Abstract
Protein–protein interactions (PPIs) are an important category of putative drug targets. Improvements in high-throughput screening (HTS) have significantly accelerated the discovery of inhibitors for some categories of PPIs. However, methods suitable for screening multiprotein complexes (e.g. those composed of three or more different components) have been slower to emerge. Here, we explored an approach that uses reconstituted multiprotein complexes (RMPCs). As a model system, we chose heat shock protein 70 (Hsp70), which is an ATP-dependent molecular chaperone that interacts with co-chaperones, including DnaJA2 and BAG2. The PPIs between Hsp70 and its co-chaperones stimulate nucleotide cycling. Thus, to re-create this ternary protein system, we combined purified human Hsp70 with DnaJA2 and BAG2 and then screened 100,000 diverse compounds for those that inhibited co-chaperone–stimulated ATPase activity. This HTS campaign yielded two compounds with promising inhibitory activity. Interestingly, one inhibited the PPI between Hsp70 and DnaJA2, whereas the other seemed to inhibit the Hsp70–BAG2 complex. Using secondary assays, we found that both compounds inhibited the PPIs through binding to allosteric sites on Hsp70, but neither affected Hsp70's intrinsic ATPase activity. Our RMPC approach expands the toolbox of biochemical HTS methods available for studying difficult-to-target PPIs in multiprotein complexes. The results may also provide a starting point for new chemical probes of the Hsp70 system.
Keywords: high-throughput screening (HTS); protein complex; protein–protein interaction; chemical biology; inhibitor; inhibition mechanism; molecular chaperone; Hsp70, heat shock protein
Introduction
Protein–protein interactions (PPIs)2 are emerging as targets for both drug discovery and chemical biology (1). Indeed, 20% of the total number of PPI inhibitors have been reported in the last 5 years alone (2). This accelerated rate of discovery has been fueled, in part, by improvements in the design of chemical libraries (e.g. fragment-based collections and macrocycles) and the development of specialized HTS methodologies that are suitable for studying PPIs (e.g. Alpha-Lisa and split-GFP). However, not all PPIs are proving amenable to these solutions. PPIs vary dramatically in their relative affinity (KD), polarity, and buried surface area (BSA) (3). Based on retrospective analyses of reported successes and failures, it seems that the subset of “difficult” PPIs are ones that (a) are of weak affinity and/or (b) occur over large, shallow interfaces (4). Protein contacts with these features seem to be relatively difficult to inhibit because they lack good binding sites for drug-like (e.g. less than 500 Da) small molecules. In addition, difficult PPIs are commonly found within multiprotein complexes, which are composed of three or more components. This feature adds to the complexity of the system because, even if one can identify an inhibitor of one difficult PPI, it is often unclear how that event might influence binding to other partners. Thus, it is becoming clear that next-generation HTS approaches are needed to tackle these issues.
To explore these questions, we focused on the molecular chaperone, heat shock protein 70 (Hsp70), as a model system. Hsp70 is composed of an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD). The hydrolysis of ATP in the NBD causes a dramatic conformational change that powers the chaperone cycle (5). This process is tightly regulated by binding of Hsp70 to its co-chaperones (6). The major classes of Hsp70 co-chaperones are the J-domain proteins (also called Hsp40s or DNAJs), which stimulate the hydrolysis step, and the nucleotide exchange factors (NEFs), which bind a distinct region of Hsp70 and promote ADP release. Together, these two co-chaperones have been observed to increase Hsp70's steady-state ATP hydrolysis by nearly 200-fold (7). For these reasons, the Hsp70 system is a well-studied multiprotein machine, and we reasoned that it would also be a good one for studying difficult PPIs. Specifically, the interaction of Hsp70 with J proteins is known to be weak (micromolar) and to involve a large BSA that is highly polar (8). Similarly, the interaction of Hsp70 with the BAG family of NEFs is relatively tight (mid-nanomolar), but the contact area is large and occurs over multiple subdomains of Hsp70's NBD (9). Thus, this ternary system is emblematic of modern, multiprotein drug targets and includes multiple categories of difficult PPIs.
We envisioned a strategy in which measuring the ATP cycling of this three-protein system might provide a readout for each of the PPIs. The intrinsic hydrolysis activity of Hsp70 in the absence of its co-chaperones is slow (5 × 10−4 s−1) (6, 7), such that most of the measured signal would be expected to emerge as a direct result of co-chaperone interactions. In support of this idea, screens performed against a combination of the prokaryotic Hsp70 (DnaK) and one of its co-chaperones (DnaJ) had previously identified PPI inhibitors (10). However, it is not clear that a binary combination replicates the full conformational cycling of this complicated system. For example, Hsp70–NEF activity contributes to the kinetics of substrate binding and release, but it is only rate-limiting in the presence of J protein activity (11). Put another way, it would be expected that only a full complement of co-chaperones would sample the full range of conformational states that normally occur under physiological conditions.
Here, we report an HTS campaign using the human Hsp70 system: Hsp72 (HSPA1A), BAG2, and DnaJA2. There are ∼13 hsp70 genes, ∼45 J proteins, and 6 BAG-domain–containing NEFs in humans. From these possibilities, we chose Hsp72, DnaJA2, and BAG2 because they have been shown to be important in cancer (12–15) and because their biochemistry has been fully characterized (16). Leveraging this existing knowledge, we generated a reconstituted multiprotein complex (RMPC) of purified Hsp72, DnaJA2, and BAG2 at a carefully chosen ratio of 5:1:2.5. Then, we screened 100,000 diverse compounds for inhibitors of steady-state nucleotide turnover in a 384-well format. To our knowledge, this is the largest HTS campaign targeting Hsp70 that has been reported. From this screen, we identified multiple compounds that inhibit ATPase activity, and we focused on two promising chemical series. Pilot medicinal chemistry campaigns, combined with secondary assays, showed that one compound targeted the Hsp70–DnaJA2 complex, and the other inhibited Hsp70–BAG2. These findings suggest that RMPCs present a tractable, biochemical approach for the discovery of inhibitors of even difficult PPIs within multiprotein systems, thus adding to the “toolbox” of methods for chemical biology.
Results
High-throughput ATPase screen identifies inhibitors of Hsp72–DnaJA2–BAG2
Our search for inhibitors of PPIs within the Hsp72–DnaJA2–BAG2 system was guided by three major design ideas as follows: (a) to avoid discovery of nucleotide-competitive molecules, we saturated the cleft using high concentrations of ATP (1 mm) in the screening buffer; (b) DnaJA2 and BAG2 were added at concentrations that produced half-maximal stimulation of ATP hydrolysis to improve the sensitivity of the assay to potential inhibitors and activators (17); and (c) we combined all three proteins (Hsp72, DnaJA2, and BAG2) into the same wells to re-create native cycling conditions. Using these principles, we then chose inorganic phosphate release as the measure of steady-state ATPase activity. Specifically, we used a modification of the malachite green (MG) assay, in which MG is replaced by quinaldine red (QR), and the measurements are made in opaque white 384-well plates. This approach has been shown to enhance sensitivity more than 8-fold in 384-well format (10). Finally, for the chemical library, we selected a 100,000 diversity set from ChemDiv that is composed of lead-like molecules.
Recombinant human Hsp72, DnaJA2, and BAG2 were each purified from Escherichia coli (>95% purity, SDS-PAGE). To estimate the quality of the Hsp72 sample, we measured its intrinsic ATPase activity (∼4–6 pmol/min/μg Hsp72) and found that it compared favorably to literature values (16). DnaJA2 and BAG2 solutions were similarly characterized by titrating them into Hsp72 and measuring their ability to stimulate hydrolysis (Fig. 1A). From this characterization, we prepared a master mix that would yield final concentrations of 0.5 μm Hsp72, 0.1 μm DnaJA2, and 0.25 μm BAG2. In 384-well plates, individual test compounds from the 100,000 ChemDiv collection were dispensed to the master mix at a final concentration of ∼20–40 μm. After a short incubation, an ATP solution (1 mm final) was added to start the reaction. After 2 h, the reaction was quenched, and the QR signal was converted to a hydrolysis rate by comparison with an inorganic phosphate standard. Each screening plate included a negative control (a sample lacking Hsp72) and a positive control containing 40 μm myricetin (19). The overall Z-factor was calculated to be 0.65, and the co-efficient of variance was 9%. Finally, it is important to note that the screen was carried out in the presence of 0.01% Triton X-100 to minimize aggregator artifacts.
