Abstract
Background
In monogastric animals, phosphorus (P) homeostasis is maintained by regulating intestinal absorption, bone mobilization, and renal excretion. Since P is a non-renewable resource, a shortage is imminent due to widespread over-usage in the farming and animal husbandry industries. As a consequence, P efficiency should be improved in pig production. We sought to characterize the transcriptional response in re−/absorbing and excreting tissues in pigs to diets varying in calcium: phosphorus ratios. Weaned piglets were assigned to one of three groups fed diets varying in digestible P content for a period of five weeks. Gene expression profiles were analyzed in jejunum, colon, and kidney.
Results
Transcriptome analysis revealed that reduced dietary P intake affects gene expression in jejunum and kidney, but not in colon. The regulation of mineral homeostasis was reflected via altered mRNA abundances of CYP24A1, CYP27A1, TRPM6, SPP1, and VDR in jejunum and kidney. Moreover, lowered abundances of transcripts associated with the classical complement system pathway were observed in the jejunum. In kidney, shifted transcripts were involved in phospholipase C, calcium signaling, and NFAT signaling, which may have immunomodulatory implications.
Conclusions
Our results revealed local transcriptional consequences of variable P intake in intestinal and renal tissues. The adaptive responses are the result of organismal efforts to maintain systemic mineral homeostasis while modulating immune features at local tissue sites. Therefore, the deviation from the currently recommended dietary P supply must be carefully considered, as the endogenous mechanisms that respond to low P diets may impact important adaptive immune responses.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4584-2) contains supplementary material, which is available to authorized users.
Keywords: Calcium: Phosphorus ratio, Pig, Gene expression, Immune response, Diet
Background
Over the last few decades, efforts to conserve the non-renewable resource phosphorus (P) have become increasingly prominent in scientific research. In mammals, the biologically active form, phosphate , is an essential structural component of nucleic acids, phospholipids, adenosine triphosphate (ATP), and hydroxyapatite in bone. P homeostasis is controlled by absorption in the small intestine (especially in the jejunum), bone remodeling, and reabsorption/excretion in the kidney [1]. In farmed pigs, a deficient P intake could lead to less bone formation and, therefore, may reduce weight gain [2, 3]. To avoid P deficiency symptoms and to ensure maximal growth, dietary P is often supplemented to levels exceeding age-specific requirements, with up to two-thirds of the consumed P being excreted [4, 5]. In agriculture, the majority of P input is the result of mineral fertilizers and animal manure, which are used to encourage crop production [6]. Unfortunately, over-application and accumulation of P results in P-laden runoff, leading to pollution and eutrophication of surface waters [7, 8].
To deal with the environmental concerns surrounding P usage, increased efficiency should be considered in pig husbandry. P recommendations should be re-evaluated considering the requirements of modern pig breeds and the mechanisms of P utilization, which are known to be affected by genetic factors and transcriptional regulation [9]. To date, only a few studies have described the molecular mechanisms that respond to varying P supplementation in pigs. Activation of vitamin D and increased P uptake from the intestine are widely accepted. Specifically, a low P diet prompted an increased mRNA abundance of 1α-hydroxylase and parathyroid hormone receptor [10], and an increased sodium-dependent phosphate uptake in the jejunum in pigs [11]. Furthermore, a previous study of our group investigated the transcriptional response of peripheral blood mononuclear cells to diets varying in calcium (Ca):P ratios, showing P as a link between bone remodeling and immune features [12]. In fact, an impact of P on the immune system has also been observed in other farm animals where e.g. dietary effects on T cell function, lymphocyte proliferation and antibody production have been reported [13–15].
The intestinal mucosa is of crucial importance to immune regulation due to its continuous contact with the external environment. In particular, the gut-associated lymphoid tissue (GALT), including Peyer’s patches, mediates the homeostatic balance between tolerance of commensal microorganisms and immune response to pathogens [16, 17]. Moreover, the mucosa-associated bacterial community can be influenced by dietary compositions [18]. The kidney also has a special role in immune tolerance against dietary antigens and hormones [19]. Furthermore, the kidney contributes to the production of hormones with immunomodulatory properties, including vitamin D [20]. Hence, several local immunological processes are present in different tissues and these may be orchestrated by systemic endocrine processes.
To evaluate the consequences of reducing dietary P supplementation in swine production, we sought to characterize the organismal response to variable P supply in pigs. The endocrine regulation of P homeostasis due to diets varying in Ca:P ratios has been previously reported [12]. Here, we describe the transcriptional effects of dietary P on tissues that are involved in P re−/absorption and excretion such as jejunum, colon, and kidney.
Methods
Animals and diets
Animals were provided by the Leibniz Institute for Farm Animal Biology (FBN). The experimental protocol was approved by the ethics committee of the federal state of Mecklenburg-Western Pomerania, Germany (Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei; LALLF MV 7221.3–1-053/15). The experimental design was described previously [12]. In short, 19 German landrace piglets from three litters of two boars were randomly assigned to one of three dietary groups (selection according to gender and litter balanced). For a period of 5 weeks (day 28–64), piglets were fed diets which differed in relation to the digestible P-content (Additional file 1). Neither phytase nor other phosphatases were added. Specifically, the achieved levels of digestible P were 0.33% (low; L), 0.51% (medium; M) and 0.74% P (high; H) on a dry matter basis. The dietary P content in group M corresponded to current recommendations [21].
Collection and preparation of tissue samples
At 64 days of age, pigs were killed by electrical stunning followed by exsanguination in the experimental slaughterhouse of FBN. Kidney, jejunum, and colon tissue were immediately collected, frozen in liquid nitrogen, and stored at − 80 °C.
RNA isolation, target preparation, and hybridization
Total RNA was isolated using TRI Reagent per manufacturer’s directions (Sigma-Aldrich, Taufkirchen, Germany), then treated with DNase and purified with the column-based NucleoSpin RNA II-Kit (Macherey-Nagel, Düren, Germany). RNA integrity was determined by visualization on a 1% agarose gel containing ethidium bromide and the concentration was measured using the NanoDrop ND-1000 spectrometer (PEQLAB, Erlangen, Germany). DNA contamination was assessed by PCR amplification of the porcine RPL32 gene (forward primer: 5’-AGCCCAAGATCGTCAAAAAG-3′; reverse primer: 5’-TGTTGCTCCCATAACCAATG-3′). All RNA samples were stored at − 80 °C.
Each RNA sample was transcribed to DNA using the Ambion WT Expression Kit (Ambion, Austin, TX, USA). The DNA preparations were fragmented and labelled with the WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, USA). DNA preparations were hybridized on genome-wide snowball arrays (Affymetrix), which were invented for genome-wide analysis of the pig transcriptome [22]. Raw data was generated with Affymetrix GCOS 1.1.1 software and deposited in a MIAME-compliant database [23], the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession number: GSE94448).
