Abstract
Cholesterol biosynthesis is tightly regulated in the cell. For example, high sterol concentrations can stimulate degradation of the rate-limiting cholesterol biosynthetic enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase, HMGCR). HMGCR is broken down by the endoplasmic reticulum membrane–associated protein complexes consisting of insulin-induced genes (Insigs) and the E3 ubiquitin ligase gp78. Here we found that HMGCR degradation is partially blunted in Chinese hamster ovary (CHO) cells lacking gp78 (gp78-KO). To identify other ubiquitin ligase(s) that may function together with gp78 in triggering HMGCR degradation, we performed a small-scale short hairpin RNA–based screening targeting endoplasmic reticulum–localized E3s. We found that knockdown of both ring finger protein 145 (Rnf145) and gp78 genes abrogates sterol-induced degradation of HMGCR in CHO cells. We also observed that RNF145 interacts with Insig-1 and -2 proteins and ubiquitinates HMGCR. Moreover, the tetrapeptide sequence YLYF in the sterol-sensing domain and the Cys-537 residue in the RING finger domain were essential for RNF145 binding to Insigs and RNF145 E3 activity, respectively. Of note, amino acid substitutions in the YLYF or of Cys-537 completely abolished RNF145-mediated HMGCR degradation. In summary, our study reveals that RNF145, along with gp78, promotes HMGCR degradation in response to elevated sterol levels and identifies residues essential for RNF145 function.
Keywords: cholesterol metabolism, ubiquitin ligase, ER-associated degradation, lipid, ubiquitylation (ubiquitination), gp78, HMG-CoA reductase, Insig, RNF145, ubiquitin ligase
Introduction
Cholesterol is the most abundant sterol in mammalian cells. It regulates membrane function and serves as the precursor for bile acids and steroid hormones. Cholesterol can either be synthesized through the mevalonate pathway (1) or taken up from diets via Niemann–Pick C1-like 1 (NPC1L1)–mediated absorption (2–5).
HMG-CoA4 reductase (HMGCR) catalyzes the rate-limiting step in cholesterol biosynthesis in which HMG-CoA is converted to mevalonate. The half-life of HMGCR varies with cellular sterol levels: more than 12 h in sterol-depleted cells and less than 1 h in sterol-overloaded cells (6, 7). HMGCR is tightly regulated by sterols at both transcriptional and posttranslational levels. High concentrations of cholesterol decrease transcription of the HMGCR gene by inhibiting the activation of sterol regulatory element–binding protein 2 (SREBP-2) (8, 9). In addition, excess levels of 24,25-dihydrolanosterol, an intermediate in the mevalonate pathway, promote ubiquitination and degradation of the HMGCR protein (10–12). Oxysterols can inhibit HMGCR transcription and stimulate HMGCR degradation (13, 14). Besides sterols, geranylgeraniol, a non-steroid product downstream of mevalonate, acts on the post-ubiquitination step to accelerate sterol-induced HMGCR degradation (7).
The sterol-induced degradation of HMGCR initiates when the endoplasmic reticulum (ER)-localized Insig-1 and -2 proteins bind to HMGCR (15) and recruit the ubiquitin ligase (E3) gp78 to catalyze ubiquitination (16). The HMGCR protein is eventually degraded in the proteasome. Ufd1 enhances the E3 activity of gp78 and accelerates the degradation of HMGCR (17). Ablation of gp78 in mouse liver increases the stability of HMGCR, Insig-1, and Insig-2 (18, 19). Elevated levels of Insigs inhibit the SREBP pathway and decrease cholesterol synthesis (18). These data suggest that gp78 is a major E3 essential for HMGCR degradation in the hepatocytes.
Besides gp78, TRC8 and MARCH6 are two other ER-localized E3s involved in HMGCR degradation (20, 21). TRC8 interacts with Insig-1 and -2 and ubiquitinates HMGCR for proteasomal degradation. In addition to sterol-regulated degradation, the basal turnover of HMGCR is mediated by Hrd1, an ER-anchored E3 homologous to gp78 (22, 23). Interestingly, sterol-induced HMGCR degradation has been found to persist in gp78-deficient primary mouse embryonic fibroblasts (24), which leads us to speculate that there might be other E3(s) compensating for the function of gp78 in cultured cells.
In this study, we identified that an ER-anchored E3 named RNF145 catalyzed sterol-induced ubiquitination of HMGCR. Knockout of gp78 or Rnf145 alone had partial or little effect on HMGCR degradation in Chinese hamster ovary (CHO) cells. However, knockout of both genes dramatically blunted sterol-induced degradation of HMGCR. The E3 activity–deficient RNF145 (C537A) failed to promote sterol-induced ubiquitination and degradation of HMGCR. Moreover, we found that Insigs were required for RNF145-catalyzed HMGCR degradation and that RNF145 interacted with Insigs constitutively through its transmembrane domains. We therefore conclude that RNF145 is a new E3 promoting sterol-induced degradation of HMGCR.
