Abstract
Replicative erosion of telomeres is naturally compensated by telomerase and studies in yeast and vertebrates show that homologous recombination can compensate for the absence of telomerase. We show that RAD51 protein, which catalyzes the key strand-invasion step of homologous recombination, is localized at Arabidopsis telomeres in absence of telomerase. Blocking the strand-transfer activity of the RAD51 in telomerase mutant plants results in a strikingly earlier onset of developmental defects, accompanied by increased numbers of end-to-end chromosome fusions. Imposing replication stress through knockout of RNaseH2 increases numbers of chromosome fusions and reduces the survival of these plants deficient for telomerase and homologous recombination. This finding suggests that RAD51-dependent homologous recombination acts as an essential backup to the telomerase for compensation of replicative telomere loss to ensure genome stability. Furthermore, we show that this positive role of RAD51 in telomere stability is dependent on the RTEL1 helicase. We propose that a RAD51 dependent break-induced replication process is activated in cells lacking telomerase activity, with RTEL1 responsible for D-loop dissolution after telomere replication.
INTRODUCTION
Telomeres are present at the end of linear chromosomes to protect them from the action of nucleases and to prevent their recognition as Double Strand Breaks (DSB). Telomere DNA is constituted of long tandem arrays of a short sequence: TTAGGG in mammals and TTTAGGG in Arabidopsis. In mammals the G-strand terminates in a single stranded 3′ overhang which can invade the double stranded telomeric DNA to form the so-called T-loop (1–3). Telomeres compensate the loss of sequences associated to incomplete DNA replication through the action of a reverse transcriptase called telomerase (4,5). Cells lacking telomerase activity show progressive telomere shortening which ultimately will induce a DNA Damage Response (DDR) leading to a permanent cell cycle arrest and senescence.
Lying at the ends of linear chromosomes, telomeres are uni-directionally replicated by forks initiated at the most distal replication origins. This means that collapse of the replication fork at the telomere cannot be compensated by a second fork coming in from the opposite direction, which makes telomeres particularly sensitive to replication fork stalling and collapse (6).Thus stalling of replication forks at telomeres will lead to rapid telomere loss and this must be compensated by telomerase or Homologous Recombination (HR) (7–9). Telomeres thus are genomic regions sensitive to replication stress and have been identified as fragile sites (10). This sensitivity to replication stress is worsened by the G-rich content of the telomeric DNA, which can give rise to the formation of highly stable G-quadruples (G4) DNA structures. The presence of G-quadruplex at human telomeres has been visualised by using a G-quadruplex specific antibody (11,12) and G4 formation during replication of telomeric repeats can cause replication fork arrest and breakage within the telomeres (13–15). Several helicases known to unwind G-quadruplexes in vitro (PIF1, Petite Integration Frequency 1; WRN, Werner syndrome ATP-dependent helicase; BLM, Bloom syndrome protein; RTEL1, Regulator of Telomere Elongation helicase 1) are thought to facilitate replication in vivo (16–18) and RTEL1, the best characterized, has been shown to interact with the replisome and facilitate telomere replication through G4-DNA unwinding (14). Furthermore, telomerase activity has been shown to be involved in G-quadruplex unfolding in vivo both in ciliates and mammals (19,20).
A second natural barrier to telomere replication is the presence of RNA–DNA hybrids (R-loops) generated during TERRA (Telomeric Repeat-containing RNA) transcription (21). TERRA is a non-coding RNA transcribed by RNA polymerase II from subtelomeric sites (22,23). To avoid replication fork stalling, cells have developed different systems to protect the genome from the effects of RNA–DNA hybrids. Among these, RNA–DNA hybrids are removed by RNAseH1 and RNAseH2 which specifically digest the RNA of the hybrid (24). As expected, R-loops accumulate at telomeres in yeast cells lacking RNAseH activity and simultaneous depletion of telomerase activity induces increased rates of HR at telomeres (23,25–28).
Cells lacking telomerase compensate replicative telomere shortening through a mechanism known as Alternative Lengthening of Telomeres (ALT), which is thought to operate through HR (2,29). Cancer cells lacking telomerase activity lengthen their short telomeres by synthesizing new telomeric DNA using telomeric DNA as the copy template (30). Different models have been proposed to describe the mechanisms of telomere maintenance through HR. Cancer ALT cells present elevated levels of Telomere Sister Chromatid Exchanges (T-SCE), which involve the elongation of one sister telomere at the expense of shortening the other one - thus explaining the presence in these cells of very long and short telomeres. Telomere synthesis can also involve Break-Induced Replication (BIR), by which the shortened end invades another chromosome or extra-chomosomal telomeric DNA and uses it as template to synthesize up to several kilobases of new telomeric DNA (7). Both of these mechanisms are forms of recombination and genes participating in the HR process are essential for telomerase negative yeast survivors and inactivation of several HR proteins in ALT human cells leads to loss of telomeric sequences (31–33).
Arabidopsis plants lacking telomerase lose 250–500bp at telomeres per generation (G) and survive up to 11 generations with increasingly severe developmental and cytological abnormalities from the sixth generation (34,35). It has been shown that plant cells can compensate the absence of telomerase activity with the activation of ALT to maintain telomeres length. The ALT mechanism participates in early plant development and its activation is ATM (Ataxia-Telangiectasia Mutated) dependent (36,37) but little is known of the mechanisms involved. Here we asked whether a RAD51 (Radiation sensitivite 51)-dependent HR process is activated in plant cells devoid of telomerase activity.
In contrast to mammals, where RAD51 knockout is lethal, Arabidopsis plants lacking RAD51 protein are viable but completely sterile due to the failure of meiosis.To study the consequences of RAD51 inactivation over multiple plant generations we took advantage of a dominant negative RAD51-GFP (RAD51-Green fluorescent protein) fusion allele, which lacks strand-invasion catalytic activity but the presence of which supports the action of DMC1 in meiosis and RAD51-GFP plants thus lack RAD51-dependent recombination but are fully fertile (38,39). Absence of RAD51 strand-transfer activity in telomerase mutant plants results in a significantly earlier onset of developmental defects and increased end to end chromosomal fusions. These fusions result from stochastic loss of telomeric repeats, indicative of a replicative origin. Accordingly, introduction of a replicative stress through inactivation of RNAseH further reduces the number of surviving plants. This study demonstrates the essential role of the RAD51 recombinase in compensating the stochastic telomere loss during replication in the absence of telomerase activity. The positive role of RAD51 is dependent on the presence of the RTEL1 helicase. In the absence of RTEL1, RAD51 induces telomere shortening suggesting a role of RTEL1 in resolving recombination intermediates through dissolution rather than endonucleolytic processing which will lead to telomere shortening.
MATERIALS AND METHODS
Plant material and growth conditions
All Arabidopsis thaliana plants used in this study were of Col-0 ecotype. The T-DNA insertion Arabidopsis mutants and PCR-based genotyping of tert (34), trd1–2 (40) and rtel1–1 (41) have been previously described. The expression vector containing the RAD51-GFP sequence has been previously described (38). Presence of the RAD51-GFP construct was verified by PCR using 5′-TGCCGTATGCTCAACAGGAGGT and 5′-GGTCTTGTAGTTGCCGTCGT primers.
Seeds were sown in soil, stratified for two days at 4°C and grown in a greenhouse with 16/8 hour light/dark cycle, temperature 23°C and 45–60% relative humidity (standard conditions). For in vitro culture, seeds were surface-sterilised with 0.05% SDS, 70% ethanol for 5 min, rinsed twice for 5 min with 95% ethanol, air dried, sown on 0.8% agar, 1% sucrose and 1× Murashige and Skoog salts (M0255; Duchefa Biochemie) medium in Petri dishes, stratified for 2 days at 4°C and grown in plant growth cabinets (SANYO MLR-351H) under standard conditions.
Immunocytology
Five days after germination, root tips were prepared as previously described (42). Briefly, seedlings were fixed for 45 min in 4% paraformaldehyde (PFA) in 1× PME (50 mM PIPES, 5 mM MgSO4, 1 mM EGTA; pH 6.9 with KOH) and then washed three times for 5 min in 1× PME. Tips were digested for 1 h in a moist chamber at 37°C in a 1% (w/v) cellulase, 0.5% (w/v) cytohelicase, 1% (w/v) pectolyase (C1794, C8274, P5936; Sigma-Aldrich) solution prepared in 1× PME and then were washed three times for 5 min in PME. These tips were squashed gently onto slides as described previously, air dried and stored at -80°C.
