Abstract
Polyglutamine (polyQ) diseases are a class of progressive neurodegenerative disorders characterized by the expression of both expanded CAG RNA and misfolded polyQ protein. We previously reported that the direct interaction between expanded CAG RNA and nucleolar protein nucleolin (NCL) impedes preribosomal RNA (pre-rRNA) transcription, and eventually triggers nucleolar stress-induced apoptosis in polyQ diseases. Here, we report that a 21-amino acid peptide, named “beta-structured inhibitor for neurodegenerative diseases” (BIND), effectively suppresses toxicity induced by expanded CAG RNA. When administered to a cell model, BIND potently inhibited cell death induced by expanded CAG RNA with an IC50 value of ∼0.7 µM. We showed that the function of BIND is dependent on Glu2, Lys13, Gly14, Ile18, Glu19, and Phe20. BIND treatment restored the subcellular localization of nucleolar marker protein and the expression level of pre-45s rRNA. Through isothermal titration calorimetry analysis, we demonstrated that BIND suppresses nucleolar stress via a direct interaction with CAG RNA in a length-dependent manner. The mean binding constants (KD) of BIND to SCA2CAG22, SCA2CAG42, SCA2CAG55, and SCA2CAG72 RNA are 17.28, 5.60, 4.83, and 0.66 µM, respectively. In vivo, BIND ameliorates retinal degeneration and climbing defects, and extends the lifespan of Drosophila expressing expanded CAG RNA. These effects suggested that BIND can suppress neurodegeneration in diverse polyQ disease models in vivo and in vitro without exerting observable cytotoxic effect. Our results collectively demonstrated that BIND is an effective inhibitor of expanded CAG RNA-induced toxicity in polyQ diseases.
Keywords: Drosophila, Huntington's disease, spinocerebellar ataxias
INTRODUCTION
Polyglutamine (PolyQ) diseases are a set of dominantly inherited neurodegenerative disorders. These diseases include spinocerebellar ataxia type 2 (SCA2), SCA3/Machado–Joseph disease (MJD), and Huntington's disease (HD) (Orr and Zoghbi 2007). These diseases are characterized by a genomic CAG trinucleotide repeat expansion in their respective affected genes. The affected genes are transcribed and translated into toxic species, specifically, the expanded CAG RNA and polyQ domain-containing protein (Williams and Paulson 2008; Fiszer and Krzyzosiak 2013; Nalavade et al. 2013). In addition to the well-studied polyQ protein-induced neurotoxicity (Sakahira et al. 2002; Michalik and Van Broeckhoven 2003; Takahashi et al. 2010), an increasing number of studies in recent years emphasized the contribution of expanded CAG RNA to the pathogenesis of polyQ diseases (Li et al. 2008; Fiszer and Krzyzosiak 2013, 2014; Martí 2016) by sequestering RNA splicing factors via expanded CAG RNA foci (de Mezer et al. 2011; Mykowska et al. 2011), activating nucleolar stress (Tsoi et al. 2012; Tsoi and Chan 2013) and generating small toxic CAG RNA via Dicer cleavage (Krol et al. 2007; Bañez-Coronel et al. 2012).
Ribosomes are complexes of ribosomal RNAs (rRNA) and ribosomal proteins essential for cellular protein synthesis. Failure in rRNA transcription hinders the synthesis and assembly of ribosomes (Boulon et al. 2010), up-regulates cellular p53 expression, and eventually leads to apoptosis (Wang et al. 2014). The nucleolus coordinates rRNA transcription (Rickards et al. 2007) and processing (Ginisty et al. 1998), as well as ribosomal assembly (Bouvet et al. 1998; Rickards et al. 2007). Upon exposure to cytotoxic species, the nucleolus rapidly loses its compact organization, and several nucleolar proteins, including nucleolin (NCL) and nucleophosmin (B23), mislocalize to the nucleoplasm (Rubbi and Milner 2003; Meng et al. 2006; Yao et al. 2010; Haeusler et al. 2014; Tao et al. 2015). This phenomenon is defined as “nucleolar stress” and is widely implicated in the pathogenesis of various neurodegenerative diseases (Parlato and Kreiner 2013; Tsoi and Chan 2014).
Our group previously demonstrated that expanded CAG RNA interacts with NCL protein. This interaction results in hypermethylation of “upstream control element (UCE) of pre-rRNA” and consequently impedes its transcription. In turn, the impeded transcription of NCL triggers nucleolar stress and eventually causes apoptosis in polyQ diseases (Tsoi et al. 2012). Pull-down studies revealed that RNA recognition motifs 2 and 3 (RRM2 and RRM3), but not RRM1 and RRM4, of NCL are crucial for the interaction of NCL protein with RNA (Tsoi et al. 2012). We screened different peptide sequences of RRM2 and RRM3 on the basis of this information and identified a peptidylic inhibitor, P3 (Zhang et al. 2016). P3 binds directly to expanded CAG RNA, thus restoring pre-rRNA transcription and suppressing neurodegeneration in polyQ diseases (Zhang et al. 2016). P3, a 13-amino acid peptide, comprises the sequence of the ribonucleoprotein domain (RNP1) motif of NCL RRM2 (Fujiwara et al. 2011; Zhang et al. 2016). Structural prediction suggested that P3 is flexible and mostly adopts coil conformation.
The above information prompted us to develop a novel peptidylic inhibitor with low flexibility to reduce entropic loss during RNA binding, and with improved potency against RNA toxicity in polyQ diseases. In this study, we performed a structure-based design and efficacy study of a 21-amino acid peptide inhibitor, which we termed “beta-structured inhibitor for neurodegenerative diseases” (BIND). BIND comprises the β2, β3 strands and the β2–β3 loop of NCL RRM2. Although BIND also contains the RNP1 motif, it folds into a partial β structure, in contrast to the flexible P3.
RESULTS
Identification of a NCL-derived inhibitor with partial β-structure
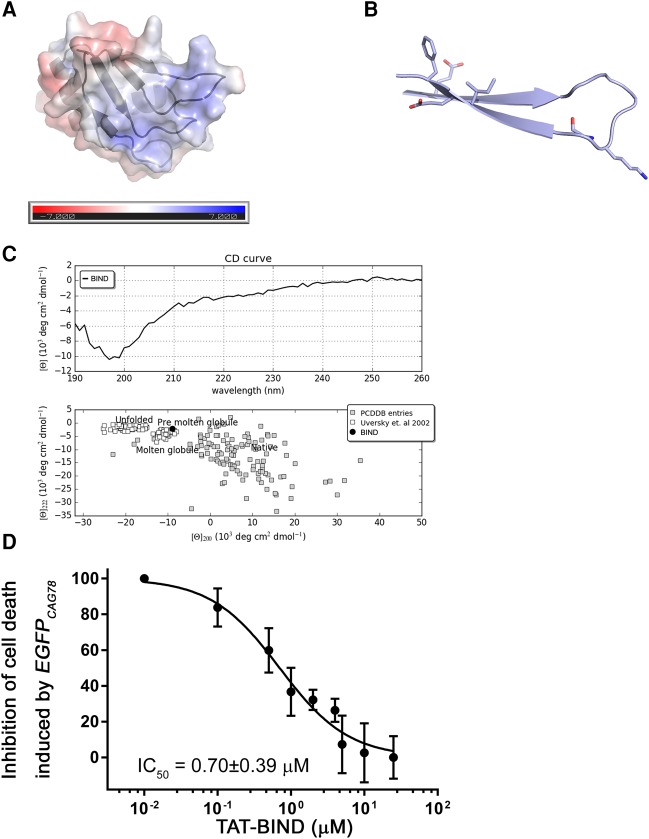
The central region of NCL contains four RNA recognition motifs (RRMs). Our previous work showed that the RRM2 and RRM3 of NCL are responsible for direct interaction with expanded CAG RNA (Tsoi et al. 2012). Both RRMs adopt the canonical RRM fold and each contains two conserved short RNP motifs. Structural studies of NCL RRM2 in complex with a nucleolin recognition element (NRE) RNA revealed that its β-sheet comprises β1, β2, and β3 strands, and that its β2–β3 loop provides an electropositive solvent-accessible surface that favorably binds with RNA (Fig. 1A; Allain et al. 2000a,b; Arumugam et al. 2010). In particular, the side chains and backbones of the β2 and β3 strands extensively interact with RNA. We speculate that the same surface is also important to the interaction between NCL and expanded CAG RNA. To interfere with this interaction, we synthesized beta-structured inhibitor for neurodegenerative diseases (BIND), a 21-amino acid peptide (AEIRLVSKDGKSKGIAYIEFK), as a competitive inhibitor. BIND comprises the RNP1 and forms β2 and β3 strands and β2–β3 loop of NCL RRM2. The secondary and tertiary structures predicted by Jpred 4 (http://www.compbio.dundee.ac.uk/jpred/) (Drozdetskiy and Cole 2015) and PEP-FOLD (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/) (Maupetit et al. 2010) suggested that this peptide adopts a β hairpin conformation (Fig. 1B; Maupetit et al. 2010; Drozdetskiy and Cole 2015).