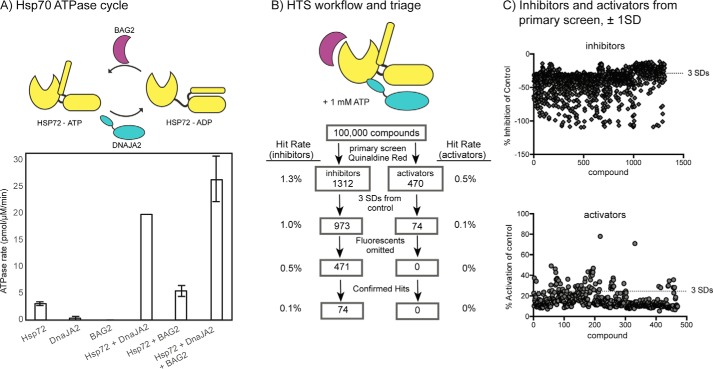
Figure 1.
High-throughput ATPase screen identifies inhibitors and activators of the Hsp72/DnaJA2/BAG2 system. A, schematic of the Hsp72 ATPase cycle, highlighting the role of the DnaJA2 and BAG2 co-chaperones. Hsp72 (0.5 μm) has a slow hydrolysis rate in the absence of DnaJA2 (0.1 μm) or BAG2 (0.25 μm). Results are the average of triplicates, and the error bars represent standard error of the mean (S.E.). B, overview of the HTS campaign. A total of 100,000 molecules were screened against the Hsp72/DnaJA2/BAG2 combination. Approximately 1.8% of the molecules had inhibitor or activator activity at ±1 S.D., and 1.1% were inhibitors or activators at ±3 S.D. After triage for fluorescent artifacts and dose-response curves (EC50 <50 μm), 74 molecules were predicted to be inhibitors. See the text and under “Materials and methods” for details. C, overview of the compounds with activity at ±1 S.D. and ±3 S.D. from the controls, highlighting the distribution of the top active molecules.
The screen revealed 1,312 molecules that appeared to inhibit the hydrolysis of ATP and 470 molecules that increased it by at least ±1 S.D. This list was further filtered to only include those compounds that showed effects at least ±3 S.D. from the control, leaving 973 inhibitors and 74 activators (Fig. 1, B and C). Although it was not the focus of this study, we were potentially interested in pursuing both inhibitors and activators. However, we were also concerned that apparent activators might be fluorescence artifacts. Accordingly, both sets of active compounds were cherry-picked and tested for intrinsic fluorescence (in the absence of protein) that might interfere with the QR assay (excitation 430 nm/emission 530 nm). At the same time, we removed any compounds that were annotated as common pan-assay hits (e.g. PAINS) and those flagged as containing reactive functionalities. Together, these triage steps reduced the number of potential inhibitors to 471 (0.5%) and removed all of the activators. The potential inhibitors were then subject to validation in eight-point dose-response curves (DRCs). Molecules with EC50 values <50 μm were then considered to be confirmed actives, resulting in 74 inhibitors (0.1% hit rate).
Compound R and compound F inhibit distinct binary combinations
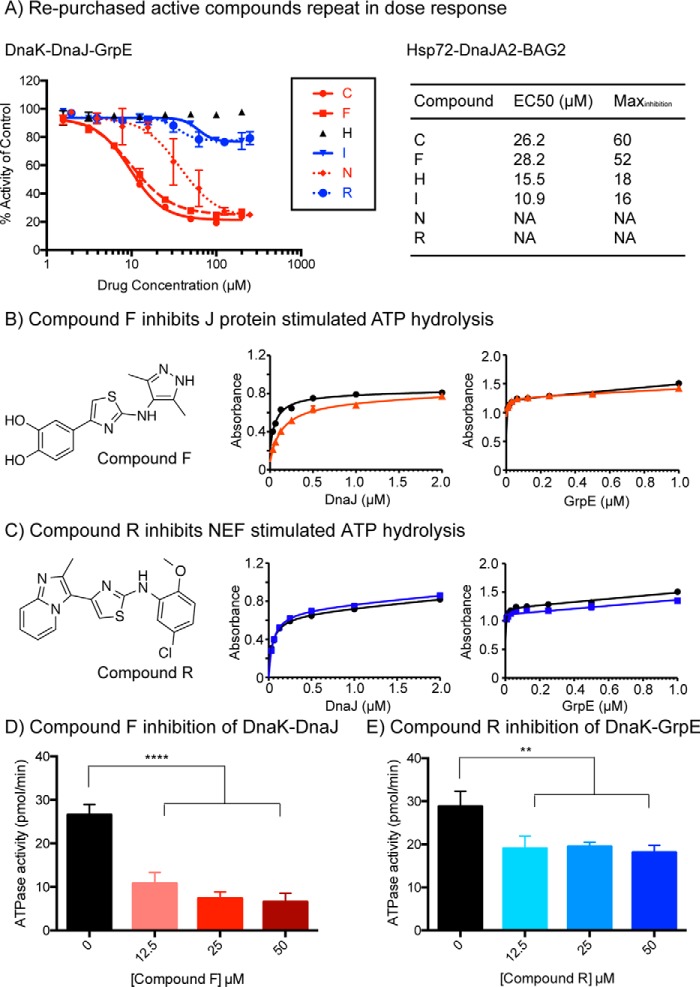
Next, the top 18 inhibitors were re-purchased from ChemDiv or ChemBridge and tested in secondary DRCs. In these assays, we used both the human Hsp72–DnaJA2–BAG2 system and the highly conserved prokaryotic orthologs DnaK–DnaJ–GrpE. We included the prokaryotic system in the secondary screening because of its faster rate of ATP turnover (20), which tends to produce more robust EC50 values. Of the 18 re-purchased compounds, five had EC50 values less than 50 μm (compounds F, C, I, N, and R) in the DnaK–DnaJ–GrpE system. Interestingly, these active molecules appeared to visually “bin” into two distinct groups, based on the shape of their inhibition curves (Fig. 2A). The first set (compounds I and R) inhibited ∼20% of the overall activity at a saturating concentration, whereas the other molecules (compounds C, F, and N) inhibited ATP turnover to a maximum of 80%. These two categories were immediately intriguing because our pre-screening characterization studies (see Fig. 1A) had shown that ∼24% (∼6 pmol/μm Hsp72/min) of the overall ATPase activity was due to stimulation by BAG2, whereas 80% (∼20 pmol/μm Hsp72/min) is produced by DnaJA2's contribution. Thus, it was compelling to imagine that the two categories of inhibitors might be targeting the Hsp70–NEF or Hsp70–J protein pairs. Qualitatively similar results were obtained with the human system (Fig. 2A).
Figure 2.
Confirmation of active molecules reveals that they are either inhibitors of J protein or NEF co-chaperones. A, top 18 active molecules were re-purchased and subject to DRCs in the prokaryotic and human Hsp70 systems. Compounds not shown were inactive or not sufficiently soluble for testing. B, compound F inhibits DnaJ-mediated stimulation but has little effect on GrpE-mediated stimulation in ATPase assay. Absorbance is measured at 620 nm. C, compound R has no activity against DnaK–DnaJ but modestly inhibits DnaK–GrpE. Results in B and C are the average of at least three experiments performed in triplicate each, and the error bars represent S.E. D, compound F inhibits a combination of DnaK and DnaJ dose-dependently at low micromolar concentrations (****, p < 0.0001). E, compound R showed significant inhibition of a combination of DnaK, DnaJ, and GrpE at low micromolar concentrations (**, p = 0.005). Results are the average of triplicate values and error bars represent S.E.
To explore this idea, we selected two representative compounds for further mechanistic studies. From the available compounds, F and R were particularly intriguing because they are both 2-aminothiazoles, yet they had very different activity profiles. Specifically, compound R inhibited 21% of the total activity of DnaK–DnaJ–GrpE, with an EC50 of 35 μm, whereas compound F inhibited 73% of the total ATP turnover, with an EC50 of 10 μm. To understand whether these molecules might target distinct binary combinations, we tested them against mixtures of either DnaK–DnaJ or DnaK–GrpE alone. Consistent with the speculation, we found that compound F inhibited the combination of DnaK and DnaJ but that it had no activity against the DnaK–GrpE system (Fig. 2B). Conversely, compound R had no activity against the DnaK–DnaJ pair but had modest activity against DnaK–GrpE (Fig. 2C). Thus, screening against the ternary complex seemed to yield at least one inhibitor of each difficult PPI.
