Abstract
Summary
Networks have become ubiquitous in systems biology. Visualization is a crucial component in their analysis. However, collaborations within research teams in network biology are hampered by software systems that are either specific to a computational algorithm, create visualizations that are not biologically meaningful, or have limited features for sharing networks and visualizations. We present GraphSpace, a web-based platform that fosters team science by allowing collaborating research groups to easily store, interact with, layout and share networks.
Availability and implementation
Anyone can upload and share networks at http://graphspace.org. In addition, the GraphSpace code is available at http://github.com/Murali-group/graphspace if a user wants to run his or her own server.
Contact
murali@cs.vt.edu
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Computational analysis of molecular interaction networks has become pervasive in systems biology. Nearly every publication that uses network analysis includes a layout of a graph in which the nodes and edges are laid out in two dimensions. Several systems implement methods for network analysis and visualization. Despite these advances, interdisciplinary research teams in network biology face several challenges in sharing, exploring and interpreting computed networks in their collaborations:
Sharing. Computational biologists may generate hundreds or thousands of networks that they seek to share without manual overhead.
Organization. A research group working on multiple projects may seek to keep the networks from different collaborations private from each other.
Searching and Filtering. Collaborators may search within and across these networks for proteins and interactions of interest. Collaborators may also want to examine subgraphs of a network.
Layouts. Automatic layout algorithms incorporate almost no knowledge of the biological information underlying the networks. Thus, researchers need layout tools that can help them readily use their knowledge and intuition to modify node positions manually to bring out salient features.
Release. Upon the publication of a project, researchers may seek to make a subset of networks publicly available for access.
We are not aware of any existing systems that address all these challenges (see Supplementary Section S1 for a comparison).
2 GraphSpace features
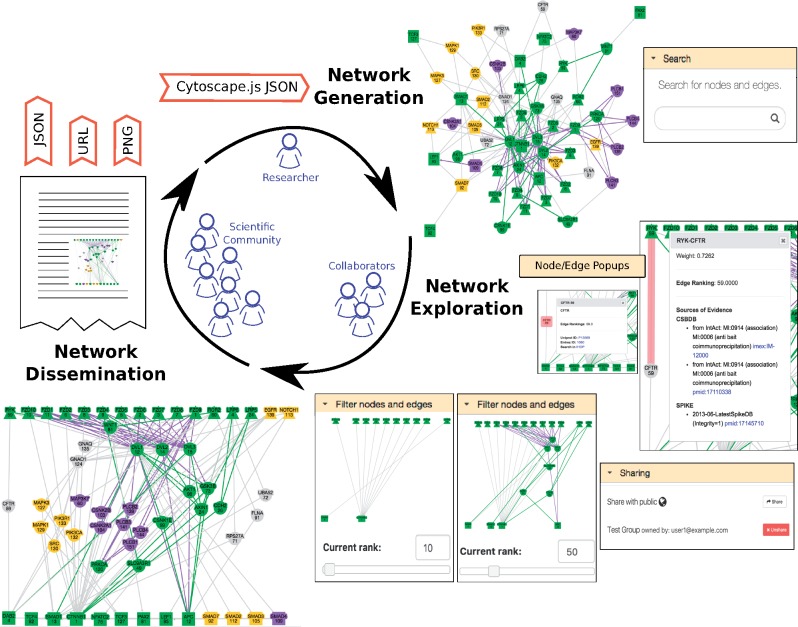
We developed GraphSpace, a web-based system that addresses the challenges described earlier using a rich set of user-friendly features designed to enhance network-based collaboration (Fig. 1). Users can upload richly annotated networks, irrespective of the algorithms or software used to generate them. Users can create private groups, invite other users to join groups and share networks with group members. A user may search for networks for specific attributes and for the presence of a specific node or nodes. A powerful layout editor allows users to efficiently modify node positions, edit node and edge styles, save new layouts and share them with other users (Supplementary Fig. S1). Users may make networks public and provide a persistent URL in a publication, enabling others to explore these networks. Supplementary Figures S2–S32 illustrate a user’s interaction with the GraphSpace web interface. While users can view and interact with GraphSpace networks anonymously, several features become available after a user creates an account and logs in. We describe the main features of GraphSpace here and in Supplementary Section S2. Supplementary Section S3 discusses use cases for GraphSpace.
Fig. 1.
A typical GraphSpace workflow. A user may upload networks in the Cytoscape.js-supported JSON format into GraphSpace. The user and collaborators can explore networks using search, sharing, annotation and filtering capabilities. Users may also create manual layouts of the networks (also shown in Supplementary Fig. S1A). Upon publication, networks may be shared with the scientific community via URLs. These networks may be used by others for their research, and re-imported into GraphSpace for further collaborations
2.1 Sharing and organization
Groups lie at the core of GraphSpace’s functionality. Any GraphSpace user may create a group and add other GraphSpace users as members of the group using their email address. Group owners can also invite other users by sharing a signup link for the group. Groups are private by design. A member of a group may share any network he or she owns with the group (‘Sharing panel’ in Fig. 1). Only members of the group can view the shared network. Groups provide a level of privacy that is intermediate between the default (a network is visible only to its owner) and public networks (a network is visible to anyone; see ‘Public Networks’ below).
