Abstract
Thioredoxin 1 (Trx1) and glutaredoxin 1 (Grx1) are two ubiquitous redox enzymes that are central for redox homeostasis, but also are implicated in many other processes, including stress sensing, inflammation, and apoptosis. In addition to their enzymatic redox activity, there is increasing evidence showing that Trx1 and Grx1 play regulatory roles via protein-protein interactions with specific proteins, including Ask1. The currently available inhibitors of Trx1 and Grx1 are thiol-reactive electrophiles or disulfides that may suffer from low selectivity due to their thiol reactivity. In this report, we used a phage peptide library to identify a 7-mer peptide, 2GTP1, that binds to both Trx1 and Grx1. We further showed that a cell-permeable derivative of 2GTP1, TAT-2GTP1, disrupts the Trx1-Ask1 interaction, which induces Ask1 phosphorylation with subsequent activation of JNK, stabilization of p53, and reduced viability of cancer cells. Notably, as opposed to a disulfide-derived Trx1 inhibitor (PX-12), TAT-2GTP1 was selective for activating the Ask1 pathway without affecting other stress signaling pathways, such as ER stress and AMPK activation. Overall, 2GTP1 will serve as a useful probe for investigating protein interactions of Trx1.
INTRODUCTION
Thioredoxin 1 (Trx1) and glutaredoxin 1 (Grx1) are two ubiquitous redox enzymes that belong to a thioredoxin-fold family with one or two conserved cysteine residues in the active site for redox activity.1, 2 These two small oxidoreductases play pleiotropic functions that can be overlapping, but also distinct in cellular protection. For example, both Trx1 and Grx1 provide the reducing power to central antioxidant enzymes to maintain redox homeostasis.1 In addition, Trx1 and Grx1 directly modulate the redox status of a broad range of signaling proteins, transcription factors and others,2 including p53,3, 4 Nf-kB,5, 6 and ribonucleotide reductase (RNR),7 by several distinct mechanisms, including disulfide reduction, de-nitrosylation, and de-glutathionylation.2 Moreover, Trx1 and Grx1 are secreted to the extracellular matrix where Trx1 or its truncated form acts as cytokines for activating immune cells,8, 9 whereas the exact role of Grx1 is unclear. Overall, Trx1 and Grx1 are implicated in diverse processes, e.g., enhancing DNA synthesis, activating inflammation, and suppressing apoptosis.10, 11 Indeed, a high level of Trx1 was found in many cancer cell lines and in patient tumors.12–15 Extensive data support that a high level of Trx1 strongly correlates with tumor proliferation, drug resistance, tumor invasion and metastasis, and poor clinical prognosis.16–20 A Trx1 knockout is lethal in mice.21 The exact role of Grx1 in cancer is less understood, but an increased level of Grx1 was also found in pancreatic carcinoma tissue,22 and murine allergic airway disease,23 but at low levels in alveolar macrophages of sarcoidosis and allergic alveolitis.24 Grx1 knockdown decreases lipopolysaccharide-induced inflammation and alveolar macrophage activation.25
The redox activity of Trx1 for disulfide reduction and Grx1 for de-glutathionylation is essential for their biological activity in various processes. However, there is increasing evidence to show that Trx1 and Grx1 play regulatory functions via non-covalent or covalent protein-protein interactions with specific partners, including apoptosis signal-regulating kinase 1 (Ask1),26–29 thioredoxin-interacting protein,30, 31 RNR,32 and murine protein Ser/Thr kinase 38 (MPK38).33 One example is suppression of apoptosis in cancer cells by inhibiting Ask1.34 Ask1 is a stress-sensing kinase that is activated by a variety of stimuli, including reactive oxygen species (ROS) and cytokines.34, 35 Trx1 and Grx1 negatively inhibit activation of Ask1 by binding to an N-terminal Trx-binding domain and a C-terminal oligomerization domain of Ask1, respectively.29, 36 At a molecular level, Trx1 binds to Ask1 via non-covalent interactions26, 27 or disulfide formation.37, 38 In response to ROS, Trx1 and Grx1 are oxidized and dissociate from Ask1,29, 36 which allows for Ask1 activation by autophosphorylation39 or an upstream kinase, such as MPK38.40 Ask1 activation induces a cascade of the MAPK pathway, phosphorylating MKK 3/6 and 4/7 with subsequent activation of JNK and p38.35 The sustained activation of JNK and p38 is associated with apoptosis; thus, up-regulation of Trx1 and/or Grx1 contributes to inhibiting apoptosis in tumor cells.
The strong correlation of Trx1 with cancer proliferation led to the development of Trx1 inhibitors,41–43 including sec-butyl 2-imidazoylyl disulfide known as PX-12,44 which was evaluated in clinical trials.45, 46 The currently available inhibitors are thiol-reactive electrophiles or disulfides in which inhibition is primarily mediated by covalent modification of the redox-active cysteine in Trx1. Similarly, Grx1 inhibitors, including cadmium, primarily rely on cysteine reactivity in Grx1.47–49 Many of these electrophilic or reactive inhibitors have low selectivity due to their thiol reactivity. Here, we employed phage display to develop a redox-inactive peptide-based inhibitor (2GTP1) that has high binding affinity to both Trx1 and Grx1. We further show that a cell-permeable derivative of 2GTP1, TAT-2GTP1, selectively disrupts the Trx1-Ask1 interaction, thus activating Ask1 and JNK.
MATERIAL AND METHODS
Phage display selection
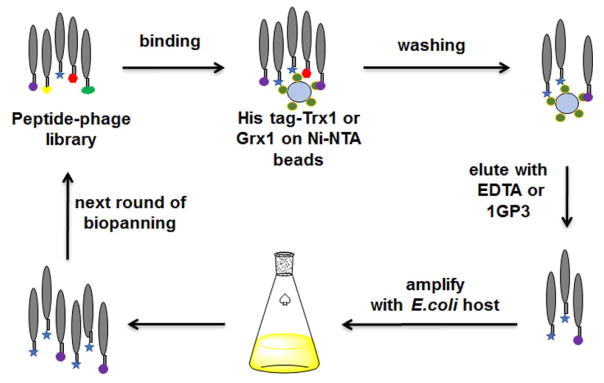
The initial phage library (New England Biolabs, phage titer 4 × 1011 pfu/μl) was incubated with Ni-NTA beads overnight to remove non-selective phage binding to beads, and the non-bound phage were used for selection against Grx1 or Trx1. Human His-tagged Grx1 or Trx1 (1 μM) was immobilized on fresh Ni-NTA magnetic beads, which were then incubated with the pre-cleared phage library. All bio-panning procedures were conducted in a TBST buffer containing DTT (1 mM) to exclude phage that form disulfide bond with a bait protein. EDTA (50 mM) or 1GP3 (50 μM) was used to elute the bound phage. Eluted phage were used to infect bacteria (E. coli ER2738) for amplification. After growing bacterial cells overnight, the amplified phage in supernatant were precipitated by addition of 20% polyethyleneglycol-8000 and 2.5 M NaCl. The collected phage were used in the next round of bio-panning. After three or four rounds of bio-panning, bacteria infected with eluted phage were cultured on IPTG/X-gal plates. The activity of β-galactosidase in bacterial cells, which is produced by combination of a lacZα gene of phage and a lacZ omega-gene of E. coli, was detected by addition of IPTG that induces expression of β-galactosidase, and X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) that is hydrolyzed to a blue product by β-galactosidase. The blue bacterial colonies were randomly selected. The DNA sequences were amplified by colony PCR, and subjected to DNA sequencing.
