Abstract
Fibrotic disorders of the renal, pulmonary, cardiac, and hepatic systems are associated with significant morbidity and mortality. Effective therapies to prevent or curtail the advancement to organ failure, however, remain a major clinical challenge. Chronic kidney disease, in particular, constitutes an increasing medical burden affecting >15% of the US population. Regardless of etiology (diabetes, hypertension, ischemia, acute injury, urologic obstruction), persistently elevated TGF-β1 levels are causatively linked to the activation of profibrotic signaling networks and disease progression. TGF-β1 is the principal driver of renal fibrogenesis, a dynamic pathophysiologic process that involves tubular cell injury/apoptosis, infiltration of inflammatory cells, interstitial fibroblast activation and excess extracellular matrix synthesis/deposition leading to impaired kidney function and, eventually, to chronic and endstage disease. TGF-β1 activates the ALK5 type I receptor (which phosphorylates SMAD2/3) as well as non-canonical (e.g., src kinase, EGFR, JAK/STAT, p53) pathways that collectively drive the fibrotic genomic program. Such multiplexed signal integration has pathophysiological consequences. Indeed, TGF-β1 stimulates the activation and assembly of p53-SMAD3 complexes required for transcription of the renal fibrotic genes plasminogen activator inhibitor-1, connective tissue growth factor and TGF-β1. Tubular-specific ablation of p53 in mice or pifithrin-α-mediated inactivation of p53 prevents epithelial G2/M arrest, reduces the secretion of fibrotic effectors and attenuates the transition from acute to chronic renal injury, further supporting the involvement of p53 in disease progression. This review focuses on the pathophysiology of TGF- β1-initiated renal fibrogenesis and the role of p53 as a regulator of profibrotic gene expression.
Keywords: Fibrosis, p53, Kidney, TGF-β1, Plasminogen Activator Inhibitor-1
Introduction
Sustained inflammation and repeated cycles of kidney injury/repair (or incomplete recovery) leads to tubular atrophy, progressive fibrosis, functional decline and, ultimately, organ failure [1–7]. Episodic acute injury (AKI) to the proximal tubular (largely S3 segment) epithelium is a major factor in the transition to chronic kidney disease (CKD) [e.g., 6–8]; patients who survive AKI have an increased risk of development of CKD [8]. Excessive accumulation of extracellular matrix (ECM; e.g., the fibrillar collagens, fibronectin) in the glomerular, interstitial and vascular compartments is accompanied by a significant decline in glomerular filtration rate and impaired epithelial regeneration [1]. In this regard, interstitial fibrosis is both a pathophysiologic hallmark feature and prognostic biomarker of end-stage renal disease (ESRD) [1,9–12]. The primary sources of ECM synthesis during interstitial fibrogenesis are activated fibroblasts or myofibroblasts [13,14]. Although controversial in origin, recent biomarker analysis and lineage-tracing studies suggest that this cell type-predictor of disease progression likely derives from FoxD1+ mesenchymal precursors (i.e., vascular pericytes and tissue-resident fibroblasts) with perhaps minor varying contributions from endothelial cells, completely or partially transdifferentiated tubular epithelia, and bone marrow fibrocytes [15–21]. The persistence of such activated fibroblasts is a critical factor in the initiation and development of renal disease where they likely participate in the silent scarring phase prior to development of significant organ dysfunction [22].
Signaling Transducers of the Renal Fibrotic Phenotype
Transforming growth factor-β1 (TGF-β1) drives the myofibroblastic phenotype, particularly in the context of a stiff microenvironment such as a fibrosing tissue. Signals generated from an increasingly non-compliant stroma, moreover, distort the latency constraints on TGF-β1 releasing the active TGF-β1 dimer facilitating interaction with its receptor complex to promote myofibroblast differentiation and/or retention [23] while activating cellular pathways that impact chromatin architecture and transcription of disease-relevant genes [24,25]. Elevated levels of TGF-β1 in the injured kidney, moreover, orchestrates a program of pathologic renal ECM synthesis and advancing fibrosis in response to diabetes, hypertension, ischemic or repeat tubular injury and urinary tract obstruction [e.g., 12,26–31]. Within hours after ureteral occlusion, for example, the affected kidney exhibits changes in hydrostatic forces and increased oxidative stress [32–34]. Tubular stretch, in turn, further stimulates TGF-β1 expression (>20-fold), increases the epithelial apoptotic index and leads to the development of an inflammatory inflitrate [30,35]. Non-resolving inflammation and continued interstitial ECM deposition accompanies tubular dilation and atrophy, nephron loss and scarring [9,11,13,27,37–41]. Maintenance of renal TGF-β1 expression in response to ischemic or obstructive stimuli, results in escalating tissue injury, impaired regenerative growth, and eventual loss of organ function [28,42,43].
Early findings suggested that renal disease onset and progression could be attenuated by blockade of TGF-β1 expression or function. TGF-β-neutralizing antibodies reduced trauma-initiated inflammation, tubular epithelial apoptosis, and fibrosis [30,44] while retrograde ureteral introduction of TGF-β1 antisense oligodeoxynucleotides or small interfering RNA (siRNA) blunted both collagen I mRNA expression and interstitial involvement [45,46]. Overexpression of latent TGF-β1, to minimize availability of bioactive TGF-β1, resulted in a decrease in both SMAD2/3 activation (the transcriptional effectors of canonical TGF-β1 signaling) and the number α-smooth muscle actin-positive cells (presumably myofibroblasts) in the injured kidney [47] consistent with the finding that SMAD3 knockout mice are protected from renal fibrosis [31,48]. A caveat regarding SMAD involvement in gene control, however, is that positioning of an activated SMAD complex on a target promoter requires repeat SMAD-binding elements (SBEs) and complicated, in part, by recognition of the increasing number of SMAD-interacting transcriptional partners [49]. For SMAD2/3, the most relevant SMADs in fibrotic disorders, these various co-factors contribute to the defined self-enabling, switch enhancer and derepression modes of SMAD-dependent transcription (Hill, 2016) suggesting a model of “contextual” signaling in the varied responses to TGF-β family ligands [50].
The repertoire of TGF-β-dependent non-canonical signaling contributors to normal and dysfunctional tissue repair is expanding and includes the three mitogen-activated protein kinase (MAPK) families (ERK, p38, JNK) as well as the Wnt/β-catenin, Jagged/Notch, Hedgehog, JAK/STAT, Hippo/YAP-TAZ, epidermal growth factor receptor (EGFR), p53, RhoA/ROCK/PTEN, Numb and Toll-like receptor (TLR) networks [e.g., 51–55]. There is, however, considerable pathway cross-talk [56]. Numb increases TGF-β1 expression and promotes a p53-dependent tubular epithelial G2/M arrest, a prominent profibrotic response in the injured renal epithelium [8,12,40], following ischemia/reperfusion or ureteral obstruction [57] while the Hippo/YAP-TAZ axis integrates mechanochemical and TGF-β/SMAD signaling as a function of YAP-TAZ phosphorylation [24,58]. A progressively non-compliant microenvironment, in fact, induces the YAP-TAZ, SMAD2/3-dependent expression of a subset of profibrotic TGF-β1 target genes including several collagens, plasminogen activator inhibitor-1 (PAI-1) and connective tissue growth factor (CTGF) [e.g., 24,59]. The impact of increasing biomechanical strain, as is likely encountered in a fibrosing tissue, however may well transcend just the YAP-TAZ system since mechanical stress regulates (as least in some cell types) several network “hubs” and their constituent genes by activating the TGF-β1, tumor necrosis factor α (TNFα) and p53 pathways [60,61].
Integration of TGF-β1-Activated p53 in Renal Fibrogenesis
Various species-, tissue- and cell type-specific cis-acting factors regulate the genomic program of fibrosis [62]. Recent findings, however, indicate a further layer of complexity to TGF-β1 signaling and implicate p53 in the transcriptional control of renal disease-causative genes (Figure 1A) [40,55,63]. p53 isoforms are involved in a subset of TGF-β1 responses attributable to, in part, interactions between phosphorylated p53 (p-p53) and SMADs to form transcriptionally-active multi-protein complexes [64–66]. Binding specifically involved the N-terminal MAD homology 1 (MH1) domain of SMAD2/3 and the receptor tyrosine kinase/Ras/MAPK cascade-phosphorylated N-terminus of p53 [63,66]. Increased p53S15 phosphorylation, accelerating renal damage and compromised organ function are evident following ureteral obstruction-, ischemia/reperfusion- or nephrotoxin-induced (e.g., cisplatin, aristolochic acid) injury as well as in the dysmorphic tubular epithelium and interstitial cells of renal allograft patients (Figure 1B,C) consistent with a role for p53 in promoting tubular cell apoptosis and proliferative inhibition [67,68]. Recent studies, furthermore, link epithelial growth arrest following both acute (e.g., due to ischemia/reperfusion, nephrotoxins) and more protracted (i.e., ureteral ligation) injury to the development of renal fibrosis via mechanisms involving p53 and JNK with retention of TGF-β signaling [2,40]. p53 inactivation by pifithrin-α or siRNA-directed p53 silencing suppresses p53 phosphorylation, attenuates tubular epithelial apoptosis and G2/M arrest reducing the severity of cisplatin- or ischemia-induced kidney damage and subsequent renal fibrosis [2,40,69].
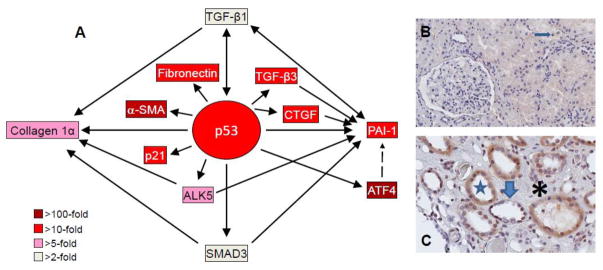
Figure 1. p53 induction and target gene expression in the UUO-injured kidney and in renal allografts.
