Abstract
Miniproteins simplify the protein-folding problem, allowing the dissection of forces that stabilize protein structures. Here we describe PPα-Tyr, a designed peptide comprising an α helix buttressed by a polyproline-II helix. PPα-Tyr is water soluble, monomeric, and unfolds cooperatively with a midpoint unfolding temperature (TM) of 39 °C. NMR structures of PPα-Tyr reveal proline residues docked between tyrosine side chains as designed. The stability of PPα is sensitive to the aromatic residue: replacing tyrosine by phenylalanine, i.e. changing three solvent-exposed hydroxyl groups to protons, reduces the TM to 20 °C. We attribute this to the loss of CH–π interactions between the aromatic and proline rings, which we probe by substituting the aromatic residues with non-proteinogenic side chains. In analyses of natural protein structures we find a preference for proline-tyrosine interactions over other proline-containing pairs, and abundant CH–π interactions in biologically important complexes between proline-rich ligands and SH3 and similar domains.
Introduction
The accumulation and cooperation of weak non-covalent interactions (NCIs) are critical for the stabilization of the folded, functional states of proteins.1 In addition to hydrogen bonds, van der Waals’ interactions and salt bridges, other NCIs are increasingly recognized as important contributors to protein stability, e.g., CH–π, cation–π and n→π* interactions.2–5 Cooperativity, interplay, and even competition between many such weak interactions further complicate computational analysis and experimental dissection of NCIs.6,7 Indeed, our current understanding of such forces and how they work together is incomplete and largely qualitative.
One route to improving our understanding of NCIs in proteins is to engineer or design smaller protein-like structures; i.e., so-called miniproteins, which are polypeptide chains shorter than 40 – 50 amino acids with stable tertiary structures.8–11 However, the requirement for optimized NCIs is even greater in these structures, where, despite the lower entropic cost of folding, the potential for NCIs is reduced because of their small size. Consequently, few miniproteins have been structurally characterized to high resolution, and of those that have many oligomerize,12 are stabilized by disulfide bonds,13–15 or depend on metal binding.16
For example, cysteine knots have two disulfide bonds that form a ring through which a third disulfide bond is threaded. This imparts exceptional stability even to enzymatic proteolysis.14 The folding of zinc-finger peptides depends entirely on the binding of zinc, which is usually coordinated by sequence and spatially conserved cysteine and histidine residues. Remarkably, this leaves the majority of the remaining sequence positions free for mutations to many other amino acids without disrupting the overall tertiary structure. Although calcium-binding EF hands comprise two short α-helices separated by the metal-binding loop these usually dimerize for additional stability.17
The folded structures of the villin headpiece,18 the tryptophan zipper,19 the Trp-cage,10 and most recently of TrpPlexus11 are notable exceptions to the above, as the stabilities of these miniproteins are not contingent on the presence of covalent crosslinks or ligand binding. As the first example, the 20-residue Trp-cage peptide is particularly noteworthy. It has a short α helix that presents a single tryptophan (Trp) residue, which is penned in by three proline (Pro) residues from an abutting irregular piece of structure. The Trp-cage has a midpoint of thermal unfolding (TM) of 42 °C, although the transition is broad and the peptide is fully folded only below 10 °C.10 This stability has been improved by rational design.20
α Helices are standard building blocks in many natural proteins and the majority of successful protein designs described to date, including miniproteins. Although examples of persistent free-standing α helices are found in nature and have been designed,21 α helices are usually stabilized through tertiary and quaternary interactions. Commonly, the α helices of natural and designed water-soluble proteins have hpphppp or similar sequence repeats of hydrophobic (h) and polar (p) residues. For example, in α-helical coiled coils the repeats usually have 7 residues, called heptads, i.e. ‘abcdefg’ where the ‘a’ and ‘d’ positions are hydrophobic.12 These repeats closely match the 3.6-residues-per-turn periodicity of the α helix, leading to amphipathic helices with distinct hydrophobic and polar faces. Stabilization is then conferred through the packing the hydrophobic faces of two or more such amphipathic helices, leaving the polar faces exposed to aqueous media. More specifically, these h-type residues project out from neighbouring helices and combine in so-called knobs-into-holes (KIH) packing, which define and stabilize the helical assemblies.22,23 Short runs of these intimate KIH interactions can lead to a variety of very stable and specific quaternary structures formed by peptides just 20 – 40 residues in length.24
Here we describe the design and characterization of a series of short peptides, PPα, which adopt a stable monomeric fold that combines an amphipathic α helix and a stretch of polyproline-II helix. This compact tertiary structure is stabilized by tight inter-digitation of proline (Pro) residues from the latter and aromatic side chains displayed by the α helix. This packing is reminiscent of KIH interactions.22,23 Our experimental studies of PPα and bioinformatics analyses of proline-aromatic side-chain contacts in protein structures more generally unveil a key role for CH–π interactions3,25,26 in these Pro-Tyr-based packing arrangements. We argue that these contribute to the global stability of PPα, as well as for the affinities of proline-based protein-protein interactions more widely.
Results
Miniprotein design and characterization
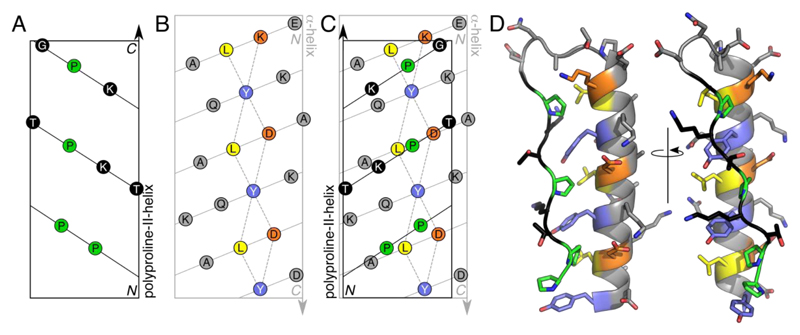
The design of PPα-Tyr borrowed from two natural proteins: a surface adhesin and antigen (AgI/II) from Streptococcus mutans, and the family of pancreatic polypeptide hormones (Supplementary Results, Supplementary Fig. 1).27,28 In both structures, a polyproline-II helix and an α helix combine to form an unusual tertiary structure in which Pro residues from the former dock into holes formed by regularly spaced aromatic residues of the latter, Figures 1A-C. In AgI/II the helices are long, resulting in an overall fibrous structure, and the smaller peptide hormones form dimers.29 To reduce this complexity—i.e., to reduce the size of the system, and to eliminate dimer formation—we combined short segments of the two helical types in silico, choosing segment lengths to match best the different helical repeats, and sequences based on fragments from AgI/II, with tyrosine (Tyr) at the aromatic ‘d’ sites, Figure 1C. Engineering loops in protein design is notoriously difficult.30,31 Therefore, we connected the two elements of secondary structure with the loop from the bovine pancreatic polypeptide hormone sequence. A full model for the resulting PPα-Tyr sequence, Table 1, with the topology polyproline-II helix—loop—α-helix, was constructed, energy minimized and found to be stable over 100 ns of molecular-dynamics (MD) simulations in water, Figure 1D.
Figure 1. Design of PPα combining polyproline-II and α helices.
A-C: 2D helical nets—i.e., projections of Cα atoms onto the surfaces of cylinders of appropriate radii—for a canonical polyproline-II helix (A), an α helix (B), and with these two overlaid showing ‘knobs-into-holes’ packing of the Pro and Tyr side chains (C). The paths of the backbones are shown as solid lines, while dashed lines outline the ‘holes’ presented by the α helix. Color key: Tyr, slate; Leu, yellow; Lys and Asp, orange; and Pro, green. D: In silico model for the designed PPα-Tyr sequence, Table 1, after 100 ns of molecular-dynamics simulation in water.