A side product of compound F is the active molecule and it inhibits the Hsp70–DnaJA2 interaction
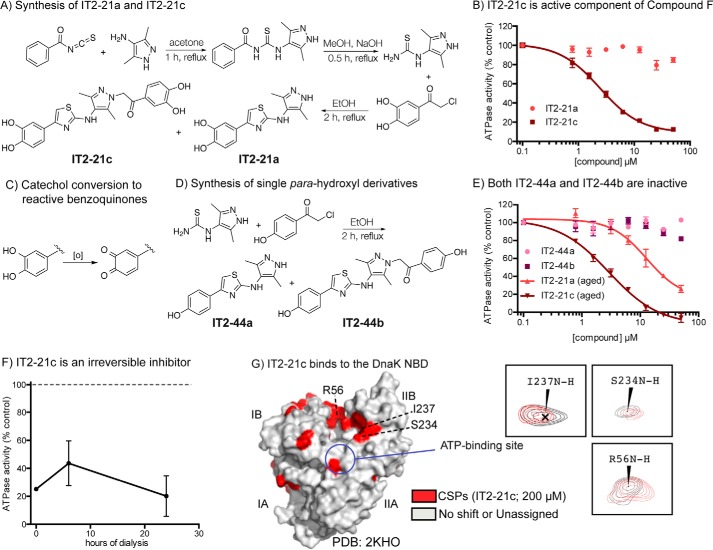
To further probe the mechanism of compound F, we first re-synthesized the molecule (now termed IT2-21a) using a known synthetic route (Fig. 3A). We were initially frustrated by the lack of activity of IT2-21a in ATPase assays, so we decided to re-evaluate the chemical identity of the authentic molecule from the screening collection. By LC-MS, we found trace amounts of a side-product featuring two catechols (IT2-21c) in the original samples (Fig. S1). We reasoned that such a side-product might be expected from the undesired nucleophilic attack of the pyrazole secondary amine onto the α carbon of the acetophenone starting material, so we separated the two products from the crude reaction mixture by HPLC. Pure (>95%) samples of both compounds were then tested in the ATPase assay. Indeed, we found that IT2-21c was the potent inhibitor, whereas the molecule that is annotated in the commercial library, IT2-21a, was inactive (Fig. 3B).
Figure 3.
Side product of compound F synthesis is an irreversible inhibitor of Hsp70 systems. A, synthesis of IT2-21a and IT2-21c by Hantzsch thiazole condensation. Compounds IT2-21c and IT2-21a were separated by HPLC. B, IT2-21c, not IT2-21a, is an inhibitor of DnaK/DnaJ/GrpE ATPase activity. Results are the average of triplicates, and error bars represent S.E. C, known oxidation of catechol rings to yield reactive benzoquinones. D, synthesis of IT2-44a and IT2-44b. Compounds were separated by HPLC. E, neither of the para-phenolic analogs were active in the ATPase assay using DnaK/DnaJ/GrpE. Results are the average of triplicates, and error bars represent S.E. “Aging” DMSO stocks of IT2-21a and IT2-21c by overnight incubation at room temperature improved their activity, consistent with an oxidative mechanism. F, IT2-21c (100 μm) was incubated with DnaK (2.5 μm) and then subjected to dialysis. Samples were removed from dialysis after 6 and 24 h, and the ATPase assay was performed with added DnaJ. Even 24 h of dialysis was not able to reverse compound activity. G, a potential binding site of IT2-21c was revealed by HSQC experiments with 15N-labeled DnaK NBD. CSPs (>0.01 ppm in 1H and/or >0.1 ppm in 15N) in the presence of IT2-21c were localized to the IB and IIB subdomains (red). See under “Materials and methods” for details. Peaks shifting for select residues are highlighted with NBD + 2% DMSO in gray and NBD + 200 μm IT2-21c in red.
With the active molecule now clear, we turned to examination of structure–activity relationships (SAR). Our initial focus was to try to eliminate the catechols, because these groups can sometimes form covalent bonds with protein nucleophiles. Briefly, it is known that catechols can be oxidized to ortho-benzoquinones, creating strong Michael acceptors (Fig. 3C). Accordingly, we synthesized IT2-44a and IT2-44b, in which the catechols of IT2-21a and IT2-21c were replaced with less reactive, para-hydroxy substitutions (Fig. 3D). We found that both IT2-44a and IT2-44b were inactive in the ATPase assay (Fig. 3E), consistent with a possible covalent mechanism. It is known that long-term storage in DMSO can exacerbate oxidation of the catechol, so we next tested “aged” DMSO stocks of IT2-21a and IT2-21c that were incubated overnight at room temperature. Consistent with the model, both samples gained activity in the ATPase assay (see Fig. 3E).
Given this information, we would expect that IT2-21c might covalently modify the target (21, 22). Indeed, IT2-21c still had an inhibitory effect on DnaK after the compound-treated protein was dialyzed for 24 h (Fig. 3F). To understand where this compound might bind, we titrated IT2-21c into 15N-labeled DnaK NBD and performed 2D NMR, HSQC experiments (Fig. S2). Plotting the chemical shift perturbations (CSPs) onto the structure of DnaK's NBD (2KHO) showed that IT2-21c seemed to bind a conserved region that is located between the IB and IIB subdomains (Fig. 3G). It is not yet clear which residue(s) is the target for covalent modification, but there are a number of possible nucleophiles, particularly lysines, in this vicinity. Interestingly, this region was previously identified as the binding site for myricetin, the positive control from our screen (19). Together, we concluded that IT2-21c was acting by a covalent mechanism and therefore not likely to be a suitable scaffold for further study. However, it binds an interesting location on the NBD, which might be exploited with structure-based drug design in the future.
Compound R analogs inhibit the Hsp70–NEF interaction
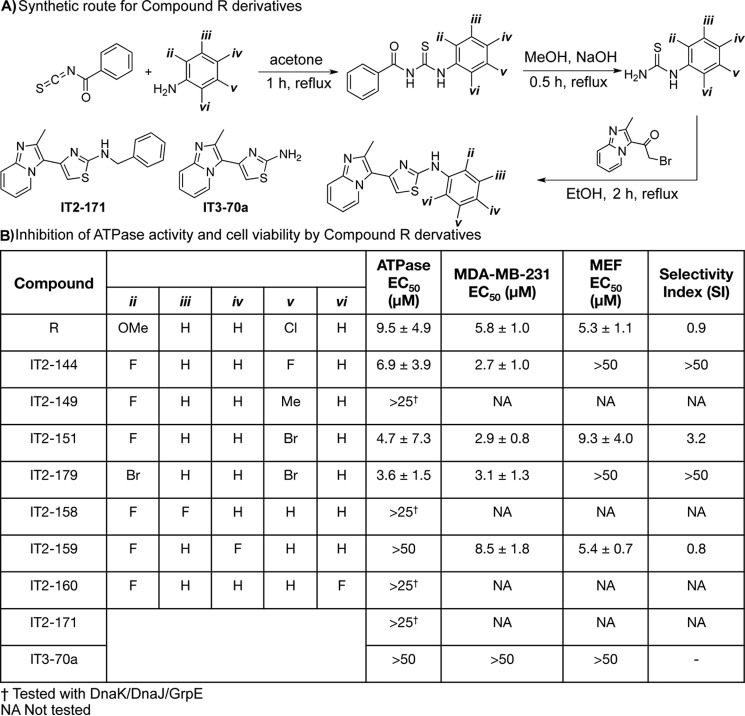
Unlike compound F, we found that compound R bound reversibly and that it lacked the chemical features to make it a potential covalent inhibitor. Thus, our exploration of compound R focused on generating analogs to better understand the pharmacophore. A total of 30 analogs were created using a previously described synthetic route that features a Hantzsch thiazole condensation (Fig. 4A) (23). These analogs largely focused on modifications of the “right” side of the molecule, as there were many commercially available anilines from which to diversify the substituted thiourea. Each of the compounds was tested for the ability to inhibit the ATPase activity of the DnaK–DnaJ–GrpE complex (Fig. S3). Then, the active molecules (<25 μm) were additionally tested against the Hsp72–DnaJA2–BAG2 combination. We did not discover any molecules that were selective for the prokaryotic or eukaryotic system, which is not surprising given the remarkably high sequence and structure conservation between these systems. From this effort, it seemed that 2,5 di-substitutions were required for activity (Fig. 4B). The importance of the 2,5 di-substitution is best exemplified by comparing IT2-144 with its regio-isomers, IT2-158, IT2-159, and IT2-160; the 2,3, 2,4, and 2,6 di-fluoro derivatives, respectively. All three of these derivatives were inactive (EC50 >25 μm), whereas the 2,5 di-fluoro regio-isomer, IT2-144, had an EC50 value of 6.9 μm. The best compound in the ATPase assay, IT2-179, replaced the fluorines for bromines, and it had an EC50 value of ∼3.6 μm against Hsp72–DnaJA2–BAG2, an approximate 3-fold improvement on compound R. Finally, removing the ring entirely (compound IT3-70a) or extending the linker to the ring in the form of a benzyl derivative (IT2-171) abolished activity, suggesting that the positioning of this ring is a key part of the pharmacophore.