Trx1 and Grx1 binding assay
Streptavidin agarose beads were washed with 0.1 M TBS (pH 7.4) and incubated with biotinylated peptides (20–50 μM) for 15 min, and washed the unbound peptides, followed by the addition of Grx1, Trx1, or Grx3 (25–50 μg). Alternatively, Grx1 or Trx1 was pre-incubated with reduced glutathione (0–5 mM), H2O2, or GSSG (10 molar equivalents to Grx1 or Trx1) for 1 h before addition to the beads. The bead mixture was incubated for 1 h at room temperature, and unbound protein was removed by washing with TBST (0.1% Tween) (10 mL). Protein bound to beads was eluted by using 0.1 M glycine-HCl (pH 2.8) and neutralized immediately by addition of 1 M Tris-HCl (pH 8.0) or eluted by addition of an SDS loading dye. Eluted proteins were analyzed on an SDS-PAGE gel by Coomassie staining or Western blotting.
Measuring the binding affinity
The bio-layer interferometry (BLI) technology (BLItz system, ForteBio) was used to measure the binding affinity of 2GTP1 to Grx1 or Trx1. The streptavidin-coated biosensor was hydrated in buffer A containing Tris-HCl (100 mM), NaCl (150 mM), EDTA (3 mM), Tween-20 (0.005%) and DTT (1 mM) for 10 min. The hydrated biosensor was transferred to the following solutions in sequence: 1) biotin-2GTP1 solution for 120 sec, 2) buffer A for 30 sec, 3) varying concentrations (0.078 – 25 μM) of Grx1 or Trx1 in buffer A for 120 sec, and 4) buffer A for 120 sec. The sensor responses were plotted versus increasing concentrations of proteins to calculate the apparent dissociation constant (Kd). The binding affinity of 1GP3 with Grx1 was measured under the similar conditions by using surface plasma resonance (SPR) (Biacore 2000, GE Healthcare) with a CM-5 biosensor chip.
Glutaredoxin enzyme assay
Grx1 activity was measured by using a glutathione reductase (GR) coupled enzyme assay.50 In this assay, Cys-SSG (Toronto Research Chemical) was used as a substrate of Grx1. Cys-SSG was reduced by Grx1, forming oxidized glutathione disulfide (GSSG). GSSG was reduced by GR with oxidation of NADPH to NADP+. The NADPH concentration was measured by UV absorbance at 340 nm. Grx1 inhibition was assayed in 0.1 M phosphate buffer (pH 7.4), containing EDTA (1 mM), BSA (0.1 mg/μL), NADPH (0.2 mM), yeast GR (4 units), GSH (0.25 mM), and Cys-SSG (100 μM) as the substrate. Grx1 (40 ng) was incubated with increasing concentrations (10–200 μM) of peptide inhibitors. The initial rates were calculated from the change of UV versus time with increasing concentrations of peptide inhibitors, which were used to calculate the percent inhibition.
Thioredoxin enzyme assay
Trx1 activity was measured as reported previously.51 Insulin was used as a substrate of Trx1. Trx1 reduces the disulfide in insulin, and the reduced insulin induces turbidity, which was measured by the absorbance at 650 nm. Trx1 activity was assayed in 0.1 M phosphate buffer (pH 7.4), containing EDTA (2 mM), insulin (35 μM), and DTT (125 μM). Trx1 (10 nM) was incubated with increasing concentrations (1–100 μM) of peptide inhibitors. Absorbance at 650 nm in the presence of peptide inhibitors was measured at the specific time point, which was used to calculate the percent inhibition.
Cell viability assay
Cell viability was measured by using a Trypan blue assay. MDA-MB-231 cells were plated in a 6-well plate. Cells were then treated with TAT-re-2GTP1, TAT-2GTP1, or PX-12 and incubated for 48 h at 37 °C. After washing with PBS once, cells were treated with trypsin to detach cells from the plates. Cells were then re-suspended in culture medium, and mixed with 0.4% trypan blue solution. The suspended cells were then loaded into the chamber on the counting slide (BioRad) and cell number was determined by using a TC20 automated cell counter (BioRad).
Detection of ROS induction
ROS induction was measured by a 2′,7′-dichlorofluorescein diacetate (DCF-DA) fluorescence assay. MDA-MB-231 cells were plated in a 96-well plate with 25,000 cells per well. After plating, cells were incubated with DCF-DA (25 μM) in a phenol red-free DMEM medium for 45 min at 37 °C. The medium containing DCF-DA was then removed, and cells were incubated with TAT-re-2GTP1, TAT-2GTP1, or PX-12 for 5 h at 37 °C. Fluorescence intensity was measured at 485 nm (excitation) and 535 nm (emission).
Western Blotting
MDA-MB-231, MCF-7 and MDA-MB-453 cells in 6 cm dishes were treated with TAT, TAT-re-2GTP1, TAT-2GTP1, and PX-12, and incubated at 37 °C for 6 h. Cells were then lysed with TBST containing Triton-X 100 (0.1%), NaF (50 mM) and an EDTA-free protease-inhibitor cocktail. The same amount of proteins in lysates were loaded and separated by SDS-PAGE, transferred to PVDF membrane, and incubated with individual primary antibodies at 4 °C overnight, followed by a secondary antibody conjugated with HRP. Individual proteins were detected by measuring chemiluminescence with a FluorChem Q imager (BioRad). Antibodies for p53, Ask1, GRP78, and Grx1 were obtained from SantaCruz, and antibodies for JNK, p-JNK (Thr183/Tyr185), p-Ask1 (Thr845), AMPK, p-AMPK (Thr172), p38, p-p38 (Thr180/Tyr182), and Trx1 were obtained from Cell Signaling.
Co-immunoprecipitation of Ask1 and Trx1
pcDNA3.1 plasmids containing FLAG-Ask1 (gift from Yardena Samuels, Addgene plasmid # 47104) or HA-Trx1 were transfected to HEK 293 cells. Cells were then treated with TAT-re-2GTP1, TAT-2GTP1, or PX-12 (50–100 μM) for 6 h. Cells were then lysed with TBST containing Triton-X 100 (0.1%), NaF (50 mM) and an EDTA-free protease inhibitor cocktail. Lysates (0.5 mg proteins) were incubated with FLAG-antibody and pre-washed protein G agarose beads, and incubated for 16 h at 4 °C. The beads were washed with TBST, and bound proteins were eluted and analyzed by Western blotting.
