Abstract
The stemness maintenance of embryonic stem cells (ESCs) requires pluripotency transcription factors, including Oct4, Nanog, and Sox2. We have previously reported that protein arginine methyltransferase 7 (PRMT7), an epigenetic modifier, is an essential pluripotency factor that maintains the stemness of mouse ESCs, at least in part, by down-regulating the expression of the anti-stemness microRNA (miRNA) miR-24-2. To gain greater insight into the molecular basis underlying PRMT7-mediated maintenance of mouse ESC stemness, we searched for new PRMT7-down-regulated anti-stemness miRNAs. Here, we show that miR-221 gene–encoded miR-221-3p and miR-221-5p are anti-stemness miRNAs whose expression levels in mouse ESCs are directly repressed by PRMT7. Notably, both miR-221-3p and miR-221-5p targeted the 3′ untranslated regions of mRNA transcripts of the major pluripotency factors Oct4, Nanog, and Sox2 to antagonize mouse ESC stemness. Moreover, miR-221-5p silenced also the expression of its own transcriptional repressor PRMT7. Transfection of miR-221-3p and miR-221-5p mimics induced spontaneous differentiation of mouse ESCs. CRISPR-mediated deletion of the miR-221 gene, as well as specific antisense inhibitors of miR-221-3p and miR-221-5p, inhibited the spontaneous differentiation of PRMT7-depleted mouse ESCs. Taken together, these findings reveal that the PRMT7-mediated repression of miR-221-3p and miR-221-5p expression plays a critical role in maintaining mouse ESC stemness. Our results also establish miR-221-3p and miR-221-5p as anti-stemness miRNAs that target Oct4, Nanog, and Sox2 mRNAs in mouse ESCs.
Keywords: embryonic stem cell, microRNA (miRNA), pluripotency, microRNA mechanism, histone methylation, miR-221, Nanog, Oct4, PRMT7, stemness, arginine methylation, Sox2
Introduction
Embryonic stem cells (ESCs)3 can be derived from inner cell masses of blastocysts (1) and are defined by two main characteristics: 1) long-term self-renewal and 2) the ability to form all three germ layers and differentiate into all kinds of different cell types (pluripotency) (2). These stemness characteristics of ESCs are maintained by pluripotency transcription factors (e.g. Oct4, Nanog, and Sox2) and their regulatory networks (3, 4). These factors co-occupy and activate their own genes and other numerous genes important for maintaining ESC pluripotency (e.g. Oct4, Nanog, Sox2, STAT3, and Zic3) while repressing lineage-specific transcription factor genes (e.g. Hox clusters, Pax6, and Meis1) to prevent ESC differentiation (5, 6).
In addition to transcription factors, microRNAs (miRNAs) regulate pluripotency (7–9). miRNAs are small single-stranded RNAs of 21–25 nucleotides that negatively regulate gene expression. The miRNA-mediated targeting of mRNAs induces post-transcriptional repression through Argonaute-2–mediated mRNA degradation, translational repression, and mRNA deadenylation (10–13). miRNA-mediated regulation of stemness often results from changes in miRNA levels between the ESC state and the differentiated state (14, 15). For example, miR-27a has been identified as a differentiation-associated miRNA that is induced during ESC differentiation and directly targets the pluripotency factor Foxo1 and signal transducers (gp130 and smad3) to inhibit ESC pluripotency (16).
The expression patterns of miRNAs can be highly cell type–specific and thus are important to regulating cellular differentiation and development (17–19). It has been shown that the expression of miRNAs is regulated by multiple different mechanisms, including transcriptional control, epigenetic modulation, and post-transcriptional regulation (20, 21). Dysregulation of miRNAs is linked to cancer and other diseases (22, 23). For instance, the expression of multiple miRNAs (e.g. miR-124, miR-34, miR-9, and miR-200 families) is silenced by DNA hypermethylation in many types of cancer (24, 25).
We have reported previously that protein arginine methyltransferase 7 (PRMT7), a transcriptional co-repressor, is essential for maintaining mouse ESC stemness. In the same study, we showed that miR-24-3p and miR-24-2-5p levels are highly up-regulated by PRMT7 knockdown and are increased during mouse ESC differentiation (26). We also characterized miR-24-3p and miR-24-2-5p as anti-stemness miRNAs that can induce mouse ESC differentiation and directly inhibit the expression of the major pluripotency factors Oct4, Nanog, Sox2, Klf4, and c-Myc (26). We further showed that PRMT7-mediated repression of the expression of the miR-24-2 gene encoding miR-24-3p and miR-24-2-5p is required for maintaining mouse ESC stemness.
To better understand how PRMT7 maintains mouse ESC stemness, we sought to identify new anti-stemness miRNAs that are repressed by PRMT7. We thus re-analyzed our previous miRNA expression profile data of control and PRMT7-depleted mouse ESCs to determine which miRNAs in mouse ESCs are highly up-regulated by PRMT7 knockdown. We found that miR-221-3p and miR-221-5p act as anti-stemness miRNAs by targeting the 3′ untranslated regions (3′UTRs) of mRNA transcripts of the major pluripotency factors Oct4, Nanog, and Sox2. Our results also revealed that negative regulation of miR-221-3p and miR-221-5p expression by PRMT7 is necessary to maintain ESC pluripotency.
Results
Expression of miR-221-3p and miR-221-5p is directly repressed by PRMT7
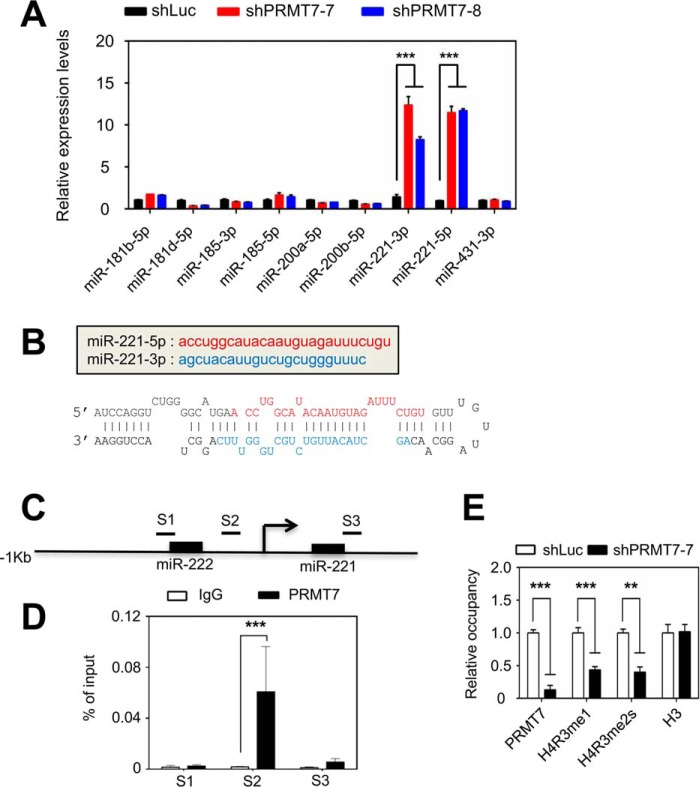
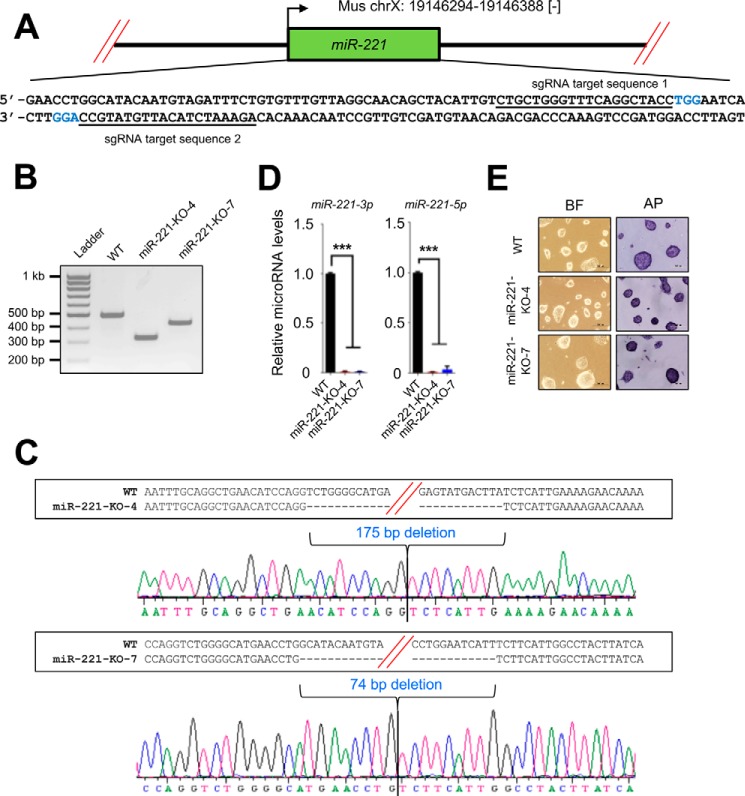
To identify PRMT7-repressed anti-stemness miRNAs, we searched miRNAs that are highly up-regulated by knockdown of the pluripotency factor PRMT7 in mouse ESCs using our previously reported microRNA microarray data (26). This search resulted in several candidate miRNAs, including miR-221-3p and miR-221-5p. Our miRNA-specific quantitative PCR results confirmed that miR-221-3p and miR-221-5p in V6.5 mouse ESCs were highly up-regulated by two independent PRMT7 shRNAs (shPRMT7-7 and shPRMT7-8) (Fig. 1A). The two mature microRNAs, miR-221-3p and miR-221-5p, are derived from the stem loop structure of the precursor miRNA miR-221 (Fig. 1B). The miR-221 gene is located on chromosome X.