Figure 4.
Synthesis and activity of compound R derivatives. A, synthesis of compound R analogs. Structures of IT2-171 and IT3-70a are shown here. B, activity of analogs in ATPase assays (Hsp72, DnaJA2, and BAG2 at 0.5, 0.1, and 0.25 μm, respectively). Results are the average of three independent experiments, each performed in triplicate ± S.D. In addition, compounds were tested for membrane permeability using cell growth assays in MDA-MB-231 breast cancer cells and normal MEFs. EC50 values in cell experiments are the result of two independent experiments, each in triplicate ± S.D. The selectivity index is the EC50 in MDA-MB-231 cells divided by the EC50 in MEF cells.
To understand possible membrane permeability, we tested the analogs in anti-proliferative MTT assays using the breast cancer cell line, MDA-MB-231. This cell line was chosen because it is known to require Hsp70 complexes for growth (13). Encouragingly, compounds active in the ATPase assays had the expected anti-proliferative activity. For example, IT2-144 had an EC50 of 2.7 μm in suppressing growth of MDA-MB-231 cells, whereas the negative control, IT3-70a, was inactive (EC50 >50 μm). As an initial test of potential selectivity for cancer cells, we also tested the analogs for effects on growth of normal mouse embryonic fibroblasts (MEFs). Multiple groups have suggested that Hsp70 is selectively required in cancer cells, creating an “addiction” to this chaperone (24). Although compound R was not selective (selectivity index ∼0.9), the more potent analogs were significantly less toxic to MEFs. For example, IT2-144 and IT2-179 had EC50 values >50 μm in these normal cells (Fig. 4B). Together, these studies suggest that compound R analogs are at least partially membrane-permeable, and they might be good starting points for Hsp70 inhibitors. It is important to note that their molecular selectivity has not been established and will require additional efforts.
Next, we wanted to understand whether compound R analogs might act on other NEFs besides BAG2. This question is important because there are six members of the BAG family in mammals, and more broadly, there are at least two other structurally distinct classes of NEFs. To explore this idea, we tested the activity of IT2-144 against a combination of Hsp70 (or DnaK) and five different, purified NEFs: human BAG1, BAG2, BAG3, Hsp105, and E. coli GrpE. The BAG family of NEFs bind to Hsp70's NBD through a conserved motif, termed the BAG domain (9). Thus, in addition to testing the three full-length BAG proteins (BAG1–3), we also measured activity against a truncated protein composed of only the BAG domain of BAG1. Hsp105 is a member of the Hsp110 class of NEFs, which share similar domain architecture with Hsp70 and bind in a “face-on” orientation (25). Finally, the prokaryotic GrpE makes contacts with DnaK in the IB and IIB subdomains, similar to the BAG proteins, but also the IA subdomain, and even parts of the SBD (26). Thus, this collection of NEFs includes members that bind Hsp70 in different locations.
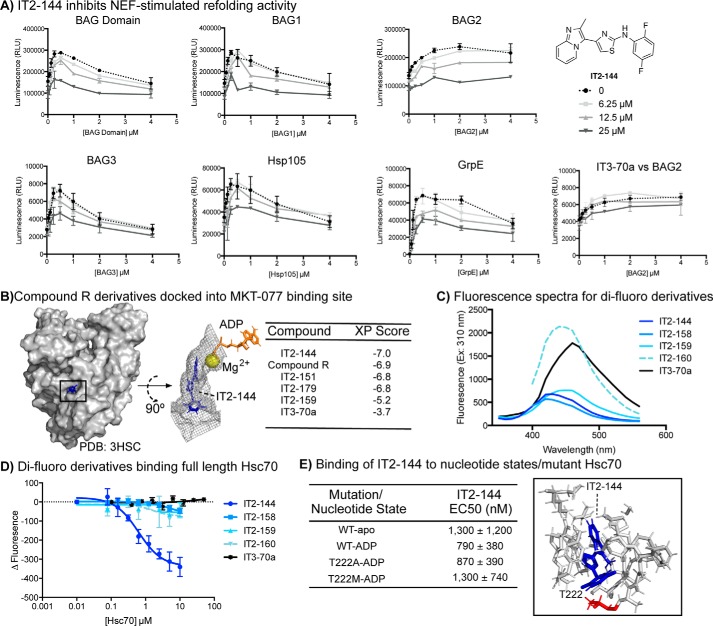
Because Hsp105 has intrinsic ATPase activity that is hard to resolve from Hsp70's in hydrolysis assays, we instead turned to a luciferase-refolding assay to estimate inhibition of the panel of NEFs. Luciferase refolding is commonly used to measure NEF activity, because the co-chaperone yields a characteristic concentration profile in which activity is first stimulated and then inhibited (16, 27). Briefly, in this experiment chemically denatured firefly luciferase is refolded into active enzyme by the action of Hsp70 complexes, and luminescence is used to track this process (28). In control experiments, we found that IT2-144 did not inhibit native luciferase (Fig. S4), allowing us to use this molecule in refolding assays without interference. Remarkably, we found that IT2-144 suppressed the refolding activity of Hsp70 with any of its NEFs at low micromolar concentrations (Fig. 5A).
Figure 5.
IT2-144 inhibits NEF-mediated activity and potentially binds in the MKT-077 binding pocket. A, IT2-144 inhibits a wide range of Hsp70–NEF combinations in luciferase refolding assays. Hsp72 (1 μm) was used with all of the NEFs except for GrpE, which used DnaK (1 μm). Likewise, DnaJA2 (0.5 μm) was used for all of the NEFs except for GrpE, which used DnaJ (0.5 μm). Denatured luciferase (0.1 μm) and potassium phosphate (10 mm) were also added. Results are the average of triplicate values, and the error bars represent S.E. B, IT2-144 docked to the MKT-077-binding site of Hsc70 (3HSC). Top view of the binding pocket (residues within 5 Å of IT2-144) highlights IT2-144 as well as ADP and magnesium in the adjacent nucleotide-binding cassette. C, scanning fluorescence spectra for all di-fluoro derivatives and the negative compound IT3-70a, with excitation wavelength set at 310 nm. D, change in fluorescence intensity with increasing Hsc70 concentration. Compounds were held at 50 μm, and fluorescence emission was read at optimal wavelengths, determined in spectral scans (see C). E, full-length Hsc70 was incubated with or without ADP and then tested for ability to bind IT2-144 (excitation 310 nm and emission 430 nm). EC50 was determined from two independent experiments in triplicate. Full-length Hsc70 with mutations at position Thr-222 was also tested for ability to bind IT2-144. Thr-222 is highlighted in the structure of IT2-144 docked to Hsc70.
