Abstract
Circadian rhythms govern immune cell function, giving rise to time-of-day variation in the recognition and clearance of bacterial or viral pathogens; to date, however, no such regulation of the host-fungal interaction has been described. In this report, we use murine models to explore circadian control of either fungal-macrophage interactions in vitro or pathogen clearance from the lung in vivo. First, we show that expression of the important fungal pattern recognition receptor Dectin-1 (clec7a), from either bone marrow–derived or peritoneum-derived macrophages, is not under circadian regulation at either the level of transcript or cell surface protein expression. Consistent with this finding, the phagocytic activity of macrophages in culture against spores of the pathogen Aspergillus fumigatus also did not vary over time. To account for the multiple cell types and processes that may be coordinated in a circadian fashion in vivo, we examined the clearance of A. fumigatus from the lungs of immunocompetent mice. Interestingly, animals inoculated at night demonstrated a 2-fold enhancement in clearance compared with animals inoculated in the morning. Taken together, our data suggest that while molecular recognition of fungi by immune cells may not be circadian, other processes in vivo may still allow for time-of-day differences in fungal clearance from the lung.
Keywords: circadian, mammalian, Aspergillus fumigatus, fungal, clec7a, Dectin-1, macrophages, phagocytosis
The probability of pathogen exposure oscillates over the circadian day, with risk generally tracking with an animal’s activity such as foraging or feeding. As these activity cycles are controlled by the circadian clock, it follows that the immune response and, hence, the consequence of the host-pathogen interaction are similarly clock-controlled. Indeed, early studies demonstrated that survival following bacterial challenge in mice is dependent on time of inoculation; for example, intraperitoneal inoculation of pneumococcus in the light phase resulted in a higher mortality rate and corresponded with higher levels of bacteremia (Feigin et al., 1969; Wongwiwat et al., 1972). Similar results were seen in flies; for example, infection with either Pseudomonas aeruginosa or Staphylococcus aureus during the light phase resulted in higher mortality rates (Lee and Edery, 2008). These observations suggest that the mammalian immune system gates its response over the circadian day depending on the likelihood of pathogen exposure. This has important implications in human health, particularly for shift-workers, whose circadian disruption leads to dysregulation of the immune system, heightening the risk of various immunopathologies, including those associated with pathogen encounter (Castanon-Cervantes et al., 2010).
Mammalian clocks are driven by a conserved transcription-translation feedback oscillatory loop using CLOCK (encoded by CLOCK) and BMAL1 (Arntl) transcription factor heterodimers driving the expression of interlocking negative regulatory PER (Per), CRY (Cry), and REV-ERBα (Nr1d1) proteins and driving clock-controlled genes as output (Antoch et al., 1997; King et al., 1997; Tei et al., 1997; Gekakis et al., 1998; Hogenesch et al., 1998; Dunlap, 1999; Ko and Takahashi, 2006). In immune cells such as macrophages, the molecular clock is cell autonomous (Keller et al., 2009) and drives up to 8% of the transcriptome. Toll-like receptor 9 (TLR9) is one of these clock-controlled genes, showing peak transcript levels at 20 to 24 h after serum synchronization of peritoneal macrophages (Silver et al., 2012). Consequently, vaccination during high TLR9 expression results in greater proliferation of T cells, and induction of bacterial sepsis during high TLR9 expression exacerbates lethality (Silver et al., 2012). Moreover, the timing of optimal host defense is not limited to bacteria; recently, control against viral pathogens (herpesvirus, influenza) and eukaryotic parasite pathogens (Leishmania) was found to be under circadian regulation (Edgar et al., 2016; Kiessling et al., 2017).
Considering the known circadian control over bacterial, viral, and parasitic immune responses, we hypothesized that the host-fungal interaction would be similarly regulated. The most well-characterized receptor in this regard is Dectin-1 (clec7a), which binds to β-glucans on the fungal cell wall. Dectin-1 is highly expressed on macrophages, and cells from clec7a-null (Dectin-1 deficient) mice are deficient in their uptake and clearance of various fungal pathogens, including Aspergillus fumigatus and Pneumocystis carinii (Taylor et al., 2002; Herre et al., 2004; Saijo et al., 2007; Taylor et al., 2007; Werner et al., 2009). Macrophages themselves represent the first line of defense against environmental fungi, and macrophage defects are an important risk factor for many fungal infections (MacMicking et al., 1997; Erwig and Gow, 2016). Accordingly, we reasoned that if fungal recognition were regulated by the circadian clock, this would manifest as rhythmic expression of Dectin-1 in macrophages.