Microarray data processing and analysis
All arrays were tested and passed quality control criteria as proposed previously [24]. The data were RMA (Robust Multiarray Average) [25] normalized (Log2) and filtered by both standard deviation (SD > 0.29) and mean (x̅> 2.5) using R version 3.2.0 [26]. Transcriptional effects were analyzed between samples from animals fed L, M, and H diets. Relative changes of transcript abundance were analyzed using a linear model (JMP Genomics 7.0), including the effects of diet, sire, sex, tissue, and interactions (Vijkl = μ + dieti + sirej + sexk + tissuel + dieti*tissuel + errorijkl). P-values were converted to a set of q-values to correct for multiple testing using the algorithm proposed by Storey & Tibshirani [27]. Significance levels were set at p ≤ 0.05 and q ≤ 0.20. The annotation data for snowball arrays were obtained from the developers [22]. Analysis of regulated pathways was performed using the web-based software Ingenuity Pathway Analysis (IPA) version 01–04 (Qiagen, Redwood City, CA, USA). The significance of association between altered transcripts and regulated pathways was set at p ≤ 0.05. Genes with shifted transcript abundance were used to identify the molecular and cellular functions affected by P supply using IPA ‘Diseases and Bio-functions’ (criteria: z-score ≥ 2 or z-score ≤ − 2). To visualize the differences in the gene expression of the investigated tissues, a non-parametric hierarchical cluster analysis of significantly different probe-sets was performed using R (hclust option ‘complete’; dist option ‘euclidean’). In total 29,013 probe-sets were used to generate the heatmap.
Quantitative real-time RT-PCR
All RNA samples were transcribed to cDNA. First-strand cDNA was synthesized from 2 μg of total RNA using random hexamer primers and oligo d(T)13VN in the presence of SuperScript III reverse transcriptase (Invitrogen, Karlsruhe, Germany). To verify the microarray experiments, total transcript levels of selected targets and reference genes (Additional file 2) were quantified by real-time quantitative PCR (qPCR) performed on a LightCycler®480 system (Roche, Mannheim, Germany). Target genes involved in observed immunoregulatory pathways (NFATC2, SPP1, C1QC, C7, C1R, C1S, and MMP2) and in P metabolism (CYP24A1, CYP27A1, PTH1R, SLC34A3 and VDR) were selected for analysis. Reactions were accomplished in a final volume of 12 μL using 6.0 μL of LightCycler 480 SYBR Green I Master (Roche, Basel, Switzerland), 2.8 μL of Aqua dest., 0.6 μL of each Primer (10 μM) and 2 μL (40 ng) cDNA. Amplifications were conducted in duplicate according to the manufacturer’s instructions. The temperature profiles of PCR were as follows: initial denaturation step at 95 °C for 10 min and 40 cycles including denaturation at 95 °C for 15 s, annealing at 60 °C for 10 s and extension/fluorescence acquisition at 72 °C for 15 s. After completion of amplification, melting curve analysis and agarose gel electrophoresis were conducted to confirm the absence of unspecific PCR products or primer dimers. Serial dilutions of external PCR-generated standards (108–102 copies) were used to convert the threshold cycles into copy numbers. The calculated copy numbers were factorial normalized on RPL32 expression values. Data were calculated using a linear model analysis (JMP Genomics 7.0; effects of diet, sire, and sex). Significance levels were set at p ≤ 0.05. The analyzed data was compared to microarray results using the Spearman’s Rank Correlation in R version 3.2.0.
Results
To characterize the effects of modulated dietary phosphorus supply, we analyzed the transcriptomic profiles of tissues responsible for re−/absorption (jejunum and colon) and excretion (kidney).
Transcriptome profiles
The snowball microarray covers 47,845 probe-sets corresponding to 17,964 annotated genes. After filtering as described above, 29,013 probe-sets (~ 61%) remained for analysis. These probe-sets corresponded to 14,064 annotated genes. The number and annotation of regulated transcripts (per diet and tissue) is shown in Additional file 3.
In jejunum, two probe-sets differed between groups M and H (2 probe-sets M > H). The comparison between groups L and H showed changed mRNA abundance of 168 probe-sets (72 probe-sets L > H). Between groups L and M, 172 probe-sets (129 probe-sets L < M) differed significantly. Of these, 48 probe-sets (41 probe-sets L < M and L < H) were differentially expressed in both comparisons, i.e. L vs. H and L vs. M.
Diet-dependent gene expression patterns in colon showed only marginal transcriptional alterations. Between groups H and M, seven probe-sets (5 probe-sets H > M) differed significantly. In group L, two probe-sets were increased compared to group M. There were no pathways found to be enriched.
In kidney, 74 probe-sets (20 probe-sets L > H) were altered between groups L and H. The comparison between groups L and M revealed 330 differentially expressed probe-sets (167 probe-sets L > M). Of these, 47 probe-sets (38 probe-sets L < M and L < H) were differentially expressed in both comparisons.
Differences in gene expression were visualized by hierarchical clustering (Additional file 4). The heat map shows three major clusters representing the different tissues, whereby the colon and jejunum showed less distance due to gene expression patterns. The kidney was separated from the intestinal tissues. Group L showed greater distance from groups M and H in kidney and jejunum, reflecting the outcome of the parametric statistical analyses.
Transcriptional regulation of signaling pathways and bio-functions
IPA analysis revealed a number of P-regulated canonical pathways and Bio-functions in jejunum and kidney. In colon, there were no pathways and Bio-functions enriched.
In the jejunum, gene expression pattern revealed the same top three regulated pathways for the comparisons L vs M and L vs H (Table 1). ‘Hepatic Fibrosis’, ‘RhoA Signaling’, and ‘VEGF Signaling’ are associated with a number of genes encoding for structural proteins like collagen and actin. Transcripts associated with the ‘Complement System’ showed decreased mRNA abundances in group L, highlighting genes of the classical complement pathway. Regarding Bio-functions, the categories ‘Cellular Movement’ and ‘Cell Morphology’ were altered in both comparisons (Additional file 5). For example, ‘Reorganization of Actin Cytoskeleton’ (L vs H: 5 genes; L vs M: 6 genes) and ‘Attachment of Cells’ (L vs H: 6 genes; L vs M: 6 genes) revealed a decreased state of activation in group L. In both comparisons, CYP24A1, which encodes for calcitriol 24-hydroxylase, revealed the highest positive fold change (FC; L vs M: + 5.19; L vs H: + 7.69). TRPM6 revealed a positive fold change of + 1.80 in group L compared to M. There were various genes shared between L vs M and L vs H with negative fold changes, namely C1S (L vs M: − 4.36; L vs H: -3.08), COL15A1 (L vs M: − 3.35; L vs H: -2.73), ACTA2 (L vs M: − 2.96; L vs H: -2.63), FN1 (L vs M: − 2.89; L vs H: − 2.45), and COL1A2 (L vs M: − 2.78; L vs H: − 2.63). There were no pathways enriched for the comparison M vs H.
Table 1.