Table 1. Peptides designed and characterized in this study.
The helical register for the α helix is indicated, 'efgabcd'. Key: AUC, molecular weight relative to monomer mass from analytical ultracentrifugation; TM, midpoint of thermal unfolding transition measured by CD spectroscopy; ϕ, non-proteinogenic amino acid based on L-phenylalanine with the para substituents given in the peptide name. Variants with ϕ = 4-trifluoromethyl-, 4-iodo-, 4-bromo- and 4-chloro-phenylalanine were also made, but these aggregated in solution.
| Peptide | Sequence efgabcd efgabcd efgabcd |
AUC (x monomer) |
TM
(°C) |
|---|---|---|---|
| PPα-Tyr | Ac-PPTKPTKP GDNAT PEKLAKY QADLAKY QKDLADY-NH2 | 0.9 | 39 |
| PPα-Phe | Ac-PPTKPTKP GDNAT PEKLAKF QADLAKF QKDLADF-NH2 | 0.9 | 20 |
| PPα-Trp | Ac-PPTKPTKP GDNAT PEKLAKW QADLAKW QKDLADW-NH2 | 0.9 | 36 |
| PPα-His | Ac-PPTKPTKP GDNAT PEKLAKH QADLAKH QKDLADH-NH2 | ND | < 0 |
| PPα-ϕNH2 | Ac-PPTKPTKP GDNAT PEKLAKϕ QADLAKϕ QKDLADϕ-NH2 | 1.0 | 19 |
| PPα-ϕOCH3 | Ac-PPTKPTKP GDNAT PEKLAKϕ QADLAKϕ QKDLADϕ-NH2 | 1.0 | 38 |
| PPα-ϕCH3 | Ac-PPTKPTKP GDNAT PEKLAKϕ QADLAKϕ QKDLADϕ-NH2 | 0.8 | 31 |
| PPα-ϕF | Ac-PPTKPTKP GDNAT PEKLAKϕ QADLAKϕ QKDLADϕ-NH2 | 1.0 | 26 |
| PPα-ϕCN | Ac-PPTKPTKP GDNAT PEKLAKϕ QADLAKϕ QKDLADϕ-NH2 | 0.9 | 22 |
| PPα-ϕNO2 | Ac-PPTKPTKP GDNAT PEKLAKϕ QADLAKϕ QKDLADϕ-NH2 | 1.1 | 17 |
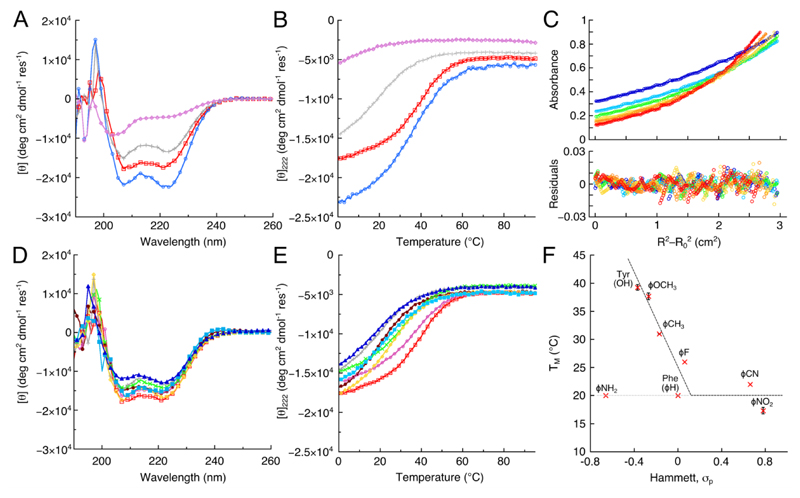
A 34-residue synthetic peptide for PPα-Tyr (Supplementary Fig. 2) was soluble in aqueous buffer at pH 7.4. As judged by circular dichroism (CD) spectroscopy and consistent with the design, PPα-Tyr was folded with approximately 50% α-helical structure at 5 °C, Figure 2 and Supplementary Figure 4. Temperature dependence of the far-UV CD signal at 222 nm, which reports directly on the secondary structure present, revealed a reversible unfolding transition with a TM of 39 °C, Figure 2B and Supplementary Figure 4. Furthermore, monitoring this transition by near-UV CD spectroscopy, which reports on the tertiary structure, gave an unfolding and refolding curve that were coincident with the far-UV CD traces (Supplementary Fig. 4). These data indicate fully cooperative unfolding and refolding behavior. Analytical ultracentrifugation (AUC) showed that PPα-Tyr was monomeric, Figure 2C and Supplementary Figure 5.
Figure 2. Folding and stability of PPα-Tyr and PPα variants.
A: CD spectra recorded at 5 °C and B: thermal unfolding curves measured through the CD signal at 222 nm for PPα-Trp (blue circles), PPα-Tyr (red squares), PPα-Phe (gray crosses) and PPα-His (lilac diamonds). C: AUC data for PPα-Tyr (circles), and fits to a monomeric single ideal species (lines, upper panel), with residuals (lower panel) at rotor speeds of 40 krpm (blue), 44 krpm (light blue), 48 krpm (green), 52 krpm (yellow), 56 krpm (orange) and 60 krpm (red). D: CD spectra recorded at 5 °C for p-substituted phenylalanine-containing peptides; and E: thermal unfolding curves for the same peptides. Color key for D&E: (listed in order of σp values for the p-substituent) PPα-NH2 (burgundy filled circles), PPα-Tyr (red squares), PPα-ϕOCH3 (pink circles), PPα-ϕCH3 (green saltires), PPα-Phe (gray crosses), PPα-ϕF (yellow diamonds), PPα-ϕCN (light blue filled squares) and PPα-ϕNO2 (blue filled triangles). F: Plot of TM values against the Hammett σp parameter for the corresponding aromatic substituent. Errors bars represent one standard deviation from the mean of at least three data sets. Dashed lines are included simply to guide the eye. In parts A, B, D&E, representative spectra from at least three replicate experiments are shown.
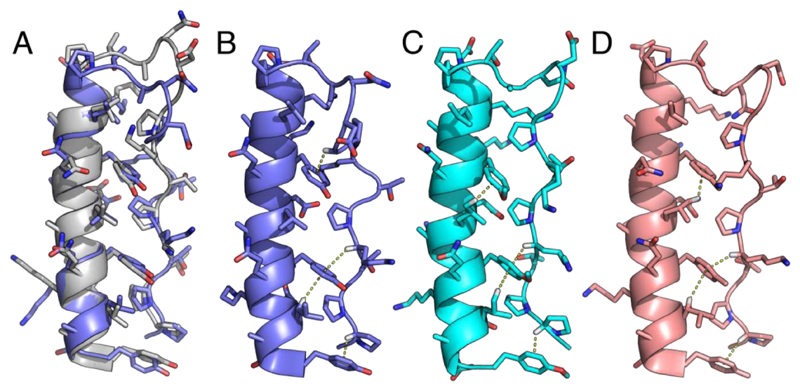
High-resolution and high-sensitivity nuclear magnetic resonance (NMR) spectroscopy was used to determine the solution structure of PPα-Tyr. This employed standard homo-nuclear experiments and natural-abundance 15N- and 13C-edited HSQC spectra. 87% of the 1H resonances were assigned, Supplementary Table 1, with the side chains of solvent-exposed lysine residues mostly accounting for the missing assignments. Consistent with the design, the PPα-Tyr structure comprised a polyproline-II helix and loop (residues 1—13) and an α helix (residues 14—33), Figures 3A&B. The core of the structure was highly defined with numerous strong NOEs between the aromatic side chains and surrounding residues. Unsurprisingly, the conformations of some of the solvent-exposed side chains were less well defined and could not be fully assigned, which resulted in some variation across the ensemble: the backbone RMSD was 0.514 Å ± 0.121 Å, and the all-atom RMSD 0.825 ± 0.122 Å, Supplementary Figure 6. A representative structure from this ensemble matched the in silico model with RMSDs of 0.7 Å and 1.3 Å measured over the backbone and all atoms, respectively. Moreover, at the helix-helix interface KIH-type packing between the tyrosine and proline residues was evident, and these side chains were in close contact, Figure 3.