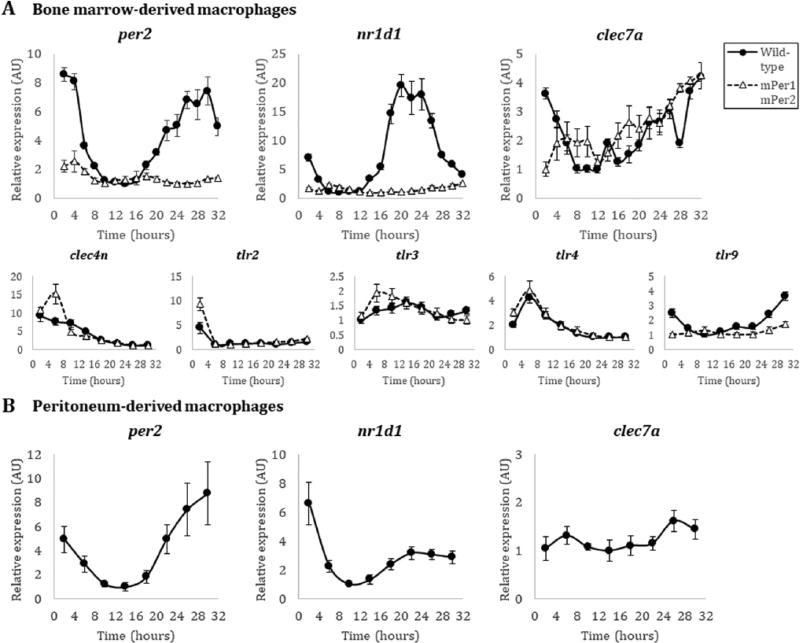
To test for rhythmicity in Dectin-1 expression, we first quantified transcript levels isolated from bone marrow–derived macrophages (BMDMs) over the course of the circadian day. Homozygous mPer2Luc/Luc knock-in mice (herein, wild-type) or mPer1/mPer2 mutant mice (Yoo et al., 2004) were used as a source of BMDMs. Briefly, cells were differentiated from a starting bone marrow population with L929 cultured supernatants as previously described (Weischenfeldt and Porse, 2008), resulting in a naïve BMDM population 90% double positive for F4/80 and CD11b markers using flow cytometry (Supplementary Fig. 1A). These cells were serum starved for 24 h in Leibovitz’s L-15 medium without serum and then synchronized with 50% horse serum. PER2LUC luminescence from macrophages was monitored in a LumiCycle (ActiMetrics, Wilmette, IL, USA) and found to be circadian (Supplementary Fig. 1A). Rhythmic cells were pulled directly from the LumiCycle and used for assay. RNA was isolated from BMDMs every 2 h after release from serum entrainment. As expected, rhythmicity could be detected by qRT-PCR for both per2 and nr1d1 transcripts from mPer2Luc/Luc macrophages, peaking between hours 28–30 and 20–22, respectively (Fig. 1A). Notably, these phases are similar to those previously described in peritoneal macrophages (Silver et al., 2012). As a control, no rhythmicity could be detected in per2 and nr1d1 transcripts in BMDMs derived from mPer1Brdm1/BrdmmPerBrdm1/Brdm (hereafter mPer1m/mmPer2m/m) mice (Zheng et al., 2001) (Fig. 1A). In contrast to per2 and nr1d1, clec7a transcripts from wild-type mPer2Luc/Luc BMDMs did not oscillate (Fig. 1A). Beyond Dectin-1, other pathogen recognition receptors (PRRs) against fungi may be important for circadian gating; for example, Dectin-1 and TLR2 colocalize during recognition of fungal zymosan (Brown et al., 2003), and Dectin-1 synergizes with TLR4 (which also detects bacterial LPS) for recognition of Candida albicans by the innate immune system (Ferwerda et al., 2008). Therefore, we tested several other PRR genes implicated in the control of fungal infections (clec4n, TLR2, TLR3, TLR4, and TLR9) from the same RNA time-course and found that Dectin-2 (clec4n), TLR2, and TLR4 transcripts did not appear to be under circadian control, whereas TLR3 and TLR9 transcripts appeared circadian in nature (Fig. 1A).