Microarray analyses of TGF-β1-stumulated proximal tubular epithelial cells illustrating the relative expression levels of target genes within the p53 transcriptome node (A). p53-dependent up-regulation of ALK5, SMAD3, TGF-β1, TGF-β3, CTGF, α-smooth muscle actin (α-SMA) and PAI-1 may constitute a complex feed-forward loop that maintains a profibrotic renal microenvironment. Compared to a normal appearing patient allograft (B) in which only infrequent p-p53S15 tubular epithelial cells were evident (arrow), a renal transplant exhibiting dysmorphic tubules (star) with a flattened and occasionally denuded epithelium (thick arrow) and a markedly expanded interstitial region (asterisk) has abundant nuclear and cytoplasmic p-p53S15 immunoreactivity (C).
p53 is a critical co-factor in the TGF-β1-initiated transcription of a subset of pro-fibrotic genes [54,55,70,71] suggesting widespread involvement in the TGF-β1-directed genomic response to tissue injury. Cluster analysis indicated, moreover, that the p53/TGF-β1 synergy specifically involves genes that regulate growth inhibition, extracellular matrix remodeling and cell substrate attachment [63,72–74]. p53 response element(s) are present in the promoters of the PAI-1, collagen 1α, smooth muscle α-actin and other TGF-β1 target pro-fibrotic genes [75,76]. Oligonucleotide mobility shift and DNase I footprinting/methylation interference analyses confirmed that p53 binds to specific motifs in the PAI-1 promoter, including the two p53 half-sites (AcACATGCCT, cAGCAAGTCC) at −224 bp to −204 bp relative to the transcription start site as well as to the upstream 4G/5G polymorphic sequence [75,77] (Figure 2A). Application of the p53MH algorithm, which identifies genome-wide p53-binding motifs, confirmed that the two PAI-1 half-site motifs meet the >90 cut-off score threshold for potential p53-responsive genes [78]. Induction is due to, in part, the formation of transcriptionally-active p-p53/SMAD multi-protein complexes [54,64–66] with DNA site occupancy reflected in both p53 sequence-driven reporter gene transcription and induced expression of the endogenous PAI-1 gene. Multiple approaches established the involvement of p53 in TGF-β1-stimulated PAI-1 gene expression [54,55,71] and revealed that: (a) TGF-β1 induced binding of p53 to the PAI-1 promoter in human proximal tubular epithelial cells, (b) p53-null fibroblasts do not express PAI-1 upon stimulation with TGF-β1, (c) PAI-1 expression “rescue” was evident in p53-null cells engineered to re-express human p53, (d) pre-treatment of a PAI-1 promoter-luciferase reporter cell line with the p53 inhibitor pifithrin-α suppressed TGF-β1-dependent PAI-1 transcription and protein synthesis, (e) transient siRNA knockdown or pharmacologic blockade of p53 in kidney epithelial cells inhibited PAI-1 induction in response to TGF-β1, and (f) the p53/SMAD2/SMAD3 complex recruits histone acetyltransferase CREB-binding protein to the PAI-1 promoter enhancing H3 acetylation and TGF-β1-stimulated PAI-1 transcriptional activation.
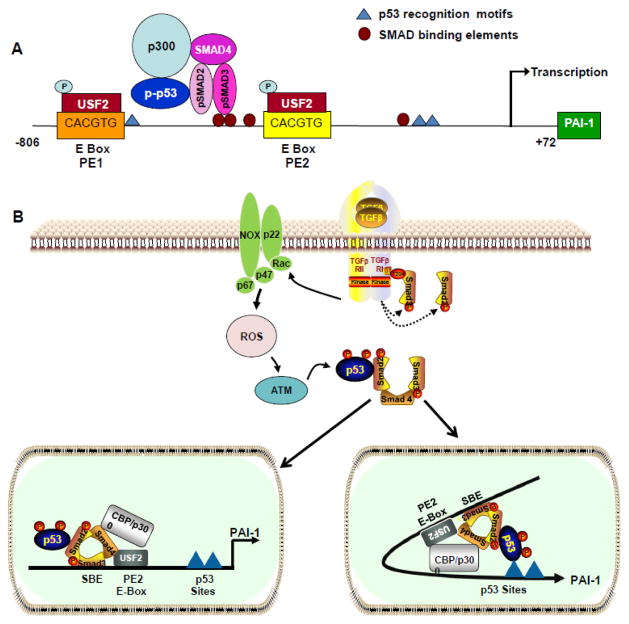
Figure 2. Topography of transcriptional motifs in the PAI-1 promoter and pathways involved in TGF-β1-induced expression.
Downstream p53 binding sites (AcACATGCCT, cAGCAAGTCC) map to nucleotides -224 to -204 relative to the transcription start site and the upstream 4G/5G polymorphic sequence (blue triangles) in the PAI-1 promoter. pSMAD2/3/p-p53 interactions, at the PE2 USF2-binding E Box site located immediately 3′ of three clustered SMAD-binding elements (SBEs), are critical for PAI-1 transcription (A). Ligand-dependent TGF-β1 receptor activation initiates SMAD2/3 phosphorylation (by the ALK5/TGF-β1 type I receptor) (B). Rapid TGF-β1-induced generation of ROS stimulates non-SMAD-mediated signaling (e.g., upon p53 phosphorylation/acetylation). The SMAD and non-SMAD pathways collectively regulate target gene expression. In one model (B, left panel), p53 interacts directly with SMAD2; such SMAD2-p53 interactions may occur independently of p53 occupancy of its consensus motif. Alternatively, certain bHLH-LZ factors (including USF) bend DNA toward the minor groove potentially promoting interactions between p53, bound to its two downstream half-site motifs, with SMAD2 tethered to SBE sites immediately upstream of the CACGTG E-Box [72] (B, right panel).
PAI-1 transcripts are short-lived (<2 hours), as is typical of unstable p53-induced mRNAs, and targeted by the microRNAs miR-143-3p and miR-145-5p both of which are also p53- responsive [79–81]. The 3′ untranslated regions (UTRs) of unstable p53 inducible transcripts are typically longer, and have a higher incidence of U-, AU- and GU-rich sequences, than 3′ UTRs from stable mRNAs [79]. Indeed, an AU-rich region is followed by an AUUUA instability pentamer in the 3′ UTR of the short-lived 3.0-kb PAI-1 transcript. This rather brief PAI-1 mRNA half-life may provide at least a partial basis for the translational utility of designed expression suppression since in vivo delivery of p53 siRNA or pifithrin-α effectively reduced cisplatin- or ischemia/reperfusion-induced renal injury and blunted advancement to CKD [40,69,82].
While the mechanism is uncertain, proximal tubular p53 deficiency down-regulated expression of specific genes involved in apoptosis, signaling and oxidative stress and attenuated ischemia-induced inflammation and interstitial fibrosis [83]. Recent findings, moreover, suggest promoter level competition among certain p53 family members, with p53 target gene control implications. Overexpression of the Δ133p53 isoform (which lacks the N-terminal transactivation domain but retains the C-terminal tetramerization sequence) in human fibroblasts represses specific p53-inducible genes involved in cellular senescence, including PAI-1, p21 and IGFBP7 and enhances reprogramming to an induced pluripotent stem cell phenotype [84]. Δ133p53 physically interacts with p53 and it appears likely that heterodimerization of p53 with Δ133p53 at p53 responsive promoters may constitute a dominant-negative mode of expression regulation. Whether such titration of key transcriptional effectors has clinical efficacy in the context of fibrotic disease remains to be determined.
The PE2 promoter motif in the PAI-1 gene may provide a unique opportunity to probe the intricacies of p53 involvement in gene control. Differential residence of upstream stimulatory factor (USF) family members (involving a USF1→USF2 switch) at the PE2 region E box (CACGTG), which is immediately juxtaposed to three 5′ SBEs, characterized the G0→G1 transition period and growth state-dependent transcriptional activation of the PAI-1 gene [85] (Figure 2A). USF2, moreover, is up-regulated in the obstructed kidney [70] and a consensus PE2 E box motif at nucleotides -566 to -561 is required for USF/E box interactions and serum-dependent PAI-1 transcription [85]. Site-directed CG→AT substitution at the two central nucleotides inhibited formation of USF/probe complexes and PAI-1 promoter-driven reporter expression, confirming the importance of this site in expression control, while Tet-OFF induction of a dominant-negative USF construct or a double-stranded PE2 “decoy” or “trap” [86] attenuated both serum- and TGF-β1-stimulated PAI-1 synthesis [87]. Phasing analysis, moreover, revealed that certain MYC family bHLH-LZ proteins (including USF) redirect DNA minor grove orientation [88] potentially promoting interactions between p53, bound to its half-site response motif, with SMAD2/3 tethered to the PE2 region SBE (Figure 2B). This conformation would facilitate direct interactions between the MH1 N-terminal domain of SMAD2/3 and the p53 N-terminus transactivation domain [63] consistent with the topographic requirement that p53 transcriptionally activates TGF-β1 target genes with both SBEs and p53 binding motifs in their promoter regions and, perhaps, between the C-terminus of p53 and the MH2 region of SMAD3 [89]. Alternatively, since p53 interacts directly with SMAD2 [65,66,72], the PE2 site (with its 3 SBEs) may also serve as a docking platform for p53/SMAD complexes [54,71] (Figure 2B). p300/CREB-binding protein, a histone acetyltransferase, acetylates SMAD2/3 in response to TGF-β1 [90] facilitating the creation of a SMADs/p53/USF2 transcriptional complex necessary for optimal PAI-1 induction [91–93]. The importance of such interactions is underscored by the finding that RAP250, a protein devoid of intrinsic enzymatic activity yet effectively recruits histone acetyltransferases and methylases to chromatin complexes, also interacts with SMAD2/3 and is essential for maximal TGF-β1-stimulated PAI-1 expression [94].
While the growing complexity of the PE2-based transcriptional control unit in the PAI-1 promoter, as well as the ability to target the involved cis-acting and epigenetic factors [86], promises to provide new opportunities to manage expression of disease-relevant fibrotic genes, p53 may also initiate a profibrotic genomic program indirectly though transcription of specific microRNAs. p53 appears to promote fibrosis following unilateral ureteral obstruction (UUO) by up-regulation of miR199a-3p that, in turn, suppresses expression of SOCS7 stimulating, thereby, STAT3 activation in proximal tubular cells [95]. SOCS1, SOCS3 and SOCS7 are potent inhibitors of STAT1, STAT3 and ERK phosphorylation [96] suggesting a model [95] whereby TGF-β1 activation of p53 stimulates p53-dependent transcription of miR-199a-3p inhibiting SOSC7 expression resulting in STAT3-induced renal fibrosis.
The ancestral p53 family member p73 [97], in particular ΔNp73, also functions in PAI-1 transcriptional control [98]. ΔNp73 transcripts are generated from the alternate P2 promoter site in intron 3 and encode a N-terminal truncated protein lacking the transactivation domain [99,100]. While ΔNp73 generally inhibits gene transcription by p53, and other p73 isoforms, ΔNp73 actually increased expression of the TGF-β1 target genes PAI-1 and collagen 1α [100]. ΔNp73 knockdown attenuated TGF-β1 signaling and reporter anayses confirmed that ΔNp73 induction of the PAI-1 gene, unlike p53, was not dependent on p53 binding motifs but on association with SMADs at the SBEs in the PAI-1 and collagen 1α promoters. DNA pull-down assays indicated, moreover, that ΔNp73 and SMADs form a complex on an SBE oligonucleotide platform and that ΔNp73 enhances SMAD3 binding to the SBE target construct. p53, however, is essential for PAI-1 transcription at least in response to TGF-β1 [54]. It remains to be determined, therefore, if ΔNp73 requires p53 to activate TGF-β1-dependent transcription of the PAI-1 gene. This possibility is supported by the observation that p73 is not sufficient to completely compensate for p53 deficiency in renal development as p53-null mice have defects in nephron differentiation [102] and that p53-p73 cooperation regulates p53 transcriptional activity and genomic impact [100]. p53 family proteins, moreover, form multimeric complexes often described as “dimers on dimers”. It appears that JNK-induced phosphorylation of p53T81 drives the formation of transcriptionally-active p53/p73 complexes [103]; it is also conceivable that p73 may, in the context of chromatin, heterodimerize with mutant p53 [104]. The various p53 members, thus, likely foster different transcriptional outcomes by competing for DNA binding sites, acting in a dominant-negative fashion or inhibiting or enhancing function via heterotetramerization or other protein-protein interactions. Addressing these issues will require individual target gene assessments.