Figure 3. NMR structures for the p-substituted phenylalanine variants of PPα.
A: NMR structure closest to the geometric mean of the ensemble (model 20) for PPα-Tyr (slate) overlaid with the in silico model after 100 ns of MD (gray). B-D: Representative NMR structures from the ensembles showing the CH–π interactions found for PPα-Tyr, model 14 (B), PPα-ϕOCH3, model 8 (C), and PPα-ϕCH3, model 5 (D). The average numbers of CH–π interactions per ensemble structure were 2.25, 2.7 and 2.5, respectively, with 1.2, 0.65 and 0.55 per structure involving Pro. Although the remaining PPα peptides were folded by NMR they gave poor-quality spectra and structure calculations were not possible, which corroborated their reduced thermal stability. PDB codes: PPα-Tyr, 5LO2; PPα-ϕOCH3, 5LO3; and PPα-ϕCH3, 5LO4.
Pro-aromatic contacts promote CH–π interactions
Because of the close contacts between the tyrosine and proximal aliphatic side chains, we searched for potential CH–π interactions in PPα-Tyr. To do this, we used an operational definition for these interactions and parameters adapted from previous studies (Supplementary Figs. 7&8).3,32 We found 24 CH–π interactions between these residues across the ensemble of 20 structures, and detected additional CH–π interactions involving 15 lysine, 4 leucine and 2 glycine residues as the CH donors, Supplementary Table 2.
On this basis, we posited that the stability of PPα might be promoted through improved CH–π interactions with the Tyr residues substituted for tryptophan (Trp), which has a more electron-rich aromatic system, and reduced when replaced by histidine (His) or phenylalanine (Phe), which have less electron-rich rings.2,32 Consistent with this, but nevertheless surprisingly, PPα-His was largely unfolded as judged by CD spectroscopy, Figs 2A&B. We took the characterization of this peptide no further. The other variants, PPα-Trp and PPα-Phe, Table 1, were soluble, cooperatively folded by CD spectroscopy (Figs. 2A&B and Supplementary Fig. 4), and monomeric in AUC, (Supplementary Fig. 5). PPα-Phe was destabilized to a significant degree with the TM reduced to 20 °C compared with 39 °C for PPα-Tyr. This is remarkable given the small chemical changes involved, i.e. three solvent-exposed hydroxyl groups in the protein were replaced by protons. Contrary to initial expectations, the stability of PPα-Trp (TM = 36 °C) was comparable to that of PPα-Tyr. Thus, whilst electron-poor His and Phe do destabilize PPα, both electron-rich aromatics (Tyr and Trp) stabilize the structure to similar extents.
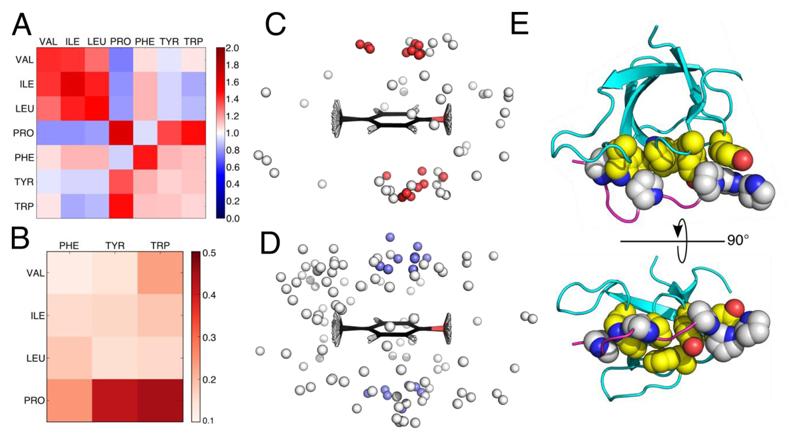
To understand this better, we analyzed interactions between Pro and aromatic side chains, and, for comparison, all aliphatic—aromatic side-chain contacts in the RCSB Protein Data Bank (PDB), Figure 4 and Supplementary Figures 7&8. We used only non-redundant (<40% sequence identity) X-ray protein crystal structures of 1 Å resolution or better, and those that had all CH protons experimentally determined. A side-chain contact map for the propensity of interactions between Val, Ile, Leu, Pro, Phe, Tyr and Trp, revealed several trends, Figure 4A, some of which have been noted by others:3,33 (1) like-with-like contacts were favored, i.e. aliphatic—aliphatic and aromatic— aromatic; (2) in general, aliphatic—aromatic contacts were neutral, i.e. they occurred at rates expected by chance; and (3) Phe was unusual in that it made more contacts than expected with all of the other hydrophobic residues except Pro. However, we found Pro broke these patterns: despite having an aliphatic cyclic side chain, it contacted the other aliphatic residues and Phe less often than expected; and, in contrast, Pro interacted with Tyr and Trp significantly more often than expected by chance, Fig. 4A and Supplementary Fig. 7.
Figure 4. Pairwise side-chain and CH–π interactions in the PDB.
A: Heat map for the propensity (observed/expected ratio) of amino-acid pairs with one or more sub-3 Å atom-atom contacts. B: Heat map of the proportion of aliphatic-aromatic close contacts that participate in CH–π interactions normalized for propensity of the pairs to be in close contact. C: Overlay of Pro-Tyr side-chain contacts within 3 Å; gray spheres represent the centers of mass of Pro side chains, with those that tested positive for CH–π interactions colored red. D: Similar to C but for Val-Tyr contacts, and CH–π positive interactions colored slate. E: Orthogonal views of human adapter protein Tuba SH3 domain (PDB: 4CC7), which has 7 CH–π interactions between its binding domain and Pro-rich peptide of N-WASP. Color key: atoms of proline side chains of the ligands, gray and blue; atoms of the interacting side chains from the SH3 domain, yellow, red and blue.
Closer examination of the Pro-aromatic pairings from the PDB showed that approximately one quarter had potential CH–π interactions, Supplementary Table 3. Moreover, Pro-Trp and Pro-Tyr pairs participated in these predicted CH–π interactions much more than Pro-Phe, Figure 4B. As a result, these pairs were highly directional compared with the more-isotropic distributions of aliphatic residues around aromatics, compare Figures 4C&D. This directionality likely arises from a combination of electrostatic and electronic interactions between the electron-rich aromatic groups, and the slightly acidic protons of the pyrrolidine ring of Pro, which are both consistent with electrostatic surface potentials, Supplementary Figure 9.
