Summary
N6-methyladenosine (m6A) is an abundant modification in eukaryotic mRNA, regulating mRNA dynamics by influencing mRNA stability, splicing, export and translation. However, the precise m6A regulating machinery still remains incompletely understood. Here we demonstrate that ZC3H13, a zinc finger protein, plays an important role in modulating RNA m6A methylation in the nucleus. We show that knockdown of Zc3h13 in mouse embryonic stem cell significantly decreases global m6A level on mRNA. Upon Zc3h13 knockdown a great majority of WTAP, Virilizer and Hakai translocate to the cytoplasm, suggesting that Zc3h13 is required for nuclear localization of the Zc3h13-WTAP-Virilizer-Hakai complex, which is important for RNA m6A methylation. Finally, Zc3h13 depletion, as does WTAP, Virilizer or Hakai, impairs self-renewal and triggers mESC differentiation. Taken together, our findings demonstrate that Zc3h13 plays a critical role in anchoring WTAP, Virilizer and Hakai in the nucleus to facilitate m6A methylation and to regulate mESC self-renewal.
Keywords: Zc3h13, m6A, mESC self-renewal, nuclear localization
eTOC blurb
Wen et al. show that Zc3h13 is a critical RNA m6A regulator that is part of an evolutionarily conserved complex containing WTAP, Virilizer, and Hakai. Zc3h13 anchors this complex in the nucleus to facilitate m6A methylation and mESC pluripotency.
Introduction
Identified in 1970s, N6-Adenosine methylation (m6A) is the most abundant chemical modification occurring on mRNA and long noncoding RNA, and is present broadly among eukaryotic species from yeast, plants, insects to mammals (Desrosiers et al., 1974; Rottman et al., 1974). Recent studies in a variety of species have revealed a role for RNA m6A modification in yeast sporulation (Bodi et al., 2010), plant development (Zhong et al., 2008), Drosophila neuronal function and sex determination (Haussmann et al., 2016; Lence et al., 2016), mouse embryonic stem cell (mESC) stemness and differentiation (Batista et al., 2014; Geula et al., 2015; Wang et al., 2014b), zebrafish embryogenesis (Zhao et al., 2017) and the DNA damage response (Xiang et al., 2017). At the molecular level, m6A has been demonstrated to regulate RNA stability, translation, splicing and export (Meyer et al., 2015; Sommer et al., 1978; Wang et al., 2014a; Wang et al., 2015; Xiao et al., 2016).
RNA m6A methylation is mediated by a core complex of three components, METTL3, METTL14 and WTAP (Liu et al., 2014). The crystal structure of the METTL3 and METTL14 complex suggests that METTL3 is the catalytic component while METTL14, which forms a heterodimer with METTL3, contributes to substrate RNA binding (Wang et al., 2016a; Wang et al., 2016b). WTAP, on the other hand, recruits METTL3 and METTL14 to nuclear speckles (Ping et al., 2014). This core complex is believed to be responsible for methylation of about 0.1–0.5% of total adenosine (A) in polyadenylated RNA (Wei et al., 1975), installing methylation on a conserved sequence motif of “RRACH” (R represents A or G, and H represents A, C or U), mainly near stop codons and 3′ UTR (Dominissini et al., 2012; Meyer et al., 2012).
In addition to the core complex, a number of other proteins have been implicated in regulating RNA m6A. For instance, Virilizer and Hakai were identified as the components associated with WTAP in mammalian cells (Horiuchi et al., 2013). The depletion of Virilizer or Hakai decreases RNA m6A level and affects Drosophila sex determination and Arabidopsis development, respectively (Haussmann et al., 2016; Lence et al., 2016; Ruzicka et al., 2017; Schwartz et al., 2014;). Indeed, purification of WTAP using different antibodies identified 26 core interacting factors among hundreds of potential WTAP binding proteins (Horiuchi et al., 2013). In addition, a separate study suggested that more than 100 proteins may bind METTL3 or METTL14 (Malovannaya et al., 2011). These findings suggest interacting proteins outside the core complex are likely to contribute to the regulation of RNA m6A methylation. Recently, Wan et al. studied endogenous protein complexes from different species in metazoan using quantitative mass spectrometry and identified Zc3h13-WTAP-Virilizer-Hakai as an evolutionarily conserved complex (Wan et al., 2015). Although WTAP, Virilizer and Hakai have been linked to m6A manipulation, the role of Zc3h13 and how the Zc3h13-WTAP-Virilizer-Hakai complex components work together to facilitate mRNA m6A processing remain unknown.
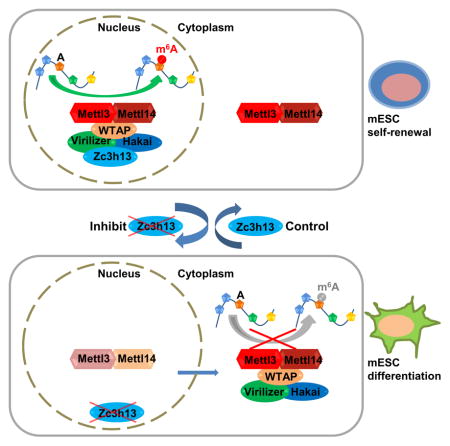
In this study, we provide evidence for the physical interaction among Zc3h13 and WTAP, Virilizer, Hakai, and identified the C-terminal region of Zc3h13 to be necessary and sufficient for its interaction with the other members of the complex. LC-MS/MS shows that Zc3h13 is critical for m6A methylation, and Zc3h13 depletion mainly affects m6A methylation at 3′ UTR of mRNA. Importantly, Zc3h13 knockdown also leads to a significant decrease of the nuclear presence of WTAP, Virilizer and Hakai, indicating that Zc3h13 is critical for nuclear localization of the other components of its associated complex, but not vice versa. Correlating with a robust decrease in m6A level, Zc3h13 depletion significantly impairs self-renewal and triggers differentiation in mESCs. Similar phenotypes were observed upon inhibition of WTAP, Virilizer or Hakai in mESCs. Our findings suggest that Zc3h13 is critical for mESC self-renewal by anchoring the other components of the complex in the nucleus for mRNA m6A methylation.
Results
Zc3h13 interacts with WTAP, Virilizer and Hakai
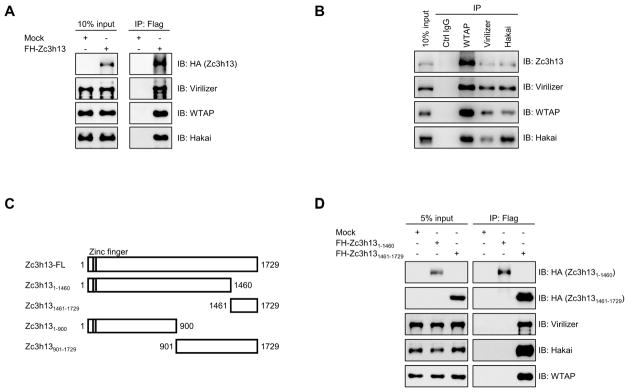
As discussed above, Zc3h13 was identified in a WTAP pull-down experiment but whether it plays a role in m6A methylation was unknown. To investigate Zc3h13 function, we first carried out co-immunoprecipitation (co-IP) using a Flag-HA-tagged Zc3h13 in mESCs. As shown in Figure 1A, we identified interactions of Zc3h13 with Virilizer, WTAP, and Hakai (Figure 1A). In the reciprocal IP, Zc3h13 was also pulled down by antibodies of Virilizer, WTAP or Hakai, respectively (Figure 1B). Moreover, treating cell lysates with RNase did not interfere with their interactions in the co-IP experiments (Figure S1A) suggesting that the interaction of Zc3h13 with WTAP-Virilizer-Hakai is likely to be independent of RNA. Our findings are consistent with the previous mass spectrometry studies of native macromolecular complex, which suggested that Zc3h13, WTAP, Virilizer and Hakai are in the same biochemical complex (Wan et al., 2015). We next aimed to identify the corresponding region of Zc3h13 for interaction. Zc3h13 was divided into four different segments (Figure 1C; Figure S1B). While the N-terminal regions of Zc3h13 (aa 1-900 or aa 1-1460) did not bind WTAP, Virilizer and Hakai, the C-terminal regions of Zc3h13 (aa 901-1729 or aa 1461-1729) interacted with WTAP, Virilizer and Hakai (Figure 1D; Figure S1C). Based on these data, we conclude that Zc3h13 physically associates with WTAP, Virilizer and Hakai, and the C-terminal domain (aa 1461-1729) is necessary and sufficient for the interactions.
Figure 1. Zc3h13 interacts with WTAP, Virilizer and Hakai.
