Abstract
Motivation: Given the abundance of genome sequencing and omics data, an opprtunity and challenge in bioinformatics relates to data mining and visualization. The majority of current bioinformatics visualizations are implemented either as multi-tier web server applications that require significant maintenance effort, or as client software that presumes technical expertise for installation. Here we present the Visual Omics Explorer (VOE), a cross-platform data visualization portal that is implemented using only HTML and Javascript code. VOE is a standalone software that can be loaded offline on the web browser from a local copy of the code, or over the internet without any dependency other than distributing the code through a file sharing service. VOE can interactively display genomics, transcriptomics, epigenomics and metagenomics data stored either locally or retrieved from cloud storage services, and runs on both desktop computers and mobile devices.
Availability and implementation: VOE is accessible at http://bcil.github.io/VOE/.
Contact: agbiotec@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Data visualization applications are key for representing, integrating and mining next-generation sequencing and other types of omics data. A common paradigm involves omics data visualizations that are computed on a remote server and displayed through Common Gateway Interface web pages (UCSC, https://genome.ucsc.edu/), or using a Java client-server architecture that provides interactive, local data visualizations (IGV, Robinson et al. 2011). Recently, a number of omics databases came online that provide rich Application Programming Interfaces (API, Dooley et al., 2012; Cerami et al. 2012; Paten et al., 2015), or which combine the API with data storage and computational capacity for rent such as Google Genomics (https://cloud.google.com/genomics/). Furthermore, a new generation of omics data visualizations (Almeida et al., 2012; Gómez et al., 2013; Gille et al., 2014), provide interactive interfaces that leverage computing within the web browser, and are completely de-coupled from remote web servers. Here we present the Visual Omics Explorer (VOE), a data visualization portal that offers dynamic, mobile-friendly omics data displays, combining the power of APIs with that of local compute within the browser using Javascript. VOE provides a diverse set of visualizations (Fig. 1, Supplementary Fig.S1) for data on the Google Genomics Cloud, genetic variants, ChIP-seq, RNA-seq and phylogenetic data, using HTML5 and, Javascript – D3 web technologies (https://github.com/mbostock/d3). VOE is standalone software that can be loaded offline on the web browser from a local copy of the code (https://github.com/BCIL/VOE/archive/master.zip, index.html file), over the internet from GitHub (http://bcil.github.io/VOE/) or online storage such as Amazon S3 (http://tinyurl.com/BioITCore), and similarly from Google Drive or Dropbox. VOE has been tested to work on all current versions of desktop or mobile web browsers, and by using the PhoneGap framework (http://phonegap.com) we packaged VOE as a touch-enabled, mobile app for the Android system (http://tinyurl.com/voe-apk).
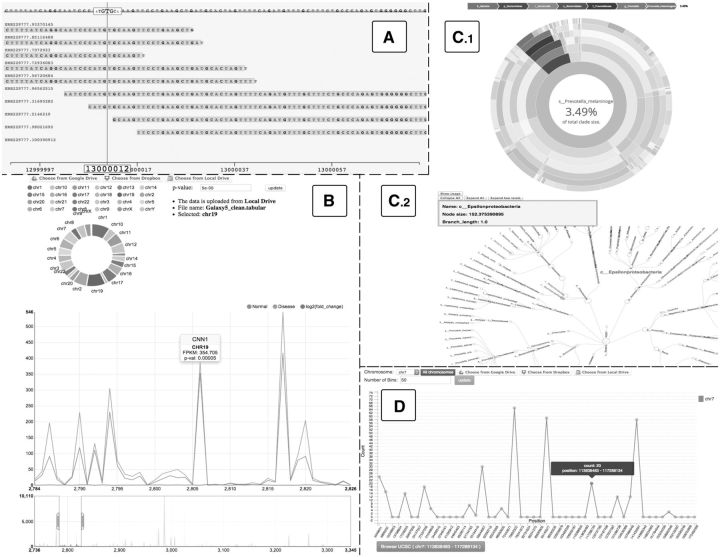
Fig. 1.