Results
Identification of Rnf145 involved in HMGCR degradation
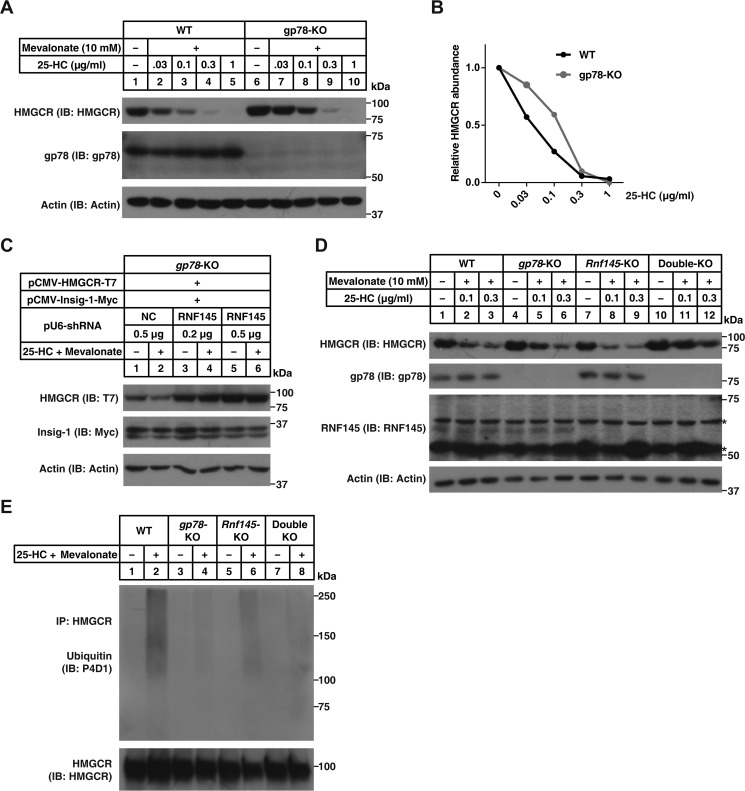
To determine whether gp78 is exclusively responsible for HMGCR degradation, we treated WT CHO and gp78 knockout (gp78-KO) cells with increasing concentrations of 25-hydroxycholesterol (25-HC). Although the degradation of HMGCR was partially impaired when cells were treated with low concentrations of 25-HC (0.03 and 0.1 μg/ml), high concentrations of 25-HC (0.3 and 1 μg/ml) almost or completely diminished HMGCR in gp78-KO cells (Fig. 1, A and B), suggesting that gp78 is not the only E3 mediating HMGCR degradation. Because previous studies identified a total of 24 ER membrane–spanning E3s (25), we next transfected plasmids expressing shRNAs targeting each E3 together with those expressing HMGCR-T7 and Insig-1-Myc into gp78-KO cells and examined sterol-induced degradation of HMGCR (Fig. S1). In contrast to control shRNA transfected cells where HMGCR degraded rapidly in response to sterols (Fig. 1C, compare lanes 1 and 2), no HMGCR degradation was detected upon Rnf145 deficiency in gp78-KO cells (Fig. 1C, lanes 3–6). These results suggest that RNF145 is involved in HMGCR degradation. We further generated Rnf145-KO and gp78 plus Rnf145 knockout (double KO) CHO cells using the CRISPR/Cas9 technique (26). Knockout of gp78 slightly affected HMGCR degradation relative to WT cells (Fig. 1D, lanes 4–6), and knockout of Rnf145 alone had little influence on HMGCR degradation (Fig. 1D, lanes 7–9). However, HMGCR degradation was largely blunted in the double KO cells (Fig. 1D, lanes 10–12). Next we measured sterol-induced ubiquitination of HMGCR in cells lacking either E3 or both. Knockout of gp78, Rnf145, or both genes decreased sterol-induced ubiquitination of HMGCR (Fig. 1E). These results suggest that RNF145 plays a critical role in sterol-induced degradation of HMGCR.
Figure 1.
RNF145 is involved in sterol-regulated HMGCR degradation. A, WT CHO and gp78-KO CHO cells were depleted of sterol in medium C for 16 h. Cells were then treated with medium C supplemented with indicated concentrations of 25-HC plus 10 mm mevalonate for 5 h. Cells were harvested and subjected to SDS-PAGE, followed by immunoblot (IB) analysis. B, quantification of the HMGCR protein in A. C, the gp78-KO CHO cells were transfected with plasmids encoding HMGCR-T7, Insig-1-Myc, and the shRNA targeting RNF145. After 48 h, cells were depleted of sterol and then treated with or without 0.3 μg/ml 25-HC plus 10 mm mevalonate for 5 h as described in A. Cells were harvested and subjected to SDS-PAGE, followed by immunoblot analysis. Results shown are representative of three independent experiments. D, WT, gp78-KO, Rnf145-KO, and double KO CHO cells were depleted of sterol and then treated with indicated concentrations of 25-HC plus 10 mm mevalonate for 5 h as described in A. Cells were harvested and subjected to SDS-PAGE, followed by immunoblot analysis. Asterisks indicate nonspecific bands. Results shown are representative of two independent experiments. E, WT, gp78-KO, Rnf145-KO, and double KO CHO cells were depleted of sterol and then treated with 10 μm MG132 in the presence or absence of 1 μg/ml 25-HC and 10 mm mevalonate for 2 h. Cells were harvested, and lysates were immunoprecipitated with protein A/G beads coupled with the anti-HMGCR antibody. Pellet fractions were immunoblotted with anti-ubiquitin (P4D1) and polyclonal anti-HMGCR antibodies.