Mechanism of p53 Activation by TGF-β1
TGF-β1 regulates p53 activity by serine site phosphorylation, in the N-terminal transactivation domain, and serine/lysine acetylation/methylation (among other post-translational modifications) in the tetramerization and regulatory domains in the C-terminus [105]. Collectively, these promote interactions with activated SMADs and subsequent binding of p53/SMAD3 to the PAI-1 promoter in human renal proximal tubular epithelial cells [54]. Phosphorylation the p53 amino-terminal serines15/20 and threonine18 residues increases the association of p53 with members of the p300/CREB binding protein (CBP) co-activator family while stimulating p53 transactivation function [106]. p300 and CBP protein/histone acetyltransferases relax chromatin structure facilitating recruitment of accessory transcriptional factors to promoter domains of target genes. The creation of a multi-component p300/p53/SMAD complex preceded optimal TGF-β1-dependent induction of the PAI-1 gene [54]. Similarly, interactions between p53 and SMAD2/3, at their respective binding sites, recruits CBP to the PAI-1 promoter increasing histone H3 acetylation and PAI-1 transcription [89]. Not all p53/SMAD interactions result in gene activation, however. Consistent with the potential opposing actions of SMAD3 (pro-fibrotic) vs. SMAD2 (anti-fibrotic) [107,108], partnering of p53 with SMAD2 in hepatic cells represses expression of the developmentally-dependent alpha fetoprotein gene (AFP). p53 DNA binding is required to anchor TGF-β1-activated SMADs as well as the transcriptional co-repressor mSin3A to the SMAD-binding motifs and the p53 response elements in the AFP promoter [109]. In this context, SnoN (an inhibitor of TGF-β1 signaling) is a critical co-factor in AFP suppression functioning to up-regulate mSin3A levels. Whether other TGF-β1 down-regulated p53-sensitive genes [54] utilize a similar mechanism of repression is unknown.
Recent findings have shed light on the mechanism of p53 activation in response to TGF-β1. One potential regulator of p53 function in the context of tissue injury is the serine/threonine kinase tumor suppressor ataxia telangiectasia mutated (ATM). Activated ATM (pATMS1981) increased significantly in the tubulointerstitial region of the UUO-injured kidney correlating with SMAD3 and p53S15 phosphorylation, elevation of the p22phox subunit of the NADP(H) oxidases, and expression of the fibrotic markers PAI-1 and fibronectin [71]. This likely reflects elevated levels of TGF-β1 in response to ureteral ligation as ATM is rapidly phosphorylated at the same site (S1981) upon TGF-β1 stimulation of cultured proximal tubular cells. Stable silencing (by lentiviral delivery of short hairpin RNAs) or pharmacological inhibition (with KU-55933) of ATM attenuated TGF-β1-induced p53 activation and markedly reduced expression of the downstream targets PAI-1, fibronectin, CTGF and p21 in human tubular epithelial cells as well as in kidney fibroblasts [71]. The participating elements in TGF-β1-induced ATM mobilization are becoming clarified. Knockdown of the NADPH oxidase (NOX) subunits, p22phox and p47phox in HK-2 cells blocked TGF-β1-stimulated phosphorylation of ATM (pATMS1981) and target gene induction via p53- dependent mechanisms. Thus, TGF-β1 promotes NOX-dependent ATM activation leading to TGF-β1-initiated p53 phosphorylation and p53-mediated fibrotic gene reprogramming (Figure 3). Depletion of ATM or p53, moreover, resulted in a bypass of TGF-β1-mediated cytostasis in HK-2 cells [71]. Furthermore, TGF-β1/ATM-stimulated secretion of paracrine factors by the dysfunctional renal epithelium promotes interstitial fibroblast growth, suggesting a role for tubular ATM in mediating epithelial-mesenchymal cross-talk highlighting the translational benefit of targeting the NOX/ATM/p53 axis in renal disease.
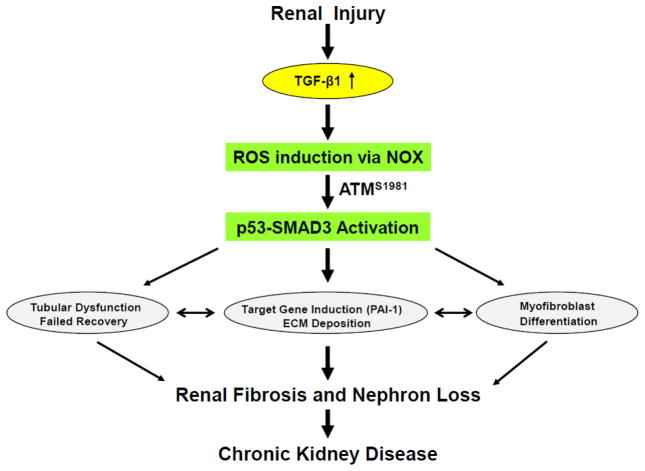
Figure 3. Events downstream of renal injury-induced TGF-β1 expression that contribute to fibrotic disease.
Activated ATM (pATMS1981) increased significantly in the tubulointerstitial region of the UUO-injured kidney, likely in response to elevated TGF-β1 levels and expression of the p22phox subunit of the NADP(H) oxidases, correlating with SMAD3 and p53S15 phosphorylation and induction of the fibrotic markers PAI-1 and fibronectin [71]. Stable silencing or pharmacological inhibition of ATM attenuated TGF-β1-induced p53 activation and expression of the downstream targets PAI-1, fibronectin, CTGF and p21. Silencing of the NADPH oxidase (NOX) subunits, p22phox and p47phox in HK-2 cells blocked TGF-β1-stimulated phosphorylation of ATM (pATMS1981) and target gene induction via p53- dependent mechanisms. Thus, TGF-β1 promotes NOX-dependent ATM activation leading to p53-mediated fibrotic gene reprogramming and growth arrest in HK-2 cells. Depletion of ATM or p53 in HK-2 cells, moreover, resulted in a bypass of TGF-β1-mediated cytostasis [71]. Furthermore, TGF-β1/ATM-initiated paracrine factor secretion by the dysfunctional renal epithelium promotes interstitial fibroblast growth, suggesting a role for tubular ATM in mediating epithelial-mesenchymal cross-talk highlighting the translational benefit of targeting the NOX/ATM/p53 axis in renal disease.
Relevant is the recent finding that SMG7 (suppressor with morphological defects in genitalia 7), a regulator of nonsense-mediated mRNA decay, binds to and stabilizes p53 under conditions of genotoxic stress [110]. The mechanism appears to involve promotion of ATM-dependent phosphorylation and subsequent inhibition of the E3 ubiquitin ligase mouse double minute 2 homolog (MDM2). This role for SMG7 may well have an impact on expression/function of p53 and recovery from AKI, particularly in patients with lupus nephitis who exhibit reduced expression of SMG7 [111]. The participation of MDM2 in fibrosis, however, is complex. MDM2 is a major regulator of p53 function, via inhibition of transcriptional activity, and stability by virtue of its role as a ligase [112]. While TGF-β1 induces p53 activation in vitro and in vivo, this growth factor also increases MDM2 expression in a p53-dependent manner establishing a feedback loop where p53 initiates expression of its negative regulator [54,112,113]. Recent findings suggest that MDM2 also mediates fibroblast activation and renal interstitial fibrosis through a p53-independent pathway perhaps involving Notch I down-regulation [113]. Nevertheless, while underlying events require clarification, it is apparent that the p53-MDM2/murine double minute X (MDMX) axis is required for normal kidney development. Germline p53 depletion results in renal anomalies (with some dependency on genetic background) while MDM2/MDMX deficiencies are associated with acute renal injury, epithelial cell death and fatal dysgenesis [114,115]. The clinical relevance of these findings is highlighted by a recent systems analysis of patients with diabetic nephropathy confirming a marked down-regulation of MDM2 expression in the glomerular and tubular compartments [116].
PAI-1 Involvement in Renal Disease: p53-Dependent/Independent Pathways
Transcriptome profiling highlights the complexity of gene expression patterns in kidney disorders [37,117–124] as well as in TGF-β1-stimulated epithelial cells [54,125,126]. Certain TGF-β1 target genes directly influence the development of the myofibroblastic phenotype and renal fibrogenesis. PAI-1, in particular, is a prominent TGF-β1 response gene in proximal tubular epithelial cells as well as interstitial fibroblasts and is involved in the TGF-β1-induced conversion of fibroblasts to myofibroblasts [71,127–129]. Among its varied functions, PAI-1 negatively regulates the plasmin-dependent pericellular proteolytic cascade effectively limiting ECM degradation and fibrinolytic activity, thereby, contributing to the initiation and/or progression of interstitial fibrosis and vascular thrombosis [62,130]. PAI-1 is a member of both the growth arrest/fibrosis genomic cluster in the diabetic rat kidney [131] and the 11-gene urine mRNA discovery set signature predictive of human renal allograft fibrosis [132]. PAI-1 null mice are, in fact, protected from excessive ECM accumulation as well as lung, liver, kidney and vascular fibrosis and PAI-1 urokinase/tissue-type plasminogen activator domain decoys reduced both UUO-initiated and established interstitial fibrosis [133].