Figure 1.
Transcript expression of fungal-recognition receptor genes in macrophages. (A) Quantitative PCR of per2, nr1d1, clec7a, clec4n, tlr2, tlr3, tlr4, and tlr9 transcripts from male wild-type mPer2Luc/Luc (black filled circle, solid line) and male mutant mPer1m/mmPer2m/m (white filled triangle, dashed line) BMDMs in the C57BL/6 background. (B) Quantitative PCR of per2, nr1d1, and clec7a transcripts from wild-type mPer2Luc/Luc peritoneum-derived macrophages in the C57BL/6 background. Time (hours) represents time after initial serum synchronization. Error bars represent ± SE from technical replicates, n=3. Methods. Because the 129 strain of mice used to create the knock-outs/knock-ins (Yoo et al., 2004; Zheng et al., 2001) is not isogenic with the C57BL/6J background, and to avoid subtle genetic background issues, we carried out crosses to the C57BL/6J background, followed by SNP mapping, to select the best progeny to speed the track to isogenicity with C57. After repeated backcrosses, mPer2Luc/Luc mice used here are >97% homozygous with the C57BL/6J background.
Because macrophages derived from different tissues exhibit diverse phenotypic behaviors (Okabe and Medzhitov, 2016), we investigated whether clec7a transcript expression would be rhythmic in peritoneal macrophages. These cells were elicited from mPer2Luc/Luc animals by thioglycolate broth injection 4 days prior to peritoneal lavage and yielded a population that was 84% double positive for F4/80 and CD11b staining (Zhang et al., 2008) (Supplementary Fig. 1B). The cells were synchronized as described above for the BMDMs, and robust luciferase rhythms were detected in the LumiCycle (Supplementary Fig. 1B). RNA was isolated from peritoneal macrophages every 4 h after serum entrainment and per2 and nr1d1 transcripts again appeared rhythmic, peaking at approximately the same times as in BMDMs; however, clec7a transcripts were again not rhythmic (Fig. 1B). Consistent with the BMDM population, these data suggest that Dectin-1 transcripts are not clock-controlled in macrophages at the level of mRNA.
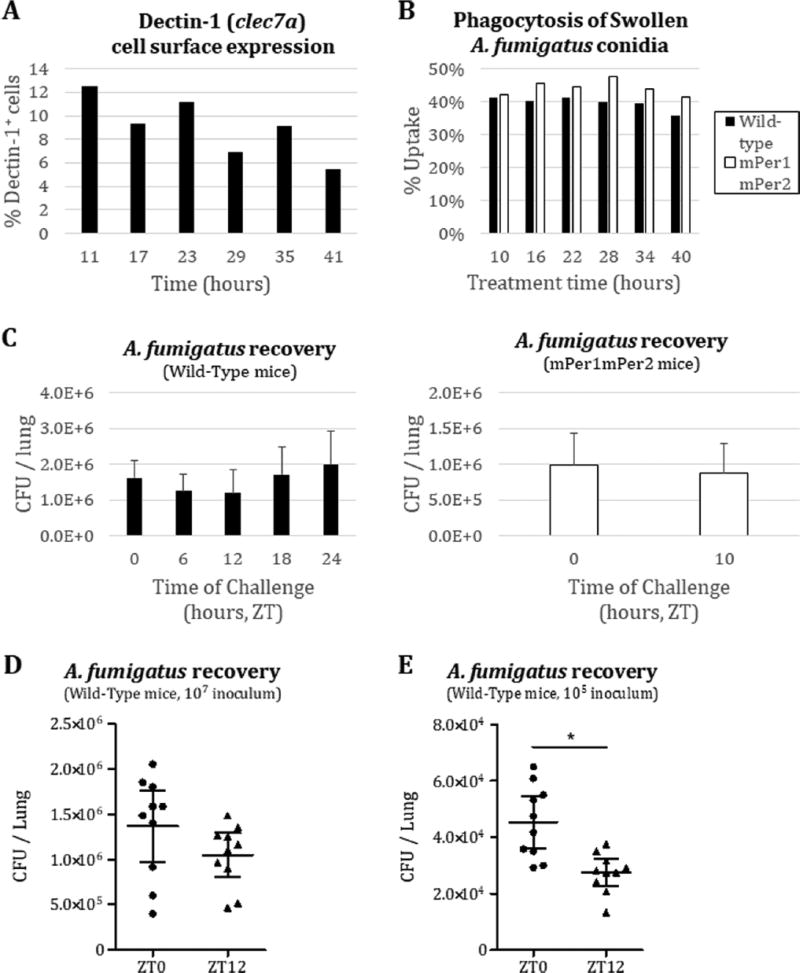
Although genes may be arrhythmic at the transcript level, gene products may still exhibit circadian rhythmicity at the protein level, as half of cycling proteins in the liver lack a corresponding cycling transcript (Reddy et al., 2006). We therefore assayed for cell surface expression of Dectin-1 in BMDMs. Flow cytometric quantification of cell surface Dectin-1 from mPer2Luc/Luc BMDMs did not vary in a circadian manner (Fig. 2A), even though the PER2 protein levels in the cells used for flow cytometry clearly were rhythmic (Supplementary Fig. 1C). These data suggest that Dectin-1 protein expression is also not under circadian control in BMDMs.
Figure 2.
