Abstract
The discovery of the microRNAs, lin-4 and let-7 as critical mediators of normal development in Caenorhabditis elegans and their conservation throughout evolution has spearheaded research towards identifying novel roles of microRNAs in other cellular processes. To accurately elucidate these fundamental functions, especially in the context of an intact organism various microRNA transgenic models have been generated and evaluated. Transgenic C. elegans (worms), Drosophila melanogaster (flies), Danio rerio (zebrafish), and Mus musculus (mouse) have contributed immensely towards uncovering the roles of multiple microRNAs in cellular processes such as proliferation, differentiation, and apoptosis, pathways that are severely altered in human diseases such as cancer. The simple model organisms, C. elegans, D. melanogaster and D. rerio do not develop cancers, but have proved to be convenient systesm in microRNA research, especially in characterizing the microRNA biogenesis machinery which is often dysregulated during human tumorigenesis. The microRNA-dependent events delineated via these simple in vivo systems have been further verified in vitro, and in more complex models of cancers, such as M. musculus. The focus of this review is to provide an overview of the important contributions made in the microRNA field using model organisms. The simple model systems provided the basis for the importance of microRNAs in normal cellular physiology, while the more complex animal systems provided evidence for the role of microRNAs dysregulation in cancers. Highlights include an overview of the various strategies used to generate transgenic organisms and a review of the use of transgenic mice for evaluating pre-clinical efficacy of microRNA-based cancer therapeutics.
Keywords: microRNA, function, mouse, model systems, zebrafish, fruit fly, in vivo, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Mus musculus, GEMMs, transgenic, cancer
1. Introduction
The central dogma of molecular biology, laid down by Francis Crick in 1958, stated that the fundamental role of RNA molecules is to transmit the genetic code into proteins1,2. However, with the characterization of the first transfer RNA (tRNA) in 19653, additional RNAs emerged that violated the central dogma. For example, ribosomal RNAs (rRNAs)4, small nuclear RNAs (snRNAs)5 and small nucleolar RNAs (snoRNAs)6 are not translated into protein products like a messenger RNAs (mRNAs), but indirectly influence the process of protein synthesis. Such RNA molecules were collectively termed “non-coding RNAs”(Review7). While the novel roles performed by some non-coding RNAs were being assimilated as imperative molecular mechanisms, a study conducted to identify genes in heterochronic signaling incidentally led to the discovery of an additional non-coding RNA, a 22 nucleotide RNA molecule, lin-48. The discovery of lin-4 further defied the central dogma of molecular biology via an unprecedented mechanism and led to the establishment of a new class of small non-coding RNAs called “microRNAs”9–11.
Lin-4 was the first microRNA (miRNA) identified, which was determined to be indispensable for the normal development of Caenorhabditis elegans8. Functionally, lin-4 interacts with the 3′-untranslated region (3′-UTR) of the mRNA transcript lin-14, resulting in a marked repression of the lin-14 protein12. Unfortunately, due to limited knowledge in RNA biology at the time, lin-4 and its peculiar role were overlooked to be a worm-specific phenomenon. Seven years later, a second C. elegans miRNA, let-7 was discovered which encouraged further miRNA investigations13. It became apparent that let-7 was not only critical for the development of C. elegans, but is also evolutionarily conserved in other organisms, including humans14. Currently ~2,500 human encoded miRNAs have been identified, which are listed in a miRNA database, miRBase (http://www.mirbase.org/, Release 21)15. In addition to their identification, biochemical and molecular studies have determined that the canonical function of miRNAs is to post-transcriptionally regulate a repertoire of protein-coding mRNA transcripts, whereas a few miRNAs perform unanticipated or “non-canonical” functions (Review16,17).
Following the identification of these first two miRNAs, lin-4 and let-7, our understanding of miRNAs in normal physiology and diseased states, such as cancer (Review18) has advanced remarkably. Advancements in the field have been possible due to state-of-the-art technologies such as high throughput screening and deep sequencing, but majorly due to the development of appropriate in vivo model systems (Review19). Therefore, the focus of this review will be on the various model systems that have been instrumental in elucidating the roles of miRNAs in cancers and the technologies that have been extensively applied to generate these animal model systems. Briefly, the utility of in vivo models in evaluating the potential of miRNAs as therapeutic agents or targets for treatment of various cancers will also be touched upon.
1.1. MicroRNA biogenesis, mechanism of action and function
1.1.1. Biogenesis
1.1.1.1 Expression of miRNA genes
The transcription of miRNA genes is regulated by multiple mechanisms eventually dictating the level of expression of a particular miRNA in normal or diseased states (Reviews20–22).
(a) Regulation mediated by availability of transcription factors
A transcription factor can enhance or repress the expression of a miRNA gene depending on the availability of the particular factor (Reviews23,24). The prominent tumor-suppressor p53 which functions as a transcription factor for several genes, also enhances the transcription of miRNA genes. Examples of miRNAs that are directly induced by p53 include mir-34a and b/c25–28 and two miR-200 subfamilies, mir-200c/141 and mir-200a/200b/42929,30. In the case of mir-15a~16-1 and mir-107 a p53-indirect effect leads to upregulation of the miRNAs through activation of the host genes DLEU228 and PANK131, respectively. Conversely, MYC, a well-studied oncogenic transcription factor, negatively regulates the tumor suppressive miRNA, let-7a-132,33.
(b) Regulation mediated by genomic location of miRNA genes
(i) Location in the epigenome
Transcriptional activation or inactivation of specific miRNA genes is largely influenced by epigenetics. Such epigenetic regulation includes the proximity of the miRNA gene promoter to a CpG island, various histone modifications to the chromatin, and availability of factors that maintain and regulate expression from the epigenome. The expression of mir-127, a miRNA located near a CpG island is dependent on the methylation status of the promoter, implying epigenetic control on the expression of miRNAs34,35. MiRNAs also undergo massive upregulation when the DNA methylatransferases 1 and 3b (DNMT1, DNMT3) are downregulated34, lending further support to the role of DNA methylation in regulating miRNA expression (Reviews23,24).
(ii) Location relative to host genes
The origin of a miRNA gene from a specific chromosomal location impacts the extent of expression of the miRNA. In the context of other genes, miRNAs genes are either intragenic where they are embedded within a host gene, or intergenic if they are located between two genes on a chromosome. Expression of an intragenic miRNA is dependent on the expression of the host gene (Reviews23,24). MiR-126 is one such miRNA whose expression is concomitantly controlled by epigenetic regulation of its host gene EGFL736. Intragenic miRNAs are also regulated by canonical mechanisms that influence host gene expression such as transcription factor occupancy at the promoter of the host gene28,31 (Review37). MiRNAs that are not directly regulated by a host gene are still subject to nearby epigenetic influence. For example, let-7a-3 and miR-129-1 expression are dependent on a nearby region of the genome that is prone to altered methylation states during the onset of cancer. The proximity to this differentially methylated region severely impacts their expression24,38,39.
(iii) Regulation by miRNA copy number
A single mature miRNA can be expressed and processed from multiple loci in the genome. For example, three individual genes encoding human mir-7 produce an identical mature miRNA product40. Conversely, miR-21 is generated from a single genomic locus41,42. The advantages of miRNAs originating from various loci relative to one originating from a single locus is discussed in a later section.
(iv) Cancer-Associated Genomic Regions (CAGRs)
Specific regions in the human genome that are prone to amplification or loss upon the onset of cancers are referred to as Cancer-Associated Genomic Regions (CAGRs). CAGRs contain amplified or deleted miRNA and/or protein-coding genes. Many of these genetic aberrations are required for tumorigenesis. MiRNAs that are lost are frequently located in either fragile sites of the genome or regions susceptible to loss of heterozygosity (LOH). For example, the mir-15a~16-1 cluster located in a fragile region of the genome at 13q14.3 is frequently deleted in Chronic Lymphocytic Leukemia (CLL) patients28,43,44. Whereas other miRNAs are commonly amplified in multiple cancers due to their location in fragile regions. For example, the 17q23-25 chromosomal region containing mir-21 gene, a commonly overexpressed miRNA in multiple cancers41 is an amplified CAGR (Reviews23,24).
1.1.1.2 Process of Biogenesis
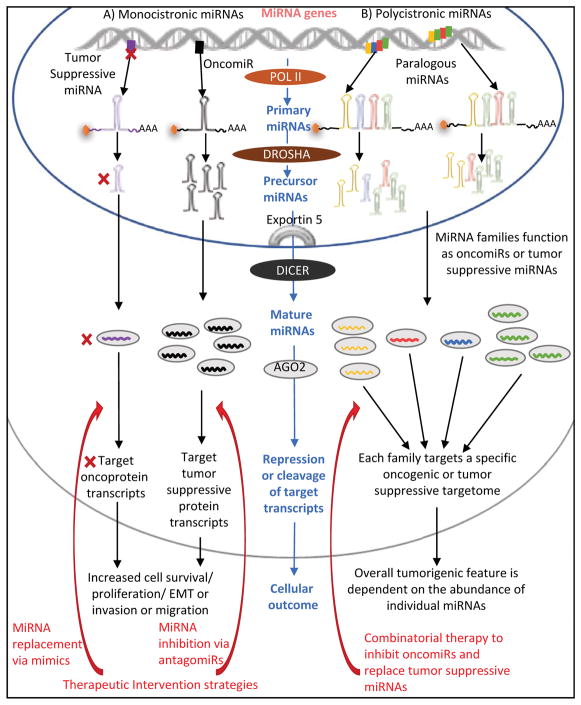
The primary miRNA (pri-miRNA) transcript produced as a result of RNA Polymerase II/III dependent transcription containing a single miRNA or as a cluster of miRNAs, produces a monocistronic or polycistronic pri-miRNA transcript, respectively45. Pri-miRNA transcripts form stem-loop structures flanked by single-stranded (ss) RNA ends. For RNA Polymearse II transcripts, the ends contain a canonical 5′ 7-methylguanosine cap and a polyadenylation signaling at the 3′-end. The size of a typical pri-miRNA can range from a hundred to a few kilobases in length and can originate from either intragenic or intergenic miRNA genes45,46 (Figure 1).
Figure 1. Overview of oncomiRs or tumor suppressive miRNAs encoded as monocistronic or polycistronic genes, their involvement in tumorigenesis, and potential use as miRNA-based cancer therapeutics.
(A) A monocistronic miRNA gene encodes a transcript containing a single primary miRNA. In cancers, one mechanism to alter the abundance of a mature miRNAs is through changes in transcription of the primary miRNA, where the expression of a tumor suppressive miRNA is downregulated, while that of an oncomiR is enhanced. A tumor-suppressive miRNA typically targets transcripts encoding oncogenic proteins, therefore miRNA replacement therapies using tumor suppressive miRNA mimics are currently being tested. OncomiRs on the other hand target tumor suppressor protein transcripts, and hence their inhibition via antagomiRs is also a potential miRNA-based therapeutic strategy. (B) Transcription of a polycistronic miRNA gene or a miRNA cluster results in a primary miRNA transcript containing multiple miRNAs. The duplication of a cluster, and expression of a more or less intact cluster from multiple genomic loci generates paralogous miRNAs. The resultant miRNAs from paralogues can be predominantly tumor-suppressive or oncogenic; however, their function is often largely context dependent – i.e. tissue-specific, temporally regulated, etc. The potential therapeutic strategy targeting miRNAs expressed from clusters depends on the abundance of the individual tumor suppressive or oncogenic miRNAs. Combinatorial miRNA therapeutics is a potential strategy currently being evaluated to combat tumorigenesis where the altered ratio between oncogenic/tumor suppressive miRNAs drives cancer development.
Processing of most pri-miRNAs begins with the association of the RNaseIII enzyme DROSHA and its cofactor Di George Syndrome Critical Region 8 (DGCR8) forming the microprocessor complex47,48. The microprocessor complex recognizes the ssRNA regions of the pri-miRNA sequence flanking the stem-loop and cleaves the ends. The resultant ~60–80 nucleotide long hairpin structure is referred to as a precursor miRNA (pre-miRNA)47,48. The pre-miRNA is translocated into the cytoplasm via Exportin-5 where another RNase III enzyme, DICER1 performs additional processing49,50. DICER1 cleaves the pre-miRNA to generate a ~22 nucleotide duplex molecule containing the guide and the passenger miRNA strands. Following cleavage, the DICER1-miRNA duplex associates with Transactivation-Responsive RNA-binding protein (TRBP) that mediates a stable transfer of the miRNA duplex into an Argonaute protein (AGO) 51,52. Selective incorporation of the miRNA duplex into either AGO1, AGO2, AGO3 or AGO4 is dictated by the presence of bulges or mismatches in nucleotides 9–12 of the duplex53,54. Incorporation of a miRNA duplex in AGO2, an AGO protein with endonuclease activity, results in selective cleavage of the passenger strand. The ssRNA guide strand is retained, and with AGO forms the mature miRNA-induced silencing complex (miRISC)55,56. On the other hand, the endonuclease activity deficient AGOs, AGO1, AGO3 and AGO4, generate a functional miRISC by binding to the guide strand and separating the passenger strand based on thermodynamic instability. The released passenger strand is shunted for further degradation57 (Biogenesis reviews20,22,46, Argonautes reviews54,58).
1.1.2. Mechanisms of action and functions of miRNAs
(a) Incorporation into miRISC and targeting
The well-established role of functionally active miRISC is to negatively regulate transcription of the target protein-coding transcripts. The canonical mechanism by which miRISC performs its function depends on the extent of complementary binding between the 5′-end “seed region” of the miRNA, the 3′-UTR of the target mRNA transcript, and the enzymatic activity of the AGO protein17,54,59,60. Perfect complementarity between the seed sequence, nucleotides 2–7 of the guide miRNA strand, and the target results in either degradation or translational repression of the target. The fate of the target transcript is dictated by whether the incorporated AGO displays catalytic activity or not, and whether additional complementarity occurs between the target and the miRNA. If a catalytically active AGO is incorporated into miRISC and the binding between the miRNA and the target are complementary between nucleotides 9–12 of the miRNA, then target cleavage will occur56,61. Translational inhibition in the absence of target degradation occurs when the miRNA binds to its target via partial complementarity (Review17,54,60) or if an endonuclease deficient AGO is included in mRISC. The partial complementarity between the miRNA and its target is highly conserved across species, providing the basis for a combinatorial interactome. A combinatorial interactome is the mechanism by which a single miRNA regulates multiple targets, thus simultaneously exerting its regulatory effects on various signaling pathways. For example, the very well-studied miRNA miR-21 simultaneously targets transcripts of proteins that regulate cell division and apoptosis, such as phosphatase and tensin homolog (PTEN)62, and programmed cell death 4 (PDCD4)63 to drive the process of tumorigenesis (Review42). Partial complementarity between the miRNA and the target also facilitates targeting of a single transcript mRNA by multiple miRNAs resulting in enhanced repression of the target. This resulting moderate-to-severe downregulation of target transcripts via a miRISC is the canonical mode of action of miRNAs (Reviews46,60,64).
(b) The role of family members in expression and targeting
The mechanism of action of miRNAs originating from a single locus, or a single mature miRNA originating from multiple loci remain largely unchanged. In these instances the same cohort of target mRNA transcripts is repressed42,62,65,66. However, miRNAs originating from several loci that contain subtle variations in their mature sequences 46,67 can exert their repressive functions on a larger repertoire of target transcripts. These miRNA families have acquired an evolutionary advantage relative to miRNAs with a single mature miRNA sequence. In addition to an increased pool of potential targets, the presence of multiple miRNA family members across the genome may allow at least one of the family members to evade transcriptional or epigenomic regulation. Therefore, the presence of multiple genetically distinct miRNA family members may prevent the depletion of an entire pool of a specific mature miRNA during the onset of a diseased state. For example, transcription of the twelve let-7 miRNA genes produces nine unique mature miRNA sequences that differ by at most three nucleotides (Reviews67,68). These minor nucleic acid changes can potentially alter the targeting affinity of the various family members (Figure 1 depicts miRNA family members). It has also become apparent that the promoter of let-7a3 resides in a heavily methylated region of the genome in normal cells resulting in low levels of let-7a3 in a normal cell. This is however not the case for the other let-7 family members which are highly expressed under normal conditions leading to a stably differentiated state of the cell. Nonetheless, upon the onset of tumorigenesis, the methylation state of cells become severely disrupted, and except for let-7a3 all the other let-7 isoforms become repressed24,38,67. Thus, the presence of multiple genetic loci encoding miRNA family members and slight variations in sequence between members adds an additional layer of complexity in the regulation of miRNAs in adverse cellular conditions.