FIGURE 1.
Identification of BIND. (A) Solvent-accessible surfaces of NCL RRM2 (PDB ID = 2KRR) (Arumugam et al. 2010) colored on the basis of electrostatic potential (−7 to +7 kT/e in red to blue) computed by APBS (Baker et al. 2001). (B) Three-dimensional structure of BIND as predicted by PEP-FOLD server (Maupetit et al. 2009). Residues crucial for interaction with RNA that contains CAG repeats are indicated. (C) CAPITO analysis of circular dichroism (CD) spectroscopy data of BIND. (Upper panel) CD curve of BIND; (lower panel) double-wavelength plot of BIND suggesting that the inhibitor adopts a partially folded structure. (D) Dose-dependent suppressive effect of TAT-BIND on the death of EGFPCAG78-expressing HEK293 cells. The IC50 value represents the concentration of TAT-BIND that decreased lactate dehydrogenase (LDH) enzyme activity by 50% relative to that in the no-peptide treatment control group. All experiments were repeated at least three times.
To assess if BIND adopts a β fold as predicted, we performed circular dichroism (CD) spectroscopy data analysis (Raussens et al. 2003) on BIND. We then used the web servers CAPITO (http://capito.nmr.leibniz-fli.de) (Wiedemann et al. 2013) and BeStSel (http://bestsel.elte.hu) (Micsonai et al. 2015) to analyze the obtained CD spectra. Both algorithms predicted that, in solution, BIND forms a partially folded structure with ∼36% to 37% β-strand content (Fig. 1C; Supplemental Table 1). In particular, BeStSel, an algorithm that can reliably predict secondary structures and distinguish the parallel/anti-parallel orientation of β-structures, predicted that BIND folds as a right-hand twisted anti-parallel β-sheet. This result supported the prediction by PEP-FOLD that BIND folds into a β-hairpin structure.
The TAT peptide (YGRKKRRQRRR) is a cell-penetrating peptide (CPP) that mediates protein translocation across the cell membrane (Frankel and Pabo 1988; Green and Loewenstein 1988). We previously demonstrated that the TAT-fusion peptide inhibitor was capable of suppressing polyQ toxicity in vitro and in vivo (Zhang et al. 2016). We synthesized the TAT-BIND fusion peptide to address its suppressive effects on neurodegeneration in polyQ diseases. Notably, the TAT peptide adopts a random coil structure and should not affect the formation of the β-hairpin (Eiriksdóttir et al. 2010). We first performed a lactate dehydrogenase (LDH) assay (Bañez-Coronel et al. 2012) to confirm that a HEK293 cell model that solely expressed EGFPCAG78 RNA had significantly higher cell death than the control cell model (Supplemental Fig. S1). TAT-BIND treatment significantly suppressed the death of EGFPCAG78-expressing cells with a calculated maximal inhibitory concentration (IC50) of 0.70 ± 0.39 µM (Fig. 1D; Supplemental Fig. S1). This value is over sixfold lower than 4.37 µM, the IC50 of TAT-P3 calculated from the same assay (Zhang et al. 2016) and strongly indicated the better in vitro efficacy of TAT-BIND than that of TAT-P3. Treatment with TAT-BIND-S (YGRKKRRQRRRGGEDIKSRVEAASILYFIKKK), a scrambled version of the TAT-fused BIND peptide, as a negative control did not suppress or elicit cell death in EGFPCAG78-expressing HEK293 cells, suggesting that the TAT peptide alone is nontoxic and does not contribute to the efficacy results of TAT-BIND (Supplemental Fig. S1).
TAT-BIND suppresses expanded CAG RNA-induced nucleolar stress
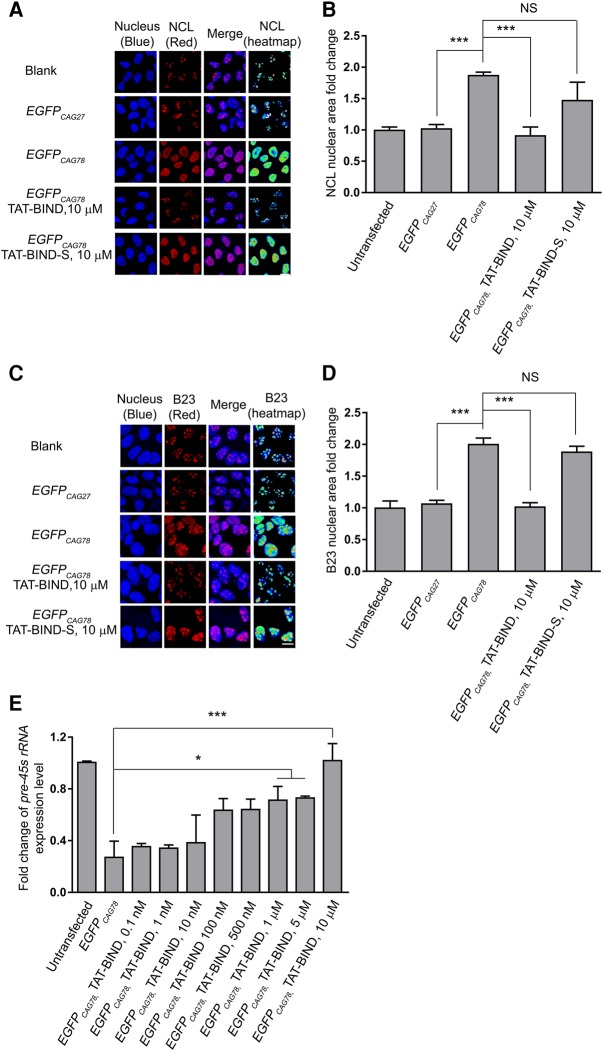
The expression of expanded CAG RNA causes nucleolar stress in polyQ diseases (Tsoi et al. 2012). The subcellular localizations of NCL and B23 are markers of nucleolar stress (Yao et al. 2010; Avitabile et al. 2011). As shown in Figure 2A and C, endogenous NCL and B23 proteins were confined to the nucleoli of blank and unexpanded EGFPCAG27-expressing cells. However, NCL is highly dispersed throughout the nuclei of EGFPCAG78-overexpressing cells, whereas B23 is localized in the nucleoplasm (Fig. 2A,C). TAT-BIND treatment restored the normal localization of NCL and B23 in the nucleoli of most EGFPCAG78-expressing cells (Fig. 2A,C). Statistical analysis of NCL and B23 nuclear fold changes were performed as reported (Haeusler et al. 2014). The fact that TAT-BIND fully rescued the mislocalization of NCL and B23 in the nucleus strongly suggested that TAT-BIND treatment suppressed nucleolar stress induced by expanded CAG RNA (Fig. 2B,D). NCL interacts with various mRNAs, including HBB mRNA (RNA encoded for β-globulin) (Jiang et al. 2006) and GADD45A mRNA (RNA encoded for the growth arrest- and DNA damage-inducible 45 protein [Gadd45α]) (Zhang et al. 2006), and decreases their stability (Abdelmohsen and Gorospe 2012). TAT-BIND/TAT-BIND-S treatments resulted in no significant change in the expression of HBB and GADD45A mRNAs in HEK293 cells (Supplemental Fig. S2), suggesting that these peptides do not affect the binding of NCL protein to other mRNAs.
FIGURE 2.
BIND suppressed nucleolar stress induced by expanded CAG RNA in vitro. (A) Increased diffusion of NCL (red) was shown in nucleus (Hoechst, blue) in EGFPCAG78-expressing HEK293 cells relative to that in the blank. TAT-BIND treatment suppressed the mislocalization of NCL protein in the disease cell model. Heat map allowed better visualization of the mislocalized cytoplasmic NCL in EGFPCAG78-expressing cells. Scales bar represents 10 µm. (B) Quantification of NCL in the nucleus. (C) B23 (red) was highly dispersed throughout the nucleus (Hoechst, blue) of EGFPCAG78-expressing HEK293 cells relative to that in the blank. TAT-BIND treatment suppressed the mislocalization of B23 protein in the disease cell model. Heat map allowed better visualization of the mislocalized cytoplasmic B23 in EGFPCAG78-expressing cells. Scale bar represents 10 µm. (D) Quantification of B23 in the nucleus. NCL and B23 measurements were normalized to that of the blank control with n = 100–150 cells calculated per treatment. (E) TAT-BIND rescued pre-45s rRNA expression level in EGFPCAG78-expressing HEK293 cells in a dose-dependent manner. Data for real-time PCR were normalized to untransfected control. All the experiments were repeated at least three times, and data were plotted as mean ± SEM. (*) P < 0.05 and (***) P < 0.001. NS, no significant difference.