Figure 1.
PRMT7 directly down-regulates the expression of the miR-221 gene. A, comparison of miRNA levels between shLuc-treated and PRMT7-depleted (shPRMT7–7 and shPRMT7–8) mouse ESCs. miRNA-specific quantitative PCR was performed. B, the sequences of mature miR-221-5p (red) and miR-221-3p (blue) and the predicted stem loop structure form of pre-mature miR-221. C, schematic representation of mouse miR-221 gene. S1, S2, and S3 indicate the PCR-amplified regions in ChIP assay. D, analysis of PRMT7 occupancy at the miR-221 promoter using quantitative ChIP. E, comparison of the occupancy of PRMT7, H4R3me1, H4R3me2s, and total H3 between shLuc-treated and PRMT7-depleted mouse ESCs at the miR-221 promoter region in V6.5 mouse ESCs. Data are presented as the mean ± S.D. of three independent experiments. **, p < 0.01 and ***, p < 0.001.
To determine whether miR-221 expression is directly repressed by PRMT7, we performed quantitative chromatin immunoprecipitation (ChIP) experiments. ChIP results showed that PRMT7 occupied the promoter region in the miR-221 gene in V6.5 mouse ESCs (Fig. 1, C and D). It has been generally accepted that PRMT7 monomethylates arginine residues, such as H4R3, for gene repression (27). We and others have also shown that PRMT7 represses gene expression by indirectly establishing symmetric dimethylation at H4R3 (H4R3me2s) (26–29). We therefore examined the effect of PRMT7 knockdown on monomethylated H4R3 (H4R3me1) and H4R3me2s levels at the miR-221 promoter. Our results showed that H4R3me1 and H4R3me2s levels at the miR-221 promoter were decreased by PRMT7 depletion (Fig. 1E). Together, these results indicate that PRMT7 represses the expression of miR-221, at least in part, by up-regulating repressive histone marks (e.g. H4R3me1 and H4R3me2s) at the miR-221 promoter in V6.5 mouse ESCs.
miR-221 has an anti-stemness function
It has been known that miR-221 has oncogenic functions (30), but little is known about the anti-stemness function of miR-221. To determine whether miR-221-3p and miR-221-5p have an anti-stemness function, we examined the effects of their mimics on mouse ESC stemness. An alkaline phosphatase (AP) staining analysis demonstrated that the transfection of miR-221-3p and miR-221-5p mimics induced spontaneous differentiation of V6.5 mouse ESCs (Fig. 2A). Consistent with this, the quantitative RT-PCR results showed that the mRNA levels of the major pluripotency factors Oct4, Nanog, Sox2, Klf4, and c-Myc were down-regulated by miR-221-3p and miR-221-5p mimics (Fig. 2B). We also compared the expression levels of miR-221-3p and miR-221-5p between V6.5 mouse ESCs and their retinoic acid (RA)–induced differentiated cells. As shown in Fig. 2C, the expression levels of miR-221-3p and miR-221-5p were increased upon RA treatment. To further validate the effect of miR-221-3p and miR-221-5p mimics on mouse ESC stemness, we used another mouse ESC line, R1. Similar to the results obtained using V6.5 mouse ESCs, the transfection of miR-221-3p and miR-221-5p induced spontaneous differentiation of R1 mouse ESCs (Fig. 2D) while down-regulating the pluripotency factors Oct4, Nanog, Sox2, and Klf4 (Fig. 2E). These results indicate that miR-221 negatively regulates ESC stemness.
Figure 2.
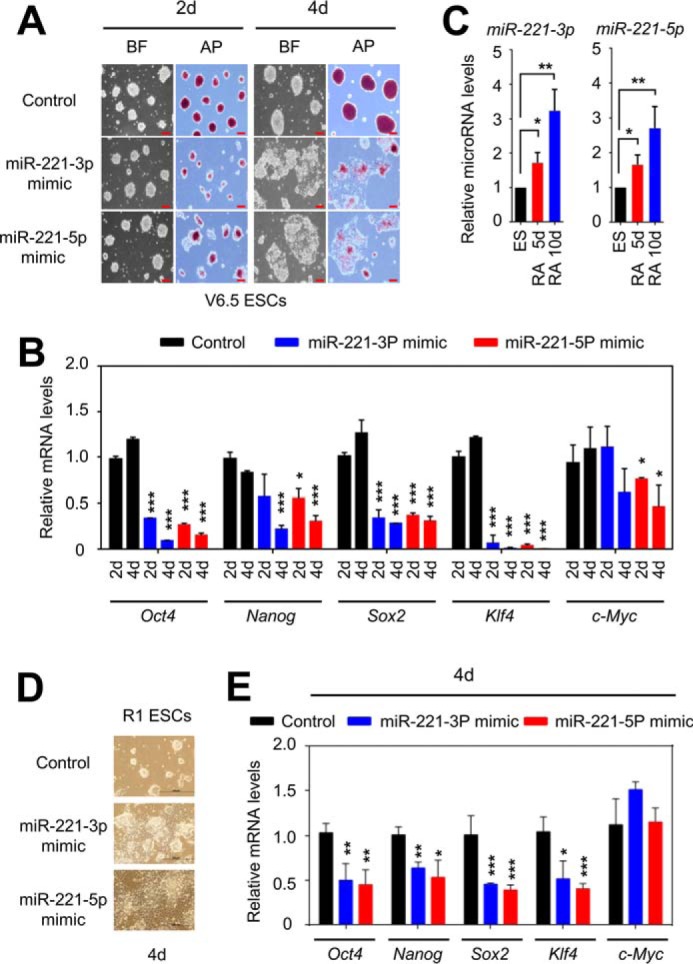
miR-221 mimics induce mouse ESC differentiation. A and D, microscopic images of V6.5 (A) and R1 (D) mouse ESCs after treatment with miR-221-3p and miR-221-5p mimics. Mouse ESCs were treated with miRNA mimics for 2 days (2d) or 4 days (4d). Red bars, 100 μm; black bars, 200 μm; BF, bright field; AP, alkaline phosphatase. B and E, analysis of relative Oct4, Nanog, Sox2, Klf4, and c-Myc mRNA levels in V6.5 (B) and R1 (E) mouse ESCs after treatment with miR-221-3p and miR-221-5p mimics. C, relative miR-221-3p and miR-221-5p levels during RA-induced V6.5 mouse ESC differentiation. RA 5d, retinoic acid treatment for 5 days; RA 10d, retinoic acid treatment for 10 days. Data are presented as the mean ± S.D. of three independent experiments. *, p < 0.05; **; p < 0.01; and ***, p < 0.001.
