Abstract
The differentiation and maturation of oligodendrocyte precursor cells (OPCs) is essential for myelination and remyelination in the CNS. The failure of OPCs to achieve terminal differentiation in demyelinating lesions often results in unsuccessful remyelination in a variety of human demyelinating diseases. However, the molecular mechanisms controlling OPC differentiation under pathological conditions remain largely unknown. Myt1L (myelin transcription factor 1-like), mainly expressed in neurons, has been associated with intellectual disability, schizophrenia, and depression. In the present study, we found that Myt1L was expressed in oligodendrocyte lineage cells during myelination and remyelination. The expression level of Myt1L in neuron/glia antigen 2-positive (NG2+) OPCs was significantly higher than that in mature CC1+ oligodendrocytes. In primary cultured OPCs, overexpression of Myt1L promoted, while knockdown inhibited OPC differentiation. Moreover, Myt1L was potently involved in promoting remyelination after lysolecithin-induced demyelination in vivo. ChIP assays showed that Myt1L bound to the promoter of Olig1 and transcriptionally regulated Olig1 expression. Taken together, our findings demonstrate that Myt1L is an essential regulator of OPC differentiation, thereby supporting Myt1L as a potential therapeutic target for demyelinating diseases.
Electronic supplementary material
The online version of this article (10.1007/s12264-018-0207-9) contains supplementary material, which is available to authorized users.
Keywords: Myt1L, Oligodendrocyte precursor cells, Remyelination, Demyelination, Olig1
Introduction
In the central nervous system (CNS), myelin, which is produced by oligodendrocytes, plays a pivotal role in accelerating the propagation of action potentials, and insulating and protecting axons. CNS demyelination occurs widely in a variety of neurological and psychiatric diseases such as multiple sclerosis (MS), periventricular leukomalacia, hypoxic ischemic encephalopathy, depression, and schizophrenia [1]. When timely and effective repair does not occur, demyelination leads to the disruption of electrical conduction in neurons and irreversible axonal injury, eventually having a serious impact on cognition and/or motor function. Currently, there is no valid treatment for CNS demyelinating diseases. Thus it is urgent to identify novel molecular targets and develop therapeutic strategies.
Remyelination in the CNS is a complicated and elaborate process which is mostly mediated by endogenous oligodendrocyte precursor cells (OPCs). In response to injury signals at sites of demyelination, OPCs in the vicinity are activated. They then proliferate and migrate into the injured area and further differentiate into oligodendrocytes, which form new compact myelin sheaths wrapping the naked axons [2, 3]. Although remyelination is usually spontaneous and efficient in many rodent models of demyelination, it fails in the chronic inactive demyelinating lesions of MS patients [4]. One of the primary reasons for remyelination failure is OPC maturation arrest [3, 5]. In addition, aging can attenuate the spontaneous remyelination capacity in the CNS, so demyelinating axons are not repaired in a timely and effective fashion, leading to conduction disorders and even the degeneration of neurons [6]. However, the molecular mechanisms regulating OPC differentiation and maturation under pathological conditions and during aging are far from fully understood.
The sequential steps in the process of myelination or remyelination involve a complex interplay among intrinsic and extrinsic factors [7]. Transcriptional regulation has been identified as a major mechanism that directs OPCs to differentiate into myelinating oligodendrocytes [8]. Myt1L (myelin transcription factor 1 like), a member of the Myt/neural zinc finger (NZF) family, encodes a neural transcription factor that has six characteristic zinc finger domains with a CX4CX4HX7HX5C (CCHHC) consensus DNA binding sequence [9]. Myt1L plays an indispensable physiological role in CNS development. It has been reported that mutation of Myt1L in chromosome band 2p25.3 can result in intellectual disability [9, 10]. Duplication of Myt1L has also recently been reported to result in schizophrenia [11–13] and depression [14, 15].
Myt1L is expressed predominantly in differentiating, post-mitotic neurons in the cerebral cortex, thalamus, hindbrain, and dorsal root ganglia [9]. It has been reported that Myt1L regulates the differentiation of neurons and induces the formation of functional mature neurons by binding to the transcription factor Brn2 [16, 17]. The pivotal role of Myt1L in regulating neuronal determination has been further underlined in a study suggesting that this molecule, in conjunction with Ascl1 and NeuroD1, can directly convert human fibroblasts into neurons [18, 19]. Moreover, Myt1L can recruit histone deacetylase to regulate neuronal transcription via its interaction with Sin3β during neurogenesis [20]. More broadly, the latest report shows that Myt1L safeguards neuronal identity by actively repressing many non-neuronal fates [21]. Unlike conventional repressors that inhibit specific lineages, such as REST and Groucho, Myt1L appears to block a multitude of differentiation programs and lineage identities, but not the neuronal lineage [21]. Myt1L is the only known transcription factor that is specifically expressed in all neurons throughout life, indicating that active repression of non-neuronal programs is critical for maintaining neuronal identity [22]. Although extensive studies have confirmed the role of Myt1L in neuronal differentiation, its function in oligodendrocyte lineage cells remains unknown. In the present study, we demonstrated that Myt1L promotes the differentiation of OPCs and remyelination after L-α-lysophosphatidylcholine (LPC)-induced demyelination.
Materials and Methods
Animal Experiments
All animal experiments were performed in adherence with the National Institutes of Health Guidelines on the Use of Laboratory Animals and approved by the Second Military Medical University Committee on Animal Care. To create the focal demyelination model, a spinal cord lesion was induced as described previously. Briefly, 1 μL of 1% LPC (Sigma Aldrich, St. Louis, MO) in 0.9% NaCl was microinjected into the dorsal or ventral column following a laminectomy at T11–T12 in C57BL/6 mice (8–10 weeks old) using a micromanipulator and a glass tip attached to a syringe (Hamilton Company, Reno, NA). The day of LPC injection was designated day 0 (0 dpi) [23]. Control mice were injected with 1 μL of saline. For transfection of shRNA lentivirus in the LPC-induced focal lesion model, a laminectomy was performed and 3 μL shRNA lentivirus was injected 0.5 cm above and below the injection site using the above method. The virus was injected immediately after the LPC injection.
Cell Cultures
OPCs and oligodendrocytes were purified and cultured as described previously [24, 25]. Mixed cortical glial cell cultures were generated from P0 rats and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) for 10 days at 37°C under 5% CO2. The culture flasks were shaken at 180 rpm for 1 h followed by an additional 20 h at 200 rpm with fresh medium at 37°C. The purified OPCs were collected from mixed glia and then the cell suspension was left to adhere in uncoated Petri dishes. To examine differentiation, OPCs were cultured in Neurobasal medium supplemented with 2% B27 (NB/B27) [26]. To expand the OPCs and keep them undifferentiated, the culture medium was supplemented with 30% B104 conditioned medium [27].
