Abstract
Background
Kyasanur Forest Disease (KFD), a tick borne flavivirus, which was earlier endemic to Karnataka state, India, has been confirmed and detected from neighboring states of Tamil Nadu, Maharashtra, Goa and Kerala states in India. Increased human and vector surveillance therefore becomes essential for the identification of KFD affected regions and control of further spread of the disease. Currently, available KFD detection assays include realtime RT-PCR and nested RT-PCR assays. Here we describe the development of a sensitive single step RT-PCR assay for the detection of KFD viral RNA. This can be easily used in any BSL-2 laboratory for screening of KFD suspected cases or for differential diagnosis of viral hemorrhagic fever panel.
Method
Three primer sets were designed and checked for sensitivity using known dilutions of KFD viral RNA (Ranging from 106 copies to 10 copies). The primer set (2) was found to be most sensitive was selected and tested for specificity for Kyasanur forest disease virus (KFDV) by testing against zika, dengue, chikungunya, crimean congo hemorrhagic fever (CCHF), yellow fever, japanese encephalitis (JE) and west nile viruses. A total of 104 samples (human, monkey and tick positive and negative samples) were tested using this assay.
Result
No false positive or false negative results were seen for human, monkey or tick samples. The assay was specific for KFD and could detect upto 100 copies of KFD viral RNA.
Discussion and conclusion
The previously published sensitive real time RT-PCR assay requires higher cost in terms of reagents and machine setup and technical expertise has been the primary reason for development of this assay. A single step RT-PCR is relatively easy to perform and more cost effective than real time RT-PCR in smaller setups in the absence of Biosafety Level-3 facility. This study reports the development and optimization of single step RT-PCR assay which is more sensitive and less time-consuming than nested RT-PCR and cost effective for rapid diagnosis of KFD viral RNA.
Keywords: Infectious disease, Molecular biology, Virology
1. Introduction
The recent years have witnessed the emergence of Kyasanur Forest Disease zoonotic disease (KFD) as an causing agent for human suffering and monkey death in India in states of Maharashtra, Kerala, Goa and Tamil Nadu [1, 2, 3, 4].
The presence of KFD was reported from; Bandipur National Park [2], Chamarajnagar district, Karnataka State [5], Mudumalai Tiger Reserve, Tamil Nadu State [5], Wayanad district, Kerala State, Malappuram district of Kerala State [1], Pali village, Sattari taluka, Goa state [6] and more than 50 cases from Sindhudurg district, Maharashtra state, India. The disease was earlier restricted to a few districts of Karnataka state, India [7, 8, 9].
KFD is transmitted by bite of KFD virus infested ticks and nymphs belonging to Haemaphysalis species. Persons visiting forest areas for collection of firewood, grass and other forest products, and those living in close proximity of forests are probable to be affected by KFDV [10]. KFD is characterized by clinical symptoms of chills, frontal headache, body ache, and high fever for 5–12 days, and a case-fatality rate >30% [11]. KFDV viremia lasts for 12–13 days of illness and probably longer after infection. The initial symptoms of KFD are overlapping with other viral diseases like dengue which are also endemic in India and therefore differential laboratory diagnosis of the disease plays a vital role in identifying the causative etiological agent and further resulting in channelizing the measures for prevention of spread of the disease. The increased region of endemicity has resulted in the need to identify field-friendly KFDV detection assays that can be used to survey for the presence of KFDV in humans, monkeys, ticks, and other probable reservoirs/hosts of the virus.
Currently the methods of laboratory diagnosis of KFDV include real time RT-PCR assay, nested RT-PCR assay, anti-KFD IgM and anti-KFD IgG ELISAs [12]. None of the diagnostic assays are currently available commercially. In Indian setting many laboratories cannot afford to have trained manpower for real-time PCR machine and reagents while RT-PCR is traditionally performed in many sectors. In the current study, we describe an indigenously developed single step RT-PCR assay for the detection of KFDV RNA. The assay is efficient, fast, sensitive and specific to KFD and can be used for any kind of clinical samples.
