36 trimethylation has also been observed in higher eukaryotes. Self-renewal of embryonic stem cells (ESCs) is partly regulated through control of histone methylation. The H3K4me3-specific demethylase KDM5B (JARID1B) is recruited to intragenic regions by interaction of its chromodomain protein MRG15 with trimethylated H3K36 nucleosomes [165]. KDM5B is important for maintaining low levels of H3K4 trimethylation over ORFs, as knock-down of either KDM5B or MRG15 results in marked increases of H3K4me3, recruitment of non-phosphorylated Pol II and increased cryptic transcription [165]. KDM5B also interacts with the Rpd3S orthologs HDAC1 and Sin3A, providing a further link to the Set2-Rpd3 pathway. In contrast to yeast, increased cryptic transcription in ES cells concurrently reduced levels of the functional full-length mRNA [165] .
4.4 Cryptic transcription and H2Bub
Other mutations known to cause cryptic transcription are bur2Δ and deletion of the PAF complex subunit Ctr9. Both are elongation factors known to play roles in histone H2B ubiquitination and early transcription elongation [166, 167]. Interestingly, mutations in both BUR2 and the PAF complex result in significant reductions of H3K36 tri- but not dimethylation [168]. While dimethylated H3K36 is sufficient to prevent cryptic transcription [162], deletion of BUR2 or PAF1 exhibited additive effects on cryptic transcription in a set2Δ background when compared to set2Δ alone. These results suggest that Bur2 and the PAF complex do not act within the Set2 pathway, but act in parallel with Set2 [168].
A recent study on the effects of H2Bub on chromatin organization concluded that the presence of H2Bub stabilizes nucleosomes. Furthermore, yeast strains with a histone H2BK123A mutation or deletion in the gene encoding the ubiquitin-conjugating enzyme Rad6 displayed reduced levels of histone H3 over coding regions genome-wide [76]. These results suggest that in both bur2Δ and ctr9Δ strains cryptic transcription may be a consequence of increased nucleosomal disassembly and/or decreased reassembly in the wake of Pol II passage, analogous to histone chaperone defects. In agreement with these data, cryptic transcription has been reported for strains with a histone H2BK123R mutation or deletions in the H2B ubiquitination pathway (rad6Δ, bre1Δ, lge1Δ) [82, 154], although another report did not detect cryptic transcripts for either bre1Δ or lge1Δ [162]. This discrepancy is presumably accounted for by different experimental approaches.
Chd1 was recently shown to play a role in the maintenance of H2Bub levels over coding regions [83], which indicates that it may be part of the same pathway (Fig. 3D). However, it is worthwhile to point out that deposition of H2Bub and H3K36me3 marks over ORFs is highly correlated, as both are enriched over long and highly transcribed genes [77]. Chd1 prevents histone exchange in yeast [130, 161]. Furthermore, Drosophila CHD1 can catalyze the transfer of histones from the histone chaperone NAP1 onto DNA in vitro [16] and it is also important for the replication-independent deposition of histone variant H3.3 in male fly pronuclei [169]. Taken together, these results suggest that nucleosomal reassembly may be impaired in a chd1Δ mutant. Genome-wide ChIP-seq experiments observed relatively small reductions in positioned nucleosomes over ORFs [83, 152], but ChIP-chip data show a clear redistribution of nucleosome occupancy from the 3′- towards the 5′-end of ORFs [130]. The mechanism by which Chd1 affects the initiation of cryptic transcription is not entirely clear at this time. Given the fact that a chd1Δ strain also exhibits a weak cryptic transcript phenotype [130, 137, 163] and seems to be involved in the Set2 pathway, it will be interesting to determine its contribution to the H2Bub pathway.
5. Future directions
Our knowledge about histone modifications and how they are established, recognized and reversed has exploded over the last decade. Yet a number of enzymes have remained elusive. How recognition of histone modifications by downstream effector molecules is influenced by other, co-existing modifications will remain an interesting area of study, especially in higher eukaryotes where several histone modifiers and remodelers often tend to be part of the same complexes.
Cryptic promoters are common throughout the yeast genome. A small number are expressed even in wildtype cells. The question remains what function they serve, especially since we know that some at least are translated into proteins. Some cryptic transcripts are present in wildtype yeast in response to stress conditions [137] although their function remains elusive. Furthermore, aberrant intragenic transcription in yeast shares some similarities with the production of alternatively spliced transcripts found in higher eukaryotes. Nucleosomal organization and H3K36 methylation play important roles in the regulation of alternative splicing. Interestingly, highly constitutive exons display higher levels of nucleosome occupancy and H3K36me3than alternative ones (reviewed in [170]). While H3K36 trimethylation is not the only histone modification to affect splicing, it may also regulate histone dynamics in metazoans and thus presumably impact alternative splicing. If so, it will be interesting to learn if lessons from yeast will be applicable.