RESULTS
Phage display to find peptides binding to Trx1 or Grx1
To develop a potential high-affinity binder to Trx1 or Grx1, we used a phage peptide library (Ph.D.™) that displays an N-terminal 7-mer peptide fused to a coat protein pIII on M13 phage. We initially used Grx1 as a bait protein immobilized on beads by N-terminal His-tagging, and performed three rounds of bio-panning while eluting with EDTA (Figure 1 and Table 1, trial 1). All bio-panning procedures were conducted in a reducing condition (DTT, 1 mM) to prevent disulfide bond between phage and a bait protein. After the last bio-panning, bacteria were infected with eluted phage and plated to find infected bacterial colonies. The sequencing of 16 selected bacterial colonies identified three peptides (1GP1, 1GP2, and 1GP3 in trial 1, Table 1). Notably, 1GP3 was found in 12 out of 16 colonies, showing its high enrichment. To confirm their binding to Grx1, we synthesized biotinylated peptides (biotin-1GPs) (Figure S1–2). In vitro binding assays showed that 1GP2 and 1GP3 bind to Grx1, whereas 1GP1 did not show significant binding with Grx1 (Figure S1C). We further evaluated whether these peptides can inhibit Grx1 enzyme activity of de-glutathionylation. While 1GP2 did not have any inhibitory activity, 1GP3 showed dose-dependent inhibition of Grx1 de-glutathionylation (IC50 = 46 μM, Figure S1D). To measure binding affinity of 1GP3 with Grx1, we used SPR with biotin-1GP3, which confirmed direct binding of 1GP3 to Grx1 (Kd = 38 ± 2 μM, Table 1 and Figure S1E).
Figure 1.
Identification of Trx1- or Grx1-binding peptides via phage library screening. Bound phage were eluted by EDTA or 1GP3.
Table 1.
Identified peptides from phage display with their sequences and apparent dissociation constants (Kd values). ND: not determined.
| Trial | Target Protein | Elution | Peptide Sequence | # of colony | Name | Kd (μM) |
|---|---|---|---|---|---|---|
| 1 | Grx1 | EDTA | SPSRSIH | 2 | 1GP1 | ND |
| SHYNYKV | 2 | 1GP2 | ND | |||
| ANHTMQV | 12 | 1GP3 | 38 ± 2 | |||
|
| ||||||
| 2 | Grx1 | 1GP3 | ETRWPNT | 23 | 2GTP1 | 1.2 ± 0.2 |
| SVGSFSM | 1 | 2GP1 | ND | |||
| GRYAIPL | 2 | 2GP2 | ND | |||
|
| ||||||
| 3 | Trx1 | 1GP3 | ETRWPNT | 26 | 2GTP1 | 2.5 ± 0.5 |
| AMYSRGT | 2 | 2TP1 | ND | |||
| RFADPRL | 1 | 2TP2 | ND | |||
| SLKTTMP | 1 | 2TP3 | ND | |||
To improve the binding affinity, we re-designed the phage display screening in which bound phage were eluted with free 1GP3 to enrich for phage that bind specifically to Grx1. During screening, both Grx1 and Trx1 were used as bait proteins independently immobilized on beads (Table 1, trial 2 and 3). Note that both Grx1 and Trx1 belong to a thioredoxin-fold family with high structural similarity.1 Thus, we envisioned that 1GP3 could be used as a competitor during bio-panning with Trx1. After three rounds of bio-panning, the sequencing results identified a relatively small number of peptide sequences (Table 1). Notably, there was significant enrichment of one peptide, 2GTP1, which was identified in both bio-panning involving Grx1 or Trx1 (23/26 colonies in the 3rd rounds with Grx1; 26/30 colonies in the 3rd rounds with Trx1). We synthesized five peptides identified from bio-panning with either Grx1 or Trx1, among which only 2GTP1 showed significant inhibitory activity to both Grx1 de-glutathionylation and Trx1 disulfide reduction (Figure S4). Therefore, 2GTP1 was selected and used for further analyses.
Biochemical analyses of 2GTP1 with Trx1 and Grx1
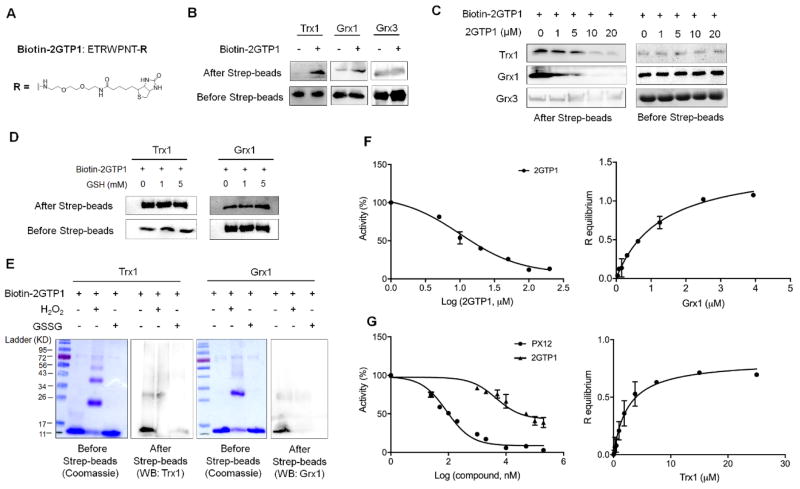
Initially, we synthesized a biotinylated 2GTP1 peptide (biotin-2GTP1) to evaluate its binding to Grx1 and Trx1 (Figure 2A and S2–3). Biotin-2GTP1 was bound to streptavidin-beads, and incubated with Grx1, Trx1, or Grx3 (also known as PICOT52). Note that Grx3 contains three thioredoxin-fold domains, including one Trx-like domain and two Grx-like domains.11 Therefore, Grx3 was used as a control protein to examine selective binding of 2GTP1 to Trx1 and Grx1. A binding assay showed that 2GTP1 binds to Grx1 and Trx1, but not Grx3 (Figure 2B). Similarly, the competition experiment with non-biotinylated 2GTP1 confirmed that 2GTP1 specifically binds to Grx1 and Trx1, but not Grx3 (Figure 2C), showing selectivity of 2GTP1 to Grx1 and Trx1 over Grx3. Importantly, Trx1 and Grx1 can be oxidized to form an intramolecular disulfide and glutathionylated disulfide (glutathionylation), respectively. Thus, we evaluated whether oxidation or the presence of glutathione can affect binding of 2GTP1 to Trx1 and Grx1. Interestingly, incubation with reduced glutathione did not change binding of 2GTP1 to either Grx1 or Trx1 (Figure 2D), which agrees with the fact that human Grx1 does not have significant binding affinity with reduced glutathione.53 In contrast, Grx1 or Trx1 incubated in oxidizing conditions decreased their binding with 2GTP1 (Figure 2E), suggesting that 2GTP1 binds to reduced forms of Trx1 and Grx1.
Figure 2.
In vitro binding and inhibitory analyses of 2GTP1 with Trx1 and Grx1. (A) The structure of biotinylated 2GTP1 (bitotin-2GTP1). (B–E) The binding analyses of 2GTP1 with Trx1 and Grx1. Biotin-2GTP1 was immobilized on streptavidin beads, and incubated with purified Trx1, Grx1, or Grx3 (B), which was repeated in the presence of non-biotinylated 2GTP1 as a competitor (C), reduced glutathione (D), or oxidants (10 molar equivalents to Trx1 or Grx1) (E). Trx1, Grx1, or Grx3 was analyzed by Western blotting before and after streptavidin beads. All Western blots represent data from two or three independent experiments. (F) Inhibitory activity of 2GTP1 for Grx1 deglutathionylation (left) and the binding affinity of 2GTP1 to Grx1 measured by BLI (right). (G) Inhibitory activity of 2GTP1 for Trx1-mediated insulin reduction (left) and the binding affinity of 2GTP1 to Trx1 measured by BLI (right). Data represent the mean ± SD, n= 3 independent experiments.