2.2 Searching across networks and within a network
A group may contain hundreds or even thousands of networks. The search interface in GraphSpace allows a user to search for networks that have a specific attribute or tag and contain one or more nodes using simple syntax (Supplementary Section S2 and Supplementary Fig. S5). When the user visits a matching network, GraphSpace highlights the nodes that match the search term. An identical search interface on the page for an individual network enables the user to refine the query. This interface also enables the user to search for specific edges within the network (Supplementary Figs S6–S8). GraphSpace allows the user to record the URL to the network with the search terms, for sharing or for further study.
2.3 Creating, saving and sharing layouts
GraphSpace exposes a subset of the layout algorithms available in Cytoscape.js (Franz et al., 2015) (the ‘Change Layout’ button in Supplementary Fig. S1(B)). These algorithms often do not create layouts that match biological intuition (e.g. the spring embedder layout at the top right of Fig. 1 and Supplementary Fig. S14). As an alternative, GraphSpace includes a powerful and intuitive layout editor that allows the user to quickly select nodes with specific properties (e.g. color and shape), to organize the selected nodes in defined shapes (e.g. a horizontal line or a filled circle) and to adjust the spacing between them (Supplementary Figs S1(A) and S18–S21). In addition, the user can manually move nodes into any configuration (Supplementary Fig. S16). The layout editor also allows users to manually change the visual properties of nodes and edges (Supplementary Figs S27–S32). Finally, the user can save the layout, make this layout appear by default and share it with other viewers of the network. Only users that have access (i.e. owned or shared via a group) can manipulate a network layout.
2.4 Filtering network nodes and edges
Graph algorithms may output networks where nodes and edges can be ordered, e.g. by path index (Ritz et al., 2016) or by edge weight/score. GraphSpace allows each node and edge to have a integer-valued attribute called ‘k’ that specifies the rank of the node or the edge (‘Filter nodes and edges’ panel in Fig. 1 and Supplementary Figs S23–S26). For any network that contains this attribute, the user can adjust a slider in the panel to view only those nodes and edges with values of k below a threshold. This interface element allows the user to unveil the network gradually in real time and gain intuition about how the network expands or contracts as this threshold changes.
2.5 Public networks
An owner may make a specific network or all networks associated with a tag public, e.g. as accompanying information for a publication. All public networks and tags can be accessed using a URL to GraphSpace. For example, several automated reconstructions of human signaling pathways (Ritz et al., 2016) appear at http://graphspace.org/graphs?tags=2015-npj-sysbio-appl-pathlinker.
2.6 Uploading networks
GraphSpace networks follow the JavaScript Object Notation (JSON) format supported by Cytoscape.js (Supplementary Section S2). GraphSpace allows a user to upload a network in three different ways:
Users may create their own JSON files and upload networks one-by-one at http://graphspace.org/upload.
Cytoscape users may export their visually coded networks from Cytoscape and import them directly into GraphSpace. This functionality works as follows. Since v3.1, Cytoscape has supported export of the structure of networks into JSON files and the visual elements of networks in JSON-based style files. GraphSpace can import both these types of files at http://graphspace.org/upload. Moreover, users can import a Cytoscape style file when they are editing the layout of a network in GraphSpace. In the future, we intend to develop a Cytoscape app to make the integration between the two systems seamless.
A comprehensive RESTful API and a Python module called ‘graphspace_python’ facilitate bulk uploads of networks. Both the API and the module are easy to integrate into software pipelines (Supplementary Section S2).
3 Conclusions
We originally created GraphSpace in order to facilitate a collaboration on budding yeast networks involving cell cycle regulation (Kraikivski et al., 2015; Poirel et al., 2013). Since then, GraphSpace has proven useful for other projects involving remote collaborators and in diverse contexts (Supplementary Section S3). Currently, GraphSpace supports more than 100 users who have stored more than 21 000 graphs (most of them private) containing a total of over 1.4 million nodes and 3.8 million edges.
Conceptually, GraphSpace serves as a bridge between visualization and analysis of individual networks supported by systems such as Cytoscape (Smoot et al., 2011) and the network indexing capabilities of NDex (Pratt et al., 2015). We anticipate that GraphSpace will find wide use in network biology projects and will assist in accelerating all aspects of collaborations among computational biologists and experimentalists, including preliminary investigations, manuscript development and dissemination of research.
Supplementary Material
Acknowledgements
We thank Craig Estep, Naveed Massjouni and Jaeil Yi for assistance with earlier versions of GraphSpace.
Funding
This work was supported by the National Science Foundation [DBI-1062380 to TMM], National Institute of General Medical Sciences of the National Institutes of Health [R01GM095955 to TMM, JP and JJT], National Cancer Institute of the National Institutes of Health [UH2CA203768 to KL and TMM], the Environmental Protection Agency [EPA-RD-83499801 to TMM], the M. J. Murdock Charitable Trust [2015288 to AR] and the National Science Foundation Graduate Research Fellowship [to CLP].
Conflict of Interest: none declared.
References
- Franz M. et al. (2015) Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics, 32, 309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraikivski P. et al. (2015) From START to FINISH: computational analysis of cell cycle control in budding yeast. NPJ Syst. Biol. Appl., 1, 15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel C.L. et al. (2013) Top-down network analysis to drive bottom-up modeling of physiological processes. J. Comput. Biol., 20, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt D. et al. (2015) NDEx, the network data exchange. Cell Syst., 1, 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz A. et al. (2016) Pathways on demand: Automated reconstruction of human signaling networks. NPJ Syst. Biol. Appl., 2, Article number 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E. et al. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics, 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.