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (PFA) for 20 min followed by three washes in phosphate buffered saline (PBS) at room temperature. The cells were next permeabilized with 0.1% Triton X-100 for 5 min. Then they were incubated overnight at 4°C with the following primary antibodies: mouse anti-myelin basic protein (MBP; Chemicon, Temecula, CA, 1:50), mouse anti-neuron/glia antigen 2 (NG2; Millipore, Temecula, CA, 1:50), mouse anti-CC1 (Abcam, Cambridge, UK, 1:50), rabbit anti-Myt1L (Abcam, 1:300), and mouse anti-caspase3 (Millipore, 1:100). The cells were then incubated in fluorescein isothiocyanate (FITC)- or tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, 1:100) containing Hoechst 33342 for 2 h at room temperature. They were counted in at least 10 randomly-selected fields from each of at least 3 coverslips from each group.
Immunohistochemistry
Frozen tissue samples of LPC lesions were used to cut cryostat sections at 14 μm for further histological analysis. Coronal sections were heated for 10 min in 10 mmol/L citrate buffer (pH 6.0) at 93°C before permeabilization with 0.6% Triton for 15 min, then placed in blocking solution, and incubated overnight at 4°C with the primary antibodies mouse anti-NG2 (Millipore, 1:50), mouse anti-CC1 (Abcam, 1:50), rabbit anti-Myt1L (Abcam, 1:300), mouse anti-Myt1L (Millipore, 1:300), goat anti-Sox10 (Santa Cruz Biotechnology, Santa Cruz, CA, 1:100), or mouse anti-Olig1 (Millipore, 1:300). The sections were then incubated in FITC- or TRITC-conjugated secondary antibodies (Jackson ImmunoResearch, 1:100) containing Hoechst 33342 for 2 h at room temperature. Five sections were selected from each sample, and 3–5 mice were tested in each experimental group.
RNA Isolation and qPCR Analysis
Total RNA was purified from cultured cells using TRIzol reagent (Invitrogen). After removal of the residual DNA with DNase I, cDNA was obtained by reverse transcription according to the manufacturer’s instructions. The mRNA level was then assessed using real-time quantitative PCR (qPCR) with SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan). The expression of GAPDH served as control. PCRs were performed under the following conditions: 95°C for 3 min followed by 45 cycles at 95°C for 10 s, 65°C for 30 s, and 72°C for 10 s, then 72°C for 10 min. The primers used were: Myt1L F: GGACCAGTCTCCAAGACAGC R: ATTTACGTGGCCGGTTCCAT; GAPDH F: ACAGCAACAGGGTGGTGGAC R: TTTGAGGGTGCAGCGAACTT; MBP F: CATTGTGACACCTCGTACACC R: TTGAATCCCTTGTGAGCCGAT; Olig1 F: CGTCCCTTCTCCCTAAACCTG R: GCTGTTGATCTTGCGCCTCA; Olig2 F: CTGGCGCGAAACTACATCC R: CCCCGTAAATCTCGCTCACC; Sox10 F: TCCGGCAAGGCAGATCCCAA R: CCAGCTCAGTCACATCAAAGGTCT; Sox2 F: AGAACTAGACTCCGGGCGA R: ACCCAGCAAGAACCCTTTCC; Ascl1 F: TCCGGTTTCGTCCTACTCCT R: CATTCCCAGTAGGGCCTGTC; NKX2.2 F: TACGACAGCAGCGACAACCC R: CTCCTTGTCATTGTCCGGTGA.
Western Blot Analysis
Western blotting used standard protocols. The primary antibodies rabbit anti-olig1 (1:300), mouse anti-MBP (1:500), and horseradish peroxidase (HRP)-conjugated anti-actin (Kangchen Biotechnology, Shanghai, China, 1:5000) were incubated overnight at 4°C. HRP-conjugated goat against rabbit and mouse secondary antibodies (Kangchen Biotechnology, 1:5000) were incubated for 2 h. The HRP-conjugated secondary antibodies were detected using chemiluminescent reagents (Pierce ECL; Thermo Scientific, Waltham, MA).
BrdU Incorporation
To assess the proliferation of OPCs, 5-bromo-2-deoxyuridine (BrdU, 10 μmol/L; Sigma) was added to the culture medium and left for 8 h before the cells were fixed in 4% PFA. Cells on coverslips were permeabilized with 0.3% Triton for 5 min, incubated in 2 N HCl for 30 min, and then in 0.1 mol/L borate buffer (pH 8.0) for 20 min. They were then blocked in PBS containing 10% goat serum for 1 h at room temperature and incubated overnight with anti-BrdU antibody (Sigma, 1:100) and anti-GFP antibody (Millipore, 1:400). Cells were then incubated in FITC- or TRITC-conjugated secondary antibodies (Jackson ImmunoResearch, 1:100) for 2 h at room temperature. The percentage of BrdU+ cells versus GFP+ cells was calculated.
Cell Migration
Migration of OPCs was assessed using 8-μm pore-sized transwell based Boyden chamber system (Corning Incorporated, NY). Both sides of the transwell membrane were precoated with 10 μg/mL poly-L-lysine at 37°C in 5% CO2 overnight, followed by DMEM containing 10% FBS for 2 h. Then transfected OPCs were planted to the upper chamber at ~ 80,000 cells/transwell in NB/B27 medium. Ara-C (5 μmol/L) was systematically included in the medium to stop cell proliferation. And the lower chamber was coated with NB/B27 medium. Cells were allowed to migrate for 12 h at 37°C in 5% CO2. Nonmigrated cells on the upper side of the filter was wiped off with a cotton swab and the cells in the lower chamber were fixed with 4% PFA for 20 min followed by three washes in PBS. Then the cells were incubated with Coomassie brilliant blue for 4 h followed by permeabilization with 0.1% Triton X-100 for 5 min. Finally, the cells were washed in PBS until the supernatant was clear [28].
Luxol Fast Blue Staining
Frozen samples of LPC lesions were cut at 14 μm on a cryostat. The sections were incubated in 95% ethanol for 5 min and with 0.1% Luxol fast blue overnight at 58°C, then differentiated with 0.05% lithium carbonate and 70% ethanol [29]. The percentage of demyelinating area in the dorsal column white matter of the spinal cord was calculated by blinded independent readers. At least five animals for each time point in different treated groups were included in one experiment, and ten sections were examined per animal for each individual analysis.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using an EZ ChIP Chromatin Immunoprecipitation kit (Millipore) according to the kit instructions. OPCs (107) were fixed in 1% formaldehyde. The chromatin was sonicated into 200 bp–500 bp fragments and immunoprecipitated using anti-Myt1L. Normal mouse IgG was used as a negative control. Quantitative PCR was conducted on an MyiQ Detection System (Bio-Rad, Hercules, CA). The primers used for Olig1, PLP and MBP were listed in supplementary Table S1.