RNF145 is an ER-localized ubiquitin ligase mediating HMGCR degradation
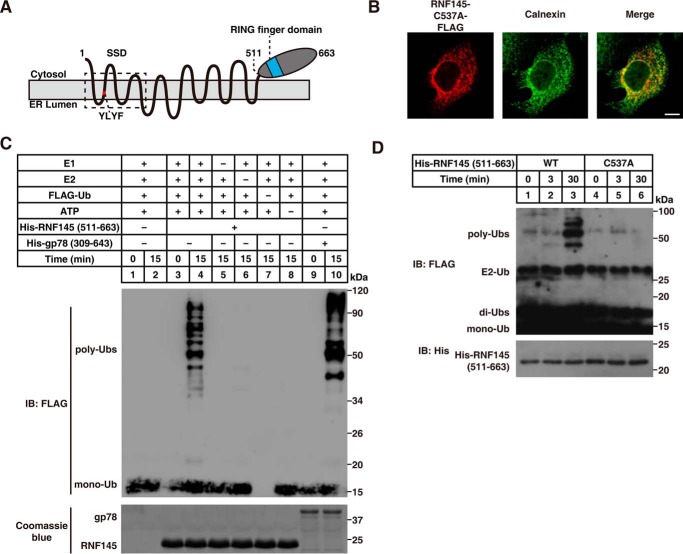
RNF145 is a putative ER transmembrane protein (Fig. 2A) (25). To confirm the subcellular location of RNF145, we performed immunofluorescence experiments by co-staining transfected RNF145 together with the endogenous ER marker Calnexin. RNF145-FLAG was largely colocalized with Calnexin, indicating that RNF145 is indeed an ER-localized protein (Fig. 2B). Next, we purified the recombinant cytosolic domain (aa 511–663) of RNF145 and performed an in vitro ubiquitination assay. The recombinant cytosolic domain of gp78 (309–643) was used as a positive control. We found that RNF145 (511–663) could efficiently catalyze the formation of polyubiquitin chains in the presence of E1, E2, FLAG-ubiquitin, and ATP (Fig. 2C). Replacement of the conserved Cys-537 residue with alanine (C537A) in the RING finger domain of RNF145 (27), however, completely abolished the E3 ligase activity of RNF145 (Fig. 2D). These results indicate that RNF145 is an ER-localized ubiquitin ligase and that the Cys-537 residue is required for its E3 activity.
Figure 2.
RNF145 is an ER-localized ubiquitin ligase. A, predicted topology of RNF145. YLYF, the amino acids from 81 to 84 of RNF145. B, subcellular localization of RNF145. HeLa cells were transfected with a plasmid encoding RNF145-FLAG and stained with anti-FLAG and anti-calnexin antibodies. Scale bar = 10 μm. C, in vitro ubiquitination assay showing that RNF145 (511–663) possesses E3 activity. Recombinant proteins, including E1, E2 (Ubc7), FLAG-ubiquitin (FLAG-Ub), RNF145 (511–663), and gp78 (309–643), were added to the reaction system as indicated. After incubation at 37 °C for 15 min, samples were subjected to SDS-PAGE, followed by immunoblot (IB) analysis. D, in vitro ubiquitination assay comparing RNF145 (511–663) and RNF145 (511–663) (C537A). Experiments were carried out as described in C.
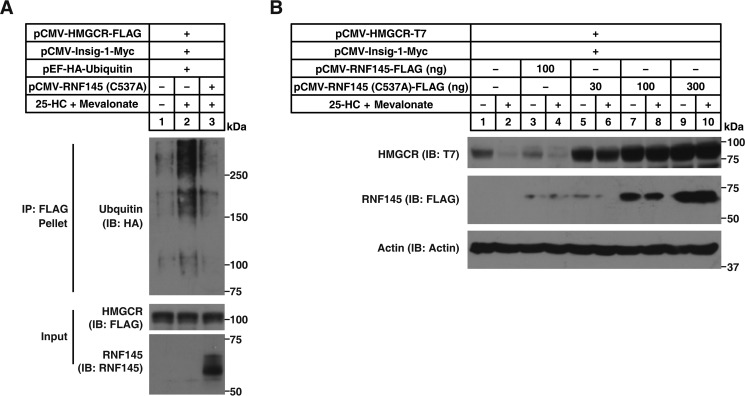
We then sought to determine whether Cys-537 is required for sterol-induced HMGCR ubiquitination. As shown in Fig. 3A, lanes 1 and 2, sterols substantially increased the ubiquitination of HMGCR, as evidenced by the high-molecular-weight smears of the immunoprecipitates. Addition of RNF145 (C537A) competitively blocked the ubiquitination of HMGCR induced by sterols (Fig. 3A, lane 3). Consistent with the ubiquitination results, the WT form of RNF145 accelerated HMGCR degradation (Fig. 3B, lanes 1–4), whereas the C537A mutant completely abrogated HMGCR degradation (Fig. 3B, lanes 5–10). These results suggest that RNF145 acts as a ubiquitin ligase promoting HMGCR degradation.
Figure 3.
The E3 activity–deficient RNF145 blocks sterol-induced ubiquitination and degradation of HMGCR. A, CHO cells were transfected with indicated plasmids, depleted of sterol, and treated with 10 μm MG132 in the presence or absence of 1 μg/ml 25-HC and 10 mm mevalonate for 3 h. Cells were harvested, and lysates were immunoprecipitated with anti-FLAG beads. Input and pellet fractions were immunoblotted (IB) with anti-HA, anti-FLAG, and polyclonal anti-RNF145 antibodies. Results shown are representative of two independent experiments. B, CHO cells were transfected with indicated plasmids, depleted of sterol, and treated with or without 1 μg/ml 25-HC plus 10 mm mevalonate for 5 h. Cells were harvested and subjected to SDS-PAGE and immunoblot analysis. Results shown are representative of two independent experiments.