Interestingly, recent work has shown that benzothiazole-rhodacyanines, such as MKT-077 and JG-48, bind to an allosteric site on Hsp70 and interrupt binding to NEFs (29). Like IT2-144, these compounds are known to inhibit multiple categories of NEFs (27). Therefore, we wondered whether IT2-144 might bind to the same site on Hsp70. This result would be important because MKT-077 and JG-48 have strong activity in multiple disease models, but they have significant liabilities (i.e. photoreactivity, poor solubility) that limit their use. In other words, the benzothiazole-rhodacyanines are good proof-of-concept compounds, but a new scaffold might expand the utility of Hsp70 inhibitors.
To test this idea, we first performed extra precision (XP) induced-fit docking experiments focused on the known binding site for MKT-077 (Fig. S5) (30). In these studies, we used the ADP-bound structure of the Hsc70 NBD (PDB 3HSC). Consistent with the biochemical studies, we found that analogs with 2,5 substitutions, such as IT2-144, had the best XP scores, whereas the 2,4 di-fluoro derivative (IT2-159) and the negative control (IT3-70a) had significantly worse XP scores (Fig. 5B). The docking pose of IT2-144 suggested that the imidazo[1,2-a]pyridine was located within a deep, narrow pocket composed of Pro-176, Val-369, Glu-175, Asp-199, Ile-197, and Val-337. Interestingly, this group approached the magnesium that is required to chelate nucleotide. The central 2-aminothiazole then seemed to position the pendant benzyl ring to make favorable contacts with a second pocket composed of Tyr-149, Gly-224, Thr-226, Ala-223, and Thr-222. The striking, overall feature of the interaction was the excellent “fit” of the molecule in a narrow channel, perhaps best exemplified by a surface rendering (Fig. 5B). This docked pose also helped explain the observed SAR from the ATPase assays. Specifically, we noted that the fluorines at the 2- and 5-positions of IT2-144 were predicted to make favorable contacts in the second pocket. Specifically, the 2-fluorine was positioned to engage in fluorine bonding (31) with the backbone of Asp-206, and the 5-fluorine was positioned near Tyr-149. In this pose, placement of a fluorine at the 3-position (as in IT2-158) would not be expected to make favorable contacts, and the 4-position (as in IT2-159) would point into solvent. Together, these docking studies supported the biochemical results and suggested possible ways of improving the molecule.
To further explore the SAR, we turned to a binding assay. Specifically, we took advantage of the intrinsic fluorescence of the 2-aminothiazoles (Fig. 5C) to directly measure an interaction. In this format, purified human Hsc70 was added to test compounds, and the binding-induced quenching of compound fluorescence was measured at the λmax. Consistent with the model, only IT2-144 had a measurable affinity for purified human Hsc70 (Fig. 5D), whereas compounds IT2-158, IT2-159, and IT2-160, as well as the negative control, IT3-70a, did not bind (Kapp >10 μm). Furthermore, the apparent affinity of IT2-144 was consistent with the functional and cellular assays (Kapp ∼1 μm).
Another prediction of the docking study is that IT2-144 should not directly compete with ADP for binding. To test this idea, we measured the affinity of IT2-144 for human Hsc70 in the apo- and ADP-bound states. If IT2-144 was competitive for nucleotide, rather than allosteric, we would anticipate that ADP would significantly reduce the apparent affinity. However, we found that addition of ADP did not inhibit binding and, in fact, might have a modestly enhanced affinity (Kapp 0.79 versus 1.3 μm). In further support, we found that IT2-144 did not compete for binding of DnaK to a fluorophore-labeled ATP analog (Fig. S6). Finally, we measured binding of IT2-144 to Hsc70 variants that have mutations in Thr-222, a residue that docking predicted to be a potential gatekeeper to the binding site (Fig. 5E). In the ADP-bound state, we found that mutation to a small amino acid (T222A) had no effect on affinity (WT Kapp ∼790 nm; T222A Kapp ∼870 nm), whereas mutation to methionine (T222M) mildly reduced affinity (Kapp ∼1.3 μm) (Fig. 5E). Attempts to mutate other residues in the putative binding site affected activity or folding, so we were not able to conclusively determine their role in binding. Although further work needs to be done, these results suggest that IT2-144 might have a mechanism-of-action that is similar to the benzothiazole-rhodacyanines, such as JG-48. Thus, IT2-144 could become an important starting point for the development of chemical probes of the Hsp70 system.
Discussion
Multiprotein complexes, especially those involving difficult PPIs, present a challenge for HTS campaigns. Cell-based approaches can sometimes recreate the dynamics of these systems, yet it is often challenging to identify which protein is the target. As an alternative, we pursued a biochemical approach that is intended to borrow some strengths of a physiological milieu, such as multiple binding partners, with the precision of a well-defined, purified system. Specifically, we explored an HTS approach in which the enzymatic activity of the “core” protein (i.e. Hsp70) is measured, and the effects of binding partners (i.e. DnaJA2 and BAG2) on turnover is used as a surrogate for their respective PPIs. We used the Hsp70 system as a model, but many other biological examples have conceptually similar architecture (e.g. Rho-Gap-GEF, Hsp90-p23-Aha1, RNA polymerase II). A key feature in these systems, including Hsp70, is that the enzyme is often weak. This allows discrimination of the effects of binding partners, such as co-chaperones, from the intrinsic activity. Indeed, a fascinating observation from our Hsp70 screen is that the potential mechanism of an inhibitor starts to become clear, even in the primary assay results. For example, compound F inhibited the Hsp72–DnaJA2 pair and blocked ∼80% of the steady-state turnover. Thus, this RMPC approach may have significant advantages for a subset of targets, specifically those with slow enzymes.
HTS campaigns, including the one described here, often result in initial inhibitors with modest efficacy values (in the range of low- to mid-micromolar). Then, the active series must be advanced through a hit-to-lead medicinal chemistry campaign to achieve high selectivity and potency. Historically, inhibitors of PPIs seem to disproportionately fail to progress through this process, likely because many PPIs have physical features, such as large BSA, which make it relatively challenging to develop competitive inhibitors. Indeed, a 2015 review of the literature found that most reported inhibitors of difficult PPIs have allosteric, not orthosteric, mechanisms (2). These allosteric inhibitors often take advantage of binding sites that are located far from the PPI contact, where the features of the pocket can be more favorable for tight binding. Thus, although there is no guarantee that PPI inhibitors from RMPC screens will progress through hit-to-lead campaigns, there is some reason to think that those with allosteric mechanisms, such as compound R, should be prioritized over orthosteric ones.
Hsp70 is an emerging drug target for a number of diseases (32). Accordingly, this work might have implications beyond HTS technology development. For example, whereas compound F itself is not a suitable starting point for probe development, its binding site might serve as a guide for future fragment-based tethering screens (33). Perhaps more interesting is the possibility that compound R and its analogs might directly serve as a starting point for the development of Hsp70–NEF inhibitors. Hsp70 and its NEFs have been shown to play important roles in cancer (34), and this PPI seems to be a particularly attractive one for controlling chaperone function (27). Thus, further medicinal chemistry campaigns and mechanistic studies on IT2-144 seem warranted. Perhaps more broadly, this RMPC approach seems to be particularly well suited to the Hsp70 system, and it is intriguing to consider primary screens that add more partners, such as “client” proteins.
Materials and methods
Compound library
The HTS campaign was carried out using the commercial ChemDiv 100K diversity set library, which is composed of “lead-like” molecules (e.g. molecular weight, logP, polar surface area, number of rotatable bonds, and the presence/absence of reactive or toxic structural motifs). Active compounds were confirmed with re-purchased samples from either ChemDiv or ChemBridge. Compound identity was confirmed using LC-MS and 1H NMR.