(A) Co-immunoprecipitation analysis showing Zc3h13 interacts with WTAP, Virilizer and Hakai in mESC. Mock, mES cells transfected with empty vector; IP, immunoprecipitation; IB, immunoblotting.
(B) Reciprocal-IP assay indicating Zc3h13 was immunoprecipitated by anti-WTAP, anti-Virilizer and anti-Hakai antibodies. IgG was used as a control.
(C) Schematic representation of Zc3h13 full-length (FL) protein and various truncated fragments. Amino acid positions are indicated.
(D) Interactions between Zc3h13 fragments (Zc3h131-1460 and Zc3h131461-1729) and Virilizer, Hakai, WTAP were determined by co-immunoprecipitation. FH-Zc3h13, Flag-HA-Zc3h13; Mock, mES cells transfected with empty vector. See also Figure S1.
Zc3h13 depletion decreases global polyadenylated RNA m6A level
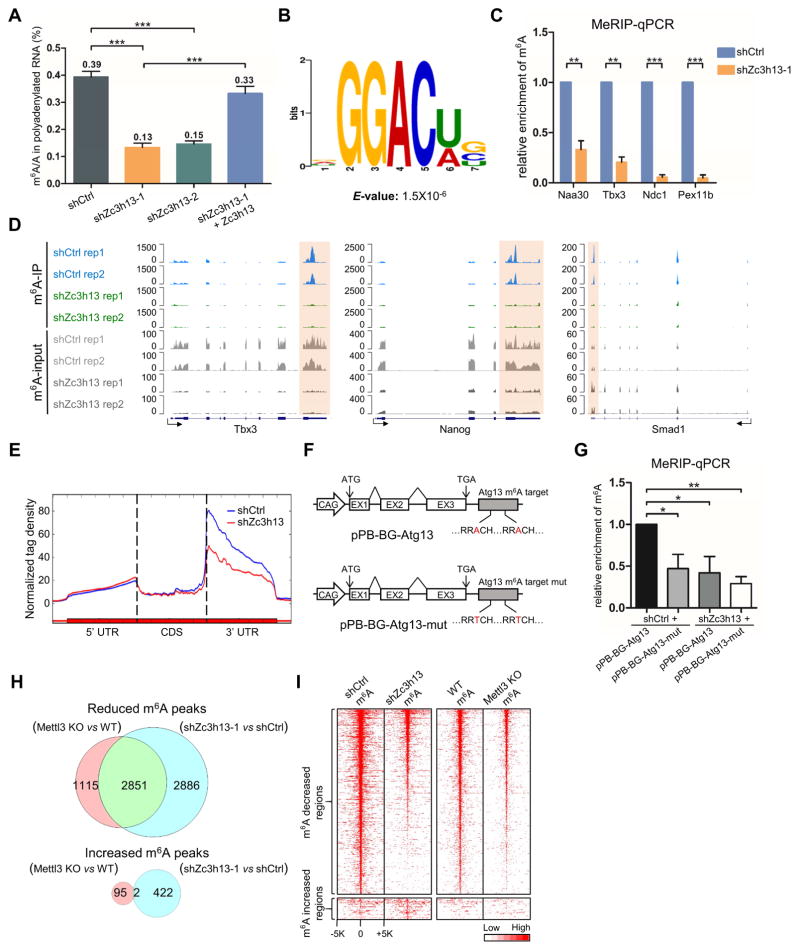
Given that Zc3h13 interacts with the other known RNA m6A regulatory proteins, WTAP and Virilizer (Wan et al., 2015) (Figure 1), we speculated that Zc3h13 might also regulate RNA m6A dynamics in vivo. To test this hypothesis, we determined the m6A/A ratio on polyadenylated RNA in mESCs treated with control or Zc3h13 shRNAs, respectively. Efficient knockdown of Zc3h13 in mESCs was shown by Western blot (Figure S2A). Using LC-MS/MS, we determined the relative level of m6A/A in control mESCs to be ~0.39%, which is comparable to the published m6A/A data in other cells including HeLa and 293T (Liu et al., 2014; Ping et al., 2014) (Figure 2A). In two different Zc3h13 knockdown cell lines, we found the global m6A/A level in mRNA reduced to about 0.13% and 0.15%, respectively (approximately 30%–40% of that of the control cells) (Figure 2A). To rule out potential shRNA off-target effects, we re-introduced Zc3h13 into the Zc3h13 knockdown cells and found the global mRNA m6A/A level restored to 0.33% (about 85% of the control cells) (Figure 2A; Figure S2A), indicating that Zc3h13 indeed modulates mRNA m6A dynamics in cells.
Figure 2. Zc3h13 regulates mRNA m6A in mESCs.
(A) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from the indicated mES cell lines. shCtrl, control.
(B) Sequence motif identified from top 1000 m6A peaks.
(C) MeRIP-qPCR analysis of m6A level in the select mRNAs in control and Zc3h13 kd mESCs. shCtrl, control.
(D) UCSC snapshots of MeRIP-seq reads along indicated mRNAs. Normalized reads density levels are shown as blue (control), green (shZc3h13) and gray (input) shades respectively. Ranges of reads are shown to the left of each track. Two replicates are shown. Transcription directions are indicated by arrows.
(E) The normalized distribution of m6A peaks across the 5′ UTR, CDS, and 3′ UTR of mRNA in control and Zc3h13 kd mESCs. shCtrl, control. (F) The reporter constructs of pPB-BG-Atg13 and pPB-BG-Atg13-mut.
(G) MeRIP-qPCR analysis of m6A modification in indicated RNAs of pPB-BG-Atg13 and pPB-BG-Atg13-mut reporters in Zc3h13 kd mESCs and control cells.
(H) Venn diagram showing overlap between Zc3h13-dependent and Mettl3-dependent m6A peaks. MeRIP-seq data of Mettl3 KO and WT mES cells were obtained from GEO database (GSE52662).
(I) Heatmap analysis of MeRIP-seq reads density in m6A modified regions with statistically significant difference in Zc3h13 kd mES cells versus control cells, and Mettl3 KO mES cells versus control cells. m6A modified regions were sorted according to m6A reads density level. MeRIP-seq data of Mettl3 KO and WT mES cells were obtained from GEO database (GSE52662).
All data are represented as mean ± SD from three biological replicates (A, C and G).
*p < 0.05; **p < 0.01; ***p < 0.001; t test. See also Figure S2.
m6A decrease mainly occurs at 3′ UTR on mRNA upon Zc3h13 loss
To gain insight into the mechanism by which Zc3h13 manipulates m6A level, we performed methylated RNA immunoprecipitation sequencing (MeRIP-seq) in control and Zc3h13 knockdown mESCs. We detected 12,368 putative m6A sites in wild-type cells (treated with control shRNA) (Figure S2B). De novo motif analysis using program MEME identified the consensus sequence “GGACU” (Figure 2B), consistent with previous findings (Dominissini et al., 2012; Meyer et al., 2012). Upon Zc3h13 knockdown, we observed a complete loss and gain of 836 and 318 m6A events, respectively, and many more m6A events (4,901) showing varying degrees of m6A signal reduction, which were confirmed by MeRIP-qPCR (Figure 2C; Figure S2B). We also performed MeRIP-seq and observed more than 80% overlap of the decreased m6A peaks between two biological repeats (84.6% for m6A-seq1, 92.2% for m6A-seq2), indicating good reproducibility (Figure S2C). An example of loss of the m6A peak signal in response to Zc3h13 knockdown is shown in Figure 2D and Figure S2D. Global m6A peak analysis also shows that normalized m6A reads density decreases significantly in Zc3h13 depleted cells (Figure S2E). Furthermore, we found that the m6A reduction in the Zc3h13 kd cells mainly occurred at 3′ UTR but not 5′ UTR or coding regions (CDS) in mRNA (Figure 2E). These findings suggest that Zc3h13 is essential for a large number of m6A methylation events in mESCs. To further investigate whether those m6A peaks are indeed controlled by Zc3h13, we generated a β-globin minigene reporter (Du et al., 2016) harboring wild type or mutant m6A sites in a Zc3h13 targeted 3′ UTR, and the reconstituted minigenes were introduced into Zc3h13 kd and control mES cells (Figure 2F; Figure S2F). We found that Zc3h13 mediated m6A is completely dependent on the existence of the methylatable A residue in the minigene (Figure 2G). This finding provides further support that Zc3h13 directly regulates RNA m6A in vivo.