Since impeded pre-45 rRNA transcription is an indicator of nucleolar stress (Rickards et al. 2007; Tsoi et al. 2012), we measured pre-45 rRNA levels in HEK293 cells expressing expanded CAG transcripts. Our result showed that TAT-BIND, but not TAT-BIND-S, restored the expression of pre-45s rRNA in EGFPCAG78-expressing HEK293 cells in a dose-dependent manner (Fig. 2E; Supplemental Fig. S3). This result provided additional evidence that TAT-BIND inhibits nucleolar stress induced by expanded CAG RNA.
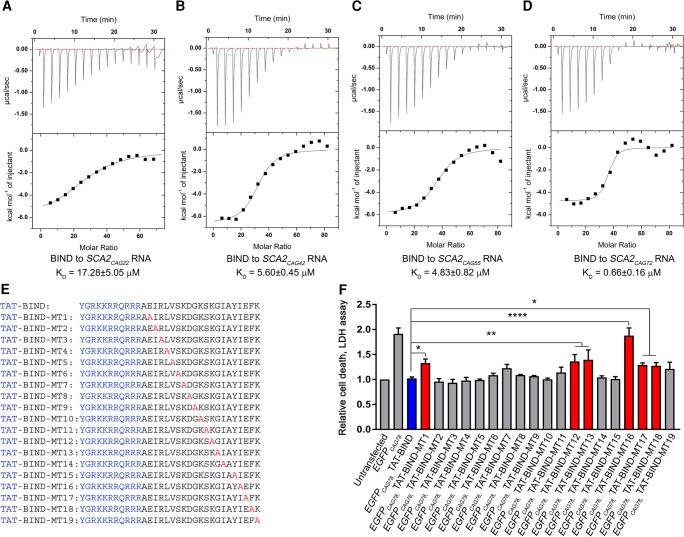
Through a pull-down assay, we demonstrated TAT-BIND and CAG78 RNA interact in EGFPCAG78-expressing HEK293 cells (Supplemental Fig. S4A). This data suggests that similar to P3, BIND also inhibits nucleolar stress through direct binding with expanded CAG RNA in polyQ diseases. We next performed a filter-binding assay to confirm whether BIND directly interacts with expanded CAG78 RNA. Since the small molecular weight of BIND is not favorable for nitrocellulose binding (data not shown), we constructed and purified a GST-BIND fusion protein. Different concentrations of GST-BIND or free GST protein as control were incubated with 32P-labeled CAG72 RNA. The mixtures were then blotted on nitrocellulose membrane filters, washed and dried. The dried filters were exposed for auto-radiography. Our results show that GST-BIND, but not GST alone, interacted with CAG72 RNA in a dose-dependent manner, indicating that the BIND peptide interacts directly with the expanded CAG RNA (Supplemental Fig. S4B). To examine the binding affinity of BIND for CAG repeats-containing RNAs of different lengths, we generated a set of in vitro transcribed RNA, including the unexpanded SCA2CAG22, the near-pathogenic threshold expanded SCA2CAG42 and SCA2CAG55, as well as the fully expanded SCA2CAG72. Isothermal titration calorimetry (ITC) data showed that P3 interacts with SCA2CAG72 RNA with a KD value of 35.55 ± 7.31 µM (Supplemental Fig. S5). However, BIND associates with SCA2CAG22 RNA, SCA2CAG42 RNA, SCA2CAG55 RNA and SCA2CAG72 RNA with KD values of 17.28 ± 5.05, 5.60 ± 0.45, 4.83 ± 0.82, and 0.66 ± 0.16 µM, respectively (Fig. 3A–D). These results demonstrated that BIND binds to expanded CAG RNA in a length-dependent manner. Furthermore, expanded CAG RNA showed a 50-fold higher binding affinity for BIND than P3. To test if the fusion of the TAT sequence affects the binding and specificity of BIND toward CAG RNA, we repeated the ITC assays using TAT-BIND. Our results show that TAT-BIND associated with SCA2CAG22/42/55/72 RNAs with KD values of 44.39 ± 1.84, 20.10 ± 4.14, 4.99 ± 0.60 and 2.22 ± 0.48 µM, respectively (Supplemental Fig. S6). Although the KD values obtained are slightly different from those of BIND alone, our results demonstrate that the conjugation of TAT sequence did not change the specificity of our inhibitor toward expanded CAG RNA.
FIGURE 3.
BIND directly interacted with RNA containing CAG repeats and structural activity relationship study of BIND. (A–D) Isothermal titration calorimetry study of the binding affinity of BIND peptide (2 mM) to SCA2CAG22/42/55/72 RNA (4 µM). The top panel shows the raw thermogram and the bottom panel shows the binding isotherm fitted to a single-site model. (E) Amino acid sequences of TAT-BIND mutants (TAT-BIND-MT1-19). (F) Effect of TAT-BIND mutants on suppressing cell death of EGFPCAG78-expressing HEK293 cells. Data for LDH assay were normalized to the untransfected control. Each experiment was repeated at least three times and data were plotted as mean ± SEM. (*) P < 0.05, (**) P < 0.01, (****) P < 0.0001.
Structural activity relationship study of TAT-BIND
We performed a structural activity relationship (SAR) study of TAT-BIND to identify the amino acids in BIND that are crucial for suppressing cell death induced by expanded CAG RNA. We synthesized 19 TAT-BIND mutants (TAT-BIND-MT1-19). Each mutant carries a single alanine substitution (Fig. 3E). Similar to the TAT-BIND peptide, the mutants TAT-BIND-MT2-11/14/15/19, inhibited the death of EGFPCAG78-expressing HEK293 cells, indicating that residues at these positions are not essential for the bioactivity of BIND (Fig. 3F). In contrast, the inhibitory effect of TAT-BIND-MT1/12/13/16/17/18 with a single Glu2Ala, Lys13Ala, Gly14Ala, Ile18Ala, Glu19Ala, and Phe20Ala mutation, respectively, significantly deviated from that of TAT-BIND (Fig. 3F). The predicted structure of BIND indicated that Glu2, Ile18, Glu19, and Phe20 cluster together at the same end of the β-hairpin, suggesting that this region is important for interaction with expanded CAG RNA. This result provided indirect evidence that the peptide likely folds into the predicted structure when bound to RNA (Fig. 1B). On the other hand, Lys13 and Gly14 are located at the hairpin loop immediately N-terminal to β2 strand. The reduced inhibitory activity of the BIND of the Gly14Ala mutant suggested that the conformational flexibility provided by Gly14 is important for peptide interaction, likely by correctly positioning Lys13, with RNA.
TAT-BIND suppresses expanded CAG RNA-induced neurodegeneration in vivo
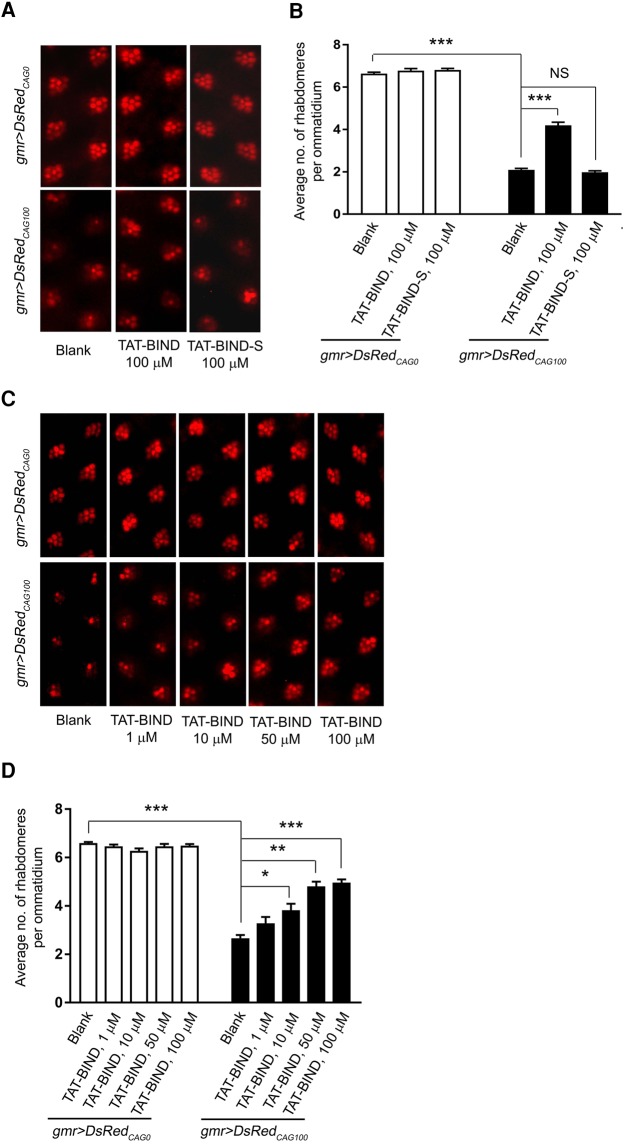
To investigate the suppressive effects of TAT-BIND on expanded CAG RNA-induced neurodegeneration in vivo, we used DsRedCAG0/100/250 Drosophila models (Li et al. 2008). The models express untranslated CAG repeats RNA due to the location of CAG repeats being in the 3′ untranslated region of the DsRed reporter gene. In Drosophila, expression of DsRedCAG100 by the gmr-GAL4 driver causes retinal degeneration that can be quantified by a pseudopupil assay (Li et al. 2008; Tsoi et al. 2011). We then performed this assay on DsRedCAG100 Drosophila to test if TAT-BIND could suppress expanded CAG RNA-induced neurodegeneration and restore the number of rhabdomeres within the ommatidium. Treatment with 10, 50 or 100 µM TAT-BIND dose-dependently suppressed retinal degeneration in DsRedCAG100 Drosophila without exerting any observable toxic effect on DsRedCAG0 Drosophila (Fig. 4A–D). TAT-BIND treatment also restored the expression level of pre-rRNA in DsRedCAG100 Drosophila (Supplemental Fig. S7), indicating that the inhibitory effect of TAT-BIND on nucleolar stress ameliorates expanded CAG RNA-induced neurodegeneration in vivo. In contrast, TAT-BIND-S, which is a scrambled control for TAT-BIND, did not mitigate retinal degeneration in the DsRedCAG100 Drosophila model (Fig. 4A,B).
