Abstract
Osteoblasts communicate both with normal cells in the bone marrow, and with tumor cells that metastasized to bone. Here we show that osteoblasts release exosomes, we termed osteosomes, which may be a novel mechanism by which osteoblasts communicate with cells in their environment. We have isolated exosomes from undifferentiated/proliferating (D0 osteosomes) and differentiated/mineralizing (D24 osteosomes) primary mouse calvarial osteoblasts. The D0 and D24 osteosomes were found to be vesicles of 130–140 nm by dynamic light scattering analysis. Proteomics profiling using tandem mass spectrometry (LC-MS/MS) identified 206 proteins in D0 osteosomes and 336 in D24 osteosomes. The proteins in osteosomes are mainly derived from the cytoplasm (~47%) and plasma membrane (~31%). About 69% of proteins in osteosomes are also found in Vesiclepedia, and these canonical exosomal proteins include tetraspanins and Rab family proteins. We found that there are differences in both protein content and levels in exosomes isolated from undifferentiated and differentiated osteoblasts. Among the proteins that are unique to osteosomes, 169 proteins are present in both D0 and D24 osteosomes, 37 are unique to D0, and 167 are unique to D24. Among those 169 proteins present in both D0 and D24 osteosomes, 10 proteins are likely present at higher levels in D24 than D0 osteosomes, based on emPAI ratios of more than 5. These results suggest that osteosomes released from different cellular state of osteoblasts may mediate distinct functions. Using live-cell imaging, we measured the uptake of PKH26-labeled osteosomes into C4-2B4 and PC3-mm2 prostate cancer cells. In addition, we showed that cadherin-11, a cell adhesion molecule, plays a role in the uptake of osteosomes into PC3-mm2 cells as osteosome uptake was delayed by neutralizing antibody against cadherin-11. Together, our studies suggest that osteosomes could have a unique role in the bone microenvironment under both physiological and pathological conditions.
Keywords: osteoblasts, exosomes, osteosomes, cadherin-11, mass spectrometry
Introduction
Under normal physiologic conditions, osteoblasts are in communication with cells in the bone marrow to maintain tissue homeostasis. Osteoblasts have been shown to be a component of hematopoietic stem cell niche1–3, in which cell-cell contact between osteoblast and hematopoietic stem cells leads to Notch activation, which is one mechanism of communication by which osteoblasts influence stem cell function1. Osteoblasts were also shown to use the paracrine factor, BMP, to regulate hematopoietic stem cells2. In pathological conditions, e.g., prostate cancer bone metastasis, osteoblasts and tumor cell communication through paracrine factors have been shown to increase the tumor growth4–8. Osteoblast secreted factors have also been shown to confer tumor cell survival, resulting in resistance to therapy9. The unique roles of osteoblasts in the bone microenvironment in both physiological and pathological conditions suggest that the methods of communication between these cell types needs to be fully understood.
In this report, we examined whether exosomes could be an additional mechanism for osteoblast communication with other cells in the bone marrow. Exosomes are extracellular vesicles that originate by the fusion of multivesicular endosomes with the plasma membrane10. Exosomes are endocytic vesicles released by cells and are enriched in specific proteins, lipids and RNAs, indicating the existence of specialized mechanisms that control the sorting of molecules into exosomes11. Recent discoveries that exosomes are a powerful way of cell-cell communication11–17 suggested new possibilities that osteoblasts may use exosomes to bring proteins and genetic modifiers, e.g. miRNAs, into target cells to modulate cell activities. For example, exosomes that are derived from breast cancer stroma have been shown to increase cell migration18 and confer therapy resistance19, suggesting a role of stromal exosomes in modulating cancer progression.
One of the unique properties of osteoblasts is their ability to undergo differentiation to form mineralized bone. Whether these differentiation-induced cellular changes may affect exosome composition and thus exosome-mediated intercellular communication remains to be determined. Recently, Ge et al.20 reported the proteomic analysis of microvesicles isolated from nonmineralized mouse MCT3T-E1 cells, a T-antigen immortalized mouse calvarial osteoblast cell line. They showed that the MC3T3-E1 exosomes contained typical exosomal markers, including TSG101 and Flot 120. Morhayim et al.21 reported the proteomic signature of extracellular vesicle (EV) from nonmineralizing and mineralizing T-antigen immortalized human osteoblasts SV-HFO. Among the proteins identified, they detected 3 and 22 osteoblast-specific proteins that were uniquely present in nonmineralizing and mineralizing osteoblasts, respectively21.
Exosomes from primary mouse osteoblasts have never been studied. It is known that primary mouse osteoblasts can be induced to differentiate more extensively then immortalized osteoblasts under differentiation conditions, which may more closely reflect normal osteoblast physiology. In this study, we isolated exosomes, which we termed osteosomes, from both undifferentiated/proliferating and differentiated/mineralizing primary mouse osteoblasts and determined the proteomics profile of these osteoblast-derived exosomes. Our study showed that the molecular compositions of osteosomes under undifferentiated and differentiated conditions are different, with 225 proteins uniquely present in osteosomes from differentiated but not undifferentiated osteoblasts. We also showed that cadherin-11 cell adhesion molecules play a role in the uptake of osteosomes into prostate cancer cells.
Experimental Section
Exosome-depleted FBS preparation
To deplete exosomes in serum, fetal bovine serum (FBS) was mixed with 50% polyethylene glycol (Fluka, polyethylene glycol 10,000) at 5:1 ratio. After incubation at 4°C for 2h, solution was centrifuged at 1,500g for 30 minutes at 4°C. Supernatant was collected and used as exosome-depleted FBS.
Osteoblast isolation and differentiation
Calvaria were isolated from 2–3 day old newborn mice. Collected bone tissue was twice digested using 0.1mg/mL collagenase in alpha-MEM with 1:40 diluted trypsin. These first two digestions were discarded and a third digestion using 0.2 mg/mL collagenase was performed and osteoblasts were collected. Along with undigested bone, osteoblasts were transferred to cell culture plates and allowed to grow to confluence with minimal disturbance for three days. Cell and bone fragments were trypsinized, washed, and passaged in fresh media containing exosome-depleted FBS. Cells were allowed to grow to confluence and conditioned media was collected (D0 conditioned media). The media was changed to differentiation media containing 10% exosome-depleted FBS, 5mM beta-glycerophosphate, and 100ug/mL ascorbic acid. Differentiation media was replenished every three days for a total of 24 days. At day 24, cell media was collected (D24 conditioned media).
Osteoblast differentiation assays
Von Kossa staining for mineralized bone matrix was performed as described elsewhere22. Alizarin Red S staining for calcium deposition was carried out as below: 2 g Alizarin Red S (C. I. 58005) was dissolved in 100 ml distilled water, and pH was adjusted to 4.1 – 4.3 with 0.1% NH4OH to prepare the Alizarin Red S staining solution. Filter the dark-brown solution and store it in the dark. The cell was taken from the incubator and the medium was carefully aspirated. Then the cells were washed with Dulbecco’s PBS, without Ca2+/Mg2+. For fixation, the neutral buffered formalin (10%) was used to cover the cellular monolayer and incubate at least 30 min. Then the formalin was carefully aspirated and the cells were washed with distilled water. Then enough Alizarin Red S staining solution was added to cover the cellular monolayer and incubated at room temperature in the dark for 45 min. Then the Alizarin Red S staining solution was carefully aspirated and was washed four times with 1 ml distilled water. Then PBS was added to cover the cellular monolayer and analyzed in light microscopy.
Reverse transcription and real-time PCR (qRT-PCR)
RNA was prepared using Trizol (InVitrogen) and further purified by RNAeasy mini kit plus DNase I treatment (Qiagen). The relative mRNA level for each gene was quantified by Real-time RT-PCR with SYBR Green (Applied Biosystems), using Gapdh as a control. The primers for RT-PCR are as follow. Alkaline phosphatase: CTCCTCCATCCCTTCCCTTC and CCCTGGGTAGACAGCCAAC; osteocalcin: GCTCTGTCTCTCTGACCTCA and TGGACATGAAGGCTTTGTCA; DMP1: CCCACGAACAGTGAGTCATC and GGTCTGTACTGGCCTCTGTC; SOST: ATCCCAGGGCTTGGAGAGTA and CTCGGACACATCTTTGGCGT; GAPDH: CCCAGAAGACTGTGGATG and GCAGGGATGATGTTCTGG.
Exosome isolation and analysis
Osteoblasts were isolated from 80 newborn mouse calvaria and grew to confluence in exosome-depleted fetal bovine serum. The conditioned medium was collected and centrifuged at 1000×g for 5 min to remove cells, followed by an initial filtration step (1μm) and a centrifugation step of 3000×g for 10 min to remove cellular debris. A total of 150 ml of conditioned medium was collected and ultracentrifuged at 100,000×g at 4°C overnight. The exosome pellet from the ultracentrifugation step was resuspended in 10 ml of PBS and a second step of ultracentrifugation was performed at 100,000×g at 4°C for 2 h. The pellet was resuspended in PBS and ultracentrifuged at 100,000×g one more time to remove fetal bovine serum. Osteosomes were isolated from day 0-CM and day 24-CM by serial centrifugation. In brief, media was centrifuged at 2,000g for 20 min, supernatant was then centrifuged again at 10,000g for 30 min. Supernatant was again collected and spun at 100,000g for 90 min, exosome pellet collected, washed with 1× PBS and spun at 100,000g for 90 min. Supernatant was discarded and pellet was resuspended in 1×PBS for further analysis.
Exosome particle size determination and transmission electron microscopy
The particle sizes of isolated D0 and D24 exosomes were measured by dynamic light scattering analysis using NanoSight LM-10 instrument (Nanosight Limited, Amesbury, UK). Transmission electron microscopy (TEM) was performed by MD Anderson Core facility. Samples were fix in the final concentration of 2% glutaraldehyde and were placed on 100 mesh carbon coated, formvar coated copper grids treated with poly-l-lysine for 1 hour. Samples were then negatively stained with Millipore-filtered aqueous 1% uranyl acetate for 1 min. Stain was blotted dry from the grids with filter paper and samples were allowed to dry. Samples were then examined in a JEM 1010 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) at an accelerating voltage of 80 Kv. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA).
Proteomics profiling
The osteosome were acetone precipitated (acetone:sample=5:1 ratio) and placed in −20°C overnight. The precipitated proteins were resuspended in 10 μl Rapigest (2 mg/ml in 100 mM ammonium bicarbonate) (Waters) plus 30 μl 50 mM ammonium bicarbonate, heated at 100°C for 10 min. The samples were cooled to room temperature and digested with 200–400 ng sequencing grade trypsin (20 ng/μl in 0.02% formic acid) (Promega) at 37°C overnight. The digested samples were dried down using Speedvac and reconstitute in 1% formic acid.
The resulting peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an Orbitrap Fusion mass spectrometer (Thermo Scientific). HPLC analyses were performed with Dionex RSCL 3000 Nano. Samples were injected into a Phenomenex core-shell C18 DB column (2.7 μm 15cm), with mobile phase compositions of A: 0.1% formic acid in water, B: 0.1% formic acid in acetonitrile and with a flow rate of 100. The gradient was held isocratic at 2% B for 2 min, ramped up to 35% at 165 min, ramped up to 80% at 166 min, maintained at 80% until 176 min, ramped down to 2% at 177 min, held at 2% until 190 min.
MS parameters and scan strategy were: (a) mass range for MS1: 400–1300; (b) mass resolution for MS1: 500,000; (c) mass window for precursor ion selection: 0.5d; (d) number or precursors selected for tandem MS in each scan cycle: Maximum in 2 sec; (e) mass analyzer for tandem-MS: MS1: Orbitrap; MS2: Iontrap. (f) charge state screening parameters: 2-4; (g) relative collision energy: 30%; (h) dynamic exclusion settings: 15sec.
Data processing of the MS results were as follows: (a) Database: SwissProt/2.3.02, SwissProt_040115.fasta, Total sequences: 548208; (b) Search engine: Mascot 2.5 via Proteome Discoverer 1.4; (c) Precursor and product ion mass tolerances: Peptide Mass Tolerance: 10, Peptide Mass Tolerance Units: ppm, Fragment Mass Tolerance: 0.8, Fragment Mass Tolerance Units: Da, Ions score cut-off: 20; (d) Enzyme specificity: Trypsin, 2 missed cleavages allowed; (e) Fixed and variable modifications: Fixed: none, Variable modifications: Oxidation (M), Gln->pyro-Glu (N-term Q), Trioxidation (C); (f) Additional search specifications: Decoy database also searched; (g) Method for FDR assessment: Decoy DB using Proteome Discoverer; (h) Criteria for acceptance of peptide assignments and protein identifications: Significance threshold: 0.05. Max. number of hits: auto. Use MudPIT protein scoring: not applicable; (i) Determination of probability of modification site location: not applicable.
Immunoblot
Proteins from osteosomes were subjected to 4–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The gel was transferred to a nitrocellulose membrane (Schleicher & Schnell) and stained with Ponceau S, followed by immunoblotting with specific antibodies as indicated. Signals were detected with a chemiluminescent detection kit (Pierce Biotechnology).
Exosome uptake and antibody blocking
Osteosomes and control liposomes were labeled with the red fluorescent lipophilic dye PKH26 (InVitrogen)23. Next, PKH26-labeled osteosomes or liposomes (3×105 particles) were added to prostate cancer cells (1×104 cells), C4-2b or PC3-mm2, in RPMI1640 containing exosome-depleted 0.1% FBS, and cells were plated on a glass-bottom dish (ibidi). Exosome or liposome uptake into cells was observed by live-cell imaging on a BioStation (Nikon), in which images were captured every 30 min over 30 h using both bright-field and red fluorescence channels24. For antibody blocking, PKH26-labeled osteosomes were preincubated with anti-Cad11 monoclonal antibody 1A525 at a final antibody concentration of 3 μg/ml before the osteosome-antibody mixture was added to prostate cancer cells for live-cell imaging analysis. PBS buffer alone and an unrelated antibody with similar IgG isotype were used as negative controls.
Results
Undifferentiated (D0) and Differentiated (D24) osteoblasts
Osteoblasts can be stimulated to undergo proliferation or differentiation, depending on the specific treatments or culture condition. It is not clear whether exosomes generated from undifferentiated or differentiated osteoblasts have different protein composition. To address this question, performed proteomics profiling of exosomes, which we term osteosomes, from undifferentiated or differentiated osteoblasts. The experimental scheme for the isolation and characterization of exosomes from primary mouse osteoblasts is shown in Fig. 1A. Osteoblasts isolated from newborn calvaria were cultured in the growth medium to confluence (undifferentiating condition) and the medium was then changed to differentiation medium and the osteoblasts further cultured for 24 days (differentiating condition). The morphologies of osteoblasts cultured in undifferentiating condition (D0 osteoblasts) and differentiating condition (D24 osteoblasts) are shown in Fig. 1B. Von Kossa (Fig. 1C) and alizarin staining (Fig. 1D) showed that D24 but not D0 osteoblasts are mineralized. qRT-PCR for the expression of markers of osteoblast differentiation in mRNA prepared from D0 and D24 osteoblasts was used to establish the differentiation status of osteoblasts. In one experiment, alkaline phosphatase and osteocalcin, markers of osteoblast differentiation, were increased by 20- and 2876-fold, respectively in Day 24 osteoblasts compared to D0 osteoblasts (Fig. 1E). In another experiment, the increases were 17- and 242-fold, respectively (Supplemental –Fig. S1). In addition, the osteocyte markers, dentin matrix acidic phosphoprotein 1 (DMP1) and sclerostin (SOST1), were also increased by 730- to 1076-fold and 1537- to 91,650-fold, respectively, in D24 osteoblasts compared to D0 osteoblasts (Fig. 1E, Supplemental Fig. S1). These results confirm that these osteoblasts have undergone differentiation after culturing in the differentiation medium for 24 days.
Figure 1.