miR-221 targets the 3′UTRs of several pluripotency factors, including Oct4, Nanog, Sox2, and PRMT7
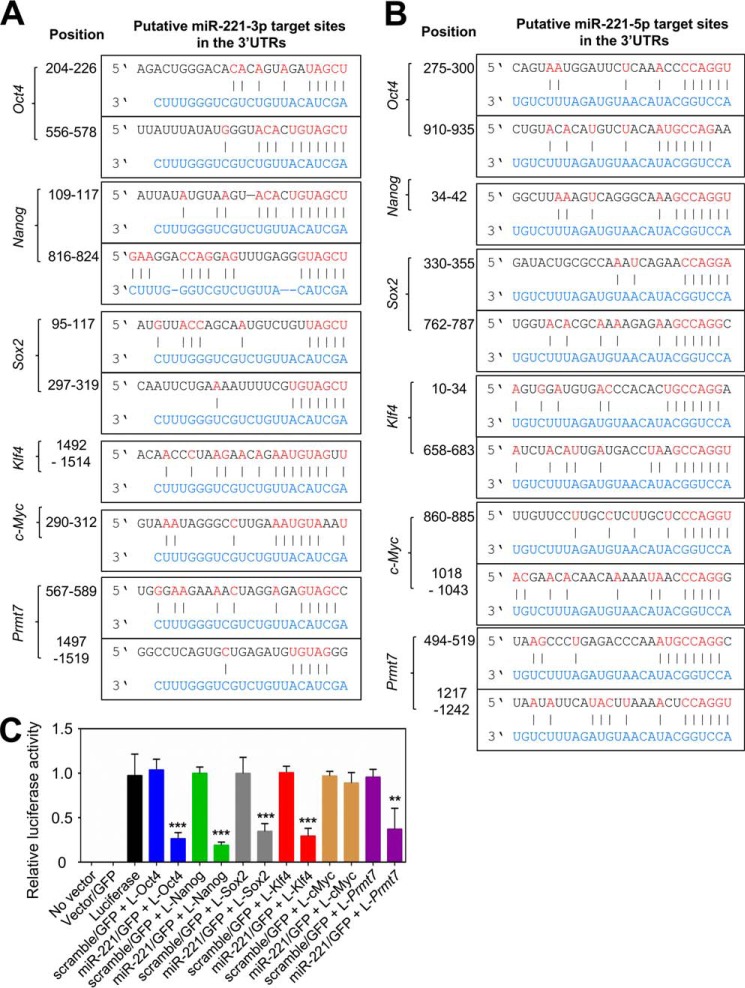
Because miR-221-3p and miR-221-5p act as anti-pluripotency miRNAs, we reasoned that their potential targets may be pluripotency factors. Specifically, we focused on determining whether miR-221 targets Oct4, Nanog, Sox2, Klf4, or c-Myc mRNAs, because their levels are down-regulated by miR-221-3p and miR-221-5p mimics and their proteins are major pluripotency factors that are critical for stemness maintenance. In addition, we examined the possibility that miR-221 silences its own transcriptional repressor PRMT7. miRNA-mediated mRNA targeting needs base pairing between an miRNA and its target mRNAs. Such base pairing is largely based on the complementarity between miRNAs' seed sequences (the nucleotide positions 2–8 in miRNAs) and their corresponding mRNA sequences. It has been known that the miRNA target sites in mRNAs are present in the 5′UTRs, open reading frames, and 3′UTRs (31). Interestingly, our analysis, based on several software and manual examinations, suggested that there are putative target sites for miR-221-3p and miR-221-5p in Oct4, Nanog, Sox2, Klf4, c-Myc, and Prmt7 3′UTRs (Fig. 3, A and B). To experimentally determine whether miR-221 can target Oct4, Nanog, Sox2, Klf4, c-Myc, and Prmt7 3′UTRs, we transfected both one of the luciferase expression plasmids containing the 3′UTRs of these pluripotency factors and the miR-221 expression plasmid encoding miR-221-3p and miR-221-5p into HEK 293T cells. Our results showed that miR-221 expression substantially reduced the luciferase activities of Oct4, Nanog, Sox2, Klf4, and Prmt7 3′UTRs but not c-Myc 3′UTR (Fig. 3C), suggesting that miR-221 can directly target Oct4, Nanog, Sox2, Klf4, and Prmt7 3′UTRs.
Figure 3.
miR-221 targets the 3′UTRs of mRNA transcripts of the pluripotency factors Oct4, Nanog, Sox2, Klf4, and PRMT7. A and B, putative target sites of miR-221-3p (A) and miR-221-5p (B) in the 3′UTRs of mouse Oct4, Nanog, Sox2, c-Myc, Klf4, and Prmt7. miR-221-3p and miR-221-5p sequences are shown in blue. C, the effect of miR-221 expression on the activity of reporter constructs containing Oct4-3′UTR (L-Oct4), Nanog-3′UTR (L-Nanog), Sox2-3′UTR (L-Sox2), Klf4-3′UTR (L-Klf4), c-Myc-3′UTR (L–c-Myc), and Prmt7-3′UTR (L-Prmt7). The miR-221 expression plasmid (pMDH1–PKG–miR-221–GFP) encoding both miR-221-3p and miR-221-5p was transfected with each luciferase reporter construct into HEK 293T cells. Firefly luciferase activity was normalized to the internal transfection control (Renilla luciferase). Data are presented as the mean ± S.D. of three independent experiments. **, p < 0.01 and ***, p < 0.001.
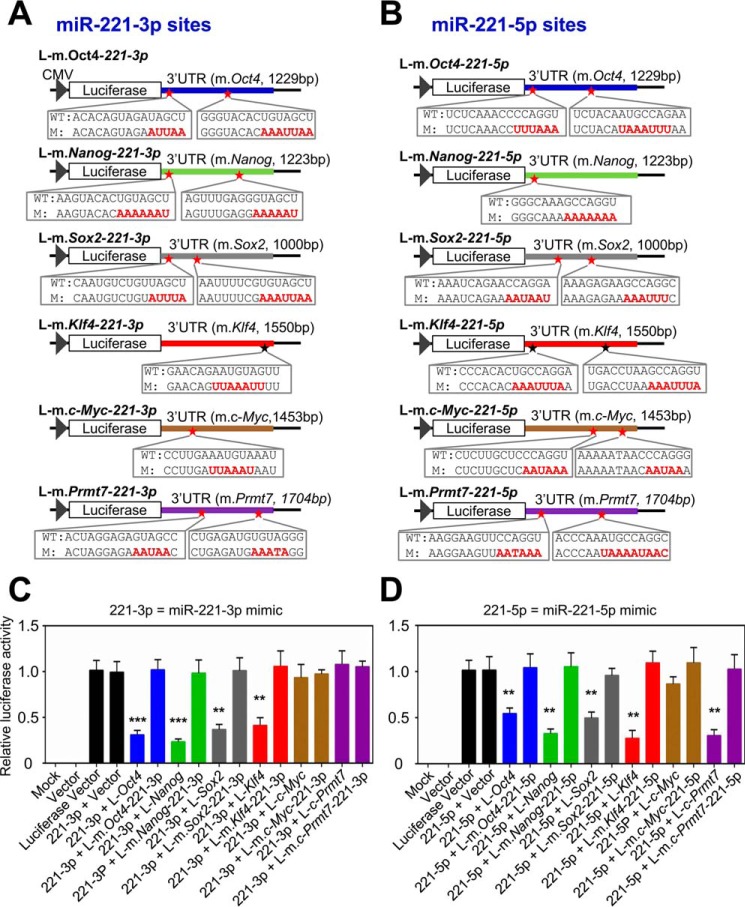
To determine specific target sites of miR-221-3p and miR-221-5p in Oct4, Nanog, Sox2, Klf4, and Prmt7 3′UTRs, we individually mutated putative target sites of miR-221-3p and miR-221-5p in the 3′UTRs in the reporter plasmids (Fig. 4, A and B). We then co-transfected WT (or mutant) 3′UTR reporter plasmids and miR-221-3p mimic (or miR-221-5p mimic) into HEK 293T cells. miR-221-3p and miR-221-5p mimics inhibited the luciferase activities of Oct4, Nanog, Sox2, and Klf4 3′UTRs while not impeding their mutant reporter plasmids (Fig. 4, C and D). Interestingly, miR-221-5p mimics, but not miR-221-3p mimics, reduced the reporter activity of Prmt7 3′UTR (Fig. 4, C and D). Together, these results indicate that target sites of miR-221-3p and miR-221-5p are located in Oct4, Nanog, Sox2, and Klf4 3′UTRs and that miR-221-5p's target site is also present in Prmt7 3′UTR.