Non-proteinogenic substitutions in PPα
To probe CH–π interactions in the PPα system, ten further variants were synthesized with para-substituted phenylalanine residues at the aromatic sites covering electron-rich p- methoxyphenylalanine through electron-poor p-nitrophenylalanine, Table 1 and Supplementary Figure 9. Six of the peptides were soluble, folded, monomeric, and gave full or near-complete thermal unfolding curves, Figures 2D&E and Supplementary Figure 5. NMR structures for the most-stable variants, p-methoxyphenylalanine and p-methylphenylalanine, again revealed intimate contacts and CH–π interactions between the pyrrolidine ring of Pro and the faces of modified aromatic rings (Figs. 3C&D). The numbers of contacts made consistent with CH–π interactions across the 20 conformers of these two ensembles were 54 and 50, respectively.
To probe the contribution of these potential CH–π interactions to PPα stability, the stabilities of the para-substituted phenylalanine variants were plotted against the corresponding Hammett constant σp, Figure 2F. Formally, the Hammett equation relates the equilibrium constant for the dissociation of substituted benzoic acids to two parameters: the substituent or Hammett constant, σ; and the reaction constant, ρ. The Hammett constant provides a measure of how much the substituent stabilizes the negative charge of the conjugate base. Traditionally, it is interpreted in terms of through-bond inductive and mesomeric effects that alter the electrostatics of the ring. Whilst we recognize that this has potential caveats,34 here we use σp as a proxy for the electron density in the aromatic ring,35 Supplementary Fig. 9, to compare the thermal unfolding reactions of the PPα variants. On this premise, we plotted the TM’s of each mutant against σp for the appropriate substituted aromatic residue Figures 2F, and we also plotted various thermodynamic parameters obtained from fitting of the full unfolding curves against σp (Supplementary Fig. 10).
The data for PPα-Tyr, PPα-ϕOMe, PPα-ϕCH3, PPα-ϕF and PPα-Phe were close to linear with a negative slope. This is strong evidence for electrostatic and electronic contributions to aromatic-Pro interactions over and above the hydrophobic effect and van der Waals’ interactions.36 Specifically, it provides evidence for CH–π interactions, which would redistribute electron density from the ring into the CH bond, and so be favored by the electron-donating groups in this series. The Hammett plot leveled off for the PPα-ϕCN and PPα-ϕNO2 variants, consistent with arguments that cyano- and nitro-functionalized benzenes have weaker interaction energies with XH groups37 and consequently, weaker CH–π interactions in PPα.
n.b. The stability of PPα-ϕNH2 was lower than expected based on the σp value of aniline. We have no clear explanation for this. We measured the pKa of the p-amino group in the peptide using a pH titration and following the UV spectrum, but we found it was unperturbed from that of the free amino acid. Thus, we assume that the lone pair of electrons of the substituent is fully available to the π-system and that the σp value is appropriate. Because of the reduced stability of this peptide compared to PPα-Tyr, a full assignment of the NMR signals was not possible nor was a structure determination.
The thermal denaturation profiles of Figure 2E were fitted by van’t Hoff analyses to determine ΔHunf, ΔSunf and ΔGunf at 5 °C where all of the peptides were close to fully folded, Supplementary Figure 10 and Supplementary Table 4. Interpreting ΔSunf and ΔHunf values for protein folding is complicated. Therefore, we focused on the free energies of unfolding, ΔGunf, which differed between the mutants, and, like the TM values (Fig. 2F), these varied linearly with σp, Supplementary Figure 10. The ΔGunf values were spread over 3.6 kJ mol-1 ≈ 0.9 kcal mol-1. With 2 – 3 CH–π interactions per structure from the NMR data, it is interesting that this energy is close to literature estimates for CH–π interactions of ≈ 1.5 – 2.8 kcal mol-1.38 Though small, energy differences on this scale shift equilibrium or binding constants by nearly an order of magnitude. Thus, the presence of even a small number of these NCIs will influence the energetics of biomolecular folding and association considerably.
Pro-aromatic interactions in ligand binding
With the potential contributions to free energies of binding in mind, we examined Pro-aromatic contacts known to be important in natural biological processes. Specifically, interactions between SH3, WW, EVH1 and profilin domains and their target proline-rich ligands were inspected, Figure 4E.39,40 Amongst other functions, these protein-peptide interactions control pathways in cell growth, transcription, cytoskeletal remodeling and other regulatory functions across all kingdoms of life. Within the 596 X-ray crystal and NMR structures containing such domains in the PDB, 135 chains had non-covalently bound polypeptide ligands with ≥ 3 contiguous residues in polyproline-II-helix conformations, Supplementary Table 5. When culled at 80% protein-sequence identity, and taking only X-ray crystal structures of ≤ 2.1 Å resolution along with NMR structures, this yielded 38 complexes. On average, the polyproline-II stretches of the ligands in the assessed structures were 4 – 5 residues long. Within this set, there were 121 CH–π inter-chain interactions, at an average of 3.18 CH–π interactions per complex. 55% of Pro, which accounted for 149 of 407 ligand residues, participated in CH–π interactions. This is significantly more than the 16% of Pro that form CH–π interactions across the entire PDB, Supplementary Tables 3&5. In other words, Pro-aromatic and CH–π interactions in the SH3 and similar domains are denser and more frequent than those generally found in proteins. Tyr was the most frequent CH–π partner for Pro in these protein-ligand interactions, followed by the two rings of Trp, and then Phe, Supplementary Figure 8.
Discussion
In conclusion, we report the fragment-based design and complete structural characterization of a new miniprotein, PPα, with a stable, monomeric polyproline-II helix—loop—α-helix fold. In the design, the lengths of the two helices were chosen to best match the different repeats of the two types of helix. This was done to promote intimate knobs-into-holes packing of Pro and Tyr side chains from the polyproline-II and the α helix, respectively. Our biophysical data and high-resolution solution-phase NMR structures validate this approach. Moreover, they reveal that, over and above the anticipated hydrophobic effect and van der Waals’ forces from the packing arrangement, PPα is stabilized by CH–π interactions between the Pro and Tyr side chains. This is supported by stability studies in a series of para-substituted phenylalanine mutants of PPα, which confirm an electrostatic/electronic component to the Pro-aromatic interactions: peptides with electron-rich aromatic π-systems are more thermally stable and have more favorable free-energies of folding than those with electron-withdrawing substituents. Of the proteinogenic aromatic amino acids, the electron-rich Tyr and Trp give more stable PPα folds and appear to make better CH–π interactions than the Phe and His mutants.
Analyses of the RCSB Protein Data Bank add considerably to these conclusions: Pro-Tyr and Pro-Trp interactions are observed much more frequently than expected by chance, and also more frequently than any other aliphatic-aromatic side-chain pairings. By contrast, Pro-Phe contacts are underrepresented. Furthermore, Pro-Tyr and Pro-Trp make many more CH–π interactions than any of the other side-chain interactions. More specifically, protein-ligand interactions involving proline-rich ligands, such as those found in SH3 domains, indicate that Pro-Tyr contacts are particularly favored and lead to unusually high densities of CH–π interactions in these complexes. This is noteworthy because the literature on protein-peptide interactions of this type focuses on the stabilizing influence of only the hydrophobic effect. Therefore, we propose that CH–π interactions also contribute to the observed affinities of these short linear-peptide ligands.