Previous MeRIP-seq identified Mettl3-dependent m6A peaks in mESC (Batista et al., 2014). We compared our Zc3h13-dependent sites with those Mettl3-dependent ones and found a significant overlap of the reduced m6A peaks between these two datasets (about 50% Zc3h13-dependent peaks and 70% Mettl3-dependent peaks, respectively, Figure 2H). Heatmap analysis also shows similar patterns of m6A peak reduction in Zc3h13 kd mESCs and Mettl3 KO mESCs (Figure 2I). These results indicate that Zc3h13-dependent m6A methylation is likely to be mediated by the known RNA m6A methyltransferase Mettl3.
Zc3h13 knockdown alters subcellular localization of WTAP, Virilizer and Hakai
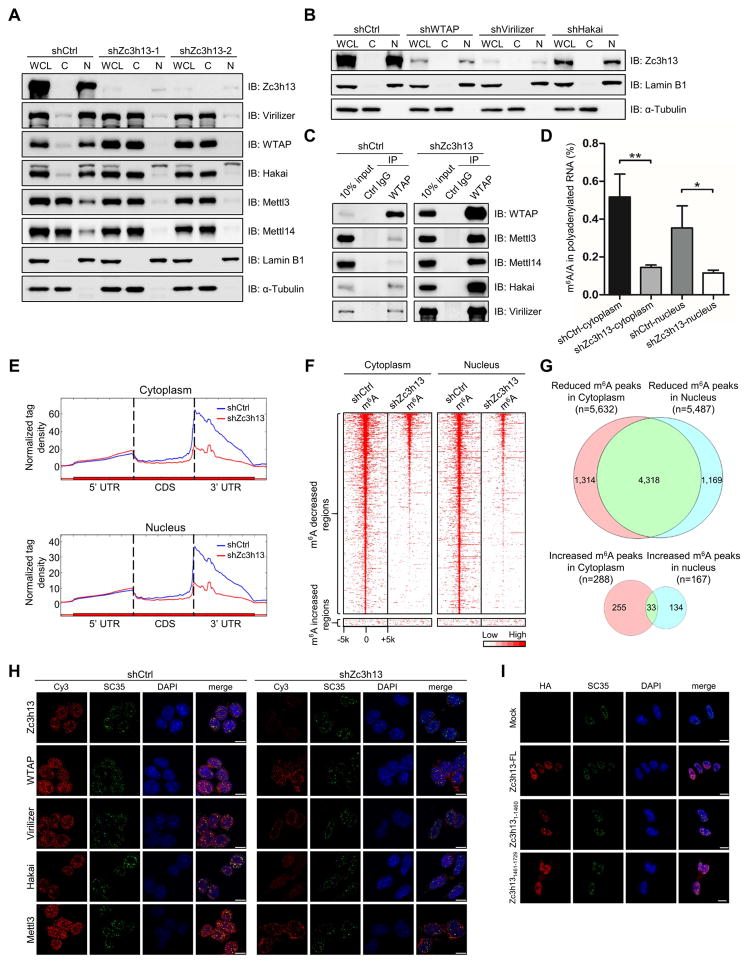
Previous studies suggest that m6A methylation takes place within the nucleus, probably in the nuclear speckles (Ping et al., 2014). Indeed, the m6A methyltransferases Mettl3, Mettl14 and demethylases FTO and ALKBH5 have all been found to co-localize with markers of nuclear speckles (Jia et al., 2011; Zheng et al., 2013). Likewise, the components of the Zc3h13 complex are also found to localize to nuclear speckles, co-localizing with the pre-mRNA splicing factor SC35 (Horiuchi et al., 2013). We therefore wished to determine whether Zc3h13 regulates m6A methylation by controlling the subcellular distribution of other members of the complex. In fractionation assays, we found that while WTAP, Virilizer and Hakai are mainly located in nucleus in the control mESCs, a significant fraction of these proteins are found in the cytoplasm in the Zc3h13 kd cells (Figure 3A; Figure S3A). Furthermore, the nuclear presence of Mettl3 and Mettl14 was also decreased in the Zc3h13 kd cells (Figure 3A; Figure S3B). As a control, we also determined the cellular localizations of several RNA processing factors including ASF/SF2, hnRNPA1, HuR and TDP-43, which showed altered cellular localization in response to different cell stress stimuli (Biamonti and Caceres, 2009; Zhang et al., 2014). We find, unlike WTAP and other components of the m6A regulatory complex, their cellular localizations are not altered in the Zc3h13 depleted mESCs (Figure S3C). Importantly, re-introducing Zc3h13 protein back into the knockdown mES cells restored cellular localizations of Zc3h13 complex components and Mettl3/Mettl14 (Figure S3D). The above results suggest that Zc3h13 is required for nuclear localization of WTAP, Virilizer and Hakai, as well as Mettl3 and Mettl14. However, WTAP, Virilizer, Hakai, Mettl3 and Mettl14 did not appear to be required for Zc3h13 nuclear localization (Figure 3B; Figures S3E and S3F).
Figure 3. Zc3h13 controls nuclear localization of WTAP, Virilizer and Hakai.
(A) Western blot analysis of Zc3h13, Virilizer, WTAP, Hakai, Mettl3 and Mettl14 in the whole cell lysate (WCL), cytoplasmic (C) and nuclear (N) fractions from Zc3h13 and control knockdown mESCs. Lamin B1 and α-Tubulin were used as nuclear and cytoplasmic markers, respectively.
(B) Western blot analysis of cytoplasmic and nuclear fractions of Zc3h13 in WTAP-depleted, Virilizer-depleted, Hakai-depleted and control mESCs.
(C) Interactions between WTAP and Virilizer, Hakai, Mettl3, Mettl14 were determined by co-immunoprecipitation using cytoplasmic fractions from control and Zc3h13 kd mESCs. IgG was used as a control. shCtrl, control.
(D) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from cytoplasmic and nuclear fractions of Zc3h13 kd and control mESCs. Data are represented as mean ± SD from three biological replicates. *p < 0.05; **p < 0.01; t test.
(E) The normalized distribution of m6A peaks across the 5′ UTR, CDS, and 3′ UTR of mRNA in cytoplasmic and nuclear fractions of control and Zc3h13 kd mESCs. shCtrl, control.
(F) Heatmap analysis of MeRIP-seq reads density in m6A modified regions with statistically significant difference in cytoplasmic or nuclear fractions of Zc3h13 kd and control mESCs. m6A modified regions were sorted according to m6A reads density level.
(G) Venn diagram showing overlap between cytoplasmic and nuclear Zc3h13- dependent m6A peaks.
(H) Immunofluorescence analysis of Zc3h13 (red), WTAP (red), Virilizer (red), Hakai (red), Mettl3 (red), SC35 (green), and DAPI (blue, cell nuclei) in Zc3h13 knockdown and control mESCs. Scale bar, 10 μm.
(I) Immunofluorescence analysis of mES cells overexpressing Flag-HA-Zc3h13-full length (FL), Flag-HA-Zc3h131-1460, or Flag-HA-Zc3h131461-1729, detected with the HA antibody (red), SC35 antibody (green) and DAPI (blue, cell nuclei). Scale bar, 10 μm. Mock, mES cells transfected with empty vector.
See also Figure S3.
We further conducted co-immunoprecipitation in the cytoplasmic fraction of Zc3h13 kd mESCs using a WTAP antibody, and found a significant fraction of Virilizer, Hakai, Mettl3 and Mettl14 proteins were pulled down (Figure 3C). These results showed that the WTAP-Mettl3-Mettl14 complex and WTAP-Hakai-Virilizer interactions were intact in the absence of Zc3h13, suggesting that Zc3h13 affects their subcellular localization but not complex formation. We next compared cytoplasmic m6A and nuclear m6A of Zc3h13 kd mES cells and control cells. m6A LC-MS/MS and MeRIP-seq analysis show that m6A level is reduced in a similar pattern both globally and gene-specifically in both subcellular locations (Figures 3D–3F; Figures S3G and S3H). Most of the decreased m6A peaks (more than 75%) are conserved between cytoplasmic and nuclear fractions (Figure 3G). These results indicate that, despite the fact that the translocated WTAP/Virilizer/Hakai in the cytoplasm still form complex with Mettl3 and Mettl14, they could not effectively mediate m6A methylation in the absence of Zc3h13. These data suggest that Zc3h13 and nuclear localization of the m6A processing machinery are essential for proper m6A methylation. Consistent with the fractionation assay results, immunofluorescence assays showed that upon Zc3h13 knockdown, the other complex components showed a significant decrease in their nuclear speckles localization (Figure 3H; Figure S3I). Our finding is consistent with a recent report demonstrating that m6A methylation occurs in the nucleus on chromatin-associated nascent RNAs (Ke et al., 2017).