Protein purification
Human Hsp72 (HSPA1A) was purified as described previously (35) using an N-terminal His6 tag and Ni-NTA column, followed by overnight TEV protease cleavage of the His tag and finally an ATP-agarose affinity column. Human DnaJA2 and E. coli DnaJ were purified as described previously (16) using an N-terminal His6 tag and Ni-NTA column, TEV protease cleavage of the His tag, a secondary Ni-NTA column to remove un-cleaved contaminants, and finally size-exclusion chromatography on a 15-ml Superdex 200 gel-filtration column (GE Healthcare). Human BAG domain, BAG1, and BAG2 were purified based on previous reports (16). Briefly, the N-terminal His6 BAG2 was subjected to a Ni-NTA column and overnight TEV protease cleavage of the His6. The NEFs were then dialyzed overnight into Buffer A (25 mm HEPES, 10 mm NaCl, 15 mm β-mercaptoethanol, 0.1 mm EDTA, pH 7.6) and separated via ion-exchange chromatography on a Mono Q HR 16/10 column (GE Healthcare). Finally, the proteins were subjected to size-exclusion chromatography on a 15-ml Superdex 200 gel-filtration column (GE Healthcare). The BAG proteins were concentrated and exchanged into BAG buffer (25 mm HEPES, 5 mm MgCl2, 150 mm KCl, pH 7.5). Hsp105 was purified on a Ni-NTA column, subjected to overnight TEV cleavage as described above, and then subjected to a second Ni-NTA column purification, followed by buffer exchange into BAG buffer. The prokaryotic chaperone proteins, DnaK, DnaJ, and GrpE, were purified following previously reported protocols (36). 15N-Labeled DnaK NBD(1–388) was expressed in M9 medium with [15N]ammonium chloride (Sigma) and purified on an ATP-agarose affinity column.
High-throughput ATPase screen with quinaldine red
The HTS method was adapted from previously reported procedures (10). A master mix of purified Hsp72, DnaJA2, and BAG2 was prepared, such that the final concentrations would be 0.5, 0.1, and 0.25 μm, respectively. Solutions were added to 384-well plates using a Multidrop dispenser (Thermo Fisher Scientific, Inc.), with the exception of compound and DMSO solutions, which were added using a Biomek HDR (Beckman). QR solution was freshly prepared each day as a 2:1:1:2 ratio of 0.05% w/v quinaldine red, 2% w/v polyvinyl alcohol, 6% w/v ammonium molybdate tetrahydrate in 6 m HCl, and water. To each well of a 384-well white, low-volume, polystyrene plate (Greiner Bio-One, Monroe, NC) was added 5 μl of a master chaperone mix. The assay buffer (100 mm Tris, 20 mm KCl, 6 mm MgCl2, pH 7.4), was supplemented with 0.01% Triton X-100 to avoid identifying aggregators as false hits. The compounds, or DMSO alone, were added as 200 nl of a 1.5 mm stock to give a final screening concentration of 43 μm, followed by 2 μl of 3.5 mm ATP to initiate the reaction. The plates were then centrifuged briefly, and subsequently incubated at 37 °C for 2 h. After incubation, 15 μl of QR solution was added, followed by 2 μl sodium citrate (32% w/v) to quench the reaction. After another 15-min incubation period at 37 °C, fluorescence (excitation 430 nm, emission 530 nm) was read in a PHERAstar plate reader.
General synthesis
Preparation of substituted thioureas
Thioureas were prepared from substituted amines according to previously described methods (23). Briefly, 1.1 molar equivalents of benzoyl isothiocyanate were added dropwise to the commercial anilines, with stirring in acetone at room temperature. The solution was then heated to reflux for 1–3 h until completion, as monitored by thin layer chromatography (TLC). Once complete, the reaction was cooled and then poured into water/ice to precipitate the benzoyl thiourea intermediate. The intermediate was isolated by vacuum filtration and dried prior to being re-dissolved in a mixture of 20 ml of methanol and 5 ml of aqueous 1 m NaOH. The hydrolysis reaction was carried out at 80 °C for 1 h, or until complete, as judged by TLC. The reaction was then cooled, added to water/ice, and neutralized by the addition of 1 m HCl. The thiourea product typically crashed out of solution upon removal of MeOH by rotary evaporation. In the cases where product did not precipitate at this stage, the product was isolated by extracting three times with 20 ml of ethyl acetate, combining the organic fractions, and removing solvent by rotary evaporation. The purified thioureas were deemed at least 95% pure by LC-MS, and carried forward to make the 2-aminothiazoles.
General preparation of 2-aminothiazoles
Thioureas were mixed with bromo- or chloro-acetophenones dissolved in 4 ml of ethanol and refluxed with stirring for ∼3 h. Over the course of the reaction, the desired product formed as an insoluble solid on the edge of the flask. After 3 h, the reaction was cooled to room temperature and then placed on ice for ∼30 min. Continued precipitation was observed, and this solid was collected by vacuum filtration. The product was further washed with hexanes and then vacuum-dried. Compound identity was determined by 1H NMR, and purity was determined by LC-MS.
Compound characterization
For IT2-144, 1H NMR (400 MHz, DMSO) δ 10.63 (s, 1H), 9.08 (d, J = 8.0 Hz, 1H), 8.34–8.29 (m, 1H), 7.97–7.88 (m, 2H), 7.54 (s, 1H), 7.47 (t, J = 12 Hz, 1H), 7.37–7.31 (m, 1H), 6.84 (t, J = 16 Hz, 1H), 2.63 (s, 3H). ESI-MS was calculated for C17H12F2N4S 342.08 and found 343.13.
For IT2-151, 1H NMR (400 MHz, DMSO) δ 10.63 (s, 1H), 9.19 (d, J = 8.0 Hz, 1H), 8.76 (d, J = 12 Hz, 1H), 7.98–7.90 (m, 2H), 7.55 (s, 1H), 7.50 (t, J = 16 Hz, 1H), 7.33–7.28 (m, 1H), 7.23–7.19 (m, 1H), 2.66 (s, 3H). ESI-MS was calculated for C17H12BrFN4S 403.99 and found 404.99.
For IT2-159, 1H NMR (400 MHz, DMSO) δ 10.30 (s, 1H), 9.08 (d, J = 8 Hz, 1H), 8.27–8.20 (q, J = 8 Hz, 1H), 7.95–7.89 (m, 2H), 7.45 (s, 1H), 7.41 (t, J = 16 Hz, 1H), 7.10 (t, J = 16 Hz, 1H), 2.62 (s, 3H).
For IT2-179, 1H NMR (400 MHz, DMSO) δ 10.06 (s, 1H), 9.19 (d, J = 8 Hz, 1H), 8.59 (s, 1H), 7.98–7.91 (m, 2H), 7.63 (d, J = 8 Hz, 1H), 7.55 (s, 1H), 7.51 (t, J = 16 Hz, 1H), 7.21 (d, J = 8 Hz, 1H), 2.65 (s, 3H). ESI-MS was calculated for C17H12Br2N4S 463.91 and found 464.94.
ATPase assays with malachite green
Active compounds from the primary screen were confirmed by subsequent ATPase assays using MG, as described previously (36). Briefly, in a clear 96-well plate, compounds were incubated with Hsp72 and co-chaperones at specified concentrations in 25-μl total volume with a final DMSO concentration of 4%. All malachite green assays were performed in an assay buffer composed of 100 mm Tris, 20 mm KCl, 6 mm MgCl2, 0.01% Triton X-100, pH 7.4. The reaction was initiated by the addition of ATP at a final concentration of 1 mm and incubated at 37 °C for 1 h. After incubation, 80 μl of MG reagent was added, followed by 10 μl of saturated sodium citrate to quench the reaction, and absorbance of 620 nm was measured on a SpectraMax M5 plate reader (Molecular Devices). ATP hydrolysis rates were calculated by comparison to a phosphate standard.
Luciferase refolding
Luciferase refolding assays followed a previously described procedure (37). All compounds were first tested by incubation with native luciferase to determine whether they interfered with the assay (Fig. S4). Compounds that reduced activity by >10% were excluded from further analysis in this platform. Briefly, native firefly luciferase (Promega) was denatured in 6 m guanidine hydrochloride for 1 h at room temperature and then diluted into assay buffer (28 mm HEPES, pH 7.6, 120 mm potassium acetate, 12 mm magnesium acetate, 2.2 mm dithiothreitol, 8.8 mm creatine phosphate, and 35 units/ml creatine kinase). Solutions were prepared of test compounds, chaperones, denatured luciferase (at 0.1 μm), 10 mm phosphate (for NEF-stimulation assays), and 1 mm ATP. Total volume was 25 μl (4% DMSO) in white 96-well plates (Corning Glass), and incubation time was 1 h at 37 °C. Steady Glo reagent was prepared fresh and added to the plate immediately prior to reading luminescence.