2. Materials and method
2.1. Ethics statement
National Institute of Virology (NIV) Pune has been involved in the investigations of viral disease outbreaks of human including zoonosis in India. Institutional Human Ethical Committee, NIV Pune was informed where the human serum samples were used and institutional animal ethical committee in case of monkey samples (Project ID: MCL-1304). All study participants provided informed consent. The consent was in written format, both in English and local language (Marathi). All the records analyzed were anonymized. Every sample was registered in the central registry of the institute and allotted a NIV number, which was used throughout the study. Samples of referred dead monkeys were in this study and therefore do not fall under the purview of ethical consent.
2.2. Preparation of viral RNA, primer designing and standardization of RT-PCR assay
KFD viral RNA extracted from tissue culture isolate (Isolate number: P9605, Genbank Accession number: EU293327) [13] was used for optimization of the One-step RT PCR assay. Viral RNA was extracted by using Trizol (Invitrogen, Life Technologies) followed by column purification by the Qiagen viral RNA isolation kit (Qiagen, USA). PCR amplification was undertaken using superscript III one-step RT-PCR system with platinum Taq high fidelity kit (Invitrogen, Life Technologies). The extracted RNA was used to set up diagnostic RT-PCR with three sets of primers from the NS5 region as this region is highly conserved among Flaviviruses [14]. The sequences of different strains of the KFDV NS-5 gene were aligned and care was taken to distinguish the primer sets from other flaviviruses, like tick borne encephalitis (TBE), japanese encephalitis (JE), west nile (WN), alkhurma and dengue (DEN) viruses.
The designed primer sets used in study are shown in Table 1. The reaction conditions used were: 50 °C for 30 minutes, 94 °C for 5 minutes followed by 20 cycles of 94 °C for 30 seconds, 48 °C for 30 seconds, 68 °C for 30 seconds, and 20 cycles of 94 °C for 30 seconds, 51 °C for 30 seconds. These conditions were followed with a final extension at 68 °C for 10 minutes. The primer concentrations and annealing temperatures were standardized according to standard procedure. Standardization of annealing temperature was done after setting up temperature gradients ranging from 45 °C to 55 °C. Primer concentrations ranging from 5 picomoles of each primer per reaction to 20 picomoles of each primer per reaction were tested. Varying concentrations of MgSO4 i.e., 0.5mM, 1mM, 1.5mM and 2mM additional MgSO4 per reaction provided along with the kit were also tested. 5μl of viral RNA was added to each reaction.
Table 1.
Primer sets used for standardization of the assay.
| Set | Primer Orientation | Primer Name | Primer position | Primer Sequence | Product Size | Reference |
|---|---|---|---|---|---|---|
| Set 1 | Forward | KFD_9090_F | 9090–9110 | CCGAGAAGCAGTGGAGGACC | 260bp | Designed during this study |
| Reverse | KFD_9350_R | 9324–9350 | GCCATCCCAGGTAGTTAAGACTGGTC | |||
| Set 2 | Forward | KFD_8610_F | 8610–8633 | ACTGGCACAGCGTGTGTGGTACT | 330bp | |
| Reverse | KFD_8940_R | 8918–8940 | CCCTCATGATGATCTTGGTTCC | |||