We next investigated the subcellular localization of the various Zc3h13 truncations using immunofluorescence assay (Figure 3I). The N-terminal domain of Zc3h13 (aa 1-1460) is found at nuclear speckles, suggesting that this domain is sufficient to direct Zc3h13 to nuclear speckles. In contrast, the C-terminal domain (aa 1461-1729), which mediates interactions with other components of the complex, exhibited a diffused expression pattern across the whole cell (Figure 3I). This suggests that Zc3h13’s localization to nuclear speckles is likely to be independent of its interactions with WTAP, Virilizer and Hakai. Based on above observations, we conclude that distinct domains of Zc3h13 direct its interactions with the other components and their nuclear speckles localization.
Zc3h13 KD impairs mESC self-renewal
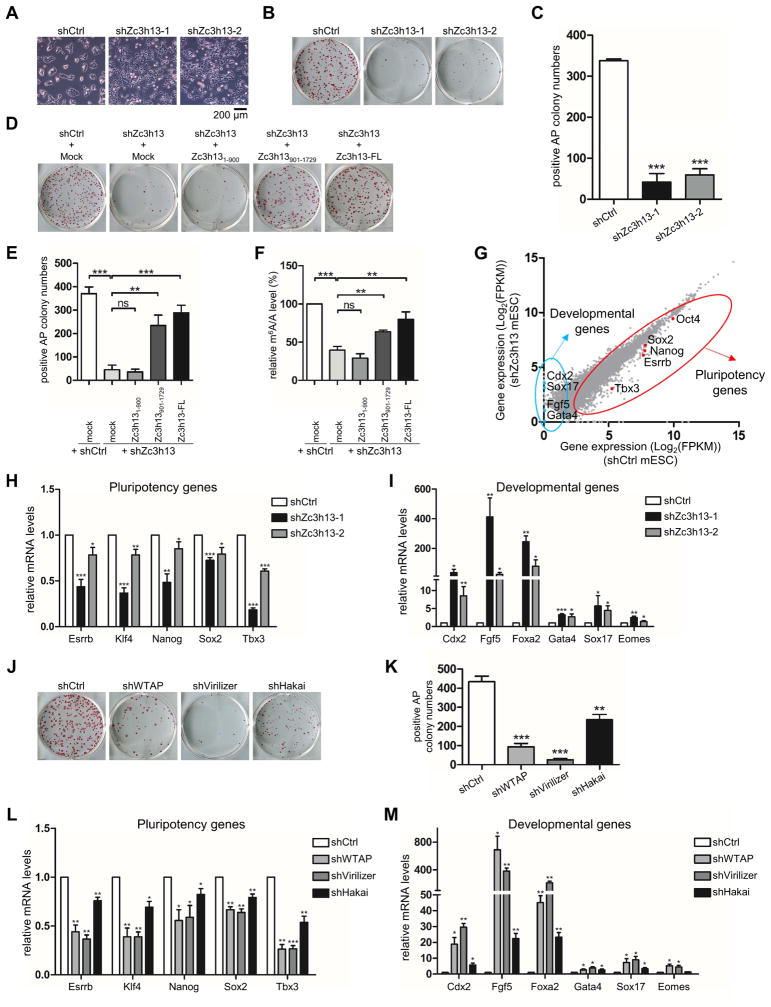
Previous explorations of m6A function suggest that Mettl3 and Mettl14 are crucial for mESC self-renewal and differentiation (Batista et al., 2014; Geula et al., 2015; Wang et al., 2014b). To determine whether Zc3h13 impacts mESC self-renewal and differentiation, we analyzed the phenotype of Zc3h13 kd mESCs. While the control cells showed typical nested and dome shape under phase contrast microscope, Zc3h13 kd cells were flatter and less compact (Figure 4A). Colony formation assays showed that the Zc3h13 kd cells displayed a significantly reduced number of AP-positive colonies (less than 20% of the control mESCs) (Figures 4B and 4C), indicating that Zc3h13 is important for mESC self-renewal. The altered cell morphology and the reduced self-renewal ability of the Zc3h13 kd mESCs are indeed due to Zc3h13 depletion because putting back a wild type copy of Zc3h13 restored the classical mESC morphology and the colony formation ability (Figures 4D and 4E).
Figure 4. Loss of Zc3h13 impairs mESC self-renewal.
(A) Phase-contrast microscopy analysis of colony morphologies of the indicated cell lines.
(B and C) Alkaline phosphatase staining (B) and quantification of AP-positive colonies (C) of control and Zc3h13 knockdown mESCs. shCtrl, control.
(D and E) Alkaline phosphatase staining (D) and quantification of AP-positive colonies (E) of control, Zc3h13 kd and indicated rescuing mES cell lines.
(F) Relative m6A/A level in polyadenylated RNAs isolated from the indicated mES cell lines. Each sample was compared with control cells transfected with empty vector.
(G) Scatter plot of up-regulated and down-regulated genes in Zc3h13-depleted mESCs compared with control cells. Pluripotency genes are highlighted by red dots and circle; developmental genes are highlighted by blue dots and circle. shCtrl, control.
(H and I) RT-qPCR analysis of pluripotency genes (H) and differentiation genes (I) in Zc3h13 kd versus control cells. Data are represented as mean ± SD from four biological replicates. *p < 0.05; **p < 0.01; ***p < 0.001; t test.
(J and K) Alkaline phosphatase staining (J) and quantification of AP-positive colonies (K) of control and the indicated knockdown mES cells. shCtrl, control.
(L and M) RT-qPCR analysis of pluripotency genes (L) and differentiation genes (M) in the indicated knockdown cells versus control cells.
Data are represented as mean ± SD from three biological replicates (C, E, F, K, L and M). *p < 0.05; **p < 0.01; ***p < 0.001; ns, no significance; t test.
See also Figures S4 and Table S1.
To explore the mechanism by which Zc3h13 regulates mESC self-renewal, we conducted colony formation assays and m6A LC-MS/MS in Zc3h13 kd mESCs rescued with different Zc3h13 fragments (Figures S4A and S4B). We found, while cells reintroduced with the N-terminal domain (aa 1-900) still exhibited dampened self-renewal phenotype with reduced AP positive colony numbers, similar to kd cells; the C-terminal domain (aa 901-1729) restored colony numbers, showing a significant rescue of self-renewal capability (Figures 4D and 4E; Figures S4C and S4D). Meanwhile, LC-MS/MS shows that overexpression of the C-terminal (aa 901-1729) but not N-terminal domain (aa 1-900) in Zc3h13 kd cells also significantly recovered m6A/A level (Figure 4F). Moreover, immunofluorescence assays showed that the C-terminal (aa 901-1729), but not the N-terminal domain (aa 1-900), robustly recovered the nuclear presence of WTAP-Virilizer-Hakai and Mettl3/Mettl14 in the Zc3h13 kd cells (Figures S4E–S4J). As discussed before, our study also showed that Zc3h13 C-terminal domain (aa 901-1729) is responsible for interaction with its complex members. Based on these data, we conclude that the interactions between Zc3h13 and WTAP, Virilizer, Hakai are essential for mRNA m6A and mESC pluripotency regulation.
To gain molecular insights into the role of Zc3h13 in mESC self-renewal, we wished to identify genes whose expression is regulated by Zc3h13. We carried out RNA-seq in control and Zc3h13 kd mESCs, and found a total of 577 genes, which displayed changes in their levels of expression upon Zc3h13 knockdown (Table S1). Among them, 330 and 247 genes showed increased or decreased expression, respectively. GO analysis shows that most of the affected genes belong to the category of genes implicated in cell development and morphogenesis change (Figure S4K). Interestingly, a group of mESC pluripotency and differentiation genes showed significant alterations in their expression in response to Zc3h13 kd (Figure 4G). Indeed, the stemness markers such as Nanog, Sox2 and Klf4 all showed decreased mRNA levels in the Zc3h13 kd cells, which were restored in the rescued cells (Figure 4H; Figure S4C). In contrast, the differentiation makers such as Fgf5, Cdx2 and Gata4 showed increased expression in the Zc3h13 kd cells (Figure 4I; Figure S4D).
In order to investigate whether Zc3h13 is directly involved in the regulation of the above gene expression, we conducted RIP-seq with a Zc3h13 antibody in mESCs. Among 577 differentially expressed genes upon Zc3h13 knockdown, we found 65 genes were likely to be directly bound by Zc3h13 based on the RIP results (Table S1). This finding was further validated by RIP-qPCR on a number of selected genes (Figures S4L and S4M). In addition, MeRIP-qPCR assays also found significant m6A reduction on these genes after Zc3h13 depletion (Figures S4L and S4N). These results suggest that Zc3h13 may directly participate in mESC gene expression regulation, possibly via regulating m6A. We next determined whether the other components of Zc3h13 complex might also play a role in mESC self-renewal and differentiation. Similar to Zc3h13 kd, knockdown of WTAP, Virilizer or Hakai in mESCs also resulted in flatter cell shape, a reduction of AP-positive colonies and altered gene expression of pluripotency and developmental regulators (Figures 4J–4M). Taken together, our findings suggest that the Zc3h13 complex plays an important role in regulating mESC self-renewal by impacting the expression of both pluripotency and differentiation genes.