Regulated canonical pathways in jejunum
| Canonical pathway | Regulation# | p-value | Involved Genes |
|---|---|---|---|
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | L<>M | 2.24E-14 | ACTA2 (− 2.96); COL15A1 (− 3.35); COL1A1 (− 2.60); COL1A2 (− 2.78); COL3A1 (− 2.48); COL4A2 (− 1.46); COL6A1 (− 1.89); COL6A3 (− 1.87); FLT1 (− 1.72); FN1 (− 2.89); IGF2 (− 2.12); IGFBP3 (− 2.02); KDR (− 1.89); MYL9 (− 1.50); PDGFRA (− 1.83); TGFBR2 (− 1.49) |
| L<>H | 2.09E-08 | ACTA2 (− 2.63); COL15A1 (− 2.73); COL1A2 (− 2.25); COL6A1 (− 1.93); EDNRB (− 2.34); FLT1 (− 1.41); FN1 (− 2.45); IGFBP3 (− 1.89); MMP2 (− 2.20); MYL9 (− 1.46); TGFBR2 (− 1.37) | |
| Complement System | L < M | 9.08E-08 | C7 (−2.48); C1QA (− 1.77); C1QB (− 2.03); C1QC (− 2.02); C1R (− 2.25); C1S (− 4.36) |
| L < H | 3.27E-06 | C1QA (− 1.71); C1QB (− 1.86); C1QC (− 1.70); C1R (− 1.85); C1S (− 3.08) | |
| RhoA Signaling | L < M | 7.72E-07 | ABL2 (+ 1.41); ACTA2 (− 2.96); ACTC1 (− 2.64); ARPC1B (− 1.43); MYL9 (− 1.50); PIP5K1A (− 1.26); ROCK2 (+ 1.71); SEPT4 (− 1.57) |
| L < H | 9.81E-04 | ACTA2 (−2.63); MYL9 (− 1.46); NEDD4 (+ 2.11); PFN2 (− 1.56); PLXNA1 (− 1.64) | |
| VEGF Signaling | L < M | 1.99E-05 | ACTA2 (− 2.96); ACTC1 (− 2.46); FLT1 (− 1.72); KDR (− 1.89); PRKCA (+ 1.35); ROCK2 (+ 1.71) |
# - L < M and L < H = z-score ≤ − 2; L<>M and L<>H = no activity pattern available. P-values were calculated using Fisher’s exact test. Fold changes in parentheses indicate positive (L > M or L > H) or negative (L < M or L < H) transcript abundance
In kidney, the comparison between groups H and M revealed no differentially expressed genes. Comparing group L to M (Table 2), the top three pathways ‘Role of NFAT in Regulation of the Immune Response’, ‘CCR5 Signaling in Macrophages’, and ‘PI3K Signaling in B Lymphocytes’ were associated with transcripts involved in the ‘Cellular Immune Response’. In addition, transcripts associated with calcium-dependent pathways (‘Phospholipase C Signaling’, ‘Calcium Signaling’) showed altered mRNA abundances. Considering the Bio-functions (Additional file 5), the categories ‘Small Molecule Biochemistry’ and ‘Molecular Transport’ were affected in both comparisons. Functional analysis predicted an increased state of activation for ‘Transport of Inorganic Cation’ (20 genes), ‘Release of Ca2+’ (9 genes), and ‘Cell Survival’ (33 genes). Simultaneously, the functions ‘Morbidity or Mortality’ (67 genes) and ‘Organismal Death’ (65 genes) were predicted to have a decreased state of activation in group L. SPP1 (osteopontin) ranked among the most up-regulated genes (+ 2.16) in group L compared to M.
Table 2.
Regulated canonical pathways in kidney
| Canonical pathway | Regulation | p-value | Involved Genes |
|---|---|---|---|
| Role of NFAT in Regulation of the Immune Response | L > M | 1.08E-05 | AKT3 (+ 1.49); CALM3 (− 1.25); CD3D (+ 1.59); CD3E (+ 1.52); FCGR3B (+ 1.64); GNAS (+ 1.33); GNG2 (+ 1.43); LCP2 (+ 1.69); NFATC2 (+ 1.51); PLCB3 (− 1.42); PLCB4 (+ 1.67) |
| CCR5 Signaling in Macrophages | L<>M | 2.33E-05 | CALM3 (− 1.25); CCR5 (+ 1.76); CD3D(+ 1.59); CD3E (+ 1.52); GNAS (+ 1.33); GNG2 (+ 1.43); PRKCB (+ 1.55) |
| PI3K Signaling in B Lymphocytes | L > M | 3.17E-05 | AKT3 (+ 1.49); CALM3 (− 1.25); CAMK2G (− 1.40); NFATC2 (+ 1.51); PLCB3 (− 1.42); PLCB4 (+ 1.67); PRKCB (+ 1.55); PTPRC (+ 1.86); VAV2 (− 1.37) |
| Phospholipase C Signaling | L > M | 4.59E-05 | CALM3 (− 1.25); CD3D (+ 1.59); CD3E (+ 1.52); GNAS (+ 1.33); GNG2 (+ 1.43); LCP2 (+ 1.69); NFATC2 (+ 1.51); PLA2G6 (− 1.39); PLCB3 (− 1.42); PLCB4 (+ 1.67); PRKCB (+ 1.55); RHOJ (+ 1.56) |
| Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis | L > M | 4.46E-04 | ADAMTS5 (+ 1.64); AKT3 (+ 1.49); CALM3 (− 1.25); CTSK (+ 1.54); FZD7 (+ 1.56); GSN (+ 1.64); IGF1 (+ 1.82); IL33 (+ 1.83); NFATC2 (+ 1.51); SPP1 (+ 2.36) |
| Calcium Signaling | L < M | 7.26E-04 | ACTC1 (− 3.24); CALM3 (− 1.25); CAMK2G (− 1.40); GRIA3 (− 1.38); LETM1 (− 1.21); NFATC2 (+ 1.51); SLC8A1 (− 1.90) |
| LXR/RXR Activation | L<>H | 3.10E-03 | ARG2 (− 1.48); SERPINF2 (− 3.08); TF (− 2.56) |
| RhoGDI Signaling | L<>H | 8.12E-03 | ACTC1 (− 2.88); CDH2 (− 2.72); ESR1 (1.87) |
# - L < M = z-score ≤ − 2; L > M = z-score ≥ 2; L<>M and L<>H = no activity pattern available. P-values were calculated using Fisher’s exact test. Fold changes in parentheses indicate positive (L > M or L > H) or negative (L < M or L < H) transcript abundance
Validation of selected transcripts by RT-qPCR
To validate the microarray experiment, transcripts involved in immunoregulatory pathways (NFATC2, SPP1, C1QC, C7, C1R, C1S, and MMP2) and in P metabolism (CYP24A1, CYP27A1, PTH1R, SLC34A3, and VDR) were analyzed by qPCR. Between microarray and qPCR data, the correlation coefficients were highly significant and ranged between 0.61 and 0.94 (Table 3). The fold changes revealed a reliable dimension. In brief, the qPCR data confirmed the selected microarray results and indicated reproducibility of the microarray analysis. It should be noted that the qPCR analysis of CYP27A1 exhibited significant alterations between the different groups (L > M; L > H; M > H).
Table 3.
