Abstract
Synthesis and turnover of phosphoinositides are tightly regulated processes mediated by a set of recently identified kinases and phosphatases. We analyzed the primary role of the phosphoinositide phosphatase Sac1p in Saccharomyces cerevisiae with the use of a temperature-sensitive allele of this gene. Our analysis demonstrates that inactivation of Sac1p leads to a specific increase in the cellular levels of phosphatidylinositol 4-phosphate (PtdIns(4)P), accompanied by changes in vacuole morphology and an accumulation of lipid droplets. We have found that the majority of Sac1p localizes to the endoplasmic reticulum, and this localization is crucial for the efficient turnover of PtdIns(4)P. By generating double mutant strains harboring the sac1ts allele and one of two temperature-sensitive PtdIns 4-kinase genes, stt4ts or pik1ts, we have demonstrated that the bulk of PtdIns(4)P that accumulates in sac1 mutant cells is generated by the Stt4 PtdIns 4-kinase, and not Pik1p. Consistent with these findings, inactivation of Sac1p partially rescued defects associated with stt4ts but not pik1ts mutant cells. To analyze potential overlapping functions between Sac1p and other homologous phosphoinositide phosphatases, sac1ts mutant cells lacking various other synaptojanin-like phosphatases were generated. These double and triple mutants exacerbated the accumulation of intracellular phosphoinositides and caused defects in Golgi function. Together, our results demonstrate that Sac1p primarily turns over Stt4p-generated PtdIns(4)P and that the membrane localization of Sac1p is important for its function in vivo. Regulation of this PtdIns(4)P pool appears to be crucial for the maintenance of vacuole morphology, regulation of lipid storage, Golgi function, and actin cytoskeleton organization.
INTRODUCTION
Phosphoinositides (PIs) are key regulators of a wide variety of cellular processes including signal transduction, cell proliferation, vesicular trafficking, apoptosis, cytoskeletal organization, and transcription (reviewed by Fruman et al., 1998; Martin, 1998; Divecha et al., 2000). The reversible phosphorylation of these lipid head groups make them ideally suited to function as temporal and spatial regulators of these processes. The synthesis and turnover of PIs are regulated by a set of kinases, phosphatases, and lipases localized to discrete membrane sites. The importance of these tightly regulated biosynthetic pathways have been highlighted by multiple studies showing that distinct PI isomers, such as phosphatidylinositol 3-phosphate (PtdIns(3)P), phosphatidylinositol (4,5)-diphosphate (PtdIns(4,5)P2), and phosphatidylinositol (3,4,5)-triphosphate, interact with and/or regulate specific downstream targets/effectors acting in a variety of cellular pathways (Fruman et al., 1998; Martin, 1998). In contrast to other PI isomers, PtdIns(4)P has been, until recently, considered mainly as a precursor for PtdIns(4,5)P2 synthesis, and its potential role as a direct mediator of cellular signaling has not been well characterized. However, the complex regulation of PtdIns(4)P biosynthesis and turnover in yeast (Balla, 1998; Guo et al., 1999) indicates that this isomer also functions as a key regulator of distinct cellular processes.
In Saccharomyces cerevisiae, synthesis of PtdIns(4)P is mediated by two PtdIns 4-kinases, Pik1p and Stt4p. Both kinases are required for cell viability, regulating distinct and essential cellular processes (Audhya et al., 2000). Pik1p is a 125-kDa soluble protein residing at the nucleus and trans-Golgi compartments (Flanagan and Thorner, 1992; Garcia-Bustos et al., 1994; Walch-Solimena and Novick, 1999). It has been demonstrated to play a direct role in late events of the secretory pathway, secretory vesicle budding at the late Golgi, integrity of Golgi structure, and cytokinesis (Garcia-Bustos et al., 1994; Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). Stt4p is a 216-kDa membrane-bound protein that is required for actin cytoskeleton organization, cell wall integrity, and maintenance of vacuole morphology (Yoshida et al., 1995; Audhya et al., 2000). Stt4p has also been implicated in the transport of an aminophospholipid from the endoplasmic reticulum (ER) to the Golgi/vacuole (Trotter et al., 1998).
Regulation of the distinct PtdIns(4)P pools generated by Stt4p and Pik1p is unclear, but recently, PI phosphatases have been proposed to inhibit or terminate PtdIns 4-kinase signaling (reviewed by Majerus et al., 1999). In particular, a novel class of PI phosphatases containing a Sac1-like phosphatase domain have been identified in yeast, mammal, and plant cells (Srinivasan et al., 1997; Guo et al., 1999). Sac1p is the prototype of this phosphatase family, and proteins including Fig4p and two members of the synaptojanin-like (Sjl) protein family, Sjl2p and Sjl3p (also called Inp52p and Inp53p), all harbor a putative functional Sac1-like phosphatase domain (Guo et al., 1999). The Sac1-like domains of Sac1p, Sjl2p, and Sjl3p have been shown to predominantly dephosphorylate PtdIns monophosphates such as PtdIns(3)P and PtdIns(4)P in vitro. A weak phosphatase activity toward phosphatidylinositol (3,5)-diphosphate (PtdIns(3,5)P2) has also been shown, preferentially against the phosphate residue at the D-3 position of the inositol ring (Guo et al., 1999; Hughes et al., 2000). Sjl proteins also can dephosphorylate PtdIns(4,5)P2 because of the presence of a second phosphatase domain that recognizes the phosphate group at the D-5 position (Guo et al., 1999). Surprisingly, single or double sjl mutants appear to alter only PtdIns(3,5)P2 and PtdIns(4,5)P2 levels in vivo (Stolz et al., 1998a, 1998b; Guo et al., 1999). In contrast, deletion of SAC1 leads to alterations in intracellular levels of several PI isomers, but the greatest effect is on PtdIns(4)P levels (8- to 10-fold; Guo et al., 1999; Rivas et al., 1999; Stock et al., 1999; Hughes et al., 2000). The alteration in PI metabolism in sac1Δ cells is accompanied by several phenotypes, including inositol auxotrophy, cold sensitivity, and hypersensitivity to different drugs (Novick et al., 1989; Whitters et al., 1993; Hughes et al., 1999). Organization of the actin cytoskeleton and chitin deposition are also affected at low temperatures (Novick et al., 1989). Genetic studies have shown that mutations in SAC1 can selectively suppress defects associated with certain actin alleles and bypass the requirement for the Sec14 PtdIns/PtdCho transfer protein (Novick et al., 1989; Whitters et al., 1993; Cockcroft, 1998). Furthermore, genetic interactions have been described between mutations in the secretory pathway (SEC genes) and sac1 mutations (Cleves et al., 1989). Sac1p has been suggested to be required for proper transport of ATP and efficient translocation of preproteins into the endoplasmic reticulum (ER; Kochendorfer et al., 1999). The stabilization of PtdIns(4)P in Sac1p-deficient cells is likely responsible for the above cellular defects because overexpression of Sjl2p or Sjl3p, which also possess PtdIns(4)P phosphatase activity, are able to rescue some of the phenotypes associated with sac1 null mutations (Hughes et al., 2000). However, all of the previous work has been based on an analysis of sac1 null mutants, leaving open the possibility that abnormal phenotypes observed in these mutants are the result of secondary defects.
Therefore, to analyze the primary function of Sac1p in PtdIns(4)P turnover, we generated alleles of sac1 that are temperature sensitive for function (sac1ts) and examined the immediate consequence of Sac1p inactivation. We found that 1Sac1p primarily turns over Stt4p-generated PtdIns(4)P and that membrane localization of Sac1p is crucial for its efficient function. High levels of PtdIns(4)P resulting from Sac1p inactivation are accompanied by changes in vacuole morphology and an accumulation of lipid droplets. Defects resulting from inactivation of Stt4p, but not Pik1p, could be partially rescued when Sac1p was also inactivated. Finally, analysis of sac1ts mutants lacking one or more of the Sjl proteins showed that Sjl3p plays an essential compensatory role in Sac1p-deficient cells.
MATERIALS AND METHODS
Strains and Media
Sources of growth media for yeast and bacterial strains have been described elsewhere (Gaynor et al., 1994). Transformation into yeast was performed by a standard lithium acetate method (Ito et al., 1983) and Escherichia coli transformations were done as previously described (Hanahan, 1983). S. cerevisiae strains used in this study are listed in Table 1.
Table 1.
Strains used in this study
| Strains | Genotypes | Reference or source |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | (Robinson et al., 1988) |
| SEY6210.1 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | (Robinson et al., 1988) |
| MFY62 | SEY6210.1; sac1Δ::TRP1 | This study |
| AAY102 | SEY6210; stt4Δ::HIS3 carrying pRS415stt4-4 (LEU2 CEN6 stt4-4) | (Audhya et al., 2000) |
| MFY55 | AAY 102; sac1Δ::TRP1 | This study |
| AAY104.5 | SEY6210; pik1Δ::HIS3 carrying pRS415pik1-83 (LEU2 CEN6 pik1-83) | (Audhya et al., 2000) |
| AAY131 | AAY104.5; sac1Δ::TRP1 | This study |
| JGY130 | SEY6210; sjl2Δ::HIS3 | This study |
| JGY131 | SEY6210; sjl3Δ::TRP1 | This study |
| JGY132 | SEY6210; sjl2Δ::HIS3 sjl3Δ::TRP1 | This study |
| MFY80 | SEY6210.1; SAC1-GFP::HIS3MX6 | This study |
| MFY107 | SEY6210.1; SAC1-MYC13::TRP1 | This study |
| MFY109 | SEY6210.1; sac1Δ522-623MYC13::TRP1 | This study |
| MFY111 | SEY6210.1; sac1Δ522-623-GFP::TRP1 | This study |
| AAY141 | SEY6210; sjl2Δ::HIS3 sac1Δ::TRP1 | This study |
| AAY142 | SEY6210; sjl3Δ::TRP1 sac1Δ::TRP1 carrying pRS416sac1-23 (URA3 CEN6 sac1-23) | This study |
| AAY143 | SEY6210; sjl2Δ::HIS3 sjl3Δ::TRP1 sac1Δ::TRP1 carrying pRS416sac1-23 (URA3 CEN6 sac1-23) | This study |
| EGY1181-10 | SEY6210; sec18-1 | (Sato et al., 1998) |
Plasmids and DNA Manipulations
Enzymes utilized for recombinant DNA techniques were purchased from commercial sources and used as recommended by the suppliers. Standard recombinant DNA techniques and standard yeast genetic methods were performed as previously described (Sherman et al., 1979; Sambrook et al., 1989).