Figure 4.
Both miR-221-3p and miR-221-5p can target the 3′UTRs of Oct4, Nanog, Sox2, and Klf4; miR-221-5p silences also the expression of Prmt7. A and B, schematic representation of luciferase reporter constructs containing Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR, c-Myc–3′UTR, and Prmt7-3′UTR. Mutations in putative target sites for miR-221-3p (A) and miR-221-5p (B) are shown in red. m, mutant. C and D, the effect of miR-221-3p and miR-221-5p mimics on the reporter activities of Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR, c-Myc–3′UTR, Prmt7-3′UTR and their mutants. Each WT or mutated reporter construct was transfected with miR-221-3p mimic (C) or miR-221-5p mimic (D) into HEK 293T cells. Firefly luciferase activity was normalized to the internal transfection control (Renilla luciferase). Data are presented as the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
PRMT7-mediated repression of miR-221-3p and miR-221-5p expression is required for maintaining mouse ESC stemness
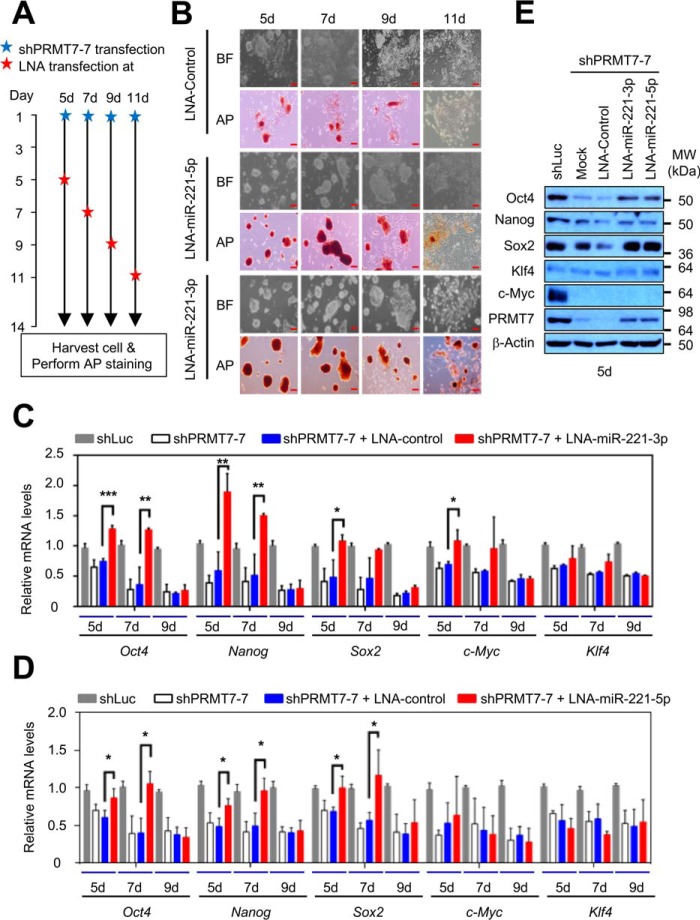
To determine whether the repression of miR-221-3p and miR-221-5p expression by PRMT7 is necessary for sustaining mouse ESC stemness, we examined the effects of locked nucleic acid (LNA) inhibitors of miR-221-3p and miR-221-5p (LNA-miR-221-3p and LNA-miR-221-5p) on the spontaneous differentiation of PRMT7-depleted mouse ESCs in which miR-221-3p and miR-221-5p levels are increased by PRMT7 depletion (LNA-miRNAs strongly bind to and inhibit their target miRNAs). Specifically, we treated cells with LNA-control, LNA-miR-221-3p, or LNA-miR-221-5p on days 5, 7, 9, and 11 after transfecting shPRMT7-7 into V6.5 mouse ESCs (Fig. 5A) and examined mouse ESC morphology, AP staining, protein levels, and mRNA expression. Our results showed that the treatment of PRMT7-depleted mouse ESCs with LNA-miR-221-3p or LNA-miR-221-5p (as compared with LNA-control) on days 5 and 7 inhibited spontaneous ESC differentiation and restored AP staining (Fig. 5B). Consistent with this, the expression levels of major pluripotency factors (e.g. Oct4 and Nanog) in shPRMT7-treated mouse ESCs were substantially recovered by treatment with LNA-miR-221-3p or LNA-miR-221-5p on days 5 and 7 (Fig. 5, C and D, red bar). The protein levels of these pluripotency factors and PRMT7 were also increased by the treatment of LNA-miR-221-3p and LNA-miR-221-5p on day 5 (Fig. 5E). In contrast, the treatment with LNA-miR-221-3p or LNA-miR-221-5p on day 9 or 11 had insignificant effects on the differentiation of PRMT7-depleted cells and barely reversed the expression of Oct4, Nanog, and Sox2, suggesting that treating miR-221 inhibitors at these two time points may be too late to inhibit the spontaneous ESC differentiation induced by PRMT7 knockdown (Fig. 5, B–D). These results indicate that transcriptional repression of miR-221-3p and miR-221-5p by PRMT7 is indispensable for maintaining Oct4, Nanog, Sox2, and PRMT7 levels and mouse ESC stemness.
Figure 5.
PRMT7-mediated down-regulation of miR-221-3p and miR-221-5p levels is indispensable for the maintenance of mouse ESC stemness. A, schematic representation of the procedure for the treatment of cells with LNA–miR-221-3p and LNA–miR-221-5p. Cells were harvested 14 days after transfection of shPRMT7-7. B, microscopic and AP staining images of PRMT7-depleted V6.5 mouse ESCs. Mouse ESCs were treated with LNA-control, LNA–miR-221-5p or LNA–miR-221-3p on days 5, 7, 9, or 11 after transfection of shPRMT7-7. Red bars, 100 μm; BF, bright field; AP, alkaline phosphatase. C and D, analysis of mRNA levels of Oct4, Nanog, Sox2, c-Myc, and Klf4 after treatment of shLuc-transfected or PRMT7-depleted V6.5 mouse ESCs with LNA–miR-221-3p (C) or LNA–miR-221-5p (D) at different time points (days 5, 7, and 9). E, Western blot analysis of Oct4, Nanog, Sox2, Klf4, c-Myc, PRMT7, and β-actin (loading control) levels after treatment of shLuc-transfected or PRMT7-depleted V6.5 mouse ESCs with LNA-control, LNA–miR-221-3p or LNA–miR-221-5p. Data are presented as the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
CRISPR-mediated deletion of the miR-221 gene impedes spontaneous differentiation of PRMT7-depleted mouse ESCs
To confirm that the repression of miR-221-3p and miR-221-5p expression by PRMT7 is required for mouse ESC stemness, we sought to determine whether miR-221 loss impedes the spontaneous differentiation of PRMT7-depleted mouse ESCs. To generate miR-221–null mouse ESCs, we used CRISPR-Cas9, which is a powerful tool for genome editing in living cells (32). Specifically, we used a double nicking strategy involving two guide RNAs and Cas9-D10A nickase (a mutant form of the RNA-guided, double-strand cleaving DNA endonuclease Cas9) because this strategy may introduce DNA double-strand breaks with significantly increased specificity around the target region (33). For this double nicking strategy, we first cloned two 20-bp–long single guide RNAs (sgRNAs) into a Cas9-D10A nickase expression plasmid that was used to target the mouse miR-221 gene (Fig. 6A). We then transfected these plasmids into V6.5 mouse ESCs and screened mouse ESC colonies to obtain miR-221–null clones. Because V6.5 mouse ESCs were derived from a male mouse embryo and the miR-221 gene is located on chromosome X, one allele of chromosome X needs to be deleted for the generation of miR-221–null mouse ESCs. Our genomic PCR results demonstrated that the miR-221 gene was deleted in two clones (miR-221–KO-4 and miR-221–KO-7) (Fig. 6B). DNA sequencing of genomic PCR products also confirmed the knockout of miR-221 (Fig. 6C). Furthermore, miRNA-specific quantitative PCR results showed that the expression of miR-221-3p and miR-221-5p was undetectable (Fig. 6D). Importantly, these two miR-221 knock-out clones (miR-221–KO-4 and miR-221–KO-7) were morphologically normal and positively stained by AP (Fig. 6E), in line with the findings of a previous report showing a minor effect of miR-221 inhibition on ESC proliferation (34).