Electron Microscopy
Mouse spinal cord tissue perfused with a phosphate buffer solution containing 2.5% glutaraldehyde and 4% PFA (pH 7.3) was fixed in 1% osmium tetroxide for 45 min, dehydrated, and embedded in Araldite resin. Sections (1 μm) were stained with toluidine blue to select regions of demyelination, and then ultra-thin (60 nm) sections were cut with uranyl acetate and lead citrate staining. The sections were observed in a transmission electron microscope (H-7650; Hitachi, Tokyo, Japan) at 100 kV. The thickness of myelin sheaths was measured by g-ratio (the ratio of the diameter of the axon itself to the diameter of axon plus its myelin sheath).
Virus Production and Infection
The Myt1L- and Olig1-overexpressing viruses were from GeneChem (Shanghai, China). The shRNAs for Myt1L (Myt1L-RNAi) or GFP alone were ligated into the pLKD-CMV-GFP-U6 plasmid. The sequences for the control shRNA were 5′-TTCTCCGAACGTGTCACGT-3′ and 5′-ACGTGACACGTTCGGAGAA-3′. The sequences for the Myt1L shRNA were 5′-CCGGTGCGAACTGAGTCTAGACTTTTCAAGAGAAAGTCTAGACTCAGTTCGCTTTTTTG-3′ and 5′-AATTCAAAAAAGCGAACTGAGTCTAGACTTTCTCTTGAAAAGTCTAGACTCAGTTCGCA-3′.
Statistical Analysis
All experiments were repeated three times and the results are shown as mean ± SEM. We used two-tailed Student’s t-test to compare 2 groups, and one-way ANOVA with Dunnett’s post hoc test for multiple groups. A value of P <0.05 was considered statistically significant.
Results
Myt1L is Expressed in Oligodendrocyte Lineage Cells and is Down-Regulated During Oligodendrocyte Maturation
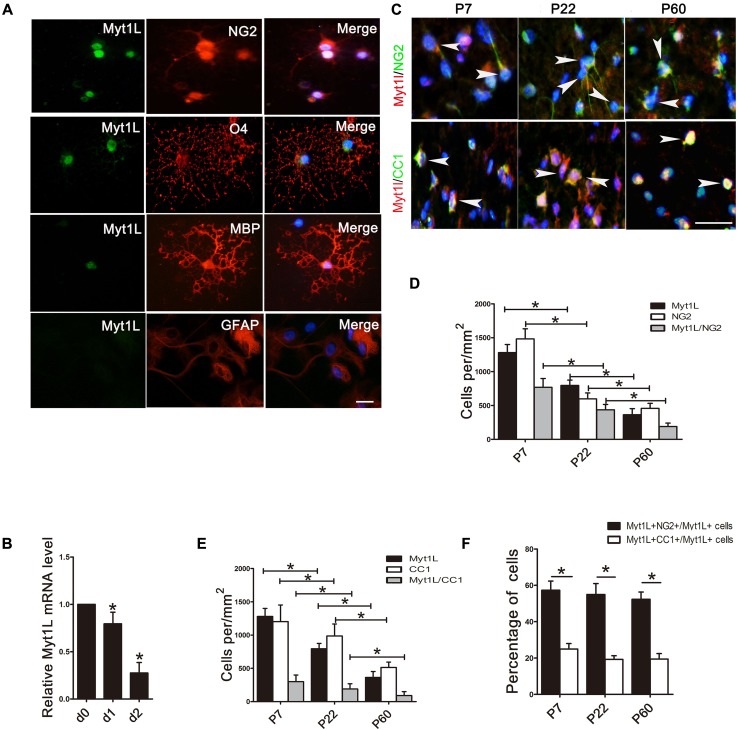
We first characterized the expression pattern of Myt1L in oligodendrocyte lineage cells. Myt1L was expressed in cultured rat NG2+ OPCs, O4+ pre-myelinating oligodendrocytes, and MBP+ mature oligodendrocytes. However, it was not expressed in GFAP-positive astrocytes (Fig. 1A). And the qPCR analysis revealed that the mRNA level of Myt1L was down-regulated during the differentiation process (Fig. 1B). These results indicated that Myt1L is consistently expressed at various developmental stages of oligodendrocytes in vitro and gradually decreases with the maturation of OPCs.
Fig. 1.
Myt1L is dynamically expressed in oligodendrocyte-lineage cells in vitro and in vivo. A Cultured oligodendrocytes at different developmental stages stained for Myt1L (green) as well as NG2, O4, and MBP (red). Cultured astrocytes were stained for Myt1L (green) as well as GFAP (red). Scale bar, 20 μm. B Quantitative PCR of mRNA levels of Myt1L during OPC differentiation in vitro. C Immunofluorescence staining for the expression of Myt1L (red) in NG2+ OPCs and mature CC1+ oligodendrocytes (green) in the spinal cord of P7, P22, and P60 mice (arrowheads indicate representative dual-labeled cells). Scale bar, 20 μm. D, E Density of different cell types in spinal cord white matter that express Myt1L throughout postnatal development as in C. F Percentages of Myt1L+/NG2+ and Myt1L+/CC1+ double-positive cells in total Myt1L+ cells. *P < 0.05, one-way ANOVA with Tukey’s post hoc test; n = 3/group.
The process of myelination in the spinal cord of rodents begins within the first week after birth and finishes at around two months of age [30]. In order to further explore the expression patterns of Myt1L during myelination in vivo, we performed immunofluorescence staining for Myt1L in the spinal white matter of neonatal to adult mice. This revealed that the density of Myt1L+ cells decreased during myelination. The density of Myt1L+ OPCs (Myt1L+/NG2+ cells) (Fig. 1C, D) and mature oligodendrocytes (Myt1L+/CC1+ cells) (Fig. 1C, E) was also down-regulated. Moreover, among the total Myt1L+ cells, the percentage of NG2+ OPCs was much higher than mature CC1+ oligodendrocytes (Fig. 1F). These results suggest that Myt1L is expressed in both OPCs and mature oligodendrocytes during CNS myelination, which is consistent with the expression pattern in vitro, and its expression in oligodendrocyte lineage cells decreased with oligodendrocyte maturation.