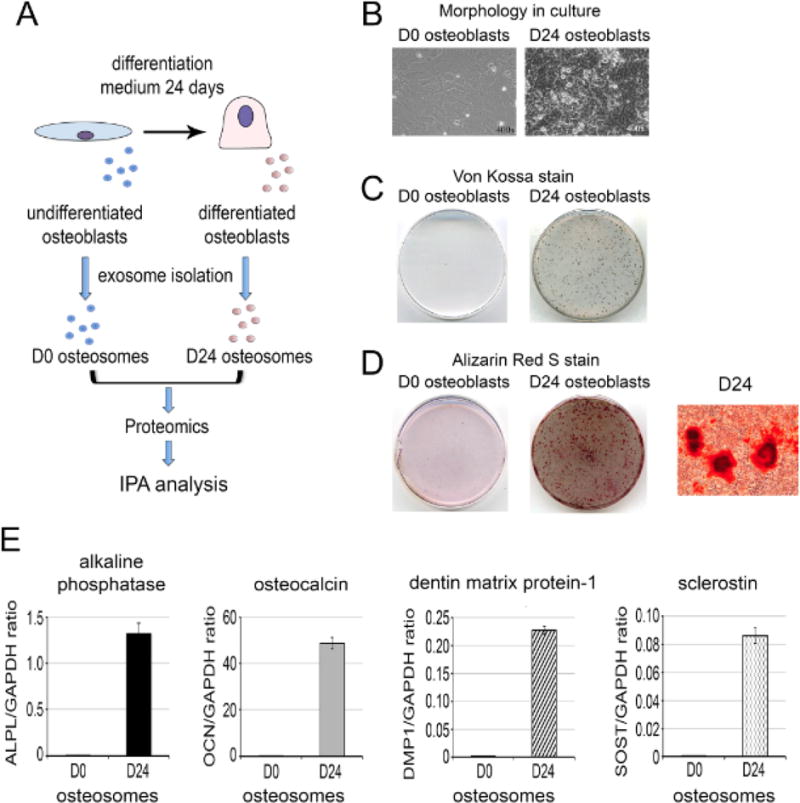
Preparation of osteosomes from undifferentiated (D0) and differentiated osteoblasts (D24). (A) Experimental scheme for the isolation and characterization of exosomes from primary mouse osteoblasts, here termed “osteosomes”. (B) Morphology of D0 undifferentiated and D24 differentiated osteoblasts in culture. (C) Von Kossa stain for the mineralization of osteoblasts cultured in the absence (D0) or presence (D24) of differentiation medium. (D) Alizarin Red stain for mineralization of osteoblasts. Right panel, enlarged image of Alizarin Red staining of D24 differentiated osteoblasts. (E) Real-time RT-PCR for the expression of osteoblast differentiation markers, including alkaline phosphatase, osteocalcin, dentin matrix phosphoprotein-1, and sclerostin in D0 and D24 osteoblasts. Real-time RT-PCRs were performed on total RNAs prepared from calvarial osteoblasts cultured in the absence (D0) or presence (D24) of osteoblast differentiation medium using gene-specific primers as indicated.
Characterization of osteosomes isolated from D0 and D24 osteoblasts
Conditioned media were collected from D0 and D24 osteoblasts and exosomes were isolated using ultracentrifugation. Exosomes prepared from the undifferentiated (D0 osteoblasts) and differentiated (D24 osteoblasts) conditions are named D0 and D24 osteosomes, respectively. When examined by light scattering spectroscopy, both D0 and D24 osteosomes have particle sizes around 100 nm (Fig. 2A, C), which is the typical size of exosomes. Transmission electron microscopy (TEM) showed that both the osteosome vesicles (D0 and D24) exhibit a cup-shaped morphology (Fig. 2B, 2D), which is the characteristic morphology of exosomes. The average sizes of D0 and D24 osteosomes from four independent experiments were 134.9 ± 12.6 and 138.9 ± 12.5, respectively (Fig 2E). We note that the number of osteosomes from primary mouse osteoblasts is very low, ~ 4000 and ~3300 particles per million cells from undifferentiated and differentiated osteoblasts, respectively. In contrast, the exosomes from C4-2B4 and PC3-mm2 PCa cells are ~184,000 and 108,000 particles per million cells. Thus, the number of exosomes from primary mouse osteoblasts is around 2–4% of those from PCa cells.
Figure 2.
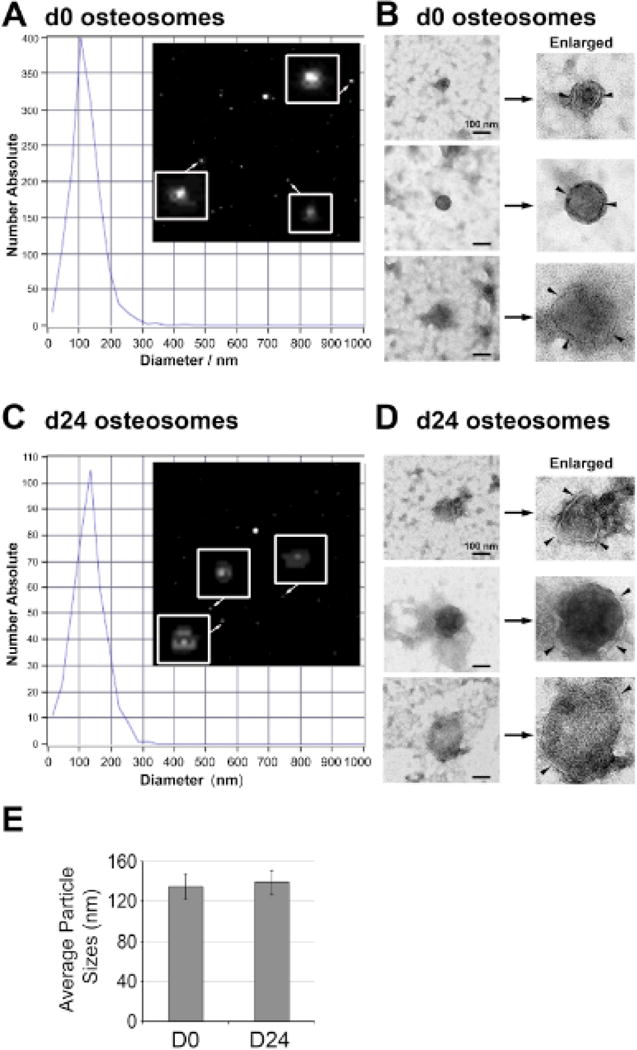
Characterization of osteosomes. (A) Particle size and images of D0 osteosomes by dynamic light scattering analysis using a Zetasizer Nano ZS instrument. Osteosomes (see enlarged in insets) were found to be mainly ~50–150 nm size particles. (B) Transmission electron microscopy images of three representative D0 osteosomes were found to exhibit cup-shaped morphology (arrowheads) characteristic of exosomes. (C) Particle size and images of D24 osteosomes by dynamic light scattering analysis as in A. (D) Three representative transmission electron microscopy images of D24 osteosomes. Scale bar, 100 nm. (E) Average sizes of osteosomes from D0 and D24 osteoblasts. N=4. Data represent average ± sem.
Comparison of osteosomal proteins with other exosomal proteins
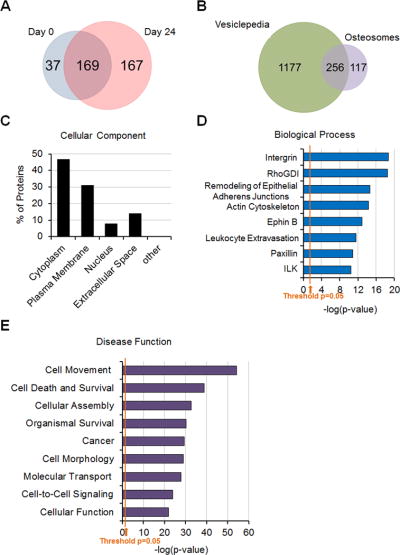
To characterize the proteins in osteosomes, D0 and D24 osteosomes were subjected to mass spectrometry analysis. Proteomics profiling by mass spectrometry identified 206 and 336 proteins with a 1% false discovery rate (FDR) from D0 and D24 osteosomes, respectively (Fig. 3A). 169 proteins were found in both D0 and D24 osteosomes, resulting in a total of 373 osteosomal proteins from combining the proteins from D0 and D24 osteosomes. A comparison of our osteosome proteomics data with a published exosome database, i.e., Vesiclepedia26, showed that 256 (69%) proteins are also found in Vesiclepedia (Fig. 3B), resulting in 117 proteins that are unique to osteosomes. The canonical exosome proteins10 found in osteosomes are shown in Supplemental Table S1. They include tetraspanins (CD9, CD81), endosomal molecules (clathrin), multivesicular body proteins (Chmp4b), membrane trafficking proteins (RAB proteins, annexins), cytoskeletal proteins (actin, tubulin, myosin), heat shock proteins (HSP90, HSP70), and adhesion proteins (integrins). The molecular composition of osteosomes reflects their origin in endosomes. These results demonstrate that osteosomes have similar characteristics as exosomes from other cell types.
Figure 3.
Proteomics analysis of osteosomes. (A) Venn diagram of proteins in D0 vs D24 osteosoms. (B) Venn diagram of proteins in osteosomes and in Vesiclepedia. (C) Ingenuity Pathway Analysis of the intracellular origin of osteosome proteins. (D) The involvement of osteosome proteins in various biological processes. (E) The involvement of osteosome proteins in disease functions. These pathways are selected based on p values (expressed as −log(p-value)). The marked thresholds in D and E represent p=0.05.
Ingenuity pathway analysis of osteosomal proteins
Analysis of the 373 osteosomal proteins from combining the proteins from D0 and D24 osteosomes using Ingenuity Pathway Analysis showed that the osteosomal proteins are originated from the cytoplasm (47%) and plasma membrane (31%) (Fig. 3C). We further analyzed these proteins based on the potential biological processes and found that these proteins are involved in integrin signaling, RhoGDI, and remodeling of epithelial adherens junctions (Fig. 3D). Importantly, the disease function analysis showed that these osteosome proteins are mainly involved in cell movement, cell death and survival, cellular assembly and cancer (Fig. 3E).
Changes in the levels of osteosomal proteins during osteoblast differentiation
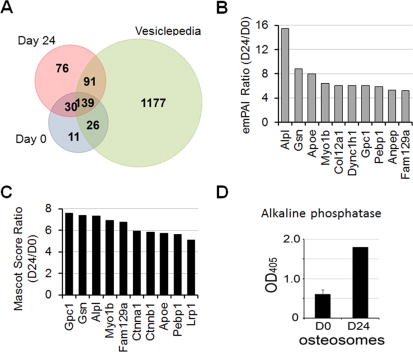
Among the 117 proteins that are unique to osteosomes (Fig. 4A), 30 proteins are common between D0 and D24 osteosomes (Table 1). This results in 11 proteins that are unique to D0 osteosomes (Fig. 4A, Table 2) and 76 proteins that are unique to D24 osteosomes (Fig. 4A, Table 3). For the 169 proteins that are common between D0 and D24 osteosomes, we compared their levels of expression under different differentiation status. Although the mass spectrometry method we used for protein identification is not quantitative, the Experimentally Modified Protein Abundance Index (emPAI) can provide an estimate for the relative levels of expression. A comparison of emPAI scores among the 169 common osteosome proteins, 10 of the 169 proteins (6%) show a greater than 5-fold increase in D24 osteosomes when compared to D0 osteosomes (Fig. 4B). Among them, the protein with the highest fold of increase is alkaline phosphatase (ALPL, 15-fold). When protein scores were used as comparison, seven of these proteins also have more than 5-fold increase (Fig. 4C). Measurement of the enzymatic activity of alkaline phosphatase in D0 and D24 osteosomes showed that there was an increase, about 3.5-fold, in D24 osteosomes compared to that in D0 osteosomes (Fig. 4D), confirming the results from mass spectrometry analysis. These observations suggest that osteosome compositions differ depending on the differentiation states of osteoblasts.
Figure 4.
Comparison of proteomics profile of D0 and D24 osteosomes. (A) Venn diagram of proteins in D0, D24 osteosoms versus those in Vesiclepedia. (B) Proteins that showed a more or equal to 5-fold increase, based on emPAI values, in D24 osteosomes when compared to D0 osteosomes. (C) Proteins that showed a more or equal to 5-fold increase, based on protein score, in D24 osteosomes when compared to D0 osteosomes. (D) Enzymatic activity of alkaline phosphatase in D0 and D24 osteosomes.
Table 1.
Proteins common to D0 and D24 osteosomes
| prot_acc | GN | prot_desc | prot_mass (Da) | prot_scor e (Day 0) | prot_scor e (Day 24) | Num. of significant matches (day 0) | Num. of significant matches (day 24) | Number of unique peptide (day 0) | Number of unique peptide (day 24) | Sequenc e coverage (day 0) | Sequenc e coverage (day 24) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A2MG_MOUSE | A2m | Alpha‐2‐macroglobulin‐P OS=Mus musculus GN=A2m PE=2 SV=2 | 164248 | 79 | 118 | 7 | 12 | 2 | 3 | 1.8 | 2.4 |
| SYAC_MOUSE | Aars | Alanine–tRNA ligase, cytoplasmic OS=Mus musculus GN=Aars PE=1 SV=1 | 106841 | 57 | 36 | 2 | 1 | 2 | 1 | 3.7 | 1 |
| ACTB_MOUSE | Actb | Actin, cytoplasmic 1 OS=Mus musculus GN=Actb PE=1 SV=1 | 41710 | 995 | 1160 | 95 | 88 | 18 | 19 | 62.9 | 62.9 |
| ACTC_MOUSE | Actc1 | Actin, alpha cardiac muscle 1 OS=Mus musculus GN=Actc1 PE=1 SV=1 | 41992 | 710 | 670 | 63 | 48 | 15 | 13 | 55.4 | 48 |
| ACTN1_MOUSE | Actn1 | Alpha‐actinin‐1 OS=Mus musculus GN=Actn1 PE=1 SV=1 | 103004 | 176 | 446 | 4 | 9 | 3 | 7 | 3.9 | 9.2 |
| ARP3_MOUSE | Actr3 | Actin‐related protein 3 OS=Mus musculus GN=Actr3 PE=1 SV=3 | 47327 | 95 | 90 | 2 | 2 | 2 | 2 | 4.8 | 4.8 |
| ALBU_MOUSE | Alb | Serum albumin OS=Mus musculus GN=Alb PE=1 SV=3 | 68648 | 128 | 121 | 10 | 10 | 1 | 1 | 2.1 | 2.1 |
| ALDOA_MOUSE | Aldoa | Fructose‐bisphosphate aldolase A OS=Mus musculus GN=Aldoa PE=1 SV=2 | 39331 | 206 | 144 | 5 | 4 | 5 | 4 | 14.3 | 14 |
| PPBT_MOUSE | Alpl | Alkaline phosphatase, tissue‐nonspecific isozyme OS=Mus musculus GN=Alpl PE=1 SV=2 | 57478 | 120 | 879 | 3 | 81 | 2 | 15 | 4.8 | 35.7 |
| AMPN_MOUSE | Anpep | Aminopeptidase N OS=Mus musculus GN=Anpep PE=1 SV=4 | 109582 | 159 | 753 | 5 | 30 | 4 | 16 | 6 | 25.6 |
| ANXA1_MOUSE | Anxa1 | Annexin A1 OS=Mus musculus GN=Anxa1 PE=1 SV=2 | 38710 | 508 | 822 | 15 | 38 | 8 | 13 | 29.5 | 41.9 |
| ANXA2_MOUSE | Anxa2 | Annexin A2 OS=Mus musculus GN=Anxa2 PE=1 SV=2 | 38652 | 683 | 834 | 47 | 63 | 12 | 14 | 36 | 46.9 |
| ANXA3_MOUSE | Anxa3 | Annexin A3 OS=Mus musculus GN=Anxa3 PE=1 SV=4 | 36362 | 65 | 50 | 2 | 1 | 2 | 1 | 7.4 | 5 |
| ANXA4_MOUSE | Anxa4 | Annexin A4 OS=Mus musculus GN=Anxa4 PE=1 SV=4 | 35893 | 214 | 684 | 7 | 23 | 4 | 8 | 16.6 | 29.5 |
| ANXA5_MOUSE | Anxa5 | Annexin A5 OS=Mus musculus GN=Anxa5 PE=1 SV=1 | 35730 | 581 | 1208 | 26 | 71 | 12 | 22 | 42 | 67.1 |
| ANXA6_MOUSE | Anxa6 | Annexin A6 OS=Mus musculus GN=Anxa6 PE=1 SV=3 | 75837 | 495 | 1026 | 18 | 47 | 11 | 22 | 23.2 | 42.1 |
| AP2A1_MOUS E | Ap2a1 | AP‐2 complex subunit alpha‐1 OS=Mus musculus GN=Ap2a1 PE=1 SV=1 | 107596 | 34 | 40 | 1 | 1 | 1 | 1 | 0.9 | 0.9 |
| APOE_MOUSE | Apoe | Apolipoprotein E OS=Mus musculus GN=Apoe PE=1 SV=2 | 35844 | 108 | 619 | 4 | 23 | 3 | 12 | 10 | 35 |