Recent data suggest a rather novel role for PAI-1 in fibrotic disorders apart from its impact on ECM turnover. While it is well established that p53 limits cellular proliferation by inducing a state of replicative senescence, little is known about the mechanisms involved in this growth-limiting response. Suppression of the p53 target gene PAI-1 by RNA interference leads to senescence escape by sustained activation of the PI(3)K-AKT-GSK3β pathway and nuclear retention of cyclin D1 [73]. Genetically-deficient PAI-1 knockout (PAI-1−/−) mouse embryonic fibroblasts (MEFs), in fact, proliferate well beyond the senescence checkpoint, albeit at a slower rate than p53−/− MEFs. Moreover, ectopic expression of PAI-1 in p53-null fibroblasts induces a phenotype displaying all the hallmarks of replicative senescence-induced growth arrest [73]. These data were the first to conclusively indicate that PAI-1 is not merely a marker of senescence, but is both necessary and sufficient for the induction of senescence downstream of p53. Similarly, TGF-β has a significant cytostatic effect on various cell types. p53 knockdown results in escape from TGF-β1-induced growth arrest in various cell types [24,89,134] which exhibit a strong growth inhibitory response to TGF-β1 including those derived from the renal proximal tubular epithelium [71]. Collectively, it appears that p53 plays an important role in TGF-β-induced cytostasis via induction of PAI-1 transcription and that loss of p53 or its target gene PAI-1 confers resistance to TGF-β1-mediated growth inhibition. These findings suggest the utility of p53 pathway disruption in renal disease, perhaps as a strategy to promote compensatory regeneration, and are underscored by very recent findings that loss of phosphatase and tensin homolog (PTEN) expression correlated with increased PAI-1 levels in the obstructed kidney [135]. PTEN knockdown in HK-2 cells, moreover, promoted fibrotic factor expression (PAI-1, fibronectin, CTGF) and G1 cell cycle arrest [135]. PTEN loss also results in p53, SMAD3, AKT activation and formation of p53/SMAD3 complexes associated with epithelial dysfunction. As is the case for TGF-β1-treated cells, growth restriction was PAI-1-dependent since silencing of PAI-1 expression in PTEN-knockdown HK-2 proximal tubular cells rescued the proliferative response. The increased population density evident in dual-deficient PTEN/PAI-1 cultures was comparable to that of cells with stable silencing of both p53 and PTEN expression. Moreover, the elevated PAI-1 levels evident in PTEN-deficient cultures significantly decreased upon p53 shRNA lentiviral transduction additionally reinforcing a role for p53 in fibrotic gene induction [135]. Furthermore, PCNA expression markedly increased in both dual PTEN+PAI-1 shRNA- and PTEN+p53 shRNA-expressing HK-2 cells compared to similarly seeded PTEN shRNA cultures confirming that depletion of p53 or PAI-1 levels leads to a bypass of cell growth inhibition triggered by PTEN loss in proximal tubular epithelial cells. Since PTEN deficiency is a common event in diabetic-, UUO-, ischemia/reperfusion- and aristocholic acid-induced renal injury and the associated failed regeneration [2,135], approaches designed to limit p53 activation and/or PAI-1 expression may promote tubular epithelial recovery and attenuate nephron loss.
Current data also confirm an unexpected involvement of PAI-1 in innate immunity. Indeed, following kidney injury, PAI-1-null mice develop an attenuated inflammatory/fibrotic response while transgenic PAI-1 over-expressing animals exhibit increased renal interstitial monocyte/macrophage density suggesting that this serine protease inhibitor may promote macrophage and T-cell infiltration and/or immune cell tissue residence time [136,137]. Monocyte adhesion to the aortic intima was significantly reduced in streptozotocin-treated PAI-1−/− mice and accompanied by decreases in the inflammatory mediators TNF-α and monocyte chemotactic protein-1 [138]. Since PAI-1 provides a “don’t eat me” signal, effectively inhibiting neutrophil efferocytosis [139,140], these findings [138–140] suggest that this serine protease inhibitor may impact cellular influx as well as the intensity and/or duration of the injury-initiated inflammatory phase. Indeed, elevated PAI-1 levels closely mirrors systemic and localized inflammation while exogenously-delivered PAI-1 stimulates expression of proinflammatory cytokines (e.g., TNFα and macrophage inflammatory protein-2) in primary bone marrow macrophages [137]. The protease inhibitory-, vitronectin- or LRP1-binding properties of PAI-1, however, are not necessary for macrophage activation but TLR4 appears to be required, at least in part, since TLR4 neutralizing antibodies or the genetic depletion of TLR4 attenuated PAI-1-induced inflammation. This response was PAI-1 dose-dependent but LPS-independent and reduced in TLR4−/− macrophages [137] suggesting that PAI-1 may function as a matricellular damage-associated molecular pattern (DAMP) TLR ligand [141,142]. There is also evidence for PAI-1 involvement in lipopolysaccharide (LPS) signaling. PAI-1 knockdown attenuated LPS-induced increases in macrophage TLR4, MD-2, MyD88, TNFα, IL-1β and NF-κB levels while vector-driven PAI-1 over-expression enhanced these responses [143,144]. Although the mechanism is unclear, data suggest that PAI-1 is involved in host inflammatory responses via TLR4, at least in macrophages [137]. This is likely to have a significant impact on fibrogenic outcomes following tissue injury as exogenous PAI-1 treatment increased TGF-β1, collagen 1α1, collagen 1α2 and MCP-1 transcripts in renal mesangial and proximal tubular epithelial cells [145–147]. The TLR4/RAGE DAMP-type ligand HMGB1 also activates a subset of genes in the TGF-β1 profibrotic signature that includes PAI-1, CTGF and TGF-β1 [148] suggesting that DAMPs and LPS utilize common and unique signaling pathways that may be exploited in the design of interventional strategies. Collectively, it appears that TLR4 may function as a molecular “switch”, activated by endogenous DAMPs to initiate repair while stimulating TGF-β1 signaling (by down-regulating the TGF-β pseudoreceptor BAMBI) promoting the persistent expression of TGF-β target genes to create and maintain a progressive fibrotic microenvironment [149,150].
Exogenous PAI-1 also activates the JAK/STAT, AKT and focal adhesion kinase (FAK) pathways via LRP1-dependent mechanisms [151–153]. PAI-1 may engage several cellular receptors (TLR4, LRP1), therefore, with differing phenotypic outcomes. Whether PAI-1 occupancy of its binding site on the somatomedin B domain of vitronectin or to urokinase plasminogen activator/urokinase plasminogen activator receptor complexes on the cell surface is required for signaling through TLR4 or LRP1 is not clear. Recombinant PAI-1, however, does mobilize the RhoA/ROCK1/MLC-P pathway stimulating amoeboid cell migration [142] and apparently modulates TGF-β1 signaling, through direct effects on TGF-β1 bioavailability, as PAI-1-null mice subjected to obstructive nephropathy have lower TGF-β1 levels compared to wild-type controls [136,154]. Exogenously-delivered PAI-1 alone, moreover, stimulates TGF-β1 synthesis which could be attenuated by pretreatment with small molecule PAI-1 functional inhibitors, suggesting the existence of a PAI-1/TGF-β1-positive feedback mechanism [145,147]; these same compounds reduced up-regulation of fibronectin, collagen 1 and PAI-1 transcripts in the kidneys of diabetic mice [146]. It appears that PAI-1 may initiate, perhaps maintain, a potential pro-fibrogenic “loop” in the context of renal disease [145,147]. It is tempting to speculate, therefore, that targeted down-modulation of PAI-1 expression or function may provide multi-level therapeutic opportunities to inhibit the onset and progression of tissue fibrosis (Figure 4).
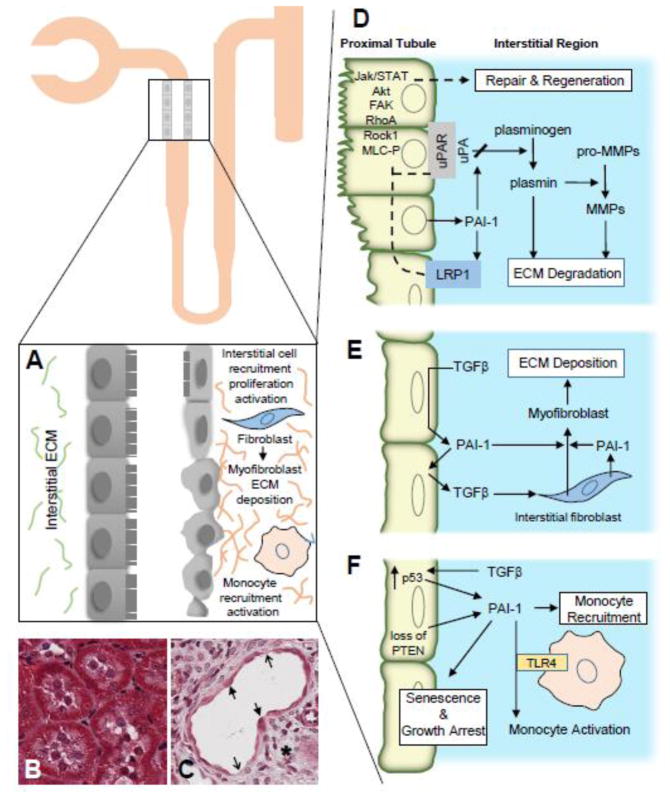
Figure 4. Multifunctional roles of PAI-1 in renal fibrosis.
Development of renal disease upon proximal tubule injury is characterized by major functional and morphological changes including epithelial dedifferentiation, ECM accumulation, cell cycle arrest and tubular dysmorphism (A left, B = normal kidney, A right, C = fibrosis). Thick and thin arrows denote flattened epithelium and denuded regions, respectively. Following tissue injury, an inter-dependent plasmin-generating/matrix metalloproteinase (MMP) pericellular proteolytic cascade is finely titered, both temporally and spatially, by local PAI-1 levels (D). Collectively, these highly integrated systems cooperate to regulate ECM degradation and stromal remodeling. Elevated PAI-1 abundance in the wounded kidney compromises ECM degradation and fibrin clearance promoting increased matrix accumulation that contributes to the initiation of the fibrotic process and eventual development of CKD. Increased TGF-β levels or activation in response to trauma facilitate the transition of pericytes and resident interstitial fibroblasts (with perhaps minor contributions from other cell types) toward a myofibroblastic phenotype (E). An increased persistence and/or density of myofibroblasts further accelerates ECM deposition and eventual loss of tissue function. In conjunction with high p53 expression and loss of PTEN, TGF-β expression in the injury microenvironment mediates epithelial growth arrest with loss of regenerative repair, exacerbating ECM accumulation, likely through elevation of local levels of profibrotic factors including PAI-1 (F). Microenvironmental cues in the injured epithelial and interstitial compartments initiate TGF-β1-dependent monocyte recruitment/activation (perhaps via PAI-1 modulation of TLR4 signaling) (E,F). Additional mechanistic details are provided in the text. Collectively, these events (D–F) illustrate the clinical significance of the collaborative effects of TGF-β1 and TGF-β1 target genes (e.g., PAI-1) on the development of renal fibrosis.
Conclusion
TGF-β1 is the principal driver of tissue scarring leading to interstitial renal fibrosis and eventual organ failure. TGF-β1 stimulates p53 phosphorylation promoting interactions with activated SMADs and subsequent binding of p53/SMAD3 to target promoters. Recent findings further suggest that PTEN deficiency, perhaps TGF-β1-mediated, is a common event in diabetic-, ischemia/reperfusion-, ureteral ligation- and aristocholic acid-induced renal injury resulting in p53 and SMAD3 activation and formation of p53/SMAD3 complexes. The details in this pathophysiologically-relevant interplay of signaling effectors are only beginning to emerge however p-p53 is required for expression of PAI-1 and CTGF, major TGF-β1 target genes and key causative factors in fibrotic disorders. One regulator of p53 function in the context of renal injury is ATM. pATMS1981 levels increase in the injured kidney, as well as in TGF-β1-stimulated tubular epithelial cells, correlating with SMAD3/p53 phosphorylation and expression of the p22phox subunit of the NADPH oxidases. Silencing or pharmacologic inhibition of ATM attenuated TGF-β1-induced p53 activation and subsequent PAI-1, fibronectin, CTGF and p21 up-regulation in human proximal tubular cells and kidney fibroblasts. Engineered reductions in the NOX subunits, p22phox and p47phox blocked TGF-β1-induced ATMS1981 phosphorylation and gene induction via p53-dependent pathways. TGF-β1, therefore, appears to promote NADPH oxidase-dependent ATM activation leading to p53-dependent profibrotic genomic reprogramming. Importantly, PAI-1 is a member of the signature gene set predictive of renal allograft fibrosis [132] as well as a prominent p53 target [73]. Increased p-p53S15 immunoreactivity is evident in the epithelial and intertubular compartments in human renal transplants with established tubular dysmorphism and interstitial involvement (Figure 1C). Administration of pifithrin-α to mice with ischemic renal injury reduced both the expression of profibrotic genes and the extent of interstitial fibrosis [40]. Pharmacologic inhibition of p53 function or the p53 activation network, if appropriately managed, may have significant clinical implications. These data collectively highlight the translational potential in targeting the TGF- β1/p53 axis in renal disease but which also may be relevant to the global problem of tissue fibrosis regardless of the involved site.