Slides were incubated overnight at 4°C with anti-γ-H2AX antiserum diluted 1:600 or/and anti-RAD51 diluted 1:750; in fresh blocking buffer (3% BSA, 0.05% Tween-20 (Sigma-Aldrich) in 1× PBS). γ-H2AX antiserum was raised and purified against a phospho-specific Arabidopsis H2AX peptide as previously described (42). The RAD51 specific antibody used was kindly provided by Peter Schlögelhofer and was raised in rat (43). Slides were washed three times for 5 min in 1× PBS solution, air dried, and then incubated for 2–3 h in a moist chamber at room temperature in blocking buffer consisting of Alexa 488-conjugated goat anti-rabbit (Molecular Probes; 1:1000) or/and Alexa 594-conjugated goat anti-rat (Molecular Probes; 1:1000) secondary antibodies. Finally, slides were washed three times for 5 min in 1× PBS and mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (2 mg/ml) (Vector Laboratories).
For EdU incorporation, 5-day-old seedlings were transferred to liquid medium containing 10 μM of EdU for 30 min and rinsed twice 5 min in fresh medium before fixation and treatment as previously described (44). EdU detection was performed at the end of immunodetection using the Click-It EdU Alexa Fluor 594 imaging kit (Invitrogen).
For immunostaining coupled with FISH, inflorescences were fixed in 4% PFA in 1× PME during at least 45 min then washed three times for 5 min in 1× PME. Tissue was cut in a 1% (w/v) cellulase, 0.5% (w/v) cytohelicase, 1% (w/v) pectolyase (Sigma-Aldrich) solution prepared in 1× PME, and incubated for 3 h at 37°C. The cell suspension was transferred to poly-lysine coated slides, spread first in 1% lipsol in 1× PME, and then in 4% PFA in 1× PME, air dried and stored at -80°C. FISH was performed with telomeric probe after immunostaining, as described above.
Fluorescent in situ hybridisation
Whole inflorescences were collected and fixed in absolute ethanol/glacial acetic acid (3:1) and stored at –20°C. Influorescences were rehydrated at room temperature in distilled water twice for 5 min followed by two washes in 1× citrate buffer (10 mM Na3 citrate, 10 mM citric acid) for 5 min. Then buds were digested for 3 h to 3 h 30 min in enzyme mixture (0.3% (w/v) cellulase, 0.3% (w/v) pectolyase, (0.3%) cytohelicase (Sigma-Aldrich) prepared in 1× citrate buffer) in a moist chamber at 37°C and washed rapidly in 1× citrate buffer. For studies of anaphase bridges, mitotic nuclei of flower pistils were squashed onto slides, air dried and stored at –80°C until hybridisation with set of subtelomeric probes. For metaphase and anaphase spreading, flower buds of appropriate size were selected under a binocular microscope and each bud was then softened in 15 ml 60% acetic acid on a microscopic slide at 45°C, fixed with ice-cold ethanol/glacial acetic acid (3:1) and air dried. Slides were then rinsed for 2 min in distilled water, 10 min in 4% PFA in 1× PBS and finally rinsed for 5 min in distilled water; air-dried and mounted in Vectashield mounting medium with DAPI (2 mg/mL) (Vector Laboratories). Slides with the desired stages were selected for further hybridisation, rinsed twice 15 min in 1× PBS and submitted to FISH using subtelomeric and telomeric probes.
Slides were heated at 60°C for 30 min and treated with RNAseA (100 μg/mL) prepared in 2× (3 M NaCl, 300 mM Na3 citrate, pH 7 HCl) for 1 h at 37°C in a moist chamber, washed twice in 2× SSC for 5 min and 1 min in 1× PBS. Then slides were fixed in 4% PFA in 1× PBS for 10 min and rinsed twice in 1× PBS for 5 min. FISH after immunostaining requires a specific post fixation step in place of this step of 4% PFA in 1× PBS for at least 1 h followed by three washes in 1× PBS for 5 min, three washes in 2× SSC for 5 min and a rapid rinse in sterile water. All slides were dehydrated by successive 2 min baths of 70%, 90%, and 100% Ethanol and air dried.
FISH hybridisations were performed with slides prepared as described above according to (45) for multiple hybridisations of the same slide using telomeric or/and subtelomeric probes. Telomeric probe was labelled by PCR (95°C 15 min; 95°C 1 min, 55°C 40 s, 72°C 2 min; *5; 95°C 1 min, 60°C 40 s, 72°C 2 min; *25) with biotin-dUTP (Roche) using specific primers 5′-(TTTAGGG)6–3′. Arabidopsis BACs from subtelomeric regions (F6F3, F23A5, F17A22, F4P13, T20O10, F6N15, T19P19, F7J8 and K9I9 (46)) were labelled with either biotin- or digoxygenin- dUTP (Roche) using a Nick-Translation kit (Roche) following the manufacturer's recommendations. The detection of probes was carried out as previously described (46). Slides were then mounted in Vectashield mounting medium with DAPI (2 µg/ml) (Vector Laboratories).
Microscopy
All images were acquired with a motorised Zeiss AxioImager Z1 epifluorescence microscope (Carl Zeiss) using an AxioCam Mrm camera (Carl Zeiss), PL Apochromat 100×/1.40 oil objective and Zeiss filter sets adapted for the fluorochromes used: (25HE (DAPI), 38HE (Alexa 488), 43HE (Alexa 596)). Images were captured as Z-stacks and collapsed Z-stack projections obtained using the Extended-focus module (projection method) of the Axiovision 4.6.2 and further processed with the Zeiss ZEN lite and Adobe Photoshop CS4 software.
Telomere length analyses
Terminal restriction fragment (TRF) analysis of telomere length in Mbo1- or Hinf1-digested genomic DNA from aerial parts of two-week-old seedlings was carried out as previously described (47). DNA was extracted in all cases from pools of 200 sibling plants, except for that used in Supplemental Figure S3 which involves analyses of individual plants.
The Primer-extension telomere repeat amplification (PETRA) assay was carried out with genomic DNA according to the published protocol (48).
Mitomycin sensitivity test
40 μM MitomycinC (Sigma-Aldrich) was included in solid media on plates during in vitro culture. Sensitivity was analysed in 2-week-old seedlings by counting the number of true leaves as previously described (49). Plants with more than three true leaves were considered to be resistant.
Statistical analysis
The software GraphPad PRISM 7 was used to perform the statistical analyses and to generate the graphs. To determine whether differences between two groups were statistically significant, groups were compared using the Mann–Whitney test. For multiple comparisons, statistical analyses were conducted by ordinary one-way ANOVA followed by Tukey's multiple comparison post-hoc test with a single pooled variance. A P-value of 0.05 or less was considered to be statistically significant. *P-value < 0.05, **P-value < 0.005, ***P- value < 0.0001.
RESULTS
RAD51 is present at deprotected telomeres in early generation tert mutants
Both the absence of telomerase leading to the gradual erosion of telomeric DNA and telomere truncation resulting from incomplete replication lead to deprotection of chromosome ends, with dramatic consequences for genome stability. Arabidopsis plants lacking telomerase activity are able to reproduce for up to eleven generations, with plants showing clear developmental defects from generation six onwards (35). To determine whether HR compensates for the loss of telomeric repeats in the absence of telomerase activity, we have analyzed the implication of RAD51 in telomere metabolism and stability in these plants.
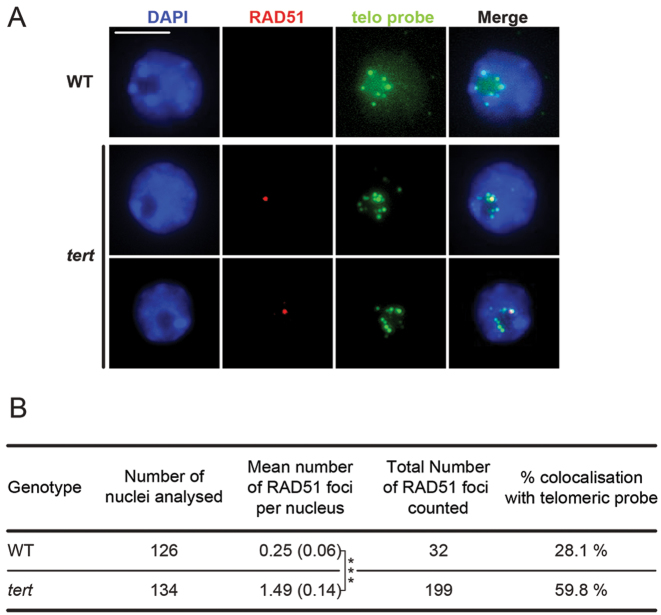
RAD51 immunostaining plus Fluorescence In Situ Hybridization (FISH) using a telomeric DNA probe, was carried out on nuclei from immature flower buds of fourth generation tert mutant plants to check for the presence of telomeric RAD51 foci. As shown in Figure 1, 6-fold more RAD51 foci were observed in tert mutant plants than in the wild-type (means of 1.49 versus 0.25 RAD51 foci/nucleus respectively). As expected, the majority of these foci (59%) in the tert plants colocalize with the telomeric sequence, while only 28% do so in WT plants—confirming that RAD51 is recruited to telomeres in cells lacking telomerase.