Cell culture and viability assays
MDA-MB-231 and MEF cells were purchased from ATCC and maintained at 37 °C and 5% CO2 in appropriate growth media (Dulbecco's modified Eagle's medium, 5% penicillin/streptomycin, 10% FBS for MDA-MB-231, and 15% FBS for MEF, respectively). Cell proliferation assays were carried out as described previously (34) using the MTT kit (ATCC number: 30-1010 K) with some modifications. Briefly, cells were plated in tissue culture-treated 96-well plates (5,000 cells per well for MDA-MB-231 and 2,000 cells per well for MEF) with compounds (1% DMSO) in 200 μl of growth medium for 72 h. Cells were washed three times with PBS, and the medium was replaced with fresh medium plus 10% MTT reagent, then subsequently incubated at 37 °C, 5% CO2 for 4 h. The medium was then removed and replaced with DMSO and then read at an absorbance of 540 nm.
HSQC with 15N-labeled DnaK NBD
2D HSQC experiments were carried out to measure binding of IT2-21c to 15N-labeled DnaK NBD(1–388). Experiments were performed on a Bruker 800 MHz NMR at 27 °C in NMR buffer (25 mm Tris, 10 mm KCl, 5 mm MgCl2, 10 mm sodium phosphate, 5 mm ADP, 0.015% NaN3, 5% D2O, pH 7.2). For determining a binding site of IT2-21c, DnaK NBD concentration was held at 100 μm, and compound was titrated in at ratios of 1:1 and 1:2 (NBD/compound) at 2% DMSO. The spectra were processed with NMRPipe (38) and displayed with Sparky (39). The chemical shift assignments used were from the triple resonance data obtained by Bertelsen et al. (18). Compound-induced shifts that are larger than 0.01 ppm in 1H and/or larger than 0.1 ppm in 15N were deemed significant and were displayed on the DnaK NBD structure (2KHO).
Docking to MKT-077-binding site
Compound R derivatives were docked to a bovine Hsc70 crystal structure (PDB code 3HSC) using both InducedFit and Glide docking software (Software Suite 2017-3, Schroedinger, LLC). The nucleotide-binding domain of Hsc70 is 100% identical between bovine and human. Prior to docking, we optimized the protein structure using Protein Preparation Wizard. During this step, water molecules, sodium, and an inorganic phosphate ion from the crystal structure were removed. Only ADP and a magnesium ion were retained in the nucleotide-binding pocket. Hydrogen atoms were added to the structure; protonation states of titratable residues were adjusted, and overall structure was energy-minimized such that heavy atoms were not allowed to move beyond 0.5 Å from their starting positions. The allosteric binding site was partially closed in the crystal structure. Therefore, we docked the allosteric inhibitor IT2-144 using an InducedFit docking algorithm. The allosteric binding pocket was defined using residues Thr-13, Lys-71, Tyr-149, Glu-175, Asp-199, Thr-204, and ADP. InducedFit docking program treats both protein and ligand flexibly during docking. The lowest energy docking pose predicted by the InducedFit docking program is shown in Fig. 5B. The methyl substituted imidazopyridine ring is deeply buried in the hydrophobic binding pocket and the hydrogen atom of the secondary amine makes a hydrogen bonding interaction with the backbone carbonyl oxygen of Val-207. The 2,5 di-fluoro phenyl ring stacks up between the β-sheet and Tyr-149 residue.
With the allosteric binding pocket optimized, the IT2-144 inhibitor-bound model was used in subsequent rigid-receptor flexible-ligand docking calculations. IT2-144 analogs were docked to the allosteric site using Glide docking with XP-docking scoring function. All inhibitors were built using the Edit/Build panel of Maestro (Schroedinger, LLC). They were subsequently energy minimized using LigPrep software (v4.3016, Schroedinger, LLC). Inhibitor structure and docking scores are reported in Fig. 5B, and their binding poses are shown in Fig. S5. The 2,5 di-substituted derivatives all have similar docking scores, whereas the 2,4 di-substituted compound (IT2-159) and the truncated negative compound (IT3-70a) have poorer scores. Docking poses also show a similar binding mode for all but IT2-159 and IT3-70a.
IT2-144 binding by fluorescence quenching
All di-fluoro compound R derivatives, as well as IT3-70a, were subjected to spectral characterization by determining a full scanning fluorescence spectrum at an excitation wavelength of 310 nm. These spectra were used to determine the optimal emission wavelength for each compound (IT2-144, 430 nm; IT2-158, 420 nm; IT2-159, 450 nm; IT2-160, 440 nm; IT3-70a, 460 nm). It is important to note that compound R did not show any fluorescence in this range and did not have an artifactual effect in the original QR assay. Only the di-fluoro derivatives appeared to give a strong enough fluorescence signature with this excitation/emission profile. Hsc70 was incubated at the indicated concentrations with nucleotide (ADP or ATP) for 30 min in a low-volume black 384-well plate (Corning Glass) at room temperature (Buffer: 200 mm Tris, 40 mm KCl, 12 mm MgCl2, 0.01% Triton X-100, pH 7.4). Following incubation with nucleotide, compounds were added to the wells at a constant concentration of 50 μm and 5% DMSO. The protein/nucleotide/compound mixture was allowed to incubate at room temperature, in the dark, for 30 min, prior to reading fluorescence at excitation 310 nm and the appropriate emission wavelength, determined from the spectral characterization of each compound.
Author contributions
I. R. T., B. M. D., T. K., and E. R. P. Z. data curation; I. R. T., B. M. D., H. S., V. A. A., C. K., and J. E. G. formal analysis; I. R. T., B. M. D., H. S., X. R., V. A. A., C. K., J. N. R., and E. R. P. Z. investigation; I. R. T. and J. N. R. methodology; I. R. T. and J. E. G. writing-original draft; I. R. T., B. M. D., H. S., X. R., E. R. P. Z., and J. E. G. writing-review and editing; B. M. D. and J. E. G. conceptualization; M. P. J. software; M. P. J., E. R. P. Z., and J. E. G. supervision; E. R. P. Z. and J. E. G. funding acquisition; J. E. G. project administration.
Supplementary Material
Acknowledgments
We thank Martha Larsen and Thomas McQuade (University of Michigan; Center for Chemical Genomics) for technical assistance with the HTS campaign. We also thank Robert Briski for help in the triage of active molecules.
This work was supported by National Institutes of Health Grant R01NS059690 (to J. E. G. and E. R. P. Z.) and Stand Up 2 Cancer (to J. E. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
- PPI
- protein–protein interaction
- HTS
- high-throughput screening
- RMPC
- reconstituted multiprotein complex
- DRC
- dose-response curve
- MG
- malachite green
- QR
- quinaldine red
- FBS
- fetal bovine serum
- MEF
- mouse embryonic fibroblast
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- NEF
- nucleotide exchange factor
- PDB
- Protein Data Bank
- BSA
- buried surface area
- SAR
- structure–activity relationship
- XP
- extra precision
- TEV
- tobacco etch virus
- CSP
- chemical shift perturbation
- ESI-MS
- electrospray ionization-mass spectrometry
- HSQC
- heteronuclear single quantum coherence.