RT-qPCR results and correlation. Values in parentheses represent Fold Changes (FC)
| Tissue | Gene name | Microarray | qPCR# | Correlation## | ||||
|---|---|---|---|---|---|---|---|---|
| H vs L | H vs M | M vs L | H vs L | H vs M | M vs L | |||
| Kidney | CYP27A1 | H < L (2.11) |
H < M (1.45) |
M < L (1.46) |
H < L*** (2.83) |
H < M** (1.60) |
M < L** (1.77) |
0.94 |
| NFATC2 | H < L (1.33) |
H > M (1.13) |
M < L** (1.51) |
H < L* (1.76) |
H > M (1.04) |
M < L* (1.82) |
0.66 | |
| PTH1R | H > L (1.14) |
H < M (1.09) |
M > L (1.26) |
H > L (1.03) |
H < M (1.20) |
M > L (1.24) |
0.62 | |
| SLC34A3 | H > L (1.04) |
H < M (1.26) |
M > L (1.31) |
H < L (1.30) |
H < M* (1.40) |
M > L (1.07) |
0.61 | |
| SPP1 | H < L (1.48) |
H > M (1.46) |
M < L*** (2.16) |
H < L* (2.02) |
H > M (1.19) |
M < L** (2.41) |
0.92 | |
| VDR | H > L*** (1.75) |
H < M (1.05) |
M > L*** (1.84) |
H > L*** (1.75) |
H < M (1.08) |
M > L*** (1.88) |
0.76 | |
| Jejunum | CYP24A1 | H < L*** (7.69) |
H < M (1.48) |
M < L*** (5.19) |
H < L** (222.14) |
H < M (2.79) |
M < L* (79.59) |
0.89 |
| C1QC | H > L** (1.70) |
H < M (1.18) |
M > L*** (2.02) |
H > L** (1.79) |
H < M (1.05) |
M > L** (1.88) |
0.83 | |
| C1R | H > L** (1.85) |
H < M (1.19) |
M > L*** (2.24) |
H > L** (2.19) |
H < M (1.15) |
M > L** (2.52) |
0.93 | |
| C1S | H > L*** (3.08) |
H < M (1.19) |
M > L*** (4.36) |
H > L** (2.48) |
H < M (1.34) |
M > L*** (3.33) |
0.89 | |
| C7 | H > L (2.02) |
H < M (1.18) |
M > L** (2.47) |
H > L* (2.64) |
H < M (1.14) |
M > L** (3.00) |
0.94 | |
| MMP2 | H > L*** (2.20) |
H > M (1.18) |
M > L** (1.88) |
H > L** (4.05) |
H > M (1.21) |
M > L** (3.35) |
0.92 | |
Significance level was set at ***p < 0.001; **p < 0.01, *p < 0.05 and q < 0.2 for results from microarray and qPCR analyses. Fold Changes are stated in parentheses. # - Values were calculated by factorial normalization on RPL32 expression values. ## - Correlation of normalized values was calculated by Spearman; the significance level for correlation was set at p < 0.01. Scatterplots of jejunal and renal transcripts are displayed in Additional file 6
Discussion
Growing piglets face the challenge of maintaining P homeostasis despite increased P requirements for body growth and weight gain. Hence, piglets have to orchestrate numerous regulatory mechanisms in different tissues, which is reflected on the endocrine [12] and transcriptional levels.
Decreased P intake provokes a transcriptional response in the jejunum, but not in the colon
The expression profiles of jejunum and colon resulting from lowered P intake exhibited considerable differences in the tissue-specific regulation of transcription. In jejunum, numerous transcripts were differentially expressed between the dietary groups, whereas the colon showed barely any transcriptional differences, revealing a variable contribution to maintain P homoeostasis in specific parts of the intestine. The jejunum is assumed to be the major site for active P absorption in monogastric mammals [28, 29]. P absorption is hormonally stimulated by calcitriol [30], whose serum levels have been demonstrated previously to be increased in the group L [12]. In fact, calcitriol is able to induce the expression of target genes involved in enteral absorption, renal excretion and bone remodeling which affect calcium absorption and renal P excretion [31]. In jejunum, CYP24A1 exhibited the highest positive fold change in group L compared to groups H and M. In fact, CYP24A1 transcription is known to be strongly regulated by calcitriol as the encoded 24-hydroxylase inactivates calcitriol. This also corresponds to our observations in a pig experiment in which the jejunal transcription of CYP24A1 varied greatly in diets with variable calcium-P ratios [32]. This feedback loop could prevent hypercalcemic effects in L-samples, but could also have effects on P-absorption in the jejunum [33]. In addition, there are a number of genes encoding for proteins responsible for P uptake in jejunum. From these genes, SLC34A2, encoding the sodium dependent phosphate cotransporter type IIb, and S100G, encoding calbindin-D9k, showed no mRNA alterations in our study. These findings are supported by Saddoris et al., who assumed a post-transcriptional regulation of SLC34A2 and S100G [11].
The jejunum represents an important site for absorption of other minerals. Our results revealed that the expression of the transient receptor potential cation channel subfamily M member 6 (TRPM6) was diet-dependently altered (L > M). In fact, TRPM6 is essential for magnesium absorption [34]. As previously presented by our group [12], the analyzed pigs exhibited diet-dependent magnesium levels in serum (L < M; H < M). This observation supports previous studies stating an interrelation between magnesium, calcium, and inorganic P [35, 36]. However, mRNA expression of TRPM6 was increased in L but not in H animals when compared to M fed animals. This pattern might reflect compensatory attempts of L animals to maintain mineral homoeostasis.
Our results indicate that the large intestine has no part in P absorption and that the differentially expressed transcripts in colon are not associated with mineral homeostasis or intestinal functions. As reviewed by Sabbagh et al., P absorption is physiologically irrelevant in the colon under normal conditions; in fact, Breves and Schröder could not detect P absorption in the large intestine of pigs [28, 37]. Therefore, our microarray experiment in the colon validated previous results and supports the finding that the transcriptional response in jejunum is specific to P diets.
Transcriptional response in kidney to lowered P intake reveals the importance of CASR for mineral homeostasis
Of the three investigated organs, the kidney exhibited the highest number of differentially expressed genes. The supporting qPCR analysis revealed that the mRNA abundance of CYP27A1 was increased in pigs with lowered P intake (L > M; L > H; M > H). The enzyme (CYP27A1) has 25-hydroxylase activity to convert cholecalciferol to calcidiol [38]. Calcidiol is the precursor of calcitriol and acts as a transport and storage form of vitamin D [39]. Since CYP27A1 is known to be primarily expressed in liver tissue, the diet-dependent CYP27A1 expression in kidney reflects that mineral homoeostasis comprises feedback mechanisms at local tissue sites. Apart from this known physiological mechanism, there were several canonical pathways associated with calcium-dependent signaling that were altered by P diet. In particular, the phospholipase C beta 4 (PLCB4; L > M) was significantly altered by P intake, implicating a change in signal transduction. The catabolic activity of phospholipase C depends on the activation of G protein-coupled receptors [40] such as the calcium sensing receptor (CASR). Consequently, CASR mRNA abundance was increased in group L (Fig. 1). In fact, CASR expression was probably induced by the elevated levels of calcium in serum observed in group L [12]. It is known from kidney that CASR is involved in the control of calcium and P homeostasis, transport of cations, urinary acidification and concentration, and renin release [41]. Activated PLCB4 catalyzes the conversion of phosphatidylinositol 4,5-bisphosphate (PIP2) to inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). IP3 and DAG are both essential as second messengers for numerous cellular and metabolic processes [42]. Functional analysis predicted an increased state of activation for ‘release of Ca2+’, which is consistent with the fact that IP3 induces the release of calcium from intracellular stores into the cytoplasm, thus activating downstream signaling cascades [43]. DAG and calcium are necessary to activate protein kinase C beta (PRKCB), which in turn has various functions in diverse cellular signaling pathways [44]. Our results showed increased mRNA abundance for PRKCB and decreased transcript levels for VDR. This is consistent with findings demonstrating that the elevation of intracellular calcium and activation of protein kinase C inhibits vitamin D receptor (VDR) gene expression [45]. The activation of protein kinase C has been shown to enhance the induction of SPP1 by calcitriol but is not correlated with VDR activation [46]. As reviewed elsewhere [47], osteopontin, encoded by SPP1, has various functions in different cell types. In kidney, the protein is synthesized in the epithelial cells and secreted into urine. There, osteopontin inhibits the urinary crystallization of calcium salts, thus avoiding the formation of renal stones and increasing urinary calcium excretion [48]. Moreover, our results showed increased mRNA abundance of STC1 in group L. STC1 encodes stanniocalcin, which enhances P re−/absorption in small intestine and kidney [49]. Circulating calcium and calcitriol [12] induce STC1 gene expression [50], whereby these effects are possibly mediated by activation of CASR [51].