Myt1L Expression is Up-Regulated in Oligodendrocyte Lineage Cells in the LPC Model
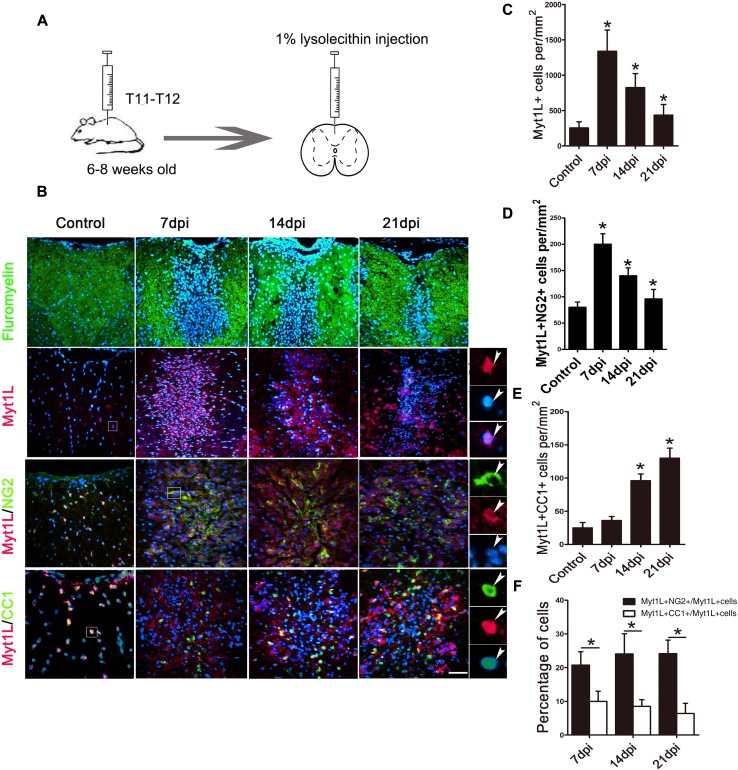
In order to validate the expression pattern of Myt1L in the acute demyelination model in vivo, we constructed a model of LPC-induced demyelination in mouse spinal cord (Fig. 2A). Generally, both the demyelinating lesion and the proliferation of OPCs reached a peak and a small number of mature oligodendrocytes emerged at 7 dpi, while at 14 dpi, a large number of OPCs had differentiated into mature oligodendrocytes accompanied by significant remyelination, and the whole process of remyelination was nearly completed at 21 dpi [2, 31]. Therefore, we selected these three time points to assess the expression of Myt1L. The extent of demyelination and repair of lesions were assessed by fluoromyelin staining (Fig. 2B). The expression of Myt1L measured in the LPC lesions was significantly increased at 7 dpi and then declined (Fig. 2B). Quantitative analysis showed that the number of Myt1L+ cells was markedly elevated in LPC lesions from 7 dpi to 21 dpi compared with controls (Fig. 2C). These data show that, in LPC-induced lesions, Myt1L expression is significantly increased, and its highest expression coincides with the peak of OPC proliferation.
Fig. 2.
Expression of Myt1L in oligodendrocyte lineage cells in LPC-induced demyelinating lesions. A Schematic of the LPC-induced spinal cord demyelination model. B Immunofluorescence staining for Myt1L in lesions. Upper row of panels: representative micrographs of fluoromyelin (green) and Hoechst (blue) staining in control and demyelinated spinal cord at 7, 14, and 21 days after LPC injection (dpi). Lower 3 rows of panels: representative micrographs of immunofluorescence staining for Myt1L (red) and NG2 or CC1 (green) in lesions and in saline-injected control spinal cord. High magnification micrographs of outlined regions (right-most panels) show co-localization of Myt1L with Hoechst and other oligodendrocyte lineage markers. C–E Density of Myt1L+ cells (C), Myt1L+/NG2+ OPCs (D), and Myt1L+/CC1+ oligodendrocytes (E) in saline-injected controls and LPC lesions. F Percentage of Myt1L+/NG2+ or Myt1L+/CC1+ double-positive cells among total Myt1L+ cells. *P < 0.05, one-way ANOVA with Tukey’s post hoc test: n = 3/group, scale bar, 20 μm.
To further define the involvement of Myt1L in repair processes, we evaluated the regulation of its expression in oligodendrocyte lineage cells and astrocytes by immunofluorescence staining. We found that Myt1L was almost exclusively expressed in Sox10+ oligodendrocyte lineage cells in LPC lesions (Fig. S1) and Myt1L barely coincided with GFAP before and after LPC-induced demyelination (Fig. S2). Moreover, the largest number of Myt1L+ NG2+ OPCs in LPC lesions occurred at 7 dpi, compared with control and LPC lesions at 14 dpi and 21 dpi (Fig. 2B, D). The number of Myt1L+/CC1+ oligodendrocytes was significantly increased at 14 dpi and 21 dpi (Fig. 2B, E). In addition, Myt1L was expressed more in NG2+ OPCs than in mature CC1+ oligodendrocytes (Fig. 2F). These results showed that Myt1L expression is up-regulated in LPC lesions and is higher in OPCs than in mature oligodendrocytes, similar to the expression pattern during development.
Myt1L Promotes OPC Differentiation In Vitro
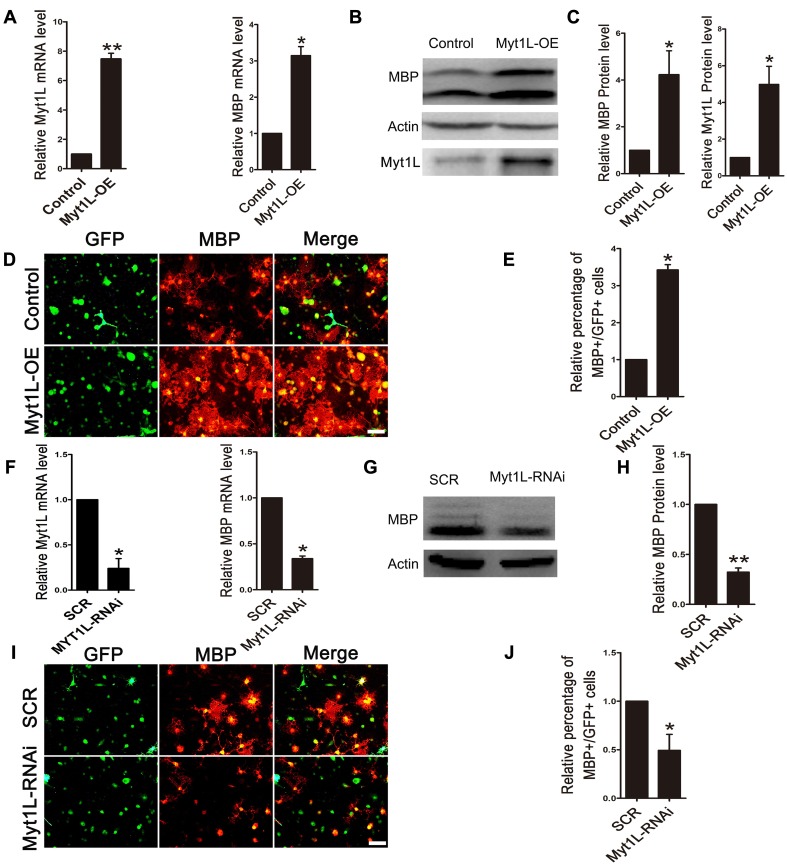
To investigate the effect of Myt1L on OPC differentiation in vitro, we transfected GFP-labeled Myt1L-overexpressing viruses (Myt1L-OE) and Myt1L interfering viruses (Myt1L-RNAi). After 2 days of differentiation, we found that the overexpression of Myt1L increased the Myt1L and MBP mRNA levels compared with the control group (Fig. 3A). Consistently, Western blot analysis also revealed that Myt1L overexpression led to an elevation of MBP and Myt1L protein expression (Fig. 3B, C). We found that overexpression of Myt1L resulted in an increase of MBP+ cells (Fig. 3D, E). In contrast, OPC cultures transfected with Myt1L-RNAi showed a marked decrease in MBP at both the mRNA (Fig. 3F) and protein levels (Fig. 3G, H). Similarly, the percentage of MBP+ cells also decreased when Myt1L was knocked down. Thus, our data indicate that Myt1L plays a positive regulatory role in OPC differentiation.