| ARF1_MOUSE | Arf1 | ADP‐ribosylation factor 1 OS=Mus musculus GN=Arf1 PE=1 SV=2 | 20684 | 265 | 203 | 11 | 9 | 6 | 4 | 43.6 | 32 |
| ARF4_MOUSE | Arf4 | ADP‐ribosylation factor 4 OS=Mus musculus GN=Arf4 PE=1 SV=2 | 20384 | 104 | 106 | 3 | 5 | 3 | 2 | 23.3 | 11.7 |
| ARF6_MOUSE | Arf6 | ADP‐ribosylation factor 6 OS=Mus musculus GN=Arf6 PE=1 SV=2 | 20069 | 50 | 82 | 2 | 4 | 1 | 2 | 5.7 | 12 |
| GDIR1_MOUSE | Arhgdia | Rho GDP‐dissociation inhibitor 1 OS=Mus musculus GN=Arhgdia PE=1 SV=3 | 23393 | 106 | 121 | 3 | 5 | 2 | 2 | 15.2 | 15.2 |
| AT1A1_MOUSE | Atp1a1 | Sodium/potassium‐transporting ATPase subunit alpha‐1 OS=Mus musculus GN=Atp1a1 PE=1 SV=1 | 112910 | 616 | 972 | 20 | 35 | 13 | 17 | 18.5 | 23 |
| AT2B1_MOUSE | Atp2b1 | Plasma membrane calcium‐transporting ATPase 1 OS=Mus musculus GN=Atp2b1 PE=1 SV=1 | 134662 | 61 | 222 | 2 | 6 | 2 | 5 | 2.3 | 5.9 |
| B2MG_MOUSE | B2m | Beta‐2‐microglobulin OS=Mus musculus GN=B2m PE=1 SV=2 | 13770 | 59 | 110 | 2 | 10 | 2 | 2 | 16 | 16 |
| BASP1_MOUSE | Basp1 | Brain acid soluble protein 1 OS=Mus musculus GN=Basp1 PE=1 SV=3 | 22074 | 364 | 321 | 11 | 7 | 8 | 6 | 51.3 | 38.1 |
| BASI_MOUSE | Bsg | Basigin OS=Mus musculus GN=Bsg PE=1 SV=2 | 42418 | 166 | 77 | 9 | 7 | 4 | 2 | 10.3 | 9.5 |
| CO3_MOUSE | C3 | Complement C3 OS=Mus musculus GN=C3 PE=1 SV=3 | 186366 | 111 | 189 | 3 | 5 | 3 | 5 | 1.5 | 3.3 |
| CALM_MOUSE | Calm1 | Calmodulin OS=Mus musculus GN=Calm1 PE=1 SV=2 | 16827 | 293 | 272 | 16 | 21 | 5 | 5 | 38.3 | 38.3 |
| CAP1_MOUSE | Cap1 | Adenylyl cyclase‐associated protein 1 OS=Mus musculus GN=Cap1 PE=1 SV=4 | 51532 | 32 | 102 | 1 | 4 | 1 | 3 | 1.7 | 12.4 |
| CSKP_MOUSE | Cask | Peripheral plasma membrane protein CASK OS=Mus musculus GN=Cask PE=1 SV=2 | 105042 | 94 | 86 | 3 | 2 | 3 | 2 | 3.7 | 1.7 |
| TCPB_MOUSE | Cct2 | T‐complex protein 1 subunit beta OS=Mus musculus GN=Cct2 PE=1 SV=4 | 57441 | 89 | 79 | 2 | 2 | 2 | 2 | 6.9 | 5.6 |
| CD44_MOUSE | Cd44 | CD44 antigen OS=Mus musculus GN=Cd44 PE=1 SV=3 | 85565 | 78 | 62 | 3 | 3 | 1 | 1 | 1.5 | 1.5 |
| CD47_MOUSE | Cd47 | Leukocyte surface antigen CD47 OS=Mus musculus GN=Cd47 PE=1 SV=2 | 33076 | 42 | 77 | 1 | 1 | 1 | 1 | 4.6 | 4.6 |
| CD81_MOUSE | Cd81 | CD81 antigen OS=Mus musculus GN=Cd81 PE=1 SV=2 | 25797 | 62 | 94 | 4 | 6 | 1 | 1 | 8.5 | 8.5 |
| CDC42_MOUSE | Cdc42 | Cell division control protein 42 homolog OS=Mus musculus GN=Cdc42 PE=1 SV=2 | 21245 | 172 | 211 | 7 | 9 | 4 | 4 | 25.7 | 25.7 |
| CAD11_MOUSE | Cdh11 | Cadherin‐11 OS=Mus musculus GN=Cdh11 PE=1 SV=1 | 88058 | 30 | 69 | 1 | 2 | 1 | 2 | 1.3 | 2.3 |
| COF1_MOUSE | Cfl1 | Cofilin‐1 OS=Mus musculus GN=Cfl1 PE=1 SV=3 | 18548 | 253 | 375 | 12 | 13 | 5 | 8 | 46.4 | 43.4 |
| CLIC1_MOUSE | Clic1 | Chloride intracellular channel protein 1 OS=Mus musculus GN=Clic1 PE=1 SV=3 | 26996 | 60 | 103 | 2 | 3 | 2 | 3 | 10.8 | 14.1 |
| CLIC4_MOUSE | Clic4 | Chloride intracellular channel protein 4 OS=Mus musculus GN=Clic4 PE=1 SV=3 | 28711 | 90 | 170 | 3 | 5 | 3 | 4 | 16.6 | 19.8 |
| CLH1_MOUSE | Cltc | Clathrin heavy chain 1 OS=Mus musculus GN=Cltc PE=1 SV=3 | 191435 | 221 | 555 | 7 | 14 | 6 | 12 | 6.1 | 10.1 |
| COCA1_MOUSE | Col12a1 | Collagen alpha‐1(XII) chain OS=Mus musculus GN=Col12a1 PE=2 SV=3 | 340004 | 45 | 115 | 1 | 4 | 1 | 4 | 0.3 | 1.3 |
| CO1A1_MOUSE | Col1a1 | Collagen alpha‐1(I) chain OS=Mus musculus GN=Col1a1 PE=1 SV=4 | 137948 | 1728 | 539 | 76 | 27 | 38 | 10 | 42 | 8.5 |
| CO1A2_MOUSE | Col1a2 | Collagen alpha‐2(I) chain OS=Mus musculus GN=Col1a2 PE=1 SV=2 | 129478 | 996 | 608 | 35 | 25 | 25 | 10 | 29.5 | 9.9 |
| CTNA1_MOUSE | Ctnna1 | Catenin alpha‐1 OS=Mus musculus GN=Ctnna1 PE=1 SV=1 | 100044 | 52 | 309 | 2 | 8 | 2 | 7 | 5 | 12.6 |
| CTNB1_MOUSE | Ctnnb1 | Catenin beta‐1 OS=Mus musculus GN=Ctnnb1 PE=1 SV=1 | 85416 | 55 | 321 | 2 | 10 | 2 | 7 | 3.3 | 12.4 |
| DDAH1_MOUSE | Ddah1 | N(G),N(G)‐dimethylarginine dimethylaminohydrol ase 1 OS=Mus musculus GN=Ddah1 PE=1 SV=3 | 31361 | 52 | 49 | 2 | 1 | 2 | 1 | 7 | 3.5 |
| DEST_MOUSE | Dstn | Destrin OS=Mus musculus GN=Dstn PE=1 SV=3 | 18509 | 61 | 79 | 2 | 2 | 2 | 2 | 10.9 | 14.5 |
| DYHC1_MOUSE | Dync1h 1 | Cytoplasmic dynein 1 heavy chain 1 OS=Mus musculus GN=Dync1h1 PE=1 SV=2 | 531710 | 44 | 200 | 1 | 6 | 1 | 6 | 0.3 | 1.8 |
| EDIL3_MOUSE | Edil3 | EGF‐like repeat and discoidin I‐like domain‐containing protein 3 OS=Mus musculus GN=Edil3 PE=1 SV=2 | 53677 | 780 | 678 | 75 | 50 | 16 | 13 | 37.9 | 34 |
| EF1A1_MOUSE | Eef1a1 | Elongation factor 1‐alpha 1 OS=Mus musculus GN=Eef1a1 PE=1 SV=3 | 50082 | 369 | 438 | 24 | 32 | 7 | 9 | 19.3 | 22.5 |
| EF2_MOUSE | Eef2 | Elongation factor 2 OS=Mus musculus GN=Eef2 PE=1 SV=2 | 95253 | 220 | 326 | 6 | 9 | 6 | 9 | 10.5 | 13.5 |
| EHD1_MOUSE | Ehd1 | EH domain‐containing protein 1 OS=Mus musculus GN=Ehd1 PE=1 SV=1 | 60565 | 105 | 147 | 2 | 5 | 2 | 3 | 7.1 | 8.4 |
| IF5A1_MOUSE | Eif5a | Eukaryotic translation initiation factor 5A‐1 OS=Mus musculus GN=Eif5a PE=1 SV=2 | 16821 | 78 | 82 | 3 | 2 | 2 | 2 | 22.7 | 22.7 |
| ENOA_MOUSE | Eno1 | Alpha‐enolase OS=Mus musculus GN=Eno1 PE=1 SV=3 | 47111 | 630 | 508 | 28 | 23 | 12 | 10 | 33.6 | 30.9 |
| EZRI_MOUSE | Ezr | Ezrin OS=Mus musculus GN=Ezr PE=1 SV=3 | 69364 | 222 | 233 | 9 | 8 | 6 | 6 | 13.1 | 11.4 |
| FABP5_MOUSE | Fabp5 | Fatty acid‐binding protein, epidermal OS=Mus musculus GN=Fabp5 PE=1 SV=3 | 15127 | 32 | 66 | 1 | 1 | 1 | 1 | 6.7 | 6.7 |
| NIBAN_MOUSE | Fam129 a | Protein Niban OS=Mus musculus GN=Fam129a PE=1 SV=2 | 102585 | 60 | 405 | 2 | 14 | 2 | 9 | 2.8 | 12.7 |
| FARP1_MOUSE | Farp1 | FERM, RhoGEF and pleckstrin domain‐containing protein 1 OS=Mus musculus GN=Farp1 PE=1 SV=1 | 118801 | 120 | 32 | 2 | 1 | 2 | 1 | 2.7 | 1 |
| FLNA_MOUSE | Flna | Filamin‐A OS=Mus musculus GN=Flna PE=1 SV=5 | 281046 | 234 | 163 | 6 | 5 | 5 | 4 | 2.3 | 1.5 |
| FINC_MOUSE | Fn1 | Fibronectin OS=Mus musculus GN=Fn1 PE=1 SV=4 | 272368 | 2549 | 2319 | 165 | 137 | 46 | 42 | 31.2 | 28.6 |
| FSCN1_MOUSE | Fscn1 | Fascin OS=Mus musculus GN=Fscn1 PE=1 SV=4 | 54474 | 51 | 96 | 2 | 3 | 2 | 3 | 6.9 | 8.7 |
| FRIL1_MOUSE | Ftl1 | Ferritin light chain 1 OS=Mus musculus GN=Ftl1 PE=1 SV=2 | 20790 | 70 | 130 | 3 | 5 | 2 | 3 | 15.8 | 30.6 |
| G3P_MOUSE | Gapdh | Glyceraldehyde‐3‐phosphate dehydrogenase OS=Mus musculus GN=Gapdh PE=1 SV=2 | 35787 | 439 | 369 | 24 | 21 | 8 | 8 | 34.5 | 42.9 |
| SYG_MOUSE | Gars | Glycine–tRNA ligase OS=Mus musculus GN=Gars PE=1 SV=1 | 81826 | 35 | 94 | 1 | 3 | 1 | 3 | 1.4 | 5.3 |
| GDIB_MOUSE | Gdi2 | Rab GDP dissociation inhibitor beta OS=Mus musculus GN=Gdi2 PE=1 SV=1 | 50505 | 215 | 447 | 6 | 15 | 5 | 9 | 13.9 | 28.3 |
| GNAI2_MOUSE | Gnai2 | Guanine nucleotide‐binding protein G(i) subunit alpha‐2 OS=Mus musculus GN=Gnai2 PE=1 SV=5 | 40463 | 321 | 556 | 14 | 32 | 6 | 9 | 24.8 | 33.5 |
| GNAS1_MOUSE | Gnas | Guanine nucleotide‐binding protein G(s) subunit alpha isoforms XLas OS=Mus musculus GN=Gnas PE=1 SV=1 | 121429 | 139 | 220 | 7 | 14 | 3 | 5 | 3.2 | 4.7 |
| GBB1_MOUSE | Gnb1 | Guanine nucleotide‐binding protein G(I)/G(S)/G(T) subunit beta‐1 OS=Mus musculus GN=Gnb1 PE=1 SV=3 | 37353 | 134 | 233 | 5 | 10 | 3 | 5 | 9.1 | 13.2 |
| GBG12_MOUSE | Gng12 | Guanine nucleotide‐binding protein G(I)/G(S)/G(O) subunit gamma‐12 OS=Mus musculus GN=Gng12 PE=1 SV=3 | 7992 | 142 | 100 | 3 | 2 | 3 | 2 | 47.2 | 25 |
| GPC1_MOUSE | Gpc1 | Glypican‐1 OS=Mus musculus GN=Gpc1 PE=1 SV=1 | 61321 | 31 | 235 | 1 | 8 | 1 | 5 | 2.5 | 14 |
| GELS_MOUSE | Gsn | Gelsolin OS=Mus musculus GN=Gsn PE=1 SV=3 | 85888 | 67 | 495 | 2 | 17 | 2 | 12 | 4.2 | 23.6 |
| GSTP1_MOUSE | Gstp1 | Glutathione S‐transferase P 1 OS=Mus musculus GN=Gstp1 PE=1 SV=2 | 23594 | 138 | 203 | 5 | 6 | 3 | 3 | 20.5 | 20.5 |
| HA1B_MOUSE | H2‐K1 | H‐2 class I histocompatibility antigen, K‐B alpha chain OS=Mus musculus GN=H2‐K1 PE=1 SV=1 | 41276 | 105 | 165 | 4 | 7 | 2 | 4 | 5.1 | 12.2 |
| HBE_MOUSE | Hbb‐y | Hemoglobin subunit epsilon‐Y2 OS=Mus musculus GN=Hbb‐y PE=1 SV=2 | 16126 | 52 | 38 | 5 | 5 | 1 | 1 | 6.8 | 6.8 |
| HS90A_MOUS E | Hsp90aa 1 | Heat shock protein HSP 90‐alpha OS=Mus musculus GN=Hsp90aa1 PE=1 SV=4 | 84735 | 230 | 355 | 7 | 8 | 6 | 8 | 10.6 | 13.2 |
| HS90B_MOUS E | Hsp90a b1 | Heat shock protein HSP 90‐beta OS=Mus musculus GN=Hsp90ab1 PE=1 SV=3 | 83229 | 292 | 357 | 11 | 11 | 7 | 8 | 10.8 | 12 |
| HSP7C_MOUS E | Hspa8 | Heat shock cognate 71 kDa protein OS=Mus musculus GN=Hspa8 PE=1 SV=1 | 70827 | 593 | 784 | 23 | 29 | 15 | 17 | 29.3 | 35.3 |
| PGBM_MOUSE | Hspg2 | Basement membrane‐specific heparan sulfate proteoglycan core protein OS=Mus musculus GN=Hspg2 PE=1 SV=1 | 398039 | 892 | 242 | 29 | 5 | 20 | 5 | 7.6 | 1.4 |
| IFM2_MOUSE | Ifitm2 | Interferon‐induced transmembrane protein 2 OS=Mus musculus GN=Ifitm2 PE=1 SV=1 | 15733 | 34 | 40 | 2 | 5 | 1 | 1 | 5.6 | 5.6 |
| IGSF8_MOUSE | Igsf8 | Immunoglobulin superfamily member 8 OS=Mus musculus GN=Igsf8 PE=1 SV=2 | 64970 | 70 | 271 | 2 | 9 | 2 | 6 | 4.7 | 15.9 |
| ILK_MOUSE | Ilk | Integrin‐linked protein kinase OS=Mus musculus GN=Ilk PE=1 SV=2 | 51340 | 35 | 32 | 1 | 1 | 1 | 1 | 2.2 | 2.2 |
| IQGA1_MOUSE | Iqgap1 | Ras GTPase‐activating‐like protein IQGAP1 OS=Mus musculus GN=Iqgap1 PE=1 SV=2 | 188624 | 226 | 407 | 5 | 9 | 5 | 9 | 4.4 | 9.2 |
| ITA11_MOUSE | Itga11 | Integrin alpha‐11 OS=Mus musculus GN=Itga11 PE=1 SV=1 | 132929 | 38 | 74 | 1 | 3 | 1 | 2 | 0.8 | 1.4 |
| ITAV_MOUSE | Itgav | Integrin alpha‐V OS=Mus musculus GN=Itgav PE=1 SV=2 | 115287 | 166 | 500 | 6 | 14 | 5 | 11 | 7.2 | 12 |
| ITB1_MOUSE | Itgb1 | Integrin beta‐1 OS=Mus musculus GN=Itgb1 PE=1 SV=1 | 88173 | 161 | 298 | 6 | 10 | 4 | 7 | 5.3 | 12.9 |
| ITIH2_MOUSE | Itih2 | Inter‐alpha‐trypsin inhibitor heavy chain H2 OS=Mus musculus GN=Itih2 PE=1 SV=1 | 105861 | 272 | 471 | 10 | 17 | 6 | 8 | 6.7 | 9.3 |
| ITIH3_MOUSE | Itih3 | Inter‐alpha‐trypsin inhibitor heavy chain H3 OS=Mus musculus GN=Itih3 PE=1 SV=3 | 99296 | 42 | 72 | 1 | 3 | 1 | 2 | 1.1 | 2.7 |
| IMB1_MOUSE | Kpnb1 | Importin subunit beta‐1 OS=Mus musculus GN=Kpnb1 PE=1 SV=2 | 97122 | 33 | 103 | 1 | 2 | 1 | 2 | 1.4 | 3.1 |
| K1C10_MOUSE | Krt10 | Keratin, type I cytoskeletal 10 OS=Mus musculus GN=Krt10 PE=1 SV=3 | 57735 | 333 | 79 | 29 | 3 | 6 | 2 | 10 | 3.7 |
| K22E_MOUSE | Krt2 | Keratin, type II cytoskeletal 2 epidermal OS=Mus musculus GN=Krt2 PE=1 SV=1 | 70880 | 125 | 104 | 9 | 2 | 2 | 2 | 3.3 | 3.3 |
| K2C5_MOUSE | Krt5 | Keratin, type II cytoskeletal 5 OS=Mus musculus GN=Krt5 PE=1 SV=1 | 61729 | 208 | 60 | 9 | 2 | 3 | 1 | 5.7 | 2.1 |
| K2C73_MOUSE | Krt73 | Keratin, type II cytoskeletal 73 OS=Mus musculus GN=Krt73 PE=1 SV=1 | 58875 | 168 | 128 | 8 | 5 | 2 | 2 | 4.3 | 4.3 |