Highlights for Review.
Fibrotic disorders of the renal, pulmonary, cardiac, and hepatic systems are associated with significant morbidity and mortality..
TGF-β1 is the principal driver of renal fibrogenesis,.
TGF-β1 activates the ALK5 type I receptor (which phosphorylates SMAD2/3) as well as non-canonical (e.g., src kinase, EGFR, Jak/Stat, p53) pathways that collectively drive the fibrotic genomic program.
TGF-β1 stimulates the activation and assembly of p53-SMAD3 complexes required for transcription of the renal fibrotic genes plasminogen activator inhibitor-1 (PAI-1, SERPINE1), connective tissue growth factor (CTGF, CCN2) and TGF-β1.
Focus on TGF-β1-initiated signaling in renal fibrogenesis and the role of p53 as a regulator of profibrotic gene expression.
Acknowledgments
This work was supported by grants from the NIH (GM057242), the Roach Family Foundation, the Graver Family Endowment, and the Friedman Cancer Research Fund.
Abbreviations
- AKT
AKT8 oncogene, alpha serine/threonine-protein kinase or protein kinase B
- bHLH-LZ
basic helix-loop-helix/leucine zipper
- CREB
cAMP response element-binding protein
- ERK
extracellular signal-regulated kinases
- JAK
Janus kinases
- JNK
c-Jun N-terminal kinases
- IGFBP7
insulin-like growth factor-binding protein 7
- MLC-P
phospho-myosin light chain
- PCNA
proliferating cell nuclear antigen
- RhoA
Ras homolog gene family, member A
- ROCK
Rho-associated protein kinase
- shRNA
short hairpin RNA
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- TAZ
transcriptional coactivator with PDZ-binding motif
- YAP
Yes-associated protein
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zeisberg M, Neilson EG. Mechanisms of tubulointestinal fibrosis. J Am Soc Neprhol. 2010;21:181901834. doi: 10.1681/ASN.2010080793. [DOI] [PubMed] [Google Scholar]
- 2.Venkatachalam MA, Weinberg JM, Kriz W, Bidani AK. Failed tubule recovery, AKI-CKD transition, and kidney disease progress. J Am Soc Neprhol. 2015;26:1765–1776. doi: 10.1681/ASN.2015010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnaper HW. The tubulointerstitial pathophysiology of progressive kidney disease. Adv Chronic Kidney Dis. 2017;24:107–116. doi: 10.1053/j.ackd.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster AC, Nagler EV, Morton RL, Masson P. Chronic kidney disease. Lancet. 2017;389:1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 5.Mei C, Zheng F. Chronic inflammation potentiates kidney aging. Semin Nephrol. 2009;29:555–568. doi: 10.1016/j.semnephrol.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Grgic I, Campanholle G, Bijol V, Wang C, Sabbisetti VS, Ischimura T, Humphreys BD, Bonventure JV. Targeted proximal tubule injury triggers interstitial fibrosis and glomerulosclerosis. Kidney Int. 2012;82:172–183. doi: 10.1038/ki.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takaori K, Nakamura J, Yamamoto S, Nakata H, Sato Y, Takase M, Nameta M, Yamamoto T, Economides AN, Kohno K, Haga H, Sharma K, Yanagita M. Severity and frequency of proximal tubule injury determines renal prognosis. J Am Soc Nephrol. 2016;27:2393–2406. doi: 10.1681/ASN.2015060647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferenbach DA, Bonventre JV. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat Rev Nephrol. 2015;11:264–276. doi: 10.1038/nrneph.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy AA. Molecular basis of renal fibrosis. Pediatr Nephrol. 2000;15:290–301. doi: 10.1007/s004670000461. [DOI] [PubMed] [Google Scholar]
- 10.Eddy AA. Progression in chronic kidney disease. Adv Chronic Kidney Dis. 2005;12:353–365. doi: 10.1053/j.ackd.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Strutz F, Nelson EG. New insights into mechanisms of fibrosis in immune renal injury. Springer Semin Immunopathol. 2003;24:459–476. doi: 10.1007/s00281-003-0123-5. [DOI] [PubMed] [Google Scholar]
- 12.Bonventre JV. Pathophysiology of AKI: injury and normal and abnormal repair. Contrib Nephrol. 2010;165:9–17. doi: 10.1159/000313738. [DOI] [PubMed] [Google Scholar]
- 13.Grande MT, Lopez-Nova JM. Fibroblast activation and myofibroblast generation in obstructure nephropathy. Nat Rev Nephrol. 2009;5:319–328. doi: 10.1038/nrneph.2009.74. [DOI] [PubMed] [Google Scholar]
- 14.Richter K, Konzack A, Pihlajaniemi T, Helijasvaara R, Kietzmann T. Redox-fibrosis: impact of TGF-β1 on ROS generators, mediators and functional consequences. Redox Biol. 2015;6:344–352. doi: 10.1016/j.redox.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vega G, Alarcon S, San Martin R. The cellular and signalling alterations conducted by TGF-β contributing to renal fibrosis. Cytokine. 2016;88:115–125. doi: 10.1016/j.cyto.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Picard N, Baum O, Vogetseder A, Kaissling B, Le Hir M. Origin of renal myofibroblasts in the model of unilateral ureter obstruction in the rat. Histochem Cell lBiol. 2008;130:1410155. doi: 10.1007/s00418-008-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook HT. The origin of renal fibroblasts and progression of kidney disease. Am J Pathol. 2010;176:22–24. doi: 10.2353/ajpath.2010.090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y. New insights into epithelial-mesenchymal transition in kidney fibrosis. J Am Soc Nephrol. 2010;21:212–222. doi: 10.1681/ASN.2008121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricardo SD, van Goor H, Eddy AA. Macrophage diversity in renal injury and repair. J Clin Invest. 2008;118:3522–3530. doi: 10.1172/JCI36150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi W, Chen X, Poronnik P, Pollock CA. The renal cortical fibroblast in renal tubulointerstitial fibrosis. Int J Biochem Cell Biol. 2006;38:1–5. doi: 10.1016/j.biocel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Leung JY, Wilson HL, Voltzke KJ, Williams LA, Lee HJ, Wobker SE, Kim WY. Sav1 loss induces senescence and Stat3 activation coinciding with tubulointerstitial fibrosis. Mol Cell Biol. 2017;37(12):e00565–16. doi: 10.1128/MCB.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong X, Zhao B, laboc RE, Zhu J, Koksal AC, Lu C, Engen JR, Springer TA. Force interacts with macromolecular structure in activation of TGF-β. Nature. 2017;542:55–59. doi: 10.1038/nature21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szeto SG, Narimatsu M, Lu M, He X, Sidiqi AM, Tolosa MF, Chan L, De Freitas K, Bialik JF, Majumder S, Boo S, Hinz B, Dan Q, Advani A, John R, Wrana JL, Kapus A, Yuen DA. YAP/TAX are mechanoregulators of TGF-β-Smad signaling and renal fibrogenesis. J Am Soc Nephrol. 2016;27:3117–3128. doi: 10.1681/ASN.2015050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Tiam B, Brasier AR. Targeting chromatin remodeling in inflammation and fibrosis. Adv Protein Chem Struct Biol. 2017;107:1–36. doi: 10.1016/bs.apcsb.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Sun YB, Qu X, Caruana G, Li J. The origin of renal fibroblasts/myofibroblasts and the signals that trigger fibrosis. Differentiation. 2016;92:102–107. doi: 10.1016/j.diff.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 28.Chevalier RL, Forbes MS, Thomhill BA. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009;75:1145–1152. doi: 10.1038/ki.2009.86. [DOI] [PubMed] [Google Scholar]
- 29.McGaraughty S, Davis-Taber RA, Zhu CZ, Cole TB, Nikkel AM, Chhaya M, Doyle KJ, Olson LM, Preston GM, Grinnell CM, Salte KM, Giamis AM, Luo Y, Sun V, Goodearl AD, Gopalakrishnan M, Lacy SE. Targeting anti-TGF-β therapy to fibrotic kidneys with a dual specificity antibody approach. J Am Soc Nephrol. 2017 Aug 21; doi: 10.1681/ASN.201710013. pii: ASN.2017010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyajima A, Chen J, Lawrence C, Ledbetter S, Soslow RA, Stern J, Jha S, Pigato J, Lemer ML, Poppas DP, Vaughan ED, Felsen D. Antibody to transforming growth factor-beta ameliorates tubular apoptosis in unilateral ureteral obstruction. Kidney Int. 2000;58:2301–2313. doi: 10.1046/j.1523-1755.2000.00414.x. [DOI] [PubMed] [Google Scholar]
- 31.Inazaki K, Kanamaru Y, Kojima Y, Sueyoshi N, Okumura K, Kaneko K, Yamashiro Y, Ogawa H, Nakao A. Smad3 deficiency attenuates renal fibrosis, inflammation, and apoptosis after unilateral ureteral obstruction. Kidney Int. 2004;66:597–604. doi: 10.1111/j.1523-1755.2004.00779.x. [DOI] [PubMed] [Google Scholar]
- 32.Schreiner GF, Harris KP, Purkerson ML, Klahr S. Immunological aspects of acute ureteral obstruction: immune cell infiltrate in the kidney. Kidney Intl. 1988;34:487–493. doi: 10.1038/ki.1988.207. [DOI] [PubMed] [Google Scholar]
- 33.Klahr S, Morrissey J. Obstructive nephropathy and renal fibrosis. Am J Physiology Renal Physiol. 2002;283:F861–F875. doi: 10.1152/ajprenal.00362.2001. [DOI] [PubMed] [Google Scholar]
- 34.Dendooven A, Ishola DA, Jr, Nguyen TQ, Van der Giezen DM, Kok RJ, Goldschmeding R, Joles JA. Oxidative stress in obstructive nephropathy. Int J Exp Pathol. 2011;92:202–210. doi: 10.1111/j.1365-2613.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohatgi R, Flores D. Intratubular hydrodynamic forces influence tubulointerstitial fibrosis in the kidney. Curr Opin Nephrol Hypertens. 2010;19:65–71. doi: 10.1097/MNH.0b013e32833327f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eddy AA, Fogo AB. Plasminogen activator inhibitor-1 in chronic kidney disease: evidence and mechanisms of action. J Am Soc Neprhol. 2006;17:2999–3012. doi: 10.1681/ASN.2006050503. [DOI] [PubMed] [Google Scholar]