Similar to the let-7 family, another well-studied family of miRNAs is the miR-34 family. The three canonical miR-34 family members include miR-34a that arises from a monocistronic locus, and miR-34b/c, which are expressed from a polycistronic transcript25,27,34 (Figure 1 depicts mono- and polycistronic miRNA genes). The function of miR-34 in normal physiology is well established as an inducer of cellular senescence and cell cycle arrest69 (Review27). Nevertheless, the advantage of multi-loci encoding miR-34 family members is that miR-34a and miR-34 b/c can be differentially regulated in tissue specific context70–72.Recent reports suggest that the tissue-specific expression of the miR-34 paralogues miR-449a/b/c add an additional level of complexity to the control of cancer cell proliferation, invasion, and migration73–75. Indeed, it was not until the paralogue mir-449a/b/c cluster was deleted in mouse models that the mir-34a, mir-34b/c double mutant displayed a phenotype76–78.
(c) The role of miRNA clusters and paralogous in targeting
Analogous to the overlapping role that miRNA family members have on gene expression, some paralogous clusters can also have overlapping roles while others have gained novel functions. A paralogous miRNA cluster is generated when a cluster undergoes duplication and translocates to another area of the genome (Figure 1 depicts miRNA paralogues). The resultant paralogue may express miRNAs similar to the parent cluster, located in relatively analogous positions79–81. One such miRNA cluster, miR-17~92 has been extensively studied due to its implication in the human developmental syndrome, Feingold disease. Loss of mir-17~92 results in severe skeletal abnormalities, and learning and developmental disabilities associated with Feingold disease82. However, similar developmental defects were not observed following the knockout of two mir-17~92 paralogous clusters– mir-106b~25 and mir-106a~36383. Additionally, the presence of a single wild-type mir-17~92 allele was capable of mitigating the deleterious effects of the loss of mir-17~92, despite the absence of its paralogues84. Collectively the three paralogous clusters encode a total of fifteen miRNAs that can be sub-classified into four miRNA families that are presumed to target analogous target transcripts. However in this case it can be inferred that alterations in certain nucleotides of the paralogues may have ceased their ability to compensate for mir-17~92 deletion80,83,84 (Figure 1 depicts miRNA paralogues). Therefore, in order to dissect the function of each miRNA in a family of miRNAs or within paralogues demands the generation of appropriate model systems to advance the field forward.
(d) Use of bioinformatics to elucidate miRNA function
Recently there has been a surge in the development of bioinformatic tools to precisely predict targets of a miRNA, or predict miRNAs that target a particular transcript19. Several computationally predicated miRNA-target pairs based on complementarity between the miRNA seed sequence and the 3′-UTR of transcripts have been experimentally validated8,46,60,67,85–87. Thus, implying that the developed algorithms are powerful in predicting relevant targets of novel miRNAs with unknown functions based on the canonical mode of action of miRNAs. However, there is increasing evidence that demonstrates some non-canonical mechanisms of actions of miRNAs17,64,88. For example, a passenger strand (also known as miRNA*) may not always be released for degradation once the miRNA: miRNA* duplex is incorporated into AGO. The miRNA* strand and may have an equal or a higher potential of becoming incorporated in an active miRISC17,89. Moreover, certain miRNAs modulate the expression of targets either positively or negatively by physically interacting with the 5′-UTR of transcripts90,91, the coding sequences (CDS) of transcripts91,92, or with gene promoters93. Mature miRNAs have also been experimentally validated to interact with other non-coding RNAs such as other miRNAs, long non-coding RNAs (lncRNAs)91,94, or circular RNAs (circRNAs)91,95. The canonical “seed-sequence” binding hypothesis has also been challenged as miRNAs can bind some targets independently of the seed sequence96,97. Data from these studies, along with the identification that certain miRNAs are contained in extracellular vesicles such as exosomes, has added an additional level of complexity in the mechanism by which miRNAs function, including non-autonomous mechanisms98.
1.2. MiRNA function and relevance in cancer
MiRNAs are important players in the normal developmental processes of animal species. As such, disruption in the normal physiological levels of certain miRNAs can lead to the development of multiple diseases, including cancers.
Detailed characterization of various miRNAs have revealed many important properties of these powerful post-transcriptional modulators in both normal and diseased states. In the context of cancers, certain miRNAs have been identified as functional “drivers of cancer”, whereas others are regarded as mere “passengers” in the tumorigenic process. A few known miRNA drivers of cancer become upregulated while others are severely downregulated or lost. The miRNAs that promote hallmarks of cancer are referred to as oncogenic miRNAs (oncomiRs). Those that prevent or reduce tumorigenesis are collectively called tumor suppressive miRNAs (Figure 1).
1.2.1. OncomiRs
OncomiR coding genes are frequently located in regions of the genome that are aberrantly amplified, or are subject to increased expression99. Increased expression of an oncomiR can be attributed to enhanced transcription of the oncomiR gene due to (i) availability of transcription factors, (ii) hypomethylation of its promoter, or (iii) its location in an intra- or intergenic region that is subject to increased expression in cancer via other mechanisms. OncomiRs can also be upregulated due to defects in biogenesis and/or stability of the mature miRNA22,23,46,64,84. The way by which an oncomiR typically functions is through targeting tumor suppressive protein-coding transcripts via canonical mechanisms, or through other less understood non-canonical mechanisms.
The first oncomiR to be validated was the miR-17~92 cluster (oncomiR-1). Overexpression of the cluster led to the development of lymphoproliferative and auto-immune diseases in mice via targeting of BIM, a pro-apoptotic protein100. Other targets of miR-17~92 that support the oncogenic role for this cluster include PTEN, E2Fs, and MYC. More detailed analysis of this cluster confirmed that the cell-type and context specific processing of individual miRNAs from the cluster adds an additional level of complexity to the function of the oncomiR80,101. Co-operatively the individual miRNAs processed from miR-17~92 functions as an oncomiR. However, miR-92 alone can antagonize an additional cluster member, miR-19 and also negatively regulates the oncogenic effects of c-Myc84,101,102. Because miR-19 alone can recapitulate the oncogenic role of the intact mir-17~92 cluster102,103, negative regulation by miR-92 suggest that miR-92 may be functioning as a tumor suppressor. The function of the miR-17~92 miRNA cluster is extremely intriguing and is currently under active investigation. Specifically, molecular roles and tissue specific effects of individual miRNAs of the miR-17~92 cluster are being determined in appropriate model systems83,84,102–105. These positive findings highlight the importance of carefully dissecting individual miRNAs from clusters so as to accurately identify the functions of each of the miRNAs contained within them.
Other miRNAs that have been well established as oncomiRs due to their implication in multiple solid tumors and hematological malignancies are miR-21 and miR-155106–108. Independent studies determined that overexpression of individual miRNAs such as miR-21 and miR-155 are sufficient to cause lymphoproliferative diseases. The mechanism by which miR-155 initiates cancer is not well understood, however, in leukemic mouse models it was determined that miR-155 promotes cancer progression, perhaps through gradual downregulation of its targets, SHIP and C/EBP109. In miR-21-dependent mouse models of lung cancer or pre-B-lymphoma, downregulation of the miR-21 targets PTEN and PDCD4 (negative regulators of cell death and cell-cycle, respectively) contributed to enhanced proliferation and growth42,62,63.
1.2.2. Tumor Suppressive miRNAs
About 50% of the miRNAs involved in repressing oncogenic protein-coding genes are located in or are close to fragile regions of the genome that are frequently deleted in cancer. Additional mechanisms elicited by cancer cells to repress tumor suppressive miRNAs include LOH, hypermethylation of the promoter, or the activation of transcriptional repressors that specifically downregulate the expression of the miRNA gene23,25,43,69. In the case of most tumor suppressive miRNA genes, identification of their role in development preceded their characterization as tumor suppressors. The most striking example of such a tumor suppressive miRNA is let-7. Let-7 was identified as a crucial differentiation factor in C. elegans prior to its identified role in tumorigenesis. Indeed, the development of cancer requires a reversal of a well-differentiated state to an undifferentiated state, thus, it is perhaps not surprising that downregulation or loss of let-7 family members is common in tumorigenesis67,68.
High levels of let-7 expressed from multiple genomic loci are expected in normal fully differentiated cells67,68. This results in repression of let-7 targets which are important oncogenes, such as KRAS, NRAS85,110, HMGA2110,111, LIN28112–115, and MYC116. A candid tumor-suppressive miRNA, such as let-7 has multiple loci of origin in order to maintain an appropriate level of the tumor suppressive miRNA as a defense mechanism against developing cancers67,117. However, since most let-7 isoforms are located in regions of the genome frequently deleted in cancer, let-7 is severely downregulated in multiple cancers43. One anomaly to this rule is the expression of let-7a-3 gene. In lung adenocarcinoma, epigenetic regulation of the gene encoding let-7-a3 results in hypomethylation of the promoter enhancing the accumulation of the pre-let-7a-3 transcript in lung cancer cells, and subsequently its potential oncogenic effects24,38.
Additional miRNAs that have been well established as tumor suppressors include miR-15a and miR-16-1, which were among the first miRNAs that demonstrated a negative correlation with the development of cancers43,118. Mir-15a~16-1 is located in 13q14.3, a region that is homozygously or hemizygously lost in over 50% of CLL cases118. Early reports correlated loss of mir-15a~16-1 with an increase in expression of BCL2, a pro-survival factor that normally prevents cell death119–122. In addition to targeting BCL-2, targets of miR-15a~16-1 include several cell-regulatory proteins, such as MCL1122,123, another BCL2-family member, CCND1121, a cell cycle regulator, and WNT3A121, a protein that induces several tumorigenic features including survival, proliferation, and invasion. Hence it is speculated that the simultaneous overexpression of these pro-survival onco-proteins, as a result of the loss of mir-15a~16-1 cluster may synergistically contribute to the development of cancers118,124. To further evaluate the tumor-suppressive potential of each miRNA in the miR-15a~16-1 cluster, modelling CLL in more sophisticated model systems is required.
1.3. Tools that advanced miRNA research
Research in the miRNA field exponentially increased following the discovery of the second miRNA in C. elegans, let-7. Let-7 was experimentally found to be homologous in a few species such as Drosophila melanogaster (fruit fly) and Danio rerio (zebrafish)14,67. However, additional computational evidence showed that let-7 was conserved further throughout evolution in all metazoans evaluated, and therefore, the discovery of let-7 was regarded as one of the most significant breakthroughs in the history of miRNA research18,67. Moreover, bioinformatic analysis corroborated with molecular studies also established that the let-7 target, lin-41 is conserved across species10,12,14,125. These studies were fundamental in establishing the proposed global mechanism of action of miRNAs, that miRNAs negatively regulate protein coding gene expression through miRNA:target interactions. The more recent utility of computational target prediction algorithms in miRNA research has further advanced the field in two ways: (i) a miRNA and its targets can be predicted in silico such that a biologically relevant target is putatively identified prior to in depth biochemical and molecular analysis, and (ii) computational methods spread awareness about similarities between various species. This knowledge can be used to identify appropriate, and perhaps simpler organisms that can be used as an adequate model system in miRNA research. In silico analysis combined with biochemical and molecular studies conducted in in vivo models, and in vitro from cells isolated from the in vivo models, or from human samples, have remarkably advanced our understanding of miRNAs in development and disease.
While the functional significance of miRNAs in various developmental stages of C. elegans was emerging, and the correlation between miRNA levels and disease such as cancer were being reported, a spontaneously occurring mouse model of CLL, the New Zealand Black (NZB) mouse was discovered. This discovery incidentally highlighted the importance of mouse models in delineating the role of miRNAs in cancers. It was determined that the endogenous loss of the mir-15a~16-1 cluster in this mouse led to the spontaneous development of CLL as the mouse aged126,127. This discovery followed by bioinformatic developments that described ~60% of miRNA loci remain conserved from mouse to humans spearheaded the use of murine models in miRNA research125,128. Indeed, from worms to mice many miRNAs and the components of the miRNA biogenesis machinery are conserved. The benefits of using simple model organisms such as worms, zebrafish and drosophila in miRNA-based biochemical studies have indirectly paved the path towards characterization of miRNAs that have relevance in cancer (Figures 4,5). Validation and clinically relevant studies become possible due to the increased use of very robust and durable mouse model systems (Figures 4,5). Therefore, in this review, a short background on the historical perspective of using various model systems will be followed by an detailed explanation on the current technology used to generate appropriate model systems. Some of the most successful models that have faithfully aided in uncovering the roles of various tumor suppressive or oncogenic miRNAs involved in cancer will be discussed. In closing, the pros and cons of the most widely used model systems in miRNA functional studies in cancer will be elaborated upon.
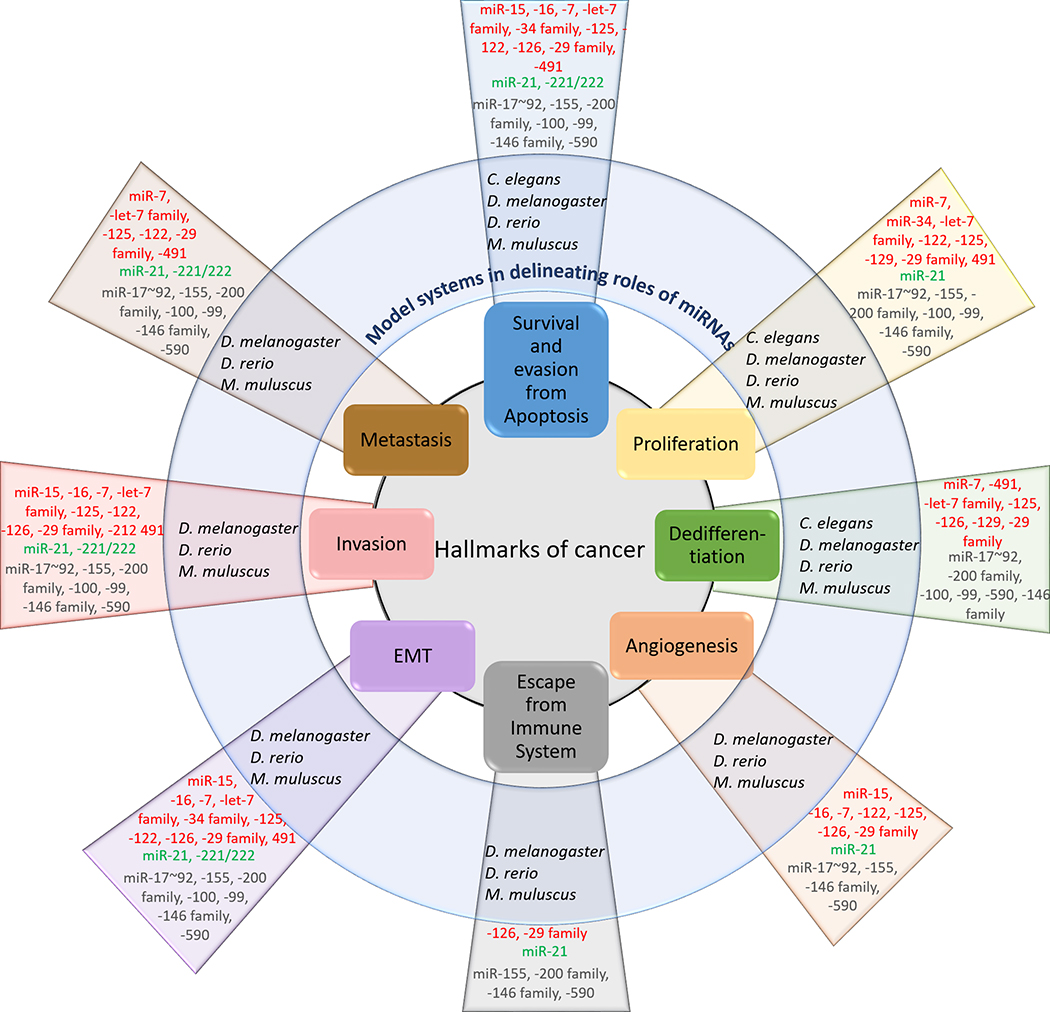
Figure 4. Functions of miRNAs in regulation of the hallmarks of cancer, identified via the use of in vivo model organisms.