| K22O_MOUSE | Krt76 | Keratin, type II cytoskeletal 2 oral OS=Mus musculus GN=Krt76 PE=1 SV=1 | 62806 | 96 | 51 | 6 | 1 | 2 | 1 | 3.4 | 1.5 |
| K2C79_MOUSE | Krt79 | Keratin, type II cytoskeletal 79 OS=Mus musculus GN=Krt79 PE=1 SV=2 | 57517 | 88 | 77 | 2 | 2 | 1 | 2 | 2.3 | 4.3 |
| LAMP1_MOUSE | Lamp1 | Lysosome‐associated membrane glycoprotein 1 OS=Mus musculus GN=Lamp1 PE=1 SV=2 | 43837 | 117 | 218 | 5 | 9 | 3 | 4 | 7.6 | 11.3 |
| LAMP2_MOUSE | Lamp2 | Lysosome‐associated membrane glycoprotein 2 OS=Mus musculus GN=Lamp2 PE=1 SV=2 | 45652 | 63 | 72 | 3 | 2 | 2 | 2 | 4.3 | 4.1 |
| LDHA_MOUSE | Ldha | L‐lactate dehydrogenase A chain OS=Mus musculus GN=Ldha PE=1 SV=3 | 36475 | 426 | 369 | 18 | 14 | 8 | 8 | 26.8 | 25.6 |
| LEG1_MOUSE | Lgals1 | Galectin‐1 OS=Mus musculus GN=Lgals1 PE=1 SV=3 | 14856 | 146 | 181 | 9 | 7 | 2 | 3 | 17.8 | 25.2 |
| LRP1_MOUSE | Lrp1 | Prolow‐density lipoprotein receptor‐related protein 1 OS=Mus musculus GN=Lrp1 PE=1 SV=1 | 504411 | 39 | 198 | 1 | 4 | 1 | 4 | 0.2 | 0.9 |
| LYZ2_MOUSE | Lyz2 | Lysozyme C‐2 OS=Mus musculus GN=Lyz2 PE=1 SV=2 | 16678 | 53 | 109 | 2 | 6 | 1 | 1 | 10.1 | 10.1 |
| MARCS_MOUSE | Marcks | Myristoylated alanine‐rich C‐kinase substrate OS=Mus musculus GN=Marcks PE=1 SV=2 | 29644 | 173 | 201 | 8 | 10 | 4 | 4 | 16.8 | 16.8 |
| MRP_MOUSE | Marcksl 1 | MARCKS‐related protein OS=Mus musculus GN=Marcksl1 PE=1 SV=2 | 20153 | 77 | 59 | 2 | 1 | 2 | 1 | 14 | 6.5 |
| MFGM_MOUS E | Mfge8 | Lactadherin OS=Mus musculus GN=Mfge8 PE=1 SV=3 | 51208 | 895 | 1287 | 125 | 331 | 17 | 21 | 42.8 | 44.7 |
| MIF_MOUSE | Mif | Macrophage migration inhibitory factor OS=Mus musculus GN=Mif PE=1 SV=2 | 12496 | 56 | 51 | 4 | 3 | 1 | 1 | 9.6 | 9.6 |
| MOES_MOUSE | Msn | Moesin OS=Mus musculus GN=Msn PE=1 SV=3 | 67725 | 771 | 1229 | 38 | 47 | 19 | 28 | 35 | 45.2 |
| MVP_MOUSE | Mvp | Major vault protein OS=Mus musculus GN=Mvp PE=1 SV=4 | 95865 | 62 | 156 | 2 | 4 | 2 | 4 | 2.7 | 6.2 |
| MYADM_MOUSE | Myadm | Myeloid‐associated differentiation marker OS=Mus musculus GN=Myadm PE=1 SV=2 | 35261 | 69 | 77 | 5 | 5 | 1 | 1 | 5.3 | 5.3 |
| MYH9_MOUSE | Myh9 | Myosin‐9 OS=Mus musculus GN=Myh9 PE=1 SV=4 | 226232 | 535 | 828 | 16 | 21 | 15 | 19 | 12.7 | 14.5 |
| MYL6_MOUSE | Myl6 | Myosin light polypeptide 6 OS=Mus musculus GN=Myl6 PE=1 SV=3 | 16919 | 57 | 138 | 2 | 4 | 2 | 4 | 15.9 | 29.1 |
| MYO1B_MOUSE | Myo1b | Unconventional myosin‐Ib OS=Mus musculus GN=Myo1b PE=1 SV=3 | 128483 | 77 | 533 | 2 | 17 | 2 | 11 | 3.5 | 16.4 |
| MYO1C_MOUSE | Myo1c | Unconventional myosin‐Ic OS=Mus musculus GN=Myo1c PE=1 SV=2 | 121868 | 338 | 595 | 12 | 20 | 9 | 15 | 13.8 | 17.7 |
| NID2_MOUSE | Nid2 | Nidogen‐2 OS=Mus musculus GN=Nid2 PE=1 SV=2 | 153816 | 167 | 75 | 4 | 2 | 4 | 2 | 4.1 | 1.6 |
| NDKA_MOUSE | Nme1 | Nucleoside diphosphate kinase A OS=Mus musculus GN=Nme1 PE=1 SV=1 | 17197 | 31 | 68 | 2 | 5 | 1 | 2 | 11.2 | 21.1 |
| PDC6I_MOUSE | Pdcd6ip | Programmed cell death 6‐interacting protein OS=Mus musculus GN=Pdcd6ip PE=1 SV=3 | 95964 | 164 | 224 | 6 | 8 | 5 | 6 | 8.2 | 8.1 |
| PEBP1_MOUSE | Pebp1 | Phosphatidylethanola mine‐binding protein 1 OS=Mus musculus GN=Pebp1 PE=1 SV=3 | 20817 | 33 | 185 | 1 | 5 | 1 | 4 | 13.9 | 32.6 |
| PROF1_MOUS E | Pfn1 | Profilin‐1 OS=Mus musculus GN=Pfn1 PE=1 SV=2 | 14948 | 232 | 275 | 8 | 11 | 4 | 4 | 42.1 | 42.1 |
| PI4KA_MOUSE | Pi4ka | Phosphatidylinositol 4‐kinase alpha OS=Mus musculus GN=Pi4ka PE=1 SV=2 | 236889 | 43 | 60 | 1 | 1 | 1 | 1 | 0.6 | 0.6 |
| KPYM_MOUSE | Pkm | Pyruvate kinase PKM OS=Mus musculus GN=Pkm PE=1 SV=4 | 57808 | 603 | 678 | 27 | 25 | 12 | 12 | 29.8 | 33 |
| PLP2_MOUSE | Plp2 | Proteolipid protein 2 OS=Mus musculus GN=Plp2 PE=1 SV=1 | 16597 | 56 | 112 | 1 | 3 | 1 | 2 | 7.9 | 24.3 |
| PPIA_MOUSE | Ppia | Peptidyl‐prolyl cistrans isomerase A OS=Mus musculus GN=Ppia PE=1 SV=2 | 17960 | 212 | 239 | 12 | 16 | 6 | 6 | 31.7 | 34.8 |
| 2AAA_MOUSE | Ppp2r1a | Serine/threonine‐protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform OS=Mus musculus GN=Ppp2r1a PE=1 SV=3 | 65281 | 61 | 57 | 1 | 1 | 1 | 1 | 1.7 | 1.7 |
| PRDX1_MOUSE | Prdx1 | Peroxiredoxin‐1 OS=Mus musculus GN=Prdx1 PE=1 SV=1 | 22162 | 108 | 288 | 4 | 12 | 3 | 8 | 16.1 | 40.2 |
| PRDX2_MOUSE | Prdx2 | Peroxiredoxin‐2 OS=Mus musculus GN=Prdx2 PE=1 SV=3 | 21765 | 38 | 74 | 1 | 2 | 1 | 2 | 4 | 17.2 |
| PRIO_MOUSE | Prnp | Major prion protein OS=Mus musculus GN=Prnp PE=1 SV=2 | 27960 | 33 | 81 | 1 | 2 | 1 | 2 | 3.5 | 7.1 |
| PTK7_MOUSE | Ptk7 | Inactive tyrosine‐protein kinase 7 OS=Mus musculus GN=Ptk7 PE=1 SV=1 | 117457 | 284 | 181 | 12 | 5 | 8 | 4 | 11.7 | 5.6 |
| RAB10_MOUSE | Rab10 | Ras‐related protein Rab‐10 OS=Mus musculus GN=Rab10 PE=1 SV=1 | 22527 | 173 | 272 | 7 | 12 | 4 | 5 | 20 | 26 |
| RAB14_MOUSE | Rab14 | Ras‐related protein Rab‐14 OS=Mus musculus GN=Rab14 PE=1 SV=3 | 23882 | 83 | 191 | 5 | 8 | 2 | 4 | 8.4 | 19.1 |
| RAP1A_MOUSE | Rap1a | Ras‐related protein Rap‐1A OS=Mus musculus GN=Rap1a PE=1 SV=1 | 20974 | 244 | 334 | 13 | 17 | 5 | 5 | 27.2 | 30.4 |
| RAP2B_MOUSE | Rap2b | Ras‐related protein Rap‐2b OS=Mus musculus GN=Rap2b PE=1 SV=1 | 20491 | 43 | 83 | 1 | 2 | 1 | 2 | 6.6 | 20.8 |
| RADI_MOUSE | Rdx | Radixin OS=Mus musculus GN=Rdx PE=1 SV=3 | 68500 | 220 | 364 | 9 | 11 | 6 | 9 | 11.3 | 18.4 |
| RHOA_MOUSE | Rhoa | Transforming protein RhoA OS=Mus musculus GN=Rhoa PE=1 SV=1 | 21768 | 118 | 201 | 2 | 11 | 2 | 4 | 13 | 31.6 |
| RS27A_MOUSE | Rps27a | Ubiquitin‐40S ribosomal protein S27a OS=Mus musculus GN=Rps27a PE=1 SV=2 | 17939 | 107 | 186 | 8 | 13 | 2 | 4 | 16 | 30.1 |
| RS8_MOUSE | Rps8 | 40S ribosomal protein S8 OS=Mus musculus GN=Rps8 PE=1 SV=2 | 24190 | 34 | 39 | 1 | 1 | 1 | 1 | 4.3 | 4.3 |
| RRAS_MOUSE | Rras | Ras‐related protein R‐Ras OS=Mus musculus GN=Rras PE=1 SV=1 | 23749 | 90 | 147 | 2 | 4 | 2 | 4 | 10.6 | 21.1 |
| RRAS2_MOUSE | Rras2 | Ras‐related protein R‐Ras2 OS=Mus musculus GN=Rras2 PE=1 SV=1 | 23385 | 120 | 173 | 2 | 4 | 2 | 4 | 13.7 | 23.5 |
| S10AA_MOUS E | S100a10 | Protein S100‐A10 OS=Mus musculus GN=S100a10 PE=1 SV=2 | 11179 | 74 | 97 | 4 | 6 | 2 | 2 | 35.1 | 35.1 |
| S10AB_MOUSE | S100a11 | Protein S100‐A11 OS=Mus musculus GN=S100a11 PE=1 SV=1 | 11075 | 66 | 111 | 2 | 3 | 1 | 1 | 16.3 | 16.3 |
| S10A4_MOUSE | S100a4 | Protein S100‐A4 OS=Mus musculus GN=S100a4 PE=1 SV=1 | 11714 | 41 | 135 | 1 | 3 | 1 | 3 | 8.9 | 17.8 |
| S10A6_MOUSE | S100a6 | Protein S100‐A6 OS=Mus musculus GN=S100a6 PE=1 SV=3 | 10044 | 107 | 147 | 4 | 3 | 3 | 3 | 47.2 | 47.2 |
| SH3L3_MOUSE | Sh3bgrl 3 | SH3 domain‐binding glutamic acid‐rich‐like protein 3 OS=Mus musculus GN=Sh3bgrl3 PE=1 SV=1 | 10470 | 43 | 40 | 1 | 1 | 1 | 1 | 10.8 | 10.8 |
| MOT1_MOUSE | Slc16a1 | Monocarboxylate transporter 1 OS=Mus musculus GN=Slc16a1 PE=1 SV=1 | 53232 | 116 | 129 | 3 | 3 | 2 | 2 | 4.3 | 4.3 |
| SATT_MOUSE | Slc1a4 | Neutral amino acid transporter A OS=Mus musculus GN=Slc1a4 PE=1 SV=1 | 56026 | 66 | 39 | 2 | 1 | 2 | 1 | 3.6 | 2.1 |
| 4F2_MOUSE | Slc3a2 | 4F2 cell‐surface antigen heavy chain OS=Mus musculus GN=Slc3a2 PE=1 SV=1 | 58300 | 454 | 325 | 13 | 9 | 9 | 8 | 20.7 | 16.9 |
| LAT1_MOUSE | Slc7a5 | Large neutral amino acids transporter small subunit 1 OS=Mus musculus GN=Slc7a5 PE=1 SV=2 | 55836 | 233 | 213 | 7 | 5 | 4 | 4 | 16 | 16 |
| TAGL_MOUSE | Tagln | Transgelin OS=Mus musculus GN=Tagln PE=1 SV=3 | 22561 | 36 | 62 | 1 | 2 | 1 | 2 | 6.5 | 10 |
| TFR1_MOUSE | Tfrc | Transferrin receptor protein 1 OS=Mus musculus GN=Tfrc PE=1 SV=1 | 85677 | 179 | 158 | 10 | 6 | 4 | 4 | 6 | 5.8 |
| THY1_MOUSE | Thy1 | Thy‐1 membrane glycoprotein OS=Mus musculus GN=Thy1 PE=1 SV=1 | 18069 | 66 | 189 | 6 | 20 | 2 | 4 | 15.4 | 23.5 |
| TLN1_MOUSE | Tln1 | Talin‐1 OS=Mus musculus GN=Tln1 PE=1 SV=2 | 269653 | 107 | 352 | 2 | 7 | 2 | 7 | 1 | 3.4 |
| TYB4_MOUSE | Tmsb4x | Thymosin beta‐4 OS=Mus musculus GN=Tmsb4x PE=1 SV=1 | 5676 | 32 | 33 | 1 | 1 | 1 | 1 | 14 | 14 |
| TENA_MOUSE | Tnc | Tenascin OS=Mus musculus GN=Tnc PE=1 SV=1 | 231659 | 209 | 84 | 5 | 2 | 5 | 2 | 3.8 | 2.5 |
| TPIS_MOUSE | Tpi1 | Triosephosphate isomerase OS=Mus musculus GN=Tpi1 PE=1 SV=4 | 32171 | 214 | 185 | 7 | 5 | 5 | 4 | 20.4 | 15.7 |
| TPM4_MOUSE | Tpm4 | Tropomyosin alpha‐4 chain OS=Mus musculus GN=Tpm4 PE=1 SV=3 | 28450 | 71 | 117 | 2 | 3 | 2 | 3 | 14.9 | 18.1 |
| TBA1A_MOUSE | Tuba1a | Tubulin alpha‐1A chain OS=Mus musculus GN=Tuba1a PE=1 SV=1 | 50104 | 391 | 323 | 11 | 9 | 7 | 5 | 22.2 | 17.3 |
| TBB5_MOUSE | Tubb5 | Tubulin beta‐5 chain OS=Mus musculus GN=Tubb5 PE=1 SV=1 | 49639 | 472 | 449 | 22 | 24 | 9 | 10 | 26.4 | 31.3 |
| UBE2N_MOUSE | Ube2n | Ubiquitin‐conjugating enzyme E2 N OS=Mus musculus GN=Ube2n PE=1 SV=1 | 17127 | 41 | 60 | 1 | 1 | 1 | 1 | 7.2 | 7.2 |
| VAMP1_MOUSE | Vamp1 | Vesicle‐associated membrane protein 1 OS=Mus musculus GN=Vamp1 PE=1 SV=1 | 12882 | 32 | 43 | 1 | 1 | 1 | 1 | 5.9 | 5.9 |
| VAT1_MOUSE | Vat1 | Synaptic vesicle membrane protein VAT‐1 homolog OS=Mus musculus GN=Vat1 PE=1 SV=3 | 43069 | 252 | 345 | 14 | 16 | 7 | 7 | 28.8 | 23.4 |
| VINC_MOUSE | Vcl | Vinculin OS=Mus musculus GN=Vcl PE=1 SV=4 | 116644 | 60 | 286 | 2 | 8 | 2 | 8 | 3.3 | 9.9 |
| TERA_MOUSE | Vcp | Transitional endoplasmic reticulum ATPase OS=Mus musculus GN=Vcp PE=1 SV=4 | 89266 | 71 | 277 | 2 | 8 | 2 | 7 | 4 | 13 |
| VIME_MOUSE | Vim | Vimentin OS=Mus musculus GN=Vim PE=1 SV=3 | 53655 | 358 | 349 | 11 | 11 | 8 | 8 | 19.1 | 16.1 |
| WDR1_MOUSE | Wdr1 | WD repeat‐containing protein 1 OS=Mus musculus GN=Wdr1 PE=1 SV=3 | 66365 | 67 | 36 | 3 | 1 | 2 | 1 | 4 | 1.3 |
| YKT6_MOUSE | Ykt6 | Synaptobrevin homolog YKT6 OS=Mus musculus GN=Ykt6 PE=1 SV=1 | 22300 | 52 | 33 | 2 | 1 | 2 | 1 | 9.1 | 4.5 |
| 1433B_MOUSE | Ywhab | 14‐3‐3 protein beta/alpha OS=Mus musculus GN=Ywhab PE=1 SV=3 | 28069 | 241 | 299 | 6 | 9 | 5 | 6 | 18.3 | 24 |
| 1433E_MOUSE | Ywhae | 14‐3‐3 protein epsilon OS=Mus musculus GN=Ywhae PE=1 SV=1 | 29155 | 214 | 555 | 9 | 20 | 5 | 10 | 21.6 | 45.5 |
| 1433G_MOUS E | Ywhag | 14‐3‐3 protein gamma OS=Mus musculus GN=Ywhag PE=1 SV=2 | 28285 | 346 | 446 | 11 | 14 | 8 | 9 | 31.2 | 35.2 |
| 1433F_MOUSE | Ywhah | 14‐3‐3 protein eta OS=Mus musculus GN=Ywhah PE=1 SV=2 | 28194 | 195 | 267 | 6 | 7 | 4 | 6 | 15.9 | 22 |
| 1433T_MOUSE | Ywhaq | 14‐3‐3 protein theta OS=Mus musculus GN=Ywhaq PE=1 SV=1 | 27761 | 226 | 249 | 6 | 6 | 4 | 4 | 15.9 | 15.9 |
| 1433Z_MOUSE | Ywhaz | 14‐3‐3 protein zeta/delta OS=Mus musculus GN=Ywhaz PE=1 SV=1 | 27754 | 338 | 415 | 12 | 14 | 7 | 7 | 40.4 | 35.9 |
Table 2.