Fig. 1.
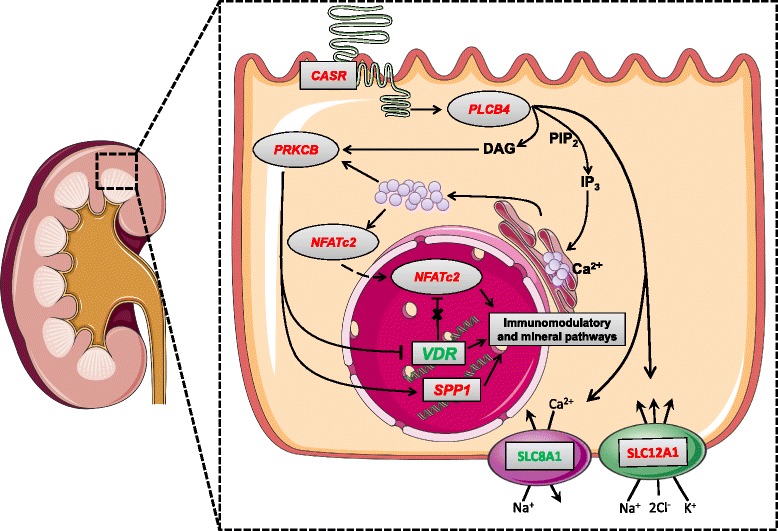
Model for signal transduction in renal cells due to lowered P intake. Solid arrows represent activation, dashed arrows represent inhibition. The data is based on a microarray comparison between groups L and M. Genes in red: L > M; genes in green: L < M. L – Low P supply; M – Medium P supply; H – High P supply
Our results also reveal a number of P-regulated genes encoding transporter molecules. The kidney-specific proteins NKCC2, encoded by SLC12A1 (L > M), and NCC, encoded by SLC12A3 (L < M), are responsible for renal sodium chloride absorption [52]. Another target transport protein is NCX1, encoded by SLC8A1 (L < M), which regulates intracellular calcium homeostasis [53]. Furthermore, the phospholipases PLCB3 (L < M) and PLA2G6 (L < M) modulate electrolyte transport by CASR signaling [41, 54, 55]. The proteins NPT4, encoded by SLC17A3 (L < M), and NPT5, encoded by SLC17A4 (L < M), mediate the transport of organic anions such as urate. Unfortunately, there is not much known about their regulation and functional physiology [56]. However, our results suggest that changes in serum P and calcium levels are associated with altered mRNA of various transporter proteins in kidney in order to maintain mineral homeostasis.
Genes involved in immune-relevant pathways are altered in jejunum by decreased P intake
The gene expression in jejunum revealed considerable shifts due to low P intake, resulting primarily in the downregulation of genes involved in several signaling pathways. The top canonical pathway ‘Hepatic fibrosis’ might state an accumulation of extracellular matrix proteins (mainly collagen) which can occur in all tissues. The pathways ‘RhoA Signaling’ and ‘VEGF Signaling’ comprise genes associated with cytoskeleton organization like actin, myosin, and Rho Kinase 2. Interestingly, Suyama et al. demonstrated an upregulation for collagen fibril organization in rat kidney due to a high P diet, confirmed by fibrosis-like regions in kidney sections [57]. During fibrogenesis, the intestinal function is restricted due to impaired motility and deposition of collagens [58]. Additionally, it is known from other diseases that deposition of collagen can impair the immune function of GALT [59]. Furthermore, Kutuzova and DeLuca demonstrated that calcitriol suppresses various matrix modeling proteins which increases the intestinal epithelial tight junction permeability [60]. Consequently, the downregulation of various genes associated with the extracellular matrix (e.g. COL1A1, COL1A2, SPARC, and MMP2) and cytoskeleton (ACTC1, ACTA2, MYL9, and KDR) indicates that a low P intake has not only anti-calcific features and counteracts fibrous changes in the small intestine, but also regulates paracellular calcium absorption through calcitriol actions. Apart from this, jejunal transcripts associated with the ‘Complement System’ showed lowered mRNA abundances due to the low P diet. The complement system acts as an immune surveillance system, discriminating between healthy tissue, apoptotic cells, and microbial intruders [61]. The C1 complex, encoded by C1QA, C1QB, and C1QC, recognizes surface structures on microbial and apoptotic cells and activates the proteases C1r and C1s, initiating the classical pathway of the complement system. All genes, which encode for the C1 complex, were downregulated in group L, indicating an inhibition of the classical pathway. It can be argued that such an inhibition could prevent the immune response to commensal microorganisms, which can be considered as an increase in immune tolerance by blocking the classical pathway and saving energy resources [62]. Taken together, our results indicate that a low P diet has an effect on immune features in jejunum and possibly supports a healthy intestinal function through calcitriol actions.
Low P results in transcriptional changes involved in mineral homeostasis and immunomodulatory signaling pathways
In kidney, the top three P-regulated pathways were found to be associated with transcripts involved in the cellular immune response. The canonical pathways ‘Role of NFAT in Regulation of the Immune Response’ and ‘Phospholipase C Signaling’ are partly associated with the same transcripts, indicating that they are integrated. Of particular interest is the nuclear factor of activated T cells 2 (NFATC2; L > M), a transcription factor that integrates calcium signaling with other signaling pathways and regulates different aspects of the immune system. In fact, the previously described CASR mediated pathway is able to induce the activation of NFATC2 [63]. Our results exhibited increased mRNA abundance for CD3D, CD3E, and PTPRC, encoding for the CD3 complex and CD45, respectively. The CD3 complex is part of the T cell receptor and CD45 is required for T cell activation, both implicating altered T cell signaling as a result of variable dietary P. Activation of the T cell receptor and subsequent involvement of NFAT proteins leads to T helper cell differentiation and induction of immune genes [64]. Furthermore, an unbalanced activation of NFAT without co-stimulatory molecules (e.g. CD28) leads to the expression of anergy-inducing genes, mediating a status of T cell unresponsiveness [65, 66]. Moreover, NFATC2 is also a molecular target for calcitriol-dependent effects mediated by VDR, which is able to inhibit the NFATC2 complex formation via AP-1 (activator protein 1) [67, 68]. Thus, the potential down-regulation of VDR by protein kinase C might enable the formation of the NFATC2 complex. Calcitriol has immunoregulatory properties, such as T cell activation [69] as well as T cell suppression [67]. These contrary regulatory effects on the immune system of vitamin D are mediated by VDR expression level [70], which in turn is modulated by the activity of protein kinase C. Also of interest is osteopontin, which is not only an inhibitor of urinary crystallization, but has also immunomodulatory properties like T cell activation and leucocyte recruitment [47]. The functions and mechanisms of osteopontin are diverse, and its expression is affected by various factors, including calcitriol, calcium and P. Thus, osteopontin might function as a further link between mineral homeostasis and the immune system, but the distinct molecular routes due to modulated P supply remain unclear. The genes NFATC2, VDR, and SPP1 are targets of pathways sensitive to mineral alterations as well as T cell receptor signaling, highlighting the interaction between P and the immune system.