Fig. 3.
Myt1L promotes OPC differentiation in vitro. A QPCR analysis of Myt1L mRNA and MBP mRNA in purified OPCs transfected with overexpressing virus (Myt1L-OE) and control virus. B, C Western blot analysis of MBP and Myt1L protein levels. D Representative images of anti-MBP immunocytofluorescence staining of OPCs transfected with Myt1L-OE. E Quantification of MBP+/GFP+ cells as in D. F QPCR analysis of Myt1L and MBP mRNAs in purified OPCs transfected with interfering virus (Myt1L-RNAi). G, H Western blot analysis of MBP protein levels. I Representative images of anti-MBP immunocytofluorescence staining of OPCs transfected with Myt1L-RNAi virus. J Quantification of MBP+/GFP+ cells as in I. Transfected cells were all cultured for 72 h in differentiation medium before analysis. *P <0.05, **P <0.01, Student’s t-test; n = 3 independent experiments; scale bars, 20 μm.
Myt1L has No Effect on OPC Proliferation, Survival, and Migration
The ability of Myt1L to increase the number of MBP+ oligodendrocytes could be due to OPC proliferation and apoptosis. To exclude these possibilities, we performed BrdU incorporation assays (Fig. 4A, B) and caspase3 staining (Fig. 4E, F) in Myt1L-overexpressing and knockdown OPCs. We found that neither overexpression nor knockdown affected OPC proliferation and survival, as demonstrated by the comparable percentages of BrdU+/GFP+ cells (Fig. 4C, D) and caspase3+/GFP+ cells in the experimental and control groups (Fig. 4G, H). Similarly, Myt1L had no effect on the migration of OPCs (Fig. 4I–L) assessed by transwell assays. Thus, our results indicate that Myt1L specifically promotes OPC differentiation without affecting their survival, proliferation, and migration in vitro.
Fig. 4.
Myt1L does not affect OPC proliferation, survival, and migration. A, B Representative images showing BrdU immunocytofluorescence staining (red) in Myt1L-OE (A) and Myt1L-RNAi (B) virus-transfected OPCs (green). C, D Percentages of BrdU+/GFP+ cells among all GFP+ cells as in A and B. Data are normalized to control. E, F Representative images showing caspase3 immunocytofluorescence staining (red) in Myt1L-OE (E) and Myt1L-RNAi (F) virus-transfected OPCs (green). G, H Percentages of caspase3+/GFP+ cells among all GFP+ cells as in E and F. Data are normalized to control; n = 3 independent experiments. I, J Representative transwell assays visualizing migration of Myt1L-OE (I) and Myt1L-RNAi (J) virus-transfected OPCs. K, L Quantification of migrating cells as in I and J. Data are normalized to control. Student’s t-test: n = 5 independent experiments; scale bars, 20 μm.
Myt1L is Involved in the Remyelination Process in LPC-Induced Demyelination in Mice
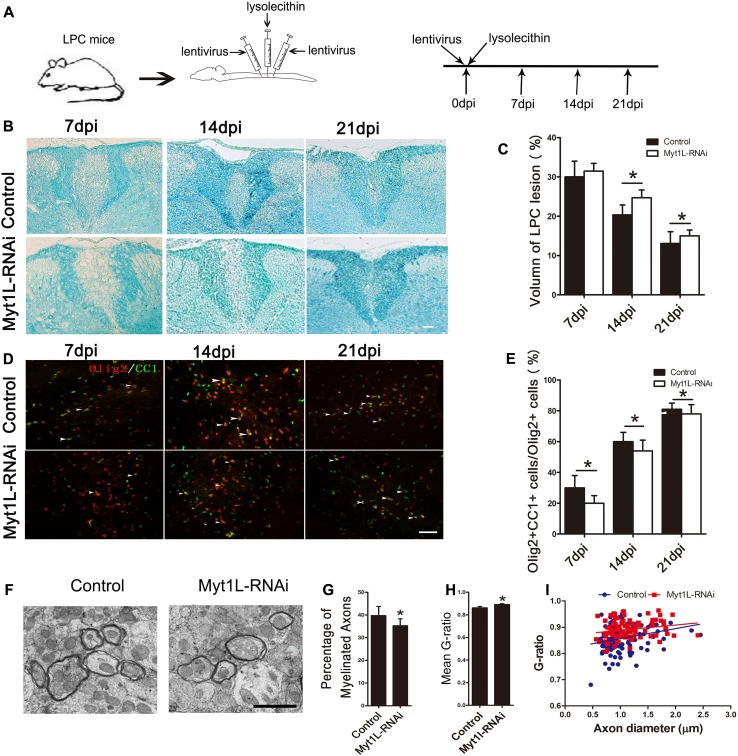
Our data showed that Myt1L significantly promoted the differentiation and maturation of OPCs in vitro, and its expression in oligodendrocyte lineage cells was remarkably up-regulated in LPC-induced lesions, suggesting that Myt1L is involved in myelin regeneration in models of demyelination. To define the effect of Myt1L on LPC-induced demyelination, lentivirus encoding Myt1L-shRNA (Myt1L-RNAi), or control virus encoding scrambled-shRNA, was injected directly into the lesions immediately after LPC injection (Fig. 5A). The changes in area of demyelination were assessed by Fast Blue staining at 7 dpi, 14 dpi, and 21 dpi (Fig. 5B). The results showed that, compared with the control group, the area of demyelination after transfection with Myt1L-RNAi was significantly enlarged, especially at 14 dpi and 21 dpi (Fig. 5C). Furthermore, Olig2/CC1 immunofluorescence double-staining was used to assess the differentiation of local oligodendrocytes (Fig. 5D). At 7 dpi, 14 dpi, and 21 dpi, compared with the control group, the proportion of mature oligodendrocytes (Olig2+/CC1+) among all oligodendrocytes lineage cells (Olig2+) was significantly lower in the lesions transfected with Myt1L-RNAi (Fig. 5E). These results showed that local interference with Myt1L expression significantly aggravates the demyelination injury and inhibits oligodendrocyte differentiation in vivo. To further determine whether Myt1L results in reduced remyelination, we examined demyelination in the spinal cord transfected with Myt1L-RNAi using electron microscopy. The results showed that Myt1L interference significantly reduced the percentage of myelinated axons (Fig. 5F, G) and increased the g-ratio compared with the control group (Fig. 5H). Therefore, interference with Myt1L inhibits the remyelination process in vivo.