| Set 3 | Forward | KFD_7800_F | 7800–7818 | CGCAGAGGAGGTGCCGAG | 360bp | |
| Reverse | KFD_8160_R | 8141–8160 | AGTCTTGGGACCTCATGGCC | |||
| Real time RT-PCR primers | Forward | KFD NS5F | 548–567 | TGGAAGCCTGGCTGAAAGAG | 63bp | Mourya et al 2012 [12] |
| Reverse | KFD NS5R | 611–592 | TCATCCCCACTGACCAGCAT | |||
| Probe | KFD NS 5P | 569–590 | ATGGAGAGGAGCGCCTGACCCG | |||
| Nested RT-PCR | Outer forward | KFDNS5 3S | 8896–8917 | GTCAGATGAACAAAATCGCTGG | 756bp | Mourya et al 2012 [12] |
| Outer reverse | KFDNS5 3R | 9632–9651 | TCATCCCCACTGACCAGCAT | |||
| Inner forward | KFDNS5 4S | 9217–9235 | GAAGAAGCTGTCCGAACTC | 355bp | ||
| Inner reverse | KFDNS5 4R | 9554–9572 | GGTCCTGTGAGTCAGATGG |
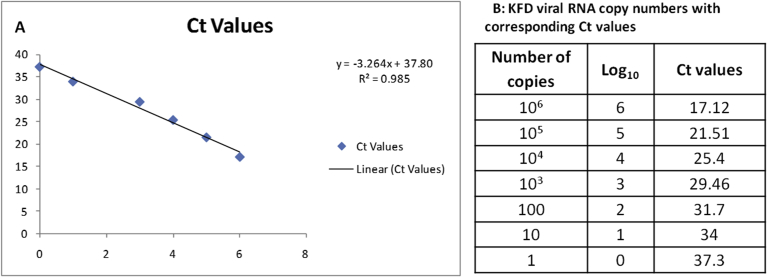
For determination of copy number, KFD viral RNA of known copy number (106 copies) was 10-fold serially diluted. The serially diluted viral RNA was used to set up real time RT-PCR assay described by Mourya et al., 2012 [12] to determine Ct values corresponding to copy numbers. The Ct values thus obtained were plotted against the log of viral RNA copy numbers (10-fold serially diluted). The graph thus obtained along with the Ct values is described in Fig. 1.
Fig. 1.
A: Standard curve for the quantitation of KFD viral RNA copy number. The RNA used for serial dilutions was quantitated using the generated equation from the standard curve. B: Table depicting copies of KFD viral RNA and their corresponding Ct values that were used to generate Fig. 1A.
2.3. Testing specificity and sensitivity of the assay
For testing sensitivity of the assay, known copy numbers KFD viral RNA stock was serially diluted from 106 copies of RNA to one copy of RNA per reaction in distilled water. The sensitivity of the designed assay was compared with that of published real time qRT-PCR and nested RT-PCR assays [12]. The copy number of KFD viral RNA was calculated based on a standard curve described in Fig. 1.
The primers were checked for specificity using zika (Strain MR766, titre: 105.5 plaque forming units (pfu)/ml), dengue (strains 16007; titre: 1.4 × 106 pfu/ml, 803347; titre: 1.4 × 105 pfu/ml, 059826; titre: 1.3 × 105 pfu/ml and 642069; titre: 4.2 × 106 pfu/ml for dengue virus strain 1, 2, 3 and 4 respectively), chikungunya (Strain: 61573; titre: 1.9 × 1012 pfu/ml), CCHF (Strain 11704, 2.5 × 105 copies of viral RNA/ml), yellow fever (strain 17D; titre: 4.2 × 109 pfu/ml), JE (Strains: 9117857; titre: 1.6 × 106 pfu/ml and 733913; titre: 1.1 × 106 pfu/ml) and WN (strains: 22672, titre: 2.1 × 108 pfu/ml and 80245, titre: 1.2 × 107 pfu/ml) viral RNAs. KFD viral RNA isolated from tissue culture was used as positive control. A total of 104 samples (36 KFDV positive samples (22 human, 9 monkey and 5 tick samples) and 68 KFDV negative samples (44 human, 3 monkey and 21 tick samples) were tested using this assay. Human and tick samples used during this study were known positive and negative samples included from Sindhudurg district of Maharashtra State, India. Monkey samples used in this study were received from Sattari taluka, Goa State, India. All the samples included in this study had been previously screened for KFD viral RNA using real-time RT-PCR described previously [12].