Proteins unique to D0 osteosomes.
| prot_acc | GN | prot_desc | prot_mass (Da) | prot_score | Number of significant matches | Number of significant unique peptide sequences | Sequence coverage |
|---|---|---|---|---|---|---|---|
| ARF2_MOU SE | Arf2 | ADP‐ribosylation factor 2 OS=Mus musculus GN=Arf2 PE=1 SV=2 | 20733 | 219 | 7 | 5 | 43.6 |
| HS71A_MO USE | Hspa1a | Heat shock 70 kDa protein 1A OS=Mus musculus GN=Hspa1a PE=1 SV=2 | 70036 | 197 | 6 | 4 | 7.6 |
| K2C1_MOU SE | Krt1 | Keratin, type II cytoskeletal 1 OS=Mus musculus GN=Krt1 PE=1 SV=4 | 65565 | 168 | 11 | 2 | 3.6 |
| GNAI3_MO USE | Gnai3 | Guanine nucleotide‐binding protein G(k) subunit alpha OS=Mus musculus GN=Gnai3 PE=1 SV=3 | 40512 | 144 | 8 | 3 | 11.6 |
| K1C17_MO USE | Krt17 | Keratin, type I cytoskeletal 17 OS=Mus musculus GN=Krt17 PE=1 SV=3 | 48132 | 113 | 10 | 3 | 6.2 |
| RALA_MOUSE | Rala | Ras‐related protein Ral‐A OS=Mus musculus GN=Rala PE=1 SV=1 | 23538 | 109 | 3 | 3 | 25.2 |
| STOM_MOUSE | Stom | Erythrocyte band 7 integral membrane protein OS=Mus musculus GN=Stom PE=1 SV=3 | 31355 | 95 | 3 | 3 | 15.1 |
| GNA12_MO USE | Gna12 | Guanine nucleotide‐binding protein subunit alpha‐12 OS=Mus musculus GN=Gna12 PE=1 SV=3 | 44067 | 85 | 5 | 2 | 5 |
| GTR1_MOU SE | Slc2a1 | Solute carrier family 2, facilitated glucose transporter member 1 OS=Mus musculus GN=Slc2a1 PE=1 SV=4 | 53949 | 81 | 4 | 2 | 3.7 |
| PGAM1_MOUSE | Pgam1 | Phosphoglycerate mutase 1 OS=Mus musculus GN=Pgam1 PE=1 SV=3 | 28814 | 78 | 1 | 1 | 7.1 |
| AKA12_MO USE | Akap12 | A‐kinase anchor protein 12 OS=Mus musculus GN=Akap12 PE=1 SV=1 | 180586 | 76 | 2 | 2 | 1.1 |
| NID1_MOU SE | Nid1 | Nidogen‐1 OS=Mus musculus GN=Nid1 PE=1 SV=2 | 136450 | 65 | 1 | 1 | 1.4 |
| RAP2C_MO USE | Rap2c | Ras‐related protein Rap‐2c OS=Mus musculus GN=Rap2c PE=1 SV=1 | 20731 | 64 | 2 | 2 | 14.8 |
| LAMA4_MOUSE | Lama4 | Laminin subunit alpha‐4 OS=Mus musculus GN=Lama4 PE=1 SV=2 | 201692 | 63 | 2 | 2 | 1.8 |
| TTYH3_MO USE | Ttyh3 | Protein tweety homolog 3 OS=Mus musculus GN=Ttyh3 PE=1 SV=1 | 57677 | 61 | 1 | 1 | 2.7 |
| ANT3_MOU SE | Serpinc 1 | Antithrombin‐III OS=Mus musculus GN=Serpinc1 PE=1 SV=1 | 51971 | 60 | 2 | 2 | 6.2 |
| APOM_MO USE | Apom | Apolipoprotein M OS=Mus musculus GN=Apom PE=1 SV=1 | 21259 | 59 | 1 | 1 | 4.2 |
| RS2_MOUSE | Rps2 | 40S ribosomal protein S2 OS=Mus musculus GN=Rps2 PE=1 SV=3 | 31212 | 51 | 1 | 1 | 4.4 |
| GSLG1_MO USE | Glg1 | Golgi apparatus protein 1 OS=Mus musculus GN=Glg1 PE=1 SV=1 | 133646 | 46 | 1 | 1 | 0.9 |
| GAPR1_MO USE | Glipr2 | Golgi‐associated plant pathogenesis‐related protein 1 OS=Mus musculus GN=Glipr2 PE=1 SV=3 | 17080 | 43 | 1 | 1 | 8.4 |
| RAB5C_MO USE | Rab5c | Ras‐related protein Rab‐5C OS=Mus musculus GN=Rab5c PE=1 SV=2 | 23398 | 43 | 1 | 1 | 6.5 |
| S38A5_MO USE | Slc38a5 | Sodium‐coupled neutral amino acid transporter 5 OS=Mus musculus GN=Slc38a5 PE=1 SV=1 | 52582 | 42 | 1 | 1 | 1.7 |
| AK1A1_MO USE | Akr1a1 | Alcohol dehydrogenase [NADP(+)] OS=Mus musculus GN=Akr1a1 PE=1 SV=3 | 36564 | 39 | 1 | 1 | 5.8 |
| SF3A3_MO USE | Sf3a3 | Splicing factor 3A subunit 3 OS=Mus musculus GN=Sf3a3 PE=1 SV=2 | 58805 | 39 | 1 | 1 | 1.4 |
| CTR1_MOU SE | Slc7a1 | High affinity cationic amino acid transporter 1 OS=Mus musculus GN=Slc7a1 PE=1 SV=1 | 67048 | 35 | 1 | 1 | 3.1 |
| NCAM1_M OUSE | Ncam1 | Neural cell adhesion molecule 1 OS=Mus musculus GN=Ncam1 PE=1 SV=3 | 119353 | 34 | 1 | 1 | 0.6 |
| RL8_MOUSE | Rpl8 | 60S ribosomal protein L8 OS=Mus musculus GN=Rpl8 PE=1 SV=2 | 28007 | 34 | 1 | 1 | 4.3 |
| VATL_MOUSE | Atp6v0c | V‐type proton ATPase 16 kDa proteolipid subunit OS=Mus musculus GN=Atp6v0c PE=1 SV=1 | 15798 | 33 | 1 | 1 | 20 |
| EHD2_MOU SE | Ehd2 | EH domain‐containing protein 2 OS=Mus musculus GN=Ehd2 PE=1 SV=1 | 61136 | 33 | 1 | 1 | 2.8 |
| H4_MOUSE | Hist1h4a | Histone H4 OS=Mus musculus GN=Hist1h4a PE=1 SV=2 | 11360 | 33 | 1 | 1 | 9.7 |
| PLAK_MOU SE | Jup | Junction plakoglobin OS=Mus musculus GN=Jup PE=1 SV=3 | 81749 | 33 | 1 | 1 | 2.4 |
| PCDAA_MOUSE | Pcdha10 | Protocadherin alpha‐10 OS=Mus musculus GN=Pcdha10 PE=2 SV=1 | 101994 | 33 | 1 | 1 | 0.8 |
| ASNS_MOUSE | Asns | Asparagine synthetase [glutamine‐hydrolyzing] OS=Mus musculus GN=Asns PE=1 SV=3 | 64241 | 31 | 1 | 1 | 1.6 |
| TCPZ_MOUSE | Cct6a | T‐complex protein 1 subunit zeta OS=Mus musculus GN=Cct6a PE=1 SV=3 | 57968 | 31 | 1 | 1 | 3.2 |
| RASH_MOUSE | Hras | GTPase HRas OS=Mus musculus GN=Hras PE=1 SV=2 | 21285 | 31 | 1 | 1 | 5.8 |
| PARVA_MOUSE | Parva | Alpha‐parvin OS=Mus musculus GN=Parva PE=1 SV=1 | 42304 | 31 | 1 | 1 | 3.8 |
| PGK1_MOUSE | Pgk1 | Phosphoglycerate kinase 1 OS=Mus musculus GN=Pgk1 PE=1 SV=4 | 44522 | 31 | 1 | 1 | 4.3 |
Table 3.