Insigs are required for RNF145-mediated HMGCR degradation
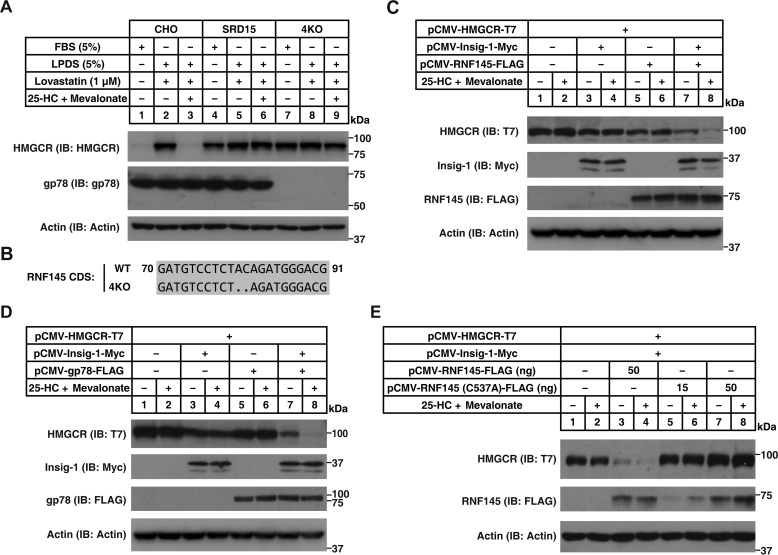
Insigs are indispensable for sterol-induced degradation of HMGCR mediated by gp78 (7, 10, 16). Indeed, SRD15 cells, a cell line lacking both Insig-1 and Insig-2 (28), failed to display sterol-regulated degradation of HMGCR (Fig. 4A, lanes 4–6). To test whether RNF145-mediated degradation of HMGCR requires Insigs, we generated an SRD15 cell line deficient in Rnf145 and gp78 (4KO) (Fig. 4B) that was also insensitive to sterol treatment (Fig. 4A, lanes 7–9). Re-expression of Insig-1 or RNF145 alone in 4KO cells did not restore sterol-induced degradation of HMGCR (Fig. 4C, lanes 3–6). Interestingly, co-expression of Insig-1 and RNF145 triggered HMGCR degradation in 4KO cells exposed to sterols (Fig. 4C, lanes 7 and 8). It was noteworthy that sterol-regulated HMGCR degradation was also detected in 4KO cells co-expressing Insig-1 and gp78 (Fig. 4D, lanes 7 and 8), suggesting that gp78 and RNF145 may function redundantly in mediating HMGCR degradation. In contrast to these findings, RNF145 (C537A) did not induce the degradation of HMGCR in the reconstitution system, even in the presence of Insig-1 (Fig. 4E, lanes 5–8).
Figure 4.
Insig is required for RNF145-mediated degradation of HMGCR. A, CHO, SRD15 (deficient in Insig-1 and -2), and 4KO (deficient in Insig-1, Insig-2, gp78, and RNF145) cells were incubated in indicated media for 16 h. Cells were harvested and subjected to SDS-PAGE, followed by immunoblot (IB) analysis. B, sequence alignment of the guide RNA targeting region showing that Rnf145 is knocked out. CDS, coding sequence. C–E, 4KO cells were transfected with indicated plasmids, depleted of sterol, and treated with or without 1 μg/ml 25-HC plus 10 mm mevalonate for 5 h. Cells were harvested and subjected to SDS-PAGE and immunoblot analysis.
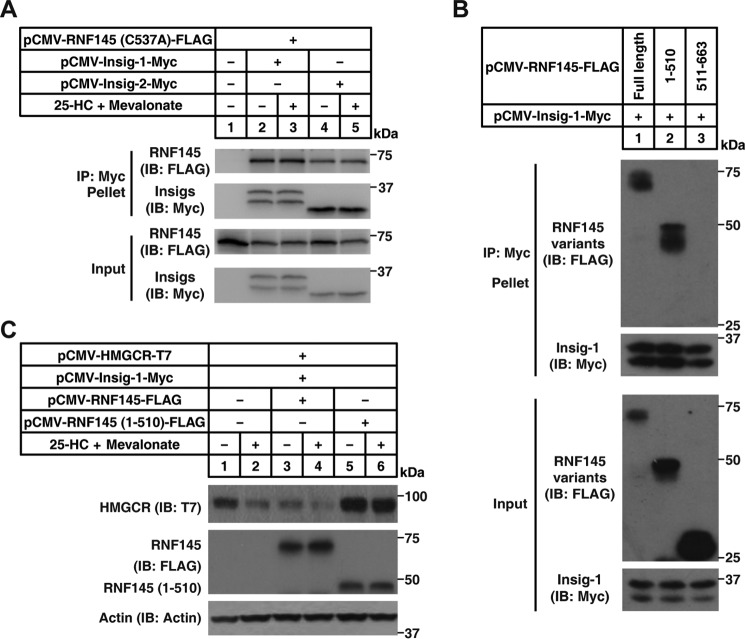
We then tested whether RNF145 could interact with Insigs. Fig. 5A shows that RNF145 co-immunoprecipitated with both Insig-1 and Insig-2 regardless of sterol levels. Specifically, it was the transmembrane domain (aa 1–510) but not the cytosolic domain (aa 511–663) of RNF145 that bound to Insig-1 (Fig. 5B). Further, overexpression of the transmembrane domain (aa 1–510) of RNF145 effectively blunted sterol-regulated HMGCR degradation (Fig. 5C).
Figure 5.
RNF145 interacts with Insigs through its transmembrane domain. A, CHO cells were transfected with indicated plasmids, depleted of sterol, and treated with or without 1 μg/ml 25-HC plus 10 mm mevalonate for 2 h. The cell lysates were immunoprecipitated with anti-Myc beads. Pellets and input were immunoblotted (IB) with indicated antibodies. Results shown are representative of two independent experiments. B, CHO cells were transfected with indicated plasmids and cultured in medium B. 48 h later, cells were harvested, and lysates were immunoprecipitated with anti-Myc beads. Pellets and input were blotted with indicated antibodies. C, CHO cells were transfected with indicated plasmids, depleted of sterol, and treated with or without 1 μg/ml 25-HC plus 10 mm mevalonate for 5 h. Cells were harvested and subjected to SDS-PAGE, followed by immunoblot analysis. Results shown are representative of two independent experiments.