(A) Read alignments on Google Genomics Cloud; (B). Differential Gene Expression using RNA-seq data; PhyloXML visualization using (C1). Sunburst and (C2). Radial Tree; (D) ChIP-Seq peak binding sites
2 Implementation
VOE Architectural Design: The VOE portal was implemented using a skeleton of HTML, with embedded Javascript-D3 code for parsing the omics data elements and rendering the visualizations, in addition to providing interactive graphs on the interface by controlling the CSS transitions. Furthermore, the D3 ‘brush’ function was utilized to support focusing, zooming and navigation to features of interest within the visualized data, while D3 parser methods were used to divide large datasets for computational efficiency. New D3 functions were also written to convert the omics datasets to Scalable Vector Graphics (SVG), and allow exporting the visualizations in SVG file format for use in publications. Finally, a new set of functions were implemented also in D3 to parse and index files with various bioinformatics data formats, and allow importing omics data to VOE from local files or cloud storage such as Google Drive and Dropbox. In addition to the user documentation in the supplementary material, we prepared video screencasts (http://tinyurl.com/bioitcore-voe) demonstrating data analysis with VOE. In summary, VOE visualizations include: (1). Google Genomics Cloud (video: http://tinyurl.com/bioitcore-viz-gg). This visualization enables viewing sequence read alignments to the reference human genome from the 1000 Genomes and other public projects hosted on Google Genomics (Fig. 1A, Supplementary Fig. S2). Users can select a specific project, genome and chromosomal region from the Google Genomics Cloud, and then navigate through the visualization by highlighting and zooming into a sequence alignment track showing variants between reads and the reference genome; (2). ChIP-Seq and genetic variants (video: http://tinyurl.com/bioitcore-viz-cv). The visualization of molecular interactions predicted by ChIP-Seq data analysis pipelines (BED output files) and genetic variants (VCF files, Danecek et al., 2011), share a similar graphical representation, but are computed and displayed separately given the different data formats. In more detail, the x-axis on the graph (Fig. 1D, Supplementary Fig. S3) represents the length of the complete genome or a specific chromosome selected by the user, while the y-axis indicates the number of molecular interaction sites for ChIP-Seq, or the number of genetic variants. By default, the data are grouped in ten bins, each represented by one point on the graph. Users have the option to specify the number of bins within a specific chromosome, and then the graph is automatically recomputed. With selection for example of 1000 bins on a single chromosome and input file with 48 000 variants (Supplementary files ‘Sample_inputFiles’ in VOE code), the visualization takes only a few seconds to render. By clicking on each point on the line graph, users can see the exact number or variants included in the bin, and a table of web links is shown under the graph that redirects to the UCSC Genome database for further information on the genomic region represented by the bin; (3). RNA-Seq Gene Expression data (http://tinyurl.com/bioitcore-viz-rna-seq). Gene expression omics data are visualized in two displays (Fig. 1B, Supplementary Fig. S4), with the first being a circular chart where each chromosome is represented as a slice with a different color and size proportional to the number of differentially expressed genes (P-value < 0.05 as default). Users can adjust the P-value, and also add or remove chromosomes to the visualization. The second display shows two lines, each for gene expression levels in a different experimental condition, and a line corresponding to expression fold change between conditions. In both displays additional information provide the gene name, chromosome, expression in FPKM and P-value; (4). PhyloXML data (video: http://tinyurl.com/bioitcore-viz-phyloxml). VOE displays Sunburst, Radial and Indented Tree visualizations (Supplementary Fig. S5, Han and Zmasek, 2009). The Radial tree algorithm (Fig. 1, C2) aligns on the outside the circle the deepest level of nodes, which users can collapse or expand further by clicking on each. Hovering with the mouse pointer over a tree node displays the node name, size and branch length. The Sunburst tree (Fig. 1, C1) displays simultaneously the hierarchical and quantitative relationship between tree levels, with the size of an outside slice for example representing the proportion of a species found under an inner slice for the genus.