References
- 1. Arkin M. R., Tang Y., and Wells J. A. (2014) Small-molecule inhibitors of protein–protein interactions: progressing toward the reality. Chem. Biol. 21, 1102–1114 10.1016/j.chembiol.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cesa L. C., Mapp A. K., and Gestwicki J. E. (2015) Direct and propagated effects of small molecules on protein–protein interaction networks. Front. Bioeng. Biotechnol. 3, 119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thompson A. D., Dugan A., Gestwicki J. E., and Mapp A. K. (2012) Fine-tuning multiprotein complexes using small molecules. ACS Chem. Biol. 7, 1311–1320 10.1021/cb300255p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scott D. E., Bayly A. R., Abell C., and Skidmore J. (2016) Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550 10.1038/nrd.2016.29 [DOI] [PubMed] [Google Scholar]
- 5. Chiappori F., Merelli I., Milanesi L., Colombo G., and Morra G. (2016) An atomistic view of Hsp70 allosteric crosstalk: from the nucleotide to the substrate binding domain and back. Sci. Rep. 6, 23474 10.1038/srep23474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zuiderweg E. R., Bertelsen E. B., Rousaki A., Mayer M. P., Gestwicki J. E., and Ahmad A. (2013) Allostery in the Hsp70 chaperone proteins. Top. Curr. Chem. 328, 99–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCarty J. S., Buchberger A., Reinstein J., and Bukau B. (1995) The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249, 126–137 10.1006/jmbi.1995.0284 [DOI] [PubMed] [Google Scholar]
- 8. Ahmad A., Bhattacharya A., McDonald R. A., Cordes M., Ellington B., Bertelsen E. B., and Zuiderweg E. R. (2011) Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl. Acad. Sci. U.S.A. 108, 18966–18971 10.1073/pnas.1111220108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., and Misra S. (2008) Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317 10.1038/nsmb.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miyata Y., Chang L., Bainor A., McQuade T. J., Walczak C. P., Zhang Y., Larsen M. J., Kirchhoff P., and Gestwicki J. E. (2010) High-throughput screen for Escherichia coli heat shock protein 70 (Hsp70/DnaK): ATPase assay in low volume by exploiting energy transfer. J. Biomol. Screen. 15, 1211–1219 10.1177/1087057110380571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mally A., and Witt S. N. (2001) GrpE accelerates peptide binding and release from the high affinity state of DnaK. Nat. Struct. Biol. 8, 254–257 10.1038/85002 [DOI] [PubMed] [Google Scholar]
- 12. Yue X., Zhao Y., Liu J., Zhang C., Yu H., Wang J., Zheng T., Liu L., Li J., Feng Z., and Hu W. (2015) BAG2 promotes tumorigenesis through enhancing mutant p53 protein levels and function. eLife 4, 10.7554/eLife.08401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kiang J. G., Gist I. D., and Tsokos G. C. (2000) Regulation of heat shock protein 72 kDa and 90 kDa in human breast cancer MDA-MB-231 cells. Mol. Cell. Biochem. 204, 169–178 10.1023/A:1007016822939 [DOI] [PubMed] [Google Scholar]
- 14. Powers M. V., Clarke P. A., and Workman P. (2008) Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14, 250–262 10.1016/j.ccr.2008.08.002 [DOI] [PubMed] [Google Scholar]
- 15. Whitmore A. (2017) Targeting human DNAJAs for sensitization to chemotherapeutic agents. FASEB J. 31, 995 [Google Scholar]
- 16. Rauch J. N., and Gestwicki J. E. (2014) Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 289, 1402–1414 10.1074/jbc.M113.521997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cesa L. C., Patury S., Komiyama T., Ahmad A., Zuiderweg E. R. P., and Gestwicki J. E. (2013) Inhibitors of difficult protein–protein interactions identified by high-throughput screening of multiprotein complexes. ACS Chem. Biol. 8, 1988–1997 10.1021/cb400356m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bertelsen E. B., Chang L., Gestwicki J. E., and Zuiderweg E. R. (2009) Solution conformation of wildtype E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 10.1073/pnas.0903503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang L., Miyata Y., Ung P. M., Bertelsen E. B., McQuade T. J., Carlson H. A., Zuiderweg E. R., and Gestwicki J. E. (2011) Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem. Biol. 18, 210–221 10.1016/j.chembiol.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonomo J., Welsh J. P., Manthiram K., and Swartz J. R. (2010) Comparing the functional properties of the Hsp70 chaperones, DnaK and BiP. Biophys. Chem. 149, 58–66 10.1016/j.bpc.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyata Y., Rauch J. N., Jinwal U. K., Thompson A. D., Srinivasan S., Dickey C. A., and Gestwicki J. E. (2012) Cysteine reactivity distinguishes redox sensing by the heat-inducible and constitutive forms of heat shock protein 70. Chem. Biol. 19, 1391–1399 10.1016/j.chembiol.2012.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang Y., Taldone T., Patel H. J., Patel P. D., Rodina A., Gozman A., Maharaj R., Clement C. C., Patel M. R., Brodsky J. L., Young J. C., and Chiosis G. (2014) Heat shock protein 70 inhibitors. 1. 2,5′-thiodipyrimidine and 5-(phenylthio)pyrimidine acrylamides as irreversible binders to an allosteric site on heat shock protein 70. J. Med. Chem. 57, 1188–1207 10.1021/jm401551n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gallardo-Godoy A., Gever J., Fife K. L., Silber B. M., Prusiner S. B., and Renslo A. R. (2011) 2-Aminothiazoles as therapeutic leads for prion diseases. J. Med. Chem. 54, 1010–1021 10.1021/jm101250y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., Srinivasan S. R., Connarn J., Ahmad A., Young Z. T., Kabza A. M., Zuiderweg E. R., Sun D., and Gestwicki J. E. (2013) Analogs of the allosteric heat shock protein 70 (Hsp70) inhibitor, MKT-077, as anti-cancer agents. ACS Med. Chem. Lett. 4, 10.1021/ml400204n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andréasson C., Fiaux J., Rampelt H., Druffel-Augustin S., and Bukau B. (2008) Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc. Natl. Acad. Sci. U.S.A. 105, 16519–16524 10.1073/pnas.0804187105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., and Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 10.1126/science.276.5311.431 [DOI] [PubMed] [Google Scholar]
- 27. Young Z. T., Rauch J. N., Assimon V. A., Jinwal U. K., Ahn M., Li X., Dunyak B. M., Ahmad A., Carlson G. A., Srinivasan S. R., Zuiderweg E. R., Dickey C. A., and Gestwicki J. E. (2016) Stabilizing the Hsp70-Tau complex promotes turnover in models of tauopathy. Cell Chem. Biol. 23, 992–1001 10.1016/j.chembiol.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levy E. J., McCarty J., Bukau B., and Chirico W. J. (1995) Conserved ATPase and luciferase refolding activities between bacteria and yeast Hsp70 chaperones and modulators. FEBS Lett. 368, 435–440 10.1016/0014-5793(95)00704-D [DOI] [PubMed] [Google Scholar]
- 29. Li X., Shao H., Taylor I. R., and Gestwicki J. E. (2016) Targeting allosteric control mechanisms in heat shock protein 70 (Hsp70). Curr. Top. Med. Chem. 16, 2729–2740 10.2174/1568026616666160413140911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rousaki A., Miyata Y., Jinwal U. K., Dickey C. A., Gestwicki J. E., and Zuiderweg E. R. (2011) Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 411, 614–632 10.1016/j.jmb.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gillis E. P., Eastman K. J., Hill M. D., Donnelly D. J., and Meanwell N. A. (2015) Applications of fluorine in medicinal chemistry. J. Med. Chem. 58, 8315–8359 10.1021/acs.jmedchem.5b00258 [DOI] [PubMed] [Google Scholar]
- 32. Assimon V. A., Gillies A. T., Rauch J. N., and Gestwicki J. E. (2013) Hsp70 protein complexes as drug targets. Curr. Pharm. Des. 19, 404–417 10.2174/138161213804143699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erlanson D. A., Braisted A. C., Raphael D. R., Randal M., Stroud R. M., Gordon E. M., and Wells J. A. (2000) Site-directed ligand discovery. Proc. Natl. Acad. Sci. U.S.A. 97, 9367–9372 10.1073/pnas.97.17.9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X., Colvin T., Rauch J. N., Acosta-Alvear D., Kampmann M., Dunyak B., Hann B., Aftab B. T., Murnane M., Cho M., Walter P., Weissman J. S., Sherman M. Y., and Gestwicki J. E. (2015) Validation of the Hsp70-Bag3 protein–protein interaction as a potential therapeutic target in cancer. Mol. Cancer Ther. 14, 642–648 10.1158/1535-7163.MCT-14-0650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang L., Thompson A. D., Ung P., Carlson H. A., and Gestwicki J. E. (2010) Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK). J. Biol. Chem. 285, 21282–21291 10.1074/jbc.M110.124149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang L., Bertelsen E. B., Wisén S., Larsen E. M., Zuiderweg E. R., and Gestwicki J. E. (2008) High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal. Biochem. 372, 167–176 10.1016/j.ab.2007.08.020 [DOI] [PubMed] [Google Scholar]
- 37. Wisén S., and Gestwicki J. E. (2008) Identification of small molecules that modify the protein folding activity of heat shock protein 70. Anal. Biochem. 374, 371–377 10.1016/j.ab.2007.12.009 [DOI] [PubMed] [Google Scholar]
- 38. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 39. Goddard T. D., and Kneller D. G. (2000) SPARKY 3. University of California, San Francisco, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.