Next, we further evaluated 2GTP1 for its enzyme inhibitory activity and binding affinity to Grx1 and Trx1. Encouragingly, a Grx1 de-glutathionylation enzyme assay showed that 2GTP1 inhibits Grx1 more potently than 1GP3 (IC50 11 μM for 2GTP1 vs. 46 μM for 1GP3) (Figure 2F, left, and S1D). The subsequent BLI binding analysis also confirmed binding interaction of 2GTP1 with Grx1 (Kd = 1.2 ± 0.2 μM) (Figure 2F, right), which is significantly higher affinity than 1GP3 (Kd = 38 ± 2 μM) (Table 1). Similarly, we evaluated 2GTP1 for potential inhibition of Trx1 activity. 2GTP1 showed dose-dependent inhibition of Trx1 activity in an insulin-reduction assay (Figure 2G, left) albeit incomplete inhibition even at high concentrations. In contrast, PX-12, a disulfide-derived Trx1 inhibitor, showed higher inhibitory potency and efficacy to Trx1 activity in this assay (Figure 2G, left). The BLI binding analysis confirmed binding interaction of 2GTP1 to Trx1 (Kd = 2.5 ± 0.5 μM) (Figure 2G, right). Interestingly, direct binding of 2GTP1 to Trx1 was decreased in the presence of PX-12 (Figure S5), which suggests overlapping binding sites of 2GTP1 and PX-12 in Trx1. Overall, despite the shallow surface in the active site of Trx1 and Grx1, 2GTP1 was found to bind to both Trx1 and Grx1.
2GTP1 disrupts the Trx1-Ask1 interaction
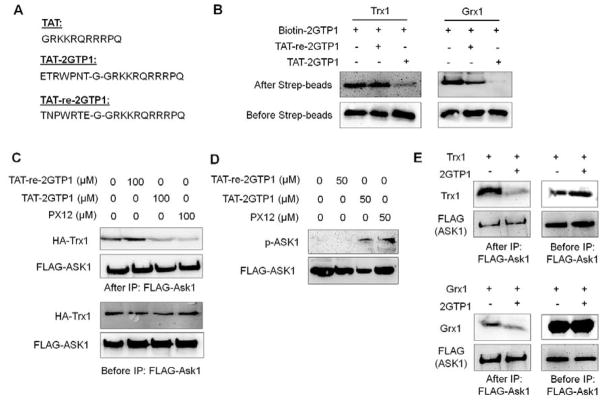
Both Trx1 and Grx1 bind to Ask1, protecting Ask1 from activation.29, 36 It was previously shown that oxidation of Trx1 and Grx1 induces their dissociation from Ask1, leading to activation and phosphorylation of Ask1.29, 36 To evaluate 2GTP1 in cells, we made a cell-permeable derivative of 2GTP1 by adding the TAT-sequence (TAT-2GTP1), along with two control peptides, TAT only and the reverse sequence of 2GTP1 with TAT (TAT-re-2GTP1) (Figure 3A and S3). An in vitro binding assay confirmed that TAT-2GTP1 binds to Trx1 and Grx1, while TAT-re-2GTP1 does not (Figure 3B).
Figure 3.
A cell-permeable derivative of 2GTP1 (TAT-2GTP1) for disruption of the Ask1-Trx1 interaction and activation of Ask1. (A) The sequences of TAT, TAT-2GTP1, and TAT-re-2GTP1 that contains a reverse sequence of 2GTP1. (B) In vitro validation of TAT-2GTP1 and TAT-re-2GTP1 for binding to Trx1 (left) or Grx1 (right). TAT-2GTP1 or TAT-re-2GTP1 was added to a solution containing Trx1 or Grx1 bound to biotin-2GTP1 on streptavidin beads. Bound Trx1 or Grx1 was analyzed by Western blotting. (C) Disruption of the Ask1-Trx1 interaction by TAT-2GTP1. Ask1-Trx1 interaction was analyzed by co-IP after incubation of TAT-2GTP1 to MDA-MB-231 cells for 6 h. (D) Ask1 activation by TAT-2GTP1. Ask1 phosphorylation was analyzed after incubation of TAT-2GTP1 to MDA-MB-231 cells for 6 h. (E) Disruption of Ask1-Trx1 and Ask1-Grx1 interaction by 2GTP1 in vitro. The transfected FLAG-Ask1 was pull-downed by FLAG-antibody, and incubated with purified Trx1 (top) or Grx1 (bottom) in vitro in the presence of 2GTP1. Bound Trx1 or Grx1 was analyzed by Western blotting. All Western blots represent data from two or three independent experiments.
TAT-2GTP1 was then examined for disrupting the Trx1-Ask1 interaction in MDA-MB-231 cells that express a high level of Trx1 (Figure 4C). After incubation of TAT-2GTP1 to cells, co-immunoprecipitation (co-IP) experiments showed that TAT-2GTP1 decreased binding of Trx1 with Ask1 (Figure 3C, lane 1 vs 3, and Figure S6A). Interestingly, treatment of TAT-2GTP1 induced phosphorylation of Ask1 (Figure 3D, lane 1 vs. 3, and Figure S6B), suggesting that Trx1 dissociation from Ask1 even without ROS stimulus induces activation of Ask1. In the control, TAT-re-2GTP1 did not induce disruption of the Trx1-Ask1 interaction (Figure 3C, lane 1 vs. 2, and Figure S6A) nor phosphorylation of Ask1 (Figure 3D, lane 1 vs. 2, and Figure S6B), showing the specific effect of TAT-2GTP1. In the same manner, PX-12 also disrupted the Trx1-Ask1 interaction (Figure 3C, lane 1 vs 4, and Figure S6A) and activated Ask1 by phosphorylation (Figure 3D, lane 1 vs 4). On the other hand, under similar conditions, we did not observe significant loss of the Ask1-Grx1 interaction upon incubation of TAT-2GTP1 or TAT-re-2GTP1 (Figure S6C).
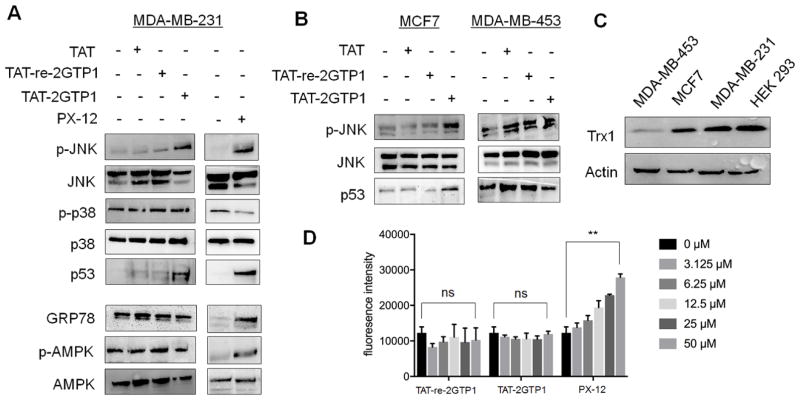
Figure 4.