Proteins unique to D24 osteosomes
| prot_acc | GN | prot_desc | prot_mass (Da) | prot_sco re | Number of significant matches | Number of significant unique peptide sequences | Sequence coverage |
|---|---|---|---|---|---|---|---|
| CO6A1_MOUSE | Col6a1 | Collagen alpha‐1(VI) chain OS=Mus musculus GN=Col6a1 PE=1 SV=1 | 108422 | 1441 | 117 | 21 | 26 |
| CO6A2_MOUSE | Col6a2 | Collagen alpha‐2(VI) chain OS=Mus musculus GN=Col6a2 PE=1 SV=3 | 110266 | 871 | 50 | 19 | 23 |
| FPRP_MOUSE | Ptgfrn | Prostaglandin F2 receptor negative regulator OS=Mus musculus GN=Ptgfrn PE=1 SV=2 | 98660 | 630 | 20 | 13 | 18.7 |
| E41L2_MOUSE | Epb41l2 | Band 4.1‐like protein 2 OS=Mus musculus GN=Epb41l2 PE=1 SV=2 | 109873 | 603 | 19 | 14 | 16.8 |
| PHEX_MOUSE | Phex | Metalloendopeptidase homolog PEX OS=Mus musculus GN=Phex PE=1 SV=1 | 86364 | 469 | 14 | 10 | 17.1 |
| MYO1D_MOUSE | Myo1d | Unconventional myosin‐Id OS=Mus musculus GN=Myo1d PE=1 SV=1 | 116007 | 393 | 12 | 11 | 12.8 |
| PPIC_MOUSE | Ppic | Peptidyl‐prolyl cis‐trans isomerase C OS=Mus musculus GN=Ppic PE=1 SV=1 | 22780 | 389 | 27 | 7 | 48.6 |
| ACTN4_MOUSE | Actn4 | Alpha‐actinin‐4 OS=Mus musculus GN=Actn4 PE=1 SV=1 | 104911 | 388 | 9 | 7 | 10.1 |
| PLCD1_MOUSE | Plcd1 | 1‐phosphatidylinositol 4,5‐bisphosphate phosphodiesterase delta‐1 OS=Mus musculus GN=Plcd1 PE=1 SV=2 | 85819 | 383 | 12 | 8 | 17.9 |
| LUM_MOUSE | Lum | Lumican OS=Mus musculus GN=Lum PE=1 SV=2 | 38241 | 369 | 13 | 7 | 23.4 |
| PHOP1_MOUSE | Phospho1 | Phosphoethanolamine/phosphocholine phosphatase OS=Mus musculus GN=Phospho1 PE=1 SV=1 | 29892 | 311 | 12 | 7 | 31.5 |
| IGSF8_MOUSE | Igsf8 | Immunoglobulin superfamily member 8 OS=Mus musculus GN=Igsf8 PE=1 SV=2 | 64970 | 271 | 9 | 6 | 15.9 |
| EMIL1_MOUSE | Emilin1 | EMILIN‐1 OS=Mus musculus GN=Emilin1 PE=1 SV=1 | 107518 | 266 | 5 | 5 | 7.3 |
| GBB2_MOUSE | Gnb2 | Guanine nucleotide‐binding protein G(I)/G(S)/G(T) subunit beta‐2 OS=Mus musculus GN=Gnb2 PE=1 SV=3 | 37307 | 253 | 10 | 5 | 13.2 |
| NEP_MOUSE | Mme | Neprilysin OS=Mus musculus GN=Mme PE=1 SV=3 | 85648 | 236 | 8 | 6 | 11.1 |
| IDHC_MOUSE | Idh1 | Isocitrate dehydrogenase [NADP] cytoplasmic OS=Mus musculus GN=Idh1 PE=1 SV=2 | 46644 | 226 | 5 | 5 | 17.1 |
| RAB35_MOUSE | Rab35 | Ras‐related protein Rab‐35 OS=Mus musculus GN=Rab35 PE=1 SV=1 | 23011 | 216 | 10 | 4 | 20.9 |
| CATB_MOUSE | Ctsb | Cathepsin B OS=Mus musculus GN=Ctsb PE=1 SV=2 | 37256 | 205 | 5 | 4 | 15.9 |
| KAD1_MOUSE | Ak1 | Adenylate kinase isoenzyme 1 OS=Mus musculus GN=Ak1 PE=1 SV=1 | 21526 | 203 | 5 | 4 | 20.6 |
| CATD_MOUSE | Ctsd | Cathepsin D OS=Mus musculus GN=Ctsd PE=1 SV=1 | 44925 | 194 | 5 | 4 | 12.4 |
| PEDF_MOUSE | Serpinf1 | Pigment epithelium‐derived factor OS=Mus musculus GN=Serpinf1 PE=1 SV=2 | 46205 | 194 | 8 | 5 | 21.3 |
| ASM3B_MOUSE | Smpdl3b | Acid sphingomyelinase‐like phosphodiesterase 3b OS=Mus musculus GN=Smpdl3b PE=1 SV=1 | 51567 | 180 | 4 | 4 | 14.7 |
| CD109_MOUSE | Cd109 | CD109 antigen OS=Mus musculus GN=Cd109 PE=1 SV=1 | 161557 | 174 | 5 | 4 | 3.6 |
| AQP1_MOUSE | Aqp1 | Aquaporin‐1 OS=Mus musculus GN=Aqp1 PE=1 SV=3 | 28775 | 172 | 7 | 4 | 24.5 |
| GNA11_MOUSE | Gna11 | Guanine nucleotide‐binding protein subunit alpha‐11 OS=Mus musculus GN=Gna11 PE=1 SV=1 | 41997 | 169 | 5 | 5 | 17.5 |
| TM119_MOUSE | Tmem119 | Transmembrane protein 119 OS=Mus musculus GN=Tmem119 PE=1 SV=1 | 29383 | 161 | 20 | 4 | 15.7 |
| AEBP1_MOUSE | Aebp1 | Adipocyte enhancer‐binding protein 1 OS=Mus musculus GN=Aebp1 PE=1 SV=1 | 128284 | 158 | 4 | 3 | 3.1 |
| SDCB1_MOUSE | Sdcbp | Syntenin‐1 OS=Mus musculus GN=Sdcbp PE=1 SV=1 | 32359 | 158 | 4 | 3 | 21.1 |
| EHD3_MOUSE | Ehd3 | EH domain‐containing protein 3 OS=Mus musculus GN=Ehd3 PE=1 SV=2 | 60783 | 150 | 6 | 4 | 9 |
| GSTM1_MOUSE | Gstm1 | Glutathione S‐transferase Mu 1 OS=Mus musculus GN=Gstm1 PE=1 SV=2 | 25953 | 142 | 4 | 3 | 17.4 |
| FLNC_MOUSE | Flnc | Filamin‐C OS=Mus musculus GN=Flnc PE=1 SV=3 | 290937 | 134 | 4 | 3 | 1.1 |
| MMP14_MOUSE | Mmp14 | Matrix metalloproteinase‐14 OS=Mus musculus GN=Mmp14 PE=2 SV=3 | 65877 | 132 | 4 | 4 | 7.6 |
| CTND1_MOUSE | Ctnnd1 | Catenin delta‐1 OS=Mus musculus GN=Ctnnd1 PE=1 SV=2 | 104860 | 126 | 3 | 3 | 5.1 |
| AT2B4_MOUSE | Atp2b4 | Plasma membrane calcium‐transporting ATPase 4 OS=Mus musculus GN=Atp2b4 PE=1 SV=1 | 132984 | 125 | 4 | 3 | 3.7 |
| NRP2_MOUSE | Nrp2 | Neuropilin‐2 OS=Mus musculus GN=Nrp2 PE=1 SV=2 | 104565 | 123 | 3 | 3 | 3.8 |
| DLG1_MOUSE | Dlg1 | Disks large homolog 1 OS=Mus musculus GN=Dlg1 PE=1 SV=1 | 100058 | 122 | 4 | 3 | 3.9 |
| PANX3_MOUSE | Panx3 | Pannexin‐3 OS=Mus musculus GN=Panx3 PE=1 SV=1 | 44899 | 122 | 6 | 3 | 12.2 |
| GDIA_MOUSE | Gdi1 | Rab GDP dissociation inhibitor alpha OS=Mus musculus GN=Gdi1 PE=1 SV=3 | 50489 | 116 | 3 | 3 | 10.5 |
| SAP_MOUSE | Psap | Prosaposin OS=Mus musculus GN=Psap PE=1 SV=2 | 61381 | 116 | 6 | 3 | 7.2 |
| DPYL2_MOUSE | Dpysl2 | Dihydropyrimidinase‐related protein 2 OS=Mus musculus GN=Dpysl2 PE=1 SV=2 | 62239 | 115 | 3 | 3 | 9.8 |
| NSMA2_MOUSE | Smpd3 | Sphingomyelin phosphodiesterase 3 OS=Mus musculus GN=Smpd3 PE=1 SV=1 | 71152 | 110 | 3 | 3 | 7.5 |
| ANO6_MOUSE | Ano6 | Anoctamin‐6 OS=Mus musculus GN=Ano6 PE=1 SV=1 | 106186 | 108 | 3 | 3 | 3.4 |
| MYOF_MOUSE | Myof | Myoferlin OS=Mus musculus GN=Myof PE=1 SV=2 | 233177 | 108 | 2 | 2 | 1 |
| PLXB2_MOUSE | Plxnb2 | Plexin‐B2 OS=Mus musculus GN=Plxnb2 PE=1 SV=1 | 206099 | 108 | 2 | 2 | 1 |
| CO3A1_MOUSE | Col3a1 | Collagen alpha‐1(III) chain OS=Mus musculus GN=Col3a1 PE=1 SV=4 | 138858 | 106 | 3 | 3 | 1.9 |
| FERM2_MOUSE | Fermt2 | Fermitin family homolog 2 OS=Mus musculus GN=Fermt2 PE=1 SV=1 | 77750 | 106 | 3 | 3 | 5.6 |
| TKT_MOUSE | Tkt | Transketolase OS=Mus musculus GN=Tkt PE=1 SV=1 | 67588 | 106 | 3 | 3 | 8.2 |
| S13A5_MOUSE | Slc13a5 | Solute carrier family 13 member 5 OS=Mus musculus GN=Slc13a5 PE=2 SV=1 | 63780 | 103 | 4 | 3 | 5.6 |
| MMP2_MOUSE | Mmp2 | 72 kDa type IV collagenase OS=Mus musculus GN=Mmp2 PE=1 SV=1 | 74055 | 102 | 3 | 3 | 8.6 |
| FMOD_MOUSE | Fmod | Fibromodulin OS=Mus musculus GN=Fmod PE=2 SV=1 | 43027 | 101 | 2 | 2 | 11.2 |
| CAPG_MOUSE | Capg | Macrophage‐capping protein OS=Mus musculus GN=Capg PE=1 SV=2 | 39216 | 97 | 2 | 2 | 8 |
| UBA1_MOUSE | Uba1 | Ubiquitin‐like modifier‐activating enzyme 1 OS=Mus musculus GN=Uba1 PE=1 SV=1 | 117734 | 97 | 2 | 2 | 1.9 |
| DDAH2_MOUSE | Ddah2 | N(G),N(G)‐dimethylarginine dimethylaminohydrolase 2 OS=Mus musculus GN=Ddah2 PE=1 SV=1 | 29627 | 96 | 2 | 2 | 12.3 |
| RB11A_MOUSE | Rab11a | Ras‐related protein Rab‐11A OS=Mus musculus GN=Rab11a PE=1 SV=3 | 24378 | 96 | 3 | 3 | 14.8 |
| CA2D1_MOUSE | Cacna2d1 | Voltage‐dependent calcium channel subunit alpha‐2/delta‐1 OS=Mus musculus GN=Cacna2d1 PE=1 SV=1 | 124551 | 93 | 2 | 2 | 3.5 |
| NUCB1_MOUSE | Nucb1 | Nucleobindin‐1 OS=Mus musculus GN=Nucb1 PE=1 SV=2 | 53376 | 93 | 2 | 2 | 4.4 |
| ARF5_MOUSE | Arf5 | ADP‐ribosylation factor 5 OS=Mus musculus GN=Arf5 PE=1 SV=2 | 20517 | 90 | 5 | 2 | 11.7 |
| PLTP_MOUSE | Pltp | Phospholipid transfer protein OS=Mus musculus GN=Pltp PE=1 SV=1 | 54419 | 89 | 2 | 2 | 6.3 |
| TSP2_MOUSE | Thbs2 | Thrombospondin‐2 OS=Mus musculus GN=Thbs2 PE=1 SV=2 | 129798 | 89 | 2 | 2 | 3.2 |
| NDKB_MOUSE | Nme2 | Nucleoside diphosphate kinase B OS=Mus musculus GN=Nme2 PE=1 SV=1 | 17352 | 84 | 6 | 2 | 23.7 |
| PCBP1_MOUSE | Pcbp1 | Poly(rC)‐binding protein 1 OS=Mus musculus GN=Pcbp1 PE=1 SV=1 | 37474 | 83 | 2 | 2 | 5.3 |
| NAC3_MOUSE | Slc8a3 | Sodium/calcium exchanger 3 OS=Mus musculus GN=Slc8a3 PE=1 SV=1 | 102917 | 79 | 2 | 2 | 1.9 |
| 5NTD_MOUSE | Nt5e | 5~‐nucleotidase OS=Mus musculus GN=Nt5e PE=1 SV=2 | 63824 | 78 | 2 | 2 | 4.2 |
| S12A2_MOUSE | Slc12a2 | Solute carrier family 12 member 2 OS=Mus musculus GN=Slc12a2 PE=1 SV=2 | 130950 | 78 | 3 | 3 | 4.8 |
| ANXA7_MOUSE | Anxa7 | Annexin A7 OS=Mus musculus GN=Anxa7 PE=1 SV=2 | 49893 | 77 | 2 | 2 | 5.6 |
| OX2G_MOUSE | Cd200 | OX‐2 membrane glycoprotein OS=Mus musculus GN=Cd200 PE=1 SV=1 | 31236 | 77 | 6 | 2 | 8.6 |
| ENPP1_MOUSE | Enpp1 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 OS=Mus musculus GN=Enpp1 PE=1 SV=4 | 103109 | 76 | 2 | 2 | 4.1 |
| FKB1A_MOUSE | Fkbp1a | Peptidyl‐prolyl cis‐trans isomerase FKBP1A OS=Mus musculus GN=Fkbp1a PE=1 SV=2 | 11915 | 76 | 1 | 1 | 13 |
| CHP1_MOUSE | Chp1 | Calcineurin B homologous protein 1 OS=Mus musculus GN=Chp1 PE=1 SV=2 | 22418 | 75 | 2 | 2 | 11.8 |
| CTL2_MOUSE | Slc44a2 | Choline transporter‐like protein 2 OS=Mus musculus GN=Slc44a2 PE=1 SV=2 | 80057 | 71 | 2 | 2 | 3.4 |
| CHM4B_MOUSE | Chmp4b | Charged multivesicular body protein 4b OS=Mus musculus GN=Chmp4b PE=1 SV=2 | 24921 | 70 | 2 | 2 | 11.2 |
| FA5_MOUSE | F5 | Coagulation factor V OS=Mus musculus GN=F5 PE=1 SV=1 | 247076 | 70 | 2 | 2 | 1 |
| K1C15_MOUSE | Krt15 | Keratin, type I cytoskeletal 15 OS=Mus musculus GN=Krt15 PE=1 SV=2 | 49107 | 70 | 3 | 2 | 3.5 |
| PLST_MOUSE | Pls3 | Plastin‐3 OS=Mus musculus GN=Pls3 PE=1 SV=3 | 70697 | 70 | 2 | 2 | 3.7 |
| MRC2_MOUSE | Mrc2 | C‐type mannose receptor 2 OS=Mus musculus GN=Mrc2 PE=1 SV=3 | 166968 | 69 | 2 | 2 | 1.4 |
| PRDX5_MOUSE | Prdx5 | Peroxiredoxin‐5, mitochondrial OS=Mus musculus GN=Prdx5 PE=1 SV=2 | 21884 | 69 | 2 | 1 | 8.1 |
| SYTC_MOUSE | Tars | Threonine‐‐tRNA ligase, cytoplasmic OS=Mus musculus GN=Tars PE=1 SV=2 | 83303 | 69 | 2 | 2 | 2.2 |
| KCY_MOUSE | Cmpk1 | UMP‐CMP kinase OS=Mus musculus GN=Cmpk1 PE=1 SV=1 | 22151 | 68 | 2 | 2 | 11.2 |
| PSA5_MOUSE | Psma5 | Proteasome subunit alpha type‐5 OS=Mus musculus GN=Psma5 PE=1 SV=1 | 26394 | 68 | 1 | 1 | 5 |
| PKHO2_MOUSE | Plekho2 | Pleckstrin homology domain‐containing family O member 2 OS=Mus musculus GN=Plekho2 PE=1 SV=1 | 53839 | 67 | 1 | 1 | 2.4 |
| CERU_MOUSE | Cp | Ceruloplasmin OS=Mus musculus GN=Cp PE=1 SV=2 | 121074 | 62 | 2 | 2 | 3.1 |
| DCTN1_MOUSE | Dctn1 | Dynactin subunit 1 OS=Mus musculus GN=Dctn1 PE=1 SV=3 | 141588 | 62 | 1 | 1 | 0.9 |
| DC1I2_MOUSE | Dync1i2 | Cytoplasmic dynein 1 intermediate chain 2 OS=Mus musculus GN=Dync1i2 PE=1 SV=1 | 68352 | 62 | 1 | 1 | 1.6 |
| HTRA1_MOUSE | Htra1 | Serine protease HTRA1 OS=Mus musculus GN=Htra1 PE=1 SV=2 | 51182 | 62 | 4 | 2 | 8.5 |
| SERC5_MOUSE | Serinc5 | Serine incorporator 5 OS=Mus musculus GN=Serinc5 PE=2 SV=1 | 51795 | 61 | 1 | 1 | 2 |
| PRS8_MOUSE | Psmc5 | 26S protease regulatory subunit 8 OS=Mus musculus GN=Psmc5 PE=1 SV=1 | 45597 | 60 | 1 | 1 | 3.2 |
| IFM5_MOUSE | Ifitm5 | Interferon‐induced transmembrane protein 5 OS=Mus musculus GN=Ifitm5 PE=1 SV=1 | 14657 | 59 | 2 | 1 | 6.7 |
| TCPQ_MOUSE | Cct8 | T‐complex protein 1 subunit theta OS=Mus musculus GN=Cct8 PE=1 SV=3 | 59518 | 56 | 1 | 1 | 1.8 |
| STEA3_MOUSE | Steap3 | Metalloreductase STEAP3 OS=Mus musculus GN=Steap3 PE=1 SV=1 | 54714 | 56 | 3 | 1 | 2.7 |
| APOB_MOUSE | Apob | Apolipoprotein B‐100 OS=Mus musculus GN=Apob PE=1 SV=1 | 509113 | 55 | 1 | 1 | 0.3 |