FIGURE 4.
TAT-BIND suppressed retinal degeneration in DsRedCAG100 Drosophila in a dose-dependent manner. (A) TAT-BIND suppressed retinal degeneration in DsRedCAG100 Drosophila. (B) Statistical analysis of panel A. (C) Dose-response effect of TAT-BIND on suppressing eye degeneration in DsRedCAG100 Drosophila. (D) Statistical analysis of panel C. Third instar larvae were treated with respective peptides dissolved in 2% sucrose solution for 2 h at room temperature. Pseudopupil assay was performed on 12-d-old adult flies. The genotypes of the flies were: w; gmr-GAL4 UAS-DsRedCAG0/+; +/+ and w; gmr-GAL4/+; UAS-DsRedCAG100/+. Data are plotted as mean ± SEM and were collected from at least three independent experiments. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. NS, no significant difference.
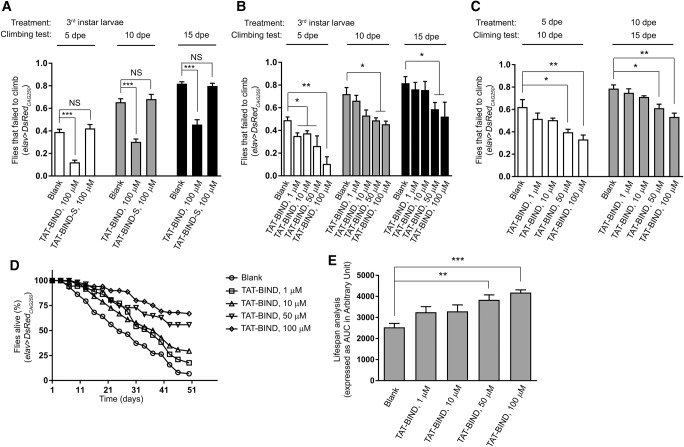
In addition to pseudopupil assay, we performed climbing ability and lifespan assays to evaluate the suppressive effect of BIND in vivo (Li et al. 2008). We first confirmed that flies expressing DsRedCAG250 in neurons using elav-GAL4 driver exhibited progressive loss of climbing ability and lethality (Fig. 5A–E). Treatment with 100 µM TAT-BIND at the third instar larval stage significantly rescued climbing defects of 5 dpe, 10 dpe, and 15 dpe adult elav>DsRedCAG250flies, whereas the scrambled TAT-BIND-S had no effect (Fig. 5A). This rescuing effect of TAT-BIND was dose-dependent (Fig. 5B). Even when TAT-BIND treatment was given as late as 5 dpe or 10 dpe adult stage, we were still able to detect a significant rescue of climbing ability within 5 d of 50 or 100 µM TAT-BIND treatment (Fig. 5C). This result demonstrated that even after the manifestation of climbing defects in adult animals, TAT-BIND is still capable of ameliorating the disease-like symptoms, highlighting the effectiveness of TAT-BIND in vivo. We further showed that the treatment with 50 or 100 µM TAT-BIND extended the lifespan of DsRedCAG250 Drosophila (Fig. 5D,E), indicating the therapeutic potential of TAT-BIND against RNA toxicity in polyQ diseases.
FIGURE 5.
TAT-BIND dose-dependently improved the climbing ability and extended the lifespan of DsRedCAG250 Drosophila. (A) TAT-BIND treatment during third instar larval stage rescued the climbing defects of adult DsRedCAG250 Drosophila, whereas TAT-BIND-S had no effect. (B) TAT-BIND treatment during third instar larval stage rescued the climbing defects of adult DsRedCAG250 Drosophila in a dose-dependent manner. (C) TAT-BIND treatment during adult stage rescued the climbing defects of adult DsRedCAG250 Drosophila in a dose-dependent manner. (D) TAT-BIND treatment extended the lifespan of DsRedCAG250 Drosophila in a dose-dependent manner. (E) Area under the curve (AUC) analysis of panel D. For TAT-BIND treatment during larval stage, third instar larvae were treated with respective peptides dissolved in 2% sucrose solution for 2 h at room temperature. For TAT-BIND treatment during adult stage, adult flies were starved for 8 h followed by overnight feeding with respective peptides in 2% sucrose solution at room temperature. The genotypes of flies were: elav-GAL4; +/+; UAS-DsRedCAG250/+. Data are plotted as mean ± SEM and were from at least six independent experiments. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. NS, no significant difference.
TAT-BIND suppresses neurodegeneration in different polyQ disease models in vitro and in vivo
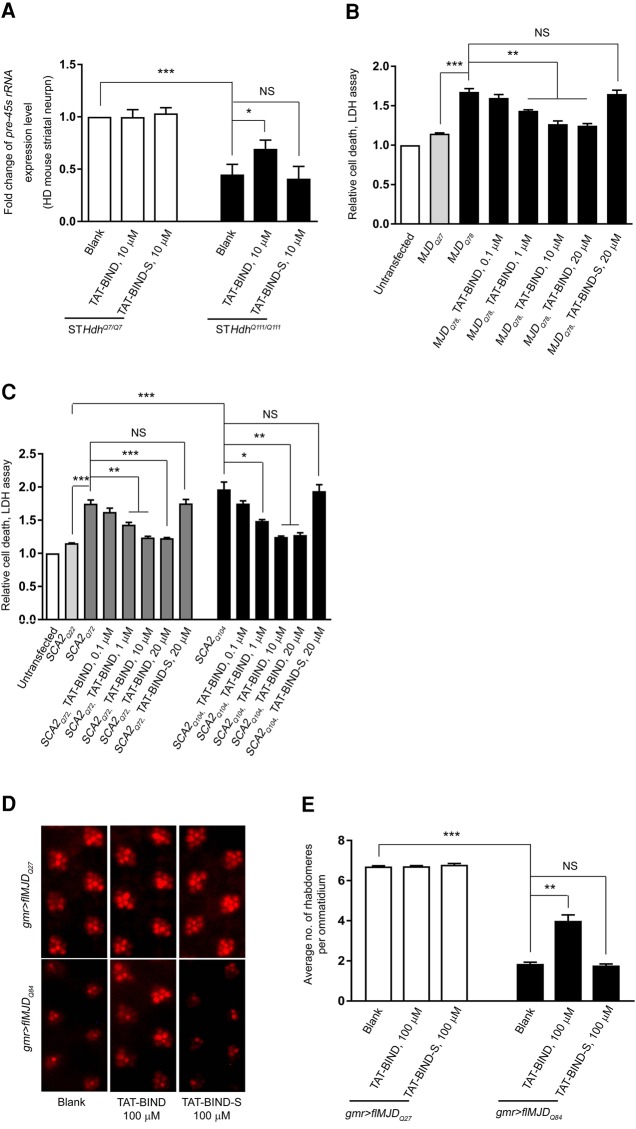
In general, expanded CAG RNA induces neurotoxicity in polyQ diseases (Li et al. 2008; Tsoi et al. 2012; Tsoi and Chan 2013). We thus investigated if TAT-BIND suppresses expanded CAG RNA-induced neurotoxicity in polyQ disease models. We selected the mouse striatal cell model of HD (STHDhQ111/Q111) expressing mutant huntingtin proteins with 111 glutamines (Trettel et al. 2000) for our investigation. pre-45s rRNA expression in STHDhQ111/Q111 cells was decreased compared to that of control STHDhQ7/Q7 cells expressing normal huntingtin (Fig. 6A). The expression level of pre-45s rRNA in STHDhQ111/Q111 cells was partially rescued with 10 µM TAT-BIND treatment (Fig. 6A), indicating that nucleolar stress was suppressed by TAT-BIND in this HD cell model.