The evaluation of TAT-2GTP1 on ROS induction and stress signaling pathways. (A–B) The effect of TAT-2GTP1 versus PX-12 on Ask1 downstream signaling pathway, ER stress, or AMPK activation in MDA-MB-231 (A), MCF7, and MDA-MB-453 cell lines (B). TAT-2GTP1, TAT-re-2GTP1, TAT, or PX-12 (all 100 μM) was incubated for 6 h. Lysates were analyzed by Western blotting. (C) The level of Trx1 in different cell lines. All Western blots represent data from two or three independent experiments. (D) The level of ROS induced by TAT-2GTP1 versus PX-12. After incubation of TAT-2GTP1, TAT-re-2GTP1, or PX-12 to MDA-MB-231 cells for 6 h, a DCF-DA assay was used to measure induction of ROS. Data represent the mean ± SD, n= 2 independent experiments. Difference is significant by two-tailed Student’s unpaired t-test with Welch’s correction, *p < 0.05, **p < 0.01, ***p < 0.001, and ns (not significant, p > 0.05).
To confirm the direct effect of 2GTP1 in disrupting Trx1-Ask1 and Grx1-Ask1, FLAG-Ask1 was transfected to HEK293 cells, isolated by using FLAG-antibody, and incubated with Grx1 or Trx1 in vitro in the presence of 2GTP1. The Trx1-Ask1 interaction was significantly decreased upon incubation of 2GTP1 (Figure 3E, top), whereas the Grx1-Ask1 interaction was decreased, but relatively at a modest level (Figure 3E, bottom), which suggests that 2GTP1 may have low potency in disrupting Grx1-Ask1, resulting in no disruption of the Grx1-Ask1 interaction by TAT-2GTP1 in cells. Overall, our data demonstrate that TAT-2GTP1 can disrupt the Trx1-Ask1 interaction in cells with subsequent activation and phosphorylation of Ask1.
2GTP1 induces selective activation of Ask1 stress signal pathway
Ask1 phosphorylation induces activation and phosphorylation of MAP kinase, such as JNK and/or p38.35 The sustained activation of JNK induces apoptosis and stabilization of p53.54 Therefore, we further evaluated the effects of TAT-2GTP1 in downstream pathways of Ask1. Incubation of TAT-2GTP1 to MDA-MB-231 cells induced phosphorylation of JNK and stabilization of p53 (Figure 4A) while TAT or TAT-re-2GTP1 did not induce phosphorylation of JNK nor stabilization of p53 (Figure 4A). On the other hand, p38 was not phosphorylated upon incubation of TAT-2GTP1 (Figure 4A). Similarly, PX-12 also induced JNK phosphorylation and p53 stabilization without p38 phosphorylation (Figure 4A). In addition to MDA-MB-231 cells, we found that Trx1 is highly expressed in MCF7 cells while it is expressed in low levels in MDA-MB-453 cells (Figure 4C). Interestingly, TAT-2GTP1 also induced JNK activation and p53 stabilization in MCF7 cells, but not in MDA-MB-453 cells (Figure 4B), suggesting the potential effect of TAT-2GTP1 targeting Trx1 for JNK activation and p53 stabilization.
TAT-2GTP1 is a non-electrophilic peptide that binds to Trx1 while PX-12 relies on disulfide reactivity that may induce ROS or activate other stress signaling pathways. Notably, an increasing dose of PX-12 induced a high level of ROS, while TAT-2GTP1 did not show any significant induction of ROS (Figure 4D). We further evaluated the effect of TAT-2GTP1 on activation of other stress-signaling pathways. Glucose-regulated protein 78 (GRP78/Bip) is induced in response to ER stress,55 and AMPK is phosphorylated in energy- or ATP-depletion, or in response to ROS.56 TAT-2GTP1 did not induce GRP78 nor phosphorylation of AMPK, while PX-12 induced expression of GRP78 and phosphorylation of AMPK (Figure 4A), suggesting that there are differential effects of TAT-2GTP1 versus PX-12. Overall, our data support that TAT-2GTP1 is selective in disrupting Trx1-Ask1 interaction and activating Ask1 downstream pathways, without significant effects on cellular redox state or other stress signaling pathways.
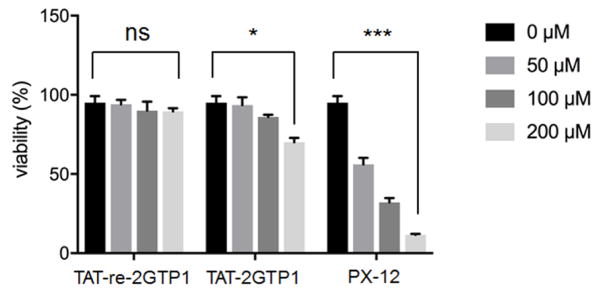
2GTP1 decreases viability of cancer cells
The sustained activation of Ask1 and JNK, induction of ER stress, and activation of AMPK are associated with apoptosis in cancer cells. Therefore, we evaluated the viability of cancer cells upon incubation of TAT-2GTP1 versus PX-12. PX-12 significantly decreased viability of MDA-MB-231 cells (Figure 5). In contrast, TAT-2GTP1 decreased viability of MDA-MB-231 cells in a dose-dependent manner, but less potently than PX-12 (Figure 5). In controls, TAT-re-2GTP1 did not show any inhibitory effect on the viability of MDA-MB-231 cells (Figure 5). The difference between TAT-2GTP1 and PX-12 in cell viability may be attributed to the selective effect of TAT-2GTP1 on Ask1 activation without affecting ER stress and AMPK activation.
Figure 5.
Cell viability induced by TAT-2GTP1. Trypan blue assay was used to measure viability of MDA-MB-231 cells after incubation of TAT-2GTP1, TAT-re-2GTP1 or PX-12 for 48 h. Data represent the mean ± SD, n= 2 independent experiments. Difference is significant by two-tailed Student’s unpaired t-test with Welch’s correction, *p < 0.05, **p < 0.01, ***p < 0.001, and ns (not significant, p > 0.05).
DISCUSSION
Trx1 is a well-appreciated target protein for anti-cancer therapy, due to its role in cancer proliferation, inflammation, and anti-apoptosis.57 While the role of Grx1 in cancer is not clearly understood, Grx1 plays a role in inflammatory responses.25 Accordingly, there have been previously many inhibitors developed for Trx1 and a few inhibitors for Grx1.41–44, 47–49 However, most of the available inhibitors of Trx1 and Grx1 rely on their thiol-reactivity, covalently conjugating to redox active cysteine residues in the active site.
In this report, we developed a non-reactive 7-mer peptide that binds to and inhibits Trx1 and Grx1 in vitro (Figure 2). Notably, 2GTP1 showed partial inhibition of Trx1 activity even at high concentrations (Figure 2G), which may explain its lack of ROS induction, and no effect on ER stress and AMPK phosphorylation in cells (Figure 4A and D). However, TAT-2GTP1 could disrupt the Ask1-Trx1 interaction and activate Ask1 (Figure 3) and its downstream pathway (Figure 4). In contrast, the disulfide-derived PX-12 induced a high level of ROS and activated other stress-associated pathways, such as ER stress and AMPK phosphorylation (Figure 4A), which could be due to inhibiting Trx1 redox activity (Figure 2G), but also may be attributed to a high level of ROS (Figure 4D). Indeed, ROS can induce AMPK phosphorylation,58, 59 whereas Trx1 was shown to suppress ER stress.60 However, it is worth noting that ROS detection by a fluorescent probe, DCF-DA, has been criticized due to its lack of selectivity to various oxidants.61 Therefore, the increased signal detected by DCF-DA in our study may result from ROS or any other oxidants in cells. Overall, these data suggest that TAT-2GTP1 selectively targets the Trx1-protein interaction without significantly altering the cellular redox state.