Fig. 5.
Down-regulation of Myt1L suppresses remyelination in LPC-induced focal demyelinating lesions. A Schematic of LPC-induced spinal demyelination model. Myt1L-RNAi virus was administered immediately after LPC injection. B Representative Luxol fast blue staining of dorsal cord sections from the Myt1L-RNAi and control virus-injected groups at 7 dpi, 14 dpi, and 21 dpi. Scale bar, 10 μm. C Quantitative analysis of lesion size in the dorsal column as in B. D Representative immunohistofluorescence staining showing Olig2 (red) and CC1 (green) expression at various time points in different groups in the demyelinating area of the spinal cord. E Quantification of Olig2+/CC1+ cells among the total Olig2+ cells in the dorsal spinal cord as in D. *P < 0.05, one-way ANOVA with Tukey’s post hoc test. Scale bar, 20 μm. F Electron micrographs of dorsal spinal cord sections from the Myt1L-RNAi and control virus-injected groups at 14 dpi. Scale bar, 2 μm. G Quantification of myelinated axons among total axons. H Mean g-ratio at 14 dpi determined using ImageJ. I Analysis of myelinated axons showed a reduction of the g-ratio in the spinal cord in the Myt1L-RNAi and control virus-injected groups at 14 dpi. *P < 0.05, Student’s t-test; n = 5/group.
Myt1L Binds to the Olig1 Promoter and Up-Regulates Olig1 Expression
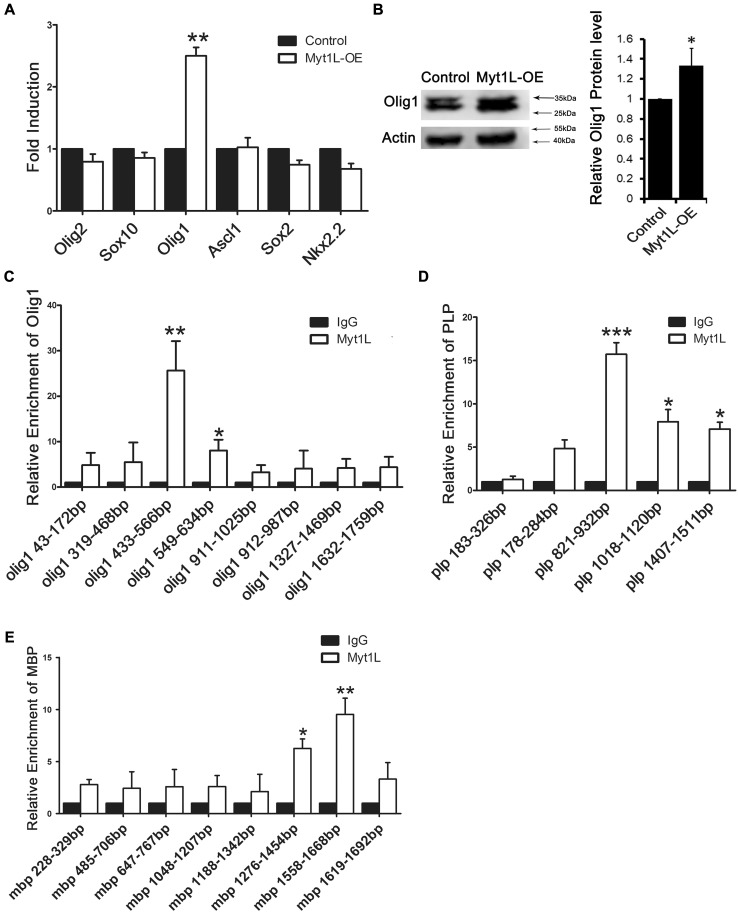
To further investigate the molecular mechanisms controlling the regulation of OPC differentiation by Myt1L, we transfected Myt1L-OE into OPCs and then used q-PCR to assess the expression of several transcription factors that have been reported to positively regulate OPC differentiation. We found that Myt1L specifically up-regulated the expression of Olig1, but had no effect on the other transcription factors Olig2, Sox10, Sox2, Nkx2.2, and Ascl1 (Fig. 6A). Consistently, Western blot analysis also revealed a remarkable increase of Olig1 protein expression when Myt1L was overexpressed (Fig. 6B). Similarly, we found that Olig1 expression was significantly reduced after interfering virus was injected into the demyelinating mouse (Fig. S3). So, we hypothesized that, as a transcription factor, Myt1L binds with the promoter region of Olig1 to enhance its expression. To test this hypothesis, we used the ChIP assay to study the binding of Myt1L to different regions of the Olig1 promoter and found that it bound to the upstream 433 bp–566 bp and 549 bp–634 bp of the Olig1 transcription initiation site (Fig. 6C). To further validate the functional relationship between Myt1L and the promoter of Olig1, we overexpressed Olig1 in Myt1L-knockdown OPCs, and found that Olig1 overexpression led to an elevation of MBP protein expression in these conditions (Fig. S4). This result indicated that overexpression of Olig1 rescues the deficits caused by Myt1l knockdown in vitro.
Fig. 6.
Myt1L binds to the promoter of Olig1 and up-regulates Olig1 expression. A Quantitative PCR analysis of the effect of Myt1L overexpression on the mRNA level of different transcription factors during OPCs differentiation. B Western blot analysis of Olig1 protein in OPCs transfected with Myt1L-OE virus and cultured in differentiation medium for 72 h. C–E Quantitative PCR analysis of different promoter regions of Olig1 (C), proteolipid protein (PLP) (D), or MBP (E) in immunoprecipitated DNA from OPCs cultured in proliferation medium for 12 h then processed for ChIP with an antibody against Myt1L. *P < 0.05, **P < 0.01, Student’s t-test; n = 3 independent experiments.
Since Myt1, which has a sequence similar to Myt1L, has been reported to bind to the promoter of proteolipid protein (PLP), we also used ChIP assays to study the binding between Myt1L and different regions of the PLP promoter and found that Myt1L bound to the upstream 821 bp–932 bp, 1018 bp–1120 bp, and 1407 bp–1511 bp of the PLP transcription initiation site (Fig. 6D). We further explored the binding between Myt1L and the promoter of MBP which is another major myelin protein, and found that Myt1L bound to the upstream 1276 bp–1454 bp and 1558 bp–1668 bp of the MBP transcription initiation site (Fig. 6E). Taken together, these results suggested that Myt1L not only acts on the PLP and MBP promoters to increase their expression, but also targets the promoter region of Olig1, thereby promoting the differentiation of OPCs by up-regulating Olig1 expression.
Discussion
OPC differentiation block is one of the most important causes of remyelination failure in demyelinating diseases such as MS. Screening and identifying the molecules affecting OPC maturation and remyelination are important strategies to elucidate the pathogenesis of demyelinating diseases and to find novel therapeutic targets. CNS demyelination also occurs in depression and schizophrenia. In the present study, we identified the roles of the transcription factor Myt1L, which is closely related to schizophrenia and depression, on OPC differentiation and remyelination.