Figure 1.
RAD51 is present at telomeres in absence of telomerase. (A) Immunofluorescence detection of RAD51 (red) combined with telomere FISH labelling (green) in immature flower bud nuclei of WT and fourth generation tert mutant plants. DNA is counter stained with DAPI (blue). Images are collapsed Z-stack projections of a three-dimensional image stack. Scale bar: 5 μm. (B) Table showing mean number of RAD51 foci per nucleus ± standard error (SEM), and percentages of RAD51 foci colocalizing with telomeric signals. ***P < 0.0001. P-value was obtained using an unpaired Mann-Whitney test.
The presence of RAD51 at telomeres should be the result of DDR pathway activation at unprotected telomeres. DDR activation involves phosphorylation of histone H2AX at sites of DNA damage and in order to verify that RAD51 is indeed present at dysfunctional telomeres, we checked whether RAD51 foci colocalize with γ-H2AX foci. Mitotic root tip nuclei of third generation tert mutant plants show three times more RAD51 foci than wild-type plants and 85% of these do colocalize with γ-H2AX foci (Supplementary Figure S1). Early-generation telomerase-mutant plants thus present unprotected telomeres that activate the DDR response and the accumulation of RAD51 at telomeres.
Absence of homologous recombination accelerates genomic instability in telomerase-minus Arabidopsis plants
The presence of RAD51 at unprotected telomeres points to HR compensating for the loss of telomeric repeats in the absence of telomerase. If this is so, plants lacking both telomerase and RAD51 would be expected to present an accelerated loss of telomeric repeats and an earlier onset of the associated developmental defects. It has not been previously possible to test this directly due to the sterility of rad51 mutants, but this problem has been solved by our recent characterization of a dominant-negative RAD51-GFP fusion protein (Supplementary Figure S2A) (38,39). Using this material we have thus been able to follow for the first time the effects of the absence of homologous recombination across multiple sexual generations.
Sibling tert RAD51-GFP and control (wild-type, tert and RAD51-GFP) lines were identified in the F2 of a tert x RAD51-GFP rad51 cross (38) (Supplementary Figure S2B) and propagated through multiple generations.
As expected (35), plants lacking telomerase (tert mutants) were indistinguishable from wild-type plants through five generations, but abnormalities became apparent from sixth generation onward, initially in vegetative organs (reduced leaf size, asymmetric and lobed leaves, deformed shoot meristems) followed by the reproductive organs (leading to reduced fertility). These phenotypes worsened in successive generations and ultimately the plants arrested at the vegetative phase without an inflorescence bolt, or with a short inflorescence bolt bearing sterile flowers. Inactivation of homologous recombination in tert mutants (tert RAD51-GFP mutants) strikingly advanced appearance of the ‘tert phenotype’, with the number of fertile plants declining significantly from the second generation (Figure 2A). RAD51-GFP and tert control plants show a wild-type phenotype, confirming that the accelerated onset of developmental defects is due to the combined absence of telomerase and the RAD51 strand-transfer activity (Figure 2A). Absence of RAD51 strand-transfer activity thus advances the appearance of developmental defects in plants lacking telomerase, suggesting that HR contributes to preserve functional telomeres. As shown in Figure 2B and C, quantification of γ-H2AX foci in mitotic root tip nuclei supports this conclusion, with mean numbers of γ-H2AX foci per nucleus being 3-fold higher in third generation tert RAD51-GFP mutants than in tert single mutant controls.
Figure 2.
Inactivation of homologous recombination induces early onset of genomic instability in the tert mutant. (A) Developmental phenotypes of four and six week-old mutants at the sixth mutant generation (left panel; scale bar: 2 cm) and percentages of fertile plants across generations. Fertility was analysed six weeks after germination on at least 60 plants (right panel) in each case. (B) Immunofluorescence for γ-H2AX (green) in mitotic root tip nuclei of fifth generation of tert and tert RAD51-GFP plants. DNA is counter stained with DAPI (blue). Scale bar: 5 μm. (C) Mean numbers of γ-H2AX foci per nucleus of wild-type, RAD51-GFP, tert, and tert RAD51-GFP plants at different generations. Error bars indicate ± SEM from at least 100 nuclei. ns means ‘not significant’. **P < 0.005. ***P < 0.0001. P-values were obtained using an ordinary one-way ANOVA. (D) Percentages of mitotic anaphases with at least one chromosome bridge of wild-type, RAD51-GFP, tert, and tert RAD51-GFP plants at different generations. Numbers of anaphase counted (n) are given beneath each column. Errors bars indicate ± SEM from at least three different plants in each case. ns means ‘not significant’. **P < 0.005. ***P < 0.0001. P-values were obtained using an ordinary one-way ANOVA. (E) (Upper panel) Images of tert RAD51-GFP mitotic anaphases from flower pistils showing chromosome bridges analysed by FISH using a pool of nine subtelomeric BAC fluorescent probes (red) merged with DAPI stain for DNA (blue). Scale bar: 5 μm. (Lower panel) Percentage of anaphase bridges containing subtelomeric signal in tert RAD51-GFP mutant at third and fifth generations. n represents number of bridges analysed by FISH from three different plants each.
Recombination of dysfunctional telomeres can result in chromosome end-to-end fusions detectable as dicentric chromosome bridges at mitotic anaphase. Absence of RAD51 activity does not result in increased numbers of anaphase bridges compared to WT controls, as seen in the analysis of generation six RAD51-GFP plants (Figure 2D). In agreement with previous work (14,35,50), 3% of anaphases have visible chromosome bridges in tert mutant plants up to generation six, where the proportion of anaphases with bridges goes up to 14%. However and in accordance with the early appearance of developmental defects, tert RAD51-GFP mutant plants show significantly earlier appearance of mitotic anaphase bridges (16% in generation three). This value increased in sucessive generations up to almost 30% in generation six, as compared to 14% in tert mutant controls. FISH using a pool of nine chromosome-specific subtelomeric BAC (Bacterial Artificial Chromosome) probes confirmed that these anaphase bridges result from chromosome end-to-end fusions: 80.6% and 78.8% of bridges observed in third and fifth generation of tert RAD51-GFP mutants respectively present a subtelomeric signal, indicating that the chromosome fusion involved at least one chromosome end (Figure 2E).
Inactivation of homologous recombination in tert mutant plants thus results in a strikingly earlier onset of developmental and telomere cytogenetic defects. RAD51 can thus compensate for the absence of telomerase, avoiding chromosomal instability associated to the generation of unprotected telomeres.