Highlights.
Histone modifications influence chromatin organization during transcription elongation.
Histone chaperones and H2B ubiquitination maintain nucleosome levels over gene bodies.
Disruption of histone H3K36 methylation leads to increased histone exchange and altered, hyperacetylated chromatin.
Perturbation of chromatin structure over gene bodies exposes cryptic promoters.
Cryptic transcripts are polyadenylated and even translated - their function is unclear though.
Acknowledgments
We would like to thank members of the Workman Lab for useful discussions, and Arnob Dutta and Swaminathan Venkatesh for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 3.Clark DJ, Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992;71:11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- 4.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 5.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 7.Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends in biochemical sciences. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 11.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 13.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 14.Adkins MW, Tyler JK. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J Biol Chem. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 15.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 17.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007;26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson KM, Schultz MC. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol Cell Biol. 2003;23:7937–7946. doi: 10.1128/MCB.23.22.7937-7946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zlatanova J, Seebart C, Tomschik M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 2007;21:1294–1310. doi: 10.1096/fj.06-7199rev. [DOI] [PubMed] [Google Scholar]
- 20.Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, Bouvet P. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 22.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 24.Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques M, Laflamme L, Gervais AL, Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010;5:267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- 26.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 27.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol Cell Biol. 2001;21:6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008 doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy S, Jacques PE, Gevry N, Forest A, Fortin ME, Laflamme L, Gaudreau L, Robert F. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS Genet. 2009;5:e1000687. doi: 10.1371/journal.pgen.1000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 40.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 41.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 42.Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 43.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gervais AL, Gaudreau L. Discriminating nucleosomes containing histone H2A.Z or H2A based on genetic and epigenetic information. BMC Mol Biol. 2009;10:18. doi: 10.1186/1471-2199-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 48.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26:5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- 53.Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 54.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 55.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci U S A. 2002;99(4):16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 59.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 60.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009 doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 64.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 67.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 68.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 69.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 71.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 72.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shieh GS, Pan CH, Wu JH, Sun YJ, Wang CC, Hsiao WC, Lin CY, Tung L, Chang TH, Fleming AB, Hillyer C, Lo YC, Berger SL, Osley MA, Kao CF. H2B ubiquitylation is part of chromatin architecture that marks exon-intron structure in budding yeast. BMC Genomics. 2011;12:627. doi: 10.1186/1471-2164-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. BMC Genomics, Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 79.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim Biophys Acta. 1994;1218:187–193. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 81.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011 doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 83.Lee JS, Garrett AS, Yen K, Takahashi YH, Hu D, Jackson J, Seidel C, Pugh BF, Shilatifard A. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012;26:914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 85.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone Crosstalk between H2B Monoubiquitination and H3 Methylation Mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 86.Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, Guccione E. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 87.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 88.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007 doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 91.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 92.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 94.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 95.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 96.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 97.Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol. 2008;15:550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 98.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 100.Oh S, Jeong K, Kim H, Kwon CS, Lee D. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun. 2010;399:512–517. doi: 10.1016/j.bbrc.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 101.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci U S A. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 108.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shah S, Henriksen MA. A novel disrupter of telomere silencing 1-like (DOT1L) interaction is required for signal transducer and activator of transcription 1 (STAT1)-activated gene expression. J Biol Chem. 2011;286:41195–41204. doi: 10.1074/jbc.M111.284190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li J, Chen T, Wang DZ, Xiao X, Zhang Y. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev. 2011;25:263–274. doi: 10.1101/gad.2018511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 113.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morris SA, Shibata Y, Noma K, Tsukamoto Y, Warren E, Temple B, Grewal SI, Strahl BD. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell. 2005;4:1446–1454. doi: 10.1128/EC.4.8.1446-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 117.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 119.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. RNA Polymerase II Carboxyl-terminal Domain Phosphorylation Regulates Protein Stability of the Set2 Methyltransferase and Histone H3 Di- and Trimethylation at Lysine 36. J Biol Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 122.