3. Results
3.1. Preparation of viral RNA, primer designing and standardization of RT-PCR assay
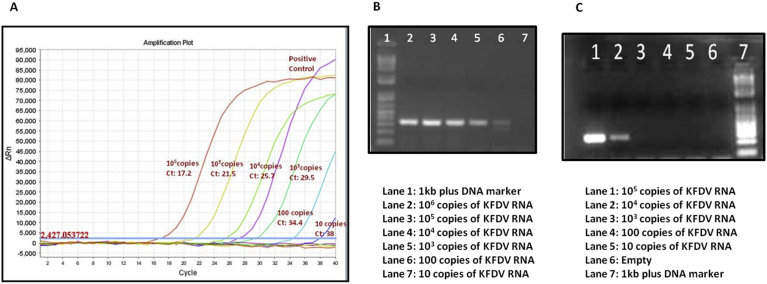
The primers from set 1, 2 and set 3 could detect down to 105, 102 and 105 copies of KFD viral RNA respectively, thereby indicating that set 2 was the most sensitive and could be used for further determining specificity of the assay. The single step RT-PCR assay using primer set 2 and was found to be more sensitive than previously published nested RT-PCR (sensitivity: Upto 104 copies of viral RNA) and 10 fold less sensitive than real time RT-PCR assay (10 copies of viral RNA) (Fig. 2). Both the nested RT-PCR assay and real time RT-PCR assay have been described by Mourya et al, 2012 [12]. The assay was found to work optimally at a primer concentration of 20 picomoles/reaction (Reaction volume: 50 μl) and an annealing temperature of 48 °C for first 20 cycles followed by 51 °C for the next 20 cycles. Additional MgSO4 other than that already present in the 2X reaction buffer provided in the kit (superscript III one-step RT-PCR system with platinum Taq high fidelity kit (Invitrogen, Life Technologies)) was not required in the optimized assay. RNA input volume was 5 μl.
Fig. 2.
Comparison of sensitivity of Real time RT-PCR (A), Single step RT-PCR (B), and nested RT-PCR (C): 106 copies of KFD viral RNA has been serially 10-fold diluted. Equal volumes of dilutions of KFDV RNA thus prepared have been used to perform the three assays.
3.2. Testing specificity and sensitivity of the assay
The assay could specifically detect KFD viral RNA and showed no non-specific PCR amplification with other representative flaviviruses namely, zika, dengue, chikungunya, CCHF, yellow fever, japanese encephalitis and west nile viral RNAs.
A total of 36 KFDV positive samples (22 human, 9 monkey tissues (Liver, heart and spleen tissues) and 5 tick samples, Ct values ranging from 22 to 34, corresponding to viral RNA copy number range of 105 to 10 copies (Fig. 2) and 68 KFDV negative samples (44 human, 3 monkey and 21 tick samples) were tested using the designed assay. The results obtained were synchronized with those obtained with previously published real-time RT-PCR assay. No false positive or false negative results were observed for human, monkey or tick samples (Table 2).
Table 2.
Comparative results of samples tested by single step RT-PCR and real time RT-PCR.
| Total number of samples tested | Number of samples concurrence by RT-PCR for positive and negative | Range Ct value of RT-PCR positive samples | ||
|---|---|---|---|---|
| Human | KFD Positive | 22 | 22 | 22–34 |
| KFD Negative | 44 | 44 | No Ct | |
| Monkey | KFD Positive | 9 | 9 | 22–34 |
| KFD Negative | 3 | 3 | No Ct | |
| Tick | KFD Positive | 5 | 5 | 22–34 |
| KFD Negative | 21 | 21 | No Ct | |
| Total number of samples tested: 36 Positive, 68 Negative (Total: 104) | ||||
4. Discussion
With increased awareness and constant support of Integrated diseases surveillance program of Ministry of Health and family welfare, Government of India, enhanced networking of laboratory capacity and improving economy in India, a focus on viral hemorrhagic fever detection is increased, especially for KFD in newer regions of Maharashtra, Karnataka, Kerala, Goa and Tamil Nadu [4] states, it becomes imperative to develop a tool for early detection of KFD infection, thereby curtailing the spread of disease. With the increasing need for surveillance in the light of detection of KFDV from five Indian states there is a necessity to develop a cost effective assay which can be easily performed even in smaller laboratories and field setups. The simultaneous presence of other viral endemic diseases like dengue and chikungunya in India with overlapping seasonality with KFD emphasizes the necessity of differential diagnosis of the disease causing etiological agent. The field friendly anti-KFDV IgM or anti-dengue IgM ELISA cannot be used for differential diagnosis in the first few days as the IgM antibodies can be detected after an interval of at least four days after the onset of these diseases. We therefore need to emphasize on a user-friendly, cost-effective and reliable assay for the detection of KFD viral RNA during the acute phase of disease.