RAD51 acts to avoid stochastic loss of telomeric repeats in the absence of telomerase
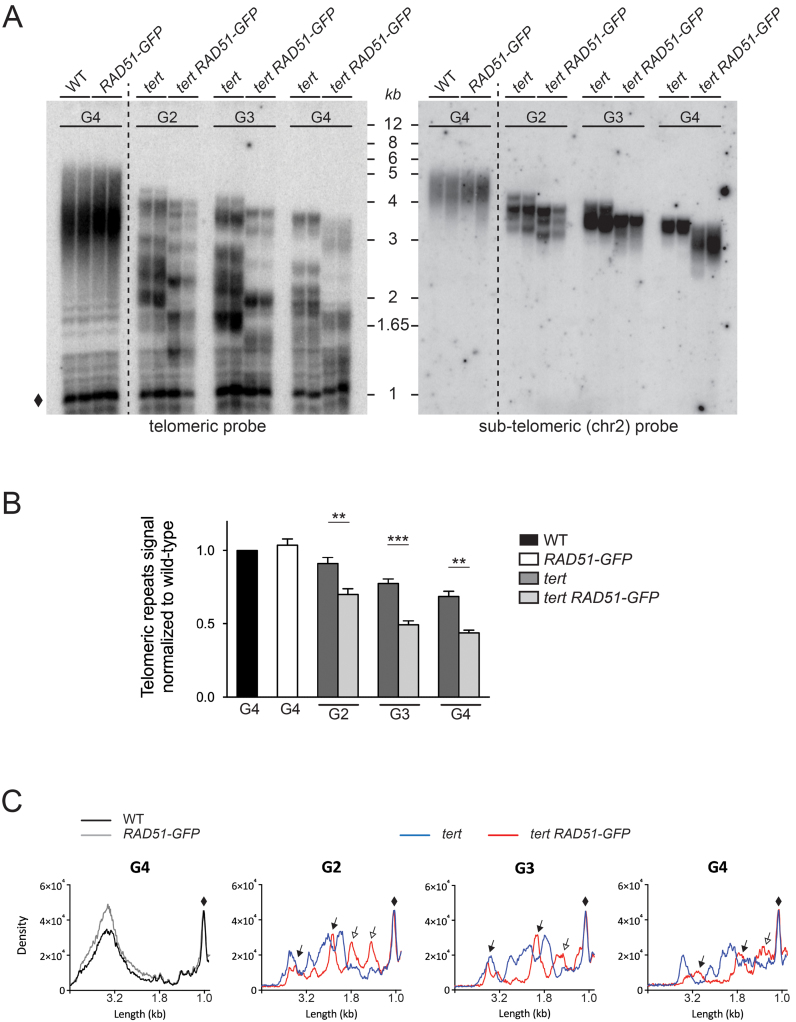
An increased rate of telomere erosion is the simplest hypothesis to explain the accelerated appearance of genomic instability in tert RAD51-GFP mutant plants. To test whether or not this is so, we carried out Telomere Restriction Fragment (TRF) analysis to determine bulk telomere length in the wild-type, RAD51-GFP, tert and tert RAD51-GFP plants (Figure 3A). Wild-type plants exhibited the expected telomere length of 2–5 kb and as expected from the absence of developmental defects and genomic instability, telomeres of RAD51-GFP mutant plants were maintained at the wild-type length at least up to generation four. As expected (35), we observed a progressive decline in telomere length over successive generations in tert mutant plants (Figure 3A). A similar heterogeneous telomere length profile is observed in tert RAD51-GFP mutant plants (Figure 3A), however the shortening of telomeres in successive generations was accelerated in plants lacking both telomerase and RAD51 activity and also confirmed in DNA from individual plants (Supplementary Figure S3A and B). Reprobing the same Southern blot with a probe specific for the end of the long arm of chromosome 2 confirmed the effect at a single chromosome arm (Figure 3A, Supplementary Figure S3A). These data thus show important telomere loss and point to sudden attrition of some telomeres in the combined absence of RAD51-dependent recombination and telomerase activity. Densitometric analysis of the TRF profiles shows a significant decrease in telomere quantity in tert RAD51-GFP mutants as compared to single tert mutant plants (Figure 3B, Supplementary Figure S3B) and the presence of shortened telomeres in tert RAD51-GFP compared to tert plants (Figure 3C). This stochastic loss of telomeric repeats is seen from the second generation of tert RAD51-GFP plants.
Figure 3.
Telomere instability in tert RAD51-GFP mutant plants. (A) Telomere restriction fragment analysis of bulk telomere length in genomic DNA from 15-day-old seedlings of indicated genotypes at different generations. Southern analysis was done with MboI-digested total DNA and using the telomeric repeat probe consisting of a hexamer of TTTAGGG repeats (left panel) and a probe to the subtelomere of the right arm of chromosome 2 (right panel). Two independent DNA preparations are presented for each genotype. The band at 1 kb marked by a diamond is derived from degenerate interstitial telomeric sequences and is used for quantification as load control. (B) Quantitative analysis of telomeric restriction fragment data carried on wild-type, RAD51-GFP, tert, and tert RAD51-GFP plants at different generations and hybridised with telomeric probe. Telomeric signal was normalised to the interstitial DNA signal (diamond) in each lane. Errors bars indicate ± SEM from four independent experiments. **P < 0.005. ***P < 0.0001 (for each value the wt was taken as 1). P-values were obtained using an ordinary one-way ANOVA. (C) Densitometric analysis of telomeric restriction fragment data (presented in A) hybridised with the telomeric probe. The diamond indicates interstitial DNA with homology to the telomeric repeat and this band was used for normalisation. Full arrows indicate telomeres found at equivalent lengths between the two genotypes, while empty arrows indicate shorter telomeres for tert RAD51-GFP.
These effects were further confirmed by realizing PETRA (Primer Extension Telomere Repeat Amplification) analysis of individual telomeres. Comparing telomere length at a given generation between tert and tert RAD51-GFP shows accelerated telomere loss in tert RAD51-GFP mutant plants for all five telomeres examined: the left arm of chromosome 1 (1L), the right and left arms of chromosome 5 (5R and 5L) and the right arms of chromosomes 2 and 4 (2R and 4R) (Supplementary Figure S4). We note that this analysis shows that the rate of telomeric loss was not uniform, with for example that of the right arm of chromosome 5 being clearly more pronounced than that of the left arm of chromosome 1. The variation in telomere length of specific telomeres in tert mutant plants has been previously observed and is most probably a stochastic effect (48).
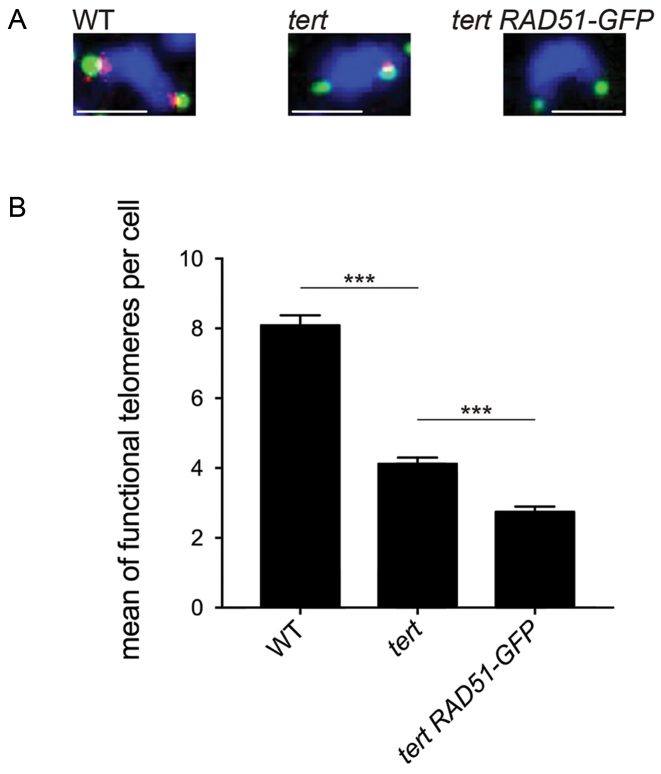
Thus sudden telomere-loss events were detectable in early generations of tert mutants lacking RAD51 activity. FISH analyses on mitotic nuclei were carried out to confirm this stochastic telomere loss at the cellular level. Wild-type Arabidopsis telomeres are short (2.5–5kb) and any further shortening in the mutants should lead to reduction or loss of telomeric DNA FISH signals. We thus used two probes: a telomeric probe (against the telomeric repeats TTTAGGG; in red) and a mixture of nine subtelomeric BAC probes (in green) (46). Coincident red and green foci thus indicate functional telomeres, while isolated green foci show the presence of a severely shortened or absent telomere. We were able to detect 8 functional telomeres per cell for wild-type plants. Inactivation of telomerase halves this number and the combined absence of telomerase and homologous recombination further decreases the fraction of functional telomeres (Figure 4). This confirms the presence in vivo of chromosome ends with very short (or absent) telomere repeat arrays in the absence of telomerase and homologous recombination and the role of RAD51 in the extension of these short telomeres.
Figure 4.
Rad51 compensates stochastic loss of telomeres in tert mutant plants. (A) DAPI-stained (blue) mitotic chromosomes analysed by FISH using telomeric repeat (red) and a pool of nine subtelomeric (green) fluorescent probes. Scale bar: 2 μm. (B) Quantification of functional telomeres per cell (mitotic metaphases or anaphases) as assessed by telomere FISH in A. All values are means ± SEM from at least 80 mitotic metaphases or anaphases of two different plants of fifth generation. ***P < 0.0001. P-values were obtained using an ordinary one-way ANOVA.