It is also worth noting that the recent result of PX-12 in clinical trials showed a low concentration of PX-12 in plasma, potentially due to its conjugation to abundant plasma proteins,45, 46 suggesting an undesired thiol reactivity. However, 2GTP1 also suffers from poor cell permeability. Indeed, a TAT-sequence was necessary to obtain favorable cellular activity of 2GTP1 at high concentrations (50–100 μM). Further structural optimization will be necessary to improve the cell-permeability and cellular potency of 2GTP1.
Trx1 has five Cys residues, including catalytic C32 and C35, and redox-sensitive C73. A thioredoxin-binding domain of Ask1 (Ask1-TBD) also has several Cys residues that can form disulfide bonds. Therefore, intermolecular disulfide bonds between Trx1 and Ask1-TBD have been observed, especially with C250 in Ask1, in non-stressed or stressed conditions.27, 37, 38 These observations are a basis for supporting covalent binding interaction between Trx1 and Ask1. However, recent biophysical analyses have also shown non-covalent interactions between Trx1 and Ask1-TBD in a reducing condition.26, 27 Structural modeling experiments predicted the potential interface between Trx1 and Ask1-TBD that includes a 31WCGPC35 motif, and a loop connecting an α3 helix and a β3 strand (loop α3-β3) around the active site of Trx1 (Figure S7).27 Note that these binding motifs in Trx1 include catalytic C32 and redox-sensitive C73, which are two major Cys residues oxidized or modified by PX-12.44 Because our data support potentially overlapping binding sites of 2GTP1 and PX-12 in Trx1 (Figure S5), we predict that 2GTP1 is likely to bind around C32 and/or C73 in Trx1, which could disrupt the Trx1-Ask1 interaction.
TAT-2GTP1 induced dissociation of the Trx1-Ask1 interaction, Ask1 phosphorylation, JNK activation (Figure 3 and 4A). In general, it is well-known that ROS induce activation and phosphorylation of Ask1.27, 29, 34, 37, 38 In contrast, we found that non-reactive TAT-2GTP1 can also induce Ask1 activation, potentially by disrupting the Trx1-Ask1 interaction, suggesting that Trx1-Ask1 dissociation without stimulus of ROS is enough to induce phosphorylation of Ask1. While it is unclear whether Ask1 phosphorylation is induced by trans-phosphorylation of Ask139 or by an upstream kinase MPK38,62 it is interesting to note that MPK38 is reported to bind non-covalently with Trx1, and it is activated upon Trx1 oxidation or dissociation.33 Thus, it is possible that TAT-2GTP1 may act on dissociation of Trx1-MPK38 interaction. Further analyses will be necessary to understand the mechanism of 2GTP1 for activating the Ask1 pathway. Overall, these data suggest that TAT-2GTP1 would be useful for probing protein interactions of Trx1 in the future.
Interestingly, both TAT-2GTP1 and PX-12 induced p53 stabilization in MDA-MB-231 and MCF7 cells (Figure 4), which have mutant p53 (R280K in a DNA-binding domain) and wild-type p53, respectively.63 It is known that JNK can induce phosphorylation and stabilization of wild-type p53.54, 64, 65 However, to our knowledge, it is unclear whether JNK can stabilize mutant p53 R280K. Despite single mutation, many hot spot mutations in a DNA-binding domain of p53, such as R280K, are known to induce significant conformational changes that confer a pro-oncogenic gain of function.66, 67 Consequently, many reports have shown that mutant p53 contributes to metastatic phenotype and survival of tumor cells.67 Accordingly, stabilization of mutant p53 in MDA-MB-231 cells was observed previously, which contributed to survival of tumor cells.68 Therefore, mutant p53 stabilization by TAT-2GTP1 in our data may compromise a cytotoxic effect of TAT-2GTP1 in MDA-MB-231 cells. Similarly, it is notable that PX-12 also shows strong stabilization of mutant p53 in MDA-MB-231 cells (Figure 4A). However, PX-12 shows potent cytotoxicity (Figure 5), which could be attributed to activation of JNK, ER stress, and AMPK (Figure 4A). Despite the cytotoxic effect of PX-12, it is important to point out low anti-tumor activity of PX-12 in clinical evaluation.45, 46 Thus, more analyses will be necessary to understand effects of PX-12 or Trx1 inhibitors on various stress signaling pathways, including mutant p53 stabilization.
Supplementary Material
Acknowledgments
Funding
This study was supported by Wayne State University Start-up funds and NIH R01 HL131740-01A1 (Ahn).
We thank all Ahn group members for providing research help and reading the manuscript, and Dr. Yardena Samuels for sharing plasmid MAP3K5.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publication website.