FIGURE 6.
TAT-BIND suppressed neurotoxicity in different polyQ disease models in vitro and in vivo. (A) TAT-BIND rescued pre-45s rRNA expression level in STHdhQ111/111 striatal cells. (B) TAT-BIND suppressed cell death in MJDQ78-expressing HEK293 cells in a dose-dependent manner. (C) TAT-BIND suppressed cell death in SCA2Q72/104-expressing HEK293 cells in a dose-dependent manner. LDH data were normalized to the untransfected control. (D) TAT-BIND suppressed neurodegeneration in flMJDQ84 Drosophila. (E) Statistical analysis of panel D. Third instar larvae were treated with respective peptides dissolved in 2% sucrose solution for 2 h at room temperature. Pseudopupil assay was performed on 12-d-old flMJDQ27/84 adult flies. The genotypes of flies were: w; gmr-GAL4 UAS-myc-flMJDQ27/+; +/+ and w; gmr-GAL4/+; UAS-myc-flMJDQ84/+. Data are plotted as mean ± SEM and were from at least three independent experiments. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. NS, no significant difference.
pcDNA3.1-MJDQ27/78 and pcDNA3.1-myc-SCA2Q22/72/104 constructs have previously been used to investigate the suppressive effects of peptide inhibitors on toxicity induced by expanded CAG RNA and polyQ protein (Tsoi et al. 2012; Li et al. 2016; Zhang et al. 2016). The expression of MJDQ78 and SCA2Q72/104 caused severe cell death when compared with unexpanded MJDQ27 and SCA2Q22 controls, respectively (Fig. 6B,C). TAT-BIND treatment suppressed the cell death of MJDQ78- or SCA2Q72/104-expressing cell models in a dose-dependent manner (Fig. 6B,C).
In addition to in vitro models, we also investigated the inhibitory effect of TAT-BIND on neurodegeneration in flMJDQ27/84 (Zhang et al. 2016) and the Htt-exon1Q93 Drosophila (Steffan et al. 2001) in vivo. The expression of flMJDQ84 (Fig. 6D) or Htt-exon1Q93 (Supplemental Fig. S8A) using gmr-GAL4 driver resulted in severe retinal degeneration. Treatment with 100 µM TAT-BIND but not TAT-BIND-S at the third instar larval stage rescued the retinal degeneration in adult flies (Fig. 6D,E; Supplemental Fig. S8A,B).
Specificity and cytotoxicity study of TAT-BIND
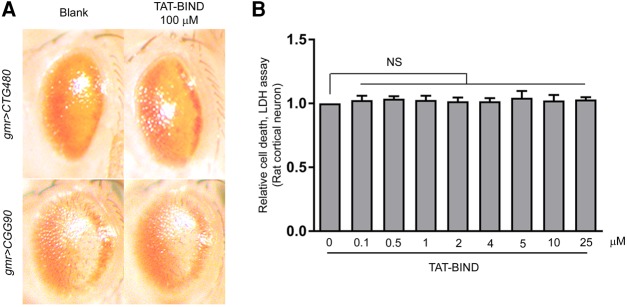
We used other repeat expansion-associated disease fly models, including CTG480 (myotonic dystrophy type 1) (Wong et al. 2014) and CGG90 (Fragile X syndrome) (Jin et al. 2003) to determine the specificity of TAT-BIND on CAG repeat expansion RNA toxicity. Consistent with previous reports, both CTG480 and CGG90-expressing flies exhibited retinal degenerative phenotypes (Fig. 7A; Jin et al. 2003; Wong et al. 2014). However, TAT-BIND treatment had no effect on these disease models (Fig. 7A). This result suggested that the TAT-BIND is a specific inhibitor toward expanded CAG repeats RNA-induced toxicity in vivo. Further, cytotoxicity tests on primary rat cortical neurons showed that the viability of wild-type primary neurons was not compromised when treated with up to 25 μM of TAT-BIND (Fig. 7B). This result clearly indicates that a concentration of 25 μM or lower of TAT-BIND does not exert a detectable toxic effect on mammalian neurons.
FIGURE 7.
Specificity and cytotoxicity study of BIND. (A) TAT-BIND did not suppress the neurodegeneration of CTG480- and CGG90-expressing fly eyes. Third instar larvae were treated with respective peptides dissolved in 2% sucrose solution for 2 h at room temperature. External eye pictures of 1 dpe CTG480 and 2 dpe CGG90 Drosophila were captured. The genotypes of flies were: w; gmr-GAL4/+; UAS-CTG480/+ and w; gmr-GAL4 UAS-EGFP-CGG90/+; +/+. (B) TAT-BIND did not exert toxic effects on primary rat cortical neurons. Data were normalized to untreated control and are plotted as mean ± SEM. All the experiments were repeated at least three times. NS, no significant difference.
DISCUSSION
Over the past decade, accumulated evidence has demonstrated that transcripts containing expanded CAG repeats contribute to the pathogenesis of polyQ diseases (Li et al. 2008; Fiszer and Krzyzosiak 2013, 2014). However, the slow development of inhibitors targeting neurotoxicity induced by expanded CAG RNA has been problematic. We identified BIND on the basis of the NCL RRM1/2:RNA complex structure. BIND is a novel peptide that could inhibit cell death induced by expanded CAG RNA at an IC50 value of 0.7 µM (Fig. 1D). It comprises residues that form the β2 and β3 strands and β2–β3 loop of NCL RNA. This novel peptide inhibitor contains the RNP1 sequence, which forms the P3 inhibitor that we have previously identified. The addition of residues from the β2 strand of NCL RRM, however, is predicted to stabilize BIND in a β-hairpin conformation (Zhang et al. 2016). In fact, our CD analysis provided evidence that BIND contains nearly 40% β-strand content, and that it likely adopts a right-hand twisted anti-parallel β-hairpin conformation.
Nucleolar stress with hindered rRNA transcription has been widely implicated in the pathogenesis of various neurodegenerative diseases (Parlato and Kreiner 2013; Tsoi and Chan 2014). Nucleolar stress induced by expanded CAG RNA is also implicated in polyQ diseases (Tsoi et al. 2012). We previously reported that the direct P3-CAG RNA interaction rescued rRNA transcription and suppressed nucleolar stress (Zhang et al. 2016). Similarly, we found that TAT-BIND restored the expression levels of pre-45s rRNA (Fig. 2E) and pre-rRNA (Supplemental Fig. S7) in the models expressing expanded CAG RNA in vitro and in vivo. TAT-BIND also suppressed the mislocalization of nucleolar proteins NCL (Fig. 2A,B) and B23 (Fig. 2C,D) in the nuclei of HEK293 cells expressing expanded CAG RNA. In the knock-in STHDhQ111/Q111 striatal cells, both CAG111-containing HTT RNAs and glutamine111-containing HTT proteins are expressed. Treatment with TAT-BIND also restored the expression level of its pre-45s rRNA relative to that in control neurons. Our biophysical study revealed that the binding affinity of BIND for expanded CAG RNA is ∼70 fold lower than that for the existing CAG RNA inhibitor P3 (Fig. 3D; Zhang et al. 2016). In summary, TAT-BIND decreases the toxicity induced by expanded CAG RNA through the same mechanism as P3 but more efficiently suppresses nucleolar stress induced by expanded CAG RNA in polyQ diseases.
P3 and BIND have different key pharmacophores despite their similar mechanisms for nuclear stress suppression. Our SAR study showed that Glu2, Lys13, Gly14, Ile18, Glu19, and Phe20 are essential for the inhibitory effects of BIND on expanded CAG RNA-induced toxicity (Fig. 3E,F). In contrast, Lys3, Lys5, Tyr9, and Phe12 are responsible for maintaining the bioactivity of P3 (Zhang et al. 2016). These data suggest that, although the two peptides have identical mechanisms of nucleolar stress suppression, the mode of recognition and interaction of expanded CAG RNA by BIND are fundamentally different from that of P3. This difference may be attributed to the partial β-structure adopted by BIND. In addition, the more stable anti-parallel β-sheet conformation of BIND than that of the flexible P3 may significantly decrease entropic energy loss upon binding to expanded CAG RNA and improve the binding affinity and suppression efficacy of BIND in polyQ diseases.
Polyglutamine diseases include at least nine disorders characterized by the pathological expansion of CAG trinucleotide repeats (Fan et al. 2014). To date, all known peptidylic inhibitors for polyQ diseases mainly target HD (Kazantsev et al. 2002; Arribat et al. 2014). Here, we demonstrated that BIND suppresses neurodegeneration in vitro and in vivo in multiple polyQ disease models, including HD, SCA2, and MJD models. We found that TAT-BIND specifically reduces expanded CAG RNA toxicity without exerting any observable toxic effect in vitro and in vivo (Figs. 4, 5, 7). Despite the conjugation of the TAT sequence to BIND slightly reduced the affinity of BIND to expanded CAG RNA in vitro, the specificity of BIND was unaffected and the in vivo activity of BIND remained effective. Thus, BIND is a specific and safe peptidylic inhibitor with potential clinical applications and its CPP-mediated delivery is a feasible therapeutic strategy for the treatment of different polyQ diseases.