The sterol-sensing domain of RNF145 is crucial for HMGCR degradation
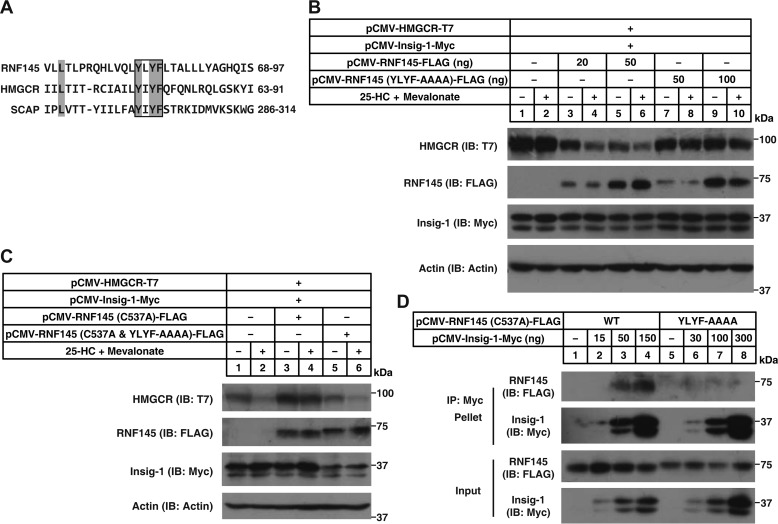
It is known that SREBP cleavage–activating protein (SCAP) and HMGCR harbor the sterol-sensing domain (SSD) for Insig binding and that the tetrapeptide YIYF is highly conserved within the SSD (7, 15). There is a putative SSD in RNF145 (Fig. 2A). Sequence alignment revealed a YLYF tetrapeptide in RNF145 corresponding to the YIYF found in HMGCR and SCAP (Fig. 6A). To test whether this motif was essential for RNF145-mediated degradation of HMGCR, we mutated YLYF to AAAA and evaluated its function in 4KO cells. Unlike WT RNF145, the RNF145 (YLYF-AAAA) mutant could not elicit the degradation of HMGCR following sterol treatment (Fig. 6B). We next combined the YLYF-AAAA mutation with E3 activity deficiency together. As shown in Fig. 6C, the RNF145 (C537A and YLYF-AAAA) mutant no longer blocked sterol-regulated degradation of HMGCR. Moreover, the RNF145 (YLYF-AAAA) mutant could not be co-immunoprecipitated with Insig-1 (Fig. 6D). Collectively, these results suggest that the YLYF motif is required for Insig binding as well as RNF145-mediated HMGCR degradation.
Figure 6.
The sterol-sensing domain of RNF145 is essential for Insig binding. A, sequence alignment of human RNF145, HMGCR, and SCAP. Invariant amino acids are shaded in gray. The conserved motif YI(L)YF is boxed. B and C, 4KO cells (B) and CHO cells (C) were transfected with indicated plasmids, depleted of sterol, and treated with or without 1 μg/ml 25-HC plus 10 mm Mevalonate for 5 h. Cells were harvested and subjected to SDS-PAGE, followed by immunoblot (IB) analysis. D, CHO cells were transfected with indicated plasmids and cultured in medium B. 48 h later, cells were harvested, and lysates were immunoprecipitated with anti-Myc beads. Pellets and input were blotted with indicated antibodies.
Discussion
In this study, we identified RNF145 as another E3 mediating the degradation of HMGCR through unbiased shRNA screening. RNF145 resides in the ER with 14 putative transmembrane segments, one to five of which constitute an SSD that is also present in SCAP, HMGCR, NPC1, NPC1L1, and TRC8 (29–31). The RING finger domain localized in the cytosolic C terminus confers RNF145 E3 activity. RNF145 and TRC8 display similar structures and sequences.
In CHO cells, knockout of gp78 partially delayed the turnover of HMGCR in response to low concentrations of sterols, and ablation of Rnf145 alone also had little effect (Fig. 1). Notably, knockout of both genes largely abolished sterol-induced degradation of HMGCR (Fig. 1D). Similar to gp78 and TRC8, Insigs are required by RNF145 for HMGCR ubiquitination. The binding between Insigs and HMGCR is regulated by sterol levels, whereas the Insig and E3 (gp78, TRC8, and RNF145) interaction is constitutive. The inactivated mutation (C537A) of RNF145 had a dominant-negative effect on HMGCR degradation. A similar C356G mutation was found in gp78 (16). Interestingly, HMGCR, SCAP, and RNF145 all bind Insigs through the SSD and require the conserved YI(L)YF tetrapeptide for binding.
The question why multiple E3s are involved in sterol-induced degradation of HMGCR is intriguing. One possibility is that different cells express different levels of E3s in response to distinct signals. In fact, recent studies have identified Rnf145 as a Liver X receptor (LXR) target gene (32, 33). We hypothesize that activation of LXR might elevate the RNF145 level and subsequently down-regulate cholesterol biosynthesis through degrading HMGCR. Another possibility is that the existence of multiple E3s for HMGCR degradation prevents saturation of specific E3(s) and ensures that ER-associated degradation functions properly when HMGCR is degraded.
The protein machineries involved in HMGCR degradation may also participate in other cholesterol-regulating processes. gp78 is the first characterized E3 catalyzing HMGCR ubiquitination (16). It is highly expressed in the liver. Knockout of gp78 in the hepatocytes largely blunted the degradation of HMGCR (18). However, gp78 deficiency also stabilizes Insigs (especially Insig-2), resulting in suppressed processing of SREBP and subsequently decreased expression of Hmgcr and other genes in the mevalonate pathway (18). As the protein levels of Insigs were dramatically increased in gp78-deficient cells (18) (Fig. S2), the effects of other E3s such as TRC8 and RNF145 might be boosted, which may explain why HMGCR is still degraded in gp78-KO cells (24). gp78 can also catalyze ubiquitination of acyl-CoA:cholesterol acyltransferase (ACAT)-2 on a cysteine residue (34). In addition to degrading HMGCR, RNF145 triggers the ubiquitination of SCAP and interferes with SCAP binding to coat protein complex II (COPII), thus inhibiting SREBP-2 maturation (33).