The synthesis and characterization of peptides, analyses of 2GTP1 in Figure S1–S7, and additional methods
References
- 1.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and Glutaredoxins: Unifying Elements in Redox Biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 2.Hanschmann EM, Godoy JR, Berndt C, Hudemann C, Lillig CH. Thioredoxins, Glutaredoxins, and Peroxiredoxins-Molecular Mechanisms and Health Significance: from Cofactors to Antioxidants to Redox Signaling. Antioxid Redox Sign. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-Binding domain during oxidative stress. Biochemistry-Us. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- 5.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EFM, Janssen-Heininger YMW. Dynamic redox control of NF-kappa B through glutaredoxin-regulated S-glutathionylation of inhibitory kappa B kinase beta. P Natl Acad Sci USA. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelleher ZT, Sha YG, Foster MW, Foster WM, Forrester MT, Marshall HE. Thioredoxin-mediated Denitrosylation Regulates Cytokine-induced Nuclear Factor kappa B (NF-kappa B) Activation. J Biol Chem. 2014;289:3066–3072. doi: 10.1074/jbc.M113.503938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avval FZ, Holmgren A. Molecular Mechanisms of Thioredoxin and Glutaredoxin as Hydrogen Donors for Mammalian S Phase Ribonucleotide Reductase. J Biol Chem. 2009;284:8233–8240. doi: 10.1074/jbc.M809338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schenk H, Vogt M, Droge W, SchulzeOsthoff K. Thioredoxin as a potent costimulus of cytokine expression. J Immunol. 1996;156:765–771. [PubMed] [Google Scholar]
- 9.Sahaf B, Soderberg A, Spyrou G, Barral AM, Pekkari K, Holmgren A, Rosen A. Thioredoxin expression and localization in human cell lines: Detection of full-length and truncated species. Exp Cell Res. 1997;236:181–192. doi: 10.1006/excr.1997.3699. [DOI] [PubMed] [Google Scholar]
- 10.Arner ESJ, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Bba-Gen Subjects. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Berggren M, Gallegos A, Gasdaska JR, Gasdaska PY, Warneke J, Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996;16:3459–3466. [PubMed] [Google Scholar]
- 13.Grogan TM, Fenoglio-Prieser C, Zeheb R, Bellamy W, Frutiger Y, Vela E, Stemmerman G, Macdonald J, Richter L, Gallegos A, Powis G. Thioredoxin, a putative oncogene product, is overexpressed in gastric carcinoma and associated with increased proliferation and increased cell survival. Hum Pathol. 2000;31:475–481. doi: 10.1053/hp.2000.6546. [DOI] [PubMed] [Google Scholar]
- 14.Raffel J, Bhattacharyya AK, Gallegos A, Cui HY, Einspahr JG, Alberts DS, Powis G. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J Lab Clin Med. 2003;142:46–51. doi: 10.1016/S0022-2143(03)00068-4. [DOI] [PubMed] [Google Scholar]
- 15.Kakolyris S, Giatromanolaki A, Koukourakis M, Powis G, Souglakos J, Sivridis E, Georgoulias V, Gatter KC, Harris AL. Thioredoxin expression is associated with lymph node status and prognosis in early operable non-small cell lung cancer. Clin Cancer Res. 2001;7:3087–3091. [PubMed] [Google Scholar]
- 16.Lin FY, Zhang PL, Zuo ZG, Wang FL, Bi RC, Shang WJ, Wu AH, Ye J, Li ST, Sun XC, Wu JB, Jiang L. Thioredoxin-1 promotes colorectal cancer invasion and metastasis through crosstalk with S100P. Cancer Lett. 2017;401:1–10. doi: 10.1016/j.canlet.2017.04.036. [DOI] [PubMed] [Google Scholar]
- 17.Bhatia M, McGrath KL, Di Trapani G, Charoentong P, Shah F, King MM, Clarke FM, Tonissen KF. The thioredoxin system in breast cancer cell invasion and migration. Redox Biol. 2016;8:68–78. doi: 10.1016/j.redox.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Feng X, Zheng L, Li SL, Ge XY, Zhang JG. Thioredoxin 1 mediates TGF-beta-induced epithelial-mesenchymal transition in salivary adenoid cystic carcinoma. Oncotarget. 2015;6:25506–25519. doi: 10.18632/oncotarget.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li CP, Thompson MA, Tamayo AT, Zuo Z, Lee J, Vega F, Ford RJ, Pham LV. Over-expression of Thioredoxin-1 mediates growth, survival, and chemoresistance and is a druggable target in diffuse large B-cell lymphoma. Oncotarget. 2012;3:314–326. doi: 10.18632/oncotarget.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heim K, Dalken B, Faust S, Rharbaoui F, Engling A, Wallmeier H, Dingermann T, Radeke HH, Schuttrumpf J, Gutscher M. High thioredoxin-1 levels in rheumatoid arthritis patients diminish binding and signalling of the monoclonal antibody Tregalizumab. Clin Transl Immunology. 2016;5:e121. doi: 10.1038/cti.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui M, Oshima M, Oshima H, Takaku K, Maruyama T, Yodoi J, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev Biol. 1996;178:179–185. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura H, Bai J, Nishinaka Y, Ueda S, Sasada T, Ohshio G, Imamura M, Takabayashi A, Yamaoka Y, Yodoi J. Expression of thioredoxin and glutaredoxin, redox-regulating proteins, in pancreatic cancer. Cancer Detect Prev. 2000;24:53–60. [PubMed] [Google Scholar]
- 23.Reynaert NL, Wouters EFM, Janssen-Heininger YMW. Modulation of glutaredoxin-1 expression in a mouse model of allergic airway disease. Am J Resp Cell Mol. 2007;36:147–151. doi: 10.1165/rcmb.2006-0259RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peltoniemi M, Kaarteenaho-Wiik R, Saily M, Sormunen R, Paakko P, Holmgren A, Soini Y, Kinnula VL. Expression of glutaredoxin is highly cell specific in human lung and is decreased by transforming growth factor-beta in vitro and in interstitial lung diseases in vivo. Hum Pathol. 2004;35:1000–1007. doi: 10.1016/j.humpath.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Aesif SW, Anathy V, Kuipers I, Guala AS, Reiss JN, Ho YS, Janssen-Heininger YMW. Ablation of Glutaredoxin-1 Attenuates Lipopolysaccharide-Induced Lung Inflammation and Alveolar Macrophage Activation. Am J Resp Cell Mol. 2011;44:491–499. doi: 10.1165/rcmb.2009-0136OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosek D, Kylarova S, Psenakova K, Rezabkova L, Herman P, Vecer J, Obsilova V, Obsil T. Biophysical and Structural Characterization of the Thioredoxin-binding Domain of Protein Kinase ASK1 and Its Interaction with Reduced Thioredoxin. J Biol Chem. 2014;289:24463–24474. doi: 10.1074/jbc.M114.583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kylarova S, Kosek D, Petrvalska O, Psenakova K, Man P, Vecer J, Herman P, Obsilova V, Obsil T. Cysteine residues mediate high-affinity binding of thioredoxin to ASK1. Febs J. 2016;283:3821–3838. doi: 10.1111/febs.13893. [DOI] [PubMed] [Google Scholar]
- 28.Weijman JF, Kumar A, Jamieson SA, King CM, Caradoc-Davies TT, Ledgerwood EC, Murphy JM, Mace PD. Structural basis of autoregulatory scaffolding by apoptosis signal-regulating kinase 1. P Natl Acad Sci USA. 2017;114:E2096–E2105. doi: 10.1073/pnas.1620813114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujino G, Noguchi T, Matsuzawa A, Yamauchi S, Saitoh M, Takeda K, Ichijo H. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol Cell Biol. 2007;27:8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang J, Suh HW, Jeon YH, Hwang E, Nguyen LT, Yeom J, Lee SG, Lee C, Kim KJ, Kang BS, Jeong JO, Oh TK, Choi I, Lee JO, Kim MH. The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein. Nat Commun. 2014;5:10–23. doi: 10.1038/ncomms3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. The interaction of thioredoxin with Txnip - Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou M, Liu Q, Ren GP, Zeng JL, Xiang XP, Ding YF, Lin QH, Zhong TT, Liu X, Zhu LJ, Qi HY, Shen J, Li HR, Shao JM. Physical interaction between human ribonucleotide reductase large subunit and thioredoxin increases colorectal cancer malignancy. J Biol Chem. 2017;292:9136–9149. doi: 10.1074/jbc.M117.783365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoharan R, Seong HA, Ha H. Thioredoxin inhibits MPK38-induced ASK1, TGF-beta, and p53 function in a phosphorylation-dependent manner. Free Radical Bio Med. 2013;63:313–324. doi: 10.1016/j.freeradbiomed.