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 124.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 125.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 126.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 127.Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, Tubingen Screen C, Kimmel CB, Schneider R, Hammerschmidt M. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development. 2008;135:1935–1946. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 129.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2312. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maltby VE, Martin BJ, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. Histone H3 Lysine 36 Methylation Targets the Isw1b Remodeling Complex to Chromatin. Mol Cell Biol. 2012 doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78:13522–13533. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, Reinberg D. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 137.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin-and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Flynn RA, Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev. 2012;22:172–178. doi: 10.1016/j.gde.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 143.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 144.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, Nicolas A, Thermes C, Morillon A. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 145.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 147.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pattenden SG, Gogol MM, Workman JL. Features of cryptic promoters and their varied reliance on bromodomain-containing factors. PLoS ONE. 2010;5:e12927. doi: 10.1371/journal.pone.0012927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Venkatesh S, Smolle M, Li H, Gogol M, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine36 suppresses histone exchange on transcribed genes. Nature. 2012 doi: 10.1038/nature11326. in the press. [DOI] [PubMed] [Google Scholar]
- 154.Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC. The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J Biol Chem. 2012;287:1709–1718. doi: 10.1074/jbc.M111.316489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stevens JR, O'Donnell AF, Perry TE, Benjamin JJ, Barnes CA, Johnston GC, Singer RA. FACT, the Bur kinase pathway, and the histone co-repressor HirC have overlapping nucleosome-related roles in yeast transcription elongation. PLoS ONE. 2011;6:e25644. doi: 10.1371/journal.pone.0025644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Imbeault D, Gamar L, Rufiange A, Paquet E, Nourani A. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J Biol Chem. 2008;283:27350–27354. doi: 10.1074/jbc.C800147200. [DOI] [PubMed] [Google Scholar]
- 158.Du HN, Briggs SD. A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 K36 methylation, histone acetylation, and repression of cryptic transcription. J Biol Chem. 2010 doi: 10.1074/jbc.M109.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Du HN, Fingerman IM, Briggs SD. Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Genes Dev. 2008;22:2786–2798. doi: 10.1101/gad.1700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hainer SJ, Martens JA. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol Cell Biol. 2011;31:3557–3568. doi: 10.1128/MCB.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Radman-Livaja M, Quan TK, Valenzuela L, Armstrong JA, van Welsem T, Kim T, Lee LJ, Buratowski S, van Leeuwen F, Rando OJ, Hartzog GA. A key role for Chd1 in histone H3 dynamics at the 3′ ends of long genes in yeast. PLoS Genetics. 2012;8:e1002811. doi: 10.1371/journal.pgen.1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 di-methylation (H3K36ME2) is sufficient to recruit the Rpd3S histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009 doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Quan TK, Hartzog GA. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics. 2010;184:321–334. doi: 10.1534/genetics.109.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 167.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 168.Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007 doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, Fyodorov DV. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 174.Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 175.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 176.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 177.Wu J, Suka N, Carlson M, Grunstein M. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 178.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 179.Kawasaki H, Schiltz L, Chiu R, Itakura K, Taira K, Nakatani Y, Yokoyama KK. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405:195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 180.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 181.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Carre C, Szymczak D, Pidoux J, Antoniewski C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol. 2005;25:8228–8238. doi: 10.1128/MCB.25.18.8228-8238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 184.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Foglietti C, Filocamo G, Cundari E, De Rinaldis E, Lahm A, Cortese R, Steinkuhler C. Dissecting the biological functions of Drosophila histone deacetylases by RNA interference and transcriptional profiling. J Biol Chem. 2006;281:17968–17976. doi: 10.1074/jbc.M511945200. [DOI] [PubMed] [Google Scholar]
- 186.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Angus-Hill ML, Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J Mol Biol. 1999;294:1311–1325. doi: 10.1006/jmbi.1999.3338. [DOI] [PubMed] [Google Scholar]
- 188.Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci U S A. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sutton A, Shia WJ, Band D, Kaufman PD, Osada S, Workman JL, Sternglanz R. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J Biol Chem. 2003;278:16887–16892. doi: 10.1074/jbc.M210709200. [DOI] [PubMed] [Google Scholar]
- 190.Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 192.Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 196.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Su D, Hu Q, Li Q, Thompson JR, Cui G, Fazly A, Davies BA, Botuyan MV, Zhang Z, Mer G. Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature. 2012;483:104–107. doi: 10.1038/nature10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 202.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 205.Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Muller CW, Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 206.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 208.Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 209.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 210.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 212.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 213.Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 214.Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426:78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- 216.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kim SM, Kee HJ, Eom GH, Choe NW, Kim JY, Kim YS, Kim SK, Kook H, Seo SB. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun. 2006;345:318–323. doi: 10.1016/j.bbrc.2006.04.095. [DOI] [PubMed] [Google Scholar]