Homologous recombination compensates stochastic replication-dependent telomere loss promoted by absence of RNAseH2
R-loops are structures in which a RNA hybridizes with its dsDNA template, thereby displacing the complementary strand as a single-stranded loop. R-loops hinder DNA replication genome-wide by causing replication fork stalling and collapse and potentially leading to stochastic loss of telomeres (51). Arabidopsis telomeres are transcribed from subtelomeric regions into TERRA (52,53), as is the case in yeast and mammalian cells where accumulation of telomere associated TERRA cause telomeric shortening in a DNA replication-dependent manner (28,54). Ribonuclease H enzymes counter this effect by degrading the RNA of RNA/DNA hybrids and their absence thus should increase replication fork stalling and collapse. Inactivation of RNAseH2 (trd1 gene for triffid 1) in tert mutant plants was thus used to increase collapse of telomeric replication forks and should induce an increase in telomeres HR. Monitoring RAD51 foci by immunofluorescence in these plants was used to quantify HR at telomeres. To identify mitotic cells in S/early G2 phase, root tips were incubated with the thymidine analog EdU (5-Ethynyl-2′-deoxyuridine), followed by immunodetection of EdU and RAD51. As shown in Supplementary Figure S5, in non replicating cells (EdU–), the number of RAD51 foci is similar in cells mutated for the RNAseH (trd1) and the double tert trd1 mutant. In contrast, we observed twice as many RAD51 foci in replicating cells (EdU+) of the double tert trd1 mutant compared to single mutant controls. This result is consistent with observations in yeast and in Arabidopsis showing that lack of RNAseH2 activity increases the global recombination frequency (25,40).
We thus expected that absence of RAD51 strand-exchange activity in tert trd1 plants (lacking telomerase and RNAseH2) would increase genomic instability as result of increased telomere loss. To test this, TERT/tert and trd1/trd1 plants were crossed and F1 heterozygotes for both genes were transformed with the RAD51-GFP expression construct. Plants expressing the RAD51-GFP fusion protein were selected and PCR genotyping identified wild-type, tert and tert trd1 plants, carrying or not RAD51-GFP (Supplementary Figure S6A). As these plants are transformants, we verified the presence of the RAD51-GFP protein through the Mitomycin C hypersensitivity conferred by the absence of RAD51 strand-exchange activity (Supplementary Figure S6B).
Plants deficient for telomerase, RNAseH2 and homologous recombination show severe developmental defects in second generation, in contrast to single or double mutant controls. Triple mutant plants are smaller, 76% are completely sterile and none are able to reproduce in G3 (Figure 5A, B). To check whether the accelerated onset of developmental defects was associated with an increase in telomere erosion we performed TRF analysis in second and third generation of single, double and triple mutant sibling plants (Figure 5C). Plants deficient for RNAseH2 present shorter telomeres compared with the wild-type plants and a similar profile was observed in plants lacking both the RNAseH2 and RAD51 catalytic activity. Telomere loss is accelerated in absence of telomerase in plants mutated for the RNAseH2, suggesting a role for the telomerase in compensating the loss associated with lack of the RNAseH2 activity. A much more pronounced loss of telomeres is observed in the triple mutant as compared with the double mutant sibling plants as shown in the densitometric analysis (Figure 5D). This observation was further confirmed by rehybridizing the same blot with a subtelomeric probe specific for the long arm of Chromosome 2 (Figure 5C). The accelerated loss of telomeric repeats in the triple mutant was confirmed by PETRA analysis of the telomeres of chromosomes 1L and 5R (Supplementary Figure S7). In accord with this telomere loss, second generation triple mutant plants show a 4-fold increase in the proportion of mitotic anaphases presenting chromosome bridges, relative to the single or double mutant sibling plants (Figure 5E). That these bridges involve at least one chromosome end was confirmed by FISH analysis using 9 subtelomeric specific BAC probes. As shown in Figure 5F, 82% of anaphase bridges present a subtelomeric signal. These results are in agreement with a role of RAD51 in compensating telomere loss due to replicative stress in the absence of telomerase activity.
Figure 5.
Telomere loss induced in absence of RNAseH2 is compensated through the combined action of Telomerase and RAD51. (A) Pictures of four and six week-old second-generation plants. In contrast to tert trd1–2 mutants, the tert trd1–2 RAD51-GFP mutants present strong developmental defects with most plants being sterile. Scale bar: 2 cm. (B) Percentage of plants with a wild-type (WT) phenotype (dark) or with developmental and sterility defects (gray) for the indicated genotypes at the second generation. The phenotypes were observed six weeks after germination on at least 60 plants. (C) Telomere restriction fragment analysis of bulk telomere length in genomic DNA from 15-day-old seedlings of indicated genotypes at different generations. Southern analysis was done with MboI-digested total DNA and using the telomeric repeat probe (left panel) and subtelomeric probe to right arm of chromosome 2 (right panel). The band at 1 kb marked by a diamond is derived from interstitial degenerated telomeric sequences. (D) Quantitative analysis of telomeric restriction fragment data (presented in C) hybridised with telomeric probe. Telomeric signal was normalised to interstitial degenerated telomeric sequences (diamond in C) used for quantification as load control. (E) Percentages of mitotic anaphases with at least one chromosome bridge obtained after cytogenetic analysis of flower pistil nuclei of third-generation plants. SEM are calculated from at least three different plants in each case. Numbers of anaphase counted are at least 300 for each genotype. (F) Analysis of tert trd1–2 RAD51-GFP mitotic anaphases from flower pistil by FISH using a pool of nine subtelomeric BAC fluorescent probes. Examples of anaphase bridges counter stained with DAPI (blue) containing a subtelomeric signal (red; upper panel) and percentage of anaphase bridges containing subtelomeric signal (lower panel). n represents number of bridges analysed by FISH from five different plants.
The telomere-stabilizing role of RAD51 in the absence of telomerase depends upon RTEL1
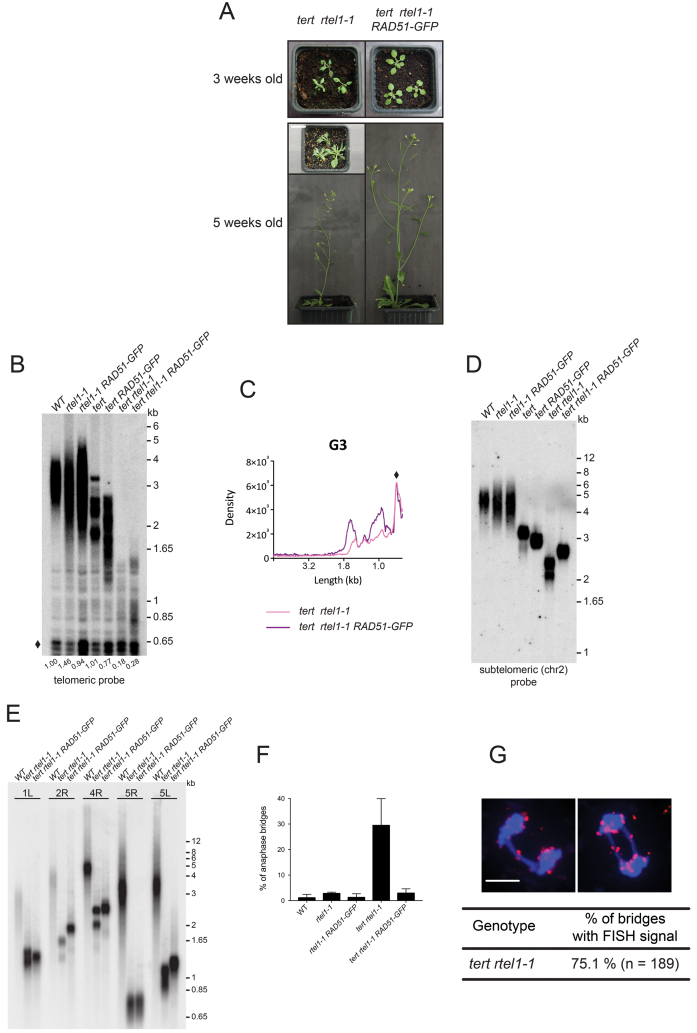
The RTEL1 helicase plays an essential role in facilitating replication and controlling homologous recombination, with depletion of RTEL1 shown to increase recombination in C. elegans, human cells and Arabidopsis (41,55). Mutation of RTEL1 induces fork stalling/collapse due to lack of resolution of secondary structures generated at the replication fork (particularly G-quadruplex) (14). Inactivation of RTEL1 in Arabidopsis plants lacking telomerase leads to rapid telomere shortening, showing a role of telomerase in compensating telomere loss generated in the absence of RTEL1 (41). Given the dual role of RTEL1 in replication and homologous recombination, we asked whether the role of RAD51 in compensating telomere loss in the absence of telomerase could be regulated by RTEL1. To test this, double heterozygote TERT/tert RTEL1/rtel1 plants were transformed with the RAD51-GFP expression construct. WT, tert, rtel1 and tert rtel1 lines were identified in the transformed progeny and RAD51-GFP homozygotes were identified in the following (T2) generation (Supplemental Figure S8A). All single and double mutant plants were hypersensitive to the crosslinking agent MMC as expected, due to the dominant-negative effect of the RAD51-GFP on HR (Supplemental Figure S8B). At the second mutant generation, single and double mutants show a wild-type phenotype, whether or not they express RAD51-GFP. However, G3 double tert rtel1 mutant plants show strong developmental defects, with 66% unable to develop past the rosette stage and the remainder reduced in size, producing small floral stems bearing sterile flowers (Figure 6A). In contrast, tert rtel1 plants of the same generation expressing the RAD51-GFP show a wild-type phenotype (Figure 6A). These tert rtel1 RAD51-GFP plants do however show severe developmental defects and reduced fertility, in the fourth generation. Thus the absence of RAD51 activity delays the appearance of developmental defects in tert rtel1 plants for two generations, with fertile plants being still present in the fifth generation.