Discussion
The evolutionarily conserved Zc3h13-WTAP-Virilizer-Hakai complex was initially identified by a biochemical/proteomics approach. Within the complex, WTAP and Virilizer were reported to control mRNA m6A level, but the role of Zc3h13 was completely unknown. Our findings identify Zc3h13 as a new RNA m6A regulator, which facilitates mESC self-renewal.
In Zc3h13 kd mESCs, the m6A/A level decreased by about 60%–70% compared with wild type cells, similar to the level of reduction when WTAP or Virilizer was knocked down in human A549 cells (Schwartz et al., 2014). A previous study identified two different types of m6A peaks: WTAP-dependent and WTAP-independent peaks (Schwartz et al., 2014). WTAP-dependent m6A peaks distribute broadly in exons, especially at the 3′ UTR of mRNA. In contrast, WTAP-independent m6A peaks are located around the first transcribed base, representing most of the m6A peaks at 5′ UTR of mRNA (Schwartz et al., 2014). Genome-wide m6A sequencing analysis shows that the m6A reduction mainly occurs at 3′ UTR, but not 5′ UTR of mRNA upon Zc3h13 knockdown. Since Zc3h13 is present in the same complex as WTAP, it’s perhaps not surprising that loss of Zc3h13 preferentially affected WTAP-dependent, 3′ UTR m6A events.
What is the role of Zc3h13 in the Zc3h13-WTAP-Hakai-Virilizer complex? We found upon Zc3h13 knockdown, the other components of the complex translocate significantly to the cytoplasm, accompanied by a decreased nuclear presence of Mettl3 and Mettl14. Similarly, WTAP, Virilizer or Hakai knockdown also decreases the nuclear accumulation of Mettl3 and Mettl14 (Figures S3J–S3L). In contrast, inhibition of WTAP, Virilizer or Hakai did not affect the nuclear localization of Zc3h13. These results suggest that Zc3h13 mainly functions to retain the Zc3h13-WTAP-Virilizer-Hakai complex in the nucleus to regulate m6A methylation. Supporting this model, the C-terminal region of Zc3h13 that mediates interactions with other members of the complex significantly restores the nuclear localization of WTAP-Virilizer-Hakai as well as Mettl3/Mettl14 and m6A methylation in the Zc3h13 kd mESCs. In contrast, the N-terminal region, which does not interact with other complex members, is not able to restore the nuclear localization of above proteins and m6A level and mESC biology. Thus, it appears that the assembly of the Zc3h13 complex in the nucleus is important for conferring m6A and mESC pluripotency.
How is Zc3h13 targeted to nuclear speckles? We noticed that 80% of Zc3h13 protein sequence is classified as low complexity (LC) regions, which are known to have the propensity to cluster together and target protein to sub-cellular “organelles” (Kato et al., 2012). Since nuclear speckles are such a sub-cellular “organelle” enriched for RNA processing enzymes and pre-mRNA splicing factors (Spector, 2001), we speculate Zc3h13 may be localized to nuclear speckles via the LC domains. Finally, we note that WTAP exists in two different complexes: Mettl3-Mettl14-WTAP m6A catalyzing complex and Zc3h13-WTAP-Virilizer-Hakai m6A regulatory complex. WTAP is likely a bridging molecule that connects Zc3h13-Virilizer-Hakai with Mettl3-Mettl14 and recruits the enzyme to nuclear speckles for m6A methylation.
Previous studies of Mettl3 or Mettl14 in mESCs indicate that regulation of mRNA m6A is crucial for mESC self-renewal and differentiation (Batista et al., 2014; Geula et al., 2015; Wang et al., 2014b). Batista et al. reported that Mettl3 knockout promotes mESC self-renewal (Batista et al., 2014). Geula et al. demonstrated that mESC pluoripotency change actually depends on the mESC state when Mettl3 was removed. Specifically, when Mettl3 was depleted under the ground state naïve condition (mESCs were cultured in N2B27 medium with 2i/LIF), mESCs were kept in a so-called hyper-naïve state. On the other hand, when Mettl3 knockout was performed in the primed state (mESCs were cultured in N2B27 medium with FGF2/Activin), the mESCs displayed minimal self-renewal and accelerated differentiation (Geula et al., 2015). In the same study, they also inhibited Mettl3 under metastable naïve condition where mESCs were cultured in the conventional FBS/LIF medium, and observed that mES C57B6 cells displayed a slight decrease in pluripotency maintenance upon Mettl3 depletion. In our study, we conducted Zc3h13 knockdown in mESCs cultured in FBS/LIF medium (metastable naïve condition) and found that Zc3h13 knockdown impaired mESC self-renewal, similar to what has been reported for Mettl3 inhibition in mESCs cultured in FBS/LIF condition (Geula et al., 2015). Consistently, our RNA-seq analysis suggests that Zc3h13 promotes pluripotency genes expression and suppresses differentiation genes in mESCs. How mRNA m6A regulates the balance between self-renewal and differentiation is not completely understood at the molecular level. Previous studies of Mettl3 suggest that mRNA m6A alters pluripotency or development gene expression by influencing their RNA stability, and the same mechanism may be operative in Zc3h13 knockdown mESCs. In addition, we also noticed changes of m6A modification and expression of key regulators in signal transduction pathways involved in ESC maintenance (such as WNT pathway, Notch pathway, unpublished data), suggesting additional regulators may also be affected by Zc3h13-mediated m6A methylation.
STAR Methods text
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yang Shi (yshi@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell cultures
E14Tg2a murine embryonic stem cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% MEM non-essential amino acid, 55μM β-Mercaptoethanol, 100 U/mL Penicillin/Streptomycin and 1000 units/mL LIF (Millipore) at 37°Cwith 5% CO2. 293T cells were cultured in DMEM supplemented with 10% FBS and 100 U/mL Penicillin/Streptomycin at 37°C with 5% CO2.
METHOD DETAILS
Stable cell lines construction
cDNA of full-length murine Zc3h13 (Zc3h13-FL, NCBI RefSeq Accession No. NM_026083.2) and several truncated fragments were cloned into the pPB-CAG-IRES-Pac plasmid with N-terminal Flag and HA tags (primer sequences were listed in Table S2). These plasmids were individually co-transfected into mESCs with pCMV-PBase plasmid in a 1:1 ratio using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Medium was replaced by fresh media with 10 μg/mL Blasticidin S at 24 hr post-infection. After continuous Blasticidin S selection for 5 days, the survived mESCs were pooled as stable overexpression cell lines.
Lentiviral shRNAs
shRNA targeting sequences against Zc3h13, WTAP, Virilizer, Hakai, Mettl3 and Mettl14 were designed, synthesized and subloned into the PLKO.1-puro vector (Addgene). shRNA targeting sequences were listed in Table S2. Lentivirus was made by co-transfection of each pLKO.1 shRNA vector with VSV-G and psPAX2 in a 3:1:1 ratio into 293T cells. Supernatant at 48 hr post-transfection was collected and passed through a 0.45 μm filter. mESCs were seeded in a 6-well plate and infected with each lentivirus supernatant in the presence of 5 μg/mL polybrene (Sigma). Medium was replaced by fresh media with 2 μg/mL puromycin at 24 hr post-infection. After continuous puromycin selection for 5 days, the survived mESCs were pooled as stable infected mESCs.
RNA m6A quantification by LC-MS/MS
RNA m6A quantification by LC-MS/MS was performed as described previously (Liu et al., 2014). In brief, total RNAs from control, Zc3h13 knockdown and rescue mES cells were isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. Polyadenylated RNAs were extracted by oligo d(T)25 magnetic beads (NEB), followed by removal of rRNA with RiboMinus Eukaryote Kit (Ambion). mRNA concentration was measured by NanoDrop. 200 ng mRNA was digested by nuclease P1 (1 U, Sigma) in 20 μl buffer containing 25 mM NaCl, 2.5 mM ZnCl2 at 37°C for 2 hr, followed by the addition of NH4HCO3 (1 M, 2.2 μl) and alkaline phophatase (1 U, Sigma). After an additional incubation at 37°C for 2 hr, the solution was centrifuged at 13000 rpm for 10 min at 4°C, and 10 μl of the solution was injected into LC-MS/MS. Quantification was performed by comparison with the standard curve obtained from pure nucleoside standards. The ratio of m6A to A was calculated based on the calculated concentrations.