Disruption of SAC1.
SAC1, including 434 bp upstream of the start codon and 134 bp downstream of the stop codon, was amplified by polymerase chain reaction (PCR) from SEY6210 genomic DNA and ligated into a SalI-SpeI–digested pBIISK(−) to generate pBIISK(−)-SAC1. The plasmid used to generate a chromosomal deletion of SAC1 was then constructed by inserting the TRP1 gene into a ClaI-BamHI–digested pBIISK(−)-SAC1 to generate pBIISK(−)-SAC1::TRP1. A SalI-SpeI fragment of pBIISK(−)-SAC1::TRP1 was then transformed into SEY6210.1 to delete 1540 bp of the SAC1-coding sequence (strain MFY62). The disruption was confirmed by PCR with the use of two different sets of primers.
SAC1 and sac1Δ522-623 Cloning.
SAC1 was subcloned from a pYEP24–based plasmid isolated from the yeast genomic library described by Carlson and Botstein (1982). Briefly, MFY62 was transformed with the pYEP24-based genomic library and plasmids containing the SAC1 open reading frame were isolated based on their ability to complement the G418 sensitivity of MFY62. To generate pRS416-SAC1, a BglII-SpeI SAC1 DNA fragment derived from a pYEP24-SAC1 was subcloned into pRS416 digested with BamHI-SpeI (Sikorski and Hieter, 1989). To generate sac1Δ522-623, PCR primers were generated to amplify the SAC1 gene lacking sequence for the last 102 amino acids (with a NcoI restriction site upstream of the second codon and an AseI restriction site 1569 bp downstream of the start codon), which was then digested with NcoI and AseI and ligated into pBluescriptSK+ (Stratagene, La Jolla, CA) containing the CPS1 promotor (pGO106). The resulting ligation product was then subcloned into either a CEN-based pRS415 vector or a 2μ-based pRS425 vector.
Generation of sac1ts (Temperature-sensitive for Function) Conditional Alleles.
A XbaI-PvuII SAC1 fragment (bp −6–1575) was amplified by error-prone PCR (Muhlrad et al., 1992) and cotransformed with HpaI-gapped pRS416-SAC1 into MFY62. Uracil auxotrophs were selected and initially screened for growth on G418 (25 μg/ml) containing YPD medium at 26 and 38°C. Mutants that grew at 26°C but not at 38°C were then tested also for growth on minimal media lacking inositol at both temperatures. From >10,000 transformants, three putative pRS416-sac1ts plasmids were isolated, retransformed into MFY62 and tested for growth phenotypes and PI phosphatase activities in vivo at the restrictive temperature. For all studies, the sac1-23 allele was analyzed.
Generation of Other sac1 Mutants.
To generate a stt4ts/sac1Δ double mutant (MFY55), AAY102 (stt4-4) was transformed with SpeI/SalI-digested pBIISK(−)-SAC1::TRP1. Tryptophan auxotrophs were then tested by PCR for deletion of SAC1 with the use of two different sets of primers. MFY55 was then transformed with pRS416-sac1-23ts. For all studies, the stt4-4 allele was analyzed.
A pik1ts/sac1Δ double mutant (AAY131) was made by mating AAY104.5 (pik1-83) with MFY62, dissecting tetrads and isolating spores by growth on selective media. AAY131 was then transformed with pRS416-sac1-23ts. For all studies, the pik1-83 allele was analyzed.
To generate sjl2Δ/sac1ts (AAY141), sjl3Δ/sac1ts (AAY142), and sjl2Δ/sjl3Δ/sac1ts (AAY143) mutants, MFY62 carrying pRS416-sac1-23ts was mated with SEY6210 sjl2Δ, SEY6210 sjl3Δ, and SEY6210 sjl2Δ/sjl3Δ (J. Gary, unpublished data), respectively. Spores resulting from tetrad dissection were then selected for the appropriate markers, and disruptions were confirmed by PCR with the use of two different sets of primers for each gene.
Generation of Green Fluorescence Protein (GFP)/myc-tagged SAC1 and sac1Δ522-623 Strains.
Tagging of the Sac1p C-terminus with GFP or a 13Myc epitope was performed as described previously (Longtine et al., 1998). In brief, PCR products containing the tags and either a TRP1 or HIS3MX6 marker, flanked by homologous regions to SAC1, were generated as described by Longtine et al. (1998) with the use of the template described therein and transformed into SEY6210. Tryptophan or histidine auxotrophs were isolated and the correct integration of the tags was confirmed by PCR with the use of two different sets of primers. In the case of sac1Δ522-623, the tag was inserted after amino acid 521, truncating Sac1p and fusing it with GFP or 13Myc.
To generate strains expressing Sec7p-GFP, the endogenous chromosomal copy of SEC7 was replaced with an SEC7-GFP fusion gene with the use of the pop-in, pop-out method as described by Seron et al. (1998).
Metabolic Labeling and Immunoprecipitation
Cell labeling and immunoprecipitations were performed as previously described (Gaynor et al., 1994) with minor modifications. In brief, log phase cultures were labeled with Tran 35S-label (DuPont NEN, Boston, MA) for 10 min and chased with cold methionine and cysteine for the indicated times; proteins were precipitated with 9% trichloroacetic acid. All temperature shifts, unless otherwise stated, were limited to 10 min at 38°C. Extracts were immunoprecipitated with antisera against carboxypeptidase Y (CPY), Hsp150p, or invertase, which have been previously characterized (Cowles et al., 1997; Gaynor and Emr, 1997). To assay internal and external fractions for the presence of invertase, cells harboring a plasmid expressing invertase (pCYI-20) were converted to spheroplasts after pulse-chase by adding a 2× buffer containing 50 mM Tris (pH 7.5), 2 M sorbitol, 40 mM NaF, 40 mM NaN3, and 10 mM dithiothreitol and incubating on ice for 10 min. Zymolyase T100 (15 μg/OD600; Seikagaku Kogyo, Tokyo, Japan) was then added to the cell suspension and incubated for 30 min at 30°C. Cells were then subjected to centrifugation at 6000 rpm for 5 min, and the supernatant was removed. Both fractions were precipitated in the presence of 9% trichloroacetic acid.
In Vivo PI Analysis
Analysis of PI levels was performed as previously described (Audhya et al., 2000). Briefly, cells from a log phase culture were shifted to the appropriate temperature for 10 min and then labeled with myo-[2-3H]inositol (Nycomed Amersham, Princeton, NJ). After 10 min excess unlabeled myo-inositol was added and cells were incubated for 30 min at the desired temperature. Cells were lysed and extracts were processed as previously described (Stack et al., 1995). Analysis of 3H-labeled glycerophosphoinositols was performed with the use of a Gold HPLC (Beckman System, Fullerton, CA) coupled to an online radiomatic detector (Packard, Meriden, CT). All phospholipid data are expressed as percentages of the total number of counts loaded onto the high-performance liquid chromatography (HPLC) column for normalization.
Fluorescence and Electron Microscopy
FM4-64 Labeling.
Labeling of vacuole membranes with the vital dye FM4-64 (Molecular Probes, Eugene, OR) was performed as previously described (Vida and Emr, 1995). Briefly, cells from a log phase culture were labeled with FM4-64 for 15 min, followed by a chase without dye for 45 min. For temperature shift experiments, cells were first pulse-chased with FM4-64 and then shifted to 38°C for the indicated time.
All the images were acquired with the use of a DeltaVision Deconvolving microscope (Applied Precision, Seattle, WA). Images were then processed with the use of Photoshop 4.0 (Adobe Systems, Mountain View, CA).
Immunofluorescence.
Cells grown to midlog phase were spheroplasted and fixed with 4% formaldehyde. Fixed cells were then washed in 50 mM Tris-HCl, pH 7.5, 1 M sorbitol, permeabilized with 0.02% Triton X-100 for 10 min, washed again, and incubated on polylysine-coated slides for 20 min. Adherent cells were incubated for 30 min with blocking buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20 containing 1% dry milk and 0.5 mg/ml bovine serum albumin) to prevent nonspecific binding of antibodies and then for 2 h with the indicated primary antibodies in WT buffer at room temperature (mouse monoclonal α-Myc 10 ng/ml OM-11-908 from Genosys [The Woodlands, TX], rabbit polyclonal α-Kar2 [1/10,000] kindly provided by Dr. M. Latterich [Salk Institute, La Jolla, CA], and rabbit polyclonal α-Kex2 (1/1000) kindly provided by Dr. R. Fuller [University of Michigan, Ann Arbor, MI]). Cells were washed three times with WT buffer and incubated again with the appropriate fluorochrome-conjugated secondary antibodies for 1 h at room temperature (Alexa Fluor568 goat anti-mouse A-11004 and Alexa Fluor488 goat anti-rabbit A-11008 from Molecular Probes). Cells were finally washed five times with phosphate-buffered saline and mounted in phosphate-buffered saline/glycerol plus 4′,6-diamidino-2-phenylindole (DAPI) for observation. For studies with the use of Sec7p-GFP as a medial Golgi marker, no antibody was required.
All the images were acquired with the use of a DeltaVision Deconvolving microscope (Applied Precision). Images were then processed with the use of Adobe Photoshop 4.0.