Hallmarks of cancer are the cellular processes that become severely dysregulated upon the onset of a cancer. The various model organisms, owing to their endogenous properties have been utilized to delineate the functions of the enlisted miRNAs that mediate the specific cancerous feature. MiRNAs represented in red are bona fide tumor suppressive miRNAs, in green are oncomiRs, while in grey represent miRNAs that, depending on their context, can function as either a tumor suppressive or oncogenic miRNA.
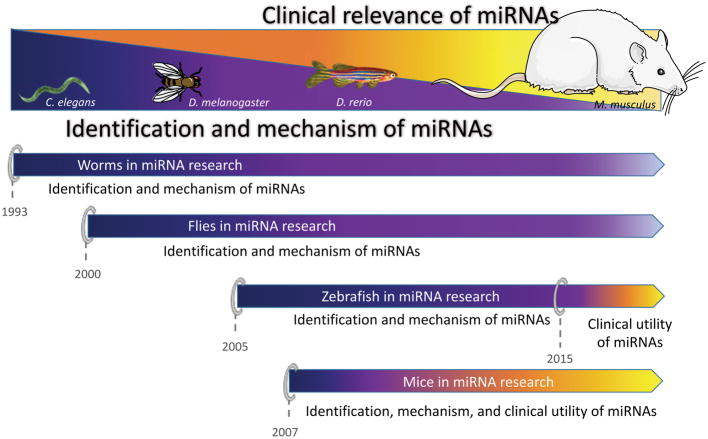
Figure 5. Utility of model systems in various aspects of miRNA research.
The model systems highlighted in this review include C. elegans (worms), D. melanogaster (flies), D. rerio (fish) and M. musculus (mice). All the enlisted model systems have contributed immensely towards the identification of evolutionarily conserved miRNAs, and delineating their mechanism of action in normal cells. The simple organisms, worms and flies do not develop cancers hence they have mainly been utilized to delineate miRNA biogenesis and function. However, the more complex systems, fish and mice, with an intact immune and angiogenic system have shown immense robustness in identifying the normal functions of miRNAs, and their roles in driving tumorigenesis. Since the discovery of the first miRNA, lin-4, in 1993, these in vivo systems have come a long way, and have demonstrated their applicability as pre-clinical model organisms to predict therapeutic relevance of certain miRNAs. Illustration created using graphics from Library of Science & Medical Illustrations (http://www.somersault1824.com/science-illustrations/) and Servier Medical Art (http://www.servier.fr/servier-medical-art).
2. Generation of model organisms and their use in miRNA functional studies
2.1. Caenorhabditis elegans
Although C. elegans do not develop cancer, they have been extensively used as model organisms to identify the functions of molecules and delineate pathways involved in normal cellular processes that are severely impaired in cancer, such as cell proliferation, differentiation, metabolism and death129–131. The completely sequenced C. elegans genome revealed that ~60% of its miRNAs have a human orthologue132. However, since C. elegans have a reduced number of miRNA family members for miRNAs that are conserved, studying miRNA function in C. elegans excludes redundancy as a hurdle to overcome. Moreover, C. elegans are self-fertilizing hermaphrodites that can produce a large number of genetically identical offspring. Additionally, their visually-traceable, well-organized transparent body make C. elegans an excellent model system129–131. Phenotypic and genetic screens, application of molecular techniques, and development of transgenic C. elegans have identified a few critical miRNAs, including their mechanism of action. Studies in C. elegans have also been an instrumental in understanding the molecular basis of miRNA biogenesis133–135. For example, the C. elegans ortholog of DICER, dcr-1, involved in RNA-mediated silencing, was identified as a critical component necessary for the processing of mature let-7 from its precursor molecule133.
2.1.1. Generation of transgenic C. elegans for use in miRNA functional studies
The most common mechanism used to generate transgenic C. elegans is transformation via either (i) microinjection or (ii) DNA bombardment. Transformation is widely used to ectopically introduce a transgene or fragment of DNA of interest into the animals to rescue a mutant phenotype or to over-express or silence a gene. The DNA is typically co-delivered with a scorable marker to determine successful transformation. For example, a scorable marker such as a promoter driven gfp::transgene allows for the selection of GFP positive worms when the promoter is positively regulated136.
Microinjection
Microinjection is a precise mechanism of introducing DNAs into the distal end of the worm gonad, which is composed of a syncytium of cells sharing cytoplasmic material. The transgene is usually a plasmid, cosmid, phage, Yeast artificial chromosome (YAC), or PCR product co-injected into the gonad with a scorable marker. Injected DNAs undergo efficient homologous recombination with each other to generate large extrachromosomal arrays. The extrachromosomal arrays contain multiple copies of the transgenic DNA that do not usually integrate into the genome, but can become inheritable by a fraction of the F1 generation. However, when integration is essential, random incorporation of the extrachromosomal DNA can be induced using radiation (gamma or UV) or through the use of a single DNA oligonucleotide that facilitates random integration and suppresses array formation 136,137.
DNA bombardment
DNA bombardment is a specialized technique where the transgene and co-injected transformation plasmid DNA mix is coated onto a gold microparticle and is bombarded into the worm using a gene-gun. The advantage of this technique is that it produces a considerable number of non-homologous integrants post-transformation136,138,139.
Apart from the above-mentioned techniques, additional newer strategies are now being applied towards generating transgenic C. elegans, such as the CRISPR-Cas9 system, discussed in a later section.
2.1.2. C. elegans as a model system for studying the function of miRNAs in cancer
The first discovered miRNA, lin-4 was identified in C. elegans via a conventional method of forward genetic mutagenesis screen, with the intent to mutate heterochronic genes resulting in phenotypic developmental defects. Lin-4 lof worms reiterate early phases of developmental fates (L1) at later stages of development, resulting in the absence of well differentiated adult phenotypes such as the adult cuticle, and a developed vulva. Development into adulthood in lin-4 null mutants was rescued by microinjection of lin-4 PCR products confirming that lin-4 lof was responsible for the mutant heterochronic phenotype. The phenotype of lin-4 lof mutants is completely opposite to that of lin-14 lof mutants attributed to the fact that the lin-4 miRNA negatively regulates the lin-14 mRNA transcript by binding to several complementary sequences in the lin-14 3′-UTR. When the lin-4 complementary sequences were mutated abnormally high lin-14 protein levels led to worms with retarded developmental phenotypes at late developmental stages8. In addition to suppressing lin-14, lin-4 also post-transcriptionally regulates the heterochronic genes, lin28 and hbl-1. Thus, lin-4 is regarded as a critical switch in nematodes for the development of well-differentiated adult structures140,141. Nevertheless, its homologs remained unidentified in higher organisms for many years, and lin-4 was presumed to have been lost during the course of evolution. But recent advances in bioinformatics have provided researchers with the tools needed to identify the human homolog for lin-4, miR-125. Analogous to the role of lin-4 in worm development, miR-125 in human cells targets LIN28 resulting in the acquisition of a differentiated state in normal cells142,143. In multiple human cancers, the two miR-125 family members, miR-125a/b are severely under-expressed, specifically in leukemia144 and melanoma145, and ovarian146, breast147, oral148 and thyroid149 carcinomas. In concert, ectopic miR-125 prevents cellular proliferation and migration in bladder cancer150, inhibits epithelial–mesenchymal transition (EMT) of triple-negative breast cancer cells151, and induces radiosensitivity and chemosensitivity in breast cancer and osteosarcoma, respectively152,153.
Seven years after the identification of lin-4 the second miRNA, let-7 was subsequently discovered, again through genetic analysis of the heterochronic pathway in C. elegans8,13. Mutants with severely retarded developmental phenotypes were identified and used to map the sequence of let-7. let-7 is expressed in the later stages of worm development, and regulates the transition of L4 larval stage worms to adulthood. A striking phenotype of let-7 lof worms is lethality as they fail to transition from larval to adults, at non-permissive temperatures. Most worms die due to bursting of the vulva. However, microinjecting the worms with PCR fragments containing the let-7 sequence rescued the progenies. Further analysis via northern blotting verified that let-7 did not encode a protein but instead encoded a 22-nucleotide RNA molecule in the rescued progenies. The offspring were selectively scored via co-injecting a GFP reporter plasmid, goa-1::GFP. The microinjected worms developed normally, supporting a role for the let-7 RNA product in inhibiting the bursting vulva phenotype. Due to the in silico finding that let-7 is complementary to the 3′-UTR of lin-41, target validations that let-7 regulates lin-41 were performed. A lacZ reporter gene was fused to the 3′-UTR of lin-41 and was co-injected with goa-1::GFP in a let-7 wild-type organism. Reduced luciferase levels and subsequent validation studies confirmed that let-7 exerts negative post-transcriptional regulation of lin-41. Moreover, overexpression of let-7 was shown to be implicated in premature development of C. elegans, therefore, acting as a critical developmental switch in worms13.
After the discovery of the first two miRNAs in C. elegans, it was speculated that nematodes had invented a novel mechanism to sequentially control their developmental course. This notion was nonetheless challenged by simple bioinformatic analysis conducted to investigate if let-7 was conserved in other organisms. Sequence analysis in Drosophila melanogaster (fruit fly), Danio rerio (zebrafish), Mus musculus (mouse), Gallus gallus (chicken), and Homo sapiens (Humans) confirmed that let-7 had been preserved throughout evolution14,125. Similarly, the let-7 target, lin-41 was also conserved14. Moreover, other detectable let-7 family members, miR-48, miR-84, and miR-241, were also identified as heterochronic miRNAs crucial for the temporal patterning of development in C. elegans 154. Both bioinformatic and biochemical analysis of let-7 family members revealed a considerable redundancy in target specificity between miRNA family members, suggesting a complex mechanism by which family members function. Although miRs-48, -84 and -241 lack human homologs, several let-7 loci are present in humans generating nine mature let-7 family members, therefore, there is a need to dissect the individual functions or functional redundancy between human encoded let-7 family members (Reviews67,68).
Despite the widespread acceptance of the role of let-7 in development, not much is known about let-7 in human embryonic development due to ethical constraints. However, shortly after the finding that miR-15a/16-1 is a bona fide tumor suppressive miRNA cluster, investigators evaluated the potential of let-7 as a tumor suppressive miRNA. Lof of let-7 in seam cells of C. elegans, leading to the inability of the seam cells to exit the cell cycle and become terminally differentiated, indicated that let-7 may play a role in maintaining the balance between cell differentiation and proliferation13,67. In human cancers let-7 expression is often reduced155, confirmed by the finding that multiple let-7 family members are located in fragile regions of the genome that are often lost in various malignancies43, signifying that let-7 is a tumor suppressive miRNA.
To delineate the molecular mechanism of let-7 in cancers, additional let-7 targets were computationally predicted in worms which identified let-60 as a putative target. Reporter plasmids containing the 3′-UTR of let-60 with or without the putative let-7 binding sites verified that let-60 was indeed a let-7 target. Moreover, let-7 mutant worms that usually die at non-permissive temperatures, when fed with silencing RNAs (RNAi) directed to let-60 survived. This was the first experimental evidence suggesting a novel role for let-7 in negatively regulating let-60 via a post-transcriptional mechanism. This breakthrough suggested that additional miRNAs may be dysregulated in cancer, other than the initially discovered mir-15~16 cluster lost in CLL. Indeed, let-60 is the human homolog of the RAS proto-oncogenes, proteins that are amplified and constitutively activated in multiple human cancers85. Multiple studies have since validated the negative regulation of RAS exerted by let-7 in various model systems and in cells derived from human patients, which has led to the advancement of let-7 family members as potential cancer therapeutics to target such oncogenes110,156–159.
With the emergence of the fact that miRNAs exist not merely as key developmental switches in nematodes, but are also crucial for normal cellular behavior in multiple species, including humans, severely dysregulated miRNAs are in the limelight for their roles in cancer initiation, development and progression. One such miRNA, miR-34, frequently lost in various cancers, has been associated with stress-response in normal cells, and with radio- and chemotherapeutic response in human cancer cells27,69,160,161. In vitro studies in wild-type and p53-mutated mouse and human cells determined that miR-34 was capable of suppressing the cell-cycle via a p53-mediated pathway25. However, the effects of miR-34 modulation in response to radio- and chemotherapies in vivo were unknown.
To determine the biochemical changes imposed by miR-34 and to record miR-34 dependent molecular observations in vivo, a mir-34-promoter::gfp transgene was constructed, and microinjected in C. elegans162. GFP signals from miR-34 transgenic animals were invariably identified in somatic tissues, including the vulval cells. Although, miR-34 expression was detected in the vulva in late larval stages and in the adult worm, miR-34 lof did not affect the development of vulva indicating that miR-34 was not a heterochronic miRNA162. Moreover, although miR-34 was upregulated in wildtype transgenic worms following exposure to radiation, miR-34 was not transcriptionally activated by the p53 worm homologue cep-1162. This observation in worms was contradictory to that predicted from human cell lines, and may be attributed to evolution of higher organisms161,162. Nevertheless, since miR-34 was upregulated in worms following exposure to radiation, the next big question was, how does miR-34 affect apoptotic and non-apoptotic pathways post-radiation exposure? C. elegans have been characterized as an excellent model to study both apoptosis in germline cells, and necrosis (non-apoptotic pathway) in vulva cells 163,164. Upon exposure of miR-34 mutant worms to radiation it was observed that the germline cells of the worms were unaffected, yet the vulva cells were radio-sensitized. Thus, miR-34 is an essential miRNA for induction of apoptosis, but is a suppressor of necrosis in nematodes. The data also provided evidence that the single miRNA, miR-34 can independently function as an apoptotic inducer, irrespective of p53 status162. Additionally, C. elegans express only one isoform of miR-34 while miR-34 in humans is encoded by four family members miR-34a/b/c and mir-449a73–75 indicating a possible evolutionary advantage towards enhancing miR-34 mediated tumor suppressive function. This notion requires further evaluation in an appropriate model system.
Conventionally, transformation has been utilized to generate transgenic worms, however, with new sophisticated techniques, such as transgenesis via the CRISPR-Cas9 system, targeting an endogenous locus has now become the preferred approach. The use of CRISPR-Cas9 system to knock out let-7 in C. elegans validated the role of let-7 in maintaining vulval integrity via the regulation of lin-41. This study exemplified the importance of ablating a miRNA to holistically understand its effects in normal physiology via reverse genetics165.
C. elegans have provided a platform for investigators to biochemically delineate the functions of some important miRNAs that are also crucial players in human cancers. However, during the time that experiments were being conducted in C. elegans, the use of computational knowledge to elucidate let-7 homology between species opened up the possibility of using additional simple organisms to identify the intricate functions of miRNAs. Conceptually, gathering functional knowledge for a single conserved miRNA in various species will aid in appropriately documenting the function of the miRNA, ultimately enhancing our understanding of the molecular biology of human diseases, such as cancer.
2.2. Drosophila melanogaster (fruit flies)
Fruit flies are a simple yet useful model system in cancer research, since cellular alterations in flies leads to the development of a few of the hallmarks of cancers, unlike in C. elegans. Although D. melanogaster lacks an intact angiogenic system, they have been very useful models to study cell survival and proliferation, cell death and apoptosis, and invasion and metastasis in the presence of normal cells. Additional advantages of using fruit flies as a model organism include i) the large brood size, one fly can lay ~100 eggs per day for about 20 days, ii) the series of conspicuous developmental stages that flies go through from embryogenesis through adulthood over the course of 10 days, and iii) the fact that genotypic changes are easily phenotypically tractable166.
2.2.1. Generation of transgenic D. melanogaster to model cancer phenotypes
Transgenesis procedures in flies rely on the flies’ endogenous transposition mechanism, P-element mediated transposition. The two-vector transposition system includes a helper vector referred to as the P-transposase vector, and the P-element transposon backbone containing the transgene and a scorable marker or a reporter vector (Figure 2). The transgene can be constitutive or inducible, or tissue-specific depending on the promoter (Figure 3). Simultaneously, the scorable marker that is expressed may be a gene that is easily detectable as a physical change – for example white eye color, small wings, GFP-wings, or lethality. The two vectors are co-injected into a generation zero (G0) embryo after which the transgene and the P-transposase are randomly incorporated into the genome. Using P-elements flies have been generated to express Gal4, a yeast derived transcription factor gene, in a tissue-specific manner. Gal4 associates with the UAS response elements to drive transcription of genes containing them. As such, UAS driven-transgenic lines have also be generated. Crossing UAS-transgene expressing lines with Gal4 expressing flies yield flies that express the transgene in the pattern of the Gal4 activator, i.e. expressed conditionally and in a tissue-specific manner (Reviews 166,167 (Figure 3).