Our observations raise the question: why does Pro interact preferably with Tyr rather than the larger π system of Trp in these cases? We suggest that the single aromatic ring of Tyr allows sufficient Pro-aromatic contacts, whereas the larger Trp makes packing more difficult and may even lead to lower solubility of the unbound states of the ligand. The last point could be important for both systems examined herein where the aromatic residues are partly exposed, as in the adhesins and pancreatic polypeptides,27,28 or exposed part of the time, as with the free SH3 and other domains.41
Our findings indicate that CH–π interactions, which are traditionally considered as weak NCIs, can have considerable impact on protein folding and stability. Moreover, these interactions could be particularly important in the design and optimization of miniproteins, protein mimics, protein-ligand interactions and possibly catalysts.42 Therefore, and as we have shown, unpicking the contributions of such NCIs to protein stability, folding and association using the subtleties of non-proteinogenic side chains will be critical in developing our understanding of and for manipulating these fundamental forces.43,44 As one of the smallest, monomeric globular protein folds described to date, PPα provides a particularly attractive model system for advancing such studies. Finally, we encourage others to consider weak NCIs, such as CH–π interactions, in the design and development of small molecules that mimic or disrupt currently undruggable natural protein-protein interactions.13,45
Accession codes
The NMR structures for PPα-Tyr, PPα-ϕOCH3, and PPα-ϕCH3 determined in this study are deposited in the RCSB PDB with accession codes 5LO2, 5LO3 and 5LO4, respectively (http://www.rcsb.org/).
Materials and Methods
Bioinformatics
All protein X-ray crystal structures in the PDB of resolution ≤ 1 Å containing at least one polypeptide chain (as of 24th August 2016) were downloaded from the PDBe.46 The number of experimentally assigned CH protons was determined for each chain in each structure, and the 123 chains which had 100% CH protons assigned were culled at 40% maximum mutual sequence identity using PISCES.47 This gave 47 non-redundant chains from 46 X-ray crystal structures. We used in-house Python scripts to find pairs of intra-chain pairs of residues with at least one inter-atomic distance of ≤ 3 Å between side chain atoms excluding covalently linked pairs. Side chains were defined as Cβ onwards, apart from Pro where Cα and N atoms were also included. Interaction propensities of amino acid pairs were calculated as the number of observed pairs in the dataset / the expected number of pairs assuming a random distribution of pairwise interactions. This approach is similar to that taken by Singh and Thornton in their previous analysis of side-chain – side-chain interactions48 and our new results using high resolution structures correlate closely.
CH–π interactions were identified using parameters based on those to find CH–π interactions involving carbohydrates in protein crystal structures32 and adapted to account for any CH protons interacting with an amino acid aromatic ring. This provides an update to a previous analysis of CH–π interactions.3 In our analysis CH–π interactions were determined between all amino acid CH bonds and the aromatic rings of Phe, Tyr, Trp and His. Trp was split into TrpA and TrpB i.e., the 5- and 6-membered rings, respectively. Interactions were classed as CH–π positive if the following conditions were met: CH–π distance (between the CH proton to center of the aromatic ring) ≤ 3.5 Å; CH–π angle (between the vector of the CH bond and the normal to the plane of the aromatic ring) ≤ 55°; H projection distance (between the projection of the CH bond to the plane of the aromatic ring and the center of the ring) ≤ 1.6 Å for the 5-membered TrpA and His rings and ≤ 2.0 Å for 6-membered rings (Phe, Tyr and TrpB), see Figure S8. A total of 742 intra-chain CH–π interactions were identified across the 55 protein chains. For Figs. 4C&D the center of mass for Pro was calculated as the center point of Cβ, Cγ and Cδ atoms, and that of Val as the center point of the side chain atoms.
We looked again to the PDB for NMR and crystal structures containing SH3 (as identified by Pfam,49 378), WW (Pfam, 163), Profilins (Pfam, 44) and EVH1 (SCOP,50 11) domains (as of 26th August 2016). Within these 596 structures, we identified 134 polypeptide chains with non-covalently bound polyproline helices with ≥ 3 contiguous residues in polyproline conformation. An 80% maximum mutual sequence identity cull using PISCES 47 gave 38 non-identical chains containing domains bound non-covalently to polyproline helix-containing polypeptides or proteins. For X-ray crystal structures where not all CH protons were assigned, these were added using REDUCE with the default settings.51 For NMR structures, the most-representative conformation was determined using OLDERADO web server (https://www.ebi.ac.uk/pdbe/nmr/olderado/), and this was used for subsequent analysis. Again we searched for CH–π interactions with the above parameters and found 121 CH–π interactions with the aromatic acceptor a residue in the binding domain and the CH donor in the polyproline helix- containing binding partner.
Electrostatic potential surfaces
Minimized conformations were generated from Density Functional Theory (B3LYP/6-31+(d)) calculations in the gas phase using Gaussian03 (Revision C.02, 2004). ESPs were then generated from Hartree–Fock (B3LYP/6-31(d)) energy calculations of these conformations at isovalue 0.002 and visualized using PyMOL (www.pymol.org).
Molecular Dynamics Simulations
Models were constructed using Accelrys (2005) and set up for molecular dynamics (MD) simulation using the Gromacs 4.6.7 suite of tools. Hydrogen atoms were added consistent with pH 7.4 using pdb3gmx, and the TIP3P water model and Amber99-ILDN forcefield were chosen. A cubic periodic boundary box was set up with dimensions 2 nm greater than the longest dimension of the model with editconf. This box was filled with water molecules using genbox and 137 mM NaCl using genion. The system was energy minimized and position-restrained MD run for 200 ps as an NPT (normal pressure and temperature) ensemble at 278 K, 1 Bar using the Verlet cut-off scheme and under PME (Particle Mesh Ewald) boundary conditions as an initial relaxation and equilibration step. The restraints were then removed and a further 100 ns of MD was performed using 1 GPU and 6 cores of an X86 workstation. Structures were saved every 10 ps.
Peptide Synthesis
Peptides were synthesized using a microwave-assisted Liberty Blue automated peptide synthesizer (CEM Corporation.) on a rink amide ChemMatrix resin™ (PCAS Biomatix Inc.) using standard Fmoc-coupling chemistry. Fmoc deprotection was via 20% morpholine (Merck Millipore), 5% formic acid (Acros Organics) in peptide-grade DMF (AGTC Bioproducts); followed by Cl-HOBt (AGTC Bioproducts, 0.5 M in DMF) and DIC (AGTC Bioproducts, 1 M in DMF) couplings for each amino acid. 5% formic acid prevented aspartimide formation. Peptides were made in three stages. The first involved single couplings under microwave conditions from residue 34 (aromatic) to residue 14 (Pro) as the peptide was synthesized C→N. The second stage was synthesis of the loop and polyproline helix i.e. residue 13 (Thr) through to residue 3 (Thr) under double coupling, non-microwave conditions (2 hours per coupling) as we found using microwave conditions for this step led to peptide degradation during synthesis. Finally, the resin was removed from the synthesizer and the N-terminal residues Pro1 and Pro2 (each at 0.5 mmoles) were coupled manually using HATU (0.49 mmoles) and DIPEA (0.6 mmoles) in DMF for 1 hour followed by Fmoc-deprotection (20 mins). Acetylations of the N-termini of peptides was achieved with pyridine (Fisher Scientific, 0.5 mL) and acetic anhydride (BDH Laboratories, 0.25 mL) in DMF for 15 mins before washing with DMF x 3 and then DCM x 3. Peptides were cleaved from the resin using TFA (Acros Organics) : H2O : TIPS (Sigma Aldrich) in 90:5:5 vol% for 2 hours under agitation. The cleavage mixtures were then filtered from the resin and the volume reduced to <5 mL under a flow of nitrogen. Diethyl ether (VWR Chemicals) was then added to precipitate the peptide over ice. The solid peptide was obtained by centrifugation and removal of the supernatant. The peptide pellet was then dissolved in H2O:MeCN (1:1 vol%) ready for lyophilisation which yielded the crude peptide as a white powder.