Actin Labeling.
To analyze actin cytoskeleton organization, cells were grown to early log phase, shifted to the appropriate temperature for 2.5 h, fixed in 3.7% formaldehyde, and stained with rhodamine-phalloidin (Molecular Probes) as previously described (Benedetti et al., 1994). Polarization of actin in small budded cells was scored on >200 cells for each condition.
Electron Microscopy.
Ultrastructural analysis was performed as previously described (Rieder et al., 1996). In brief, cells in a log phase growth were fixed with 3% glutaraldehyde for 1 h and spheroplasted before being processed for electron microscopy; >100 cells considered well preserved were examined for each analysis.
Nile Red Staining
Staining of lipid droplets with Nile red was performed on fixed cells as described by Greenspan et al. (1985) and images were acquired with the use of a DeltaVision Deconvolving microscope (Applied Precision). Images were then processed with the use of Adobe Photoshop 4.
RESULTS
sac1ts Mutant Cells Exhibit a Rapid Defect in the Turnover of Newly Synthesized PtdIns(4)P
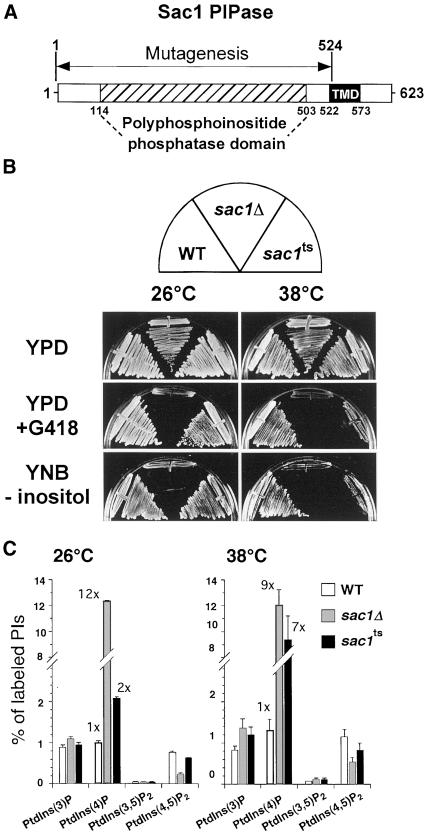
Sac1p has been identified as a PI polyphosphatase (Guo et al., 1999). To examine the primary function of Sac1p in vivo, sac1 temperature-sensitive alleles (sac1ts) were generated by PCR-mediated random mutagenesis of the SAC1 phosphatase domain (coding region corresponding to aa 1–525 as shown in Figure 1A) and introduced into haploid yeast lacking the chromosomal copy of SAC1. Selection of sac1ts mutants was facilitated by the fact that deletion of SAC1 in the SEY6210 background confers hypersensitivity to low concentrations of the aminoglycoside antibiotic geneticin (G418; 10 μg/ml) in rich media. From >10,000 transformants, three alleles were isolated that confer robust growth at 26°C, but not at the restrictive temperature of 38°C, on G418-containing rich media. Additionally, like sac1Δ cells, the sac1ts cells were found to be inositol auxotrophs at high temperature (Figure 1B). Among these three temperature-sensitive mutants, sac1-23ts exhibited the strongest defects at the restrictive temperature and was chosen for further analysis. Sequence analysis of the sac1-23 allele revealed six amino acid changes (Y111C, V190A, K223E, D336A, F366S, E416G). Interestingly, each of these mutations lie within or closely adjacent to the conserved Sac phosphatase domain of Sac1p (amino acids 114–503; reviewed by Hughes et al., 2000).
Figure 1.
sac1ts mutant cells display growth defects on selective media and generate increased levels of PtdIns(4)P at restrictive temperature. (A) The SacI PI phosphatase is a 623-amino acid protein with at least two distinct domains. The polyphosphoinositide phosphatase (PPIPase) domain (aa 114–503), also called the Sac phosphatase domain, contains seven highly conserved motifs, including the putative RXNCLDCLDRTN catalytic motif. The region between amino acids 522 and 573 encompasses two potential helical membrane-spanning domains predicted by hydropathy plot analysis according to Kyte and Doolittle. The region of Sac1p that was randomly mutagenized is indicated. (B) WT, sac1Δ, and sac1ts mutant cells were streaked on YPD-rich media with or without 10 μg/ml G418 or minimal synthetic media lacking inositol at both the permissive temperature of 26°C and the restrictive temperature of 38°C for 3 d. (C) Cells were preincubated at either 26 or 38°C for 10 min, labeled with myo-[2-3H]inositol for 10 min, and then chased with excess unlabeled myo-inositol for 30 min at the indicated temperature. Lipids were then deacylated from cellular membranes, and glycerophosphoinositols were extracted and analyzed by HPLC. Quantitative comparisons of glycerophosphoinositols generated by WT, sac1Δ, and sac1ts cells at 26 or 38°C are shown. These data represent the means ± SEM of at least three independent experiments.
Labeling of cells with myo-[2-3H]inositol demonstrated that sac1Δ mutant cells show pleiotropic alterations of intracellular PI levels as compared with WT cells (Rivas et al., 1999; Stock et al., 1999; Hughes et al., 2000). We assessed steady-state intracellular PI levels in SEY6210 lacking the SAC1 gene. Cells were labeled for 12 h with myo-[2-3H]inositol and then processed for HPLC analysis of PIs as described in MATERIALS AND METHODS. We found that sac1Δ cells accumulated PtdIns(3)P (1.7-fold increase), PtdIns(4)P (19.8-fold increase), and PtdIns(3,5)P2 (2.5-fold increase) but had decreased levels of PtdIns(4,5)P2 (4-fold decrease) as compared with WT cells. Because phospholipid metabolism is highly dependent on the cell growth phase (Paltauf et al., 1992), we also performed pulse-labeling experiments of PIs in the sac1Δ mutant. PIs were labeled with myo-[2-3H]inositol for 10 min and chased for 30 min, and the cellular levels of PIs were analyzed by HPLC. Under these conditions, where only PIs synthesized during a short pulse/chase were analyzed, the levels of PtdIns(3)P and PtdIns(3,5)P2 in sac1Δ cells were only slightly affected at 26°C, whereas PtdIns(4)P was increased by >12-fold. Similar to the steady-state labeling, PtdIns(4,5)P2 was found to be decreased by >75% (Figure 1C). When sac1ts cells were labeled at 26°C with the use of the same protocol, intracellular PI levels were only mildly affected as compared with those observed in WT cells (2-fold increase in PtdIns(4)P). However, when the labeling was performed at the restrictive temperature of 38°C, sac1ts mutant cells exhibited a dramatic increase in PtdIns(4)P levels (>7-fold), whereas the levels of PtdIns(3)P, PtdIns(3,5)P2, and PtdIns(4,5)P2 were only slightly altered as compared with WT cells (Figure 1C).
Together these results indicate that Sac1p primarily functions in the turnover of a rapidly synthesized pool of PtdIns(4)P in vivo. Changes in the intracellular levels of other PIs previously observed in sac1Δ mutant cells are likely an indirect consequence of the long-term effect of Sac1p inactivation on PI metabolism.
Membrane Localization of Sac1p Is Required for Efficient PtdIns(4)P Turnover
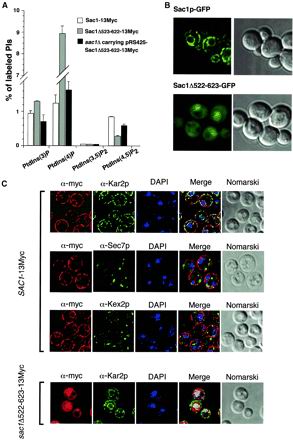
Sac1p, in contrast to the other known yeast PI phosphatases, is an integral membrane protein (Whitters et al., 1993). Additionally, in vitro, Sac1p exhibits a higher phosphatase activity against PtdIns(3)P than PtdIns(4)P, its major in vivo substrate (Hughes et al., 2000). These features suggest that the intracellular localization of Sac1p is crucial for its biological specificity. To clarify this point, the chromosomal copy of SAC1 was tagged with GFP or a 13Myc epitope at the 3′-end (Sac1p-GFP and Sac1p-13Myc, respectively) to further examine intracellular localization of Sac1p. In addition, to assess the role of the C-terminal transmembrane domain of Sac1p, GFP- or 13Myc-tagged truncated forms of the protein were also generated by integration into the chromosome as described in MATERIALS AND METHODS (Sac1Δ522-623–GFP and Sac1Δ522-623–13Myc, respectively).
Tagging of full-length Sac1p with either GFP or a 13Myc epitope did not alter the activity of Sac1p in vivo. The fusion proteins were normally expressed, as assayed by Western blot analysis, and strains harboring this fusion as their sole copy of Sac1p did not display any growth defects (Foti, Audhya, and Emr, unpublished results). Furthermore, intracellular levels of PIs in cells expressing these fusion proteins were similar to WT cells (Figure 2A). In contrast, cells expressing the Sac1Δ522-623 proteins exhibited altered intracellular PI levels and showed growth defects similar to sac1Δ cells, although the truncated proteins were well expressed (Figure 2A). Interestingly, when Sac1Δ522-623 was overexpressed in sac1Δ cells, normal growth and near WT levels of PtdIns(4)P and PtdIns(4,5)P2 were restored, indicating that this truncated version of Sac1p was active (Figure 2A). Moreover, these data suggest that the Sac1p transmembrane domain is required for the efficient turnover of PtdIns(4)P.
Figure 2.