Figure 6.
The RTEL1 helicase is essential for compensation of telomere loss by RAD51-dependent HR. (A) Pictures of three and five week-old third-generation plants. In contrast to tert rtel1–1 RAD51-GFP mutant which develop normally, the tert rtel1–1 mutants present severe developmental defects with plants being completely sterile. Scale bar: 2 cm. (B) Telomere restriction fragment analysis of bulk telomere length in genomic DNA from 15 days-old third generation seedlings of the indicated genotypes. Southern analysis was done with HinfI-digested total DNA and using the telomeric repeat probe. The band at 1 kb marked by a diamond is derived from interstitial degenerated telomeric sequences and is used for quantification as load control. Telomeric signals were normalised to the total DNA signal (diamond) in each lane. Numbers beneath telomeric blot give relative ratios of telomeric DNA compared to the wild-type. (C) Densitometric analysis of telomeric restriction fragment data (presented in C) hybridised with telomeric probe. The diamond indicates interstitial DNA with homology to the telomeric repeat and this was used for normalisation. (D) Telomere restriction fragment analysis of bulk telomere length in genomic DNA from 15 days-old seedlings of indicated genotypes at the third generation. Southern analysis was done with MboI-digested total DNA and using a subtelomeric probe to the right arm of chromosome 2. (E) PETRA of indicated genotypes (WT, tert rtel1–1, tert rtel1–1 RAD51-GFP) at the third generation. Telomere length was determined by PCR for left arm of chromosome 1 (1L), right arm of chromosome 2 (2R), right arm of chromosome 4 (4R), right arm of chromosome 5 (5R) and left arm of chromosome 5 (5R) from genomic DNA and signals were obtained by hybridisation with a telomeric probe. (F) Percentages of mitotic anaphases with at least one chromosome bridge obtained after cytogenetic analysis of flower pistil nuclei of third-generation plants. Values are calculated from at least three different plants in each case and at least 300 anaphases were counted for each genotype. (G) Analysis of tert rtel1–1 mitotic anaphases from flower pistils by FISH using a pool of nine subtelomeric BAC fluorescent probes. Examples of anaphase bridges counter stained with DAPI (blue) containing a subtelomeric signal (red; upper panel) and percentage of anaphase bridges containing subtelomeric signal (lower panel). n is the number of bridges analysed by FISH from three different plants.
To check the impact of the absence of homologous recombination on telomere length in tert rtel1 plants, we performed TRF analysis of the single and double mutants in G2 and G3 (Southern analysis of Hinf1 digested genomic DNA using the telomeric repeat probe). As previously shown, a dramatic loss of telomeric repeats was observed from G2 in tert rtel1 compared to the single mutants (Supplementary Figure S9A) (41). While the absence of RAD51 activity has no effect on telomere length in G2 plants, an increase was observed in G3 in tert rtel1 plants lacking RAD51 activity, compared to tert rtel1 (Figure 6B). That this effect is specific to the rtel tert plants is confirmed by the controls of the effect of absence of RAD51 activity. Absence of RAD51 activity in the WT has no visible effect on telomere length (RAD51-GFP. Figure 3), nor does it enhance telomere shortening in rtel1 mutants (rtel1–1 RAD51-GFP), while the expected enhancement of shortening of telomeres is seen in tert RAD51-GFP compared to tert plants. Densitometric analysis of the TRF blots clearly confirms this loss of telomeres in the presence of homologous recombination (Figure 6C). We further confirmed this observation by examining an individual telomere with a Southern of Mbo1 digested genomic DNA probed for the telomere of the long arm of chromosome 2 (Figure 6D, Supplementary Figure S9C). As expected no difference was observed between telomere lengths of second generation tert rtel1 and tert rtel1 RAD51-GFP (Supplemental Figure S9C). However G3 tert rtel1 plants show a band as short as 2kb, in contrast G3 of tert rtel1 RAD51-GFP plants show a band of 2.7 kb (Figure 6D). In Arabidopsis the minimum length of a functional telomere has been defined as 200 bp (48) and the most distal Mbo1 restriction site of the long arm of chromosome 2 is 1.9 kb from the telomeric repeats. Thus the long arm of chromosome 2 in G3 tert rtel1 plants lacks a functional telomere, while it should be functional in G3 tert rtel1 RAD51-GFP. PETRA analysis was carried out on five other chromosome ends to verify that this effect was not specific to the long arm of chromosome 2. As expected, no clear differences were observed between G2 tert rtel1 and tert rtel1 RAD51-GFP (Supplementary Figure S9D). However in G3, the ends of chromosomes 2R, 4R and 5L show longer telomeric repeats in the absence of RAD51 activity (Figure 6E), in accord with the severe developmental defects observed in tert rtel1 plants compared with the wild-type phenotype of tert rtel1 RAD51-GFP plants of the same generation.
As expected, the loss of telomeric repeats in G3 of tert rtel1 plants was accompanied by chromosomal instability. Cytogenetic analyses show 29% of mitotic anaphases with bridges in tert rtel1 plants, but only 3% in plants of the same generation lacking the RAD51 catalytic activity (Figure 6F). That these chromosome fusions involved chromosome ends was confirmed through FISH analysis using a mixture of nine subtelomeric BAC probes, showing that 75% of anaphase bridges contain a subtelomeric signal (Figure 6G).
Taken together, our results show that the role of RAD51 in telomere maintenance is dependent upon the presence of RTEL1 in the absence of telomerase, suggesting that RTEL1 is essential for the processing of recombination intermediates at telomeres in the absence of telomerase.
DISCUSSION
In this study we present an analysis of the role of RAD51-dependent homologous recombination in the maintenance of telomeres in Arabidopsis thaliana. We observe telomeric RAD51 foci in telomerase mutant plants and 85% of these colocalize with foci of phosphorylated H2AX. Thus a RAD51-dependent recombination mechanism is induced at deprotected Arabidopsis telomeres in the absence of telomerase. Through the use of a dominant-negative rad51 mutant (RAD51-GFP) which lacks strand invasion activity but remains fertile due to its ability to support DMC1 in meiosis (38,39), we were able to test whether this RAD51-dependent mechanism acts to compensate the absence of telomerase activity in telomere maintenance through the activation of homologous recombination at deprotected telomeres. Absence of RAD51-dependent HR strikingly advances the onset of developmental defects in telomerase mutant plants (by four plant generations Figure 2A). This is correlated with the early appearance of γ-H2AX foci and increased numbers of end-to-end chromosomal fusions (Figure 2C and D).
TRF analysis showed the expected loss of telomeric repeats in these plants. This is not, however, associated with bulk shortening of telomeres, suggesting that the enhanced telomere instability is due to sudden, stochastic loss of telomere repeats. FISH analyses confirmed this sudden loss of telomeres in telomerase mutant plants and that this is exacerbated in the absence of HR. RAD51-dependent HR thus, at least in part, compensates for telomerase in repair of stochastic loss of telomere repeats, which are believed to result from incomplete telomere replication. As discussed in the Introduction, telomeres are particularly subject to replicative problems due to their position at the ends of chromosomes, their transcription into TERRA RNA and their tendency to form DNA secondary structures. We thus introduced a replicative stress in these plants by increasing numbers of RNA/DNA hybrids at telomeres through mutation of RNAseH2, which normally removes them. The deletion of RNAseH2 in plants lacking telomerase and unable to perform RAD51-dependent HR advanced the onset of developmental defects, with 80% of second generation mutants being sterile (Figure 5A, B). We note that these results concord with data from yeast cells, where accumulation of RNA/DNA hybrids in the absence of telomerase and Rad52 leads to telomere loss and accelerated rates of cellular senescence (25).