In summary, we here identify that RNF145 is an E3 governing sterol-regulated degradation of HMGCR. Together with the previous findings that Rnf145 is an LXR-regulated gene and that RNF145 inhibits the SREBP pathway through ubiquitinating SCAP, RNF145 serves as an important negative regulator of cholesterol biosynthesis. Activation of RNF145 may be effective for treating hypercholesterolemia through inhibiting endogenous cholesterol synthesis.
Experimental procedures
Reagents
We obtained lovastatin, mevalonate, and 25-hydroxycholesterol from Sigma, MG132 from Calbiochem, FLAG-ubiquitin from Boston Biochem, and linear polyethyleneimine from Polysciences; lipoprotein-deficient serum was prepared from newborn calf serum as described before (35).
Antibodies
Primary antibodies used were as follows. Mouse monoclonal antibody against the T7 tag (Novagen), mouse monoclonal antibody P4D1 against ubiquitin and goat polyclonal antibody against Calnexin (Santa Cruz Biotechnology), mouse monoclonal antibody (clone 16B12) against the HA tag (Biolegend), mouse monoclonal antibody (clone AC-15) against β-actin and monoclonal antibody (clone M2) against the FLAG tag (Sigma), mouse monoclonal antibody (clone 10E2) against the His tag (Abmart), and mouse monoclonal antibody (clone 9E10) against the Myc tag and monoclonal antibody (clone A9) against HMGCR were prepared from hybridomas (ATCC). Rabbit polyclonal antibodies against gp78 and HMGCR were prepared as described before (18). Rabbit polyclonal antibody against RNF145 (aa 511–663) was generated by immunizing rabbits followed by affinity purification with antigens.
Plasmids
The following plasmids were described in the indicated references or constructed by standard molecular cloning technics. pCMV-HMGCR-T7 encodes full-length hamster HMGCR followed by the T7 tag epitope. pCMV-Insig-1-Myc encodes human Insig-1 followed by the Myc epitope, and pCMV-Insig-2-Myc encodes human Insig-2 followed by the Myc epitope. pEF-HA-ubiquitin encodes human ubiquitin preceded by the HA tag epitope (7). pCMV-gp78-FLAG encodes human gp78 followed by the 3× FLAG epitope. pCMV-RNF145-FLAG encodes mouse RNF145 followed by the 3× FLAG epitope. pCMV-RNF145 (1–510)-FLAG encodes the N-terminal mouse RNF145 (aa 1–510) followed by the 3× FLAG epitope. pCMV-RNF145 (511–663)-FLAG encodes the C-terminal mouse RNF145 (aa 511–663) followed by the 3× FLAG epitope. pCMV-RNF145 (C537A)-FLAG, in which the cysteine 537 was mutated into alanine; pCMV-RNF145 (YLYF-AAAA)-FLAG, in which the Y81LYF was mutated into AAAA; and pCMV-RNF145 (C537A & YLYF-AAAA)-FLAG, in which the cysteine 537 was mutated into alanine and the Y81LYF was mutated into AAAA, were constructed with standard methods.
Cell culture
Medium A contained a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate. Medium B contained medium A supplemented with 5% fetal bovine serum. Medium C contained medium A supplemented with 5% lipoprotein-deficient serum, 1 μm lovastatin, and 50 μm mevalonate. All cells were kept in medium B at 37 °C and 5% CO2 and treated as indicated in the figure legends.
Transient transfection of CHO cells
Cells were set up on day 0 at 8 × 105 cells per 60-mm dish. On day 1, a total of 3 μg of DNA and 6 μg of linear polyethyleneimine were mixed and transfected to each dish. The medium was refreshed 6 h after transfection.
Generating of knockout cells
CHO cells deficient in gp78 were generated by transcription activator-like effector nuclease (TALEN) technology as described before (34). CHO cells deficient in Rnf145 and Rnf145 plus gp78 were generated by CRISPR/Cas9 (26). The guide RNA sequences were as follows: Cricetulus griseus gp78, CTTATCCAGTGTATTGTGTT; C. griseus Rnf145, GCTGACGTCCCATCTGTAG.
Immunoblot analysis
Cells were lysed with 200 μl of radioimmune precipitation assay buffer and mixed with the loading buffer (23.4 mm Tris-HCl, 5.625% (w/v) SDS, 1 m urea, 3.75% (v/v) glycerol, and 37.5 mm DTT in final concentrations) and incubated at 37 °C for 30 min. The protein concentration of each lysate was quantified by Pierce BCA protein assay, and equal amounts of total proteins were loaded for SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. Membranes were blocked by TBS–Tween (1‰) supplemented with 5% skim milk for 1 h at room temperature and then incubated with the indicated primary antibodies overnight at 4 °C. Membranes were washed three times with TBS–Tween and incubated with secondary antibodies (1:5000) diluted in TBS–Tween supplemented with 5% skim milk for 1 h at room temperature, followed by at least three washes with TBS–Tween. Quantification of the immunoblot was performed with ImageJ.
Immunoprecipitation
Cells were harvested and lysed in 1 ml of immunoprecipitation (IP) buffer (1 × PBS, 0.5% (v/v) Nonidet P-40, 5 mm EDTA, 5 mm EGTA, and protease inhibitors), followed by centrifugation at 12,000 × g for 10 min at 4 °C. Supernatants were immunoprecipitated with anti-Myc beads for 4–6 h at 4 °C. Beads were washed three times with IP buffer at 4 °C and boiled at 95 °C for 10 min. Aliquots were subjected to immunoblot analysis.