Localization of Sac1p to the ER is crucial for the efficient turnover of PtdIns(4)P. (A) Cells were labeled with myo-[2-3H]inositol for 10 min at 26°C and then chased with excess unlabeled myo-inositol for 30 min. Lipids were then deacylated from cellular membranes, and the resulting soluble glycerophosphoinositols were extracted and analyzed by HPLC. Quantitative comparisons of glycerophosphoinositols generated by cells expressing the Sac1-13Myc or the Sac1Δ522-623–13Myc–tagged protein are shown. Additionally, glycerophosphoinositols from sac1Δ cells overexpressing the Sac1Δ522-623 truncation protein are shown. These data represent the means ± SEM of at least three independent experiments. (B) Full-length GFP-tagged Sac1p (Sac1p-GFP) or a truncated version of Sac1p lacking the last 102 amino acids, including the potential transmembrane domain (Sac1Δ522-623-GFP), were generated and observed by fluorescence microscopy in living cells. On the right, identical fields were observed with Nomarski optics. Pictures shown are representative of >95% of the cells observed. (C) Indirect immunofluorescence microscopy of Sac1-13Myc and Sac1Δ522-623–13Myc on paraformaldehyde-fixed cells. Cells expressing the Sac1-13Myc or Sac1Δ522-623–13Myc–tagged proteins were fixed, permeabilized, and incubated with α-Myc, α-Kar2p (ER marker), or α-Kex2p (Golgi/endosomes marker) antibodies as described in MATERIALS AND METHODS. DAPI was used to stain DNA before mounting. For cells expressing Sec7p-GFP, no antibodies were required. On the right, identical fields were observed with Nomarski optics. Pictures shown are representative of >95% of the cells observed.
Intracellular localization of GFP-tagged constructs (Sac1p-GFP and Sac1Δ522-623-GFP) were examined in living cells by fluorescence microscopy. Consistent with other studies in mammalian cells (Nemoto et al., 2000), the Sac1p-GFP fusion protein exhibited a fluorescence pattern typical of ER resident proteins, concentrating in a perinuclear ring and in a discontinuous juxtamembrane staining around the cell periphery (Figure 2B). Strikingly, the Sac1Δ522-623–GFP fusion protein was mislocalized and redistributed throughout the cytosol and the nucleus, with no evident membrane association (Figure 2B).
Identical results were obtained with the 13Myc-tagged Sac1p fusion proteins (Sac1p-13Myc and Sac1Δ522-623–13Myc) by immunofluorescence on fixed cells. Immunofluorescence detection of the full-length Sac1-13Myc protein produced an ER-staining pattern similar to that of Kar2p, an ER resident protein (Figure 2C). Localization of Sec7p, a medial Golgi marker, yielded a completely distinct pattern of staining as compared with Sac1-13Myc, and we failed to observed any colocalization of these proteins (Figure 2C). Furthermore, Sac1-13Myc failed to colocalize with the late Golgi/endosome marker, Kex2p, again indicating that little, if any, Sac1p localizes to the Golgi. Similar to the Sac1Δ522-623–GFP localization, the Sac1Δ522-623–13Myc fusion protein showed a completely different and distinct pattern of distribution as compared with the full-length Sac1p-13Myc. Coimmunofluorescence studies with the use of Kar2p showed that Sac1Δ522-623 was no longer restricted to the ER, confirming that the C-terminal 102 amino acids of Sac1p are required for normal localization of Sac1p (Figure 2C).
Together these data demonstrate that the majority of Sac1p is localized to the ER and this localization is required for efficient turnover of PtdIns(4)P.
The Stt4 PtdIns 4-Kinase Generates the Bulk of PtdIns(4)P That Accumulates in sac1ts Mutant Cells
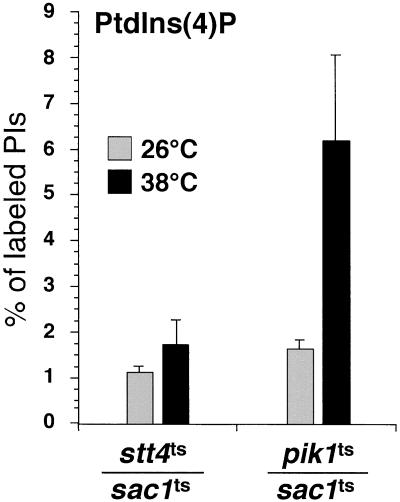
Previous studies have suggested that the Sac1p phosphatase regulates the pool of PtdIns(4)P generated by the PtdIns 4-kinase Pik1p (reviewed by Hughes et al., 2000; Huijbregts et al., 2000). To define the PI kinase that synthesizes the pool of PtdIns(4)P that accumulates in sac1 mutant cells, double mutants harboring the temperature-sensitive sac1-23 allele together with a temperature-sensitive allele of the two known PtdIns 4-kinases in yeast, pik1-83 and stt4-4, were generated. We have previously shown that each kinase accounts for approximately half of the total PtdIns(4)P generated in cells (Audhya et al., 2000). We analyzed the intracellular levels of PtdIns(4)P in these double mutants at both the permissive and restrictive temperatures. At the permissive temperature, intracellular PtdIns(4)P levels were similar to sac1ts cells, in both double mutants, stt4ts/sac1ts and pik1ts/sac1ts (compare Figure 1C with Figure 3). Surprisingly, simultaneous inactivation of Sac1p and Stt4p results in few, if any, changes in PtdIns(4)P levels after the shift to restrictive temperature (1.1 ± 0.3% at 26°C vs. 1.7 ± 0.5% at 38°C), whereas inactivation of Sac1p and Pik1p results in a dramatic increase in PtdIns(4)P that is typically observed in sac1 mutant cells (1.6 ± 0.2% at 26°C vs. 6.2 ± 1.8% at 38°C). These results indicate that Stt4p, and not Pik1p, generates the bulk of PtdIns(4)P, which accumulates upon Sac1p inactivation.
Figure 3.
Stt4p, but not Pik1p, generates PtdIns(4)P which accumulates in sac1 mutants. Cells were grown to midlog phase, shifted to the indicated temperature for 10 min, labeled with myo-[2-3H]inositol for 10 min, and chased in the presence of excess unlabeled myo-inositol for 30 min. Cellular lipids were recovered, deacylated, and separated by HPLC. Levels of glycerophosphoinositols derived from PtdIns(4)P in stt4ts/sac1ts and pik1ts/sac1ts double mutant cells at 26 and 38°C are shown. These data represent the means ± SEM of at least three independent experiments.
sac1ts Suppresses Phenotypes Associated with stt4ts But Not pik1ts Cells
To further investigate the role of Sac1p in PtdIns(4)P turnover, we assessed whether the phenotypes observed in PtdIns 4-kinase stt4ts and pik1ts mutant cells could be rescued by elimination of Sac1p activity. Although we could not detect any defects in actin cytoskeleton organization in pik1ts cells (Audhya et al., 2000), stt4ts cells fail to appropriately organize their actin cytoskeleton at restrictive temperature. Specifically, stt4ts cells display random cortical actin patches throughout both mother and daughter cells, instead of restricting these patches to the bud and septum as observed in WT cells (Audhya et al., 2000). To determine whether stabilization of the Stt4p-dependent PtdIns(4)P pool by Sac1p inactivation could prevent the defects in actin cytoskeleton organization displayed by stt4ts cells, stt4ts/sac1ts double mutant cells were incubated at the permissive or restrictive temperature, fixed, and then labeled with rhodamine-conjugated phalloidin. The distribution of actin patches and cables were then analyzed and quantified as described in MATERIALS AND METHODS. Consistent with a role for Sac1p in the turnover of PtdIns(4)P generated by Stt4p, we observed suppression of the defect in actin cytoskeleton organization in double stt4ts/sac1ts mutant cells as compared with stt4ts single mutant cells (Table 2), thus supporting a functional connection in vivo between Stt4p and Sac1p.
Table 2.
Actin cytoskeleton organization in stt4ts and sac1ts mutants
| Yeast strain | Cells with normal actin morphology
(%)
|
|
|---|---|---|
| 26°C | 38°C | |
| Wild type | >95 | >95 |
| stt4ts | >90 | <10 |
| sac1ts | >90 | >90 |
| stt4ts/sac1ts | >90 | >60 |
Percentages are based on >200 cells observed for each strain at each condition. Wild type (SEY6210), stt4ts (AAY102), sac1Δ (MFY62) carrying URA3 CEN6 sac1-23, and double mutant stt4ts/sac1Δ (MFY55) carrying URA3 CEN6 sac1-23 cells at 26°C and 38°C are described. Comparison of stt4ts and stt4ts/sac1ts cells is highlighted at 38°C.
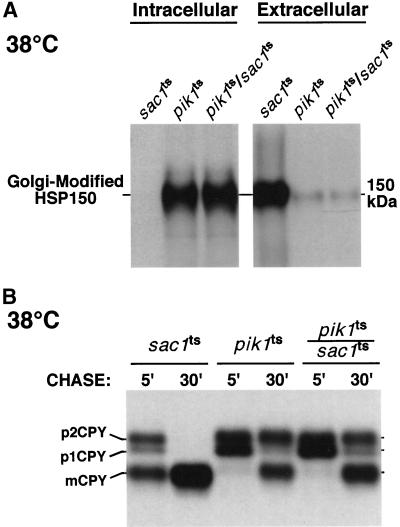
We also assessed whether the rapid inactivation of Sac1p, which prevents turnover of a pool of PtdIns(4)P generated by Stt4p, could rescue defects associated with Pik1p inactivation. We have previously shown that secretion of Hsp150p, a high-molecular-weight glycoprotein is impaired in pik1ts mutant cells at the restrictive temperature but not at the permissive temperature (Audhya et al., 2000). As shown in Figure 4A, inactivation of Sac1p neither affected protein secretion by itself nor rescued the Pik1p-dependent defect in Hsp150p secretion at the restrictive temperature. Furthermore, we previously described that pik1ts cells exhibit a kinetic delay of CPY maturation at the nonpermissive temperature, whereas at 26°C, CPY is processed normally (Audhya et al., 2000). CPY is converted from an ER-modified p1 precursor form to a Golgi-modified p2 precursor form and then transported to the vacuole where it is cleaved to generate the mature, active form of CPY (mCPY). In pik1ts cells, CPY transport to the vacuole is delayed, resulting in a significant accumulation of the p2CPY precursor form. As shown in Figure 4B, sac1ts cells displayed normal processing of CPY at the restrictive temperature, and inactivation of Sac1p in pik1ts cells could not relieve the CPY maturation defect exhibited at the nonpermissive temperature. Interestingly, in contrast to sac1ts cells, we observed a kinetic delay of CPY maturation in sac1Δ cells, as has also been shown by others (Mayinger et al., 1995; Foti, Audhya, and Emr, unpublished results), suggesting again that deletion of SAC1 results in phenotypes indirectly related to the loss of Sac1p function.