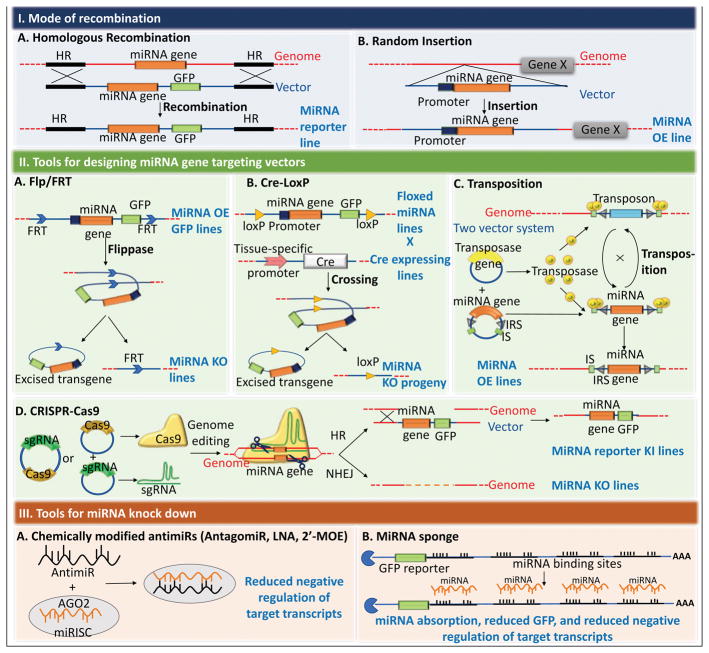
Figure 2. Strategies for the generation of targeting vector to knock-out miRNAs, and tools employed to knock-down miRNAs.
(I) Modes of recombination that govern the genomic location of incorporation of the transgene. (A) Homologous recombination (HR) allows a site-specific insertion of the transgene via crossing-over of the specific HR sites between the genomic site and the vector, in the presence of the enzyme recombinase. (B) Random insertion results in incorporation of the transgene at a random site in the genome. (II) Various gene-editing used for the generation of gene-targeting vectors (A) Flp/FRT system: Exogenously added or endogenously expressed Flippase (Flp) recombinase allows site-specific recombination with Flp recombinase target (FRT) sites flanking the transgenic miRNA gene. This results in knocking-out the targeted miRNA. (B) Cre-LoxP system: This system is analogous to the Flp/FRT system. The Cre-recombinase catalyzes site-specific recombination between two-LoxP sites flanking the miRNA of interest, resulting in its excision and miRNA knock-out. (C) Transposition is the mode of transgenic vector incorporation widely used in D. melanogaster. This mode of insertion of transgene utilizes multiple mechanisms to insert the transgene at a specific transposon site on the genome. The mechanism here explains a two-vector system. Transposase, the enzyme that facilitates transposition is encoded by the transposase vector, and the miRNA gene to be transposed at the transposon site in the genome is encoded by the second vector, flanked by inverted repeat sequences (IRS) and insertional sites (IS), that mediate the transgene exchange with a random transposon. (D) CRISPR-Cas9 system: Cas9-vector and an sgRNA vector are expressed in cells. The Cas9 endonuclease associates with the expressed sgRNA, which guides Cas9 to a homologous region in the genome to generate a double strand (ds) break. The ds break is repaired in an error-prone manner using Non-Homologous End Joining (NHEJ) or a targeting vector is inserted at the breakpoint via Homologous Recombination to generate either a miRNA knock-out or a knock-in of a miRNA and/or a reporter vector, respectively. (III) Strategies to exogenously or endogenously knock-down a mature miRNA (A–B). (A) Various chemical modifications on small miRNA-complementary oligonucleotides, double stranded or single stranded, have successfully been generated, to sequester functional mature miRNAs and inhibit their function. AntagomiRs are ssRNAs conjugated with cholesterol. Locked-Nucleotide Acid (LNA) are generated via the formation of a 2′, 4′methylene bridge in the ribose resulting in a stable bicyclic nucleotide. 2′-MOE are 2′-O-methoxyethyl phosphorothioate modified oligonucleotides. (B) A miRNA sponge depicted here contains multiple binding sites (6–8) for a specific miRNA in the 3′-UTR of a reporter vector. Sequestration of the miRNA results in negative regulation of the reporter and reduced regulation of the endogenous miRNA targets.
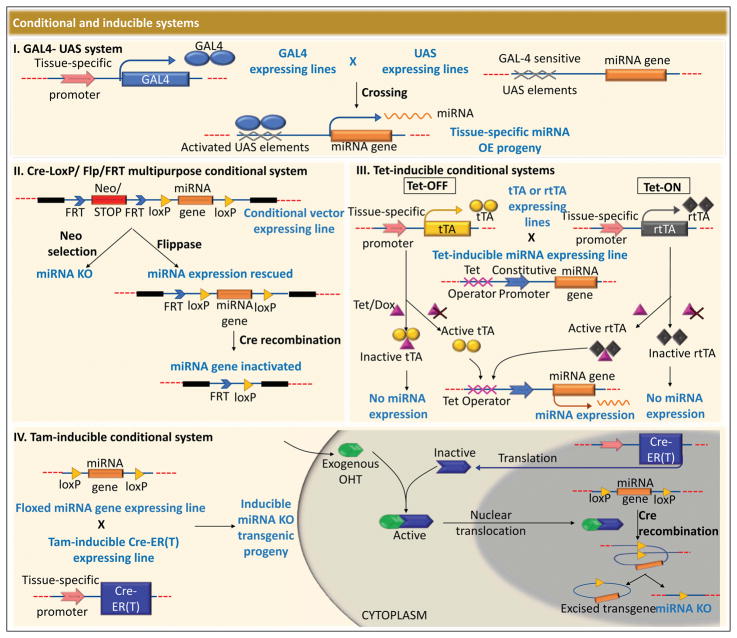
Figure 3. Conditional and inducible systems.
(I) The GAL4/upstream activating sequence (UAS) system (GAL4-UAS) is an inducible system has been utilized in the generation of transgenic flies and zebrafish models. Tissue-specifically expressed GAL4 lines are crossed with a line constitutively expressing the transgene encoded downstream of a UAS element, allowing GAL4 mediated activation of UAS in a tissue-specific manner. Specific binding of GAL4 to UAS element allows the transcription of the miRNA gene, resulting in a tissue-specific overexpression (OE) of the miRNA in the offspring. (II) A combination of Cre-LoxP and Flp/FRT is a powerful tool to generate a multi-purpose conditional and inducible targeting vector. In this case, expression of the Neomycin (Neo)/STOP cassette generates a knock-out first vector, inhibiting the expression of the downstream miRNA gene. However, expression of Flp leads to excision of the STOP cassette through recombination of the two FRT sites, rescuing the miRNA gene expression. This system allows miRNA functional studies first in the absence of miRNA expression, following which the effects of rescuing the miRNA can be evaluated. Finally, the effects of the loss of miRNA can be confirmed upon complete inactivation of the miRNA gene, achieved via expression of Cre. (III) The Tetracycline-inducible systems (Tet-OFF and Tet-ON) have proved to be very versatile in the generation of transgenic model systems. Tet-OFF is mediated via the expression of the Tet transactivator (tTA), whereas the Tet-ON system is dependent on the expression of the reverse tTA (rtTA). Lines expressing tTa or rtTA in a tissue-specific manner are crossed with a transgenic strain expressing the miRNA gene under the control of a constitutive promoter incorporated downstream of a Tetracycline activated element, the Tet Operator (TetO). tTa binds to the TetO in the absence of Tetracycline (Tet) or Doxycycline (Dox), leading to the constitutive expression of the transgenic miRNA gene, while the rtTA remains inactive and unable to bind to TetO in the absence of Tet/Dox inhibiting the expression of the miRNA gene. Upon the addition of Tet/Dox to the Tet-OFF system, tTa binds to Tet/Dox and the miRNA gene expression is turned off, whereas in the case of Tet-ON system, Tet/Dox binds to rtTa enabling it to induce the expression of the miRNA gene via direct interaction with TetO. (IV) Tamoxifen (Tam)-inducible conditional system is an extensively used inducible system in the generation of transgenic model organisms. A strain containing a floxed miRNA gene is crossed to a Tam-inducible Cre-ER(T) expressing line, generating an inducible miRNA knock-out strain. Cre-ER(T) is the Estrogen receptor (ER)-ligand binding domain fused to Cre recombinase, which remains inactive due to sequestration in the cytoplasm. However, upon exogenous addition of hydroxytamoxifen (OHT), the OHT-Cre-ER(T) complex translocates into the nucleus, and actively allows Cre-mediated recombination of the two LoxP sites to occur. The resulting Cre-LoxP recombination knocks-out the miRNA gene from the specific tissue expressing the Tam-inducible Cre-ER(T) vector.
Although the conventional method for generating transgenic flies via transposition has not become obsolete, newer strategies for more efficient transgenesis are gaining attention, such as the Flp-FRT, Cre-loxP166 and CRISPR-Cas9 systems168 (Figure 2). Such innovative and novel technologies have significantly advanced the development of transgenic animal models in miRNA functional studies, especially in the context of elucidating the function of miRNAs in various cancer-related events.
2.2.2. D. melanogaster as a model for studying the functions of miRNAs in cancer
After the discovery of let-7 in C. elegans, let-7 research was extended to D. melanogaster since flies only express a single let-7. In flies, let-7 is produced as a polycistronic pri-miRNA encoding miR-100, let-7, and miR-125, also conserved in humans148,169–171. To evaluate if the progression of juvenile flies to adulthood is spatiotemporally controlled via the expression of let-7, Gal4 was cloned into the let-7 locus removing let-7 and putting Gal4 was under the regulation of the let-7 promoter. Mutant pupae underwent normal morphogenesis into adult flies. However, the mutant adults displayed severe flight, motility and fertility defects170. This study demonstrated that although loss of let-7 in juvenile flies did not phenocopy the dramatic larval lethality observed in C. elegans, let-7 lof resulted in severe detrimental effects in the developing flies.
An important contribution to the miRNA field made through the use of D. melanogaster was achieved using the yeast Flp-FRT system to delineate the consequence of knocking-out a miRNA via homologous end recombination170,171. Using this system, the individual miRNAs from the let-7/mir100/mir-125 cluster were knocked out, resulting in data that supported that let-7 was sufficient for the normal development of D. melanogaster, whereas miR-100 and miR-125 were dispensable170. In humans the cluster exists as three paralogs, miR-100/let-7a-2/miR-125b-1, miR-99a/let-7c/miR-125b-2, and miR-99b/let-7e/miR-125a. These paralogues exist on chromosomal regions that are frequently lost in multiple cancers43,67. In a few recent studies, one or more of the individual miRNAs in each cluster has been confirmed to be negatively associated with cancers due to their potent tumor-suppressive functions67,172–174.
Until recently, transgenic D. melanogaster generation was accomplished using technologies such as the Flp-FRT or the GAL4-UAS systems that mediated successful knock-in or knock-out of miRNA genes in a spatio-temporal pattern. The transgenic D. melanogaster models so generated have contributed immensely towards identifying the roles of novel miRNAs in cellular functions of flies, and miRNAs that have a potential role in human cancers. However, the state-of-the-art technology for modeling human cancers in vivo is the CRISPR-Cas9 system, which in a recent study was successfully used to knock-out miR-219 and miR-315 in D. melanogaster168. Although the roles of miR-219 and miR-315 were not evaluated biochemically in the mutant flies, other investigations have reported that miR-219 is an essential neurodifferentiation factor175–177, and is suppressed in several human cancers, of which the reduced expression ultimately drives the acquisition of tumorigenic properties via diverse mechanisms178–181. Regardless, this pioneering study was the first to highlight the power of the CRISPR-Cas9 technology in developing transgenic D. melanogaster models to study miRNA lof, which will likely lead to new and innovative miRNA functional studies.
D. melanogaster have proven to be an instrumental models in understanding the fundamentals of miRNA biogenesis including studies identifying the molecular mechanisms of components such as dicer182 and locquacious (TRBP homologue)183. Indeed, studies in D. melanogaster validated that certain miRNAs remained conserved across evolutionary history, such as let-7184. However, due to the failure to find orthologous miRNAs for bantam185 and miR-14186, and the lack of common targets or overlapping pathways regulated by miRNAs such as miR-7, current miRNA studies are mostly being conducted in other model systems discussed in this review. This difference in miRNA conservation among flies and humans may be as a result of evolutionary canalization. Evolutionary canalization suggests that D. melanogaster encoded miRNAs are constantly evolving, acquiring distinct properties, and gaining robustness in their conspicuous functionalities40,65,66,187,188 (Review189).
2.3. Danio rerio (Zebrafish)
A model system that has been, and continues to be a major contributor in elucidating the functional role of miRNAs in both normal and cancerous cells, is zebrafish. Ever since in silico predictions identified let-7 as a conserved miRNA across species, additional miRNAs initially identified in zebrafish have displayed a striking homology in composition and function to miRNAs encoded by humans and other vertebrates14,125. D. rerio has been a useful model to conduct miRNA-based studies because of the homology with human-encoded miRNAs and additional features of zebrafish such as i) the small size, an adult zebrafish is about 2–3 cm in length, ii) the large brood size, a female lays about 100 eggs every 2–3 days, iii) the ability to easily visualize the forming embryos since fertilization is external, and iv) the short 3 month generation time of the progeny. Additionally, due to an intact angiogenic and immune system, and a well-developed organ system, successful engraftment of human cancer cells into zebrafish embryos has enabled tumor growth in the host microenvironment. Thus, zebrafish have also demonstrated to be successful model systems to assess tumoral response to anti-cancer treatments in vivo190,191. Taken together, although zebrafish is a simplistic model system, the compelling features of D. rerio have contributed towards the use of this organism to better understand the effects of aberrantly altering the endogenous levels of certain miRNAs190,191. Without question zebrafish has proven to be a successful model system that has led to the identification of multiple miRNAs via basic biochemical and molecular studies and holds immense potential to be utilized as a model to identify clinically relevant miRNAs (Figure 5).
2.3.1. Generation of transgenic D. rerio for use in miRNA functional research
There are multiple ways to generate transgenic zebrafish via microinjection at the one or two cell stage. In addition to its use in flies, the previously described Gal4-UAS system via P-element mediated transposition has been used to conditionally express transgenes in zebrafish166,167,191,192 (Figure 2 and 3). With regard to miRNA overexpressing lines, injection of a plasmid or a linearized transgene inclusive of the miRNA driven by a constitutive or conditional promoter192, or injection of miRNA mimics193 are common strategies used to overexpress a miRNA.
However, most of the zebrafish lines generated for studying the function of a miRNA have been created using a reverse genetics approach via knock-out or knock-down strategies. The tools used to knock-out miRNAs include i) Transcription Activator-Like Effector Nuclease (TALENs)194 or ii) the CRISPR-Cas9 system195. Whereas, most reported knock-down studies make use of i) synthetic anti-sense RNA-analogues called morpholinos196, or ii) heavily modified anti-sense RNA oligonucleotides, Locked-Nucleic Acids (LNA)197 (Review191). Additional approaches include combinations of the above-mentioned strategies incorporating inducible vectors, or the use of Cre-LoxP and Flp-FRT systems (Figure 2 and 3).