Ubiquitination of HMGCR
CHO cells were transfected and treated as described in the figure legends. Cells were lysed in HMG-IP buffer (1 × PBS containing 1% (v/v) Nonidet P-40, 1% (w/v) deoxycholate, 5 mm EDTA, 5 mm EGTA, 0.1 mm leupeptin, protease inhibitors, 10 μm MG132, and 10 mm N-ethylmaleimide). Lysates were first immunoprecipitated at 4 °C with 3 μg of polyclonal antibody against GFP plus 40 μl of protein A/G–agarose beads for 1 h. Beads were removed by centrifugation, and supernatants were then immunoprecipitated with 40 μl of anti-FLAG–agarose beads at 4 °C for 5 h. After incubation, beads were washed three times with HMG-IP buffer at 4 °C and boiled at 95 °C for 10 min. Aliquots were subjected to immunoblot analysis.
For detecting ubiquitination of endogenous HMGCR, CHO cells were set up on day 0 at 2.5 × 106 per 100-mm dish. Duplicate dishes were treated as described in the figure legends. 6 μg of polyclonal antibody against HMGCR plus 100 μl of protein A/G–agarose beads were used to precipitate HMGCR. Aliquots were subjected to immunoblot analysis.
In vitro ubiquitination
Ubiquitination experiments were carried out with 60 nm E1, 50 nm Ubc7, 10 μm FLAG-tagged ubiquitin and 1 μm RNF145 (511–663), 1 μm RNF145 (511–663) (C537A), or 300 nm gp78 (309–643) at 37 °C in the buffers containing 25 mm Tris-HCl (pH 7.4), 2 mm magnesium/ATP, and 0.1 mm DTT. Reactions were stopped by directly adding SDS loading buffer followed by incubation at 95 °C for 10 min.
Immunostaining
Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 1× PBS containing 0.1% Triton X-100 for 5 min. Cells were blocked with 1% BSA for 30 min, followed by incubation with primary and secondary antibodies diluted in 1% BSA for 1 h at room temperature (36). Cells were analyzed with a Leica confocal scanning laser microscope (Leica SP8) with a ×63 oil immersive objective.
Author contributions
L.-Y.J., W.J., J.W., J. Luo, X.-J.S., and B.-L.S. conceptualization; L.-Y.J. and W.J. data curation; L.-Y.J. and W.J. validation; L.-Y.J., W.J., N.T., Y.-N.X., J. Liu, and K.-Y.W. investigation; L.-Y.J. and B.-L.S. writing-original draft; L.-Y.J., W.J., and B.-L.S. project administration; L.-Y.J., J. Luo, and B.-L.S. writing-review and editing; B.-L.S. resources; B.-L.S. supervision; B.-L.S. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dan Liang, Bi-Yu Xiang, and Ying-Ying Liu for technical assistance.
This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0500100), the National Natural Science Foundation of China (31600651, 31430044, 31690102, 31771568, and 31701030), the China Postdoctoral Science Foundation (2016M592380), the 111 Project of the Ministry of Education of China (B16036), and the Natural Science Foundation of Hubei Province (2016CFA012 and 2017CFB617). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
- HMG-CoA
- 3-hydroxy-3-methylglutaryl-CoA
- HMGCR
- 3-hydroxy-3-methylglutaryl-CoA reductase
- RNF145
- ring finger protein 145
- ER
- endoplasmic reticulum
- Insig
- insulin-induced gene
- SREBP
- sterol regulatory element–binding protein
- SCAP
- SREBP cleavage–activating protein
- CHO
- Chinese hamster ovary
- KO
- knockout
- 25-HC
- 25-hydroxycholesterol
- shRNA
- short hairpin RNA
- aa
- amino acids
- SSD
- sterol-sensing domain
- LXR
- liver X receptor
- HA
- hemagglutinin
- TBS
- Tris-buffered saline
- IP
- immunoprecipitation.