Figure 4.
Inactivation of Sac1p does not rescue transport defects associated with pik1ts mutant cells. (A) Hsp150p secretion in WT, sac1ts, pik1ts, and pik1ts/sac1ts cells. Cells were preincubated at 38°C for 10 min, metabolically labeled with an 35S-protein–labeling mixture during a 10-min pulse, and then chased in the presence of excess unlabeled methionine and cysteine for 30 min. Labeled cells were subjected to centrifugation and Hsp150p was immunoprecipitated from both the media (external) fraction and the cellular (internal) fraction and resolved by SDS-PAGE. (B) CPY processing in sac1ts, pik1ts, and pik1ts/sac1ts cells. Cells were preincubated at 38°C for 10 min, metabolically labeled with a 35S-protein–labeling mixture during a 10-min pulse, and then chased in the presence of excess of unlabeled methionine and cysteine for the indicated times. CPY was then immunoprecipitated and resolved by SDS-PAGE. The migration positions of precursor and mature forms of CPY are indicated on the left. All data are representative of multiple experiments.
Together, these results demonstrate that the rapid inactivation of Sac1p can rescue the actin defect associated with the stt4ts mutant. In contrast, Sac1p inactivation cannot rescue secretory defects exhibited by pik1ts mutant cells, suggesting that the pool of PtdIns(4)P which accumulates in sac1 mutants cannot substitute for PtdIns(4)P generated by Pik1p to regulate secretion.
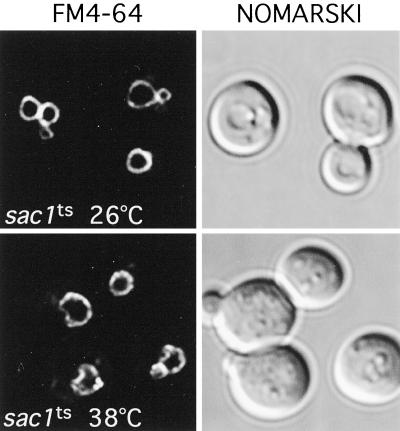
sac1ts Cells Exhibit Altered Vacuole Morphology and Accumulate Lipid Droplets at the Restrictive Temperature
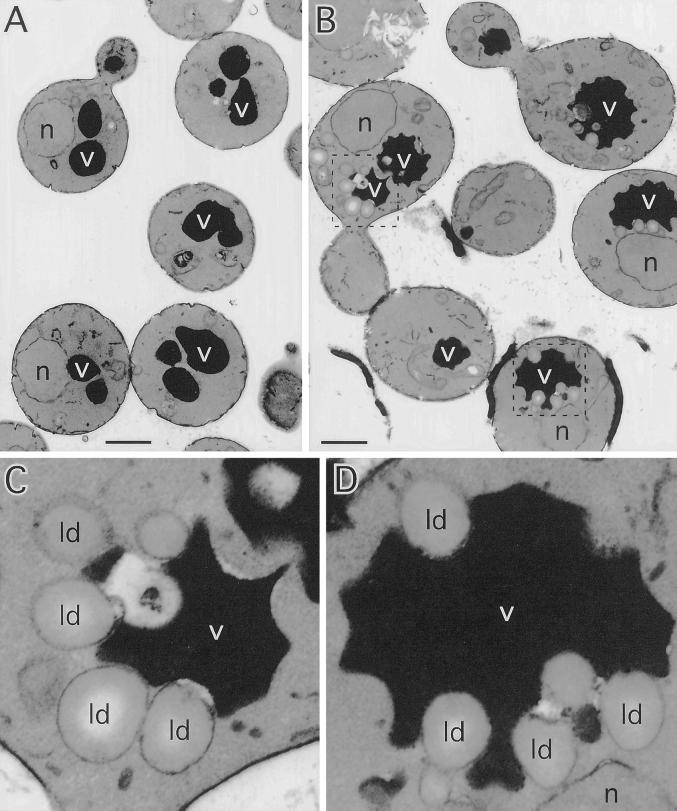
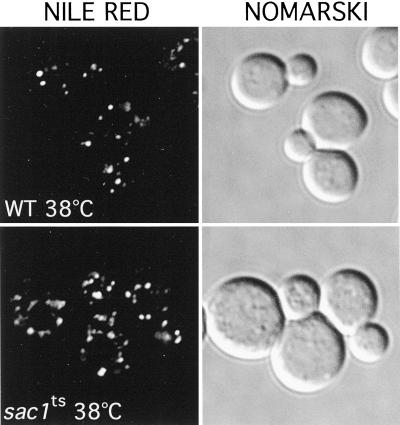
Inactivation of the two identified PtdIns 4-kinases in yeast, Stt4p and Pik1p, result in distinct alterations in vacuole morphology, suggesting that PtdIns(4)P plays a role in the maintenance of vacuole size and shape (Audhya et al., 2000). To determine whether high levels of PtdIns(4)P generated after Sac1p inactivation affect vacuole morphology, sac1ts mutant cells were labeled with the vacuolar vital dye FM4-64 (described in MATERIALS AND METHODS). At the permissive temperature, FM4-64 staining of sac1ts cells was similar to that observed in WT cells, highlighted by one to three well defined, smooth vacuolar lobes. In contrast, FM4-64 staining of sac1ts cells shifted to the restrictive temperature (2 h) revealed an abnormal vacuolar membrane morphology. In general, cells contained only one vacuole with several invaginations, creating a crenated contour (Figure 5). To more closely analyze this phenotype, sac1ts mutant cells were examined at the ultrastructural level by transmission electron microscopy at the permissive and restrictive temperatures. sac1ts cells grown at the permissive temperature were similar in appearance to WT cells with multiple round electron-dense vacuoles of regular shape and size as previously described (Figure 6A). However, when Sac1p was inactivated for 2 h by shifting cells to 38°C, a dramatic alteration in vacuole morphology occurred. Consistent with the FM4-64 staining, vacuoles in sac1ts cells were irregularly shaped with numerous invaginations of the membrane. In addition, numerous electron-lucent vesicular structures, ranging from 0.2–0.5 μm in diameter, were present. In sac1ts cells, these structures (typically 4–10/cell section in sac1ts as compared with 2–4/cell section in WT cells at 38°C) were commonly found tightly associated with the vacuolar invaginations (Figure 6, B–D; Foti, Audhya, and Emr, unpublished results). The morphology of these structures together with the lack of any apparent limiting membrane bilayer, indicated that they are lipid droplets (Zweytick et al., 2000). To further characterize these structures, cells were stained with Nile red, a fluorescent lysochrome previously demonstrated to stain cytoplasmic lipid droplets. As illustrated in Figure 7, cells presented a dotted pattern of staining consistent with the incorporation of the dye into lipid droplets. Direct observation of the stained cells on the microscope suggested, as in the above ultrastructural analysis, an increase in the number of these structures in sac1ts mutants at the restrictive temperature. In stt4ts/sac1ts double mutant cells, however, we failed to see a large increase in lipid droplets after 2 h at 38°C by electron microscopy. Furthermore, by staining with FM4-64, we observed that only a minority of these cells contained collapsed vacuoles as seen in stt4ts cells, but most did contain fragmented vacuoles, a phenotype not seen in either single mutant during the same time course (Foti, Audhya, and Emr, unpublished results). From these data, we conclude that loss of Sac1p function correlates with striking morphological alterations of the vacuole that can be, at least in part, ameliorated by elimination of Stt4p activity.
Figure 5.
SacIp phosphatase activity is required for normal vacuole morphology. sac1ts cells were grown to early log phase at 26°C, and vacuoles were labeled with the vital dye FM4-64 for 15 min (see MATERIALS AND METHODS). After labeling, cells were chased for 45 min and shifted to the indicated temperature for 2 h. On the right, cells were observed by Nomarski optics. On the left, identical fields are shown under fluorescent illumination (rhodamine channel). sac1ts cells at 26°C are shown on top, and sac1ts cells after a 2-h shift to 38°C are shown below. These cells are representative of >90% of cells observed.
Figure 6.
sac1ts cells display morphologically abnormal vacuoles surrounded by vesicular structures reminiscent of lipid droplets at the restrictive temperature. sac1ts cells were incubated at either at 26 or 38°C for 2 h, fixed in 3% gluteraldehyde, and processed for electron microscopy (see MATERIALS AND METHODS). (A) sac1ts cells at 26°C. Bar, 1.0 μm. (B) sac1ts cells at 38°C. Bar, 1.0 μm. (C and D) Fivefold enlarged view of the framed vacuoles surrounded by lipid droplets in sac1ts cells at 38°C shown in B. All cells shown are representative of >90% of cells observed. v, vacuoles; ld, lipid droplets; n, nuclei.
Figure 7.
sac1ts cells accumulate lipid droplets at the restrictive temperature. WT and sac1ts cells were incubated at 38°C for 2 h, fixed in 3% gluteraldehyde, and stained with Nile red (see MATERIALS AND METHODS). On the right, identical fields were observed with Nomarski optics. All cells shown are representative of >95% of cells observed.