Thus both telomerase and HR are implicated in repairing telomere loss caused by collapsed or stalled replication forks in Arabidopsis. Our results thus suggest a role of telomerase, not only in compensating for incomplete DNA replication, but to resolve damage from natural replicative stress at telomeres. Recent evidence from yeast supports a role of both telomerase and HR in facilitating telomeres replication. Haploid yeast cells lacking telomerase show Rad52 foci at telomeres many doublings before the appearance of senescence and inactivation of Rad52 advances the appearance of this senescence, suggesting a role of HR very early after the loss of telomerase (56). Yeast cells lacking telomerase also show DDR before critical telomere shortening and the disruption of the DNA damage response in the absence of telomerase causes severe growth defects. These effects are alleviated by increasing intracellular dNTP pools, again pointing to both telomerase and the DDR acting to deal with replicative failure occurring normally during telomeric DNA replication (57,58). Studies of individual yeast cell lineages upon telomerase loss show heterogeneity of senescing cells derived from the same cell. Some cells show transient cell cycle arrest and resumption of divisions is Rad51-dependent, suggesting that Rad51-dependent HR plays an essential role in cell proliferation early after telomerase inactivation (59). Mouse Embryonic Fibroblasts (MEFs) depleted of RAD51 also show increased telomere fragility and shorter telomeres suggesting that HR is required to restart frequent stalled replication forks at telomeres in mammalian cells lacking telomerase activity (60).
Whether HR is activated at telomeres in the presence of telomerase remains an open question. Plants cells mutated for the RNAseH2 present shorter telomeres than the wild type plants, however they remain functional at least up to the third mutant generation and no effect is observed on simultaneous depletion of RAD51 activity, suggesting that telomerase activity alone can compensate the replication stress associated to the lack of RNAseH2. In mammals, direct evidence that normal somatic cells activate HR and that this results in copying telomeric DNA templates is based on the observation in mouse cells that a DNA tag inserted in one telomere is transferred to other telomeres (61). Thus, HR is functional in normal somatic cells, but is not sufficient to fully compensate telomere shortening in the absence of telomerase. As is the case for telomerase mutant plants, telomerase mutant mice survive only a limited number of generations (35,62).
Recently, it has been shown that depletion of RAD51 in cells selected as survivors from TERC gene knockout led to an increase of fragile telomeres due to defects in DNA replication. They show that replication stress induce mitotic DNA synthesis at telomeres which is RAD51 independent (63). The authors suggest that telomeric synthesis is mediated by a RAD51-independent BIR mechanism. This is based on their observations as well as previous work in ALT cancer cells showing activation of BIR telomere synthesis independent of RAD51 (64–66). The precise mechanism of activation of RAD51-independent BIR to stabilise telomeres in cells surviving the absence of telomerase remains, however an open question. In both yeast Type II survivors and mammalian ALT cells, the presence of telomeric extrachromosomal circular DNA is believed to supply the template for rapid and heterogeneous telomere elongation and thus the presence of these circles may be the limiting factor for the appearance of RAD51-independent survivors (67–70).
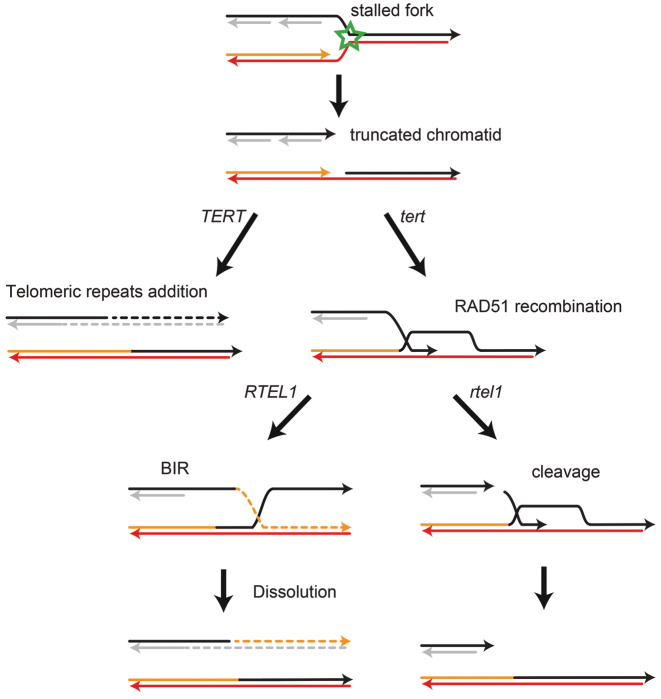
Our data show activation of RAD51 dependent HR early after telomerase deletion to facilitate telomeres replication probably through BIR. The RAD51 recombinase forms nucleofilaments on 3′ single-stranded DNA to initiate the search of a homologous recombination target sequence. The apparent absence of detectable extra-chromosomal telomeric DNA circles in Arabidopsis telomerase mutant plants suggests the use of telomeric sequences of another chromatid or chromosome as target substrate (71). We propose that the HR pathway activated relies on BIR mechanism, involved in the repair of collapsed DNA replication forks and one ended DSB (72,73). In BIR, a 3′ single-stranded DNA invades a homologous double-stranded DNA segment and the invading strand serves as a primer to initiate DNA replication (74). The resolution of the generated structure could imply the action of a helicase activity or nuclease and interestingly we found that the positive role of RAD51 in compensating the absence of telomerase is suppressed in cells lacking the helicase RTEL1. RTEL1 is able to dismantle D-loops generated during recombination (55) and we speculate that it is needed for telomere dissolution after long tract telomeric synthesis. This activity would avoid the action of endonucleases which will inhibit telomeres extension through cleavage of the D-loop (Figure 7). During the preparation of this article, an analogous model has been proposed by Sobinoff et.al. for the helicase BLM being required in a RAD51 dependent manner for ALT-mediated telomeres synthesis. Overexpression of the nuclease SLX4 result in telomeres crossing over without telomeres extension (75).
Figure 7.
Schematic model for telomeric actions of RAD51 and RTEL1. Stalling and collapse of a replication fork can lead to incomplete replication of one chromatid. Addition of telomeric repeats by telomerase can solve this problem. In the absence of telomerase, RAD51 can catalyse the invasion of the intact chromatid, thus providing a template for extension of the truncated chromatid. Recombinational repair requires the presence of the RTEL1 helicase for successful completion.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lieven de Veylder and Holger Puchta for providing trd1 and the double heterozygote TERT/tert RTEL1-1/rtel1-1 mutant seeds, respectively. We thank Nathalie Grandin for critical reading of the manuscript. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions: M.O., M.E.G. and C.I.W. conceived and designed the experiments. M.O., C.C. and S.A. performed the experiments. M.O., M.E.G. and C.I.W. analysed the data. M.O., M.E.G. and C.I.W. wrote the article.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Centre National de la Recherche Scientifique; Institut National de la Sante et de la Recherche Medicale; Université Clermont Auvergne; Ministère de l’Enseignement Supérieur et de la Recherche (to M.O.). Funding for open access charge: Lab running budget.
Conflict of interest statement. None declared.