Figure 6.
Generation of miR-221–null V6. 5 mouse ESCs using a CRISPR-Cas9 strategy. A, CRISPR-Cas9 targeting sites for generating miR-221–null V6.5 mouse ESCs. The two guide RNA (sgRNA) sequences are underlined. The PAM sequences are labeled in blue. B, PCR analysis of genomic DNA from WT and miR-221–null V6.5 mouse ESCs using primers flanking the deletion region in the miR-221 gene. The PCR band size for WT miR-221 is predicted to be 500 bp. C, DNA sequencing chromatograms for miR-221–KO-4 and miR-221–KO-7. D, comparison of miRNA levels between WT and miR-221–null V6.5 mouse ESCs (clones no. 4 and no. 7). E, microscopic and AP staining images of WT, miR-221–KO-4, and miR-221–KO-7 mouse ESCs. Black bars, 200 μm; BF, bright field; AP, alkaline phosphatase. Data are presented as the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
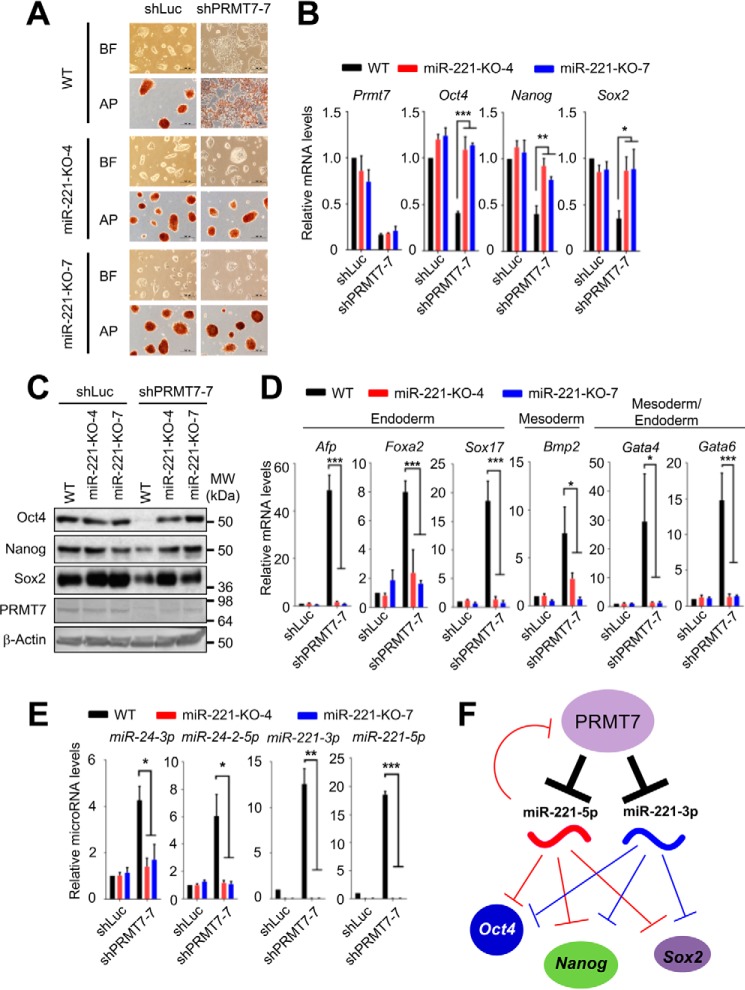
To determine whether miR-221 loss blocks the spontaneous differentiation of PRMT7-depleted mouse ESCs, we examined the effect of PRMT7 knockdown on the stemness of miR-221–null ESCs (miR-221–KO-4 and miR-221–KO-7). As evident by mouse ESC morphology as well as positive AP staining (Fig. 7A), miR-221 loss inhibited the spontaneous differentiation of PRMT7-depleted mouse ESCs. Consistent with this, miR-221 loss restored Oct4, Nanog, and Sox2 levels in PRMT7-depleted mouse ESCs (Fig. 7, B and C). Because we previously showed that PRMT7 knockdown induced endoderm (e.g. Afp, Foxa2, and Sox17), mesoderm (e.g. Bmp2), and mesoderm/endoderm (e.g. Gata4 and Gata6) markers (26), we determined whether miR-221 loss prevents the induction of these markers by PRMT7 knockdown. In fact, miR-221 loss inhibited the shPRMT7-mediated induction of Afp, Foxa2, Sox17, Bmp2, Gata4, and Gata6 (Fig. 7D). As mentioned earlier, we have previously reported that PRMT7 represses the expression of the miR-24-2 gene encoding miR-24-3p and miR-24-2-5p and that PRMT7-mediated repression of these miRNAs is necessary for sustaining mouse ESC stemness (26). Therefore, we examined whether miR-221 loss affects the expression levels of miR-24-3p and miR-24-2-5p. Interestingly, our results showed that miR-24-3p and miR-24-2-5p levels were highly reduced by miR-221 loss, suggesting that miR-221 positively regulates miR-24-3p and miR-24-2-5p levels (Fig. 7E). Together, these results further demonstrate that PRMT7-mediated repression of miR-221-3p and miR-221-5p expression is critical for maintaining mouse ESC stemness.
Figure 7.
miR-221 loss blocks spontaneous differentiation of PRMT7-depleted mouse ESCs. A, microscopic and AP staining images of WT and miR-221–null V6.5 mouse ESCs after treatment with shLuc or shPRMT7-7. Black bars, 200 μm; BF, bright field; AP, alkaline phosphatase. B and C, comparison of mRNA (B) and protein (C) levels of Prmt7, Oct4, Nanog, and Sox2 between WT and miR-221–null V6.5 mouse ESCs after treatment with shLuc or shPRMT7-7. D, comparison of mRNA levels of Afp, Foxa2, Sox17, Bmp2, Gata4, and Gata6 between WT and miR-221–null V6.5 mouse ESCs after treatment with shLuc or shPRMT7-7. E, comparison of miRNA levels between shLuc treatment and PRMT7 knockdown (shPRMT7-7) of WT mouse ESCs or miR-221–null V6.5 mouse ESCs (clones no. 4 and no. 7). F, a schematic illustration for PRMT7-mediated repression of miR-221-3p and miR-221-5p to maintain mouse ESC stemness. Data are presented as the mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Discussion
In the present study, the anti-stemness functions of miR-221-3p and miR-221-5p are supported by several lines of evidence. We showed that transfection of miR-221-3p or miR-221-5p mimics into two different ESC lines V6.5 and R1 caused the stemness loss of these ESC lines and that the loss of the miR-221 gene inhibited the spontaneous differentiation of PRMT7-depleted mouse ESCs. We also found that the expression levels of miR-221-3p and miR-221-5p were lower in mouse ESCs than in differentiated somatic cells (3T3 fibroblasts) (data not shown) and were increased during RA-induced differentiation. The results of our reporter assay, in combination with mutagenesis, showed that miR-221 miRNAs can target the 3′UTRs of the major pluripotency factors Oct4, Nanog, Sox2, Klf4, and Prmt7, indicating that miR-221 acts as an anti-stemness miRNA by targeting the mRNAs of multiple pluripotency genes, including Oct4, Nanog, Sox2, and Prmt7 (Fig. 4). In line with this, LNA-miRNA–mediated inhibition or CRISPR-mediated deletion of miR-221-3p and miR-221-5p restored the expression of Oct4, Nanog, Sox2, and Prmt7 in PRMT7-depleted mouse ESCs (Figs. 5 and 7).