Other PI Phosphatases Can Partially Compensate for the Loss of Sac1p Function
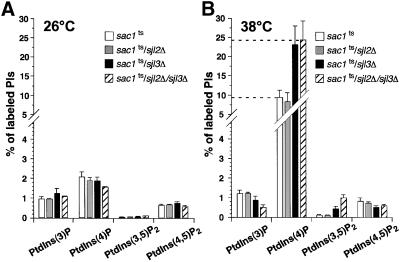
Sequence analysis of the yeast genome indicates the existence of other PI phosphatases that contain a conserved domain highly homologous to the Sac1p phosphatase domain. Of particular interest are the Sjl proteins, two of which have been shown to have similar enzymatic specificity to Sac1p in vitro (Guo et al., 1999). Notably, Sjl2p and Sjl3p have been identified as multicopy suppressors of sac1 mutant cells, suggesting that these phosphatases can partially compensate for the loss of Sac1p function (Hughes et al., 2000). Consistent with these findings, overexpression of either Sjl2p or Sjl3p suppresses the vacuole morphology defect we observe in sac1 mutant cells (Foti, Audhya, and Emr, unpublished results). Furthermore, after dissection of more than 24 tetrads generated from diploid strains harboring single chromosomal deletions of SJL3 and SAC1, we failed to isolate a single sac1Δ/sjl3Δ double mutant strain, indicating that deletion of both genes results in lethality. To address whether Sac1p, Sjl2p, and Sjl3p may function in related pathways, sjl2Δ, sjl3Δ, and sjl2Δ/sjl3Δ mutant cells harboring the sac1ts allele were generated. These mutant strains exhibited surprising growth phenotypes as follows. sac1ts/sjl3Δ cells grew well on YPD-rich medium at 26°C but poorly at 38°C, a temperature at which both single mutants grow normally. sac1ts/sjl2Δ mutant cells were not temperature sensitive for growth, consistent with our ability to isolate sjl2Δ/sac1Δ double mutant cells. However, the sac1ts/sjl2Δ/sjl3Δ triple mutant cells showed a clear growth defect, highlighted by slow growth at 26°C and cell death at 38°C. However, by performing a kill curve, we found that >90% of sac1ts/sjl2Δ/sjl3Δ mutant cells were viable after a 3-h shift to nonpermissive temperature. Therefore, to ensure cell viability, experiments with these cells were limited to conditions where they were incubated at nonpermissive temperature for <1 h.
To determine how these different phosphatases cooperate in controlling PI metabolism, intracellular PI levels were analyzed by HPLC in these double and triple mutant cells. At the permissive temperature, PI levels in all mutant strains were similar to those observed in sac1ts cells (Figure 8A). At the restrictive temperature, we could not detect any changes in the intracellular PI levels of sac1ts/sjl2Δ cells as compared with sac1ts cells. Interestingly, sac1ts/sjl3Δ and sac1ts/sjl2Δ/sjl3Δ mutant cells exhibited a dramatic accumulation of PtdIns(4)P, even when compared with sac1ts cells at the nonpermissive temperature. In these mutants, PtdIns(4)P represented 23–24% of the total newly synthesized PI, >2.5-fold greater than the level of PtdIns(4)P in sac1ts cells at the nonpermissive temperature. This incredibly high amount of PtdIns(4)P was also accompanied by a significant increase in PtdIns(3,5)P2 which represented ∼1% of the total newly synthesized PIs in sac1ts/sjl2Δ/sjl3Δ cells (Figure 8B).
Figure 8.
Defects in PI metabolism subsequent to Sac1p inactivation are exacerbated in other phosphatase mutant cells. sac1ts, sac1ts/sjl2Δ, sac1ts/sjl3Δ, and sac1ts/sjl2Δ/sjl3Δ cells were preincubated at either 26 or 38°C for 10 min, labeled with myo-[2-3H]inositol for 10 min, and then chased with excess unlabeled myo-inositol for 30 min at the appropriate temperature. Lipids were then deacylated from cellular membranes, and the glycerophosphoinositols were extracted and analyzed by HPLC. (A) Quantitative comparisons of glycerophosphoinositols generated by sac1ts, sac1ts/sjl2Δ, sac1ts/sjl3Δ, and sac1ts/sjl2Δ/sjl3Δ cells at 26°C. (B) Quantitative HPLC analysis of glycerophosphoinositols in the strains described at 38°C. These data represent the means ± SEM of at least three independent experiments.
Together, our data demonstrate that Sjl2p and Sjl3p can partially compensate for the loss of Sac1p function, likely by dephosphorylating the PtdIns(4)P that accumulates in sac1ts mutants.
Lack of Sjl2p and Sjl3p Activity in sac1 Mutant Cells Leads to Defects in Early Golgi Function
PtdIns(4)P synthesis has been shown to regulate intracellular trafficking along the secretory pathway (Hama et al., 1998; Matsuoka et al., 1998; Walch-Solimena and Novick, 1999; Audhya et al., 2000). Because deletion of SJL genes in sac1ts cells profoundly alters intracellular levels of PtdIns(4)P, we assessed whether protein sorting also was altered in these mutant cells.
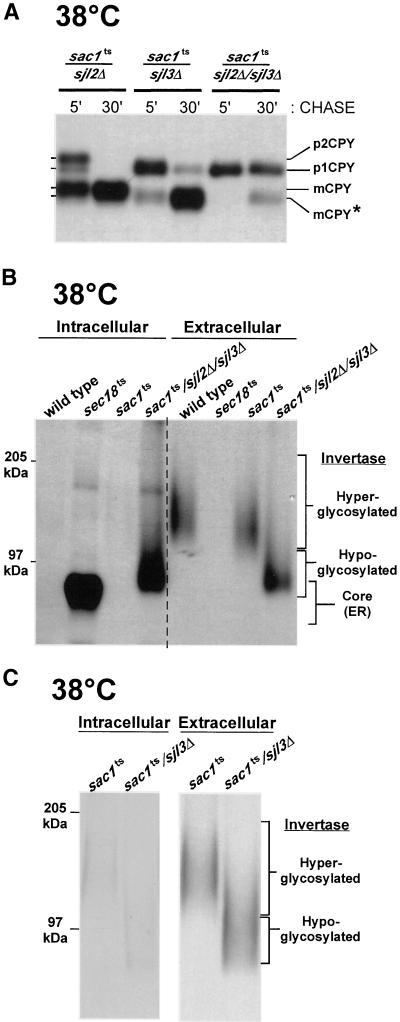
Protein glycosylation and transport were monitored by following the biosynthetic processing of CPY. At 26°C, CPY maturation was normal in sac1ts/sjl2Δ and sac1ts/sjl3Δ cells, whereas sac1ts/sjl2Δ/sjl3Δ cells displayed a minor kinetic delay in processing of this hydrolase (Foti, Audhya, and Emr, unpublished results). However, at the restrictive temperature, both sac1ts/sjl3Δ double and sac1ts/sjl2Δ/sjl3Δ triple mutant cells displayed defects in Golgi glycosylation, illustrated by the lack of p2CPY after 5 min, whereas sac1ts/sjl2Δ cells processed CPY normally (Figure 9A). After 30 min of chase, >90% of the labeled CPY remained in a ER-modified p1 form in sac1ts/sjl2Δ/sjl3Δ cells, whereas sac1ts/sjl3Δ accumulated a hypoglycosylated form of mature CPY.
Figure 9.
The dramatic accumulation of PtdIns(4)P in sac1ts/sjl2Δ/sjl3Δ mutant cells alters Golgi-dependent glycosylation pathways. (A) The indicated strains were preincubated at 38°C for 10 min, metabolically labeled with an 35S-protein–labeling mixture during a 10-min pulse, and then chased in the presence of excess unlabeled methionine and cysteine for the indicated times. CPY was then immunoprecipitated and resolved by SDS-PAGE. The migration positions of precursor and mature forms of CPY are indicated on the left. The star indicates the migration position of underglycosylated mature CPY. (B) WT, sac1ts, and sac1ts/sjl2Δ/sjl3Δ mutant cells were preincubated at 38°C for 10 min, metabolically labeled with an 35S-protein–labeling mixture during a 10-min pulse, and then chased in the presence of excess unlabeled methionine and cysteine for 30 min. Cellular transport was stopped by the addition of NaN3 and NaF after the pulse-chase, and the cells were converted to spheroplasts. Internal and external fractions were separated by centrifugation and analyzed for the presence of invertase by immunoprecipitation. (C) sac1ts and sac1ts/sjl3Δ mutant cells were preincubated at 38°C for 10 min, metabolically labeled with an 35S-protein–labeling mixture during a 10-min pulse, and then chased in the presence of excess unlabeled methionine and cysteine for 30 min. Cells were then treated as in B. All data are representative of multiple experiments.
The integrity of the secretory pathway was also assessed in sac1ts and sac1ts/sjl2Δ/sjl3Δ mutant cells by monitoring the secretion of invertase. At the permissive temperature, invertase was normally processed and secreted in both strains (Foti, Audhya, and Emr, unpublished results). Strikingly, sac1ts/sjl2Δ/sjl3Δ mutant cells exhibited a significant defect in the secretion of invertase at the nonpermissive temperature. This defect was not due to the retention of invertase in the ER as is the case for sec18ts mutant cells, which accumulate invertase exclusively in the ER core-modified form. Instead, sac1ts/sjl2Δ/sjl3Δ mutant cells accumulated a hypoglycosylated Golgi-modified form of invertase, consistent with a defect in Golgi-dependent modification of this protein (Figure 9B). In sac1ts/sjl3Δ mutant cells, invertase was secreted, but similar glycosylation defects were observed (Figure 9C).
In summary, these data indicate that, in the absence of Sac1p activity, the Sjl proteins become essential for normal Golgi function. Protein transport and glycosylation in the Golgi are impaired in the sac1ts/sjl2Δ/sjl3Δ triple mutant.