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. Embo J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichijo H, Nishida E, Irie K, tenDijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 36.Song JJ, Lee YJ. Differential role of glutaredoxin and thioredoxin in metabolic oxidative stress-induced activation of apoptosis signal-regulating kinase 1. Biochem J. 2003;373:845–853. doi: 10.1042/BJ20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadeau PJ, Charette SJ, Toledano MB, Landry J. Disulfide bond-mediated multimerization of ask1 and its reduction by thioredoxin-1 regulate H2O2-induced c-jun NH2-terminal kinase activation and apoptosis. Mol Biol Cell. 2007;18:3903–3913. doi: 10.1091/mbc.E07-05-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadeau PJ, Charette SJ, Landry J. REDOX Reaction at ASK1-Cys250 Is Essential for Activation of JNK and Induction of Apoptosis. Mol Biol Cell. 2009;20:3628–3637. doi: 10.1091/mbc.E09-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kutuzov MA, Andreeva AV, Voyno-Yasenetskaya TA. Regulation of apoptosis signal-regulating kinase 1 (ASK1) by polyamine levels via protein phosphatase 5. J Biol Chem. 2005;280:25388–25395. doi: 10.1074/jbc.M413202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung H, Seong HA, Ha H. Murine Protein Serine/Threonine Kinase 38 Activates Apoptosis Signal-regulating Kinase 1 via Thr(838) Phosphorylation. J Biol Chem. 2008;283:34541–34553. doi: 10.1074/jbc.M807219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke-Gaffney A, Callister MEJ, Nakamura H. Thioredoxin: friend or foe in human disease? Trends Pharmacol Sci. 2005;26:398–404. doi: 10.1016/j.tips.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Sun XY, Wang WG, Chen J, Cai XT, Yang J, Yang Y, Yan HJ, Cheng XL, Ye J, Lu WG, Hu CP, Sun HD, Pu JX, Cao P. The Natural Diterpenoid Isoforretin A Inhibits Thioredoxin-1 and Triggers Potent ROS-Mediated Antitumor Effects. Cancer Res. 2017;77:926–936. doi: 10.1158/0008-5472.CAN-16-0987. [DOI] [PubMed] [Google Scholar]
- 43.DiRaimondo TR, Plugis NM, Jin X, Khosla C. Selective Inhibition of Extracellular Thioredoxin by Asymmetric Disulfides. J Med Chem. 2013;56:1301–1310. doi: 10.1021/jm301775s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirkpatrick DL, Kuperus M, Dowdeswell M, Potier N, Donald LJ, Kunkel M, Berggren M, Angulo M, Powis G. Mechanisms of inhibition of the thioredoxin growth factor system by antitumor 2-imidazolyl disulfides. Biochem Pharmacol. 1998;55:987–994. doi: 10.1016/s0006-2952(97)00597-2. [DOI] [PubMed] [Google Scholar]
- 45.Ramanathan RK, Abbruzzese J, Dragovich T, Kirkpatrick L, Guillen JM, Baker AF, Pestano LA, Green S, Von Hoff DD. A randomized phase II study of PX-12, an inhibitor of thioredoxin in patients with advanced cancer of the pancreas following progression after a gemcitabine-containing combination. Cancer Chemoth Pharm. 2011;67:503–509. doi: 10.1007/s00280-010-1343-8. [DOI] [PubMed] [Google Scholar]
- 46.Baker AF, Adab KN, Raghunand N, Chow HHS, Stratton SP, Squire SW, Boice M, Pestano LA, Kirkpatrick DL, Dragovich T. A phase IB trial of 24-hour intravenous PX-12, a thioredoxin-1 inhibitor, in patients with advanced gastrointestinal cancers. Invest New Drug. 2013;31:631–641. doi: 10.1007/s10637-012-9846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chrestensen CA, Starke DW, Mieyal JJ. Acute cadmium exposure inactivates thioltransferase (glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. J Biol Chem. 2000;275:26556–26565. doi: 10.1074/jbc.M004097200. [DOI] [PubMed] [Google Scholar]
- 48.Srinivasan U, Bala A, Jao SC, Starke DW, Jordan TW, Mieyal JJ. Selective inactivation of glutaredoxin by sporidesmin and other epidithiopiperazinediones. Biochemistry-Us. 2006;45:8978–8987. doi: 10.1021/bi060440o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadhu SS, Callegari E, Zhao Y, Guan XM, Seefeldt T. Evaluation of a dithiocarbamate derivative as an inhibitor of human glutaredoxin-1. J Enzym Inhib Med Ch. 2013;28:456–462. doi: 10.3109/14756366.2011.649267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samarasinghe KTG, Godage DNPM, Zhou YN, Ndombera FT, Weerapana E, Ahn YH. A clickable glutathione approach for identification of protein glutathionylation in response to glucose metabolism. Mol Biosyst. 2016;12:2471–2480. doi: 10.1039/c6mb00175k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holmgren A. Thioredoxin Catalyzes the Reduction of Insulin Disulfides by Dithiothreitol and Dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 52.Witte S, Villalba M, Bi K, Liu YH, Isakov N, Altman A. Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-kappa B pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J Biol Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 53.Gallogly MM, Starke DW, Mieyal JJ. Mechanistic and Kinetic Details of Catalysis of Thiol-Disulfide Exchange by Glutaredoxins and Potential Mechanisms of Regulation. Antioxid Redox Sign. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuchs SY, VA, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. P Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Bio. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Powis G, Kirkpatrick DL. Thioredoxin signaling as a target for cancer therapy. Curr Opin Pharmacol. 2007;7:392–397. doi: 10.1016/j.coph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. Exposure to Hydrogen Peroxide Induces Oxidation and Activation of AMP-activated Protein Kinase. J Biol Chem. 2010;285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auciello FR, Ross FA, Ikematsu N, Hardie DG. Oxidative stress activates AMPK in cultured cells primarily by increasing cellular AMP and/or ADP. Febs Lett. 2014;588:3361–3366. doi: 10.1016/j.febslet.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng XS, Ji JJ, Kwon Y, Wang SD, Bai J. The role of thioredoxin-1 in suppression of endoplasmic reticulum stress in Parkinson disease. Free Radical Bio Med. 2014;67:10–18. doi: 10.1016/j.freeradbiomed.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 61.Kalyanaraman B, Darley-Usmar V, Davies KJA, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radical Bio Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ha H, Jung H, Seong HA. Murine proteine serine/threonine kinase 38 (MPK38) positively regulates apoptosis signal-regulating kinase 1 in a kinase-dependent manner through physical interaction. Faseb J. 2008:22. [Google Scholar]
- 63.Lacroix M, Toillon RA, Leclercq G. P53 and breast cancer, an update. Endocr-Relat Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 64.Adler V, Pincus MR, Minamoto T, Fuchs SY, Bluth MJ, BrandtRauf PW, Friedman FK, Robinson RC, Chen JM, Wang XW, Harris CC, Ronai Z. Conformation-dependent phosphorylation of p53. P Natl Acad Sci USA. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buschmann T, Potapova O, Bar-Shira A, Ivanov VN, Fuchs SY, Henderson S, Fried VA, Minamoto T, Alarcon-Vargas D, Pincus MR, Gaarde WA, Holbrook NJ, Shiloh Y, Ronai Z. Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol Cell Biol. 2001;21:2743–2754. doi: 10.1128/MCB.21.8.2743-2754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong KB, DeDecker BS, Freund SMV, Proctor MR, Bycroft M, Fersht AR. Hot-spot mutants of p53 core domain evince characteristic local structural changes. P Natl Acad Sci USA. 1999;96:8438–8442. doi: 10.1073/pnas.96.15.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Gene Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hui L, Zheng Y, Yan Y, Bargonetti J, Foster DA. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene. 2006;25:7305–7310. doi: 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.