The differentiation of ESCs requires both the down-regulation of pluripotency factors and the up-regulation of lineage-specific markers (35, 36). In this respect, our results showed that miR-221 loss in PRMT7-depleted mouse ESCs not only recovers the expression of pluripotency markers (e.g. Oct4, Nanog, and Sox2) but also down-regulates the expression of mesoderm and endoderm markers (Fig. 7). Interestingly, miR-221 is up-regulated in fully differentiated neurons (37) and plays a role in neuron differentiation (38). Therefore, it is likely that miR-221 not only antagonizes the stemness of mouse ESCs but also positively regulates terminal differentiation of certain types of stem cells. Similar to miR-221, there are many other anti-stemness miRNAs that inhibit ESC stemness and facilitate cell differentiation. For example, human miR-145 is an anti-stemness miRNA that promotes endoderm and ectoderm differentiation by targeting pluripotency factors, such as Oct4, Sox2, and Klf4 (39). In addition, miR-9 promotes the differentiation of neural stem cells while inhibiting their proliferation (40).
It has been shown that the miR-221 gene is transcriptionally regulated by several transcription factors. Estrogen receptor-α binds to the miR-221 promoter to repress miR-221 expression (41). The transcription factor FOSL1 activates miR-221 expression through its interaction with the miR-221 promoter in breast cancer cells (42). Fornari et al. (43) showed that p53 can regulate miR-221 levels in hepatocellular carcinoma. In the current study, our results indicate that PRMT7 binds to the miR-221 promoter and increases the repressive epigenetic marks H4R3me1 and H4R3me2s to down-regulate miR-221 expression. Thus, our findings provide a new miR-221 regulatory mechanism in which the expression of miR-221 is epigenetically repressed by the arginine methyltransferase PRMT7 in mouse ESCs.
Our results showed that miR-221-5p targeted its own repressor PRMT7 in addition to the well-known pluripotency factors Oct4, Nanog, and Sox2 (Fig. 4D), whereas PRMT7 directly repressed the miR-221 gene (Fig. 1). These results indicate a mutually antagonistic relationship between miR-221-5p and PRMT7. Interestingly, we have previously reported an additional antagonistic relationship between PRMT7 and miR-24-2 (i.e. miR-24-3p and miR-24-2-5p) (26). In the same study, we have also demonstrated that mouse ESC stemness requires PRMT7-mediated repression of miR-24-2 expression (26). Unexpectedly, our results in the current study showed that the CRISPR-mediated deletion of the miR-221 gene alone was sufficient to block the spontaneous differentiation of PRMT7-depleted mouse ESCs (Fig. 7A), suggesting the possibility that the PRMT7-mediated repression of the miR-221 gene plays a more predominant role in maintaining mouse ESC stemness than does the PRMT7-mediated repression of the miR-24-2 gene. However, miR-24-3p and miR-24-2-5p levels in PRMT7-depleted mouse ESCs were highly decreased by miR-221 loss (Fig. 7E). In addition, increased levels of miR-24-3p or miR-24-2-5p via transfection of miR-24-3p or miR-24-2-5p mimics induced spontaneous differentiation of mouse ESCs, as shown in our previous study (26). Therefore, it is likely that the repressed states of both miR-24-2 and miR-221 genes are critical for maintenance of mouse ESC stemness.
Interestingly, miR-221 is up-regulated in many types of tumors, including breast cancer, prostate cancer, lung cancer, and colorectal cancer (30). Tumor suppressor targets of miR-221 have been identified. For example, miR-221 directly targets the cell cycle inhibitor p27 (Kip1) and positively affects proliferation potential in prostate cancer (44). miR-221 also targets the tumor suppressor and cell cycle inhibitor p57 (CDKN1C) in hepatocarcinoma (45). Garofalo et al. (46) showed that miR-221 targets the tumor suppressors PTEN and TIMP3 to enhance the tumorigenicity of non–small cell lung cancer and hepatocellular carcinoma. Overexpression of miR-221 in several cancers is linked to resistance to various cancer therapies, in addition to a growth advantage in cancer cells (30). For instance, the expression of miR-221 is increased in several chemoresistant cancer cells (47, 48). For these reasons, miR-221 is considered an oncomiR that plays an important role in cancer development, and the inhibition of miR-221 in combination with other cancer treatments may be relevant to a new therapeutic strategy for cancer treatment (49, 50). Distinct from these studies, results reported here showed that miR-221 has an anti-stemness function to enhance the differentiation of mouse ESCs by down-regulating Oct4, Nanog, Sox2, and Prmt7 levels.
In summary, our results showed that miR-221-3p and miR-221-5p target the 3′UTRs of the major pluripotency factors Oct4, Nanog, and Sox2 in mouse ESCs, indicating an anti-stemness and pro-differentiation role for miR-221-3p and miR-221-5p. Because our results also uncovered that PRMT7 epigenetically represses the expression of miR-221-3p and miR-221-5p in mouse ESCs and that miR-221-5p silences the expression of Prmt7, it is possible that miR-221-5p and Prmt7 form a negative feedback loop (Fig. 7F). Finally, we provide evidence that the PRMT7-mediated repression of miR-221-3p and miR-221-5p expression is necessary for maintaining mouse ESC stemness.
Experimental procedures
Antibodies, plasmids, and other reagents
Anti-PRMT7 antibody was purchased from Santa Cruz Biotechnology (SC9882). Anti-Nanog (no. 61419), anti-Sox2 (no. 39823), and anti-H4R3me2s (no. 61187) antibodies were from Active Motif. Anti-Oct4 (no. 2840), anti-c-Myc (no. 5605), and anti-Klf4 (no. 4038) antibodies were from Cell Signaling Technology. Anti-β-actin antibody (A5441) was from Sigma-Aldrich. Anti-H4R3me1 antibody (PA5–27065) was from Thermo Fisher Scientific. Anti-H3 antibody (ab1791) was from Abcam. Mouse shPRMT7s (shPRMT7–7, TRCN0000097477; shPRMT7–8, TRCN0000097478) in the puromycin-resistant PLKO.1 vector were previously reported from this laboratory (26). Oligonucleotides used for site-directed mutagenesis, RT-PCR, ChIP-PCR, and CRISPR-Cas9 sgRNAs are listed in Table S1.
Mouse ESC culture
V6.5 mouse ESCs were cultured on gelatin-coated plates in complete knock-out Dulbecco's modified Eagle's medium (Life Technology), supplemented with 20% ESC grade fetal bovine serum (GenDEPOT), 2 mm l-glutamine, 50 μg/ml of penicillin, 50 μg/ml of streptomycin (Life Technology), 0.1 mm β-mercaptoethanol, 0.1 mm nonessential amino acid, and 1000 units/ml leukemia inhibitory factor (LIF). Mouse ESCs were trypsinized and split every 3 days, and the medium was changed daily.
RNA interference in mouse ESCs
The shRNA plasmids (30 μg) were transfected into mouse ESCs (5 × 105 cells in 0.8 ml) using the Gene Pulser Xcell electroporation system (500 microfarads and 250 V; Bio-Rad) according to the manufacturer's instructions. We have reported previously that the transfection in this electroporation condition was efficient (26). The cells were plated on a 6-cm dish and treated with puromycin (1.0 μg/ml). These cells were cultured for 14 days and then harvested for further analysis.
Quantitative PCR for miRNA and mRNA expression
Total RNAs were isolated using TRIzol RNA isolation reagents (Life Technology). To measure mRNA levels, cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Then, quantitative PCR was performed using the CFX384 real-time PCR detection system (Bio-Rad). GAPDH levels were used as the internal control.
For miRNA measurement, qScript microRNA cDNA synthesis kit (Quantabio) was used to synthesize microRNA cDNA. In brief, total RNAs (1 μg) were used in a poly(A) polymerase reaction that adds a poly(A) tail to miRNA, and polyadenylated miRNAs were further reverse-transcribed with qScript reverse transcriptase to synthesize miRNA cDNA. Quantitative PCR was performed. PCR data were normalized to sno66 to determine relative miRNA levels.
Chromatin immunoprecipitation assay
A ChIP assay was performed according to a previously described protocol with minor modifications (28, 51, 52). Mouse ESCs were first fixed with 1% formaldehyde. Cell pellets were then lysed with ChIP lysis buffer and sonicated for 15 min (30 s on and 30 s off for 15 cycles) to shear DNA using Bioruptor (Diagenode). Antibodies were added and incubated overnight at 4 °C. Preblocked protein A beads were added and incubated for 1–2 h to capture the antibody-DNA complex. The beads were then washed once with the following buffers: low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer. ChIP DNA was then eluted by the ChIP elution buffer (1% SDS and 0.1 m NaHCO3). The eluate was reverse cross-linked, and ChIP DNA was purified by the phenol/chloroform extraction method.