DISCUSSION
In this study, we generated a temperature-sensitive allele of the SAC1 gene to characterize the primary function of the Sac1 PI phosphatase. Our data demonstrate that Sac1p primarily metabolizes PtdIns(4)P synthesized by the Stt4p PtdIns 4-kinase and that membrane localization of Sac1p is crucial for its efficient function. The rapid inactivation of Sac1p leads to a dramatic alteration in vacuole morphology and an accumulation of lipid droplets. Furthermore, loss of Sac1p function rescues defects associated with stt4ts but not pik1ts mutant cells. Additionally, function of Sjl3p, an Sjl phosphatase homologous to Sac1p, was found to be essential for viability in Sac1p-deficient cells. We suggest that Sjl3p partially compensates for the loss of Sac1p PtdIns(4)P phosphatase activity. Our results indicate that deficiency in both Sac1p and Sjl3p activities leads to Golgi dysfunction and cell death. Together our data support a model in which Stt4p and Sac1p control the synthesis and turnover of a pool of PtdIns(4)P spatially and functionally distinct from the pool of PtdIns(4)P generated by Pik1p (Figure 10). Inactivation of Sac1p induces an accumulation of PtdIns(4)P, which may rapidly traffic to more distal compartments in the secretory pathway like the Golgi and vacuole. To prevent improper signaling in sac1 mutant cells, PtdIns(4)P must be metabolized/inactivated by other phosphatases such as Sjl3p. Consistent with this hypothesis, deletion of SJL3 in sac1ts cells results in a dramatic accumulation of PtdIns(4)P at the restrictive temperature, accompanied by defects in cell growth and Golgi function.
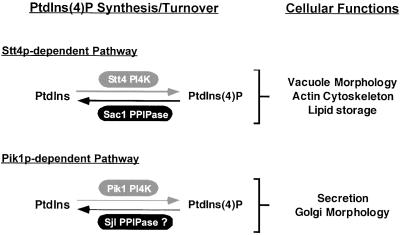
Figure 10.
Stt4p- or Pik1p-dependent phosphorylation of PtdIns leads to the production of distinct pools of PtdIns(4)P. Sac1p specifically turns over a pool of PtdIns(4)P pool generated by Stt4p, whereas Sjl proteins are potential candidate phosphatases for Pik1p-generated PtdIns(4)P turnover in the Golgi. The proposed roles for the different PtdIns(4)P pools are indicated. The Stt4p/Sac1p-dependent PtdIns(4)P pool is required for vacuole morphology, actin cytoskeleton organization, and regulation of neutral lipids storage, whereas the Pik1p-dependent PtdIns(4)P pool is essential for secretory vesicle formation from the Golgi and maintenance of Golgi/endosome morphology.
PtdIns(4)P Is the Primary Substrate for Sac1p
Analysis of PI levels in sac1ts cells has led us to conclude that the main substrate of Sac1p in vivo is PtdIns(4)P. Two of the six mutations in the sac1-23 allele lie within a conserved motif found in all Sac phosphatase domains. However, the highly conserved CX5R(T/S) motif remains intact. This is not surprising, because mutations in this motif result in steady-state defects in Sac1p function (reviewed by Hughes et al., 2000). Previous studies have clearly shown that Sac1p exhibits phosphatase activity toward all monophosphorylated PIs in vitro, especially PtdIns(3)P (Guo et al., 1999; Hughes et al., 2000). These observations support the idea that the in vivo specificity of Sac1p for PtdIns(4)P is related to the accessibility of its substrate. Our studies indicate that the majority of Sac1p localizes to the ER, where levels of PtdIns(3)P and PtdIns(3,5)P2 are likely to be very low. Indeed, PtdIns(3)P synthesis is essential for transport from the Golgi to the endosome/vacuole (Schu et al., 1993), suggesting that PtdIns(3)P is generated at the Golgi/endosome. PtdIns(3)P localization studies in yeast with the use of FYVE domain-GFP fusion proteins as a reporter also revealed a distribution pattern for PtdIns(3)P reminiscent of the Golgi/endosome (Burd and Emr, 1998). One explanation for the accumulation of PtdIns(3)P or PtdIns(3,5)P2 in sac1Δ mutants is that the high amount of PtdIns(4)P in these cells can compete as a substrate for PtdIns(3)P- or PtdIns(3,5)P2-specific phosphatases, thereby altering the normal turnover of these lipids.
The discrepancy between our data showing the localization of Sac1p to the ER and previous results from Whitters et al. (1993) reporting a localization of Sac1p to both the Golgi and the ER is still unresolved. Our data are consistent with the exclusive localization of the rat homologue of Sac1p to the ER in different types of mammalian cells, but we cannot rule out the existence of a small pool of Sac1p in the yeast Golgi (Nemoto et al., 2000). If a small Golgi pool does exist, it may rapidly recycle back to the ER.
Localization of tagged Sac1p lacking the last 102 amino acids (Sac1Δ522-623) revealed a striking loss of ER association in contrast to the full-length protein, demonstrating that the C-terminal transmembrane domain of Sac1p is required for its ER localization. Mislocalization of truncated Sac1Δ522-623 resulted in alterations in the metabolism of PIs and a growth phenotype similar to that observed in sac1Δ cells, suggesting that localization of Sac1p to ER membranes is crucial for its efficient activity. However, when overexpressed, the Sac1Δ522-623 fusion protein is capable of complementing defects exhibited by sac1Δ mutant cells. Thus, the Sac1Δ522-623 truncation protein possesses phosphatase activity in vivo, which is not unexpected because its Sac phosphatase domain remains intact, but its inappropriate localization likely prevents accessibility to a pool of PtdIns(4)P that Sac1p normally regulates.
Sac1p Acts on a Pool of PtdIns(4)P Synthesized by the Stt4 Kinase
Our analysis of PI levels in stt4ts and pik1ts mutants carrying the sac1ts allele demonstrates that Stt4p, but not Pik1p, generates the pool of PtdIns(4)P that is metabolized by Sac1p. Previously, Pik1p was thought to function as the kinase that produces high levels of PtdIns(4)P in sac1 mutant cells (Hama et al., 1999; Rivas et al., 1999). Indeed, Pik1p activity has been shown to be crucial for protein secretion out of the Golgi (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000) and overexpression of Pik1p can weakly suppress the growth defect exhibited by the sec14ts secretory mutant cells at semirestrictive temperature (Hama et al., 1999). Because mutations in the SAC1 gene can bypass the requirement for Sec14p (Cleves et al., 1989; Stock et al., 1999), it was assumed that stabilization of PtdIns(4)P generated by Pik1p in the Golgi could suppress secretory defects associated with sec14ts mutant cells (Hughes et al., 2000; Huijbregts et al., 2000). In contrast to these assumptions, our data establish a functional link between Sac1p and Stt4p, both biochemically and genetically. Strikingly, loss of Sac1p function partially rescues the actin cytoskeleton defect exhibited by stt4ts cells at the nonpermissive temperature. Consistent with this finding, Sac1p was initially identified in a screen for “suppressor of actin” mutants (Novick et al., 1989), but a mechanism for this suppression has been elusive. Together with our data, we suggest that loss of Sac1p function may rescue specific mutant alleles of act1 by affecting levels of PtdIns(4)P generated by Stt4 PtdIns 4-kinase.
Although, the intracellular location of Stt4p is unknown, our results would predict that Stt4p could localize, at least in part, to Sac1p-containing membranes like the ER. Consistent with this, Stt4p has been shown to play a role in the trafficking of aminophospholipids from the ER to the Golgi/vacuole (Trotter et al., 1998), and PtdIns 4-kinase α, the mammalian homologue of Stt4p, is associated with ER membranes (Wong et al., 1997). However, it is possible that PtdIns(4)P generated by Stt4p may be produced elsewhere and be rapidly transported to Sac1p-containing membranes for turnover. Further studies are underway to determine the localization of Stt4p.
Role of Sac1p in the Control of Vacuole Morphology and Neutral Lipid Storage
We previously demonstrated that stt4ts and pik1ts mutant cells both exhibit distinct, but dramatic, alterations in vacuole morphology, suggesting that PtdIns(4)P plays a crucial role in the appearance and/or function of this organelle (Audhya et al., 2000). The rapid inactivation of Sac1p in sac1ts mutant cells is also accompanied by dramatic changes in vacuole shape. Thus, strict control of distinct pools of PtdIns(4)P appears to be crucial to maintain normal morphology and function of the vacuole. We also observed an accumulation of lipid droplets in sac1ts cells, which often appeared to cluster around the vacuole and associate with vacuolar invaginations. These lipid droplets, consisting of a hydrophobic core of neutral lipids (steryl esters and triglycerides) surrounded by a phospholipid monolayer, are thought to be derived from ER membranes. It has been suggested that formation of these lipid droplets could provide a transport route of steryl esters to the plasma membrane, and it is feasible that Sac1p plays a role in this process (reviewed by Zweytick et al., 2000).