MATERIALS AND METHODS
Construction of plasmids
The pEGFPCAG27/78, pcDNA3.1-MJDQ27/78, and pcDNA3.1-myc-SCA2Q22/42/55/72/104 constructs were reported previously (Li et al. 2008, 2016; Tsoi et al. 2012; Zhang et al. 2016). The double-strand oligonucleotides TAT-BIND-myc and TAT-BIND-S-myc were generated using oligos: TAT-BIND-myc-F: 5′-AGCTTATGTATGGCCGCAAAAAACGCCGCCAGCGCCGCCGCGCGGAAATTCGCCTG GTGAGCAAAGATGGCAAAAGCAAAGGCATTGCGTATATTGAATTTAAAGAACAAAAACTCATCTCAGAAGAGGATCTGTGAG-3′; TAT-BIND-myc-R: 5′-GATCCTCACAGATCCTCTTCTGAGATGAGTTTTTGTTCTTTAAATTCAATATACGCAATGCCTTTGCTTTTGCCATCTTTGCTCACCAGGCGAATTTCCGCGCGGCGGCGCTGGCGGCGTTTTTTGCGGCCATACATA-3′; TAT-BIND-S-myc-F: 5′-AGCTTATGTATGGCCGCAAAAAACGCCGCCAGCGCCGCCGCGGCGGCGAAGATATTAAAAGCCGCGTGGAAGCGGCGAGCATTCTGTATTTTATTAAAAAAAAAGAACAAAAACTCATCTCAGAAGAGGATCTGTGAG-3′ and TAT-BIND-S-myc-R: 5′-GATCCTCACAGATCCTCTTCTGAGATGAGTTTTTGTTCTTTTTTTTTAATAAAATACAGAATGCTCGCCGCTTCCACGCGGCTTTTAATATCTTCGCCGCCGCGGCGGCGCTGGCGGCGTTTTTTGCGGCCATACATA-3′. The resultant fragments were then ligated with pcDNA3.1(+) using HindIII and BamHI restriction sites.
Synthesis of peptides and CAG RNAs
All peptides were purchased from GenScript USA, Inc. The TAT cell penetrating peptide used in this study was YGRKKRRQRRR (Popiel et al. 2007) and it was attached to the N terminus of all the peptides investigated. The sequences of TAT-BIND and 19 TAT-BIND mutants were shown in Figure 3E. The scrambled control peptide, BIND-S, with a sequence of GGEDIKSRVEAASILYFIKKK, was also synthesized with TAT-fused at the N terminus. The purity of peptides used in cell experiments was over 90%. Desalted peptides were used in Drosophila feeding assays. All RNAs were synthesized using the MEGAscript kit (Ambion) as previously described (Tsoi et al. 2012). The SCA2CAG22/42/55/72 RNAs were transcribed from EcoRI linearized pcDNA3.1-myc-SCA2Q22/42/55/72 constructs (Zhang et al. 2016). The concentration of RNA was determined with appropriate extinction coefficients at 260 nm on a Nanodrop 2000 spectrophotometer (Thermo Scientific).
Cell culture, plasmid transfection, and cellular peptide treatment
Human embryonic kidney 293 (HEK293) cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. Primary rat cortical neurons were isolated and cultured as previously described (Lau et al. 2008). The control striatal cells STHDhQ7/Q7 expressing normal huntingtin, and HD striatal cells STHDhQ111/Q111 expressing mutant huntingtin with 111 glutamines (kind gifts of Professor Hyokeun PARK, Hong Kong University of Science and Technology) were described previously (Trettel et al. 2000). These cells were cultured at 33°C in DMEM supplemented with 10% FBS, 1% penicillin–streptomycin and 0.4 mg/mL G418 (Takara). Transient transfection of HEK293 cells was performed using Lipofectamine 2000 (Thermo Fisher Scientific). Treatment of TAT-fusion peptides was performed immediately after plasmid transfection. At least two batches of independently synthesized peptides were used in the experiments.
Circular dichroism spectroscopy measurements of BIND
The circular dichroism (CD) spectrum for the BIND was recorded in ddH2O at 25°C on a JASCO J-810 spectropolarimeter. The JASCO J-810 parameters were set to the following: scan range 190–260 nm, scan speed 20 nm/min, 1 sec response time and 1 nm bandwidth. Data were calculated by subtracting CD spectra of ddH2O. The final CD spectrum was the result of three accumulated scans. The percentages of secondary structures were estimated using the online servers CAPITO (Wiedemann et al. 2013) and BeStSel (Micsonai et al. 2015).
Isothermal titration calorimetry (ITC) binding assay
Experiments were performed using a MicroCal iTC200 isothermal titration calorimeter (GE Healthcare) at 25°C. Data were analyzed using the Origin scientific plotting software version 7 (Microcal Software Inc.). All RNAs and peptides were dissolved in binding buffer (20 mM MOPS, pH 7.0; 300 mM NaCl). Two millimolar of respective peptide was titrated to 4 µM in vitro transcribed SCA2CAG22/42/55/72 RNA. A reference power of 8 µcal/sec was used with an initial 0.5 µL of injection of peptide followed by 2 µL for all subsequent titrations points with 60 sec initial equilibrium delay and 120 sec pause between injections. The samples were stirred at a speed of 1000 rpm. throughout the experiment. The thermal titration data were fitted to the “one binding site model” to determine the dissociation constant (KD).
RNA extraction, reverse-transcription, and real-time PCR
RNA was extracted from cells or 12-d-old fly heads by TRIzol reagent (Thermo Fisher Scientific), and 1 µg of purified RNA was then used for reverse-transcription using the ImPromII Reverse Transcription System (Promega). Random hexamer (Roche) was used as primers in reverse transcription. Taqman gene expression assays were performed on a BIO-RAD CFX96 Real-time PCR system and data were analyzed as previously described (Tsoi et al. 2012). The following probes were used: pre-45s rRNA (Assay ID: AILJIZM), Human actin (Assay ID: Hs99999903_m1), GADD45A (Assay ID: Hs00169255_m1), HBB (Assay ID: Hs00758889_s1), pre-rRNA (Assay ID: AIMSG5U), and GAPDH for Drosophila (Assay ID: Dm01841186).
Lactate dehydrogenase (LDH) cytotoxicity assay
HEK293 cells were seeded on a 24-well plate at a density of 1 × 105 and LDH enzyme activity in the cell culture medium was measured 72 h post plasmid transfection and/or drug treatment using the Cytotox 96 nonradioactive cytotoxicity assay (Promega). The experimental data were all normalized to respective controls.
For IC50 determination, various amount of TAT-BIND, 0.1, 0.5, 1, 2, 4, 5, 10, and 25 μM were added to individual culture wells after pEGFPCAG78 transfection. After normalization, data were analyzed using the dose–response inhibition curve (nonlinear regression-variable slope) to determine the IC50 value (Prism6 software, GraphPad Software, Inc.). For the cytotoxicity study, the same amounts of TAT-BIND were tested in rat cortical neuron. For the efficacy study in diverse disease models, 0.1, 1, 10, and 20 µM of TAT-BIND were added to cells transfected with pcDNA3.1-MJDQ27/78 or pcDNA3.1-myc-SCA2Q22/72/104.
Immunostaining
HEK293 cells were seeded and grown on glass coverslips at a density of 1 × 105 in a 24-well plate. Forty-eight hours after pEGFPCAG78 transfection and peptide treatment, cells were fixed with 4% paraformaldehyde followed by three washes with 1× PBS. Cells were then permeabilized in 0.5% Triton X-100 for 10 min at room temperature followed by three washes with 1× PBS. Cells were next blocked with 5% BSA in 1× PBS for 1 h at room temperature. The NCL (1:500, Abcam) or B23 (1:500, Abcam) antibody was then applied with 5% BSA (1:500) for 2 h at room temperature. Once the primary antibody was removed, cells were washed three times with 1× PBS and incubated with an anti-rabbit Cy3 or an anti-mouse Cy3 secondary antibody (1:400, Jackson Labs) for 1 h at room temperature. After washing with 1× PBS, nuclei were counterstained with Hoechst 33342 (1 µg/mL, Thermo Fisher Scientific) prior to mounting coverslips. Images were obtained on an OLYMPUS FV1000 IX81-TIRF confocal microscope. A single focal plan was obtained through the center of the nucleus. The method of quantification of NCL and B23 nuclear area fold change was described previously (Haeusler et al. 2014). To quantify both dispersed NCL/B23 and dense nucleolar NCL/B23, a threshold setting in ImageJ ranging from 25–100 was used to measure the pixel area of NCL/B23 relative to the area of the nucleus outlined by the Hoechst staining. Of note, 100–150 cells were measured per treatment, and data were normalized to untransfected control.