Conclusion
We characterized the transcriptional response in re−/absorbing and excreting tissues in pigs after variations in dietary P intake. Our analyses in jejunum, colon, and kidney revealed changes in the expression of various genes involved in adaptive responses to maintaining mineral homeostasis. The altered serum calcium, PTH and calcitriol levels observed previously [12] may trigger cellular signaling responses involving genes identified by this study. Specifically, transcripts associated with the complement system and NFAT signaling are likely to represent the involvement of P in the immune system. Furthermore, the P-dependent expression of osteopontin might link immune modulation and mineral homeostasis. However, the influence of a varying P intake during an immune challenge remains to be investigated. Taken together, our results suggest that the impact of reduced P supply during the growth period of piglets should be considered in terms of immune function, growth and performance. Possible phased feeding regimes covering age-related mineral requirements should therefore be studied in order to reconcile aspects of livestock production and animal welfare in order to derive a relevant P policy/governance.
Additional files
Composition of the experimental diets (XLSX 11 kb)
Primer used for verification of microarray by qRT-PCR (XLSX 11 kb)
Probe-sets showing significantly altered mRNA abundances (per diet and tissue). The Venn diagram depicting the number of probe-sets differentially expressed in a) colon, b) jejunum, and c) kidney. Number of up-regulated probe-sets is shown in red. Number of down-regulated probe-sets is shown in green. L – Low P supply; M – Medium P supply; H – High P supply (XLSX 254 kb)
The hierarchical clustering of filtered probe-sets is represented by log2 transformed intensity values obtained (i) from the variance component diet*tissue (least square means) and (ii) from data obtained from individual animals. L – Low P supply; M – Medium P supply; H – High P supply (PDF 2172 kb)
Molecular and cellular functions affected by P supply (XLSX 9 kb)
Scatterplots of jejunal and renal transcripts (PDF 20 kb)
Acknowledgements
The authors would like to thank Heike Riese, Dirk Ameling, and the farm stuff for assistance with pig management. We thank Hannelore Tychsen, Angela Garve, Janine Wetzel, Annette Jugert, and Joana Bittner for their excellent technical help.
Funding
This work was partly funded by the Leibniz Science Campus Phosphorus Research Rostock and the European Union Seventh Framework Programme (FP7/2007–2013) as part of the ECO-FCE project under grant agreement No. 311794. The Leibniz Institute for Farm Animal Biology provided its own matched funding. The publication of this article was funded by the Open Access Fund of the Leibniz Association and the Open Access Fund of the Leibniz Institute for Farm Animal Biology (FBN).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Raw data are deposited in a MIAME-compliant database, the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession number: GSE94448).
Abbreviations
- ATP
Adenosine triphosphate
- Ca
Calcium
- FC
Fold change
- GALT
Gut-associated lymphoid tissue
- H
Group high
- IPA
Ingenuity Pathway Analysis
- L
Group low
- M
Group medium
- P
Phosphorus
Phospate
- qPCR
Real-time quantitative PCR
- RMA
Robust Multiarray Average
- SD
Standard deviation
- x̅
Mean
Authors’ contributions
MO, KB and KW made the conception and design of research; KW supervised the experiment; FJ, MO, KB and KW performed experiments; FJ, MO and KW analyzed data; FJ, MO, LB, EM, SP, PW and KW interpreted results of experiments; FJ drafted the manuscript; FJ prepared figures; EM, SP and PW contributed reagents/materials/analysis tools; MO, LB, EM, SP, PW and KW edited and revised manuscript; FJ, MO, KB, LB, EM, SP, PW and KW approved final version of manuscript;
Ethics approval
Animals were provided by the Leibniz Institute for Farm Animal Biology (FBN). The experimental protocol was approved by the ethics committee of the federal state of Mecklenburg-Western Pomerania, Germany (Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei; LALLF MV 7221.3–1-053/15).
Consent for publication
Not applicable.
Competing interests
No conflicts of interest, financial or otherwise, are declared by the authors
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4584-2) contains supplementary material, which is available to authorized users.
Contributor Information
Franziska Just, Email: just@fbn-dummerstorf.de.
Michael Oster, Email: oster@fbn-dummerstorf.de.
Kirsten Büsing, Email: kirsten.buesing@uni-rostock.de.
Luisa Borgelt, Email: luisa.borgelt@uni-rostock.de.
Eduard Murani, Email: murani@fbn-dummerstorf.de.
Siriluck Ponsuksili, Email: ponsuksili@fbn-dummerstorf.de.
Petra Wolf, Email: petra.wolf@uni-rostock.de.
Klaus Wimmers, Email: wimmers@fbn-dummerstorf.de.
References
- 1.Prasad N, Bhadauria D. Renal phosphate handling: physiology. Indian J Endocrinol Metab. 2013;17:620–627. doi: 10.4103/2230-8210.113752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Partanen K, Siljander-Rasi H, Karhapää M, Ylivainio K, Tupasela T. Responses of growing pigs to different levels of dietary phosphorus - performance, bone characteristics, and solubility of faecal phosphorus. Livest Sci. 2010;134:109–112. doi: 10.1016/j.livsci.2010.06.113. [DOI] [Google Scholar]
- 3.Alexander LS, Mahajan A, Odle J, Flann KL, Rhoads RP, Stahl CH. Dietary phosphate restriction decreases stem cell proliferation and subsequent growth potential in neonatal pigs. J Nutr. 2010;140:477–482. doi: 10.3945/jn.109.117390. [DOI] [PubMed] [Google Scholar]
- 4.Poulsen HD, Jongbloed AW, Latimier P, Fernández JA. Phosphorus consumption, utilisation and losses in pig production in France, the Netherlands and Denmark. Livest Prod Sci. 1999;58:251–259. doi: 10.1016/S0301-6226(99)00013-5. [DOI] [Google Scholar]
- 5.Dourmad JY, Guingand N, Latimier P, Sève B. Nitrogen and phosphorus consumption, utilisation and losses in pig production: France. Livest Prod Sci. 1999;58:199–211. doi: 10.1016/S0301-6226(99)00009-3. [DOI] [Google Scholar]
- 6.Cordell D, White S. Life’s bottleneck: sustaining the World’s phosphorus for a food secure future. Annu Rev Environ Resour. 2014;39:161–188. doi: 10.1146/annurev-environ-010213-113300. [DOI] [Google Scholar]
- 7.Kruse J, Abraham M, Amelung W, Baum C, Bol R, Kühn O, et al. Innovative methods in soil phosphorus research: a review. J Plant Nutr Soil Sci. 2015;178:43–88. doi: 10.1002/jpln.201400327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Graedel TE. A half-century of global phosphorus flows, stocks, production, consumption, recycling, and environmental impacts. Glob Environ Chang Elsevier Ltd. 2016;36:139–152. doi: 10.1016/j.gloenvcha.2015.12.005. [DOI] [Google Scholar]