Transfection of miRNA mimics and LNA oligonucleotides
Mouse ESCs (5 × 105 cells) were trypsinized and transferred to 0.4-cm Gene Pulser electroporation cuvettes (Bio-Rad). Mouse miR-221-3p mimic, miR-221-5p mimic (Ambion), mouse LNA–miR-221-3p or LNA–miR-221-5p (Exiqon) (150 pmol) was added to the cuvettes. Electroporation (250 V and 500 microfarads) was performed using Gene Pulser Xcell Electroporation Systems (Bio-Rad). After electroporation, cells were rested at room temperature for 10 min and seeded in 6-cm dishes.
RA-induced differentiation of mouse ESCs
RA-induced differentiation of mouse ESCs was performed as described previously (26). In brief, mouse ESCs were trypsinized and cultured in a 10-cm Petri dish (Fisher) with ESC media, without adding leukemia inhibitory factor, to generate embryoid body (EB). After 5 days, embryoid bodies were transferred to a gelatin-coated tissue culture dish, and 0.5 μm RA was added to induce differentiation.
Luciferase reporter assays
The 3′UTRs of mouse Oct4, Nanog, Sox2, Klf4, c-Myc, and Prmt7 genes in the pMIR-REPORT (Ambion) vector have been previously described (26). The predicted miR-221-3p and miR-221-5p target sites in the 3′UTRs were mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The pre-mature miR-221 sequence was synthesized and cloned into an miRNA expression vector (pMDH1–PKG–miR-221–GFP). To perform the luciferase assay, a pMIR-REPORT vector containing WT or mutant 3′UTRs of Oct4, Nanog, Sox2, Klf4, c-Myc, and Prmt7, together with pMDH1–PKG–miR-221–GFP and Renilla vector (Promega), was transfected into HEK 293T cells. After 48 h of incubation, the transfected cells were harvested. Luciferase activity was measured using the Dual-Luciferase Reporter assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Western blot analysis
V6.5 ESCs were lysed using mammalian lysis buffer (20 mm Tris-HCl, 137 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 10% glycerol, 1% Triton X-100, and 0.2 mm PMSF) to obtain total cell lysates. The protein concentration was determined using the Bradford protein assay (Bio-Rad). Total proteins (20 μg) were subjected to a standard Western blot analysis. Antibodies against PRMT7, Oct4, Nanog, Sox2, c-Myc, or Klf4 were used for immunoblotting.
CRISPR-Cas9 gene editing
pSpCas9n(BB)-2A-GFP (PX461) plasmid with Cas9 (D10A mutant) nickase was obtained from Addgene. To generate miR-221–null V6.5 mouse ESCs, two sgRNA sequences that target miR-221 (sgRNA target sequence 1: CTGCTGGGTTTCAGGCTACC; sgRNA target sequence 2: AGAAATCTACATTGTATGCC) were separately cloned into pSpCas9n(BB)-2A-GFP using Cas9–miR-221-1 and Cas9–miR-221-2 primers (Table S1). sgRNA-containing plasmids were transfected into V6.5 mouse ESCs using Lipofectamine 3000 (Life Technology) according to the manufacturer's instructions. After 48 h of incubation, mouse ESCs were trypsinized and sorted by GFP signals. GFP-positive mouse ESCs were plated into a 96-well plate (one cell per well) to obtain a single clone. To verify miR-221 deletion, DNA purified from cells was PCR amplified using Cas9–miR-221-seq (Table S1) and PCR products were sequenced.
Statistical analysis
The statistical significance between the two groups was analyzed by Student's t test using Prism software (GraphPad Software, Inc.). Data are presented as the mean ± standard deviation (S.D.) of at least three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 indicate statistically significant changes.
Author contributions
T.-Y. C., S.-H. L., and M. G. L. conceptualization; M. G. L. resources; T.-Y. C., S.-H. L., S. S. D., and M. G. L. data curation; T.-Y. C., S.-H. L., S. S. D., and M. G. L. formal analysis; M. G. L. supervision; M. G. L. funding acquisition; T.-Y. C. and S.-H. L. validation; T.-Y. C., S.-H. L., S. S. D., and M. G. L. investigation; T.-Y. C., S.-H. L., S. S. D., and M. G. L. methodology; T.-Y. C. and M. G. L. writing-original draft; T.-Y. C. and M. G. L. writing-review and editing; S.-H. L. and S. S. D. software.
Supplementary Material
Acknowledgments
We thank the research staff in the Flow Cytometry & Cellular Imaging Facility, which is supported in part by the National Institutes of Health through M. D. Anderson's Cancer Center Support Grant CA016672. We also thank Ann Sutton (Department of Scientific Publications, M. D. Anderson) for the editorial assistance.
This study was supported by National Institutes of Health Grants R01 CA207098, R01 CA207109, and R01 GM095659 (to M.G.L.); the Cancer Prevention and Research Institute of Texas Grant RP140271 (to M. G. L.); and the Center for Cancer Epigenetics at M. D. Anderson Grant (to M. G. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
- ESC
- embryonic stem cell
- miRNA
- microRNA
- AP
- alkaline phosphatase
- LNA
- locked nucleic acid
- CRISPR
- clustered regularly interspaced short palindromic repeats
- sgRNA
- single guide RNA.
References
- 1. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., and Jones J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 2. Donovan P. J., and Gearhart J. (2001) The end of the beginning for pluripotent stem cells. Nature 414, 92–97 10.1038/35102154 [DOI] [PubMed] [Google Scholar]
- 3. Loh K. M., and Lim B. (2011) A precarious balance: Pluripotency factors as lineage specifiers. Cell Stem Cell 8, 363–369 10.1016/j.stem.2011.03.013 [DOI] [PubMed] [Google Scholar]
- 4. Thomson M., Liu S. J., Zou L.-N., Smith Z., Meissner A., and Ramanathan S. (2011) Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875–889 10.1016/j.cell.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loh Y.-H., Wu Q., Chew J.-L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K.-Y., Sung K. W., Lee C. W. H., Zhao X.-D., Chiu K.-P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C.-L., Ruan Y., Lim B., and Ng H.-H. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 10.1038/ng1760 [DOI] [PubMed] [Google Scholar]
- 6. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., and Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanellopoulou C., Muljo S. A., Kung A. L., Ganesan S., Drapkin R., Jenuwein T., Livingston D. M., and Rajewsky K. (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 10.1101/gad.1248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y., Medvid R., Melton C., Jaenisch R., and Blelloch R. (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385 10.1038/ng1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li M. A., and He L. (2012) MicroRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. BioEssays 34, 670–680 10.1002/bies.201200019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gregory R. I., Chendrimada T. P., Cooch N., and Shiekhattar R. (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 10.1016/j.cell.2005.10.022 [DOI] [PubMed] [Google Scholar]
- 11. Hutvágner G., and Zamore P. D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 10.1126/science.1073827 [DOI] [PubMed] [Google Scholar]
- 12. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., and Tuschl T. (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 13. Wu L., Fan J., and Belasco J. G. (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 4034–4039 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morin R. D., O'Connor M. D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.-L., Zhao Y., McDonald H., Zeng T., Hirst M., Eaves C. J., and Marra M. A. (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610–621 10.1101/gr.7179508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stadler B., Ivanovska I., Mehta K., Song S., Nelson A., Tan Y., Mathieu J., Darby C., Blau C. A., Ware C., Peters G., Miller D. G., Shen L., Cleary M. A., and Ruohola-Baker H. (2010) Characterization of microRNAs Involved in embryonic stem cell states. Stem Cells Dev. 19, 935–950 10.1089/scd.2009.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma Y., Yao N., Liu G., Dong L., Liu Y., Zhang M., Wang F., Wang B., Wei X., Dong H., Wang L., Ji S., Zhang J., Wang Y., Huang Y., and Yu J. (2015) Functional screen reveals essential roles of miR-27a/24 in differentiation of embryonic stem cells. EMBO J. 34, 361–378 10.15252/embj.201489957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H. R., Kauppinen S., and Plasterk R. H. A. (2005) MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- 18. Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stähler C., Meese E., and Keller A. (2016) Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alvarez-Garcia I., and Miska E. A. (2005) MicroRNA functions in animal development and human disease. Development 132, 4653–4662 10.1242/dev.02073 [DOI] [PubMed] [Google Scholar]