Circadian gating of Aspergillus fumigatus clearance from the mouse lung. (A) Flow cytometric analysis of Dectin-1 cell surface expression (BioLegend, #144303) on BMDMs, gated on CD11b+ cells (n=1). (B) Flow cytometric analysis of dsRed+ signal (A. fumigatus), gated on F4/80+ macrophages, after one-hour co-incubation with A. fumigatus in culture. Serum-synchronized BMDM from either wild-type (black) or mPer1m/mmPer2m/m (white) background were treated with swollen dsRed-expressing A. fumigatus Af293 conidia at a multiplicity of infection of one (n=1). Conidia were swollen by prior incubation in DMEM media at 37 °C with shaking for six hours. (C) Time course of intratracheal inoculation of 107 Af293 conidia into wild-type C57BL/6J (two-month-old male C57BL/6J mice from Jackson Laboratories were fed rodent chow ad libitum, maintained under constant environmental conditions, and entrained to a 12 hr light/dark cycle, light period from 7am to 7pm) or mPer1m/mmPer2m/m C57BL/6 immunocompetent mice (n=4 per group) and subsequent lung harvest after 16-hour incubation for A. fumigatus colony-forming unit (CFU) count on YPD (yeast extract, peptone, and dextrose) agar plates. (D) Intratracheal inoculation of 107 Af293 conidia into wild-type C57BL/6J mice (n=10 per group) at ZT0/ZT12 and lung harvest after 16-hour incubation for CFU count on YPD. (E) Intratracheal inoculation of 105 Af293 conidia into wild-type C57BL/6J mice (n=10 per group) at ZT0/ZT12 and lung harvest after 14-hour incubation for CFU count on YPD. Error bars represent 95% CI. *p<0.05 by Student’s two-tailed t-test.
Even though fungal recognition by Dectin-1 may not be under clock control, the functional responses leading to fungal clearance, such as phagocytosis and other immune machinery involved, might be under clock control. For example, macrophages harvested from mice appear to phagocytose zymosan in a circadian manner (Hayashi et al., 2007), and serum-entrained macrophages also appear to engulf foreign particles much more robustly during a specific time in culture (Oliva-Ramirez et al., 2014). To address the possibility that the clock controls fungal clearance, we tested for a time-of-day variability in the engulfment activity of macrophages against spores (conidia) of the predominant mold pathogen A. fumigatus. Briefly, BMDMs from clock wild-type mPer2Luc/Luc or mPer1m/mmPer2m/m mice were exposed at various times across the circadian day to dsRed-expressing A. fumigatus spores (Jhingran et al., 2012) that had been partially germinated (swollen) in order to expose surface β-glucan levels. Uptake of the swollen conidia, as assessed by flow cytometry of double-positive dsRed and F4/80 signals, from either wild-type or mPer1m/mmPer2m/m mice, did not vary significantly across the time course (Fig. 2B). As with the expression analyses, the mPER2LUC levels were followed in the macrophage cultures throughout the experiment to ensure that the population was circadian (Supplementary Fig. 1D).