- 37.Higgins DF, Lappin DW, Kieran NE, Anders HJ, Watson RW, Strutz F, Schlondorff D, Haase VH, Fitzpatrick JM, Godson C, Brady HR. DNA oligonucleotide microarray technology identifies fisp-12 among other potential fibrogenic genes following murine unilateral ureteral obstruction (UUO): modulation during epithelial-mesenchymal transition. Kidney Int. 2003;64:2079–2091. doi: 10.1046/j.1523-1755.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 38.Yabuki A, Maeda M, Matsumoto M, Kamimura R, Masuyama T, Suzuki S. SAMP1/Sku as a murine model for tubulointerstitial nephritis: a study using unilateral ureteral obstruction. Exp Anim. 2005;54:53–60. doi: 10.1538/expanim.54.53. [DOI] [PubMed] [Google Scholar]
- 39.Manucha W. Biochemical-molecular marker in unilateral ureteral obstruction. Biocell. 2007;31:1–12. [PubMed] [Google Scholar]
- 40.Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med. 2010;16:535–543. doi: 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Truong LD, Gaber L, Eknoyan G. Obstructive uropathy. Contrib Nephrol. 2011;169:311–326. doi: 10.1159/000314578. [DOI] [PubMed] [Google Scholar]
- 42.Ko GJ, Boo C-S, Jo S-K, Cho WY, Kim HK. Macrophages contribute to the development of renal fibrosis following ischemia/reperfusion-induced acute injury. Nephrol Dial Transplant. 23:842–852. doi: 10.1093/ndt/gfm694. [DOI] [PubMed] [Google Scholar]
- 43.Chevalier RL, Thornhill BA, Forbes MS, Kiley SC. Mechanisms of renal injury and progression of renal disease in congenital obstructive nephropathy. Pediatr Nephrol. 2010;25:687–697. doi: 10.1007/s00467-009-1316-5. [DOI] [PubMed] [Google Scholar]
- 44.Gagliardini E, Benigni A. Role of anti-TGF-beta antibodies in the treatment of renal injury. Cytokine Growth Factor Rev. 2006;17:89–96. doi: 10.1016/j.cytogfr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Isaka Y, Tsujie M, Ando Y, Nakamura H, Kaneda Y, Imai E, Hori M. Transforming growth factor-beta 1 antisense oligodeoxynucleotides block interstitial fibrosis in unilateral ureteral obstruction. Kidney Int. 2000;58:1885–1892. doi: 10.1111/j.1523-1755.2000.00360.x. [DOI] [PubMed] [Google Scholar]
- 46.Hwang M, Kim HJ, Noh HJ, Chang YC, Chae YM, Kim KH, Jeon JP, Lee TS, Oh HK, Lee YS, Park KK. TGF-beta1 siRNA suppresses the tubulointerstitial fibrosis in the kidney of ureteral obstruction. Exp Mol Pathol. 2006;81:48–54. doi: 10.1016/j.yexmp.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y, Border WA, Noble NA. Perspectives on blockade of TGFbeta overexpression. Kidney Int. 2006;69:1713–1714. doi: 10.1038/sj.ki.5000260. [DOI] [PubMed] [Google Scholar]
- 48.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. Targeted disruption of TGF-beta1/Smade3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest. 2003;112:1486–1494. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hill CS. Transcriptional control by SMADs. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a022079. pii: a022079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ayyaz A, Attisano L, Wrana JL. Recent advances in understanding contextual TGFβ signaling. F1000Res. 2017;6:749. doi: 10.12688/f1000research.11295.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bottinger EP, Bitzer M. TGF-beta signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610. doi: 10.1097/01.asn.0000033611.79556.ae. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat Rev Nephrol. 2011;7:684–696. doi: 10.1038/nrneph.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edeling M, Ragi G, Huang S, Pavenstadt H, Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol. 2016;12:426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Overstreet JM, Samarakoon R, Meldrum KK, Higgins PJ. Redox control of p53 in the transcriptional regulation of TGF-β1 target genes through SMAD cooperativity. Cell Signal. 2014;26:1427–1436. doi: 10.1016/j.cellsig.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samarakoon R, Dobberfuhl AD, Cooley C, Overstreet JM, Patel S, Goldschmeding R, Meldrum KK, Higgins PJ. Induction of renal fibrotic genes by TGF-β1 requires EGFR activation, p53 and reactive oxygen species. Cell Signal. 2013;25:198–209. doi: 10.1016/j.cellsig.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Luo K. Signaling cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb Perspect Biol. 3 doi: 10.1101/cshperspect.a022137. pii: a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu F, Liu W, Li T, Wan J, Tian J, Zhou Z, Li H, Liu Y, Hou FF, Nei J. Numb contributes to renal fibrosis by promoting tubular epithelial cell cycle arrest at G2/M. Oncotarget. 2016;7:25604–25619. doi: 10.18632/oncotarget.8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akhurst RJ, Padgett RW. Matters of context guide future research in TGFβ superfamily signaling. Sci Signal. 2015;8:re10. doi: 10.1126/scisignal.aad0416. 1126/scisignal.aa0416. [DOI] [PubMed] [Google Scholar]
- 59.Liang M, Yu M, Xia R, Song K, Wang J, Luo J, Chen G, Cheng J. Yap/Taz deletion in Gli+ cell-derived myofibroblasts attenuates fibrosis. J Am Soc Nephrol. 2017 doi: 10.1681/ASN.2015121354. pii: ASN2015121354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rogers R, Dharsee M, Ackloo S, Flanagan JG. Proteomics analyses of activated human optic nerve head lamina cribrosa cells following biomechanical strain. Invest Ophthalmol Vis Sci. 2012;53:3806–3816. doi: 10.1167/iovs.11-8480. [DOI] [PubMed] [Google Scholar]
- 61.Mayr M, Hu Y, Hainaut H, Xu Q. Mechanical stress-induced DNA damage and rac-p38MAPK signal pathways mediated p53-dependent apoptosis in vascular smooth muscle cells. FASEB J. 2002;16:1423–1425. doi: 10.1096/fj.02-0042fje. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. J Cell Physiol. 2012;227:493–507. doi: 10.1002/jcp.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elston R, Inman GJ. Crosstalk between p53 and TGF-β signalling. J Signal Transduct. 2012:294097. doi: 10.1155/2012/294097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piccolo S. p53 regulation orchestrates the TGF-beta response. Cell. 2008;133:767–769. doi: 10.1016/j.cell.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 65.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 66.Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science. 2007;315:840–843. doi: 10.1126/science.1135961. [DOI] [PubMed] [Google Scholar]
- 67.Wei Q, Dong G, Yang T, Megyesi J, Price PM, Dong Z. Activation and involvement of p53 in cisplatin-induced nephrotoxicity. Am J Physiol Renal Physiol. 2007;293:F1282–F1291. doi: 10.1152/ajprenal.00230.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou L, Fu P, Huang XR, Liu F, Lai KN, Lan HY. Activation of p53 promotes renal injury in acute aristolochic acid nephrology. J Am Soc Nephrol. 2010;21:31–41. doi: 10.1681/ASN.2008111133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molitoris BA, Dagher PC, Sandoval RM, Campos SB, Ashush H, Fridman E, Brafman A, Faerman A, Atkinson SJ, Thompson JD, Kalinski H, Skaliter R, Erlich S, Feinstein E. siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J Am Soc Nephrol. 2009;20:1754–1764. doi: 10.1681/ASN.2008111204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samarakoon R, Overstreet JM, Higgins SP, Higgins PJ. TGF-β1 → SMAD/p53/USF2 → PAI-1 transcriptional axis in ureteral obstruction-induced renal fibrosis. Cell Tissue Res. 2012;347:117–128. doi: 10.1007/s00441-011-1181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Overstreet JM, Samarakoon R, Cardona-Grau D, Goldschmeding R, Higgins PJ. Tumor suppressor ataxia telangiectasia mutated functions downstream of TGF-β1 in orchestrating profibrotic responses. FASEB J. 2015;29:1258–1968. doi: 10.1096/fj.14-262527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dupont S, Zacchigna L, Adorno M, Soligo S, Volpin D, Piccolo S, Cordenonsi M. Convergence of p53 and TGF-beta signaling networks. Cancer Lett. 2004;213:129–138. doi: 10.1016/j.canlet.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 73.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kortlever RM, Nijwening JH, Bernards R. Transforming growth factor-beta requires its target plasminogen activator inhibitor-1 for cytostatic activity. J Biol Chem. 2008;283:24308–24313. doi: 10.1074/jbc.M803341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53- regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 76.Chang GS, Chen XA, Park B, Rhee HS, Li P, Han KH, Mishra T, Chan-Salis KY, Li Y, Hardison RC, Wang Y, Pugh BF. A comprehensive and high-resolution genomewide response of p53 to stress. Cell Rep. 2014;8:514–527. doi: 10.1016/j.celrep.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kunz C, Pebler S, Otte J, von der Ahe D. Differential regulation of plasminogen activator and inhibitor gene transcription by the tumor suppressor p53. Nucleic Acids Res. 1995;23:3710–3717. doi: 10.1093/nar/23.18.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Melanson BD, Bose R, Hamill JD, Marcellus KA, Pan EF, McKay BC. The role of mRNA decay in p53-induced gene expression. RNA. 2011;17:2222–2234. doi: 10.1261/rna.030122.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cabrita MA, Bose R, Vanzyl EJ, Pastic A, Marcellus KA, Pan E, Hamill JD, McKay BC. The p53 protein induces stable miRNAs that have the potential to modify subsequent p53 responses. Gene. 2017;608:86–94. doi: 10.1016/j.gene.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 81.White LA, Bruzdzinski C, Kutz SM, Gelehrter TD, Higgins PJ. Growth state-dependent binding of USF-1 to a proximal promoter E box element in the rat plasminogen activator inhibitor type 1 gene. Exp Cell Res. 2000;260:127–135. doi: 10.1006/excr.2000.5001. [DOI] [PubMed] [Google Scholar]
- 82.Kelly KJ, Plotkin Z, Vulgamott SL, Dagher PC. P53 mediates the apoptotic response to GTP depletion after renal ischemia-reperfusion: protective role of a p53 inhibitor. J Am Soc Nephrol. 2003;14:128–138. doi: 10.1097/01.asn.0000040596.23073.01. [DOI] [PubMed] [Google Scholar]