| CAD13_MOUSE | Cdh13 | Cadherin‐13 OS=Mus musculus GN=Cdh13 PE=1 SV=2 | 78137 | 54 | 1 | 1 | 1.7 |
| MDHC_MOUSE | Mdh1 | Malate dehydrogenase, cytoplasmic OS=Mus musculus GN=Mdh1 PE=1 SV=3 | 36488 | 53 | 1 | 1 | 3.9 |
| CR1L_MOUSE | Cr1l | Complement component receptor 1‐like protein OS=Mus musculus GN=Cr1l PE=1 SV=1 | 53728 | 52 | 1 | 1 | 2.3 |
| TSN4_MOUSE | Tspan4 | Tetraspanin‐4 OS=Mus musculus GN=Tspan4 PE=1 SV=1 | 26036 | 52 | 2 | 1 | 10.5 |
| ARPC2_MOUSE | Arpc2 | Actin‐related protein 2/3 complex subunit 2 OS=Mus musculus GN=Arpc2 PE=1 SV=3 | 34336 | 50 | 1 | 1 | 3.3 |
| MUG1_MOUSE | Mug1 | Murinoglobulin‐1 OS=Mus musculus GN=Mug1 PE=1 SV=3 | 165193 | 50 | 2 | 1 | 0.5 |
| DDAH1_MOUSE | Ddah1 | N(G),N(G)‐dimethylarginine dimethylaminohydrolase 1 OS=Mus musculus GN=Ddah1 PE=1 SV=3 | 31361 | 49 | 1 | 1 | 3.5 |
| H2B1B_MOUSE | Hist1h2bb | Histone H2B type 1‐B OS=Mus musculus GN=Hist1h2bb PE=1 SV=3 | 13944 | 49 | 1 | 1 | 11.9 |
| NDRG1_MOUSE | Ndrg1 | Protein NDRG1 OS=Mus musculus GN=Ndrg1 PE=1 SV=1 | 42981 | 49 | 1 | 1 | 3.6 |
| CPNE1_MOUSE | Cpne1 | Copine‐1 OS=Mus musculus GN=Cpne1 PE=1 SV=1 | 58849 | 48 | 1 | 1 | 1.7 |
| PACN2_MOUSE | Pacsin2 | Protein kinase C and casein kinase substrate in neurons protein 2 OS=Mus musculus GN=Pacsin2 PE=1 SV=1 | 55798 | 48 | 1 | 1 | 1.9 |
| PSA2_MOUSE | Psma2 | Proteasome subunit alpha type‐2 OS=Mus musculus GN=Psma2 PE=1 SV=3 | 25910 | 46 | 1 | 1 | 9 |
| RAB21_MOUSE | Rab21 | Ras‐related protein Rab‐21 OS=Mus musculus GN=Rab21 PE=1 SV=4 | 24091 | 46 | 1 | 1 | 4.5 |
| CLCA_MOUSE | Clta | Clathrin light chain A OS=Mus musculus GN=Clta PE=1 SV=2 | 25588 | 45 | 1 | 1 | 3.8 |
| PCOC1_MOUSE | Pcolce | Procollagen C‐endopeptidase enhancer 1 OS=Mus musculus GN=Pcolce PE=1 SV=2 | 50136 | 45 | 1 | 1 | 2.1 |
| S29A1_MOUSE | Slc29a1 | Equilibrative nucleoside transporter 1 OS=Mus musculus GN=Slc29a1 PE=1 SV=3 | 50159 | 45 | 2 | 1 | 2.6 |
| CYTC_MOUSE | Cst3 | Cystatin‐C OS=Mus musculus GN=Cst3 PE=1 SV=2 | 15521 | 44 | 2 | 1 | 7.9 |
| SYDC_MOUSE | Dars | Aspartate‐‐tRNA ligase, cytoplasmic OS=Mus musculus GN=Dars PE=1 SV=2 | 57111 | 44 | 1 | 1 | 2 |
| MAOX_MOUSE | Me1 | NADP‐dependent malic enzyme OS=Mus musculus GN=Me1 PE=1 SV=2 | 63913 | 44 | 1 | 1 | 3.1 |
| RASA3_MOUSE | Rasa3 | Ras GTPase‐activating protein 3 OS=Mus musculus GN=Rasa3 PE=1 SV=2 | 95926 | 44 | 1 | 1 | 0.8 |
| CD9_MOUSE | Cd9 | CD9 antigen OS=Mus musculus GN=Cd9 PE=1 SV=2 | 25241 | 43 | 6 | 1 | 3.1 |
| GBG5_MOUSE | Gng5 | Guanine nucleotide‐binding protein G(I)/G(S)/G(O) subunit gamma‐5 OS=Mus musculus GN=Gng5 PE=1 SV=2 | 7314 | 43 | 1 | 1 | 13.2 |
| PSD12_MOUSE | Psmd12 | 26S proteasome non‐ATPase regulatory subunit 12 OS=Mus musculus GN=Psmd12 PE=1 SV=4 | 52861 | 43 | 1 | 1 | 2.2 |
| PRS7_MOUSE | Psmc2 | 26S protease regulatory subunit 7 OS=Mus musculus GN=Psmc2 PE=1 SV=5 | 48617 | 42 | 1 | 1 | 3 |
| RL15_MOUSE | Rpl15 | 60S ribosomal protein L15 OS=Mus musculus GN=Rpl15 PE=2 SV=4 | 24131 | 42 | 2 | 1 | 5.9 |
| TENN_MOUSE | Tnn | Tenascin‐N OS=Mus musculus GN=Tnn PE=1 SV=2 | 172983 | 42 | 1 | 1 | 0.6 |
| PP2AA_MOUSE | Ppp2ca | Serine/threonine‐protein phosphatase 2A catalytic subunit alpha isoform OS=Mus musculus GN=Ppp2ca PE=1 SV=1 | 35585 | 41 | 1 | 1 | 2.6 |
| PSB6_MOUSE | Psmb6 | Proteasome subunit beta type‐6 OS=Mus musculus GN=Psmb6 PE=1 SV=3 | 25362 | 41 | 1 | 1 | 4.2 |
| ROR1_MOUSE | Ror1 | Inactive tyrosine‐protein kinase transmembrane receptor ROR1 OS=Mus musculus GN=Ror1 PE=2 SV=2 | 104021 | 41 | 1 | 1 | 1.1 |
| NHRF1_MOUSE | Slc9a3r1 | Na(+)/H(+) exchange regulatory cofactor NHE‐RF1 OS=Mus musculus GN=Slc9a3r1 PE=1 SV=3 | 38577 | 41 | 1 | 1 | 2.8 |
| ANX11_MOUSE | Anxa11 | Annexin A11 OS=Mus musculus GN=Anxa11 PE=1 SV=2 | 54045 | 40 | 1 | 1 | 2.4 |
| CAPZB_MOUSE | Capzb | F‐actin‐capping protein subunit beta OS=Mus musculus GN=Capzb PE=1 SV=3 | 31326 | 40 | 1 | 1 | 2.9 |
| CHM1A_MOUSE | Chmp1a | Charged multivesicular body protein 1a OS=Mus musculus GN=Chmp1a PE=1 SV=1 | 21594 | 40 | 1 | 1 | 4.1 |
| LIN7A_MOUSE | Lin7a | Protein lin‐7 homolog A OS=Mus musculus GN=Lin7a PE=1 SV=2 | 25977 | 40 | 1 | 1 | 4.3 |
| VNN1_MOUSE | Vnn1 | Pantetheinase OS=Mus musculus GN=Vnn1 PE=1 SV=3 | 57054 | 40 | 2 | 1 | 2.5 |
| CAN2_MOUSE | Capn2 | Calpain‐2 catalytic subunit OS=Mus musculus GN=Capn2 PE=1 SV=4 | 79822 | 39 | 1 | 1 | 1.1 |
| EFR3A_MOUSE | Efr3a | Protein EFR3 homolog A OS=Mus musculus GN=Efr3a PE=1 SV=1 | 92554 | 39 | 1 | 1 | 1.3 |
| FAS_MOUSE | Fasn | Fatty acid synthase OS=Mus musculus GN=Fasn PE=1 SV=2 | 272257 | 39 | 1 | 1 | 0.4 |
| S39AA_MOUSE | Slc39a10 | Zinc transporter ZIP10 OS=Mus musculus GN=Slc39a10 PE=1 SV=1 | 94335 | 39 | 1 | 1 | 1.2 |
| VPS35_MOUSE | Vps35 | Vacuolar protein sorting‐associated protein 35 OS=Mus musculus GN=Vps35 PE=1 SV=1 | 91655 | 39 | 2 | 1 | 1.4 |
| APOA1_MOUSE | Apoa1 | Apolipoprotein A‐I OS=Mus musculus GN=Apoa1 PE=1 SV=2 | 30597 | 38 | 2 | 1 | 3.8 |
| F234A_MOUSE | Fam234a | Protein FAM234A OS=Mus musculus GN=Fam234a PE=1 SV=1 | 60538 | 38 | 1 | 1 | 1.8 |
| SEPR_MOUSE | Fap | Prolyl endopeptidase FAP OS=Mus musculus GN=Fap PE=1 SV=1 | 87889 | 38 | 1 | 1 | 1.3 |
| PTPRA_MOUSE | Ptpra | Receptor‐type tyrosine‐protein phosphatase alpha OS=Mus musculus GN=Ptpra PE=1 SV=3 | 93638 | 38 | 1 | 1 | 2.2 |
| TCTP_MOUSE | Tpt1 | Translationally‐controlled tumor protein OS=Mus musculus GN=Tpt1 PE=1 SV=1 | 19450 | 38 | 2 | 1 | 8.1 |
| VA0D1_MOUSE | Atp6v0d1 | V‐type proton ATPase subunit d 1 OS=Mus musculus GN=Atp6v0d1 PE=1 SV=2 | 40275 | 37 | 1 | 1 | 2.3 |
| KCRB_MOUSE | Ckb | Creatine kinase B‐type OS=Mus musculus GN=Ckb PE=1 SV=1 | 42686 | 37 | 1 | 1 | 5.5 |
| EGLN_MOUSE | Eng | Endoglin OS=Mus musculus GN=Eng PE=1 SV=2 | 69976 | 37 | 1 | 1 | 1.5 |
| EPHB2_MOUSE | Ephb2 | Ephrin type‐B receptor 2 OS=Mus musculus GN=Ephb2 PE=1 SV=3 | 109828 | 37 | 1 | 1 | 1.8 |
| MATN4_MOUSE | Matn4 | Matrilin‐4 OS=Mus musculus GN=Matn4 PE=1 SV=1 | 68874 | 37 | 1 | 1 | 1.4 |
| MRP1_MOUSE | Abcc1 | Multidrug resistance‐associated protein 1 OS=Mus musculus GN=Abcc1 PE=1 SV=1 | 171075 | 36 | 1 | 1 | 0.6 |
| CPNE2_MOUSE | Cpne2 | Copine‐2 OS=Mus musculus GN=Cpne2 PE=1 SV=1 | 60997 | 36 | 1 | 1 | 1.6 |
| FBLN1_MOUSE | Fbln1 | Fibulin‐1 OS=Mus musculus GN=Fbln1 PE=1 SV=2 | 77981 | 36 | 1 | 1 | 1.7 |
| AP2M1_MOUSE | Ap2m1 | AP‐2 complex subunit mu OS=Mus musculus GN=Ap2m1 PE=1 SV=1 | 49623 | 35 | 1 | 1 | 1.8 |
| FRMD8_MOUSE | Frmd8 | FERM domain‐containing protein 8 OS=Mus musculus GN=Frmd8 PE=1 SV=2 | 51795 | 35 | 1 | 1 | 6.7 |
| MTPN_MOUSE | Mtpn | Myotrophin OS=Mus musculus GN=Mtpn PE=1 SV=2 | 12853 | 35 | 1 | 1 | 14.4 |
| MYO9B_MOUSE | Myo9b | Unconventional myosin‐IXb OS=Mus musculus GN=Myo9b PE=1 SV=2 | 238685 | 35 | 1 | 1 | 0.4 |
| NPTN_MOUSE | Nptn | Neuroplastin OS=Mus musculus GN=Nptn PE=1 SV=3 | 44345 | 35 | 2 | 1 | 2.5 |
| PSMD2_MOUSE | Psmd2 | 26S proteasome non‐ATPase regulatory subunit 2 OS=Mus musculus GN=Psmd2 PE=1 SV=1 | 100139 | 35 | 1 | 1 | 0.9 |
| RAC1_MOUSE | Rac1 | Ras‐related C3 botulinum toxin substrate 1 OS=Mus musculus GN=Rac1 PE=1 SV=1 | 21436 | 35 | 2 | 1 | 7.3 |
| REXO1_MOUSE | Rexo1 | RNA exonuclease 1 homolog OS=Mus musculus GN=Rexo1 PE=1 SV=1 | 130709 | 35 | 1 | 1 | 0.7 |
| GPC5C_MOUSE | Gprc5c | G‐protein coupled receptor family C group 5 member C OS=Mus musculus GN=Gprc5c PE=1 SV=2 | 48390 | 34 | 1 | 1 | 3 |
| RL4_MOUSE | Rpl4 | 60S ribosomal protein L4 OS=Mus musculus GN=Rpl4 PE=1 SV=3 | 47124 | 34 | 1 | 1 | 1.9 |
| SCRN1_MOUSE | Scrn1 | Secernin‐1 OS=Mus musculus GN=Scrn1 PE=1 SV=1 | 46297 | 34 | 1 | 1 | 2.7 |
| TAGL2_MOUSE | Tagln2 | Transgelin‐2 OS=Mus musculus GN=Tagln2 PE=1 SV=4 | 22381 | 34 | 1 | 1 | 5.5 |
| AT1B3_MOUSE | Atp1b3 | Sodium/potassium‐transporting ATPase subunit beta‐3 OS=Mus musculus GN=Atp1b3 PE=1 SV=1 | 31755 | 33 | 1 | 1 | 5 |
| CAND1_MOUSE | Cand1 | Cullin‐associated NEDD8‐dissociated protein 1 OS=Mus musculus GN=Cand1 PE=1 SV=2 | 136245 | 33 | 1 | 1 | 0.7 |
| CO5A1_MOUSE | Col5a1 | Collagen alpha‐1(V) chain OS=Mus musculus GN=Col5a1 PE=1 SV=2 | 183564 | 33 | 1 | 1 | 0.5 |
| PSB5_MOUSE | Psmb5 | Proteasome subunit beta type‐5 OS=Mus musculus GN=Psmb5 PE=1 SV=3 | 28514 | 33 | 1 | 1 | 3.4 |
| RTN4_MOUSE | Rtn4 | Reticulon‐4 OS=Mus musculus GN=Rtn4 PE=1 SV=2 | 126535 | 33 | 1 | 1 | 1.1 |
| TFAM_MOUSE | Tfam | Transcription factor A, mitochondrial OS=Mus musculus GN=Tfam PE=1 SV=2 | 27970 | 33 | 1 | 1 | 2.9 |
| TCPE_MOUSE | Cct5 | T‐complex protein 1 subunit epsilon OS=Mus musculus GN=Cct5 PE=1 SV=1 | 59586 | 32 | 1 | 1 | 3.1 |
| DCTN2_MOUSE | Dctn2 | Dynactin subunit 2 OS=Mus musculus GN=Dctn2 PE=1 SV=3 | 44090 | 32 | 1 | 1 | 4.7 |
| GLOD4_MOUSE | Glod4 | Glyoxalase domain‐containing protein 4 OS=Mus musculus GN=Glod4 PE=1 SV=1 | 33296 | 32 | 1 | 1 | 3.4 |
| S39AE_MOUSE | Slc39a14 | Zinc transporter ZIP14 OS=Mus musculus GN=Slc39a14 PE=1 SV=1 | 53927 | 32 | 1 | 1 | 1.8 |
| SODC_MOUSE | Sod1 | Superoxide dismutase [Cu‐Zn] OS=Mus musculus GN=Sod1 PE=1 SV=2 | 15933 | 32 | 1 | 1 | 7.8 |
| UBP5_MOUSE | Usp5 | Ubiquitin carboxyl‐terminal hydrolase 5 OS=Mus musculus GN=Usp5 PE=1 SV=1 | 95772 | 31 | 1 | 1 | 1 |
Abbreviations used for protein and peptide identification summary tables
prot_acc: Accesion number according to protein family or pyrosequencing conread
GN: Gene name
prot_desc: Description
prot_mass: Molecular weight of translated sequence
Osteosome proteins that mediate osteosome uptake into prostate cancer cells
Uptake of exosomes has been shown to be the mechanism by which exosomes modulate their target cells. It has been reported that vesicle targeting depends on the type and activation status of recipient cells27, 28. To assess if prostate cancer cells take up released osteosomes, we investigated the uptake of osteosomes by different prostate cancer cell lines. Osteosomes or control liposomes were labeled with PKH26 dye. PKH26-labeled osteosomes or control liposomes were then co-cultured with C4-2b prostate cancer cells for various times and monitored by live-cell imaging to detect the time course of osteosome transfer into C4-2b cells. We observed an increase in osteosome uptake in C4-2b cells, with close to 60% of cells showing osteosome uptake by 10 h and 100% by 30 h (Fig. 5A). In contrast, during the same time frame, little uptake of control liposomes was detected in C4-2b cells at 10 h, and only ~20% C4-2b cells took up liposomes by 30 h. In PC3-mm2 cells, more than 60% of cells showed osteosome uptake by 10 h and 100% by 30 h (Fig. 5B). In contrast, uptake of control liposomes in PC3-mm2 cells was very low at 10 h, reaching ~ 50% by 30 h. The results using two different prostate cancer cell lines show that prostate cancer cells take up osteosomes more readily than control liposomes. These findings raise the possibility that osteosomes may contain cell surface molecules that facilitate their uptake into PCa cells.
Figure 5.