Myt1L is a transcription factor with a neural zinc finger structure. Three types of Myt/NZF family proteins have been reported in mammals: Myt1 (NZF-2 and NZF-2b) [22], Myt1L (NZF-1 or png-1) [32, 33], and ST18 (NZF-3 or Myt3) [34, 35]. Myt1 is one of the essential transcription factors regulating CNS development [22]. It can combine with the retinoic acid receptor to regulate the development of neurons and promote neurogenesis [36, 37]. Myt1 also specifically binds to the promoter of the myelin lipoprotein PLP and affects OPC proliferation and differentiation, as well as the transcription of genes associated with myelination [9]. Myt1L has 80% similarity in sequence and 62% homology at the protein level with Myt1 [10, 37]. However, previous research has reported that Myt1L is markedly different from Myt1 in tissue-specific expression, protein binding, and molecular function [9, 38]. Unlike Myt1, Myt1L is reported to be expressed mainly in post-mitotic neurons [22], with a role in promoting neuronal lineage determination and maturation. In this study, we demonstrated that Myt1L was expressed in oligodendrocyte lineage cells both in vitro and in vivo, regardless of the physiological and pathological conditions. Interestingly, we noted that Myt1L was localized in both the nucleus and cytosol of oligodendrocyte lineage cells. Further study is needed to further explore whether Myt1L translocates from nucleus to cytosol. Moreover, in the LPC-induced demyelination model, the expression of Myt1L increased significantly after injury, reaching a peak at 7 dpi, consistent with the time point when OPCs after injury reached the maximum number. At 14 dpi, when apparent remyelination can be detected, the expression of Myt1L was decreased, in accordance with the decreased number of OPCs and the generation of quite a few mature oligodendrocytes. This is similar to the reported expression pattern of Myt1 in virus-induced demyelinating lesions, in which the number of Myt1-positive cells increases markedly after injury [39].
Whether Myt1L affects the function of oligodendrocyte lineage cells has not been reported. Our experiments showed that overexpression of Myt1L in OPCs specifically enhanced OPC differentiation without affecting apoptosis, proliferation, and migration. This is distinct from Myt1, since Myt1 not only promotes the differentiation of OPCs, but also regulates their proliferation [40]. In addition, in order to exclude a redundancy effect between Myt1L and Myt1, we designed interfering viruses against different fragments of Myt1 and Myt1L in an OPC differentiation model in vitro and in the LPC-induced demyelination model in vivo. We found that specific interference with Myt1L expression significantly inhibited OPC differentiation in vivo and in vitro, and delayed the remyelination of lesions. These results suggest that Myt1L plays a positive regulatory role in OPC differentiation, and the effect cannot be compensated by Myt1 after its knockdown.
As for the molecular mechanism by which Myt1L up-regulates OPC differentiation and maturation, we hypothesized that Myt1L, as a transcription factor, may control the differentiation and maturation of OPCs by regulating the transcription of target genes. We found that Myt1L specifically up-regulated the mRNA and protein levels of Olig1 by screening a number of known transcription factors expressed by oligodendrocyte lineage cells (Olig1, Olig2, Ascl1, Sox10, and Sox2). In order to further explore the mechanism by which Myt1L improved Olig1 expression, we performed ChIP assays and found that Myt1L bound to the Olig1 promoter. As a basic helix-loop-helix transcription factor, Olig1 is essential for the fate determination and maturation of oligodendrocytes [41, 42] and its deletion would result in the delayed generation of OPCs in the spinal cord. In addition, Olig1-knockout only blocked OPC differentiation with no effect on proliferation, consistent with our findings on the effect of Myt1L on OPCs. Since Olig1 initiates the transcription of many myelin-associated genes (MBP, PLP, and myelin-associated glycoprotein) [43, 44], we hypothesized that Myt1L promotes OPC differentiation by initiating Olig1 transcriptional synergies.
It has been reported that Myt1 binds to the PLP-specific promoter to facilitate the maturation and differentiation of OPCs. Since Myt1L has 62% homology [37] with Myt1 at the protein level, we also examined the binding of Myt1L to the PLP promoter region through ChIP assays, and found that Myt1L also bound to the PLP promoter. This suggests that Myt1L promotes OPC differentiation and MAP expression through a variety of transcriptional regulation mechanisms and plays a pivotal role in OPC differentiation. Although Myt1 has been reported to bind to the promoter of transcription factor Sox2 in neural progenitor cells [36], our study found that Myt1L had no effect on Sox2 expression. As Sox2 is closely associated with the proliferation of neural progenitor cells, this result also explains our previous finding that Myt1L specifically regulates OPC differentiation which is different from Myt1. Therefore, despite their similar structures, molecular mechanisms, and functions, Myt1 and Myt1L do not act identically in oligodendrocyte lineage cells.
In summary, Myt1L is expressed in oligodendrocyte linage cells during myelination and remyelination and plays an indispensable role in OPC differentiation, suggesting a potential therapeutic target for myelin repair.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the International Cooperation and Exchange Program of the National Natural Science Foundation of China (81461138035), the National Natural Science Foundation of China (81371326, 31571066, and 31371068), the National Basic Research Development Program of China (2016YFA0100802), the UK Medical Research Council (MR/M010503/1), and the UK Multiple Sclerosis Society (33).
Footnotes
Yanqing Shi, Qi Shao and Zhenghao Li have contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1007/s12264-018-0207-9) contains supplementary material, which is available to authorized users.
Contributor Information
Cheng He, Email: chenghe@smmu.edu.cn.
Li Cao, Email: caoli@smmu.edu.cn.