Appropriate immunological clearing of A. fumigatus from the lung requires several steps beyond the initial encounter with resident macrophages, including chemokine secretion to attract immune cells such as neutrophils to the site of infection, adhesion molecule expression on recruited cells, and activation of antifungal programs. As any of these steps could be under circadian control, we wanted to take all such phenomena into account by looking at A. fumigatus clearance in a murine model. We challenged immunocompetent mice, wild-type (C57BL/6J; Jackson Laboratories, Bar Harbor, ME, USA) or mPer1m/mmPer2m/m, with 1 × 107 Aspergillus fumigatus (strain Af293) conidia through an intratracheal route at different circadian times. At 16 h after inoculation, lungs were harvested from the mice, and homogenates were plated to determine the colony-forming units (CFUs) as a marker for fungal burden. Fungal burden from the lung did not show significant time-of-day differences in either the wild-type or mPer1m/mmPer2m/m background with 4 mice per group (Fig. 2C). To eliminate the possibility that our high inoculum was perhaps hiding a subtler circadian phenotype, and to increase the power of our analysis, we tested both a 1 × 107 and a 1 × 105 conidial inoculum at ZT0 and ZT12 using 10 mice per time-point, harvesting lungs after 14 h of incubation. Although there remained no significant time-of-day effect for the higher inoculum (Fig. 2D), we found that there was a significant time-of-inoculation influence on fungal clearance for the latter dose (Fig. 2E), although the effect was less than a 2-fold difference.
In summary, we have identified a time-of-day influence on A. fumigatus clearance from the murine lung, although the basis for this effect remains unclear. Transcript levels of the Dectin and Dectin-associated pattern recognition receptors were not found to be rhythmic. The macrophage surface expression of Dectin-1 also did not appear to be under circadian control. Thus, we conclude that Dectin is not under circadian regulation in macrophages, although other fungal pattern recognition receptors might be subject to such regulation. In our study, phagocytosis of swollen A. fumigatus, presumably mediated by Dectin-1, did not appear to be under circadian regulation. Although phagocytosis has been observed to be circadian in peritoneum-derived macrophages taken directly from the mouse prior to assay (Hayashi et al., 2007), our macrophages were serum-synchronized in culture and therefore did not receive macrophage-independent biological signals from the gut immediately before phagocytosis. Therefore, circadian control of phagocytosis may not be cell-intrinsic. Furthermore, we tested only macrophages in our in vitro studies. Other innate cell types, namely neutrophils, are known to be the dominant cell type recruited to A. fumigatus–infected lungs, presumably after an initial encounter with resident macrophages (Blanco and Garcia, 2008; Hasenberg et al., 2011). However, the direct assessment of neutrophilic circadian behavior is difficult since neutrophils have short life spans in culture. Neutrophils may therefore play a role in the observed time-of-day differences of A. fumigatus clearance from the murine lung. It will also be interesting to see whether similar results are obtained with other fungal pathogens, particularly those that infect other sites in the body (i.e., the gut or central nervous system) and whose immunological response may differ markedly from A. fumigatus (Blanco and Garcia, 2008). Taken together, our data suggest that the host-Aspergillus interaction may be under subtle circadian control but through a mechanism that is independent of Dectin-1 on macrophages.
Supplementary Material
Acknowledgments
The dsRed-expressing Aspergillus strain was graciously provided by Dr. Robert A. Cramer at Dartmouth. We thank Dr. Sarah Beattie (Cramer Lab) as well as Dr. Brent L. Berwin for excellent technical support for animal experiments. We also thank the staff of the Center for Comparative Medicine and Research and the Flow Core at Dartmouth for their technical guidance. This work was supported by grants from the National Institute of General Medical Sciences of the National Institutes of Health to J.J.L. (grant R35 GM118022) and J.C.D. (grant R35 GM118021).