- 83.Ying Y, Kim J, Westphal SN, Long KE, Padanilam BJ. Targeted deletion of p53 in the proximal tubule prevents ischemic renal injury. J Am Soc Neprhol. 2014;25:2707–2716. doi: 10.1681/ASN.2013121270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horikawa I, Park KY, Isogaya K, Hiyoshi Y, Li H, Anami K, Robles AI, Mondal AM, Fujita K, Serrano M, Harris CC. Δ133p53 represses p53-inducible senescence genes and enhances the generation of human induced pluripotent stem cells. Cell Death Differ. 2017;24:1017–1028. doi: 10.1038/cdd.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qi L, Allen RR, Lu Q, Higgins CE, Garone R, Staiano-Coico L, Higgins PJ. PAI-1 transcriptional regulation during the GO → G1 transition in human epidermal keratinocytes. J Cell Biochem. 2006;99:495–507. doi: 10.1002/jcb.20885. [DOI] [PubMed] [Google Scholar]
- 86.Yan C, Higgins PJ. Drugging the undruggable: transcription therapy for cancer. Biochim Biophys Acta. 2013;1835:76–85. doi: 10.1016/j.bbcan.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qi L, Higgins CE, Higgins SP, Law BK, Simone TM, Higgins PJ. The basic helix-loop- helix/leucine zipper transcription factor USF2 integrates serum-induced PAI-1 expression and keratinocyte growth. J Cell Biochem. 2014;115:1840–1847. doi: 10.1002/jcb.24861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fisher DE, Parent LA, Sharp PA. Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove. Proc Natl Acad Sci USA. 1992;89:11770–11783. doi: 10.1073/pnas.89.24.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawarada Y, Inoue Y, Kawasaki F, Fukuura K, Sato K, Tanaka T, Itoh Y, Hayashi H. TGF-β induces p53/Smads complex formation in the PAI-1 promoter to activate transcription. Sci Rep. 2016;6:35483. doi: 10.1038/srep35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tu AW, Luo K. Acetylation of Smad2 by the co-activator p300 regulates activin and transforming growth factor beta response. J Biol Chem. 2007;282:21187–21196. doi: 10.1074/jbc.M700085200. [DOI] [PubMed] [Google Scholar]
- 92.Simonsson M, Kanduri M, Gronroos E, Heldin CH, Ericsson J. The DNA binding activities of Smad2 and Smad3 are regulates by coactivator-mediated acetylation. J Biol Chem. 2006;281:39870–39880. doi: 10.1074/jbc.M607868200. [DOI] [PubMed] [Google Scholar]
- 93.Das F, Ghosh-Choudhury N, Venkatesan B, Li X, Mahimainathan L, Choudhury GG. Akt kinase targets association of CBP with SMAD 3 to regulate TGFbeta-induced expression of plasminogen activator inhibitor-1. J Cell Physiol. 2008;214:513–527. doi: 10.1002/jcp.21236. [DOI] [PubMed] [Google Scholar]
- 94.Antonson P, Jakobsson T, Almlof T, Guldevall K, Stefffensen KR, Gustafsson JA. RAP250 is a coactivator in the transforming growth factor beta signaling pathway that interacts with Smad2 and Smad 3. J Biol Chem. 2008;283:8995–9001. doi: 10.1074/jbc.M707203200. [DOI] [PubMed] [Google Scholar]
- 95.Yang R, Xu X, Li H, Chen J, Xiang X, Dong Z, Zhang D. p53 induces miR199a-3p to suppress SOCS7 for STAT3 activation and renal fibrosis in UUO. Sci Rep. 2017;7:43409. doi: 10.1038/srep43409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sasi W, Sharma AK, Mokbel K. The role of suppressors of cytokine signalling in human neoplasms. Mol Biol Int. 2014;2014:630797. doi: 10.1155/2014/630797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Si H, Lu H, Yang X, Mattox A, Jang M, Bian Y, Sano E, Viadiu H, Yan B, Yau C, Ng S, Lee SK, Romano RA, Davis S, Walker RL, Xiao W, Sun H, Wei L, Sinha S, Benz CC, Stuart JM, Meltzer PS, Van Waes C, Chen Z. TNF-α modulates genome-wide redistribution of ΔNp63α/TAp73 and NK-κB cREL interactive binding on TP53 and AP-1 motifs to promote an oncogenic gene program in squamous cancer. Oncogene. 2016;35:5781–5794. doi: 10.1038/onc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mikulenkova E, Neradil J, Zitterbart K, Sterba J, Veselska R. Overexpression of the ΔNp73 isoform is associated with centrosome amplification in brain tumor cells lines. Tumour Biol. 2015;36:7493–7491. doi: 10.1007/s13277-015-3474-3. [DOI] [PubMed] [Google Scholar]
- 100.Osaki T, Nakagawara A. p73, a sophisticated p53 family member in the cancer world. Cancer Sci. 2005;96:729–737. doi: 10.1111/j.1349-7006.2005.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Niemantsverdriet M, Nagle P, Chiu RK, Langendijk JA, Kampinga HH, Coppes RP. ΔNp73 enhances promoter activity of TGF-β induced genes. PLos One. 2012;7:e50815. doi: 10.1371/journal.pone.0050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.El-Dahr S, Hilliard S, Aboudenhen K, Saifudeen Z. The MDM2-p53 pathway: multiple roles in kidney development. Pediatr Nephrol. 2014;29:621–627. doi: 10.1007/s00467-013-2629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bredhold K, Klein A, Mayo LD. JNK phosphorylation of p53 results in a p53-p73 complex to induce apoptosis. Proceedings of the Indiana University-Purdue University Indianapolis Research Day. 2015 [Google Scholar]
- 104.Zawacka-Pankau J, Kostecka A, Sznarkowska A, Helstrom E, Kawiak A. p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cyle. 2010;9:720–728. doi: 10.4161/cc.9.4.10668. [DOI] [PubMed] [Google Scholar]
- 105.Maclaine NJ, Hupp TR. The regulation of p53 by phosphorylation: a model for how distinct signals integrate into the p53 pathway. Aging (Albany NY) 2009;1:490–502. doi: 10.18632/aging.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duan WJ, Yu X, Huang XR, Yu JW, Lan HY. Opposing roles for Smad2 and Smad3 in peritoneal fibrosis in vivo and in vitro. Am J Pathol. 2014;184:2275–2284. doi: 10.1016/j.ajpath.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 108.Meng XM, Huang XR, Chung AC, Qin W, Shao X, Igarashi P, Ju W, Bottinger EP, Lan HY. Smad2 protects against TGF-beta/Smade3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21:1477–1487. doi: 10.1681/ASN.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor- beta-mediated transcription repression. Mol Cell Biol. 2008;28:1988–1998. doi: 10.1128/MCB.01442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo H, Cowen L, Yu G, Jiang W, Tang Y. SMG7 is a critical regulator of p53 stability and function in DNA damage stress response. Cell Discov. 2016;2:15042. doi: 10.1038/celldisc.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deng YK, Zhao J, Sakura D, Sestak AL, Osadchiy V, Langefeld CD, Kaufman KM, Kelley JA, James JA, Petri MA, Bae SC, Alarcon-Riquelme ME, Alarcon GS, Anaya JM, Criswell LA, Freedman BI, Kamen DL, Gilkeson GS, Jacob CO, Merrill JT, Gaffney PM, Sivils KM, Neiwold TB, Ramsey-Goldman R, Reveille JD, Scofield RH, Stevens AM, Boackle SA, Vila LM, Sohn II, Lee S, Chang DM, Song YW, Vyse TJ, Harley JB, Brown EE, Edberg JC, Kimberly RP, Cantor RM, Hahn BH, Grossman JM, Tsao BP. Decreased SMG7 expression associates with lupus-risk variants and elevated antinuclear antibody production. Ann Rheum Dis. 2016;75:2007–2013. doi: 10.1136/annrheumdis-2015-208441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.El-Dahr S, Hilliard S, Saifudeen Z. Regulation of kidney development by the Mdm2/Mdm4-p53 axis. J Mol Cell Biol. 2017;9:26–33. doi: 10.1093/jmcb/mjx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lei CT, Tang H, Ye C, You CQ, Zhang J, Zhang CY, Xiong W, Su H, Zhang C. MDM2 contributes to high glucose-induced glomerular mesangial cell proliferation and extracellular matrix accumulation via Notch1. Sci Rep. 2017;7:10393. doi: 10.1038/s41598-017-10927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hilliard SA, Yao X, El-Dahr SS. Mdm2 is required for maintenance of the nephrogenic niche. Dev Biol. 2014;387:1–14. doi: 10.1016/j.ydbio.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thomasova D, Ebrahim M, Fleckinger K, Li M, Molnar J, Popper B, Liapis H, Kotb AM, Siegerist F, Endlich N, Anders HJ. MDM2 prevents spontaneous tubular epithelial cell death and acute kidney injury. Cell Death Dis. 2016;7:e2482. doi: 10.1038/cddis.2016.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saito R, Rocanin-Arjo A, You YH, Darshi M, Van Espen B, Miyamoto S, Pham J, Pu M, Romoli S, Natarajan L, Ju W, Kretzler M, Nelson R, Ono K, Thomasova D, Mulay SR, Ideker T, D’Agati V, Beyret E, Belmonte JC, Anders HJ, Sharma K. JCI Insight. 2016;1:e87877. doi: 10.1172/jci.insight.87877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Murphy M, Crean J, Brazil DP, Sadlier D, Martin F, Godson C. Regulation and consequences of differential gene expression in diabetic kidney disease. Biochem Soc Trans. 2008;36:941–945. doi: 10.1042/BST0360941. [DOI] [PubMed] [Google Scholar]
- 118.Ju W, Eichinger F, Bitzer M, Oh J, McWeeney S, Berthier CC, Shedden K, Cohen DD, Henger A, Krick S, Kopp JB, Stoeckert CJ, Jr, Dikman S, Schroppel B, Thomas DB, Schlondorff D, Kretzler M, Bottinger EP. Renal gene and protein expression signatures for prediction of kidney disease progression. Am J Pathol. 2009;174:2073–2085. doi: 10.2353/ajpath.2009.080888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsuo S, Lopez-Guisa JM, Cai X, Okamura DM, Alpers CE, Bumgarner RE, Peters MA, Zhang G, Eddy AA. Multifunctionality of PAI-1 in fibrogenesis: evidence form obstructive nephropathology in PAI-1-overexperessing mice. Kidney Int. 2005;67:2221–2238. doi: 10.1111/j.1523-1755.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 120.Silverstein DM, Travis BR, Thornhill BA, Schurr JS, Kolls JK, Leung JC, Chevalier RL. Kidney Int. 2003;64:25–35. doi: 10.1046/j.1523-1755.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 121.Higgins DF, Lappin DW, Kieran NE, Anders HJ, Watson RW, Strutz F, Schlondorff D, Haase VH, Fitzpatrick JM, Godson C, Brady HR. DNA oligonucletoide microarray technology identifies fisp-12 among other potential fibrogenic genes following murine unilateral ureteral obstruction (UUO): modulation during epithelial-mesenchymal transition. Kidney Int. 2003;64:2079–2091. doi: 10.1046/j.1523-1755.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 122.Eikmans M, Baelde JJ, de Heer E, Bruijn JA. ECM homeostasis in renal diseases: a genomic approach. J Pathol. 2003;200:526–536. doi: 10.1002/path.1417. [DOI] [PubMed] [Google Scholar]