Role of PI Phosphatases containing a SacI-like Domain in Golgi Function
Elimination of Sjl3p activity, in contrast to Sjl2p activity, in sac1Δ cells results in lethality. Analysis of PI metabolism in sac1ts/sjl3Δ suggests that this growth defect is likely due to the additional increase in PtdIns(4)P levels in the double mutant as compared with sac1ts single mutant cells. The double mutant also exhibits defects in secretion and Golgi-specific glycosylation. Consistent with the observed defect in protein glycosylation, Sjl3p has been shown to play a role in trafficking from the Golgi (Luo and Chang, 1997; Bensen et al., 2000) and thus may localize to the Golgi where it can metabolize PtdIns(4)P. Recent studies of Sjl2p and Sjl3p show the proteins to be diffusely distributed throughout the cell under normal conditions, leaving open the possibility that these phosphatases regulate PI pools at the Golgi (Ooms et al., 2000). Triple sac1ts/sjl2Δ/sjl3Δ mutant cells exhibited additional defects in Golgi glycosylation when compared with sac1ts/sjl3Δ double mutant cells. Thus, even though deletion of SJL2 in sac1ts cells has no observable effect on PI metabolism as compared with sac1ts cells alone, expression of the Sjl2 phosphatase became essential in the sac1ts/sjl3Δ background at elevated temperatures. Additionally, we observed an increase in PtdIns(3,5)P2 levels at the restrictive temperature in sac1ts/sjl2Δ/sjl3Δ triple mutant cells. Although the defect in invertase secretion observed in these cells correlates with the increase in PtdIns(3,5)P2, we do not favor a role for this isomer in the alteration of Golgi secretory function. Indeed, several lines of evidence argue against this possibility. First, synthesis of PtdIns(3)P and PtdIns(3,5)P2 have been shown to be involved in the regulation of Golgi to vacuole-trafficking events but not in Golgi glycosylation or secretion (Schu et al., 1993; Odorizzi et al., 1998). Second, Fab1p, the only known yeast PtdIns(3)P 5-kinase, has been localized to prevacuolar and vacuolar compartments (Gary et al., 1998). Third, the bulk of PtdIns(3)P in yeast was shown to be localized to endosomes, multivesicular bodies, and vacuoles (Stenmark et al., 1996; Gillooly et al., 2000), and finally, sac1ts/sjl2Δ/sjl3Δ cells do not display defects in secretion of other secretory cargo such as Hsp150p. Therefore, it is likely that the dramatic increase of PtdIns(4)P levels subsequent to Sac1p inactivation in sac1ts/sjl2Δ/sjl3Δ cells causes the observed Golgi glycosylation defect, perhaps by altering the localization/trafficking of Golgi-specific glycosyltransferases. These results suggest that Stt4p-generated PtdIns(4)P that escapes degradation by Sac1p activity is, at least partially, turned over by Sjl proteins, presumably in the Golgi. Moreover, when sac1 mutant cells also lack the phosphatase activities of Sjl2p and Sjl3p, increased PtdIns(4)P levels negatively impact on normal Golgi function, and this results in a loss in cell viability. However, under normal conditions, Sjl3p does not turnover Stt4p-generated PtdIns(4)P because of the efficient activity of Sac1p. Consistent with this, deletion of SJL3 in stt4ts cells fails to stabilize PtdIns(4)P produced by this lipid kinase and fails to rescue any of the phenotypes associated with stt4ts cells (Foti, Audhya, and Emr, unpublished results).
The importance of tight regulation of both PI synthesis and turnover by PI kinases and phosphatases for proper signaling is clear. However, very little is known about the downstream effectors for PtdIns(4)P. The identification of downstream effectors of PtdIns(4)P as well as additional genetic and biochemical analysis of enzymes involved in the metabolism of PtdIns(4)P should help to elucidate the role of this particular isomer and its derivatives in protein and lipid trafficking, as well as in membrane dynamics and cytoskeletal organization.
ACKNOWLEDGMENTS
We thank Chris Hofeditz and Tammie Mcquistan for assisting with the electron microscopy analysis (Immunoelectron microscopy Core B of Program Project grant CA58689 headed by M. Farquhar). We would also like to thank Patricie Burda, Chris Stefan, and Andrew Wurmser for critical reading of the manuscript and Perla Arcaira for technical assistance. We thank Jonathan Gary for generating the yeast strains JGY130, JGY131, and JGY132 and Greg Odorizzi for providing pGO106. This work was supported by a grant from the National Institutes of Health (CA58689 to S.D.E.). S.D.E. is supported as an investigator of the Howard Hughes Medical Institute. M.F. is supported by the Swiss National Funds for the Scientific Research (fellowship 823A-053440).
Abbreviations used:
- CPY
carboxypeptidase Y
- DAPI
4′,6-diamidino-2-phenylindole
- ER
endoplasmic reticulum
- GFP
green fluorescence protein
- HPLC
high-performance liquid chromatography
- PCR
polymerase chain reaction
- PI
phosphoinositide
- PtdIns
phosphatidylinositol
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- PtdIns(4)P
phosphatidylinositol 4-phosphate
- PtdIns(3,5)P2
phosphatidylinositol (3,5)-diphosphate
- PtdIns(4,5)P2
phosphatidylinositol (4,5)-diphosphate
- Sjl
synaptojanin-like
- WT
wild-type
REFERENCES
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, stt4p and pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphatidylinositol 4-kinases. Biochim Biophys Acta. 1998;1436:69–85. doi: 10.1016/s0005-2760(98)00134-9. [DOI] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Costaguta G, Payne GS. Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae: roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S. Phosphatidylinositol transfer proteins: a requirement in signal transduction and vesicle traffic. Bioessays. 1998;20:423–432. doi: 10.1002/(SICI)1521-1878(199805)20:5<423::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Divecha N, Clarke JH, Roefs M, Halstead JR, D'Santos C. Nuclear inositides: inconsistent consistencies. Cell Mol Life Sci. 2000;57:379–393. doi: 10.1007/PL00000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA, Thorner J. Purification and characterization of a soluble phosphatidylinositol 4-kinase from the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:24117–24125. [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos JF, Marini F, Stevenson I, Frei C, Hall MN. PIK1, an essential phosphatidylinositol 4-kinase associated with the yeast nucleus. EMBO J. 1994;13:2352–2361. doi: 10.1002/j.1460-2075.1994.tb06519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hughes WE, Cooke FT, Parker PJ. Sac phosphatase domain proteins. Biochem J. 2000;350:337–352. [PMC free article] [PubMed] [Google Scholar]
- Hughes WE, Pocklington MJ, Orr E, Paddon CJ. Mutations in the Saccharomyces cerevisiae gene SAC1 cause multiple drug sensitivity. Yeast. 1999;15:1111–1124. doi: 10.1002/(SICI)1097-0061(199908)15:11<1111::AID-YEA440>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hughes WE, Woscholski R, Cooke FT, Patrick RS, Dove SK, McDonald NQ, Parker PJ. SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J Biol Chem. 2000;275:801–808. doi: 10.1074/jbc.275.2.801. [DOI] [PubMed] [Google Scholar]
- Huijbregts RPH, Topalof L, Bankaitis VA. Lipid metabolism and regulation of membrane trafficking. Traffic. 2000;1:195–202. doi: 10.1034/j.1600-0854.2000.010301.x. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochendorfer KU, Then AR, Kearns BG, Bankaitis VA, Mayinger P. Sac1p plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. EMBO J. 1999;18:1506–1515. doi: 10.1093/emboj/18.6.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luo W-J, Chang A. Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J Cell Biol. 1997;138:731–746. doi: 10.1083/jcb.138.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, Kisseleva MV, Norris FA. The role of phosphatases in inositol signaling reactions. J Biol Chem. 1999;274:10669–10672. doi: 10.1074/jbc.274.16.10669. [DOI] [PubMed] [Google Scholar]
- Martin TF. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu Rev Cell Dev Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Mayinger P, Bankaitis VA, Meyer DI. Sac1p mediates the adenosine triphosphate transport into yeast endoplasmic reticulum that is required for protein translocation. J Cell Biol. 1995;131:1377–1386. doi: 10.1083/jcb.131.6.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Kearns BG, Wenk MR, Chen H, Mori K, Alb J, De Camilli P, Bankaitis VA. Functional characterization of a mammalian SacI and mutants exhibiting substrate specific defects in phosphoinositide phosphatase activity. J Biol Chem. 2000;275:34293–34305. doi: 10.1074/jbc.M003923200. [DOI] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Ooms LM, McColl BK, Wiradjaja F, Wijayaratnam APW, Gleeson P, Gething MJ, Sambrook J, Mitchell CA. The yeast inositol polyphosphate 5-phosphatases Inp52p and Inp53p translocate to actin patches following hyperosmotic stress: mechanism for regulating phosphatidylinositol 4,5-bisphosphate at plasma membrane invaginations. Mol Cell Biol. 2000;20:9376–9390. doi: 10.1128/mcb.20.24.9376-9390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltauf F, Kohlwein SD, Henry SA. Biology of the Yeast Saccharomyces, E. W. Jones, J. R. Pringle, and J.R, Broach. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. Regulation and compartmentalization of lipid synthesis in yeast; pp. 415–500. [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p”and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Seron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel M, Guillaud P, Devilliers G, Rossanese OW, Glick BS, Riezman H, Keränen S, Haguenauer-Tsapis R. A yeast t-SNARE involved in endocytosis. Mol Cell Biol. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence LW. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr S, De Camilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis [published erratum appears in Eur. J. Cell Biol. 1998 Mar;75(3):246] Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh BH, D'Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc- binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- Stock SD, Hama H, DeWald DB, Takemoto JY. SEC14-dependent secretion in Saccharomyces cerevisiae: nondependence on sphingolipid synthesis-coupled diacylglycerol production. J Biol Chem. 1999;274:12979–12983. doi: 10.1074/jbc.274.19.12979. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Huynh CV, Thorner J, York JD. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998a;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998b;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Wu WI, Pedretti J, Yates R, Voelker DR. A genetic screen for aminophospholipid transport mutants identifies the phosphatidylinositol 4-kinase, STT4p, as an essential component in phosphatidylserine metabolism. J Biol Chem. 1998;273:13189–13196. doi: 10.1074/jbc.273.21.13189. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol 4-kinase Pik1p regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Meyers dd R, Cantley LC. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J Biol Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ohya Y, Hirose R, Nakano A, Anraku Y. STT10, a novel class-D VPS yeast gene required for osmotic integrity related to the PKC1/STT1 protein kinase pathway. Gene. 1995;160:117–122. doi: 10.1016/0378-1119(95)00214-q. [DOI] [PubMed] [Google Scholar]
- Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta. 2000;1469:101–120. doi: 10.1016/s0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]