Footnotes
Conflict of Interest Statement
The author(s) have no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary material for this article is available online.
References
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco JL, Garcia ME. Immune response to fungal infections. Vet Immunol Immunopathol. 2008;125:47–70. doi: 10.1016/j.vetimm.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, Besing RC, Menaker M, Gewirtz AT, Davidson AJ. Dysregulation of inflammatory responses by chronic circadian disruption. J Immunol. 2010;185:5796–5805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Edgar RS, Stangherlin A, Nagy AD, Nicoll MP, Efstathiou S, O’Neill JS, Reddy AB. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci U S A. 2016;113:10085–10090. doi: 10.1073/pnas.1601895113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig LP, Gow NA. Interactions of fungal pathogens with phagocytes. Nat Rev Microbiol. 2016;14:163–176. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- Feigin RD, San Joaquin VH, Haymond MW, Wyatt RG. Daily periodicity of susceptibility of mice to pneumococcal infection. Nature. 1969;224:379–380. doi: 10.1038/224379a0. [DOI] [PubMed] [Google Scholar]
- Ferwerda G, Meyer-Wentrup F, Kullberg BJ, Netea MG, Adema GJ. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol. 2008;10:2058–2066. doi: 10.1111/j.1462-5822.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Hasenberg M, Behnsen J, Krappmann S, Brakhage A, Gunzer M. Phagocyte responses towards Aspergillus fumigatus. Int J Med Microbiol. 2011;301:436–444. doi: 10.1016/j.ijmm.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30:621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- Herre J, Marshall AS, Caron E, Edwards AD, Williams DL, Schweighoffer E, Tybulewicz V, Reis e Sousa C, Gordon S, Brown GD. Dectin-1 uses novel mechanisms for yeast phagocytosis in macrophages. Blood. 2004;104:4038–4045. doi: 10.1182/blood-2004-03-1140. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhingran A, Mar KB, Kumasaka DK, Knoblaugh SE, Ngo LY, Segal BH, Iwakura Y, Lowell CA, Hamerman JA, Lin X, et al. Tracing conidial fate and measuring host cell antifungal activity using a reporter of microbial viability in the lung. Cell Rep. 2012;2:1762–1773. doi: 10.1016/j.celrep.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling S, Dubeau-Laramee G, Ohm H, Labrecque N, Olivier M, Cermakian N. The circadian clock in immune cells controls the magnitude of Leishmania parasite infection. Sci Rep. 2017;7:10892. doi: 10.1038/s41598-017-11297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Lee JE, Edery I. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol. 2008;18:195–199. doi: 10.1016/j.cub.2007.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- Oliva-Ramirez J, Moreno-Altamirano MM, Pineda-Olvera B, Cauich-Sanchez P, Sanchez-Garcia FJ. Crosstalk between circadian rhythmicity, mitochondrial dynamics and macrophage bactericidal activity. Immunology. 2014;143:490–497. doi: 10.1111/imm.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, Sudo K, Akira S, Adachi Y, Ohno N, et al. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007;8:39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36:251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PR, Brown GD, Reid DM, Willment JA, Martinez-Pomares L, Gordon S, Wong SY. The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol. 2002;169:3876–3882. doi: 10.4049/jimmunol.169.7.3876. [DOI] [PubMed] [Google Scholar]
- Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Porse B. Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5080. pdb prot5080. [DOI] [PubMed] [Google Scholar]
- Werner JL, Metz AE, Horn D, Schoeb TR, Hewitt MM, Schwiebert LM, Faro-Trindade I, Brown GD, Steele C. Requisite role for the dectin-1 beta-glucan receptor in pulmonary defense against Aspergillus fumigatus. J Immunol. 2009;182:4938–4946. doi: 10.4049/jimmunol.0804250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongwiwat M, Sukapanit S, Triyanond C, Sawyer WD. Circadian rhythm of the resistance of mice to acute pneumococcal infection. Infect Immun. 1972;5:442–448. doi: 10.1128/iai.5.4.442-448.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14(Unit 14):11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.