Though KFD is currently endemic in many states of India its spread indicates that susceptible human and tick populations from other countries also need to be surveyed. Isolation of KFDV from a febrile patient in Yuanan province of China supports this concern [15]. The concern is strengthened by isolation of Alkhurma virus, a close variant of KFDV from Makkah region of Saudi Arabia during 1994–1995 [16, 17, 18].
The present assay (detecting upto 100 copies of viral RNA) is sensitive compared to nested RT-PCR assay (104 copies of viral RNA) and nearly as sensitive as real-time RT-PCR assay (10 copies of viral RNA). Due to its single step nature contamination issue will be avoided in the laboratory and large number of samples can be simultaneously processed. The availability of a sensitive real-time RT-PCR assay has not led to its implementation in wide range of laboratories due to its greater cost in terms of reagents and machine setup. An RT-PCR is relatively easy to perform and cost effective than real time RT-PCR in smaller setups where quantitative estimation of viral RNA copy number is not required. Another advantage of the RT-PCR assay in comparison to realtime RT-PCR assay is that the amplicon obtained after RT-PCR amplification can be used for sequencing and Phylogenetic analysis for conclusive confirmation of positivity of the clinical sample. In the absence of Biosafety Level 3 facility in smaller laboratories, detection of KFD viral RNA can be performed after inactivating the patient/monkey/ticks sample with phenol, or its variants like TRIzol LS Reagent (Invitrogen Corp., Carlsbad, CA) [19]. The current assay would be user friendly, sensitive, specific, cost effective and less time consuming than nested RT-PCR. This assay would be useful not only in India, but also in adjoining areas and other countries where doubt of tick borne viral hemorrhagic fever exist and can be used for differential diagnosis.
Declarations
Author contribution statement
Gouri Chaubal: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Prasad Sarkale: Contributed reagents, materials, analysis tools or data.
Pravin Kore: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Pragya Yadav: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work is funded by Indian Council of Medical Research, National Institute of Virology, Pune, Maharashtra, India under the Intramural project Reagent preparation for highly infectious diseases.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to acknowledge the valuable contribution of staff of Maximum Containment Laboratory, NIV, Pune. In particular, we would like to thank Dr. Anita Shete, Scientist D, Dr. Deepak Patil, Scientist B, Mr. Rajen Lakra, Technican Mr. Kumar Baghmare, Technician and Mrs. Divya Bhattad, Technician for providing for the excellent technical support during the study.
References
- 1.Tandale B.V., Balakrishnan A., Yadav P.D., Marja N., Mourya D.T. New focus of Kyasanur Forest disease virus activity in a tribal area in Kerala, India, in 2014. Infect. Dis. Poverty. 2015;4:13. doi: 10.1186/s40249-015-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mourya D.T., Yadav P.D., Sandhya V.K., Reddy S. Spread of Kyasanur forest disease, Bandipur tiger Reserve, India, 2012–2013. Emerg. Infect. Dis. 2013;19(9) doi: 10.3201/eid1909.121884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awate P., Yadav P., Patil D., Shete A., Kumar V., Kore P., Dolare J., Deshpande M., Bagde S., Sapkal G., Gurav Y., Mourya D.T. Outbreak of Kyasanur forest disease (monkey fever) in Sindhudurg, Maharashtra state, India. J. Infect. 2016;72(6):759–761. doi: 10.1016/j.jinf.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Murhekar M.V., Kasabi G.S., Mehendale S.M., Mourya D.T., Yadav P.D., Tandale B.V. On the transmission pattern of Kyasanur Forest disease (KFD) in India. Infect. Dis. Poverty. 2015;4:37. doi: 10.1186/s40249-015-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mourya D.T., Yadav P.D., Patil D.Y. Highly infectious tick borne viral diseases: Kyasanur forest disease and Crimean–Congo hemorrhagic fever in India. WHO South–East Asia J. Public Health. 2014;3(1):8–21. doi: 10.4103/2224-3151.206890. 8. [DOI] [PubMed] [Google Scholar]
- 6.Department of Information & Technology, Government of Goa. Fever Cases at Pali, Confirmed as Kyasanur Forest Disease. Available at: https://www.goa.gov.in/pdf/FEVER%20CASES%20AT%20PALI.pdf.