3 Discussion
We evaluated VOE with data processed using bioinformatics pipelines (http://galaxy.hunter.cuny.edu:8080/workflow/list_published), running on our local instance of the Galaxy server. For the ChIP-Seq pipeline (based on Liu et al., 2010) data are for the H3K4 histone interactions on the MCF breast cancer line (http://www.ebi.ac.uk/ena/data/view/SRP007976). We visualized the BED output of the pipeline and identified a large number of interactions on chromosome 17 (Supplementary Fig. S3.1), where histone modifications have been associated with breast cancer (Zhang and Yu, 2011). We also visualized an RNA-Seq pipeline (Trapnell et al., 2012) output using prostate cancer data (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-567/), and found overexpression of the MYC and TSPAN13 genes (Supplementary Fig. S4.3), known to be involved in this type of cancer (Arencibia et al., 2009; Koh et al., 2010). VOE provides an interactive, mobile friendly omics visualization platform in the form of self-contained code, running within the web browser. With release of this software, we aim to present a new paradigm for bioinformatics visualizations, in contrast to the complex software stacks and monolithic web server setups. Our implementation uses purely Javascript, and enabled us to easily bundle VOE as a mobile application. As sequencing is becoming a commodity and with widespread use of powerful, yet portable computational platforms such as tablets as smartphones, software following a similar paradigm with VOE running on mobile platforms can find applications in the clinic and doctor’s offices. Furthermore, new HTML5 standards (http://www.w3.org/TR/2015/WD-workers-20150924/) including web workers, allow development of multi-threaded web browser applications that utilize multiple CPU cores for scalability. We plan to further develop VOE using these standards, and also support other developers utilizing our open source code in this respect.
Supplementary Material
Acknowledgements
The authors would like to thank all members of the Bioinformatics Core Infrastructures and Krampis’ Lab for their feedback during manuscript preparation.
Funding
Supported by the Center for Translational and Basic Research grant from National Institute on Minority Health and Health Disparities (G12 MD007599) and Weill Cornell Medical College - Clinical and Translational Science Center (2UL1TR000457-06).
Conflict of Interest: none declared.
References
- Almeida J.S. et al. (2012) ImageJS: personalized, participated, pervasive, and reproducible image bioinformatics in the web browser. J. Pathol. Inf., 3, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arencibia J.M. et al. (2009) Gene expression profiling reveals overexpression of TSPAN13 in prostate cancer. Int. J. Oncol., 34, 457–463. [PubMed] [Google Scholar]
- Cerami E. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley R. et al. (2012, November) Software-as-a-service: the iPlant foundation API. In: 5th IEEE Workshop on Many-Task Computing on Grids and Supercomputers (MTAGS). [Google Scholar]
- Gille C. et al. (2014) Sequence alignment visualization in HTML5 without java. Bioinformatics, 30, 121–122. [DOI] [PubMed] [Google Scholar]
- Gómez J. et al. (2013) BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics, 29, 1103–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.V., Zmasek C.M. (2009) phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinf., 10, 356. doi:10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C.M. et al. (2010) MYC and prostate cancer. Genes Cancer, 1, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. et al. (2010) Q&A: ChIP-seq technologies and the study of gene regulation. BMC Biology, 8, 56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B. et al. (2015) The NIH BD2K center for big data in translational genomics. J. Am. Med. Inf. Assoc., 22, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T. et al. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R. et al. (2015) The UCSC genome browser database: 2015 update. Nucleic Acids Res., 43, D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc., 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yu Y. (2011) The important molecular markers on chromosome 17 and their clinical impact in breast cancer. Int. J. Mol. Sci., 12, 5672–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.