2.3.2. D. rerio as a model for studying the function of miRNA in cancer
Zebrafish serve as a model that is simple for understanding the basic mechanisms of miRNAs in human diseases such as cancer14,67, similar to C. elegans and D. melanogaster. However, D. rerio is evolutionarily closer to humans14,67. One of the founding miRNAs, let-7, is absent in zebrafish embryos during the first ~48 hours post fertilization. Overexpressing let-7 during this critical time in zebrafish embryos causes severe developmental defects. However, upon countering the overexpressed let-7 with morpholinos, the defects are reversed. The absence of let-7 during the first ~48 hours of development, but continuous expression until adulthood describes the heterochronic nature of let-7198. This observation made in invertebrate models is also typical in the case of vertebrates197,198. However, due to ethical concerns, similar experiments cannot be conducted in human embryos, therefore, the temporal nature of let-7 in humans has yet to be validated. Importantly, let-7 family members are highly conserved between zebrafish and mammals. There are eleven mature let-7 miRNAs expressed in zebrafish while in humans there are nine. Hence zebrafish is an excellent model to begin to dissect the individual contributions of miRNA family members. This observed conservation also suggests that let-7 family members may display redundancy in activities during vertebral development14,67. Although let-7 family members are implicated as tumor-suppressors, functional redundancy due to the presence and expression of other family members presents a challenge towards precisely discerning the function of each miRNA in the let-7 family. Currently, researchers using the CRISPR-Cas9 system have successfully generated knockouts of each of the let-7 family members in zebrafish. Since the knockouts are viable, these zebrafish transgenics may be useful to uncover the functions of individual members of the let-7 family. However, detailed biochemical characterization of the individual knockouts has yet to be performed in vivo199.
The finding that let-7 is conserved in zebrafish, but that let-7 does not have a role in early zebrafish development has prompted researchers to identify other miRNAs that contribute to zebrafish developmental. One groundbreaking contribution to the miRNA field using zebrafish was the elucidation that miRNAs are dispensable for cell-fate determination, despite their indispensable roles in highly related cell-fate specification, tissue, and organ-formation197. Using microarray analysis conducted on whole organisms at different stages of development, it was determined that most miRNAs are not expressed during the first 12 hours post fertilization; however, heightened expression is observed post organogenesis at 96 hours. More detailed in situ analysis showed that during development many miRNAs are expressed in a tissue-specific manner197. For example, in Dicer mutant fish with global miNRA downregulation, the importance of miR-430 in brain morphogenesis was demonstrated. Mimics of the miR-430 family were injected into one-cell stage Dicer mutant embryos, which successfully rescued the defective brain morphology of Dicer mutant fish193. This investigation also highlighted that mature miRNA mimics can form functional miRISC complexes in the absence of active Dicer, shedding light on an unknown mechanism of miRNAs biogenesis with immense applicability in therapeutics. Another significant conclusion drawn from this study was that miRNA expression can be discriminated based on specialized cell-types within a specific organ. For example, miR-217 and miR-7 are highly expressed in exocrine and endocrine cells of the pancreas, respectively197,200,201. This observation solidified the notion that miRNAs are not always required for cell-fate determination, but may also be crucial for cellular differentiation, tissue formation, and maintenance of tissue-identity in a whole organism.
With the findings from zebrafish research that miRNAs are involved in differentiation, tissue formation, and maintenance of tissue identity, zebrafish have indirectly contributed to the characterization of tumor-suppressive miRNAs that are frequently lost in poorly differentiated human cancer cells originating from a specific organ. For example, developmental studies performed in zebrafish demonstrated that miR-122 is a liver-specific miRNA that is only expressed during organogenesis197. Analysis of miR-122 in mouse models of liver cancer and in human hepatocellular carcinoma (HCC) patients verified that miR-122 is liver-specific and functions as a tumor suppressor; loss of miR-122 correlates with aggressive HCC and poor prognosis202. Similar, although slightly more controversial results were found for miR-126. MiR-126 was shown to be specifically expressed in differentiating endothelial cells during zebrafish organogenesis197. To dissect the function of miR-126 in endothelial cell biology, morpholinos knockdown of miR-126 confirmed that miR-126 is essential for several aspects of endothelial cell biology including cell survival, migration, tissue organization, and vascular integrity and stability203. This study suggests that enhancing the expression of miR-126 in endothelial cells may inhibit migration and invasion of tumor cells through a well-integrated endothelium by enhancing endothelial cell-differentiation. However, additional contradictory reports in multiple human cancers also suggest that overexpression of miR-126 in cancer cells may induce proliferation of tumor cells via increased vascularization of tumors204. Therefore, more comprehensive studies are required to further delineate the role of miR-126 in human cancers.
An additional miRNA identified in zebrafish that showed tissue specificity was miR-200197. MiR-200 was determined to be involved in the development of sensory organs of epithelial origin in both zebrafish and mouse205. Functionally, loss of miR-200 in zebrafish during organogenesis results in the generation of embryos with underdeveloped olfactory neurons, due to terminal differentiation of olfactory progenitor cells. This finding identified that expression of miR-200 is critical for the development of sensory epithelial in zebrafish through preventing differentiation205. Consequently, several studies confirmed that loss of expression of miR-200 family members is responsible for epithelial-to-mesenchymal transition (EMT) of multiple human cancer cells206.
In a more clinical context, a recent study used zebrafish to assess the emerging role of exosomes as vehicles for drugs delivery. Exosomes are an integral mode of cellular communication, and a mechanism that is often hijacked by cancer cells. Resent literature suggests that one of the major macromolecules contained within exosomes that mediate the cancerous phenotypes that exosomes promote are miRNAs207,208. Therefore, current preclinical research is being directed towards exploring the potential use of exosomes as in vivo drug delivery vehicles for conventional therapeutics and for delivery of therapeutic miRNAs. In this pioneering study, DiD labelled human brain cancer cells were xenotransplanted in the zebrafish brain ventricle to generate a model for primary glioblastoma-astrocytoma. Following which, exosomes derived from mouse brain endothelial cells were loaded with a fluorescently labelled drug –doxorubicin, and the loaded exosomes were injected into the cardinal vein of zebrafish embryos. In vivo fluorescent imaging confirmed penetration of the blood-brain barrier (BBB). Moreover, the therapeutic effects of doxorubicin were confirmed by a reduction in vascular endothelial growth factor (VEGF) mRNA levels, and a dramatic reduction in size of the xenotransplanted brain cancer cells209. These studies provide evidence that exosomes can be used as efficient drug delivery systems, at least in a simple model system and that the exogenously added non-self-exosomes do not produce an inflammatory response . This research also opens avenues to assess exosome-mediated delivery of miRNAs that have therapeutic potential, specifically as anti-cancer drugs using zebrafish as a model system.
Zebrafish is by far the simplest model system that contains a well-developed blood circulatory system that can help to recapitulate the presence of the tumor microenvironment. Therefore, D. rerio has an immense potential to contribute towards a better understanding of the role of miRNAs and exosomes in influencing the tumor microenvironment and their potential altered behavior in a model with an intact immune system. Thus, based on the contributions that zebrafish research had in unfolding the functions of a few crucial miRNAs in cancer, it can be accepted that zebrafish is a powerful model organism and further studies using zebrafish will likely result in more breakthroughs in the field.
2.4. Mus musculus (mouse)
Considerable progress in miRNA research can be attributed to the use of the previously described in vivo models. However, the ability to closely recapitulate human cancers in mouse models, and the conservation between humans and mice suggest that studies in mice provide the most meaningful insights on the role of miRNAs in the molecular pathogenesis of human cancers. Evolutionarily, ~60% of mouse miRNA loci are conserved between mouse and humans125,128. Additionally, mice are widely used because i) they are smaller in size than other mammals that are closer in evolutionary history to humans, ii) are relatively inexpensive and easy to maintain, and iii) they produce a fairly large number of offspring in a reasonable amount of time. The use of transgenic mouse models to evaluate the contribution of miRNAs in cancers has proved to be a robust and experimentally tractable system. Recent developments in the field have resulted in the generation of new mouse models that better recapitulate the clinical outcomes of patients treated with various therapeutics than previously used conventional mouse models (Reviews 210,211), and therefore current efforts are also being directed towards the development of useful mouse models to evaluate miRNAs therapeutics pre-clinically.
2.4.1. Generation of transgenic M. musculus for use in miRNA functional research
(i) Genetically Engineered Mouse Models (GEMMs) generated via transgenesis
The most simple and straightforward method to generate a genetically engineered mouse model (GEMM) entails microinjecting a transgene into the male pronucleus of a fertilized egg, followed by transplanting the fertilized egg into a pseudopregnant female to generate offspring expressing the randomly incorporated transgene, at variable copy numbers. Littermates that have successfully incorporated the transgene into the germline are screened and crossed to generate homozygous mice. Generation of transgenic mice using this method represents the first generation of GEMMs expressing a transgene that is expressed from an exogenous promoter or an enhancer element, resulting in constitutive or tissue-specific overexpression of the transgene212. Knockout models using this method are accomplished using DNA cassettes, or knockdown of an endogenous protein-coding or miRNA gene via transgenesis of shRNAs or miRNA sponges, respectively (Reviews210,213,214) (Figure 2).
(ii) GEMMs generated via homologous recombination
To exchange an endogenous gene via site-specific homologous recombination gene-targeting vectors containing the gene of interest and a selectable marker, flanked by homologous DNA sequences of insertion, are transfected in vitro into embryonic stem (ES) cells isolated from a blastocyst. ES cells are utilized to generate GEMMs because they are pluripotent and thus retain the capacity to generate into any cell type, including cells of the germline. Additionally, ES cells maintain a normal karyotype in culture post in vitro gene manipulation, and exhibit a higher rate of homologous recombination. The genetically engineered ES cells are then re-implanted into the blastocyst of a surrogate female to generate chimeric mice. Chimeric appearance of the animals’ coat is indicative of successful incorporation of the gene-targeting vector. Chimeras are bred to generate germline transmitted GEMMs. GEMMs with knock-in, knock-out, or conditionally overexpressed genes are successfully generated with this method (Reviews210,213,215)
2.4.2. Strategies utilized to generate gene-targeting vectors
A variety of strategies have been employed to fine-tune the design of gene-targeting vectors that are used to recapitulate human cancers in multiple model systems. The application of gene-targeting vectors used to uncover the in vivo functions of miRNAs involved in the development of multiple human cancers have undeniably advanced this field of research. The most common and current technologies used to design gene-targeting vectors for generating GEMMs via homologous-recombination of these vectors into ES cells, are described in this section.
i) Cre-LoxP system
The Cre-LoxP system is derived from the bacteriophage, Coliphase P1. Cre, cyclization recombinase is a 38-kDa site-specific DNA recombinase that specifically recognizes the 34-bp sites of LoxP, locus of X-over of P1, enabling site-specific recombination. The interaction of Cre with paired LoxP sites results in excision or inversion of a DNA fragment, depending on the same or opposite orientations of the LoxP sites, respectively. LoxP sites can be located in cis surrounding a specific gene, or can be located in trans, where the two LoxP sites are located in separate areas of the genome. One of the advantages of using the Cre-LoxP system is the ability to manipulate the transgene in vivo. The Cre-LoxP system can be utilized to knock-out or knock-in a transgene constitutively, or conditionally with both spatial and temporal control. Incorporation or excision of a reporter gene cassette, such as LacZ or GFP can be included to indicate successful loss or gain of function of the gene. To generate a tissue-specific conditionally expressing transgenic model either a tissue-specific Cre expressing line is crossed with a constitutively floxed line, i.e. LoxP-transgene-LoxP line, or Cre is expressed in the specific tissue of the floxed line via administration of Cre expressing lentivirus or adenovirus (Reviews192,210,213,215,216) (Figure 3).
ii) Flp-FRT system
The Flp-FRT system uses the yeast Saccharomyces cerevisiae derived flippase (Flp) recombinase that allows site-specific recombination with a pair of 34-bp Flp recombinase target (FRT) sites that flank the transgene or a reporter gene cassette. This system is analogous to the Cre-LoxP system, and has made marked contributions towards generation of GEMMs (Reviews214,215,217)(Figure 3).
iii) Inducible systems
An inducible system is a tremendously powerful technology that enables a researcher to precisely control the expression of a transgene that can allow for a more accurate recapitulation of human cancer development and progression. Moreover, knock-out GEMMs generated using inducible systems as opposed to the conventional knock-out of a developmentally essential gene, bypass the potential lethal consequences that may be observed at early developmental stages. Inducible systems also allow for evaluating oncogene addiction in tumor maintenance, and effects of oncogene ablation on tumor progression. An inducible transgene can be generated by multiple mechanisms, however, in this review, a few of the widely used inducible systems to generate GEMMs will be focused on.
a) Tet ON/OFF system
The Tetracycline (Tet)-inducible system is typically used to turn a transgene on or off. The Tet technology is a binary system that includes tetracycline controlled transcription factors (Tet transactivator (tTA) or reverse tTA (rtTA)) and an operator sequences of the bacterial Tet Operon (TetO). TetO is fused upstream of the transgene, and when crossed with a tTA expressing line, a Tet-OFF circuit is generated such that the tTA is unable to bind TetO in the presence of tetracycline (Tet), or the less toxic derivative doxycycline (Dox). On the contrary, the Tet-ON system is generated when a TetO-regulated transgene expressing line is crossed with a rtTA line. In this case the transgene function only occurs in the presence of Tet or Dox. Dox bound rTtA binds to the TetO inducing transgene expression. When Tet or Dox is withdrawn from the diet rtTA ceases to bind to TetO, terminating the downstream transgene expression (a similar but converse mechanism occurs with the Tet-OFF system). Therefore, the reversible nature of the Tet-ON/OFF systems under the control of a tissue-specific promoter, and/or in conjugation with Cre-LoxP or Flp-FRT has been especially instrumental in modeling spatially and temporally controlled gene expression in various model organisms (Review213,215) (Figure 3).
b) Cre-ER(T) system
Another powerful tool that has wide applications in reversibly controlling transgene expression is the Cre-ER(T) system. The Cre-ER(T) system uses a mutated Estrogen receptor (ER)-ligand binding domain fused to Cre recombinase. The Cre-ER(T) fusion protein is expressed constitutively, but remains sequestered in the cytoplasm unless hydroxytamoxifen (OHT) is added. OTH allows Cre-ER(T) to translocate into the nucleus where it acts on the transgenes. Thus, the Cre-ER(T) system can allow for gene expression in either a tissue-specific manner or constitutively via local or systemic administration of OTH, respectively. When a Cre-ER(T) line is bred with a line containing a LoxP flanked gene of interest, the Cre-ER(T) fusion protein can be temporally induced via OHT resulting in translocation of Cre-ER(T) into the nucleus to exert homologous recombination at that specific site (Review213,215) (Figure 3).
The above mentioned Cre-LoxP and Flp-FRT systems have been created in complex combinations with the inducible systems to generate gene-editing vectors that are not only spatially and temporally controlled, but also enable reversible expression of the gene of interest (Figure 3). A few examples of mouse models that have been used to study the function of miRNAs in cancers have successfully been generated using these technologies. Specific use of these systems are highlighted in the following section.
2.4.3. Using M. musculus for miRNA functional studies in cancer
The first evidence of a mouse that spontaneously developed a cancer homologous to humans was the New Zealand Black (NZB) mouse strain that developed CLL. Similar to the molecular alterations occurring in human CLL patients, these NZB mice had lost the mir-15~16 cluster43,124,126,218. This discovery was the founding premise of miRNA involvement in cancer and demonstrated the power in using murine models to gain a better understanding of the contribution of miRNAs in carcinogenesis. Moreover, using this naturally occurring model of CLL, it was determined that exogenous delivery of miR-15~16 to NZB derived malignant CLL cell lines could reverse CLL phenotypes through the induction of apoptosis43,124,126,127,218. This investigation shed light on the importance of modeling cancers in appropriate organisms and on the use of mouse models to evaluate miRNAs with potential therapeutic application.
Discovering that loss of the miR-15~16 miRNA cluster in both mice and human patients was driving CLL, resulted in an exponential increase in the identification of miRNAs that are misregulated in cancer. This fueled in vivo studies to determine if these misregulated miRNAs had a significant role in promoting or maintaining disease. Initially focusing on the miR-15~16 cluster, both constitutive and conditional knock-out mouse models for mir-15a~16-1 were generated. A GEMM containing a floxed mir-15a~16-1 locus was crossed to mice constitutively expressing Cre to generate mir-15a~16-1+/−chimeric mice. To conditionally knock-out mir-15a~16-1 exclusively from B-cells, mir-15a~16-1flox/+ mice were crossed with CD19-Cre transgenic mice. Intercrossing the F1 chimeras generated constitutively null mice, or mice with mir-15a~16-1 deleted only in B-cells218. Both models demonstrated manifestations of CLL phenotypes, and cellular and molecular alterations in the models displayed a striking resemblance to human CLL, depicting the accuracy of transgenic mouse models to study the development and stage of miRNA-mediated cancer progression.