Peptide Purification
Peptides were purified using a JASCO HPLC system with a reverse phase Luna® C18 column (Phenomenex, 5 μm particle size, 100 Å pore size, 150 x 10 mm). A linear gradient of 10 – 60% buffer B (0.1 vol% TFA in MeCN, VWR Chemicals) vs. buffer A (0.1 vol% TFA in H2O) was typically used. The identities of the peptides (with the exception of PPα-ϕNO2) was confirmed by MALDI-TOF mass spectrometry using a Bruker ultrafleXtreme II instrument in reflector mode. Peptides were co-crystallized with dihydroxybenzoic acid matrix (Sigma Aldrich) on a ground steel plate. The identity of PPα-NO2 was confirmed by infusing the sample from an Advion Nanomate Triverser (Nanospray source) at 1.4 kV into a Waters Synapt G2S IMS Q-TOF mass spectrometer. Peptide purities were determined to be >95% by a JASCO analytical HPLC system equipped with a reverse phase Kinetex® C18 analytical column (Phenomenex, 5 μm particles size, 100 Å pore size, 100 x 4.6 mm).
Peptide Concentration
Peptide concentrations for PPα-Tyr and PPα-Trp were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific) using the known extinction coefficients for Trp (ε280 = 5690 M-1 cm-1) and Tyr (ε280 = 1280 M-1 cm-1). For p-nitro-Phe, p-cyano-Phe, p-amino-Phe, and p-methoxy-Phe the extinction coefficient was determined at 280 nm by measuring the absorbance of the free amino acid in solution at known concentrations (see Fig. S3). p-nitro-Phe was dissolved in DMSO:H2O, 50:50 vol%. p-amino-Phe was dissolved in PBS and the rest were dissolved in H2O. For the remaining p-substituted phenylalanine derivatives (p-fluoro-Phe and p-methyl-Phe) the extinction coefficient was determined at 214 nm in H2O.
Once the extinction coefficient at 214 nm for these substituted amino acids had been determined the extinction coefficient of each corresponding PPα peptide and also that of PPα-Phe was determined using a literature protocol.52
Circular Dichroism Spectroscopy
Peptides were prepared at 100 μM concentration (250 μL) in phosphate-buffered saline (PBS) comprising Na2HPO4 (8.2 mM), KH2PO4 (1.8 mM), NaCl (137 mM) and KCl (2.7 mM). CD spectra were baseline corrected and recorded as the average of 5 scans from 260 – 190 nm at 5°C using a JASCO 815 spectropolorimeter fitted with a peltier temperature controller, a 1 mm pathlength quartz cuvette (Starna), a scanning speed of 100 nm min-1, and a bandwidth of 1 nm. Thermal denaturation curves were obtained from 0 – 95°C (temperature slope = 40°C hr-1) by monitoring the absorbance at 222 nm (1 nm bandwidth) at 1 °C intervals with 16 sec delay and 16 sec response times. Measurements were performed on at least 3 separately prepared samples of each peptide. The midpoint of the denaturation curve (TM) was determined by taking the maximum value from the first derivative of the thermal transition. This is the TM value quoted in the manuscript unless otherwise stated.
Curve Fitting of Thermal Denaturations
The thermal denaturation data was fit to a Generalised Logistic Function (GLF): f(x) = A + ((K – A)/(1 + Qe-B(x-M))(1/v)) + c using the curve fit function from the SciPy library in Python (www.scipy.org). f(x) is the CD signal as a function of temperature. The GLF is a more complicated version of the sigmoid equation: f(x) = 1 + (1/(1 + e(x-k)). Fitting to the GLF gave the parameters A, K, Q, B, M, v, and c which were then used to extrapolate the curve to temperatures above and below those measured i.e. the upper and lower baselines (see Fig. S4). The fraction folded (α) was calculated based on the lower base line representing the fully folded peptides (α=1) and the upper baseline the fully unfolded peptide (α=0).53 We fitted the resulting fraction folded curve to the GLF of α as a function of temperature. The TM was determined as the temperature at which α = 0.5.
Van’t Hoff Analysis
A van’t Hoff analysis is a plot of ln(K) vs. 1/T where, in this case, K is the unfolding constant and T is the temperature in Kelvin. Such a plot is useful because we can extract the thermodynamic parameters associated with the thermal denaturations. To calculate K, we used the following relationship: K = ((CT / n)n-1(1-α)n) / α where CT is the total strand concentration and n is the oligomeric state, thus for a monomer this equation reduces to K = (1-α)/α. We employed a threshold, β, so that we only used values of α in the range β ≤ α ≤ 1 – β as the natural logarithm close to 0 and 1 is very sensitive. We initially fit the resulting van’t Hoff plots using a linear regression where ln(K) = -ΔH/R + ΔS/R where ΔH is the enthalpy of unfolding, ΔS is the entropy of unfolding and R is the ideal gas constant (8.314 J mol-1 K-1), however for our data the van’t Hoff plots were not completely linear. This is because a van’t Hoff analysis done in this way assumes that ΔH and ΔS are independent of temperature and therefore that the heat capacity, ΔCp, is 0. We therefore included a ΔCp term to fit the van’t Hoff plot better: ΔG(T) = ΔHr – TΔSr + ΔCp[T – Tr – Tln(T/Tr)] and extracted ΔH, ΔS, ΔG and ΔCp for any given reference temperature, Tr.54
Analytical Ultracentrifugation
AUC sedimentation experiments were performed once for each peptide at 130 μM in PBS (110 μL) at 20 °C using a Beckman XLA Analytical Ultracentrifuge with cells comprising a 2-channel aluminum centerpiece and sapphire windows in a 4-place An-60 Ti rotor. The reference channel contained 120 μL of PBS buffer. The samples were centrifuged from 44 krpm – 60 krpm in increments of 4 krpm. The absorbance was measured across the cell at a radial distance of 5.8 – 7.3 cm at each speed after 8 hours and then again after a further hour to check the samples had reached equilibrium before moving onto the next speed. The data were fitted to a single ideal species with Ultrascan II (http://ultrascan2.uthscsa.edu/) and 99% confidence limits obtained by Monte Carlo analysis of the fits.
Protein Structure Determination by NMR
Peptides were prepared once at 1 mM concentration in phosphate-buffered saline (PBS). The pH was adjusted to pH 7.4 with 10 mM NaOH and the sample freeze-dried before being reconstituted in the appropriate volume of D2O (10%) in H2O (90%). The pH and concentration were confirmed.
NMR data were acquired at 278 K on a Bruker Ascend™ spectrometer operating at 700 MHz equipped with a 1.7 mm micro-cryoprobe (BrisSynBio, University of Bristol). The peptides were assigned using standard 2D homonuclear spectra: TOCSY (60 ms mixing time) and NOESY (100 and 250 ms mixing times). Both were acquired with spectral widths of 9375 Hz, 4096 complex points in f2 and 1024 complex points in f1. To help with backbone and side chain assignment, natural abundance 15N- (96 (t1) × 1792 (t2) complex points) and 13C-edited (124 (t1) × 2048 (t2) complex points) HSQC spectra were also acquired.