Quantification of cytoplasmic and nuclear RNA m6A of Zc3h13 kd and control cells was performed using RNAs extracted from cytoplasmic or nuclear fractions of cells according to the same procedure described above.
MeRIP-seq
Polyadenylated RNAs from Zc3h13 knockdown and control mESCs were prepared as described above, and sonicated to 100–200 nt fragments by Bioruptor Plus sonicator device (Diagenode). A small portion (10%) of the RNA fragments was left aside to be used as input sample. MeRIP was performed as previously described with minor modifications (Dominissini et al., 2012). Briefly, 4 μg fragmented polyadenylated RNA was incubated with 2 μg anti-m6A antibody (Synaptic Systems) in 1 x IP buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% NP-40) for 2 hr at 4°C. The m6A-IP mixture was then incubated with Dynabeads protein A (Life Technologies) for an additional 2 hr at 4°C on a rotating wheel. After washing 3 times with 1 x IP buffer, the bound RNA was eluted by competition with N6-methyladenosine (Santa Cruz Biotechnology) and then purified using an RNA cleanup kit (Zymo Research). The purified RNA fragments from MeRIP were used for library construction using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Cat#E7420) following manufacturer’s instructions and sequenced with Illumina HiSeq 2000 or Illumina HiSeq X10.
For MeRIP-seq of cytoplasmic and nuclear fraction samples, RNAs were extracted from cytoplasmic or nuclear fractions of Zc3h13 kd mESCs and control cells. MeRIP was then performed according to the same procedure above.
MeRIP-qPCR
MeRIP was performed according to the procedure described above. Immunoprecipitated m6A RNAs were reverse transcribed into cDNA using PrimeScript RT reagent kit (Takara Bio) following manufacturer’s instruction. The primers for MeRIP-qPCR were listed in Table S2.
RNA-seq
Total RNAs were extracted from control, Zc3h13 knockdown and rescue mESCs using TRIzol reagent (Invitrogen), respectively. RNA-seq libraries were prepared and sequenced by WuXi NextCODE on an Illumina HiSeq X10 platform to produce 40–50M non-strand-specific pair-end reads of 151bp uniform length per sample.
RT-qPCR
Total RNA was isolated using TRIzol Reagent (Invitrogen) according to manufacturer’s protocol. cDNA was synthesized with PrimeScript RT reagent kit (Takara Bio) containing random primers using 1 μg of extracted RNA per sample. RT-qPCR was performed using SYBR Green Master Mix (Roche) with the Roche LightCycler 480 Instrument II system. Gapdh was used as an internal control for normalization. All primers for RT-qPCR were listed in Table S2.
RIP-seq
Zc3h13 overexpressed mESCs were washed twice by PBS and lysed in lysis buffer of 50 mM Tris, pH7.4, 150 mM NaCl, 5 mM MgCl2, 0.5% NP40, 1 mM DTT with 1x Protease Inhibitor Cocktail (Roche) and Murine RNase Inhibitor (New England Biolabs) for 30min at 4°C. Zc3h13 antibody or rabbit IgG (as control, Santa Cruz Biotechnology) was incubated with Dynabeads Protein A magnetic beads (Life Technologies) in lysis buffer for 2 hr at 4°C and washed twice by lysis buffer. The cell lysates were centrifuged and the supernatant was transferred to antibody-conjugated magnetic beads. The mixtures were rotated for another 2 hr at 4°C, and then washed 3 times with lysis buffer and twice with wash buffer (50 mM Tris, 150 mM NaCl, 1 mM MgCl2, 0.5% NP40). Then the beads were suspended in wash buffer containing 0.1% SDS and 10 μl proteinase K, and incubated at 55°C for 30 min. The elution was collected and purified by an RNA cleanup kit (Zymo Research). The purified RNA samples were used for library preparation (NEBNext Ultra Directional RNA Library Prep Kit for Illumina) and sequencing.
RIP-qPCR
RNA-IP was performed with Zc3h13 antibody or Rabbit control IgG as above. Immunoprecipitated RNAs were reverse transcribed into cDNA using PrimeScript RT reagent kit (Takara Bio) according to manufacturer’s instruction. The primers for RIP-qPCR were listed in Table S2.
Cell fractionation and immunoblotting
mES cells were lysed in hypotonic buffer (10 mM HEPES, pH7.5, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1x Protease Inhibitor Cocktail (Roche) and 1 mM PMSF) on ice for 15 min, and then NP-40 was added to a final concentration of 0.25% for another 5 min. Samples were centrifuged for 3 min at 2000 rpm at 4°C, and the supernatant was saved as cytoplasmic fraction. The nuclear pellet obtained from the low speed centrifugation was washed with hypotonic buffer once and re-suspended in RIPA buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1x Protease Inhibitor Cocktail and 1 mM PMSF) and incubated on ice for 20 min. This sample was saved as nuclear fraction. SDS loading buffer was added in the samples and boiled for 10 min. The samples were loaded on 7.5% SDS-PAGE gels and subjected to immunoblotting with different antibodies. The intensity of the band was measured by Bio-Rad Image Lab software (Bio-rad).
Primary antibodies concentrations used in immunoblotting are as below: anti-Zc3h13 (1:3000, Bethyl); anti-WTAP (1:3000, Proteintech); anti-Virilizer (1:3000, Bethyl); anti-Hakai (1:3000, Bethyl); anti Mettl3 (1:3000, Abcam); anti Mettl14 (1:2000, Sigma); anti-Lamin B1 (1:5000, Proteintech); anti-α-Tubulin (1:5000, Proteintech).
Co-immunoprecipitation
mES cells overexpressing Zc3h13 with Flag and HA tag were washed once with PBS and lysed in buffer A (10 mM HEPES pH7.5, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1x Protease Inhibitor Cocktail and 1 mM PMSF) on ice for 15 min, then NP-40 was added to a final concentration of 0.25% for another 5 min. Nuclei were collected by centrifugation (2000 rpm, 3min, 4°C) and re-suspended in buffer C (20 mM HEPES, pH 7.5, 10% Glycerol, 0.42 M KCl, 4 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF and 1x Protease Inhibitor Cocktail). After 30 min incubation on ice, insoluble chromatin fraction was removed from the nuclear extract by high-speed centrifugation (13000 rpm, 15 min, 4°C). The soluble nuclear fraction was then incubated with anti-Flag affinity gel for 4 hours at 4°C. The beads were washed for 4 times. SDS buffer was directly added in the beads and boiled for 10 min. The samples were loaded on 7.5% SDS-PAGE gels and subjected to immunoblotting using indicated antibodies.
Co-immunoprecipitation with RNase treatment was done as follows: Zc3h13 overexpressing mES cells and mock cells were lysed in buffer of 50 mM Tris, pH7.4, 150 mM NaCl, 1% NP-40, 1 x Protease Inhibitor on ice for 20 min following by sonication using Bioruptor (Diagenode). The lysates were treated with RNase I (Ambion) at 37°C for 7 min. The lysates were centrifuged and the soluble fractions were used for immunoprecipitation with anti-Flag affinity gel.
Immunofluorescence
Cultured mESCs were rinsed briefly in PBS and then fixed and permeabilized with pre-chilled methanol:acetone (1:1, v/v) for 10 min at −20°C, or cells were fixed with PBS containing 4% paraformaldehyde for 10 min at room temperature and then permeabilized with PBS containing 0.1% Triton X-100 for 10 min. Cells were subsequently washed with PBS for three times. Cells were blocked for 30 min with 1% BSA in PBS at room temperature. Primary antibodies were diluted in blocking buffer at different concentrations (see blow) and incubated overnight at 4°C. Washed twice with PBS, cells were incubated with DAPI (dilution 1:2000, Solarbio) and fluorescent dye-conjugated secondary antibodies diluted in blocking buffer for one hour at room temperature and then visualized.
Primary antibodies concentrations: anti-Zc3h13 (1:200, Bethyl); anti-WTAP (1:200, Proteintech); anti-Virilizer (1:200, Bethyl); anti-Hakai (1:200, Bethyl); anti Mettl3 (1:200, Abcam); anti Mettl14 (1:200, Sigma-Aldrich); anti SC35 (1:500, Abcam); anti HA (1:1200, Cell Signaling, Cat#3724); anti HA (1:100, Cell Signaling, Cat#2367).