REFERENCES
- 1. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 2. Cesare A.J., Quinney N., Willcox S., Subramanian D., Griffith J.D.. Telomere looping in P. sativum (common garden pea). Plant J. 2003; 36:271–279. [DOI] [PubMed] [Google Scholar]
- 3. Munoz-Jordan J.L., Cross G.A., de Lange T., Griffith J.D.. t-loops at trypanosome telomeres. EMBO J. 2001; 20:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 5. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilson E., Geli V.. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007; 8:825–838. [DOI] [PubMed] [Google Scholar]
- 7. Doksani Y., de Lange T.. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb. Perspect. Biol. 2014; 6:a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arlt M.F., Xu B., Durkin S.G., Casper A.M., Kastan M.B., Glover T.W.. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 2004; 24:6701–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller K.M., Rog O., Cooper J.P.. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006; 440:824–828. [DOI] [PubMed] [Google Scholar]
- 10. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tarsounas M., Tijsterman M.. Genomes and G-quadruplexes: for better or for worse. J. Mol. Biol. 2013; 425:4782–4789. [DOI] [PubMed] [Google Scholar]
- 13. Lipps H.J., Rhodes D.. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009; 19:414–422. [DOI] [PubMed] [Google Scholar]
- 14. Vannier J.B., Pavicic-Kaltenbrunner V., Petalcorin M.I., Ding H., Boulton S.J.. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012; 149:795–806. [DOI] [PubMed] [Google Scholar]
- 15. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huber M.D., Duquette M.L., Shiels J.C., Maizels N.. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006; 358:1071–1080. [DOI] [PubMed] [Google Scholar]
- 17. Opresko P.L., Cheng W.H., von Kobbe C., Harrigan J.A., Bohr V.A.. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003; 24:791–802. [DOI] [PubMed] [Google Scholar]
- 18. Ribeyre C., Lopes J., Boule J.B., Piazza A., Guedin A., Zakian V.A., Mergny J.L., Nicolas A.. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009; 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paeschke K., Juranek S., Simonsson T., Hempel A., Rhodes D., Lipps H.J.. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008; 15:598–604. [DOI] [PubMed] [Google Scholar]
- 20. Moye A.L., Porter K.C., Cohen S.B., Phan T., Zyner K.G., Sasaki N., Lovrecz G.O., Beck J.L., Bryan T.M.. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015; 6:7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfeiffer V., Crittin J., Grolimund L., Lingner J.. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013; 32:2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 23. Maicher A., Kastner L., Dees M., Luke B.. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012; 40:6649–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cerritelli S.M., Crouch R.J.. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009; 276:1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K., Luke B.. Telomeric RNA–DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013; 20:1199–1205. [DOI] [PubMed] [Google Scholar]
- 26. Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M.. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014; 5:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu T.Y., Kao Y.W., Lin J.J.. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arora R., Azzalin C.M.. Telomere elongation chooses TERRA ALTernatives. RNA Biol. 2015; 12:938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Draskovic I., Londono Vallejo A.. Telomere recombination and alternative telomere lengthening mechanisms. Front. Biosci. 2013; 18:1–20. [DOI] [PubMed] [Google Scholar]
- 30. Cesare A.J., Reddel R.R.. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010; 11:319–330. [DOI] [PubMed] [Google Scholar]
- 31. Pickett H.A., Reddel R.R.. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat. Struct. Mol. Biol. 2015; 22:875–880. [DOI] [PubMed] [Google Scholar]
- 32. Tacconi E.M., Tarsounas M.. How homologous recombination maintains telomere integrity. Chromosoma. 2015; 124:119–130. [DOI] [PubMed] [Google Scholar]
- 33. Le S., Moore J.K., Haber J.E., Greider C.W.. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999; 152:143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fitzgerald M.S., Riha K., Gao F., Ren S., McKnight T.D., Shippen D.E.. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riha K., McKnight T.D., Griffing L.R., Shippen D.E.. Living with genome instability: plant responses to telomere dysfunction. Science. 2001; 291:1797–1800. [DOI] [PubMed] [Google Scholar]
- 36. Ruckova E., Friml J., Prochazkova Schrumpfova P., Fajkus J.. Role of alternative telomere lengthening unmasked in telomerase knock-out mutant plants. Plant Mol. Biol. 2008; 66:637–646. [DOI] [PubMed] [Google Scholar]
- 37. Vespa L., Warrington R.T., Mokros P., Siroky J., Shippen D.E.. ATM regulates the length of individual telomere tracts in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:18145–18150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Da Ines O., Degroote F., Goubely C., Amiard S., Gallego M.E., White C.I.. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 2013; 9:e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kobayashi W., Sekine S., Machida S., Kurumizaka H.. Green fluorescent protein fused to the C terminus of RAD51 specifically interferes with secondary DNA binding by the RAD51-ssDNA complex. Genes Genet. Syst. 2014; 89:169–179. [DOI] [PubMed] [Google Scholar]
- 40. Kalhorzadeh P., Hu Z., Cools T., Amiard S., Willing E.M., De Winne N., Gevaert K., De Jaeger G., Schneeberger K., White C.I. et al. . Arabidopsis thaliana RNase H2 deficiency counteracts the needs for the WEE1 checkpoint kinase but triggers genome instability. Plant Cell. 2014; 26:3680–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Recker J., Knoll A., Puchta H.. The Arabidopsis thaliana homolog of the helicase RTEL1 plays multiple roles in preserving genome stability. Plant Cell. 2014; 26:4889–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Charbonnel C., Gallego M.E., White C.I.. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010; 64:280–290. [DOI] [PubMed] [Google Scholar]
- 43. Kurzbauer M.T., Uanschou C., Chen D., Schlogelhofer P.. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell. 2012; 24:2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amiard S., Charbonnel C., Allain E., Depeiges A., White C.I., Gallego M.E.. Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell. 2010; 22:3020–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schubert I., Fransz P.F., Fuchs J., de Jong J.H.. Chromosome painting in plants. Methods Cell Sci. 2001; 23:57–69. [PubMed] [Google Scholar]
- 46. Vannier J.B., Depeiges A., White C., Gallego M.E.. Two roles for Rad50 in telomere maintenance. EMBO J. 2006; 25:4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gallego M.E., White C.I.. RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heacock M., Spangler E., Riha K., Puizina J., Shippen D.E.. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004; 23:2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bleuyard J.Y., White C.I.. The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 2004; 23:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vannier J.B., Depeiges A., White C., Gallego M.E.. ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 2009; 5:e1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gaillard H., Aguilera A.. Transcription as a threat to genome integrity. Annu. Rev. Biochem. 2016; 85:291–317. [DOI] [PubMed] [Google Scholar]
- 52. Majerova E., Fojtova M., Mozgova I., Bittova M., Fajkus J.. Hypomethylating drugs efficiently decrease cytosine methylation in telomeric DNA and activate telomerase without affecting telomere lengths in tobacco cells. Plant Mol. Biol. 2011; 77:371–380. [DOI] [PubMed] [Google Scholar]
- 53. Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K.. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010; 6:e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rippe K., Luke B.. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015; 22:853–858. [DOI] [PubMed] [Google Scholar]
- 55. Barber L.J., Youds J.L., Ward J.D., McIlwraith M.J., O’Neil N.J., Petalcorin M.I., Martin J.S., Collis S.J., Cantor S.B., Auclair M. et al. . RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008; 135:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Khadaroo B., Teixeira M.T., Luciano P., Eckert-Boulet N., Germann S.M., Simon M.N., Gallina I., Abdallah P., Gilson E., Geli V. et al. . The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 2009; 11:980–987. [DOI] [PubMed] [Google Scholar]
- 57. Xie Z., Jay K.A., Smith D.L., Zhang Y., Liu Z., Zheng J., Tian R., Li H., Blackburn E.H.. Early telomerase inactivation accelerates aging independently of telomere length. Cell. 2015; 160:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jay K.A., Smith D.L., Blackburn E.H.. Early loss of telomerase action in yeast creates a dependence on the DNA damage response adaptor proteins. Mol. Cell. Biol. 2016; 36:1908–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu Z., Fallet E., Paoletti C., Fehrmann S., Charvin G., Teixeira M.T.. Two routes to senescence revealed by real-time analysis of telomerase-negative single lineages. Nat. Commun. 2015; 6:7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Badie S., Escandell J.M., Bouwman P., Carlos A.R., Thanasoula M., Gallardo M.M., Suram A., Jaco I., Benitez J., Herbig U. et al. . BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat. Struct. Mol. Biol. 2010; 17:1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Neumann A.A., Watson C.M., Noble J.R., Pickett H.A., Tam P.P., Reddel R.R.. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev. 2013; 27:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W.. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997; 91:25–34. [DOI] [PubMed] [Google Scholar]
- 63. Min J., Wright W.E., Shay J.W.. Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol. Cell. Biol. 2017; 37:e00226-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roumelioti F.M., Sotiriou S.K., Katsini V., Chiourea M., Halazonetis T.D., Gagos S.. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep. 2016; 17:1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cho N.W., Dilley R.L., Lampson M.A., Greenberg R.A.. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014; 159:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dilley R.L., Verma P., Cho N.W., Winters H.D., Wondisford A.R., Greenberg R.A.. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016; 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin C.Y., Chang H.H., Wu K.J., Tseng S.F., Lin C.C., Lin C.P., Teng S.C.. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell. 2005; 4:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Larrivee M., Wellinger R.J.. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 2006; 8:741–747. [DOI] [PubMed] [Google Scholar]
- 69. Basenko E.Y., Cesare A.J., Iyer S., Griffith J.D., McEachern M.J.. Telomeric circles are abundant in the stn1-M1 mutant that maintains its telomeres through recombination. Nucleic Acids Res. 2010; 38:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Henson J.D., Cao Y., Huschtscha L.I., Chang A.C., Au A.Y., Pickett H.A., Reddel R.R.. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009; 27:1181–1185. [DOI] [PubMed] [Google Scholar]
- 71. Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K.. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell. 2007; 27:163–169. [DOI] [PubMed] [Google Scholar]
- 72. Nugent C.I., Bosco G., Ross L.O., Evans S.K., Salinger A.P., Moore J.K., Haber J.E., Lundblad V.. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 1998; 8:657–660. [DOI] [PubMed] [Google Scholar]
- 73. McEachern M.J., Haber J.E.. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006; 75:111–135. [DOI] [PubMed] [Google Scholar]
- 74. Sakofsky C.J., Malkova A.. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017; 52:395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sobinoff A.P., Allen J.A., Neumann A.A., Yang S.F., Walsh M.E., Henson J.D., Reddel R.R., Pickett H.A.. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J. 2017; 36:2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.