Following the generation of robust models used to evaluate the loss of expression of tumor suppressive miRNAs, the oncogenic potential of the first speculated oncomiR, oncomiR-1 was explored. OncomiR-1 is a polycistronic gene that gives rise to a single transcript containing seven miRNA precursors, commonly known as the miR-17~92 cluster101. In patients suffering from various hematologic cancers, the genomic locus of mir-17~92 was observed to be amplified, and has since been extensively investigated via bioinformatic and biochemical approaches to confirm its oncogenic property84,100,219–221. Although initially identified as a oncomiR in hematopoietic malignancies, miR-17~92 is also involved in the development of solid cancers such medulloblastoma222, and hepatocellular223 and malignancies of the lung224 and breast225. Due to its pleotropic role in various malignancies, modeling the cluster in mouse models was a high priority. Since the knock-out model of mir-17~92 produced via crossing floxed mir-17~92 with Actin-Cre resulted in mice that suffered post-natal lethality due to severe birth defects83, multiple conditional knock-outs and overexpression GEMMs were generated to investigate the oncogenic role of miR-17~9284,100,104. Targeted overexpression of the entire miR-17~92 cluster, specifically in B-cells using B-cell specific Ig-heavy chain promoter Eμ-enhancer, resulted in severe B-cell lymphomas and leukemias104. To elucidate roles for the individual miRNAs in the cluster, transgenic mice were generated that overexpressed each individual miRNA in the miR-17~92 polycistron. The results revealed that miR-19 is sufficient to exert the oncogenic potential of oncomiR-184. MiR-92 overexpression, on the other hand, counters the effects of miR-19 in oncogenesis via a feedback mechanism102,103. Through the extensive use of mouse models, it is now evident that there exists a complex but delicate balance between miRNAs that are simultaneously expressed, but that may function individually as either potent oncomiRs or tumor-suppressive miRNAs. The balance in expression of such antagonistically functioning miRNAs likely plays a very critical role in maintenance of normal cellular physiology.
Very few phenotypes have been observed in mouse models following altered expression of a single miRNA. Indeed, overexpression of the miR-17~92 cluster or loss of the miR-15~16 cluster can both promote tumorigenesis, but in both cases, multiple miRNAs were altered. The miRNA field was further revolutionized following evidence that overexpression of either miR-21 or miR-155 is sufficient to induce tumorigenesis without the contribution of other oncogenic alterations106,107. For the first time, it was proven that hematologic cancer maintenance is dependent on a single oncomiRs, such that reducing expression of the miRNA results in cancer regression. In these studies, a transgenic vector expressing the pre-miRNA was placed downstream of a floxed STOP cassette under the control of a Tetracycline promoter, i.e. miR-21LSLtTA. In the absence of doxycycline, the miRNA is not expressed. To conditionally express the transgene in the lymphoid tissues the miRNA lines were crossed to Nestin-Cre mice resulting in STOP cassette excision, overexpression of the miRNAs, and development of pre-B cell lymphomas. However, upon impregnating the mouse chow with doxycycline, there was a rapid regression of pre-B cell lymphomas and an increase in survival. While the miR-21 study was the first to show that tumors can be addicted to changes in miRNA expression, the miR-155 report reconfirmed the role of miR-155 in lymphoma as initially reported by the Croce group106–108.
While these aforementioned studies focused on overexpressing or knocking out miRNAs independently of other genetic alterations, additional in vivo evidence has determined that miRNAs cooperate with both protein-coding genes and with each other and thus, suggests that miRNA-based therapeutics might have a place in the clinic. For example, miR-21 was found to enhance lung cancer in the inducible autochthonous model driven by the proto-oncogene, Kras, KrasLox-Stop-Lox(LSL)-G12D/+. In the absence of KrasG12D expression, miR-21 was insufficient to induce oncogenesis in the mice. However, following KrasG12D transgene expression, significantly more tumors were observed, compared to the KrasG12D/+ control mice226. Similarly, in the pancreatic autochthonous KRASG12D/+ model it was suggested that miR-21 may be involved in pancreatic cancer development via a multi-step process227. Confirmation that miR-21 was essential for pancreatic cancer maintenance was verified in animals that were orthotopically transplanted with pancreatic ductal adenocarcinoma cells (PDACs). A single dose of intratumoral administration of lentiviral vectors expressing antisense-miR-21 severely impaired tumor cell growth via onset of necrosis due to miR-21 depletion228.
The use of protein-coding transgenic models have also contributed to the miRNA field, specifically to evaluate miRNA-based therapeutics. For example, the KRASG12D/+ autochthonous lung cancer model was used to evaluate the tumor-suppressive roles of miR-34a and let-7b, and to assess their therapeutic efficacies. To this end, KrasG12D/+ mice were tail-vein injected with synthetic formulations of each of the miRNAs (miR-34a or let-7b), or orthotopically administered adenoviral-encoded let-7a. The resulting regressed tumors demonstrated that each of the individual tumor-suppressive miRNAs could act as potential therapeutic agents157,159,229. Following this, additional autochthonous NSCLC mouse models have confirmed that combinatorial RNA therapeutics produce even greater effects than treatment with an individual miRNA, some of these studies include the use of miR-34 combined with let-7b156, or miR-34 and an siRNA targeting Kras230.
The above studies proved the tumor suppressive role of let-7 through the use of exogenous let-7, however, the endogenous tumor suppressive activity of the let-7 family in a transgenic model was only recently demonstrated. To delineate if let-7 suppresses the Myc-driven tumorigenesis process, let-7g and Myc were simultaneously overexpressed using a triple transgenic, liver-specific, tet-off system. Let-7g was successfully overexpressed despite the high levels of Lin28 by innovatively cloning the mature let-7g sequence into a miR-21 stem-loop, retaining the miR-21 loop, which prevents Lin28B-mediated processing inhibition of let-7 family members. Myc overexpression led to tumorigenesis in the absence of let-7g expression; however, proliferation and growth were markedly reduced when let-7g was overexpressed. This study also showed that let-7 is transiently repressed in tissues undergoing repair and regeneration as the cells require enhanced proliferation. Further, through evaluating a conditional liver-specific knock-out of let-7b and let-7c2, investigators proved that reduced let-7 levels resulted in higher liver mass relative to control, due to increased mTOR signaling activity. This study demonstrates that let-7 is expressed in liver tissue, likely to suppress the development of liver cancer, but that controlled balance of let-7 levels is the key to maintaining the regenerative capacity of liver231.
Multiple studies have identified let-7 as a potent tumor suppressor, and the consequences of lof of let-7 results in loss of cellular differentiation, increased proliferation, and tumorigenesis. However, with the growing evidence that let-7 is an important tumor suppressor, the mechanisms involved in lof of let-7 has become a subject of interest in the field. Early studies evaluating let-7 led to the intriguing finding that loss of mature let-7 is not always associated with changes in transcription of the let-7 gene, which encouraged characterizing let-7 at the post-transcriptional level. With the ground-breaking finding that RNA-binding proteins such as LIN28A and B selectively inhibit let-7 miRNA biogenesis112–115, in vivo studies were conducted to delineate the role of LIN28B in let-7 mediated tumorigenesis. Colon cancer cells constitutively expressing LIN28B were implanted into immunocompromised mice. Biochemical evaluation of the tumors indicated increased levels of endogenous Lin28B, which strikingly negatively correlated with let-7 levels in tumor cells relative to surrounding normal cells232. This in vivo correlative study, along with additional cell-based and molecular assays confirmed the negative feedback loop that exists between let-7 family members and Lin28A/B112,113,232–234. In transgenic mouse models expressing Lin28B under the Vil1 promoter, which drives expression specifically in the intestines, tumorigenesis was directly dependent on the loss of mature let-7233. High Lin28B negatively correlated with the levels of let-7, whereas rescuing mature let-7 levels led to reversion of tumorigenic phenotypes233. This study, followed by others suggests that miRNAs are downregulated by various mechanisms in the process of tumorigenesis25–27,33,233. For example, Myc, an important oncoprotein that upregulates the oncomiR-1 cluster (mir-17~92) is predominantly associated with widespread depression of miRNA expression in humans and in cells obtained from mouse models of lymphoma through direct interaction with miRNA promoters 33.
While both MYC and LIN28 are involved in downregulation of a subset of miRNAs, global miRNA downregulation was identified as a common feature in human tumors, which could not simply be explained by MYC and LIN28. Thus, experimental designs turned to evaluating major components of the miRNA processing machinery. In mouse models where the Dicer1 locus was floxed, Dicer1 was identified as a haploinsufficient tumor suppressor235. A single copy of Dicer1 was necessary for tumorigenesis. Interestingly DICER is also lost in human cancers, and similar to the mouse model, only one allele is deleted236. Prior to this work multiple studies suggested that genetic mutations in components of the biogenesis machinery, such as DROSHA, DICER, and XPO5 may severely dysregulate miRNAs leading to cancer237,238. However, with the use of robust murine models, the anticipated pathophysiological consequences of a disrupted biogenesis pathway on global miRNA depression resulting in tumorigenesis is now regarded as a hallmark of cancer. With the growing understanding that miRNAs are globally downregulated in cancers, innovative targeting vectors have been generated to directly ablate several evolutionarily conserved miRNAs in mouse models, and to clone in reporter constructs downstream of the endogenous promoter to identify both temporal and spatial expression patterns. The targeting vectors contained floxed pre-miRNA sequences that were placed downstream of a FRT flanked promoter-less LacZ reporter to generate LacZ-STOP(Neo)-floxed miRNA transgenic vectors. Embryos generated by crossing LacZ-Neo-flox mice with Actin-Cre animals were evaluated for LacZ expression patterns in various tissues. Data suggest that approximately one third of miRNAs exhibit a global expression pattern, whereas about two thirds of miRNAs demonstrate developmental stage or tissue-specific expression patterns. For example, miR-210 and miR-146a were undetected in earlier stages, but were expressed in the adult mouse, sub-compartmentalized in immune cells. This investigation re-confirms the spatio-temporal nature of miRNAs, as suggested by the conventional model organisms, C. elegans and D. melanogaster. The observations made from these reporter studies can also be extrapolated to appropriately select a Cre-mouse for tissue-specific ablation/expression of specific miRNAs. This idea was verified using mice expressing the transgene and crossing them to animals that temporally or constitutively expressed Cre, and phenotypic and developmental alterations were observed for a subset of miRNAs239,240. Overall, this strategy holds promise for uncovering the roles of independent miRNA family members in various cancers, demanding further characterization.
The advantage of using a multi-purpose targeting vector comes from the power to generate a conditional knock-out model of the miRNA of interest (Figure 2). The expression of the upstream STOP cassette in the LacZ-STOP(Neo)-floxed miRNA transgenic mouse post-embryonic stages, via Neomycin selection allows bypassing of embryonic lethality in the absence of the miRNA, without physically excising the miRNA transgene. Nevertheless, crossing the parental-transgene expressing strain with a constitutively expressed Flp-strain leads to miRNA-rescue in the progeny by excision of the FRT-flanked LacZ-STOP regions upstream of the miRNA transgene. Although these murine models have been successfully generated, they remain to be characterized, which may shed additional light on the loss of miRNAs and their re-expression in various tissues, and may aid in the quest to better understand the role of various miRNAs in tumorigenesis239,240.
To understand the role of miRNAs in promoting metastatic potential of cancer cells in vivo, in a model with an intact immune system, investigators have relied on the syngeneic mouse model. In a recent study, primary and metastatic tumors from the KrasLA1/+;p53R172HΔG autochthonous lung cancer model were harvested, cultured, and subcutaneously injected back into the same immunocompetent host. When xenografts of cells derived from metastasized tumors were implanted in the syngeneic mice the cells metastasized, whereas animals xenografted with cells from the primary tumor only produced localized subcutaneous tumors. Upon miRNA profiling, it was observed that the metastatic tumors expressed low levels of miR-200 family members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429), relative to the localized tumors. However, when the miR-200b~200a~429 cluster was overexpressed in the metastatically derived cells, only primary tumors were capable of growing as compared to control cells that metastasized to the lung, heart, liver, and kidneys241.
3. Conclusion
Since the discovery of lin-14, scientists have come a long way in delineating the roles of miRNAs in higher vertebrates, including humans. Although the biogenesis of miRNAs and their biological role in maintaining normal physiology of cells have been fairly well established, this is a rapidly growing yet constantly evolving field of research. Currently, efforts are being made to identify miRNAs that drive aberrant cellular events responsible for abnormal behavior of a cell, leading to pathogenesis of diseases such as cancer.
Initial insights into the miRNA world were furnished through the use of a simple, yet elegant model system, C. elegans. The miRNAs lin-4 and let-7 were identified through forward-genetic screens, that determined these miRNAs are temporally expressed and required for normal development of C. elegans8,12,13,67. Nevertheless, with the advent of transgenesis and the use of more complex systems with closer homology to humans, such as D. melanogaster, D. rerio and M. musculus, a greater understanding of miRNA biogenesis, function, and misregulation in disease has become evident. Advancements in bioinformatic, biochemical and molecular approaches have revealed that miRNAs are aberrantly expressed in multiple cancers, and that in many instances they function to promote and/or maintain the tumorigenic phenotype. The first miRNAs discovered to be involved in human disease, miR-15a/16-1, were determined to be downregulated in human CLL patients. Consequently, the discovery of NZB mouse that naturally developed CLL, and displayed severe downregulation of miR-15/16 expression resulted in the recognition that mice have the potential to serve as a robust model systems for miRNA functional studies in cancer118,126,127.
Soon after this novel discovery and the realization that the mouse genome is ~90% homologous to that of humans, potent methods to generate GEMM prospered. These pioneering studies in the miRNA field led to implementing intriguing strategies to generate efficient gene targeting vectors that can not only replace an endogenous gene of interest (in this case, a miRNA gene), but also control its expression spatially and temporally. Various GEMMs have since been generated that have made immense contributions in uncovering the functions of numerous miRNAs as global and specific tumor-associated miRNAs, and have also supported the transition of miRNAs into the clinic. Models with intact immune systems are also be extensively evaluated as they more faithfully recapitulate human tumorigenesis and thus, are more accurate for studying cancer and therapeutic response, especially for agents targeted to cells of the microenvironment (Reviews247,249–251).
D. rerio or zebrafish is a vertebrate that has benefitted the research community through experiments that have helped to define the role of miRNAs as developmental factors, supported by biochemical experiments conducted in these visually traceable optically transparent embryos. Moreover, embryonic lethality, due to altered expression of critical heterochronic miRNAs is easily observed as fertilization of the egg is external, whereas in mice, lethal embryos are rapidly reabsorbed, thereby obstructing evaluation of miRNAs involved in development (Review252). Although zebrafish have proven to be convenient model organisms for identifying miRNA involvement in development, they have also indirectly contributed to the understanding of various aspects of cancer, including the role of miRNAs in the development of cancers. While zebrafish do not develop cancer, many of the hallmarks of tumorigenesis such as proliferation, migration, differentiation and apoptosis are conserved. As such, miRNAs ascertained through zebrafish studies have been validated and characterized in human cancer cell lines and/or in mouse models that recapitulate human cancer more closely. Mouse models, like zebrafish have gained immense recognition in modeling various human cancers, since they belong to the class of vertebrates. However, the contributions made by invertebrates such as C. elegans and D. melanogaster have undeniably influenced this revolutionary area of research. C. elegans and D. melanogaster have been useful in dissecting the molecular machinery of miRNA biogenesis, and contributed towards the identification of miRNA-mediated alterations of cellular pathways. Such developments achieved in simple model systems have since been extrapolated to more complex in vitro and in vivo systems to understand the effect of disturbed expression patterns of critical miRNAs, leading to cancer initiation, development and progression.
Generation of transgenic mice have come a long way since using the conventional method of random integration of an exogenous DNA into the mouse genome, via non-homologous recombination. One major drawback with this method is the lack of specificity at the site of transgene incorporation and high propensity of off-targeting effects. On the contrary, the primary advantage of generating GEMMs via the contemporary methods of gene targeting vectors is that the endogenous gene becomes replaced by the transgene (contained in the targeting vector) via site-specific homologous recombination (Figure 2). This ensures that the transgene is incorporated precisely at a specific genomic location. This same method also allows for evaluating tissue-specific effects of the miRNA through integration of tissue-specific promoters in the targeting vector.