- 9.Hittmeier LJ, Grapes L, Lensing RL, Rothschild MF, Stahl CH. Genetic background influences metabolic response to dietary phosphorus restriction. J Nutr Biochem. 2006;17:385–395. doi: 10.1016/j.jnutbio.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Alexander LS, Qu A, Cutler SA, Mahajan A, Lonergan SM, Rothschild MF, et al. Response to dietary phosphorus deficiency is affected by genetic background in growing pigs. J Anim Sci American Society of Animal Science. 2008;86:2585–2595. doi: 10.2527/jas.2007-0692. [DOI] [PubMed] [Google Scholar]
- 11.Saddoris KL, Fleet JC, Radcliffe JS. Sodium-dependent phosphate uptake in the jejunum is post-transcriptionally regulated in pigs fed a low-phosphorus diet and is independent of dietary calcium concentration. J Nutr. 2010;140:731–736. doi: 10.3945/jn.109.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oster M, Just F, Büsing K, Wolf P, Polley C, Vollmar B, et al. Towards improved phosphorus efficiency in monogastrics - interplay of serum, minerals, bone and immune system after divergent dietary phosphorus supply in swine. Am J Physiol Regul Integr Comp Physiol. 2016;310:R917–R925. doi: 10.1152/ajpregu.00215.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kegley EB, Spears JW, Auman SK. Dietary phosphorus and an inflammatory challenge affect performance and immune function of weanling pigs. J Anim Sci. 2001;79:413–419. doi: 10.2527/2001.792413x. [DOI] [PubMed] [Google Scholar]
- 14.Liu N, Ru YJ, Cowieson AJ, Li FD, Cheng XCH. Effects of Phytate and Phytase on the performance and immune function of broilers fed nutritionally marginal diets. Poult Sci. 2008;87:1105–1111. doi: 10.3382/ps.2007-00517. [DOI] [PubMed] [Google Scholar]
- 15.Heyer CME, Weiss E, Schmucker S, Rodehutscord M, Hoelzle LE, Mosenthin R, et al. The impact of phosphorus on the immune system and the intestinal microbiota with special focus on the pig. Nutr Res Rev Cambridge University Press. 2015;28:67–82. doi: 10.1017/S0954422415000049. [DOI] [PubMed] [Google Scholar]
- 16.Turner JR. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. Nat Publ Group. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 17.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 18.Mann E, Schmitz-Esser S, Zebeli Q, Wagner M, Ritzmann M, Metzler-Zebeli BU. Mucosa-associated bacterial microbiome of the gastrointestinal tract of weaned pigs and dynamics linked to dietary calcium-phosphorus. PLoS One. 2014;9(1):e86950. [DOI] [PMC free article] [PubMed]
- 19.Lukacs-Kornek V, Burgdorf S, Diehl L, Specht S, Kornek M, Kurts C. The kidney-renal lymph node-system contributes to cross-tolerance against innocuous circulating antigen. J Immunol. 2008;180:706–715. doi: 10.4049/jimmunol.180.2.706. [DOI] [PubMed] [Google Scholar]
- 20.Kurts C, Panzer U, Anders H-J, Rees AJ. The immune system and kidney disease: basic concepts and clinical implications. Nat Rev Immunol. 2013;13:738–753. doi: 10.1038/nri3523. [DOI] [PubMed] [Google Scholar]
- 21.GfE [Ausschuss für Bedarfsnormen der Gesellschaft für Ernährungsphysiologie] Empfehlungen zur Energie- und Nährstoffversorgung von Schweinen. Frankfurt am Main: DLG-Verlag; 2006. [Google Scholar]
- 22.Freeman TC, Ivens A, Baillie JK, Beraldi D, Barnett MW, Dorward D, et al. A gene expression atlas of the domestic pig. BMC Biol. 2012;10:90. doi: 10.1186/1741-7007-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics - a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.R Core Team. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria; 2015. Available from: http://www.r-project.org/.
- 27.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breves G, Schröder B. Comparative aspects of gastrointestinal phosphorus metabolism. Nutr Res Rev. 1991;4:125–140. doi: 10.1079/NRR19910011. [DOI] [PubMed] [Google Scholar]
- 29.Partridge IG. Studies on digestion and absorption in the intestines of growing pigs. Br J Nutr. 1978;39:539–545. doi: 10.1079/BJN19780069. [DOI] [PubMed] [Google Scholar]
- 30.Berndt T, Kumar R. Novel mechanisms in the regulation of phosphorus homeostasis. Physiology. 2008;24:17–25. doi: 10.1152/physiol.00034.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JG, D a B. Calcium and phosphorus homeostasis. Blood Purif. 2009;27:387–394. doi: 10.1159/000209740. [DOI] [PubMed] [Google Scholar]
- 32.Oster M, Gerlinger C, Heide K, Just F, Borgelt L, Wolf P, et al. Lower dietary phosphorus supply in pigs match both animal welfare aspects and resource efficiency. Ambio. 2017;47(Suppl 1):20–29. doi: 10.1007/s13280-017-0969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barletta F, Dhawan P, Christakos S. Integration of hormone signaling in the regulation of human 25(OH)D3 24-hydroxylase transcription. Am J Physiol Endocrinol Metab. 2004;286:E598–E608. doi: 10.1152/ajpendo.00214.2003. [DOI] [PubMed] [Google Scholar]
- 34.Robak P, Ozgo M, Michałek K, Kolasa-Wołosiuk A, Taciak M, Barszcz M, et al. Identification of TRPM6 and TRPM7 expression changes in response to a diet supplemented with inulin in porcine kidney. Arch Tierzucht. 2016;59:267–274. [Google Scholar]
- 35.Clark I. Effects of magnesium ions on calcium and phosphorus metabolism. Am J Phys. 1968;214:348–356. doi: 10.1152/ajplegacy.1968.214.2.348. [DOI] [PubMed] [Google Scholar]
- 36.Clark I. The effects of alterations in dietary magnesium on calcium, phosphate and skeletal metabolism. Calcif Tissue Res. 1967;1:204–218. doi: 10.1007/BF02008092. [DOI] [PubMed] [Google Scholar]
- 37.Sabbagh Y, Giral H, Caldas Y, Levi M, Schiavi SC. Intestinal phosphate transport. Adv Chronic Kidney. 2011;18:85–90. doi: 10.1053/j.ackd.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones G, Prosser DE, Kaufmann M. Cytochrome P450-mediated metabolism of vitamin D. J Lipid Res. 2014;55:13–31. doi: 10.1194/jlr.R031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver CM, Fleet JC. Vitamin D requirements: current and future 1 – 3. Am J Clin Nutr. 2004;80(6 Suppl.):1735S-9S. [DOI] [PubMed]
- 40.Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 41.Riccardi D, Brown EM. Physiology and Pathophysiology of the calcium-sensing receptor in the kidney. Am J Physiol Ren Physiol. 2010;298:F485–F499. doi: 10.1152/ajprenal.00608.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadamur G, Ross EM. Mammalian phospholipase C. Annu Rev Physiol. 2013;75:127–154. doi: 10.1146/annurev-physiol-030212-183750. [DOI] [PubMed] [Google Scholar]
- 43.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat. Rev Mol Cell Biol. 2000;1:11–21. [DOI] [PubMed]
- 44.Systems EA, Analogs A. Protein kinase c. Online. 1991;51:71–95. [Google Scholar]
- 45.Krishnan AV, Feldman D. Activation of protein kinase-C inhibits vitamin D receptor gene expression. Mol Endocrinol. 1991;5:605–612. doi: 10.1210/mend-5-4-605. [DOI] [PubMed] [Google Scholar]