Protein/RNA interaction
HEK293 cells were transfected with EGFPCAG78 simultaneously with pcDNA3.1(+)-TAT-BIND-myc or pcDNA3.1(+)-TAT-BIND-S-myc construct. Twenty-four hours after transfection, cells were fixed with 1% formaldehyde for 10 min at 37°C. Ice cold 1× PBS was used to wash cells twice. Binding buffer (10 mM HEPES, pH 7.5, 10% glycerol, 5 mM MgCl2, 142.5 mM KCl, 1 mM EDTA, 1% Triton X-100) supplemented with protease inhibitor cocktail (Sigma-Aldrich) and 40 U of RNAsin (Promega) was then added to the samples. Sonication was applied to lyse the cells (Duty Cycle 30, Output Control 3, Timer 20 sec, Sonifier 450, Branson Ultrasonics), and the samples were incubated at 4°C for 1 h with rotation. The samples were centrifuged at 4°C for 30 min at 18,606 r.c.f. Ten percent of supernatant was saved as “Input.” Forty microliters of protein G agarose beads, together with anti-myc antibody (1:200; Cell signaling) was quickly added to the remaining samples, and the mixture was incubated at 4°C overnight with gentle rotation. For no-antibody control, the same amount of protein G agarose beads was added to the reaction followed by gentle rotation at 4°C for overnight. Protein/RNA complex was collected by centrifuging at 4°C for 10 sec at 18,506 r.c.f., followed by three washes of binding buffer. Fifty microliters of binding buffer was used to resuspend the protein/RNA complex and the reaction was incubated for 45 min at 65°C to reverse the cross-links. RNA extraction and reverse-transcription PCR (RT-PCR) were subsequently performed. CAG78 cDNA was amplified using primers: CAG78-F: 5′-GCATGGACGAGCTGTACAAG-3′, and CAG78-R: 5′-CGCACCGGTTCTGTCCTGATAGGTCC-3′. TAT-BIND-myc or TAT-BIND-S-myc cDNA was amplified using primers: TAT-F: 5′-ATGTATGGCCGCAAAAAAC-3′ and myc-R: 5′-CAGATCCTCTTCTGAGATGAG-3′.
Protein expression and purification
SCA2CAG72 RNA was synthesized using the MEGAscript kit (Ambion) as previously described (Tsoi et al. 2012) and was PAGE-purified. BIND was subcloned into the pGEX4T2 vector. Both GST-BIND and free GST expression plasmids were expressed in BL21 (DE3) strain Escherichia coli cells. Large scale culture was grown in LB broth with 200 µg/mL ampicillin at 37°C. Protein expression was induced with 0.2 mM IPTG with shaking overnight at 16°C. Pelleted cells were lysed by sonication in 25 mM Tris 8.0, 150 mM NaCl, and 2 mM DTT, and 1 mM PMSF. The soluble fraction was loaded onto a Glutathione Sepharose 4B column (GE Healthcare) and washed with 90 mL lysis buffer. The protein was eluted with buffer containing 10 mM glutathione. The eluate was further purified by Mono-Q GL column (GE Healthcare) and the peak eluted fractions were combined, concentrated, separated in aliquots and flash frozen.
Filter-binding assay
The filter-binding assay was performed as described previously (Rio 2012). Of note, 62.4 pmol of purified RNA was 5′-end labeled with 5 µCi [γ-32P] ATP by T4 polynucleotide kinase (New England Biolabs) in nuclease-free buffer at 37°C for 1 h. The reaction was stopped by incubating at 70°C for 10 min. Labelled RNA was purified by mini Quick Spin Oligo Columns (Roche). The purified oligonucleotides were then heated at 95°C for 10 min and cooled down for 30 min. Different concentrations of protein were titrated to a constant amount of 50 nM CAG72 RNA in a final volume of 50 µL and incubated at room temperature for 1 h in binding buffer consisting of 10 mM Tris 7.5, 50 mM KCl, 750 µM MgCl2, 100 µM EDTA, 5% glycerol, 600 µM dithiothreitol, 0.1 mg/mL tRNA, and 40 µg/mL BSA. Nitrocellulose (0.45 µm pore size, Bio-Rad) and nylon filter (Roche) were presoaked in wash buffer 10 mM Tris 7.5, 50 mM KCl, and 1 mM dithiothreitol for 2 h. Both filters, nitrocellulose on top and nylon filter at the bottom, were placed into a dot-blot apparatus (Bio-Rad). The wells were washed once and vacuum was applied before the samples were loaded. The wells were washed eight times with 100 µL washing buffer after samples were loaded. The nitrocellulose filters were analyzed using auto-radiography to measure the retained radiolabeled RNA on the nitrocellulose filter.
Drosophila genetics, peptide feeding and assays
Flies were raised at 21.5°C or 25°C on cornmeal medium supplemented with dry yeast for different assays. The fly lines UAS-DsRedCAG0/100/250 (Li et al. 2008) and UAS-flMJDQ27/84 (Warrick et al. 2005) were gifts of Professor Nancy Bonini (University of Pennsylvania). The UAS-EGFP-CGG90 (Jin et al. 2003) and UAS-CTG480 (Wong et al. 2014) fly lines were obtained from Professors Stephen Warren (Emory University) and Rubén Artero Allepuz (Universitat de València, Estudi General), respectively. The gmr-GAL4, elav-GAL4, and UAS-Htt-exon1Q93 (Chan et al. 2011) fly lines were obtained from Bloomington Drosophila Stock Center. For the feeding assay, all peptides were dissolved in 2% sucrose solution, and third instar larvae were fed for 2 h at room temperature unless otherwise stated.
For the pseudopupil assay (Li et al. 2008), flies expressing UAS-DsRedCAG0/100/250, UAS-flMJDQ27/84, and UAS-Htt-exon1Q93 were raised at 21.5°C. Besides Htt-exon1Q93, in which 1 d post eclosion (dpe) adult flies were used, 12 dpe adult flies of other fly lines were used in this assay. For the external eye assay, flies were raised at 25°C and images were taken on 1 dpe CTG480 flies and 2 dpe CGG90 flies using an Olympus SZX-12 stereomicroscope. All eye images were captured using a SPOT Insight CCD camera (Diagnostic instruments Inc.). Image processing was performed using the Adobe Photoshop CS software (Adobe). Each experiment was repeated at least three times (n = 10 fly heads) and consistent results were obtained.
For climbing ability and lifespan analyses (Li et al. 2008), flies expressing DsRedCAG250 were raised at 25°C in standard fly food. For larvae treatment, the climbing ability of adult flies was investigated at 5, 10, and 15 dpe, respectively. For adult treatment, 5 or 10 dpe adult flies were starved for 8 h followed by overnight feeding with respective peptides in 2% sucrose solution soaked in filter papers. Flies were then put back into standard fly food and assayed at 10 or 15 dpe. Climbing ability was analyzed by negative geotaxis. Groups of 10–12 flies of the same gender were anesthetized and placed in a 15 mL transparent tube. After 1 h of recovery, flies were banged to the bottom, and then the climbing fail rate was calculated as the number of flies remaining at the bottom divided by the total number of flies (<2cm) at 25 sec. Three trials were performed at 3-min intervals in each experiment. Eighty to 120 flies were tested per treatment for the climbing ability assay. Of note, 120–150 flies were tested per treatment for lifespan analysis.
Statistical analyses
Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (*), (**), (***), and (****) represent P < 0.05, P < 0.01, P < 0.001, and P < 0.0001, respectively, which are considered statistically significant. NS indicates no significant difference was observed.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the ANR/RGC Joint Research Scheme (A-CUHK401/14) and Collaborative Research Fund (CUHK1/CRF/13G) of the Hong Kong Research Grants Council; Direct Grant for Research from the Research Committee of CUHK (4053148); CUHK Lui Che Woo Institute of Innovative Medicine BRAIN Initiative (8303404); CUHK Group Research Scheme (3110102); CUHK Vice-Chancellor's One-Off Discretionary Fund (VCF2014011); CUHK One-off Funding for Joint Lab/Research Collaboration (3132980); CUHK Gerald Choa Neuroscience Centre (7105306); CUHK Faculty of Science Strategic Development Fund; and donations from Chow Tai Fook Charity Foundation (6903898) and Hong Kong Spinocerebellar Ataxia Association (6903291).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062703.117.