References
- 1.He L, Lu QR. Coordinated control of oligodendrocyte development by extrinsic and intrinsic signaling cues. Neurosci Bull. 2013;29:129–143. doi: 10.1007/s12264-013-1318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moll NM, Hong E, Fauveau M, Naruse M, Kerninon C, Tepavcevic V, et al. SOX17 is expressed in regenerating oligodendrocytes in experimental models of demyelination and in multiple sclerosis. Glia. 2013;61:1659–1672. doi: 10.1002/glia.22547. [DOI] [PubMed] [Google Scholar]
- 3.Franklin RJ, Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 4.Trapp BD, Peterson J, Ransohoff RM, Rudick R, Mork S, Bo L. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 5.Lubetzki C, Williams A, Stankoff B. Promoting repair in multiple sclerosis: problems and prospects. Curr Opin Neurol. 2005;18:237–244. doi: 10.1097/01.wco.0000169739.83793.e0. [DOI] [PubMed] [Google Scholar]
- 6.Ruckh JM, Zhao JW, Shadrach JL, van Wijngaarden P, Rao TN, Wagers AJ, et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10:96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- 8.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 9.Kim JG, Armstrong RC, v Agoston D, Robinsky A, Wiese C, Nagle J, et al. Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J Neurosci Res 1997, 50: 272–290. [DOI] [PubMed]
- 10.Stevens SJ, van Ravenswaaij-Arts CM, Janssen JW, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T, et al. MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet A. 2011;155A:2739–2745. doi: 10.1002/ajmg.a.34274. [DOI] [PubMed] [Google Scholar]
- 11.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM. Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr Genet. 2012;22:206–209. doi: 10.1097/YPG.0b013e328353ae3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12. doi: 10.1007/s00439-009-0703-0. [DOI] [PubMed] [Google Scholar]
- 15.Regenold WT, Phatak P, Marano CM, Gearhart L, Viens CH, Hisley KC. Myelin staining of deep white matter in the dorsolateral prefrontal cortex in schizophrenia, bipolar disorder, and unipolar major depression. Psychiatry Res. 2007;151:179–188. doi: 10.1016/j.psychres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas CR, Kriegstein AR. Regenerative medicine: Cell reprogramming gets direct. Nature. 2010;463:1031–1032. doi: 10.1038/4631031a. [DOI] [PubMed] [Google Scholar]
- 17.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mall M, Kareta MS, Chanda S, Ahlenius H, Perotti N, Zhou B, et al. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 2017;544:245–249. doi: 10.1038/nature21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushita F, Kameyama T, Kadokawa Y, Marunouchi T. Spatiotemporal expression pattern of Myt/NZF family zinc finger transcription factors during mouse nervous system development. Dev Dyn. 2014;243:588–600. doi: 10.1002/dvdy.24091. [DOI] [PubMed] [Google Scholar]
- 23.Yu Z, Sun D, Feng J, Tan W, Fang X, Zhao M, et al. MSX3 Switches Microglia Polarization and Protects from Inflammation-Induced Demyelination. J Neurosci. 2015;35:6350–6365. doi: 10.1523/JNEUROSCI.2468-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Xiao L, Liu X, Yang W, Shen W, Hu C, et al. A functional role of NMDA receptor in regulating the differentiation of oligodendrocyte precursor cells and remyelination. Glia. 2013;61:732–749. doi: 10.1002/glia.22469. [DOI] [PubMed] [Google Scholar]
- 25.Xiao L, Guo D, Hu C, Shen W, Shan L, Li C, et al. Diosgenin promotes oligodendrocyte progenitor cell differentiation through estrogen receptor-mediated ERK1/2 activation to accelerate remyelination. Glia. 2012;60:1037–1052. doi: 10.1002/glia.22333. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Watanabe M, Nishiyama A. Optimization of oligodendrocyte progenitor cell culture method for enhanced survival. J Neurosci Methods. 2005;149:50–56. doi: 10.1016/j.jneumeth.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Bottenstein JE, Hunter SF, Seidel M. CNS neuronal cell line-derived factors regulate gliogenesis in neonatal rat brain cultures. J Neurosci Res. 1988;20:291–303. doi: 10.1002/jnr.490200303. [DOI] [PubMed] [Google Scholar]
- 28.Feng JF, Gao XF, Pu YY, Burnstock G, Xiang Z, He C. P2X7 receptors and Fyn kinase mediate ATP-induced oligodendrocyte progenitor cell migration. Purinergic Signal. 2015;11:361–369. doi: 10.1007/s11302-015-9458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang X, Sun D, Wang Z, Yu Z, Liu W, Pu Y, et al. MiR-30a positively regulates the inflammatory response of microglia in experimental autoimmune encephalomyelitis. Neurosci Bull. 2017;33:603–615. doi: 10.1007/s12264-017-0153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–467. doi: 10.1016/S0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhao C, Ma D, Zawadzka M, Fancy SP, Elis-Williams L, Bouvier G, et al. Sox2 Sustains Recruitment of Oligodendrocyte Progenitor Cells following CNS Demyelination and Primes Them for Differentiation during Remyelination. J Neurosci. 2015;35:11482–11499. doi: 10.1523/JNEUROSCI.3655-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Y, Yu VC, Buchholz F, O’Connell S, Rhodes SJ, Candeloro C, et al. A novel family of Cys-Cys, His-Cys zinc finger transcription factors expressed in developing nervous system and pituitary gland. J Biol Chem. 1996;271:10723–10730. doi: 10.1074/jbc.271.18.10723. [DOI] [PubMed] [Google Scholar]
- 33.Weiner JA, Chun J. Png-1, a nervous system-specific zinc finger gene, identifies regions containing postmitotic neurons during mammalian embryonic development. J Comp Neurol. 1997;381:130–142. doi: 10.1002/(SICI)1096-9861(19970505)381:2<130::AID-CNE2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Yee KS, Yu VC. Isolation and characterization of a novel member of the neural zinc finger factor/myelin transcription factor family with transcriptional repression activity. J Biol Chem. 1998;273:5366–5374. doi: 10.1074/jbc.273.9.5366. [DOI] [PubMed] [Google Scholar]
- 35.Jandrig B, Seitz S, Hinzmann B, Arnold W, Micheel B, Koelble K, et al. ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2. Oncogene. 2004;23:9295–9302. doi: 10.1038/sj.onc.1208131. [DOI] [PubMed] [Google Scholar]
- 36.Vasconcelos FF, Sessa A, Laranjeira C, Raposo AA, Teixeira V, Hagey DW, et al. MyT1 Counteracts the Neural Progenitor Program to Promote Vertebrate Neurogenesis. Cell Rep. 2016;17:469–483. doi: 10.1016/j.celrep.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Besold AN, Amick DL, Michel SL. A role for hydrogen bonding in DNA recognition by the non-classical CCHHC type zinc finger, NZF-1. Mol Biosyst. 2014;10:1753–1756. doi: 10.1039/C4MB00246F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JG, Hudson LD. Novel member of the zinc finger superfamily: A C2-HC finger that recognizes a glia-specific gene. Mol Cell Biol. 1992;12:5632–5639. doi: 10.1128/MCB.12.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vana AC, Lucchinetti CF, Le TQ, Armstrong RC. Myelin transcription factor 1 (Myt1) expression in demyelinated lesions of rodent and human CNS. Glia. 2007;55:687–697. doi: 10.1002/glia.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol Cell Neurosci. 2004;25:111–123. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Yang W, Xiao L, Li C, Liu X, Liu M, Shao Q, et al. TIP30 inhibits oligodendrocyte precursor cell differentiation via cytoplasmic sequestration of Olig1. Glia. 2015;63:684–698. doi: 10.1002/glia.22778. [DOI] [PubMed] [Google Scholar]
- 42.Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- 43.Kitada M, Rowitch DH. Transcription factor co-expression patterns indicate heterogeneity of oligodendroglial subpopulations in adult spinal cord. Glia. 2006;54:35–46. doi: 10.1002/glia.20354. [DOI] [PubMed] [Google Scholar]
- 44.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.