- 7.Pavri K. Clinical, clinicopathologic, and hematologic features of Kyasanur forest disease. Rev. Infect. Dis. 1989;11:S854–S859. doi: 10.1093/clinids/11.supplement_4.s854. [DOI] [PubMed] [Google Scholar]
- 8.Pattnaik P. Kyasanur forest disease: an epidemiological view in India. Rev. Med. Virol. 2006;16:151–165. doi: 10.1002/rmv.495. [DOI] [PubMed] [Google Scholar]
- 9.Holbrook M.R. Kyasanur forest disease. Antivir. Res. 2012;96:353–362. doi: 10.1016/j.antiviral.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Work T.H., Trapido H. Summary of preliminary report of investigations of the virus research centre on an epidemic disease affecting forest villagers and wild monkeys in Shimoga district, Mysore. Indian J. Med. Sci. 1957;11:340–341. [PubMed] [Google Scholar]
- 11.Banerjee K. Kyasanur forest disease. In: Monath T.P., editor. Arboviruses Epidemiology and Ecology. Boca Raton (FL) CRC Press; 1988. pp. 93–116. [Google Scholar]
- 12.Mourya D.T., Yadav P.D., Mehla R., Barde P.V., Yergolkar P.N., Kumar S.R.P. Diagnosis of Kyasanur forest disease by nested RT-PCR, real-time RT-PCR and IgM Capture ELISA. J. Virol. Method. 2012;186:49–54. doi: 10.1016/j.jviromet.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Mehla R., Kumar S.R.P., Yadav P., Barde P.V., Yergolkar P.N., Erickson B.R., Carroll S.A., Mishra A.C., Nichol S.T., Mourya D.T. Recent ancestry of Kyasanur forest disease virus. Emerg. Infect. Dis. 2009;15(9):1431–1437. doi: 10.3201/eid1509.080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiel H.-J., Collett M.S., Gould E.A., Heinz F.X., Meyers G., Purcell R.H., Rice C.M., Houghton M. Flaviviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy—Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego: 2005. pp. 981–998. [Google Scholar]
- 15.Wang J., Zhang H., Fu S., Wang H., Ni D., Nasci R., Tang Q., Liang G. Isolation of Kyasanur forest disease virus from febrile patient, Yunnan, China. Emerg. Infect. Dis. 2009;15:326–328. doi: 10.3201/eid1502.080979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blow J.A., Dohm D.J., Negley D.L., Mores C.N. Virus inactivation by nucleic acid extraction reagents. J. Virol. Methods. 2004;119(2):195–198. doi: 10.1016/j.jviromet.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Zaki A.M. Isolation of a flavivirus related to the tick-borne encephalitis complex from human cases in Saudi Arabia. Trans. R. Soc. Trop. Med. Hyg. 1997;91:179–181. doi: 10.1016/s0035-9203(97)90215-7. [DOI] [PubMed] [Google Scholar]
- 18.Qattan I., Akbar N., Afif H., Abu Azmah S., Al-Khateeb T., Zaki Al. A novel flavivirus: Makkah Region, 1994–1996. Saudi Epidemiol. Bull. 1996;3:1–3. [Google Scholar]
- 19.Charrel R.N., Zaki A.M., Attoui H., Fakeeh M., Billoir F., Yousef A.I. Complete coding sequence of the Alkhurma virus, a tick-borne flavivirus causing severe hemorrhagic fever in humans in Saudi Arabia. Biochem. Biophys. Res. Commun. 2001;287:455–461. doi: 10.1006/bbrc.2001.5610. [DOI] [PubMed] [Google Scholar]