Minigene reporter assay
A 99-nt fragment from Atg13 gene, which contains a Zc3h13-dependent m6A peak, was inserted into the 3′ UTR of the minigene reporter pTBG between NheI and XbaI sites (pTBG is a kind gift from Dr. Ligang Wu’ lab) (Du et al., 2016). The reporter genes were then subcloned into pPB-CAG-IRES-Pac plasmid (pPB-BG-Atg13) using AgeI and XhoI sites. The construction of minigene reporter pPB-BG-Atg13-mut is identical to pPB-BG-Atg13, except that the adenosines within the fragment were mutated to thymidines. The reconstituted pPB-BG-ATG13 and pPB-BG-Atg13-mut were transfected into Zc3h13 kd and control mES cells respectively. After 24 hr, total RNAs were extracted and MeRIP-qPCR was performed as described above. Targeted Atg13 m6A sequence and its mutant sequence inserted into the minigene were listed in Table S2, together with the MeRIP-qPCR primers.
Colony formation assay
After trypsinization and cell counting, 500 mES cells were seeded per well in 6-well plates and cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 1% MEM non-essential amino acid, 55μM β-Mercaptoethanol, 100 U/mL Penicillin/Streptomycin and 1000 units/mL LIF for 7 days. Cells were rinsed with PBS once and stained using Alkaline Phosphatase Detection Kit (Sigma-Aldrich), according to manufacturer’s instruction.
QUANTIFICATION AND STATISTICAL ANALYSIS
MeRIP-seq data processing
MeRIP and Input sequencing data were sent to trim-galore to remove low quality reads and adapter sequence contaminants under default parameters except for “--length 35”, remaining reads were then aligned to the mouse Ensemble genes (version NCBIM37.65) transcriptome annotation using TopHat2 aligner (v2.0.12b) (Trapnell et al., 2009) under parameters: “--max-multihits 1 --prefilter-multihits” and “--transcriptome-index”. m6A peaks were identified using exomePeak R/Bioconductor package with default parameters (Meng et al., 2014).
Differential m6A peak calling
Overall regions with differential m6A methylation values were identified using bedtools and a home-made script as follows: we firstly merged all the m6A peaks identified from MeRIP-seq datas in Zc3h13 kd and control mESCs using mergeBed in bedtools toolset, then we calculated the respective m6A enriched scores in Zc3h13 kd and control mESCs on every merged m6A peaks with a home-made script, using which m6A enrichment score on every merged m6A peaks was calculated by adding up all the value column within the merged m6A peaks in bedgraph files, bedgraph files were firstly transferred from bam files, and then normalized based on the gene expression change in Zc3h13 kd and control mESCs. m6A peaks were identified as differential m6A peaks as m6A enrichment score fold change (FC) ≥2 and an m6A score larger than 60. Mettl3 KO dependent differential m6A peaks were identified with the same methods used in MeRIP-seq data analysis in Zc3h13 kd and control mESCs. The m6A differential peaks in Zc3h13 kd and Mettl3KO mESCs were regarded as Zc3h13-dependant m6A peaks and Mettl3 dependant m6A peaks respectively. m6A enrichment score distribution on 5′UTR, CDS and 3′UTR regions in Zc3h13 kd and control mESCs were also generated by a home-made script, all the 5′UTR, CDS and 3′UTR elements were normalized to the same length unit. Heatmap analysis on Zc3h13 kd dependent increased and decreased m6A peaks was performed by cluster and Treeview. MeRIP-seq datas analysis in cytoplasmic or nuclear fractions of Zc3h13 kd and control mESCs were done using the same methods above.
MeRIP-seq data overlap analysis
Overlap between Zc3h13 dependant m6A peaks and Mettl3 dependant m6A peaks was identified by intersectBed in bedtools toolset. The same analysis method was also applied to m6A peaks in cytoplasmic and nuclear fractions of Zc3h13 kd and control mESCs.
Motif search
Identified m6A peaks were sorted according to the p-value from the lowest to the highest, then the top 1000 m6A peaks were chosen for the de novo motif analysis. 101-nucleotide-long sequences derived from the sense strand and centered on the peak summit were used as input for MEME-ChIP (Bailey et al., 2009).
RNA-Seq data processing
Polyadenylated RNAs were sequenced for a control and two Zc3h13 knockdown mouse embryonic stem cell lines. Firstly, we trimmed the low quality sequences and retain paired reads using Trim-galore under --length 100 --paired parameters. The paired-end reads were aligned to mouse genome version mm9 with TopHat2 aligner (v2.0.12b) (Trapnell et al., 2009), using TopHat2 default parameters and the Ensemble genes (version NCBIM37.65) transcriptome annotation. Then the expression of gene transcripts were assembled and quantified using cufflinks, and differentially expressed genes were identified by cuffdiff. Genes were considered as differentially expressed if gene expression is FC≥2 and with a p-value≤0.05. Heatmap analysis for up and down regulated expressed genes in Zc3h13 kd and Mettl3 kd mESCs was performed by pheatmap package in R.
RIP-seq data processing
RIP-seq data were analyzed as follows: Firstly, adapter sequence contaminants and poor-quality sequence from RIP-seq data were filtered with trim-galore software. Then RIP-Seq reads were align to the mouse reference genome (mm9, NCBI Build 37) using TopHat v2.0.12 software (Trapnell et al., 2009). The genes were annotated with Gene Transfer Format (GTF) of Ensembl (Ensembl.NCBIM37.65). The Cufflinks/Cuffdiff suite (Trapnell et al., 2009) was used to compute the fragments per kilo base of coding exon per million fragments mapped (FPKM) values as means of normalizing for gene length and depth of sequencing, and the fold-change difference of gene expression in terms of FPKM between the RIP and control (input) libraries. Gene transcripts were considered to be bound by Zc3h13 protein if their FPKM expression is > 1.0 and the ratio of FPKM (Zc3h13 RIP)/FPKM (Input) is ≥1.5.
Gene Ontology (GO) Analysis
Gene ontology analysis was performed using the web tool:The Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/).
Image quantification
All immunofluorescence images were analyzed using Image-Pro Plus software (Media Cybernetics).
For quantification of proteins’ nuclear speckles localization in Figure 3H and Figure S3I, mES cells were immunostained with SC35 to label nuclear speckles. Images were thresholded, and a mask was then generated to define the nuclear speckles region. Fluorescence intensity of indicated proteins (WTAP/Virilizer/Hakai/Mettl3) in the defined nuclear speckles region in corresponding channel was measured and ratio-ed over SC35 area. For proteins’ nuclear localization quantification in Figures S4E–S4J, images taken in the DAPI channel were first thresholded and a mask was then generated to define the nuclear region. Fluorescence intensity of indicated proteins (WTAP/Virilizer/Hakai/Mettl3) in the defined nuclear region in corresponding channel was measured and ratio-ed over DAPI area.
The statistical data are presented as mean ± SD or mean ± SEM, as described in the corresponding figure legends. The statistical significance of differences was determined using Student’s t test with GraphPad Prism 6 (Graphpad Software). p < 0.05 was considered to be statistically significant.