- 218.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 219.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 220.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 221.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 223.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, Guccione E, Gozani O. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS ONE. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, Kigawa T, Takahashi M, Kuwasako K, Tsuda K, Matsuda T, Aoki M, Seki E, Kobayashi N, Guntert P, Yokoyama S, Muto Y. Structural insight into the zinc finger CW domain as a histone modification reader. Structure. 2010;18:1127–1139. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 226.Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 227.Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- 228.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 229.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 230.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 232.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 233.Mis J, Ner SS, Grigliatti TA. Identification of three histone methyltransferases in Drosophila: dG9a is a suppressor of PEV and is required for gene silencing. Mol Genet Genomics. 2006;275:513–526. doi: 10.1007/s00438-006-0116-x. [DOI] [PubMed] [Google Scholar]
- 234.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 235.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 236.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kim KC, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003;63:7619–7623. [PubMed] [Google Scholar]
- 238.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 239.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 240.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol Cell Biol. 2007;27:3951–3961. doi: 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 245.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 246.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 247.Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–437. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Quinn AM, Bedford MT, Espejo A, Spannhoff A, Austin CP, Oppermann U, Simeonov A. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res. 2010;38:e11. doi: 10.1093/nar/gkp899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 251.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Stabell M, Larsson J, Aalen RB, Lambertsson A. Drosophila dSet2 functions in H3-K36 methylation and is required for development. Biochem Biophys Res Commun. 2007;359:784–789. doi: 10.1016/j.bbrc.2007.05.189. [DOI] [PubMed] [Google Scholar]
- 254.Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397:161–168. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 255.Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, Macalpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schubeler D. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007 doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 257.Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Molecular cancer. 2006;5:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Xu C, Cui G, Botuyan MV, Mer G. Structural Basis for the Recognition of Methylated Histone H3K36 by the Eaf3 Subunit of Histone Deacetylase Complex Rpd3S. Structure. 2008 doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 261.Fang J, Hogan GJ, Liang G, Lieb JD, Zhang Y. The Saccharomyces cerevisiae histone demethylase Jhd1 fine-tunes the distribution of H3K36me2. Mol Cell Biol. 2007;27:5055–5065. doi: 10.1128/MCB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, Freitas MA, Tsai MD. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 264.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 265.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 266.Sakaguchi A, Karachentsev D, Seth-Pasricha M, Druzhinina M, Steward R. Functional characterization of the Drosophila Hmt4-20/Suv4-20 histone methyltransferase. Genetics. 2008;179:317–322. doi: 10.1534/genetics.108.087650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, Martinez-Martinez Y, Chesi M, Bergsagel PL, Zhou MM, Waxman S, Leibovitch BA, Walsh MJ, Licht JD. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR, 3rd, Jia S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33:428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Edwards CR, Dang W, Berger SL. Histone H4 lysine 20 of Saccharomyces cerevisiae is monomethylated and functions in subtelomeric silencing. Biochemistry. 2011;50:10473–10483. doi: 10.1021/bi201120q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 272.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 273.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 275.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 Is an Effector Molecule for Arginine-Methylated Histone Marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 277.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 278.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang S. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood. 2010;115:2028–2037. doi: 10.1182/blood-2009-07-236059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 281.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 282.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 283.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, Allis CD. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 284.Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell. 2001;105:433–443. doi: 10.1016/s0092-8674(01)00325-7. [DOI] [PubMed] [Google Scholar]
- 285.Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 288.Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 289.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 290.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schubeler D, Beisel C. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet. 2012;44:94–100. doi: 10.1038/ng.1036. [DOI] [PubMed] [Google Scholar]
- 292.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 293.Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 294.Nowak SJ, Pai CY, Corces VG. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol Cell Biol. 2003;23:6129–6138. doi: 10.1128/MCB.23.17.6129-6138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 296.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 297.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 300.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 301.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 303.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 305.Chen S, Wei HM, Lv WW, Wang DL, Sun FL. E2 ligase dRad6 regulates DMP53 turnover in Drosophila. J Biol Chem. 2011;286:9020–9030. doi: 10.1074/jbc.M110.190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 307.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 309.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 311.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 313.Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, Berger SL. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]