To help resolve ambiguity in the crowded regions high-resolution data were acquired on the peptides in 5 mm tubes at 900 MHz using a Varian INOVA spectrometer equipped with a Bruker TCI 5 mm z-PFG cryogenic probe (Biomolecular NMR Facility, University of Birmingham). 2D NOESY spectra (100 and 250 ms mixing times) were acquired with spectral widths of 12019 Hz, 4096 complex points in f2 and 1330 complex points in f1. DQF-COSY spectra were acquired with spectral widths of 12019 Hz, 4096 complex points in f2 and 1024 complex points in f1.
NMR data were processed with NMRPipe and qMDD. Peak picking and assignment were carried out in CCPNMR Analysis 2.4.1.55 NOEs, peak assignment and structure calculation were carried out with ARIA 2.3.156 and CNS v 1.2.57 The final structures were water refined using the standard ARIA protocol. Dihedral restraints for the α-helix were generated using DANGLE58 and validated with the NOE spectra before inclusion into the structure calculation. The structure calculation was supplemented with several backbone ϕ angles derived from 3JNH-Hα coupling constants extracted from a high-resolution DQF-COSY and a number of 1 dihedral restraints for the core residues derived from a 50 ms NOESY and high-resolution DQF-COSY recorded at 900 MHz.
CNS topology and parameter files for the para-substituted phenylalanine residues were generated using ProDrg2 server (http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg). The final refined ensemble was composed of 20 structures with the lowest energy and no violations (>5°) and validated using PSVS v1.5.24 Protein structures were rendered with Pymol (www.pymol.org).
Secondary structure assignment
PPα secondary structure was determined using DSSP59 with an additional protocol implemented to find polyproline II structure using in-house Python scripts. Polyproline II helices were defined as at least two consecutive residues with backbone dihedral angles in the range (ϕ ± e and ψ ± e) where ϕ = -75° and ψ = +145° and e = 29°.60
Supplementary Material
One sentence summary.
Design and mutagenesis of a monomeric miniprotein provides insight into weak non-covalent interactions that help define and maintain folded proteins and protein-ligand interactions.
Acknowledgments
EGB and DNW are supported by a BBSRC/ERASynBio grant (BB/M005615/1); KLH, GJB, JWH and DNW are supported by the ERC (340764); DNW is a Royal Society Wolfson Research Merit Award holder (WM140008); and KLPG is supported by the EPSRC-funded Bristol Chemical Synthesis Centre for Doctoral Training (EP/G036764/1). We thank BrisSynBio for access to the BBSRC/EPSRC-funded 700 MHz NMR spectrometer (BB/L01386X/1), S. Whittaker and the Henry Wellcome Building NMR Facility at the University of Birmingham for access to the Wellcome Trust–funded 900 MHz spectrometer (099185/Z/12/Z), and R. Alder and members of the Woolfson group for helpful discussions.
Footnotes
Author contributions:
EGB and DNW designed the research. EGB made the synthetic peptides and performed the CD spectroscopy and AUC experiments. CW and MPC collected the NMR data. CW, KLPG and EGB analyzed the NMR data, and CW solved the NMR structures. KLH, GJB, DNW conducted the bioinformatics. EGB and RBS carried out the MD studies. JWH and EGB performed the van’t Hoff analyses. EGB and DNW wrote the paper. All authors reviewed and contributed to the manuscript.
The authors declare no competing financial interests.
Code Availability
In-house Python scripts used for interrogating the PDB for CH–π interactions, and for analyzing thermal denaturation curves is available upon request.
Data Availability
Three NMR structures have been determined for PPα-Tyr, PPα-ϕOCH3 and PPα-ϕCH3 and coordinates deposited in the RCSB PDB with accession codes 5LO2, 5LO3, and 5LO4, respectively.
References
- 1.Pace NC, Scholtz JM, Grimsley GR. Forces Stabilizing Proteins. FEBS Lett. 2014;588:2177–2184. doi: 10.1016/j.febslet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dougherty DA. Cation-π Interactions in Chemistry and Biology: A New View of Benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 3.Brandl M, Weiss MS, Jabs A, Sühnel J, Hilgenfeld R. C-H…π-Interactions in Proteins. J Mol Biol. 2001;307:357–377. doi: 10.1006/jmbi.2000.4473. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett GJ, Choudhary A, Raines RT, Woolfson DN. n → π * Interactions in Proteins. Nat Chem Biol. 2010;6:615–620. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yesselman JD, Horowitz S, Brooks CL, III, Trievel RC. Frequent Side Chain Methyl Carbon-Oxygen Hydrogen Bonding in Proteins Revealed by Computational and Stereochemical Analysis of Neutron Structures. Proteins: Struct Funct Bioinf. 2015;83:403–410. doi: 10.1002/prot.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya A, Tejero R, Montelione GT. Evaluating Protein Structures Determined by Structural Genomics Consortia. Proteins: Struct Funct Bioinf. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett GJ, Newberry RW, VanVeller B, Raines RT, Woolfson DN. Interplay of Hydrogen Bonds and n→π* Interactions in Proteins. Journal of the American Chemical Society. 2013;135:18682–18688. doi: 10.1021/ja4106122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zondlo NJ, Schepartz A. Highly Specific DNA Recognition by a Designed Miniature Protein. J Am Chem Soc. 1999;121:6938–6939. [Google Scholar]
- 9.Gellman SH, Woolfson DN. Mini-Proteins Trp the Light Fantastic. Nat Struct Biol. 2002;9:408–410. doi: 10.1038/nsb0602-408. [DOI] [PubMed] [Google Scholar]
- 10.Neidigh JW, Fesinmeyer RM, Andersen NH. Designing a 20-Residue Protein. Nat Struct Mol Biol. 2002;9:425–430. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 11.Craven TW, Cho M-K, Traaseth NJ, Bonneau R, Kirshenbaum K. A Miniature Protein Stabilized by a Cation-π Interaction Network. J Am Chem Soc. 2016;138:1543–1550. doi: 10.1021/jacs.5b10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolfson DN. The Design of Coiled-Coil Structures and Assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 13.Golemi-Kotra D, et al. High Affinity, Paralog-Specific Recognition of the Mena EVH1 Domain by a Miniature Protein. J Am Chem Soc. 2004;126:4–5. doi: 10.1021/ja037954k. [DOI] [PubMed] [Google Scholar]
- 14.Daly NL, Craik DJ. Bioactive Cystine Knot Proteins. Curr Opin Chem Biol. 2011;15:362–368. doi: 10.1016/j.cbpa.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Bhardwaj G, et al. Accurate de novo Design of Hyperstable Constrained Peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pabo CO, Peisach E, Grant RA. Design and Selection of Novel Cys(2)His(2) Zinc Finger Proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 17.Gifford JL, Walsh MP, Vogel HJ. Structures and Metal-Ion-Binding Properties of Ca2+-Binding Helix-Loop-Helix EF Hand Motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 18.McKnight CJ, Matsudaira PT, Kim PS. NMR Structure of the 35-Residue Villin Headpiece Subdomain. Nat Struct Biol. 1997;4:180–184. doi: 10.1038/nsb0397-180. [DOI] [PubMed] [Google Scholar]