DATA AND SOFRWARE AVAILABILITY
All softwares used in this study are listed in the Key Resources Table. The MeRIP-seq, RNA-seq and RIP-seq data generated by this study have been deposited to GEO database under accession number GSE94148. MeRIP-seq data of Mettl3 KO and WT mES cells were obtained from GEO database under accession number GSE52662 (Batista et al., 2014).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Zc3h13 | Bethyl | Cat#A300-748A; RRID: AB_2273126 |
| Rabbit polyclonal anti-WTAP | Proteintech | Cat#10200-1-AP; RRID: AB_2216349 |
| Rabbit polyclonal anti-Virilizer | Bethyl | Cat#A302-124A; RRID: AB_1720422 |
| Rabbit polyclonal anti-Hakai | Bethyl | Cat#A302-969A; RRID: AB_10752587 |
| Rabbit monoclonal anti-Mettl3 | Abcam | Cat#ab195352; RRID: AB_2721254 |
| Rabbit polyclonal anti-Mettl14 | Sigma-Aldrich | Cat#HPA038002; RRID: AB_10672401 |
| Mouse monoclonal anti-HA | Cell Signaling Technology | Cat#2367; RRID: AB_331789 |
| Rabbit monoclonal anti-HA | Cell Signaling Technology | Cat#3724; RRID: AB_1549585 |
| Rabbit polyclonal anti-ASF/SF2 | Proteintech | Cat#12929-2-AP; RRID: AB_2187211 |
| Rabbit polyclonal anti-HuR | Proteintech | Cat#11910-1-AP; RRID: AB_11182183 |
| Rabbit polyclonal anti hnRNP A1 | Proteintech | Cat#11176-1-AP; RRID: AB_2117177 |
| Rabbit polyclonal anti-TDP-43 | Proteintech | Cat#10782-2-AP; RRID: AB_615042 |
| Mouse monoclonal anti-Lamin B1 | Proteintech | Cat#66095-1-lg; RRID: AB_2721256 |
| Mouse monoclonal anti-α-Tubulin | Proteintech | Cat#66031-1-Ig; RRID: AB_11042766 |
| Mouse monoclonal anti-Gapdh | Proteintech | Cat#60004-1-Ig; RRID: AB_2107436 |
| Mouse monoclonal anti-β-Actin | Proteintech | Cat#60008-1-Ig; RRID: AB_2289225 |
| Mouse monoclonal anti-SC35 | Abcam | Cat#ab11826; RRID: AB_298608 |
| Rabbit polyclonal anti-m6A | Synaptic Systems | Cat#202003; RRID: AB_2279214 |
| Mouse monoclonal anti-Flag | Abmart | Cat#M20008; RRID: AB_2713960 |
| Normal rabbit IgG | Santa Cruz Biotechnology | Cat#sc-2027 |
| Dynabeads protein A | Life Technologies | Cat#10002D |
| Anti-FLAG M2 affinity gel | Sigma-Aldrich | Cat#A2220 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease Inhibitor Cocktail | Roche | Cat#4693159001 |
| RNase I | Ambion | Cat#AM2294 |
| DAPI | Solarbio | Cat#C0060 |
| Lipofectamine 2000 | Invitrogen | Cat#11668019 |
| TRIzol reagent | Invitrogen | Cat#15596018 |
| Nuclease P1 from Penicillium citrinum | Sigma-Aldrich | Cat#N8630- |
| Phosphatase, Alkaline from Escherichia coli | Sigma-Aldrich | Cat#P5931 |
| N6-methyladenosine | Santa Cruz Biotechnology | Cat#SC215524 |
| RNase Inhibitor, Murine | New England Biolabs | Cat#M1304L |
| Proteinase K | New England Biolabs | Cat# P8107S |
| Recombinant Mouse LIF Protein | Merck Millipore | Cat#ESG1107 |
| Critical Commercial Assays | ||
| PrimeScript RT reagent kit | Takara Bio | Cat#RR047A |
| Alkaline Phosphatase Detection Kit | Sigma-Aldrich | Cat#85L3R-1 |
| oligo d(T)25 magnetic beads | New England Biolabs | Cat#S1419S |
| RiboMinus Eukaryote Kit v2 | Ambion | Cat#A15026 |
| RNA clean and concentrator kit | Zymo Research | Cat#R1015 |
| NEBNext Ultra Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7420 |
| FastStart Universal SYBR Green Master (Rox) | Roche | Cat#4913914001 |
| Deposited Data | ||
| RNA-seq data | This paper | GSE94148 |
| MeRIP-seq data | This paper | GSE94148 |
| RIP-seq data | This paper | GSE94148 |
| Experimental Models: Cell Lines | ||
| E14Tg2a murine embryonic stem cells | Laboratory of Qi-Long Ying | N/A |
| 293T cells | Laboratory of Charlie Degui Chen | N/A |
| Oligonucleotides | ||
| Primers for Zc3h13 cloning | This paper | See Table S2 |
| shRNA targeting sequences | This paper | See Table S2 |
| Primers for MeRIP-qPCR, RT-qPCR and RIP-qPCR | This paper | See Table S2 |
| Zc3h13 target m6A sequence for minigene reporters | This paper | See Table S2 |
| Recombinant DNA | ||
| plasmid: pPB-Flag/HA-Zc3h13-FL | This paper | N/A |
| plasmid: pPB-Flag/HA-Zc3h13-(1-900) | This paper | N/A |
| plasmid: pPB-Flag/HA-Zc3h13-(901-1729) | This paper | N/A |
| plasmid: pPB-Flag/HA-Zc3h13-(1-1460) | This paper | N/A |
| plasmid: pPB-Flag/HA-Zc3h13-(1461-1729) | This paper | N/A |
| plasmid: pTBG | Laboratory of Li-Gang Wu (Du et al., 2016) | N/A |
| pPB-BG-Atg13 | This paper | N/A |
| pPB-BG-Atg13-mut | This paper | N/A |
| Software and Algorithms | ||
| Tophat2 | (Trapnell et al., 2009) | https://ccb.jhu.edu/software/tophat/index.shtml |
| Bowtie | (Langmead et al., 2009) | http://bowtie-bio.sourceforge.net/index.shtml |
| Samtools | (Li et al., 2009) | http://www.htslib.org/doc/samtools.html |
| Cufflinks | (Trapnell et al., 2010) | http://cole-trapnell-lab.github.io/cufflinks |
| Signalplot | This paper | N/A |
| Rseqc | (Wang et al., 2012) | http://rseqc.sourceforge.net/ |
| exomePeak | (Meng et al., 2014) | http://www.bioconductor.org/packages/release/bioc/html/exomePeak.html |
| bedtools | (Quinlan and Hall, 2010) | http://bedtools.readthedocs.io/en/latest/ |
| MEME-ChIP | (Machanick and Bailey, 2011) | http://meme-suite.org/tools/meme-chip |
| DAVID | (Huang da et al., 2009) | https://david.ncifcrf.gov/ |
| pheatmap | Raivo Kolde | https://github.com/raivokolde/pheatmap |
| Bio-Rad Image Lab | Bio-Rad | http://www.bio-rad.com/zh-cn/product/image-lab-software |
| GraphPad Prism 6.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Image-Pro Plus 6.0 | Media Cybernetics | N/A |
| Other | ||
| Bioruptor Plus sonicator | Diagenode | CAT#B01020001 |
Supplementary Material
Table S1. Differentially expressed gene list between Zc3h13 knockdown mESCs and control mESCs. Related to Figure 4.
Highlight.
Zc3h13 is a critical regulator of RNA m6A methylation
Zc3h13-WTAP-Virilizer-Hakai complex is an important regulatory complex of RNA m6A
Zc3h13 plays a role in anchoring the m6A regulatory complex in the nucleus
Zc3h13 controls mESC pluripotency by regulating m6A methylation
Acknowledgments
We thank Guoquan Yan and Lei Zhang for help with the mass spectrometry. We thank Ligang Wu for providing the minigene reporter, pTBG. We thank Xi Li and Jingyi Hui for Zc3h13 functional exploration and helpful discussions. F. L. was supported by “The national key research and development program of China” (2016YFA0101800), China “Thousand Youth Talents” (KHH1340001), NSFC (91419306), “973” State Key Development Program (2014CB943103), and ISTC (2014DFB30020). L. T. was supported by “The national key research and development program of China” (2016YFA0101800). J. D. was supported by Shanghai Pujiang Program (10PJ1400400). We thank National Institutes of Health R01 HG008688 (C. H.). C. H. is an investigator of the Howard Hughes Medical Institute. Y. S. is an American Cancer Society Research Professor.
Footnotes
Author contributions
J. W. and J. D. carried out most of the experiments and assembled the figures; R. L. analyzed MeRIP-seq data, RNA-seq data and RIP-seq data; H. M. and Chuan. H. helped with MeRIP-seq and m6A LC-MS/MS determination; H. L. conducted Hiseq2500 sequencing; Chenxi. H., Jiahua. W., F. J., and H. S. were involved in Zc3h13 constructs preparation; L. T. provided suggestions to mESC assays. P. Y. was responsible for Mass Spectrometry determination; C. H., F. L. and Y. G. S. provided advice and suggestions to the experimental designs. J. W., F. L., J. D., and Y. S., co-wrote the paper; J. D. and Y. S. supervised the project.
Declaration of Interests
Y. S. is a co-founder of Constellation Pharmaceuticals, Inc. and a member of its scientific advisory board. F. L. is a share holder of Constellation Pharmaceuticals, Inc. C. H. is a co-founder of Accent Therapeutics, Inc. and a member of its scientific advisory board.
Accession Numbers
All seq data have been deposited to GEO database (GSE94148).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34:146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nature communications. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, Hanna JH, Black DL, Darnell JE, Jr, Darnell RB. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Lu Z, Liu H, Zhang L, Zhang S, Chen Y, Rao MK, Huang Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman F, Shatkin AJ, Perry RP. Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: possible implications for processing. Cell. 1974;3:197–199. doi: 10.1016/0092-8674(74)90131-7. [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Lavi U, Darnell JE., Jr The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J Mol Biol. 1978;124:487–499. doi: 10.1016/0022-2836(78)90183-3. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Molecular cell. 2016a;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016b;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Molecular cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Zhang T, Baldie G, Periz G, Wang J. RNA-processing protein TDP-43 regulates FOXO-dependent protein quality control in stress response. PLoS genetics. 2014;10:e1004693. doi: 10.1371/journal.pgen.1004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially expressed gene list between Zc3h13 knockdown mESCs and control mESCs. Related to Figure 4.