REFERENCES
- Abdelmohsen K, Gorospe M. 2012. RNA-binding protein nucleolin in disease. RNA Biol 9: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Bouvet P, Dieckmann T, Feigon J. 2000a. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J 19: 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Gilbert DE, Bouvet P, Feigon J. 2000b. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J Mol Biol 303: 227–241. [DOI] [PubMed] [Google Scholar]
- Arribat Y, Talmat-Amar Y, Paucard A, Lesport P, Bonneaud N, Bauer C, Bec N, Parmentier ML, Benigno L, Larroque C, et al. 2014. Systemic delivery of P42 peptide: a new weapon to fight Huntington's disease. Acta Neuropathol Commun 2: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam S, Miller MC, Maliekal J, Bates PJ, Trent JO, Lane AN. 2010. Solution structure of the RBD1,2 domains from human nucleolin. J Biomol NMR 47: 79–83. [DOI] [PubMed] [Google Scholar]
- Avitabile D, Bailey B, Cottage CT, Sundararaman B, Joyo A, McGregor M, Gude N, Truffa S, Zarrabi A, Konstandin M, et al. 2011. Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc Natl Acad Sci 108: 6145–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañez-Coronel M, Porta S, Kagerbauer B, Mateu-Huertas E, Pantano L, Ferrer I, Guzmán M, Estivill X, Martí E. 2012. A pathogenic mechanism in Huntington's disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet 8: e1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. 1998. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem 273: 19025–19029. [DOI] [PubMed] [Google Scholar]
- Chan WM, Tsoi H, Wu CC, Wong CH, Cheng TC, Li HY, Lau KF, Shaw PC, Perrimon N, Chan HY. 2011. Expanded polyglutamine domain possesses nuclear export activity which modulates subcellular localization and toxicity of polyQ disease protein via exportin-1. Hum Mol Genet 20: 1738–1750. [DOI] [PubMed] [Google Scholar]
- de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. 2011. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res 39: 3852–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43: W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiríksdóttir E, Konate K, Langel U, Divita G, Deshayes S. 2010. Secondary structure of cell-penetrating peptides controls membrane interaction and insertion. Biochim Biophys Acta 1798: 1119–1128. [DOI] [PubMed] [Google Scholar]
- Fan HC, Ho LI, Chi CS, Chen SJ, Peng GS, Chan TM, Lin SZ, Harn HJ. 2014. Polyglutamine (PolyQ) diseases: genetics to treatments. Cell Transplant 23: 441–458. [DOI] [PubMed] [Google Scholar]
- Fiszer A, Krzyzosiak WJ. 2013. RNA toxicity in polyglutamine disorders: concepts, models, and progress of research. J Mol Med (Berl) 91: 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszer A, Krzyzosiak WJ. 2014. Oligonucleotide-based strategies to combat polyglutamine diseases. Nucleic Acids Res 42: 6787–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO. 1988. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55: 1189–1193. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Fujiwara K, Goda N, Iwaya N, Tenno T, Shirakawa M, Hiroaki H. 2011. Structure and function of the N-terminal nucleolin binding domain of nuclear valosin-containing protein-like 2 (NVL2) harboring a nucleolar localization signal. J Biol Chem 286: 21732–21741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J 17: 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M, Loewenstein PM. 1988. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 55: 1179–1188. [DOI] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. 2014. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Xu XS, Russell JE. 2006. A nucleolin-binding 3′ untranslated region element stabilizes β-globin mRNA in vivo. Mol Cell Biol 26: 2419–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. 2003. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 39: 739–747. [DOI] [PubMed] [Google Scholar]
- Kazantsev A, Walker HA, Slepko N, Bear JE, Preisinger E, Steffan JS, Zhu YZ, Gertler FB, Housman DE, Marsh JL, et al. 2002. A bivalent Huntingtin binding peptide suppresses polyglutamine aggregation and pathogenesis in Drosophila. Nat Genet 30: 367–376. [DOI] [PubMed] [Google Scholar]
- Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. 2007. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell 25: 575–586. [DOI] [PubMed] [Google Scholar]
- Lau KF, Chan WM, Perkinton MS, Tudor EL, Chang RC, Chan HY, McLoughlin DM, Miller CC. 2008. Dexras1 interacts with FE65 to regulate FE65-amyloid precursor protein-dependent transcription. J Biol Chem 283: 34728–34737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM. 2008. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PP, Sun X, Xia G, Arbez N, Paul S, Zhu S, Peng HB, Ross CA, Koeppen AH, Margolis RL, et al. 2016. ATXN2-AS, a gene antisense to ATXN2, is associated with spinocerebellar ataxia type 2 and amyotrophic lateral sclerosis. Ann Neurol 80: 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E. 2016. RNA toxicity induced by expanded CAG repeats in Huntington's disease. Brain Pathol 26: 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J, Derreumaux P, Tuffery P. 2009. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res 37: W498–W503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J, Derreumaux P, Tufféry P. 2010. A fast method for large-scale de novo peptide and miniprotein structure prediction. J Comput Chem 31: 726–738. [DOI] [PubMed] [Google Scholar]
- Meng L, Yasumoto H, Tsai RY. 2006. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci 119: 5124–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik A, Van Broeckhoven C. 2003. Pathogenesis of polyglutamine disorders: aggregation revisited. Hum Mol Genet 12 Spec No 2: R173–R186. [DOI] [PubMed] [Google Scholar]
- Micsonai A, Wien F, Kernya L, Lee YH, Goto Y, Refregiers M, Kardos J. 2015. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci 112: E3095–E3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. 2011. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res 39: 8938–8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalavade R, Griesche N, Ryan DP, Hildebrand S, Krauß S. 2013. Mechanisms of RNA-induced toxicity in CAG repeat disorders. Cell Death Dis 4: e752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. 2007. Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621. [DOI] [PubMed] [Google Scholar]
- Parlato R, Kreiner G. 2013. Nucleolar activity in neurodegenerative diseases: a missing piece of the puzzle? J Mol Med (Berl) 91: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popiel HA, Nagai Y, Fujikake N, Toda T. 2007. Protein transduction domain-mediated delivery of QBP1 suppresses polyglutamine-induced neurodegeneration in vivo. Mol Ther 15: 303–309. [DOI] [PubMed] [Google Scholar]
- Raussens V, Ruysschaert JM, Goormaghtigh E. 2003. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: a new scaling method. Anal Biochem 319: 114–121. [DOI] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G. 2007. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol 27: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio DC. 2012. Filter-binding assay for analysis of RNA-protein interactions. Cold Spring Harb Protoc 2012: 1078–1081. [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU. 2002. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci 99: 16412–16418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413: 739–743. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Katada S, Onodera O. 2010. Polyglutamine diseases: where does toxicity come from? what is toxicity? where are we going? J Mol Cell Biol 2: 180–191. [DOI] [PubMed] [Google Scholar]
- Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, Ying Z. 2015. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24: 2426–2441. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. 2000. Dominant phenotypes produced by the HD mutation in STHdh Q111 striatal cells. Hum Mol Genet 9: 2799–2809. [DOI] [PubMed] [Google Scholar]
- Tsoi H, Chan HY. 2013. Expression of expanded CAG transcripts triggers nucleolar stress in Huntington's disease. Cerebellum 12: 310–312. [DOI] [PubMed] [Google Scholar]
- Tsoi H, Chan HY. 2014. Roles of the nucleolus in the CAG RNA-mediated toxicity. Biochim Biophys Acta 1842: 779–784. [DOI] [PubMed] [Google Scholar]
- Tsoi H, Lau CK, Lau KF, Chan HY. 2011. Perturbation of U2AF65/NXF1-mediated RNA nuclear export enhances RNA toxicity in polyQ diseases. Hum Mol Genet 20: 3787–3797. [DOI] [PubMed] [Google Scholar]
- Tsoi H, Lau TC, Tsang SY, Lau KF, Chan HY. 2012. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc Natl Acad Sci 109: 13428–13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci 11: 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DB, Kinoshita C, Kinoshita Y, Morrison RS. 2014. p53 and mitochondrial function in neurons. Biochim Biophys Acta 1842: 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. 2005. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell 18: 37–48. [DOI] [PubMed] [Google Scholar]
- Wiedemann C, Bellstedt P, Görlach M. 2013. CAPITO—a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics 29: 1750–1757. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. 2008. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci 31: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CH, Nguyen L, Peh J, Luu LM, Sanchez JS, Richardson SL, Tuccinardi T, Tsoi H, Chan WY, Chan HY, et al. 2014. Targeting toxic RNAs that cause myotonic dystrophy type 1 (DM1) with a bisamidinium inhibitor. J Am Chem Soc 136: 6355–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Duan S, Hou D, Wang W, Wang G, Liu Y, Wen L, Wu M. 2010. B23 acts as a nucleolar stress sensor and promotes cell survival through its dynamic interaction with hnRNPU and hnRNPA1. Oncogene 29: 1821–1834. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bhatia D, Xia H, Castranova V, Shi X, Chen F. 2006. Nucleolin links to arsenic-induced stabilization of GADD45α mRNA. Nucleic Acids Res 34: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Tsoi H, Peng S, Li PP, Lau KF, Rudnicki DD, Ngo JC, Chan HY. 2016. Assessing a peptidylic inhibitor-based therapeutic approach that simultaneously suppresses polyglutamine RNA- and protein-mediated toxicities in patient cells and Drosophila. Dis Model Mech 9: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.