- 19.Cochran AG, Skelton NJ, Starovasnik MA. Tryptophan Zippers: Stable, Monomeric β-Hairpins. Proc Natl Acad Sci USA. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barua B, et al. The Trp-Cage: Optimizing the Stability of a Globular Miniprotein. Prot Eng Des Sel. 2008;21:171–185. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker EG, et al. Local and Macroscopic Electrostatic Interactions in Single α-Helices. Nat Chem Biol. 2015;11:221–228. doi: 10.1038/nchembio.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crick FHC. The Packing of α-Helices: Simple Coiled-Coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 23.Walshaw J, Woolfson DN. SOCKET: A Program for Identifying and Analysing Coiled-Coil Motifs Within Protein Structures. J Mol Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 24.Woolfson DN, et al. De novo Protein Design: How Do We Expand into the Universe of Possible Protein Structures? Curr Opin Struct Biol. 2015;33:16–26. doi: 10.1016/j.sbi.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Plevin MJ, Bryce DL, Boisbouvier J. Direct Detection of CH/π Interactions in Proteins. Nat Chem. 2010;2:466–471. doi: 10.1038/nchem.650. [DOI] [PubMed] [Google Scholar]
- 26.Nishio M, Umezawa Y, Fantini J, Weiss MS, Chakrabarti P. CH-π Hydrogen Bonds in Biological Macromolecules. Phys Chem Chem Phys. 2014;16:12648–12683. doi: 10.1039/c4cp00099d. [DOI] [PubMed] [Google Scholar]
- 27.Blundell TL, Pitts JE, Tickle IJ, Wood SP, Wu C-W. X-Ray-Analysis (1.4-Å Resolution) of Avian Pancreatic-Polypeptide: Small Globular Protein Hormone. Proc Natl Acad Sci USA. 1981;78:4175–4179. doi: 10.1073/pnas.78.7.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson MR, et al. Elongated Fibrillar Structure of a Streptococcal Adhesin Assembled by the High-Affinity Association of α-and PPII-Helices. Proc Natl Acad Sci USA. 2010;107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noelken ME, Chang PJ, Kimmel JR. Conformation and Association of Pancreatic-Polypeptide from 3 Species. Biochemistry. 1980;19:1838–1843. doi: 10.1021/bi00550a017. [DOI] [PubMed] [Google Scholar]
- 30.Fiser A, Do RKG, Šali A. Modeling of Loops in Protein Structures. Prot Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu XZ, Wang HC, Ke HM, Kuhlman B. High-Resolution Design of a Protein Loop. Proc Natl Acad Sci USA. 2007;104:17668–17673. doi: 10.1073/pnas.0707977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson KL, et al. Carbohydrate-Aromatic Interactions in Proteins. J Am Chem Soc. 2015;137:15152–15160. doi: 10.1021/jacs.5b08424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha RP, Bhattacharyya R, Chakrabarti P. Interaction Geometry Involving Planar Groups in Protein-Protein Interfaces. Proteins: Struct Funct Bioinf. 2007;67:84–97. doi: 10.1002/prot.21244. [DOI] [PubMed] [Google Scholar]
- 34.Wheeler SE, Houk KN. Through-Space Effects of Substituents Dominate Molecular Electrostatic Potentials of Substituted Arenes. J Chem Theory Comput. 2009;5:2301–2312. doi: 10.1021/ct900344g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carver FJ, Hunter CA, Seward EM. Structure-Activity Relationship for Quantifying Aromatic Interactions. Chem Commun. 1998:775–776. [Google Scholar]
- 36.Zondlo NJ. Aromatic-Proline Interactions: Electronically Tunable CH/π Interactions. Acc Chem Res. 2013;46:1039–1049. doi: 10.1021/ar300087y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bloom JWG, Raju RK, Wheeler SE. Physical Nature of Substituent Effects in XH/π Interactions. J Chem Theory Comput. 2012;8:3167–3174. doi: 10.1021/ct300520n. [DOI] [PubMed] [Google Scholar]
- 38.Tsuzuki S, Honda K, Uchimaru T, Mikami M, Tanabe K. The Magnitude of the CH/π Interaction between Benzene and Some Model Hydrocarbons. J Am Chem Soc. 2000;122:3746–3753. [Google Scholar]
- 39.Kay BK, Williamson MP, Sudol P. The Importance of Being Proline: The Interaction of Proline-Rich Motifs in Signaling Proteins with Their Cognate Domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 40.Kay BK. SH3 Domains Come of Age. FEBS Lett. 2012;586:2606–2608. doi: 10.1016/j.febslet.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ball LJ, Kuhne R, Schneider-Mergener J, Oschkinat H. Recognition of Proline-Rich Motifs by Protein-Protein-Interaction Domains. Angew Chem Int Ed. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 42.Parsons ZD, Bland JM, Mullins EA, Eichman BF. A Catalytic Role for C–H/π Interactions in Base Excision Repair by Bacillus cereus DNA Glycosylase AlkD. J Am Chem Soc. 2016;138:11485–11488. doi: 10.1021/jacs.6b07399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang YT, Malamakal RM, Chenoweth DM. Aza-Glycine Induces Collagen Hyperstability. J Am Chem Soc. 2015;137:12422–12425. doi: 10.1021/jacs.5b04590. [DOI] [PubMed] [Google Scholar]
- 44.Arnold U, Raines RT. Replacing a Single Atom Accelerates the Folding of a Protein and Increases its Thermostability. Org Biomol Chem. 2016;14:6780–6785. doi: 10.1039/c6ob00980h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobos ES, et al. A Miniprotein Scaffold Used to Assemble the Polyproline II Binding Epitope Recognized by SH3 Domains. J Mol Biol. 2004;342:355–365. doi: 10.1016/j.jmb.2004.06.078. [DOI] [PubMed] [Google Scholar]
- 46.Velankar S, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang GL, Dunbrack RL. PISCES: Recent Improvements to a PDB Sequence Culling Server. Nucleic Acids Res. 2005;33:W94–W98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh J, Thornton JM. Atlas of Protein Side-Chain Interactions. Vol. 1. Oxford University Press; 1992. [Google Scholar]
- 49.Finn RD, et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox NK, Brenner SE, Chandonia JM. SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL Data and Classification of New Structures. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and Glutamine: Using Hydrogen Atom Contacts in the Choice of Side-Chain Amide Orientation. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 52.Kuipers BJH, Gruppen H. Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm to Enable Quantitative Reverse Phase High-Performance Liquid Chromatography–Mass Spectrometry Analysis. J Agric Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 53.Marky LA, Breslauer KJ. Calculating Thermodynamic Data for Transitions of any Molecularity from Equilibrium Melting Curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 54.LiCata VJ, Liu CC. In: Methods in Enzymology. Johnson ML, Holt JM, Ackers GK, editors. Vol. 488. 2011. pp. 219–238. Biothermodynamics, Pt C Methods in Enzymology. [DOI] [PubMed] [Google Scholar]
- 55.Vranken WF, et al. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins: Struct Funct Bioinf. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 56.Rieping W, et al. ARIA2: Automated NOE Assignment and Data Integration in NMR Structure Calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 57.Brunger AT, et al. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 58.Cheung MS, Maguire ML, Stevens TJ, Broadhurst RW. DANGLE: A Bayesian Inferential Method for Predicting Protein Backbone Dihedral Angles and Secondary Structure. J Magn Resonance. 2010;202:223–233. doi: 10.1016/j.jmr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 59.Touw WG, et al. A Series of PDB-Related Databanks for Everyday Needs. Nucleic Acids Res. 2015;43:D364–D368. doi: 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mansiaux Y, Joseph AP, Gelly JC, de Brevern AG. Assignment of PolyProline II Conformation and Analysis of Sequence - Structure Relationship. Plos One. 2011;6:e18401. doi: 10.1371/journal.pone.0018401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.