References
- 1. Goldstein J. L., and Brown M. S. (1990) Regulation of the mevalonate pathway. Nature 343, 425–430 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- 2. Altmann S. W., Davis H. R. Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., and Graziano M. P. (2004) Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science 303, 1201–1204 10.1126/science.1093131 [DOI] [PubMed] [Google Scholar]
- 3. Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., and Song B. L. (2008) The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 7, 508–519 10.1016/j.cmet.2008.04.001 [DOI] [PubMed] [Google Scholar]
- 4. Ge L., Qi W., Wang L. J., Miao H. H., Qu Y. X., Li B. L., and Song B. L. (2011) Flotillins play an essential role in Niemann-Pick C1-like 1-mediated cholesterol uptake. Proc. Natl. Acad. Sci. U.S.A. 108, 551–556 10.1073/pnas.1014434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li P. S., Fu Z. Y., Zhang Y. Y., Zhang J. H., Xu C. Q., Ma Y. T., Li B. L., and Song B. L. (2014) The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1. Nat. Med. 20, 80–86 10.1038/nm.3417 [DOI] [PubMed] [Google Scholar]
- 6. Goldstein J. L., DeBose-Boyd R. A., and Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- 7. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., and DeBose-Boyd R. A. (2003) Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278, 52479–52490 10.1074/jbc.M310053200 [DOI] [PubMed] [Google Scholar]
- 8. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 10.1172/JCI0215593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown A. J., Sun L., Feramisco J. D., Brown M. S., and Goldstein J. L. (2002) Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 10, 237–245 10.1016/S1097-2765(02)00591-9 [DOI] [PubMed] [Google Scholar]
- 10. Song B. L., and DeBose-Boyd R. A. (2004) Ubiquitination of 3-hydroxy-3-methylglutaryl-CoA reductase in permeabilized cells mediated by cytosolic E1 and a putative membrane-bound ubiquitin ligase. J. Biol. Chem. 279, 28798–28806 10.1074/jbc.M402442200 [DOI] [PubMed] [Google Scholar]
- 11. Song B. L., Javitt N. B., and DeBose-Boyd R. A. (2005) Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189 10.1016/j.cmet.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 12. Lange Y., Ory D. S., Ye J., Lanier M. H., Hsu F. F., and Steck T. L. (2008) Effectors of rapid homeostatic responses of endoplasmic reticulum cholesterol and 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 283, 1445–1455 10.1074/jbc.M706967200 [DOI] [PubMed] [Google Scholar]
- 13. Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., and Goldstein J. L. (2007) Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 10.1073/pnas.0700899104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faust J. R., Luskey K. L., Chin D. J., Goldstein J. L., and Brown M. S. (1982) Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc. Natl. Acad. Sci. U.S.A. 79, 5205–5209 10.1073/pnas.79.17.5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A. (2003) Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 10.1016/S1097-2765(02)00822-5 [DOI] [PubMed] [Google Scholar]
- 16. Song B. L., Sever N., and DeBose-Boyd R. A. (2005) Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 10.1016/j.molcel.2005.08.009 [DOI] [PubMed] [Google Scholar]
- 17. Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., DeBose-Boyd R. A., Tang J. J., Li B. L., and Song B. L. (2007) Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 6, 115–128 10.1016/j.cmet.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 18. Liu T. F., Tang J. J., Li P. S., Shen Y., Li J. G., Miao H. H., Li B. L., and Song B. L. (2012) Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab. 16, 213–225 10.1016/j.cmet.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. N., Song B., DeBose-Boyd R. A., and Ye J. (2006) Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 281, 39308–39315 10.1074/jbc.M608999200 [DOI] [PubMed] [Google Scholar]
- 20. Jo Y., Lee P. C., Sguigna P. V., and DeBose-Boyd R. A. (2011) Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. U.S.A. 108, 20503–20508 10.1073/pnas.1112831108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zelcer N., Sharpe L. J., Loregger A., Kristiana I., Cook E. C., Phan L., Stevenson J., and Brown A. J. (2014) The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol. Cell. Biol. 34, 1262–1270 10.1128/MCB.01140-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang W., and Song B. L. (2014) Ubiquitin ligases in cholesterol metabolism. Diabetes Metab. J. 38, 171–180 10.4093/dmj.2014.38.3.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., and Wiertz E. (2004) Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279, 3525–3534 10.1074/jbc.M307453200 [DOI] [PubMed] [Google Scholar]
- 24. Tsai Y. C., Leichner G. S., Pearce M. M., Wilson G. L., Wojcikiewicz R. J., Roitelman J., and Weissman A. M. (2012) Differential regulation of HMG-CoA reductase and Insig-1 by enzymes of the ubiquitin-proteasome system. Mol. Biol. Cell 23, 4484–4494 10.1091/mbc.E12-08-0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neutzner A., Neutzner M., Benischke A. S., Ryu S. W., Frank S., Youle R. J., and Karbowski M. (2011) A systematic search for endoplasmic reticulum (ER) membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J. Biol. Chem. 286, 8633–8643 10.1074/jbc.M110.197459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lipkowitz S., and Weissman A. M. (2011) RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 11, 629–643 10.1038/nrc3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee P. C., Sever N., and Debose-Boyd R. A. (2005) Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J. Biol. Chem. 280, 25242–25249 10.1074/jbc.M502989200 [DOI] [PubMed] [Google Scholar]
- 29. Kuwabara P. E., and Labouesse M. (2002) The sterol-sensing domain: multiple families, a unique role? Trends Genet. 18, 193–201 10.1016/S0168-9525(02)02640-9 [DOI] [PubMed] [Google Scholar]
- 30. Davies J. P., Levy B., and Ioannou Y. A. (2000) Evidence for a Niemann-pick C (NPC) gene family: identification and characterization of NPC1L1. Genomics 65, 137–145 10.1006/geno.2000.6151 [DOI] [PubMed] [Google Scholar]
- 31. Gemmill R. M., West J. D., Boldog F., Tanaka N., Robinson L. J., Smith D. I., Li F., and Drabkin H. A. (1998) The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc. Natl. Acad. Sci. U.S.A. 95, 9572–9577 10.1073/pnas.95.16.9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cook E. C., Nelson J. K., Sorrentino V., Koenis D., Moeton M., Scheij S., Ottenhoff R., Bleijlevens B., Loregger A., and Zelcer N. (2017) Identification of the ER-resident E3 ubiquitin ligase RNF145 as a novel LXR-regulated gene. PLoS ONE 12, e0172721 10.1371/journal.pone.0172721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L., Rajbhandari P., Priest C., Sandhu J., Wu X., Temel R., Castrillo A., de Aguiar Vallim T. Q., Sallam T., and Tontonoz P. (2017) Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP. eLife 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y. J., Bian Y., Luo J., Lu M., Xiong Y., Guo S. Y., Yin H. Y., Lin X., Li Q., Chang C. C. Y., Chang T. Y., Li B. L., and Song B. L. (2017) Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat. Cell Biol. 19, 808–819 10.1038/ncb3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goldstein J. L., Basu S. K., and Brown M. S. (1983) Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98, 241–260 10.1016/0076-6879(83)98152-1 [DOI] [PubMed] [Google Scholar]
- 36. Chu B. B., Liao Y. C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H. H., Li B. L., and Song B. L. (2015) Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 161, 291–306 10.1016/j.cell.2015.02.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.