Spatially and/or temporally controlling the expression of a transgenic-miRNA has led to the discovery of miRNAs with tissue-specific or stage-specific roles in the development of cancers. A powerful contribution of GEMMs is the generation of autochthonous mice that spontaneously develop cancers at the true anatomical location once the conditional-mutant allele of a known oncogene is activated. These models, as well as multiple other GEMM and xenograft models have been utilized to uncover the contribution that miRNAs make in the presence of other genetic lesions, for example miRNA involvement in potentiating the process of tumor generation or regressing tumors growth. Thus, these pre-clinical models have applications in segregating miRNAs that may have potential therapeutic applications.
The importance of the immune system on tumorigenesis can only be appreciated using GEMM or syngeneic models. Injecting tumor cells derived from autochthonous mice into another mouse of the same genetic background has established that the ability of a tumor cell to become invasive and migrate requires a complete and efficient microenvironment, including an intact and functional immune system. Therefore, the advantage of using immunocompetent mouse models such as the autochthonous and syngeneic GEMMs over the immune-deficient xenograft models is that the former possesses an intact immune system recapitulating the natural scenario of tumor initiation, development, and progression. Each of the stages of tumor development typically require cancer cells to cross-talk with surrounding stromal cells, normal cells, and the immune cells. This is the possible reason why cancer cells show metastatic potential in syngeneic models, but often not in sub-cutaneous, or PDX xenograft mouse models (Table 1).
Table 1.
The various mouse models that have been used to identify miRNAs that contribute to tumorigenesis.
| Mouse Model | Immune status of mouse background | Generation Method | Applications |
|---|---|---|---|
| Spontaneous Models | Immune competent | Spontaneously occurring tumors127. | To identify miRNA genes that are lost or amplified in nature, resulting in the development of cancers. |
| Xenograft Models | Immune deficient mice – SCID/NOD-SCID, NOG/NSG, RAG | Sub-cutaneous injection of human tumor-cell lines106. | To evaluate in vivo oncogenic or tumor-suppressive potential of miRNAs. To determine delivery and efficacy of miRNAs that have therapeutic potential. |
| Orthotopic implantation of human tumor-derived cells or cell lines228. | The above applications. And, orthotopic xenograft mice also serve as a model for metastasis of tumor cells, therefore, they can be utilized to delineate miRNA functions in invasion and metastasis. | ||
| Patient-Derived Xenograft (PDX) Models | Immune deficient NOD-SCID mice | Primary tumor engraftment242,243 | To maintain tumor heterogeneity, and allow personalization of treatment. Additionally, evaluate exosomal miRNAs released from tumors, and evaluate miRNAs that may have therapeutic potential. |
| Genetically Engineered Mouse Models (GEMMs) | Immune competent | Autochthonous mouse156,230 – conditionally activated mutations in transgenic oncogenes that result in endogenous tumors evolving spontaneously from normal cells in the correct anatomical location | Evaluation of spontaneous tumors to identify miRNAs that drive tumorigenesis in normal cells. Evaluation of miRNAs with therapeutic potential in tumors arising in endogenous microenvironments. |
| Transgenic mouse – Random incorporation of transgene, to overexpress or knock-down/out a miRNA, in the genome. | To analyze the in vivo roles of oncogenic or tumor-suppressive miRNAs. | ||
| Conditional or inducible expression of gene targeting vectors, via homologous recombination106,240. | To analyze the tissue-specific or cancer-stage specific roles of oncogenic or tumor-suppressive miRNAs. | ||
| Site-specific recombination157,240 Can also be virus mediated – retrovirus, lentivirus, adenovirus |
The above, and to determine delivery and efficacy of miRNAs that have therapeutic potential. | ||
| CRISPR-Cas9 mediated knock-in or knock out244,245 | Has the potential to analyze the tissue-specific or cancer-stage specific roles of oncogenic or tumor-suppressive miRNAs, with greater efficacy relative to conventional site-specific recombination. | ||
| Syngeneic mouse models | Immune competent | Implantation of tumors cells derived from mice from the same strain of origin241 | Intact immune system, and microenvironment support the development of a tumor in a bona fide cancerous environment. This model aids in the identification of miRNAs that potentiate metastasis of tumors. |
| Humanized Mouse Models | Immuno deficient mice – NSG or NOG | Engraftment of human immune cells (peripheral blood mononuclear cell (PBMCs) or CD34+ cells)246,247 | Possible immune response in patients against a treatment can be evaluated in mice. Not utilized in pre-clinical studies of miRNA-based cancer therapies yet. |
| GEMMs expressing a human gene in mouse genome248 | Compensates for a gene that is missing in the mouse genome, to further study its role in oncology. MiRNAs that are not conserved can be evaluated. Has potential in human-specific miRNA functional studies. |
The most common strategy in the field to predict the function of a miRNA in cancer development is to first utilize the straight-forward and basic sub-cutaneous (SC) xenograft mouse model. Advantages of modeling human cancer in SC xenograft models include the ability of cells to grow inadvertently, develop into primary tumors in vivo, and the ease of tracking and measuring the tumors. Although the associated disadvantage is that the genetic, histological, and therapeutic responsiveness of the xenografted tumors is not comparable to the source human tumor. Moreover, the reduced propensity of SC xenografted cancer cell lines to metastasize in vivo owing to the lack of a conducive environment for the implanted cancer cells is perhaps another disadvantage that provides evidence that this model does not completely recapitulate an endogenous human cancer. Such models have immensely contributed towards a collection of preliminary evidence in understanding the effects of oncomiRs such as miR-21106, or tumor suppressive miRNAs such as let-7158 in tumorigenesis. Despite the fact that SC xenograft models demonstrate a response to therapeutic anti-sense oncomiRs or tumor-suppressive miRNA mimics, follow-up studies in more advanced models with intact immune systems are essential prior to clinical advancement 156,230,245.
On the contrary, an orthotopic model is generated when cancer cells derived from a specific anatomical location are implanted into the same location to develop primary tumors that are genetically and histologically more representative of human tumors. Orthotopic models also lack an intact immune system, similar to xenograft model systems; however, the advantage here is that the cancer cells are introduced in vivo in their original anatomical location where the tumor was harvested from. Thus, the interaction between cells of similar origin, in the correct anatomical location may provide a moderately conducive environment for the orthotopically developed cancer to metastasize, and respond to miRNA based therapeutics. Additionally, another xenograft mouse model system, the PDX model, when treated with certain drugs have shown to favor predictability of patient response to the same. PDX models stably maintain the heterogeneity of the engrafted human tumor in the mice for multiple passages, owing to the animals’ deficient immune system. Therefore, the potential of PDX models in pre-clinical evaluation of therapeutics, personalized-treatment development and useful biomarker identification is being extensively explored. The use of PDX models has recently been extended to the miRNA field. Exosomal miRNAs such as miR-21 and miR-1246 identified in PDX models of breast cancer are predicted as bona fide circulating miRNA biomarkers indicative of breast cancer in patients243. Moreover, the therapeutic potential of replacement of miR-100 has recently been evaluated in PDX models via targeted therapy of the oncogene, fibroblast growth gFactor receptor 3 (FGFR3), in FGFR3 driven PDX tumors242. Therefore, although it is becoming accepted that PDX models may be applicable to developing personalized miRNA-based cancer therapies, this model is at its infancy and requires more explicit evidence to be recognized as a faithful pre-clinical model system. With the recently accepted notion that accurate tumor progression, and therapeutic response requires an intact immune system, an emerging concept of using model systems to perform pre-clinical evaluations of miRNA-based therapies or other drugs is on the cutting edge. To this end, another mouse model that deserves a mention here is the humanized mouse model. A nude mouse is humanized by engraftment of human-donor derived immune cells. This model system is not yet in practice in miRNA functional research in cancers per se, but it may be beneficial in pre-clinical evaluation of miRNA-based therapeutics, which may eventually positively impact the time-span required for translating miRNA-based cancer treatments into the clinic.
Although advancements are being made to models for use in evaluating miRNA-based cancer therapeutics at the pre-clinical stage, many contributions in the field related to uncovering miRNAs that maintain normal archetype of a cell or miRNAs that drive the process of tumorigenesis emerged from the extensive use of GEMMs. Additionally, GEMMs have also made unparalleled contributions towards the discovery of miRNAs that confer therapeutic potential as replacement therapies or as therapeutic targets. While the technologies used to generate transgenic mice has advanced remarkably innovative strategies to generate genetically engineered pre-clinical models for miRNA-based therapeutic research requires attention. It is anticipated that additional use of the CRISPR-Cas9 system244 will support miRNA research by rapidly accelerating the process of generating GEMMs by robustly overexpressing or knocking-out a miRNA gene of interest. This will not only aid in the precise knock-out of a miRNA gene of interest to evaluate replacement therapies, but will also facilitate knock-in/out of discrete miRNA family members to finally dissect the functions of individual miRNAs that was not as easily achievable using conventional methods. The CRISPR-Cas9 system is superior in many ways to the now more traditional methods of generating GEMMs (Reviews244,245,250,253–255). Genetic manipulation of multiple genes can be simultaneously achieved in a single-embryonic cell, not requiring the laborious selection of embryonic stem cells post transgenic vector injection. The CRISPR-Cas9 system is capable of producing miRNA knock-in/out mouse models in four weeks, and therefore, has immense potential in miRNA functional delineation in multiple human cancers. Efforts towards generating model systems utilizing the CRISPR-Cas9 system targeting miRNA genes in vivo in the correct anatomical location of existing autochthonous or syngeneic models is required, which can further accelerate the fate of modeling human cancer efficiently and robustly in model organisms.
References
- 1.Crick FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 2.CRICK F. Central Dogma of Molecular Biology. Nature. 1970;227(5258):561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 3.Holley RW, Apgar J, Everett GA, et al. Structure of a Ribonucleic Acid. 1965 doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 4.Fox GE, Woese CR. The architecture of 5S rRNA and its relation to function. J Mol Evol. 1975;6(1):61–76. doi: 10.1007/BF01732674. [DOI] [PubMed] [Google Scholar]
- 5.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979;76(11):5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipowicz W, Kiss T. Structure and function of nucleolar snRNPs. Mol Biol Rep. 1993;18(2):149–156. doi: 10.1007/BF00986770. [DOI] [PubMed] [Google Scholar]
- 7.Cech TR, Steitz JA. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 9.Lau NC, Lim LP, Weinstein EG, Bartel DP. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. 2001 doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Ambros V. An Extensive Class of Small RNAs in Caenorhabditis elegans. 2001 doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of Novel Genes Coding for Small Expressed RNAs. 2001 doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 12.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 14.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 15.miRBase. 2015. [Google Scholar]
- 16.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions [mdash] beyond repression of gene expression. Nature Reviews Genetics. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 17.Cipolla GA. A non-canonical landscape of the microRNA system. Front Genet. 2014;5:337. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orellana EA, Kasinski AL. MicroRNAs in Cancer: A Historical Perspective on the Path from Discovery to Therapy. Cancers (Basel) 2015;7(3):1388–1405. doi: 10.3390/cancers7030842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinkraus BR, Toegel M, Fulga TA Radcliffe Department of Medicine UoOWIoMMOU, Radcliffe Department of Medicine UoOWIoMMOU, Radcliffe Department of Medicine UoOWIoMMOU. Tiny giants of gene regulation: experimental strategies for microRNA functional studies. Wiley Interdisciplinary Reviews: Developmental Biology. 2016;5(3):311–362. doi: 10.1002/wdev.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 21.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nature Reviews Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, Fabbri M. 4 – Epigenetics and miRNAs in Human Cancer. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- 24.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. Febs j. 2011;278(10):1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 25.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124(1):207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 27.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12(5):414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Fabbri M, Bottoni A, Shimizu M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. Jama. 2011;305(1):59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208(5):875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CJ, Chao CH, Xia W, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13(3):317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohlig L, Friedrich M, Engeland K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res. 2011;39(2):440–453. doi: 10.1093/nar/gkq796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Lin S, Li JJ, et al. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem. 2011;286(46):39703–39714. doi: 10.1074/jbc.M111.293126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 35.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379(3):726–731. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Zeng Y. Regulation of mammalian microRNA expression. J Cardiovasc Transl Res. 2010;3(3):197–203. doi: 10.1007/s12265-010-9166-x. [DOI] [PubMed] [Google Scholar]
- 38.Brueckner B, Stresemann C, Kuner R, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67(4):1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 39.Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Carthew RW. A microRNA Mediates EGF Receptor Signaling and Promotes Photoreceptor Differentiation in the Drosophila Eye. Cell. 2005;123(7):1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 41.Hirata Y, Murai N, Yanaihara N, et al. MicroRNA-21 is a candidate driver gene for 17q23–25 amplification in ovarian clear cell carcinoma. BMC Cancer. 2014;14:799. doi: 10.1186/1471-2407-14-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer. 2011;30(6):371–380. doi: 10.5732/cjc.011.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lerner M, Harada M, Loven J, et al. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16–1. Exp Cell Res. 2009;315(17):2941–2952. doi: 10.1016/j.yexcr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo j. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 47.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 48.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 49.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JE, Heo I, Tian Y, et al. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475(7355):201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawamata T, Yoda M, Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 2011;12:944–949. doi: 10.1038/embor.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Niu D, Carbonell A, et al. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nature Communications. 2014 doi: 10.1038/ncomms6468. Published online: 19 November 2014. doi:101038/ncomms6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cenik ES, Zamore PD. Argonaute proteins. Curr Biol. 2011;21(12):R446–449. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 55.Burroughs AM, Ando Y, de Hoon MJ, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8(1):158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 57.Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40(19):9850–9862. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakanishi K. Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip Rev RNA. 2016;7:637–660. doi: 10.1002/wrna.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee I, Ajay SS, Yook JI, et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19(7):1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17(4):438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411(11–12):846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 63.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27(31):4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 64.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 65.Zhao J, Tao Y, Zhou Y, et al. MicroRNA-7: a promising new target in cancer therapy. Cancer Cell Int. 2015;15:103. doi: 10.1186/s12935-015-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kefas B, Godlewski J, Comeau L, et al. microRNA-7 Inhibits the Epidermal Growth Factor Receptor and the Akt Pathway and Is Down-regulated in Glioblastoma. 2008 doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 67.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 68.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016;7(2):100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogt M, Munding J, Gruner M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458(3):313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 70.Balca-Silva J, Sousa Neves S, Goncalves AC, et al. Effect of miR-34b overexpression on the radiosensitivity of non-small cell lung cancer cell lines. Anticancer Res. 2012;32(5):1603–1609. [PubMed] [Google Scholar]
- 71.Nadal E, Chen G, Gallegos M, et al. Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res. 2013;19(24):6842–6852. doi: 10.1158/1078-0432.CCR-13-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanaka N, Toyooka S, Soh J, et al. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer. 2012;76(1):32–38. doi: 10.1016/j.lungcan.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 73.Lize M, Herr C, Klimke A, Bals R, Dobbelstein M. MicroRNA-449a levels increase by several orders of magnitude during mucociliary differentiation of airway epithelia. Cell Cycle. 2010;9(22):4579–4583. doi: 10.4161/cc.9.22.13870. [DOI] [PubMed] [Google Scholar]
- 74.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao A, Zhao Q, Zhou X, et al. MicroRNA-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells. Sci Rep. 2016;6:27346. doi: 10.1038/srep27346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Concepcion CP, Han YC, Mu P, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8(7):e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Comazzetto S, Di Giacomo M, Rasmussen KD, et al. Oligoasthenoteratozoospermia and infertility in mice deficient for miR-34b/c and miR-449 loci. PLoS Genet. 2014;10(10):e1004597. doi: 10.1371/journal.pgen.1004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song R, Walentek P, Sponer N, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510(7503):115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun J, Gao B, Zhou M, et al. Comparative genomic analysis reveals evolutionary characteristics and patterns of microRNA clusters in vertebrates. Gene. 2013;512(2):383–391. doi: 10.1016/j.gene.2012.09.102. [DOI] [PubMed] [Google Scholar]
- 80.Khuu C, Utheim TP, Sehic A. The Three Paralogous MicroRNA Clusters in Development and Disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica (Cairo) 2016 doi: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hertel J, Lindemeyer M, Missal K, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7(1):25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mogilyansky E, Rigoutsos I. The miR-17|[sol]|92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death & Differentiation. 2013;20(12):1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 86.Cho S, Jang I, Jun Y, et al. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41(Database issue):D252–257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.TargetScanHuman 5.2 Custom. 2015. [Google Scholar]