- 123.Sadlier DM, Connolly SB, Kieran NE, Roxburgh S, Brazil DP, Kairaitis L, Wang Y, Harris DC, Doran P, Brady HR. Sequential extracellular matrix-focused and baited-global cluster analysis of serial transcriptomic profiles identifies candidate modulators of renal tubulointerstitial fibrosis in murine adriamycin-induced nephropathy. J Biol Chem. 2004;279:29670–29680. doi: 10.1074/jbc.M313408200. [DOI] [PubMed] [Google Scholar]
- 124.Seseke F, Thelen P, Ringert RH. Characterization of an animal model of spontaneous congenital unilateral obstructive uropathy by cDNA microarray analysis. Eur Urol. 2004;45:374–381. doi: 10.1016/j.eururo.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 125.Freytag J, Wilkins-Port CE, Higgins CE, Carlson JA, Noel A, Foldart JM, Higgins SP, Samarakoon R, Higgins PJ. PAI-1 regulates the invasive phenotype in human cutaneous squamous cell carcinoma. J Oncol. 2009;2009:963209. doi: 10.1155/2009/963209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Freytag J, Wilkins-Port CE, Higgins CE, Higgins SP, Samarakoon R, Higgins PJ. PAI-1 mediates the TGF-beta1+EGF-induced “scatter” response in transformed human keratinocytes. J Invest Dermatol. 2010;130:2179–2190. doi: 10.1038/jid.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boehm JR, Kutz SM, Sage EH, Staiano-Coico L, Higgins PJ. Growth state-dependent regulation of plasminogen activator inhibitor type-1 gene expression during epithelial cell stimulation by serum and transforming growth factor-beta1. J Cell Physiol. 1999;181:96–106. doi: 10.1002/(SICI)1097-4652(199910)181:1<96::AID-JCP10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 128.Brennan EP, Morine MJ, Walsh DW, Roxburgh SA, Lindenmeyer MT, Brazil DP, Gaora PO, Roche HM, Sadlier DM, Cohen CD, Godson C, Martin F GENIE Consortium. Next-generation sequencing identifies TGF-β1-associated gene expression profiles in renal epithelial cells reiterated in human diabetic nephropathy. Biochim Biophys Acta. 2012;1822:589–599. doi: 10.1016/j.bbadis.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Omori K, Hattori N, Senoo T, Takayama Y, Masuda T, Nakashima T, Iwamoto H, Fujitaka K, Hamada H, Kohno N. Inhibition of plasminogen activator inhibitor-1 attenuates transforming growth factor-β-dependent epithelial mesenchymal transition and differentiation of fibroblasts to myofibroblasts. PLos One. 2016;11:e0148969. doi: 10.1371/journal.pone.0148969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Flevaris P, Vaughan D. The role of plasminogen activator inhibitor Type-1 in fibrosis. Semin Thromb Hemost. 2017;43:169–177. doi: 10.1055/s-0036-1586228. [DOI] [PubMed] [Google Scholar]
- 131.Kelley KJ, Liu Y, Zhang J, Goswami C, Lin H, Dominguez JH. Comprehensive genomic profiling in diabetic nephropathy reveals the predominance of proinflammatory pathways. Physiol Genomics. 2013;45:710–719. doi: 10.1152/physiolgenomics.00028.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anglicheau D, Muthukumar T, Hummel A, Ding R, Sharma VK, Dadhania D, Seshan SV, Schwartz JE, Suthanthiran M. Discovery and validation of a molecular signature for the noninvasive diagnosis of human renal allograft fibrosis. Transplantation. 2012;93:1136–1146. doi: 10.1097/TP.0b013e31824ef181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gonzalez J, Klein J, Chauhan SD, Neau E, Calise D, Nevoit C, Chaaya R, Miravete M, Delage C, Bascands JL, Schanstra JP, Buffin-Meyer B. Delayed treatment with plasminogen activator inhibitor-1 decoys reduces tubulointerstitial fibrosis. Exp Biol Med (Maywood) 2009;234:1511–1518. doi: 10.3181/0903-RM-105. [DOI] [PubMed] [Google Scholar]
- 134.Higgins SP, Higgins CE, Higgins PJ. PAI-1 expression is required for HDACi-induced proliferative arrest in ras-transformed renal epithelial cells. Int J Cell Biol. 2011;2011:710974. doi: 10.1155/2011/710974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Samarakoon R, Helo S, Dobberfuhl AD, Khakoo NS, Falke L, Overstreet MJ, Goldschmeding R, Higgins PJ. Loss of tumor suppressor PTEN expression in renal injury initiates SMAD3- and p53-dependent fibrotic responses. J Pathol. 2015;236:421–432. doi: 10.1002/path.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Oda T, Jung YO, Kim HS, Cai X, Lopez-Guisa JM, Ikeda Y, Eddy AA. PAI-1 deficiency attenuates the fibrogenic response to ureteral obstruction. Kidney Int. 2001;60:587–596. doi: 10.1046/j.1523-1755.2001.030002587.x. [DOI] [PubMed] [Google Scholar]
- 137.Gupta KK, Xu Z, Castellino FJ, Ploplis VA. Plasminogen activator inhibitor-1 stimulates macrophage activation through Toll-like receptor 4. Biochem Biophys Res Commun. 2016;477:503–508. doi: 10.1016/j.bbrc.2016.06.065. [DOI] [PubMed] [Google Scholar]
- 138.Zhao R, Le K, Moghadasian MH, Shen GX. Reduced monocyte adhesion to aortae of diabetic plasminogen activator inhibitor-1 knockout mice. Inflamm Res. 2017;66:783–792. doi: 10.1007/s00011-017-1057-z. [DOI] [PubMed] [Google Scholar]
- 139.Park YJ, Liu G, Lorne EF, Zhao X, Wang J, Tsuruta Y, Zmijewski J, Abraham E. PAI-1 inhibits neutrophil efferocytosis. Proc Natl Acad Sci USA. 2008;105:11784–11789. doi: 10.1073/pnas.0801394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2011;12:58–67. doi: 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
- 141.Marquerlot F, Galiacy S, Malo M, Guignabert C, Lawrence DA, d’Ortho MP, Barlovatz-Meimon G. Dual role for plasminogen activator inhibitor type 1 as soluble and as matricellular regulator of epithelial alveolar cell wound healing. Am J Pathol. 2006;169:1624–1632. doi: 10.2353/ajpath.2006.051053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cartier-Michaud A, Malo M, Charriere-Bertrand C, Gadea G, Anquille C, Supiramaniam A, Lesne A, Delaplace F, Hutzler G, Roux P, Lawrence DA, Barlovatz-Meimon G. Matrix-bound PAI-1 supports cell blebbing via RhoA/ROCK1 signaling. PLos One. 2012;7:e32204. doi: 10.1371/journal.pone.0032204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ren W, Wang Z, Hua F, Zhu L. Plasminogen activator inhibitor-1 regulates LPS-induced TLR4/MD-1 pathway activation and inflammation in alveolar macrophages. Inflammation. 2015;38:384–393. doi: 10.1007/s10753-014-0042-8. [DOI] [PubMed] [Google Scholar]
- 144.Wang ZH, Ren WY, Zhu L, Hu LJ. In recognition for her outstanding contribution to ovarian cancer research. Plasminogen activator inhibitor-1 regulates LPS induced inflammation in rat macrophages through autophagy activation. ScientificWorldJournal. 2014;204:189168. doi: 10.1155/2014/189168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nicholas SB, Aguiniga E, Ren Y, Kim J, Wong J, Govindarajan N, Noda M, Wang W, Kawano Y, Collins A, Hsueh WA. Plasminogen activator inhibitor-1 deficiency retards diabetic nephropathy. Kidney Int. 2005;67:1297–1307. doi: 10.1111/j.1523-1755.2005.00207.x. [DOI] [PubMed] [Google Scholar]
- 146.Jeong BY, Uddin MJ, Park JH, Lee JH, Lee HB, Miyata T, Ha H. Novel plasminogen activator inhibitor-1 inhibitors prevent diabetic kidney injury in a mouse model. PLoS One. 2016;11:e0157012. doi: 10.1371/journal.pone.0157012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seo JY, Park J, Yu MR, Kim YS, Ha H, Lee HB. Positive feedback loop between plasminogen activator inhibitor-1 and transforming growth factor-beta1 during renal fibrosis in diabetes. Am J Nephrol. 30:481–490. doi: 10.1159/000242477. 21009. [DOI] [PubMed] [Google Scholar]
- 148.Cheng M, Liu H, Zhang D, Liu Y, Wang C, Liu F, Chen J. HMGB1 enhances the AGE-induced expression of CTGF and TGF-β via RAGE-dependent signaling in renal tubular epithelial cells. Am J Nephrol. 2015;41:257–266. doi: 10.1159/000381464. [DOI] [PubMed] [Google Scholar]
- 149.Bhattacharyya S, Varga J. Emerging role of innate immune signaling and toll-like receptors in fibrosis and systemic sclerosis. Curr Rheumatol Rep. 2015;17:474. doi: 10.1007/s11926-014-0474-z. [DOI] [PubMed] [Google Scholar]
- 150.Bhattacharyya S, Kelley K, Melichian DS, Tamaki Z, Fang F, Su Y, Feng G, Pope RM, Budinger GR, Mutlu GM, Lafyatis R, Radstake T, Feghali-Bostwick C, Varga J. Toll-like receptor 4 signaling augments transforming growth factor-β responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am J Physiol. 2013;182:192–205. doi: 10.1016/j.ajpath.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Degryse B, Neels JG, Czekay RP, Aertgeerts K, Kamikubo Y, Loskutoff DJ. The low density lipoprotein receptor-related protein is a motogenic receptor for plasminogen activator inhibitor-1. J Biol Chem. 2004;279:22595–22604. doi: 10.1074/jbc.M313004200. [DOI] [PubMed] [Google Scholar]
- 152.Simone TM, Higgins SP, Archambeault J, Higgins CE, Ginnan RG, Singer H, Higgins PJ. A small molecule PAI-1 functional inhibitor attenuates neointimal hyperplasia and vascular smooth muscle cell survival by promoting PAI-1 cleavage. Cell Signal. 2015;27:923–933. doi: 10.1016/j.cellsig.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Thapa B, Koo BH, Kim YH, Kwon HJ, Kim DS. Plasminogen activator inhibitor-1 regulates infiltration of macrophages into melanoma via phosphorylation of FAK-Tyr. Biochem Biophys Res Commun. 2014;450:1696–1701. doi: 10.1016/j.bbrc.2014.07.070. [DOI] [PubMed] [Google Scholar]
- 154.Krag S, Danielsen CC, Carmeliet P, Nyengaard J, Wogensen L. Plasminogen activator inhibitor-1 gene deficiency attenuates TGF-beta1-induced kidney disease. Kidney Int. 2005;68:2651–2666. doi: 10.1111/j.1523-1755.2005.00737.x. [DOI] [PubMed] [Google Scholar]