- 46.Yang W, Hyllner SJ, Christakos S. Interrelationship between signal transduction pathways and 1,25(OH)2D3 in UMR106 osteoblastic cells. Am J Physiol Endocrinol Metab. 2001;281:E162–E170. doi: 10.1152/ajpendo.2001.281.1.E162. [DOI] [PubMed] [Google Scholar]
- 47.Mazzali M, Kipari T, Ophascharoensuk V, Wesson JA, Johnson R, Hughes J. Osteopontin--a molecule for all seasons. QJM. 2002;95:3–13. doi: 10.1093/qjmed/95.1.3. [DOI] [PubMed] [Google Scholar]
- 48.Kleinman JG, Beshensky A, Worcester EM, Brown D. Expression of osteopontin, a urinary inhibitor of stone mineral crystal growth, in rat kidney. Kidney Int Elsevier Masson SAS. 1995;47:1585–1596. doi: 10.1038/ki.1995.222. [DOI] [PubMed] [Google Scholar]
- 49.Madsen KL, Tavernini MM, Yachimec C, Mendrick DL, Alfonso PJ, Buergin M, et al. Stanniocalcin: a novel protein regulating calcium and phosphate transport across mammalian intestine. Am J Physiol Gastrointest Liver Physiol. 1998;274:G96–102. doi: 10.1152/ajpgi.1998.274.1.G96. [DOI] [PubMed] [Google Scholar]
- 50.Yahata K, Mori K, Mukoyama M, Sugawara A, Suganami T, Makino H, et al. Regulation of stanniocalcin 1 and 2 expression in the kidney by klotho gene. Biochem Biophys Res Commun. 2003;310:128–134. doi: 10.1016/j.bbrc.2003.08.131. [DOI] [PubMed] [Google Scholar]
- 51.Lin CH, Su CH, Hwang PP. Calcium-sensing receptor mediates Ca2+ homeostasis by modulating expression of PTH and stanniocalcin. Endocrinology. 2014;155:56–67. doi: 10.1210/en.2013-1608. [DOI] [PubMed] [Google Scholar]
- 52.Hebert SC, Mount DB, Gamba G. Molecular physiology of cation-coupled cl- cotransport: the SLC12 family. Pflugers Arch Eur J Physiol. 2004;447:580–593. doi: 10.1007/s00424-003-1066-3. [DOI] [PubMed] [Google Scholar]
- 53.DiPolo R, Beaugé L. Sodium/calcium exchanger: influence of metabolic regulation on ion carrier interactions. Physiol Rev. 2006;86:155–203. doi: 10.1152/physrev.00018.2005. [DOI] [PubMed] [Google Scholar]
- 54.Hebert S, Brown E, Harris H. Role of the Ca2+−sensing receptor in divalent mineral ion homeostasis. J Exp Biol. 1997;302:295–302. doi: 10.1242/jeb.200.2.295. [DOI] [PubMed] [Google Scholar]
- 55.Desfleur E, Wittner M, Simeone S, Pajaud S, Moine G, Rajerison R, et al. Calcium-sensing receptor : regulation of electrolyte transport in the thick ascending limb of Henle ’ s loop. Kidney Blood Press Res. 1998;21:401–412. doi: 10.1159/000025892. [DOI] [PubMed] [Google Scholar]
- 56.Reimer RJ, Edwards RH. Organic anion transport is the primary function of the SLC17/type I phosphate transporter family. Pflugers Arch. Eur. J. Physiol. 2004;447:629–635. doi: 10.1007/s00424-003-1087-y. [DOI] [PubMed] [Google Scholar]
- 57.Suyama T, Okada S, Ishijima T, Iida K, Abe K, Nakai Y. High phosphorus diet-induced changes in NaPi-IIb phosphate transporter expression in the rat kidney: DNA microarray analysis. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0029483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourgier C, Haydont V, Milliat F, François A, Holler V, Lasser P, et al. Inhibition of rho kinase modulates radiation induced fibrogenic phenotype in intestinal smooth muscle cells through alteration of the cytoskeleton and connective tissue growth factor expression. Gut. 2005;54:336–343. doi: 10.1136/gut.2004.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Estes J, Baker JV, Brenchley JM, Khoruts A, Barthold JL, Bantle A, et al. Collagen deposition limits immune reconstitution in the gut. J Infect Dis. 2008;198:456–464. doi: 10.1086/590112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kutuzova GD, DeLuca HF. Gene expression profiles in rat intestine identify pathways for 1,25-dihydroxyvitamin D 3 stimulated calcium absorption and clarify its immunomodulatory properties. Arch Biochem Biophys. 2004;432:152–166. doi: 10.1016/j.abb.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ricklin D, Hajishengallis G, Yang K, Complement LJD. A key system for immune surveillance and homeostasis. Nat. Immunol. Nat Publ Group. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rauw WM. Immune response from a resource allocation perspective. Front Genet. 2012;3:1–14. doi: 10.3389/fgene.2012.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abdullah HI, Pedraza PL, McGiff JC, Ferreri NR. CaR activation increases TNF production by mTAL cells via a Gi-dependent mechanism. Am J Physiol Renal Physiol. 2008;294:F345–F354. doi: 10.1152/ajprenal.00509.2006. [DOI] [PubMed] [Google Scholar]
- 64.Macián F. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 65.Macián F, García-Cózar F, Im SH, Horton HF, Byrne MC, Rao A. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/S0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 66.Feske S, Okamura H, Hogan PG, Rao A. Ca2+/calcineurin signalling in cells of the immune system. Biochem Biophys Res Commun. 2003;311:1117–1132. doi: 10.1016/j.bbrc.2003.09.174. [DOI] [PubMed] [Google Scholar]
- 67.Alroy I, Towers TL, Freedman LP. Transcriptional repression of the interleukin-2 gene by vitamin D3: direct inhibition of NFATp/AP-1 complex formation by a nuclear hormone receptor. Mol Cell Biol. 1995;15:5789–5799. doi: 10.1128/MCB.15.10.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takeuchi A, Reddy GS, Kobayashi T, Okano T, Park J, Sharma S. Nuclear factor of activated T cells (NFAT) as a molecular target for 1alpha,25-dihydroxyvitamin D3-mediated effects. J Immunol. 1998;160:209–218. [PubMed] [Google Scholar]
- 69.von Essen MR, Kongsbak M, Schjerling P, Olgaard K, Ødum N, Geisler C. Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat Immunol. 2010;11:344–349. doi: 10.1038/ni.1851. [DOI] [PubMed] [Google Scholar]
- 70.Kongsbak M, Levring TB, Geisler C, von Essen MR. The vitamin D receptor and T cell function. Front Immunol. 2013;4:1–10. doi: 10.3389/fimmu.2013.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composition of the experimental diets (XLSX 11 kb)
Primer used for verification of microarray by qRT-PCR (XLSX 11 kb)
Probe-sets showing significantly altered mRNA abundances (per diet and tissue). The Venn diagram depicting the number of probe-sets differentially expressed in a) colon, b) jejunum, and c) kidney. Number of up-regulated probe-sets is shown in red. Number of down-regulated probe-sets is shown in green. L – Low P supply; M – Medium P supply; H – High P supply (XLSX 254 kb)
The hierarchical clustering of filtered probe-sets is represented by log2 transformed intensity values obtained (i) from the variance component diet*tissue (least square means) and (ii) from data obtained from individual animals. L – Low P supply; M – Medium P supply; H – High P supply (PDF 2172 kb)
Molecular and cellular functions affected by P supply (XLSX 9 kb)
Scatterplots of jejunal and renal transcripts (PDF 20 kb)
Data Availability Statement
Raw data are deposited in a MIAME-compliant database, the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession number: GSE94448).