- 88.Seok H, Ham J, Jang E-S, Chi aSW. MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions. Mol Cells. 2016;39(5):375–381. doi: 10.14348/molcells.2016.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang KW, Kao SY, Wu YH, et al. Passenger strand miRNA miR-31* regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013;49(1):27–33. doi: 10.1016/j.oraloncology.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 90.Lee I, Ajay SS, Yook JI, et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. 2009 doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. 2008 doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 96.Loeb GB, Khan AA, Canner D, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48(5):760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin HC, Wani S, Steptoe AL, et al. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 2014;15(3):R51. doi: 10.1186/gb-2014-15-3-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fabbri M. TLRs as miRNA Receptors. 2012 doi: 10.1158/0008-5472.CAN-12-3229. [DOI] [PubMed] [Google Scholar]
- 99.Esquela-Kerscher A, Slack FJ. Oncomirs |[mdash]| microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 100.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olive V, Sabio E, Bennett MJ, et al. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. Elife. 2013;2:e00822. doi: 10.7554/eLife.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sandhu SK, Fassan M, Volinia S, et al. B-cell malignancies in microRNA Eμ-miR-17~92 transgenic mice. Proc Natl Acad Sci U S A. 2013;110:18208–18213. doi: 10.1073/pnas.1315365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Du P, Wang L, Sliz P, Gregory RI. A Biogenesis Step Upstream of Microprocessor Controls miR-17 approximately 92 Expression. Cell. 2015;162(4):885–899. doi: 10.1016/j.cell.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–U119. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 107.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(26):E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Costinean S, Sandhu SK, Pedersen IM, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114(7):1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 112.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 114.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283(31):21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 116.Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67(20):9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 117.Misso G, Di Martino MT, De Rosa G, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226(4678):1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 120.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 122.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105(13):5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ. 2015;22(1):6–11. doi: 10.1038/cdd.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 125.Lagos-Quintana M, Rauhut R, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raveche ES, Salerno E, Scaglione BJ, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109(12):5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scaglione BJ, Salerno E, Balan M, et al. Murine models of chronic lymphocytic leukaemia: role of microRNA-16 in the New Zealand Black mouse model. Br J Haematol. 2007;139(5):645–657. doi: 10.1111/j.1365-2141.2007.06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kato M, Slack FJ. microRNAs: small molecules with big roles –C. elegans to human cancer. Biology of the Cell. 2008;100(2):71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 130.Kirienko NV, Mani K, Fay DS. Cancer models in Caenorhabditis elegans. Dev Dyn. 2010;239(5):1413–1448. doi: 10.1002/dvdy.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kyriakakis E, Markaki M, Tavernarakis N. Caenorhabditis elegans as a model for cancer research. Mol Cell Oncol. 2015;2 doi: 10.4161/23723556.2014.975027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ibáñez-Ventoso C, Vora M, Driscoll M. Sequence Relationships among C. elegans, D. melanogaster and Human microRNAs Highlight the Extensive Conservation of microRNAs in Biology. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 135.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. Rna. 2004;10(10):1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Evans TC. Transformation and microinjection. 2006 [Google Scholar]
- 137.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo j. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157(3):1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Berezikov E, Bargmann CI, Plasterk RH. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 2004;32(4):e40. doi: 10.1093/nar/gnh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88(5):637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 141.Lin SY, Johnson SM, Abraham M, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4(5):639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 142.Liang L, Wong CM, Ying Q, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52(5):1731–1740. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 143.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25(21):9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sonoki T, Iwanaga E, Mitsuya H, Asou N. Leukemia. Vol. 19. England: 2005. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia; pp. 2009–2010. [DOI] [PubMed] [Google Scholar]
- 145.Zhang J, Na S, Liu C, Pan S, Cai J, Qiu J. MicroRNA-125b suppresses the epithelial-mesenchymal transition and cell invasion by targeting ITGA9 in melanoma. Tumour Biol. 2016;37(5):5941–5949. doi: 10.1007/s13277-015-4409-8. [DOI] [PubMed] [Google Scholar]
- 146.Lee M, Kim EJ, Jeon MJ. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. 2016;7 doi: 10.18632/oncotarget.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 148.Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased Expression of miR-125b and miR-100 in Oral Cancer Cells Contributes to Malignancy. Genes Chromosomes Cancer. 2009;48(7):569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Visone R, Pallante P, Vecchione A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26(54):7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 150.Zhao X, He W, Li J, et al. MiRNA-125b inhibits proliferation and migration by targeting SphK1 in bladder cancer. Am J Transl Res. 2015;7(11):2346–2354. [PMC free article] [PubMed] [Google Scholar]
- 151.Hong L, Pan F, Jiang H, et al. miR-125b inhibited epithelial-mesenchymal transition of triple-negative breast cancer by targeting MAP2K7. Onco Targets Ther. 2016;9:2639–2648. doi: 10.2147/OTT.S102713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang F, Yu D, Liu Z, et al. MiR-125b Functions as a Tumor Suppressor and Enhances Chemosensitivity to Cisplatin in Osteosarcoma. Technol Cancer Res Treat. 2016;15(6):Np105–np112. doi: 10.1177/1533034615618849. [DOI] [PubMed] [Google Scholar]
- 153.Metheetrairut C, Adams BD, Nallur S, Weidhaas JB, Slack FJ. cel-mir-237 and its homologue, hsa-miR-125b, modulate the cellular response to ionizing radiation. Oncogene. 2017;36(4):512–524. doi: 10.1038/onc.2016.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Abbott AL, Alvarez-Saavedra E, Miska EA, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9(3):403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 156.Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34(27):3547–3555. doi: 10.1038/onc.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 158.Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46(2):298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 161.Lacombe J, Zenhausern F. Emergence of miR-34a in radiation therapy. Crit Rev Oncol Hematol. 2017;109:69–78. doi: 10.1016/j.critrevonc.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kato M, Paranjape T, Muller RU, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28(25):2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weidhaas JB, Eisenmann DM, Holub JM, Nallur SV. A Caenorhabditis elegans tissue model of radiation-induced reproductive cell death. Proc Natl Acad Sci U S A. 2006;103(26):9946–9951. doi: 10.1073/pnas.0603791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5(3):435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 165.Ecsedi M, Rausch M, Grosshans H. The let-7 microRNA directs vulval development through a single target. Dev Cell. 2015;32(3):335–344. doi: 10.1016/j.devcel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 166.HJ VKaB. Transgenesis upgrades for Drosophila melanogaster. 2007. [DOI] [PubMed] [Google Scholar]
- 167.Brumby A, Richardson H. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005;5(8):626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- 168.RKSaU Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195(3):715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chawla G, Deosthale P, Childress S, Wu YC, Sokol NS. A let-7-to-miR-125 MicroRNA Switch Regulates Neuronal Integrity and Lifespan in Drosophila. PLoS Genet. 2016;12(8):e1006247. doi: 10.1371/journal.pgen.1006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22(12):1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.EEC, LAJ . Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang J, Yu M, Guan S, Zhang G, Cheng Y. Prognostic significance of microRNA-100 in solid tumors: an updated meta-analysis. Onco Targets Ther. 2017;10:493–502. doi: 10.2147/OTT.S122774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sun X, Zhang S, Ma X. Prognostic Value of MicroRNA-125 in Various Human Malignant Neoplasms: a Meta-Analysis. Clin Lab. 2015;61(11):1667–1674. doi: 10.7754/clin.lab.2015.150408. [DOI] [PubMed] [Google Scholar]
- 174.Rane JK, Erb HH, Nappo G, et al. Inhibition of the glucocorticoid receptor results in an enhanced miR-99a/100-mediated radiation response in stem-like cells from human prostate cancers. Oncotarget. 2016;7(32):51965–51980. doi: 10.18632/oncotarget.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hudish LI, Blasky AJ, Appel B. miR-219 regulates neural precursor differentiation by direct inhibition of apical par polarity proteins. Dev Cell. 2013;27(4):387–398. doi: 10.1016/j.devcel.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dugas JC, Cuellar TL, Scholze A, et al. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhao X, He X, Han X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65(5):612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lahdaoui F, Delpu Y, Vincent A, et al. miR-219-1-3p is a negative regulator of the mucin MUC4 expression and is a tumor suppressor in pancreatic cancer. Oncogene. 2015;34(6):780–788. doi: 10.1038/onc.2014.11. [DOI] [PubMed] [Google Scholar]
- 179.Shi JA, Lu DL, Huang X, Tan W. miR-219 inhibits the proliferation, migration and invasion of medulloblastoma cells by targeting CD164. Int J Mol Med. 2014;34(1):237–243. doi: 10.3892/ijmm.2014.1749. [DOI] [PubMed] [Google Scholar]
- 180.Lei H, Zou D, Li Z, et al. MicroRNA-219-2-3p functions as a tumor suppressor in gastric cancer and is regulated by DNA methylation. PLoS One. 2013;8(4):e60369. doi: 10.1371/journal.pone.0060369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 181.Xiong GB, Zhang GN, Xiao Y, et al. MicroRNA-219-5p functions as a tumor suppressor partially by targeting platelet-derived growth factor receptor alpha in colorectal cancer. Neoplasma. 2015;62(6):855–863. doi: 10.4149/neo_2015_104. [DOI] [PubMed] [Google Scholar]
- 182.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 183.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3(7):e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 186.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 187.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kalinowski FC, Brown RA, Ganda C, et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. 2014;54:312–317. doi: 10.1016/j.biocel.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 189.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 190.Geiger GA, Fu W, Kao GD. Temozolomide-Mediated Radiosensitization of Human Glioma Cells in a Zebrafish Embryonic System. Cancer Res. 2008;68(9):3396–3404. doi: 10.1158/0008-5472.CAN-07-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu S, Leach SD. Zebrafish models for cancer. Annu Rev Pathol. 2011;6:71–93. doi: 10.1146/annurev-pathol-011110-130330. [DOI] [PubMed] [Google Scholar]
- 192.Xiong F, Wei ZQ, Zhu ZY, Sun YH. Targeted expression in zebrafish primordial germ cells by Cre/loxP and Gal4/UAS systems. Mar Biotechnol (NY) 2013;15(5):526–539. doi: 10.1007/s10126-013-9505-4. [DOI] [PubMed] [Google Scholar]
- 193.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 194.Liu Y, Luo D, Zhao H, Zhu Z, Hu W, Cheng CH. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS One. 2013;8(10):e76387. doi: 10.1371/journal.pone.0076387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xiao A, Wang Z, Hu Y, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41(14):e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 197.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 198.Kloosterman WP, Wienholds E, et al. The Hubrecht Laboratory CfBG, 3584 CT Utrecht, The Netherlands. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Research. 2004;32(21):6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Narayanan A, Hill-Teran G, Moro A, et al. In vivo mutagenesis of miRNA gene families using a scalable multiplexed CRISPR/Cas9 nuclease system. Sci Rep. 2016;6:32386. doi: 10.1038/srep32386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Correa-Medina M, Bravo-Egana V, Rosero S, et al. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns. 2009;9(4):193–199. doi: 10.1016/j.gep.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 201.Goodwin D, Rosenzweig B, Zhang J, et al. Evaluation of miR-216a and miR-217 as potential biomarkers of acute pancreatic injury in rats and mice. Biomarkers. 2014;19(6):517–529. doi: 10.3109/1354750X.2014.944217. [DOI] [PubMed] [Google Scholar]
- 202.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48(4):648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 203.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. ScientificWorldJournal. 2010;10:2090–2100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Choi PS, Zakhary L, Choi WY, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57(1):41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5(3):115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.An T, Qin S, Xu Y, et al. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 2015;4:27522. doi: 10.3402/jev.v4.27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015 doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 209.Yang T, Martin P, Fogarty B, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32(6):2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kersten K, Visser KEd, Miltenburg MHv, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. 2016 doi: 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hidalgo M, Amant F, Biankin AV, et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. 2014 doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hanahan D, Wagner EF, Palmiter RD. The origins of oncomice: a history of the first transgenic mice genetically engineered to develop cancer. 2007 doi: 10.1101/gad.1583307. [DOI] [PubMed] [Google Scholar]
- 213.Cheon DJ, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95–119. doi: 10.1146/annurev.pathol.3.121806.154244. [DOI] [PubMed] [Google Scholar]
- 214.Coumoul X, Deng CX. RNAi in mice: a promising approach to decipher gene functions in vivo. Biochimie. 2006;88(6):637–643. doi: 10.1016/j.biochi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 215.Zhang J, Zhao J, Jiang WJ, Shan XW, Yang XM, Gao JG. Conditional gene manipulation: Cre-ating a new biological era. J Zhejiang Univ Sci B. 2012;13(7):511–524. doi: 10.1631/jzus.B1200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bouabe H, Okkenhaug K. Gene Targeting in Mice: a Review. Methods Mol Biol. 2013;1064:315–336. doi: 10.1007/978-1-62703-601-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Deng C-X. Conditional Knockout Mouse Models of Cancer. 2014 doi: 10.1101/pdb.top074393. [DOI] [PubMed] [Google Scholar]
- 218.Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 219.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 220.Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64(9):3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 221.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Murphy BL, Obad S, Bihannic L, et al. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013;73(23):7068–7078. doi: 10.1158/0008-5472.CAN-13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhu H, Han C, Wu T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis. 2015;36(10):1213–1222. doi: 10.1093/carcin/bgv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 225.Kim K, Chadalapaka G, Lee SO, et al. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2012;31(8):1034–1044. doi: 10.1038/onc.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.du Rieu MC, Torrisani J, Selves J, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56(4):603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 228.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21(5):986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72(21):5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wu L, Nguyen LH, Zhou K, et al. Precise let-7 expression levels balance organ regeneration against tumor suppression. Elife. 2015;4:e09431. doi: 10.7554/eLife.09431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee J-S, Rustgi AK. LIN28B Promotes Colon Cancer Progression and Metastasis. 2011 doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Madison BB, Liu Q, Zhong X, et al. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27(20):2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 235.Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23(23):2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lambertz I, Nittner D, Mestdagh P, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17(4):633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A. 2016;113(13):E1881–1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rakheja D, Chen KS, Liu Y, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19(R2):R169–175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Park CY, Jeker LT, Carver-Moore K, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1(4):385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gibbons DL, Lin W, Creighton CJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sun S, Wang Y, Zhou R, et al. Targeting and Regulating of an Oncogene via Nanovector Delivery of MicroRNA using Patient-Derived Xenografts. Theranostics. 2017;7(3):677–693. doi: 10.7150/thno.16357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18(1):90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc. 2014;9(8):1956–1968. doi: 10.1038/nprot.2014.134. [DOI] [PubMed] [Google Scholar]
- 245.Humphrey SE, Kasinski AL. RNA-guided CRISPR-Cas technologies for genome-scale investigation of disease processes. J Hematol Oncol. 2015;8:31. doi: 10.1186/s13045-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Humanized Mice in Tumor Studies. Charles River; 2017. [Google Scholar]
- 247.Ito R, Takahashi T, Katano I, Ito M. Current advances in humanized mouse models. Cell Mol Immunol. 2012;9(3):208–214. doi: 10.1038/cmi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Takehara S, Schulz TC, Abe S, et al. A novel transchromosomic system: stable maintenance of an engineered Mb-sized human genomic fragment translocated to a mouse chromosome terminal region. Transgenic Res. 2014;23(3):441–453. doi: 10.1007/s11248-014-9781-4. [DOI] [PubMed] [Google Scholar]
- 249.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 250.Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2017;9(2):137–153. doi: 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zhao S, Huang J, Ye J. A fresh look at zebrafish from the perspective of cancer research. J Exp Clin Cancer Res. 2015;34:80. doi: 10.1186/s13046-015-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579(26):5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 253.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wen WS, Yuan ZM, Ma SJ, Xu J, Yuan DT. CRISPR-Cas9 systems: versatile cancer modelling platforms and promising therapeutic strategies. Int J Cancer. 2016;138(6):1328–1336. doi: 10.1002/ijc.29626. [DOI] [PubMed] [Google Scholar]
- 255.Guernet A, Grumolato L. CRISPR/Cas9 editing of the genome for cancer modeling. Methods. 2017 doi: 10.1016/j.ymeth.2017.03.007. [DOI] [PubMed] [Google Scholar]