- 20. Obernosterer G., Leuschner P. J. F., Alenius M., and Martinez J. (2006) Post-transcriptional regulation of microRNA expression. RNA 12, 1161–1167 10.1261/rna.2322506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gulyaeva L. F., and Kushlinskiy N. E. (2016) Regulatory mechanisms of microRNA expression. J. Trans. Med. 14, 143 10.1186/s12967-016-0893-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kloosterman W. P., and Plasterk R. H. A. (2006) The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 23. Sayed D., and Abdellatif M. (2011) MicroRNAs in development and disease. Physiol. Rev. 91, 827–887 10.1152/physrev.00006.2010 [DOI] [PubMed] [Google Scholar]
- 24. Croce C. M. (2009) Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuki H., Maruyama R., Yamamoto E., and Kai M. (2012) DNA methylation and microRNA dysregulation in cancer. Mol. Oncol. 6, 567–578 10.1016/j.molonc.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee S.-H., Chen T.-Y., Dhar S. S., Gu B., Chen K., Kim Y. Z., Li W., and Lee M. G. (2016) A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness. Nucleic Acids Res. 44, 10603–10618 10.1093/nar/gkw788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zurita-Lopez C. I., Sandberg T., Kelly R., and Clarke S. G. (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J. Biol. Chem. 287, 7859–7870 10.1074/jbc.M111.336271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dhar S. S., Lee S.-H., Kan P.-Y., Voigt P., Ma L., Shi X., Reinberg D., and Lee M. G. (2012) Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 26, 2749–2762 10.1101/gad.203356.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee J.-H., Cook J. R., Yang Z.-H., Mirochnitchenko O., Gunderson S. I., Felix A. M., Herth N., Hoffmann R., and Pestka S. (2005) PRMT7, a new protein arginine Methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 280, 3656–3664 10.1074/jbc.M405295200 [DOI] [PubMed] [Google Scholar]
- 30. Howe E. N., Cochrane D. R., and Richer J. K. (2012) The miR-200 and miR-221/222 microRNA families: Opposing effects on epithelial identity. J. Mammary Gland Biol. Neoplasia 17, 65–77 10.1007/s10911-012-9244-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Filipowicz W., Bhattacharyya S. N., and Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9, 102–114 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- 32. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran F. A., Hsu P. D., Lin C.-Y., Gootenberg J. S., Konermann S., Trevino A. E., Scott D. A., Inoue A., Matoba S., Zhang Y., and Zhang F. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li J., Bei Y., Liu Q., Lv D., Xu T., He Y., Chen P., and Xiao J. (2015) MicroRNA-221 is required for proliferation of mouse embryonic stem cells via p57 targeting. Stem Cell Rev. Rep. 11, 39–49 10.1007/s12015-014-9543-y [DOI] [PubMed] [Google Scholar]
- 35. Young R. A. (2011) Control of the embryonic stem cell state. Cell 144, 940–954 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivey K. N., and Srivastava D. (2010) MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7, 36–41 10.1016/j.stem.2010.06.012 [DOI] [PubMed] [Google Scholar]
- 37. Pandey A., Singh P., Jauhari A., Singh T., Khan F., Pant A. B., Parmar D., and Yadav S. (2015) Critical role of the miR-200 family in regulating differentiation and proliferation of neurons. J. Neurochem. 133, 640–652 10.1111/jnc.13089 [DOI] [PubMed] [Google Scholar]
- 38. Hamada N., Fujita Y., Kojima T., Kitamoto A., Akao Y., Nozawa Y., and Ito M. (2012) MicroRNA expression profiling of NGF-treated PC12 cells revealed a critical role for miR-221 in neuronal differentiation. Neurochem. Int. 60, 743–750 10.1016/j.neuint.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 39. Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., and Kosik K. S. (2009) MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- 40. Zhao C., Sun G., Li S., and Shi Y. (2009) A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Leva G., Gasparini P., Piovan C., Ngankeu A., Garofalo M., Taccioli C., Iorio M. V., Li M., Volinia S., Alder H., Nakamura T., Nuovo G., Liu Y., Nephew K. P., and Croce C. M. (2010) MicroRNA cluster 221–222 and estrogen receptor α interactions in breast cancer. J. Natl. Cancer Inst. 102, 706–721 10.1093/jnci/djq102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stinson S., Lackner M. R., Adai A. T., Yu N., Kim H.-J., O'Brien C., Spoerke J., Jhunjhunwala S., Boyd Z., Januario T., Newman R. J., Yue P., Bourgon R., Modrusan Z., Stern H. M., Warming S., de Sauvage F. J., Amler L., Yeh R.-F., and Dornan D. (2011) TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 4, ra41 10.1126/scisignal.2001538 [DOI] [PubMed] [Google Scholar]
- 43. Fornari F., Milazzo M., Galassi M., Callegari E., Veronese A., Miyaaki H., Sabbioni S., Mantovani V., Marasco E., Chieco P., Negrini M., Bolondi L., and Gramantieri L. (2014) p53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol. Cancer Res. 12, 203–216 10.1158/1541-7786.MCR-13-0312-T [DOI] [PubMed] [Google Scholar]
- 44. Galardi S., Mercatelli N., Giorda E., Massalini S., Frajese G. V., Ciafrè S. A., and Farace M. G. (2007) miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 282, 23716–23724 10.1074/jbc.M701805200 [DOI] [PubMed] [Google Scholar]
- 45. Fornari F., Gramantieri L., Ferracin M., Veronese A., Sabbioni S., Calin G. A., Grazi G. L., Giovannini C., Croce C. M., Bolondi L., and Negrini M. (2008) miR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27, 5651–5661 10.1038/onc.2008.178 [DOI] [PubMed] [Google Scholar]
- 46. Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S.-S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P., Gasparini P., Gonelli A., Costinean S., Acunzo M., Condorelli G., and Croce C. M. (2009) miR-221&222 regulate TRAIL-resistance and enhance tumorigenicity through PTEN and TIMP3 down-regulation. Cancer Cell 16, 498–509 10.1016/j.ccr.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Pogribny I. P., Filkowski J. N., Tryndyak V. P., Golubov A., Shpyleva S. I., and Kovalchuk O. (2010) Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 127, 1785–1794 10.1002/ijc.25191 [DOI] [PubMed] [Google Scholar]
- 48. Zhou M., Liu Z., Zhao Y., Ding Y., Liu H., Xi Y., Xiong W., Li G., Lu J., Fodstad O., Riker A. I., and Tan M. (2010) MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285, 21496–21507 10.1074/jbc.M109.083337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park J.-K., Lee E. J., Esau C., and Schmittgen T. D. (2009) Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 38, e190–e199 10.1097/MPA.0b013e3181ba82e1 [DOI] [PubMed] [Google Scholar]
- 50. Gullà A., Di Martino M. T., Gallo Cantafio M. E., Morelli E., Amodio N., Botta C., Pitari M. R., Lio S. G., Britti D., Stamato M. A., Hideshima T., Munshi N. C., Anderson K. C., Tagliaferri P., and Tassone P. (2016) A 13 mer LNA-i-miR-221 inhibitor restores drug-sensitivity in melphalan-refractory multiple myeloma cells. Clin. Cancer Res. 22, 1222–1233 10.1158/1078-0432.CCR-15-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li N., Li Y., Lv J., Zheng X., Wen H., Shen H., Zhu G., Chen T. Y., Dhar S. S., Kan P. Y., Wang Z., Shiekhattar R., Shi X., Lan F., Chen K., Li W., Li H., and Lee M. G. (2016) ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell 63, 470–484 10.1016/j.molcel.2016.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dhar S. S., Lee S. H., Chen K., Zhu G., Oh W., Allton K., Gafni O., Kim Y. Z., Tomoiga A. S., Barton M. C., Hanna J. H., Wang Z., Li W., and Lee M. G. (2016) An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 44, 3659–3674 10.1093/nar/gkv1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.