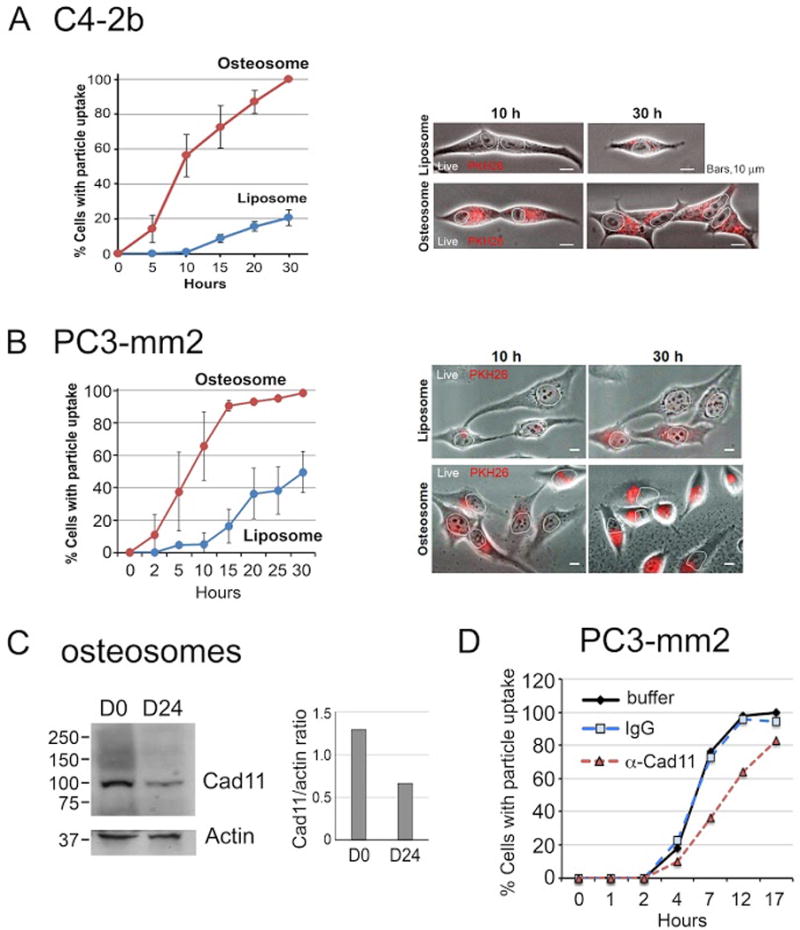
Osteosome uptake into C4-2b and PC3-mm2 cells. Live-cell imaging of osteosome uptake in (A) C4-2b cells and (B) PC3-mm2 cells. Cells (1×104) were incubated with PKH26-labeled D24 osteosomes or PKH26-labeled control liposomes (3×105 particles). Live-cell imaging was recorded at 30 min intervals over 30 h on a Nikon Biostation. Number of cells imaged live: C4-2b with osteosome (n=157) or liposome (n=48); PC3-mm2 with osteosome (n=118) or liposome (n=100) in two independent experiments. Error bars, mean ± s.d. Right panels, representative bright field images merged with PKH26 red fluorescence of cells treated with PKH26-labeled liposomes or PKH26-labeled osteosomes. Nuclei are outlined; dash line separates two cells. Bars, 10 μm. (C) Western blot of adhesion molecule cadherin-11 (Cad11) in D0 and D24 osteosomes. Right panel, quantification of Cad11 level. (D) Live-cell imaging of PC3-mm2 was performed as in B, except that PKH26-labeled osteosomes were preincubated with either anti-Cad11 mAb 1A5, isotype-matched irrelevant mAb (IgG), or PBS buffer, prior to their addition to cells. The final antibody concentration was 3 μg/ml. Number of cells imaged live following osteosome pre-incubation with: PBS (n=55), IgG (n = 52), and Cad11 mAb (n=81).
Cad11 contributes to the uptake of osteosomes into PC3-mm2 cells
We next examined whether osteosomes may contain specific membrane proteins that facilitate interaction with PC3-mm2 cells through cell surface adhesion molecules and/or receptors to favor their capture by PC3-mm2 cells. Our previous studies have shown that the osteoblast cadherin, cadherin11 (Cad11, also known as OB-cadherin) plays a role in the homing of PC3-mm2 cells, which express Cad11, to bone through interacting with Cad11 expressed on osteoblasts25, 29. We found that Cad 11 is a common osteosomal protein in both D0 and D24 osteosomes (Table 1). Cad11 is a homophilic cell adhesion molecule. Thus, Cad11 on osteosomes may enhance the uptake of osteosomes into PC3-mm2 cells through interaction with Cad11 on PC3-mm2 cells. The emPAI values of Cad11 in D0 vs D24 osteosomes were 0.06 and 0.11, respectively, and the mascot score were 30 and 54, respectively (Table 1). Western blot for the levels of Cad11 in osteosomes showed that the level of Cad11 were similar, although D24 seemed to be slightly lower when compared to D0 osteosomes (Fig. 5C).
To examine the role of Cad11 in osteosome uptake into PC3-mm2 cells, we used a Cad11 adhesion-blocking antibody mAb1A525 in live-cell imaging analysis. PKH26-labeled osteosomes were preincubated for 30 min with Cad11 mAb1A5, a control antibody with matching isotype (IgG) or buffer alone, and the antibody-osteosome mixture was added to PC3-mm2 cells at a final concentration of 3μg/ml mAb. As shown in Fig. 5D, the control cells treated with either buffer alone or an irrelevant mAb showed a similar time course of osteosome uptake, with about 50% of PC3-mm2 cells positive with osteosomes at 5 h. Treatment of osteosomes with Cad11 mAb 1A5 delayed osteosome uptake into PC3-mm2 cells, with 50% cell uptake reached at 9 h. These results suggest that Cad11 contributes to the uptake of osteosomes into PC3-mm2 cells, likely mediated through homophilic Cad11 adhesion interactions. We note that C4-2b cells do not express detectable levels of Cad1129, yet osteosomes can still be efficiently taken up relative to liposomes (Fig. 5A), suggesting that interactions of other membrane components between osteosomes and C4-2b cells likely mediate osteosome uptake into C4-2b cells. Together, these observations suggest that osteosome uptake is dependent on the expression of cell surface adhesion molecules, and that diverse membrane components of different PCa cells might be involved in osteosome uptake into different PCa cells.
Discussion
We have identified a unique set of proteins in exosomes derived from primary mouse osteoblasts termed “osteosomes”. In addition, we showed that there are significant differences in the levels and content of proteins in osteosomes isolated from undifferentiated versus differentiated osteoblasts, with 167 proteins uniquely present in osteosomes from differentiated but not undifferentiated osteoblasts. Our studies expand the list of exosome proteins differentially expressed in mineralized osteoblasts and suggest that osteosomes may mediate different functions depending on their cellular state. Furthermore, we showed that the adhesion molecules, such as cadherin-11, on the osteosome surface play a role in osteosome uptake into PCa cells. As PC3-mm2 is a highly metastatic PCa cell line, it is possible that uptake of osteosomes through cadherin-11 contributes to the metastastic potential of PC3-mm2 cells. This is the first report on the isolation and proteomics profiling of exosomes from primary mouse osteoblasts. Our study offers an additional mechanism, besides cell-cell contact and paracrine factors, by which osteoblasts may be used to communicate with cells in the bone marrow microenvironment in both physiological and pathological conditions.
The low number of osteosomes from primary mouse osteoblasts has limited our ability to examine the functional roles of osteosomes on PCa cells. Despite these challenges, our studies raise the possibility that osteosomes play a role in modulating the activities of tumor cells that have metastasized to bone. Exosomes derived from tumor-associated stroma have been shown to increase tumor cell migration through Wnt-PCP signaling18, and stromal exosomes have been shown to confer therapy resistance to breast cancer cells19. PCa is a unique malignancy with a special affinity for the bone and a remarkable capacity to develop osteoblastic metastasis4. We recently demonstrated that PCa-induced aberrant bone overgrowth promotes tumor growth in bone7. While factors secreted from osteoblasts, such as osteonectin, osteopontin, osteocalcin and bone sialoprotein, have been shown to affect different PCa cell functions30–33, a role of osteosomes in PCa progression in bone has never been studied. Morhayim et al.21 showed that upon incubating the extracellular vesicles, isolated from differentiated osteoblastic cell line SV-immortalized human osteoblasts, with PC3 PCa cells, a 2-fold increase in cell growth compare to medium control was observed. Whether such an effect also occurs in vivo awaits further studies.
Exosomes contain specific repertoires of proteins as well as RNAs, indicating the existence of mechanisms that control the sorting of molecules into exosomes. During osteoblast differentiation, there is a significant change in the expression of proteins, as reflected in the dramatic increases in osteoblast differentiation markers alkaline phosphatase, osteocalcin, DMP1 and sclerostin (Fig. 1E). Among the osteoblast differentiation markers examined, only alkaline phosphatase was found in D24 osteoblasts. In addition, the differentiation status of osteoblasts also affects the composition of exosomes. We found that several proteins are uniquely present in D24 but not D0 osteosomes. How proteins and RNAs are selected and sorted into exosomes is not clear34. The differential expression of proteins, and likely RNAs, between D0 and D24 osteosomes will likely affect the outcome of the communication between the osteoblasts (exosome-producer) and the recipient cells. This issue is under intense investigation. Isolation of osteosomes and the identification of components in osteosomes opened new possibilities that osteoblasts may use osteosomes to modulate cells in the bone marrow. Previous studies by Calvi et al.1 and Zhang et al.2 showed that osteoblasts regulate hematopoietic stem cell activity through cell-cell contact, leading to Notch activation and paracrine BMP signaling, respectively. It is intriguing to consider that osteosomes may be an additional mechanism by which osteoblasts regulate hematopoietic stem cells. Because osteosomes can bring genetic modifiers, in addition to proteins, to hematopoietic stem cells, osteosomes may represent a novel mechanism to regulate hematopoiesis. Further studies on the osteosome RNA contents and their effects on resident bone marrow cells and metastatic cancer cells are warranted.
Conclusions
Our studies suggest that osteosomes may play a role in the interaction between osteoblasts and cells in the bone marrow microenvironment in both physiological and pathological conditions.
Supplementary Material
2) Supplemental Table S1. Proteins common to osteosomes and other exosomes.
1) Supplemental Figure S1. Real-time RT-PCR for the expression of osteoblast differentiation markers, in a second set of D0 and D24 osteoblasts.
Acknowledgments
This work was supported by grants from the NIH including CA174798, 5P50 CA140388 and P30CA16672, the Prostate Cancer Foundation, Cancer Prevention and Research Institute of Texas (CPRIT RP110327, CPRIT RP150179, CPRIT RP150282), funds from the University Cancer Foundation via the Sister Institute Network Fund at the MD Anderson Cancer Center.
Footnotes
SUPPORTING INFORMATION:
The following files are available free of charge at ACS website http://pubs.acs.org:
3) Annotated tandem mass spectra for proteins identified on the basis of a single peptide assignment that are unique in D0 and in D24 osteosomes.
References
- 1.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–6. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, Harris S, Wiedemann LM, Mishina Y, Li L. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425(6960):836–41. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 3.Coskun S, Chao H, Vasavada H, Heydari K, Gonzales N, Zhou X, de Crombrugghe B, Hirschi KK. Development of the fetal bone marrow niche and regulation of HSC quiescence and homing ability by emerging osteolineage cells. Cell Rep. 2014;9(2):581–90. doi: 10.1016/j.celrep.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logothetis C, Lin S-H. Osteoblasts in prostate cancer metastasis to bone. Nature Reviews Cancer. 2005;5:21–28. doi: 10.1038/nrc1528. [DOI] [PubMed] [Google Scholar]
- 5.Dai J, Keller J, Zhang J, Lu Y, Yao Z, Keller ET. Bone morphogenetic protein-6 promotes osteoblastic prostate cancer bone metastases through a dual mechanism. Cancer Res. 2005;65:8274–85. doi: 10.1158/0008-5472.CAN-05-1891. [DOI] [PubMed] [Google Scholar]
- 6.Dai J, Kitagawa Y, Zhang J, Yao Z, Mizokami A, Cheng S, Nor J, McCauley LK, Taichman RS, Keller ET. Vascular endothelial growth factor contributes to the prostate cancer-induced osteoblast differentiation mediated by bone morphogenetic protein. Cancer Res. 2004;64:994–999. doi: 10.1158/0008-5472.can-03-1382. [DOI] [PubMed] [Google Scholar]
- 7.Lee YC, Cheng CJ, Bilen MA, Lu JF, Satcher RL, Yu-Lee LY, Gallick GE, Maity SN, Lin SH. BMP4 promotes prostate tumor growth in bone through osteogenesis. Cancer Res. 2011;71(15):5194–203. doi: 10.1158/0008-5472.CAN-10-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Sikes RA, Malaeb BS, Yeung F, Law A, Graham SE, Pei M, Kao C, Nelson J, Koeneman KS, Chung LW. Osteoblasts can stimulate prostate cancer growth and transcriptionally down-regulate PSA expression in cell line models. Urol Oncol. 2011;29(6):802–8. doi: 10.1016/j.urolonc.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Lee YC, Lin SC, Yu G, Cheng CJ, Liu B, Liu HC, Hawke DH, Parikh NU, Varkaris A, Corn P, Logothetis C, Satcher RL, Yu-Lee LY, Gallick GE, Lin SH. Identification of Bone-Derived Factors Conferring De Novo Therapeutic Resistance in Metastatic Prostate Cancer. Cancer Res. 2015;75(22):4949–59. doi: 10.1158/0008-5472.CAN-15-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 11.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18(5):199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 13.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–35. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 15.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16(4):415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Nieuwland R, Sturk A. Why do cells release vesicles? Thromb Res. 2010;125(Suppl 1):S49–51. doi: 10.1016/j.thromres.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 17.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–56. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, Azzam DJ, Twyman-Saint Victor C, Wiemann BZ, Ishwaran H, Ter Brugge PJ, Jonkers J, Slingerland J, Minn AJ. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge M, Ke R, Cai T, Yang J, Mu X. Identification and proteomic analysis of osteoblast-derived exosomes. Biochem Biophys Res Commun. 2015;467(1):27–32. doi: 10.1016/j.bbrc.2015.09.135. [DOI] [PubMed] [Google Scholar]
- 21.Morhayim J, van de Peppel J, Demmers JA, Kocer G, Nigg AL, van Driel M, Chiba H, van Leeuwen JP. Proteomic signatures of extracellular vesicles secreted by nonmineralizing and mineralizing human osteoblasts and stimulation of tumor cell growth. FASEB J. 2015;29(1):274–85. doi: 10.1096/fj.14-261404. [DOI] [PubMed] [Google Scholar]
- 22.Bhargava U, Bar-Lev M, Bellows CG, Aubin JE. Ultrastructural analysis of bone nodules formed in vitro by isolated fetal rat calvaria cells. Bone. 1988;9:155–163. doi: 10.1016/8756-3282(88)90005-1. [DOI] [PubMed] [Google Scholar]
- 23.Medina A, Ghahary A. Transdifferentiated circulating monocytes release exosomes containing 14-3-3 proteins with matrix metalloproteinase-1 stimulating effect for dermal fibroblasts. Wound Repair Regen. 2010;18(2):245–53. doi: 10.1111/j.1524-475X.2010.00580.x. [DOI] [PubMed] [Google Scholar]
- 24.Deeraksa A, Pan J, Sha Y, Liu XD, Eissa NT, Lin SH, Yu-Lee LY. Plk1 is upregulated in androgen-insensitive prostate cancer cells and its inhibition leads to necroptosis. Oncogene. 2013;32(24):2973–83. doi: 10.1038/onc.2012.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YC, Bilen MA, Yu G, Lin SC, Huang CF, Ortiz A, Cho H, Song JH, Satcher RL, Kuang J, Gallick GE, Yu-Lee LY, Huang W, Lin SH. Inhibition of cell adhesion by a cadherin-11 antibody thwarts bone metastasis. Mol Cancer Res. 2013;11(11):1401–11. doi: 10.1158/1541-7786.MCR-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Kramer-Albers EM, Lim SK, Llorente A, Lotvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-’t Hoen EN, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sanchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BW, Vazquez J, Vidal M, Wauben MH, Yanez-Mo M, Zoeller M, Mathivanan S. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segura E, Guerin C, Hogg N, Amigorena S, Thery C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179(3):1489–96. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 28.Nolte-’t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113(9):1977–81. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 29.Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ, Chen DT, Yu-Lee LY, Zhang S, Yeh ET, Hu MC, Logothetis CJ, Lin SH. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6(8):1259–67. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen N, Ye XC, Chu K, Navone NM, Sage EH, Yu-Lee LY, Logothetis CJ, Lin SH. A secreted isoform of ErbB3 promotes osteonectin expression in bone and enhances the invasiveness of prostate cancer cells. Cancer Res. 2007;67:6544–8. doi: 10.1158/0008-5472.CAN-07-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon JA, Sodek J, Hunter GK, Goldberg HA. Bone sialoprotein stimulates focal adhesion-related signaling pathways: role in migration and survival of breast and prostate cancer cells. J Cell Biochem. 2009;107(6):1118–28. doi: 10.1002/jcb.22211. [DOI] [PubMed] [Google Scholar]
- 32.Jacob K, Webber M, Benayahu D, Kleinman HK. Osteonectin promotes prostate cancer cell migration and invasion: a possible mechanism for metastasis to bone. Cancer Res. 1999;59:4453–4457. [PubMed] [Google Scholar]
- 33.Khodavirdi AC, Song Z, Yang S, Zhong C, Wang S, Wu H, Pritchard C, Nelson PS, Roy-Burman P. Increased expression of osteopontin contributes to the progression of prostate cancer. Cancer Res. 2006;66:883–8. doi: 10.1158/0008-5472.CAN-05-2816. [DOI] [PubMed] [Google Scholar]
- 34.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2) Supplemental Table S1. Proteins common to osteosomes and other exosomes.
1) Supplemental Figure S1. Real-time RT-PCR for the expression of osteoblast differentiation markers, in a second set of D0 and D24 osteoblasts.