Abstract
Massive high-throughput sequencing techniques allowed the identification of thousands of noncoding RNAs (ncRNAs) and a plethora of different mRNA processing events occurring in higher organisms. Long ncRNAs can act directly as long transcripts or can be processed into active small si/miRNAs. They can modulate mRNA cleavage, translational repression or the epigenetic landscape of their target genes. Recently, certain long ncRNAs have been shown to play a crucial role in the regulation of alternative splicing in response to several stimuli or during disease. In this review, we focus on recent discoveries linking gene regulation by alternative splicing and its modulation by long and small ncRNAs.
INTRODUCTION
Eukaryotic genes are often discontinuous with noncoding (intronic sequences) and coding DNA (exonic sequences). Therefore, in order to produce a mature mRNA that can be translated into a protein, introns need to be removed and exons joined by a process called splicing. In certain circumstances, splicing reactions can be modulated generating two or more mRNA isoforms from a single pre-mRNA by a process called alternative splicing (AS). In recent years, improvements in our ability to sequence entire genomes and complete pools of transcripts allowed the identification of a plethora of different mRNA isoforms in higher organisms. More than 90% of intron-containing genes in humans and over 60% in plants, are alternatively spliced in controlled conditions (1–4). The significant diversity in the number of transcripts in comparison to the number of genes emphasizes the extensive regulation occurring at transcriptional and post-transcriptional levels (5). Many genes have been found to produce tissue or condition-dependent isoforms (6). In humans, numerous studies point out a link between RNA splicing misregulation and several diseases (7–10). In plants, under stress conditions, AS plays an important role in the control of gene expression for an adequate response (11–14). Interestingly, the identification of alternatively spliced transcripts revealed a high number of genes encoding multiple proteins which may perform relevant functions in gene expression, as splicing regulators and transcription factors (15–20). In fact, some AS features seemed to be conserved across species and indeed genes homologous to common splicing regulators are also conserved (20). Growing evidence indicates that AS is an important mechanism that controls gene expression by (i) increasing gene-coding capacities, thus proteome complexity through the generation of different mRNA isoforms, and/or (ii) facilitating mRNA degradation through the introduction of a premature termination codon in specific isoforms that would lead to non-sense mediated decay (NMD).
Recognition of the splicing site can be modulated by cis-regulatory sequences, known as splicing enhancers or silencers, which contribute to the generation of two or more alternatively-spliced mRNAs from the same pre-mRNA. Computational analyses on genome-wide RNA sequencing have allowed the identification of splicing patterns in different organisms under various specific conditions, showing the dynamic nature of AS regulation in gene expression (21–23). AS defects are also related to various diseases in mammals (for review see, (24–26)), and to perturbations of plant responses to environmental cues (11,17,27,28).
Besides the identification of numerous AS events on mRNAs, next generation sequencing technologies have facilitated the identification of thousands of RNAs with no or low coding potential (the so-called noncoding RNAs, ncRNAs). Globally, ncRNAs are classified by their size. The long ncRNAs (lncRNAs, over 200 nt) act directly in a long form by lncRNA-protein interactions, whereas the small ncRNAs (smRNAs) act by smRNA-protein interactions and base paring recognition of their mRNA targets. They were found to be modulated by different stimuli, exerting regulatory roles in gene expression by their implied involvement in mRNA cleavage, translational repression or epigenetic DNA/chromatin modification of their targets, hence leading to a wide range of biological outputs required for cell viability and function (29–31). It has been known for a long time that splicing mechanisms are regulated by small ncRNAs named nuclear uridine (U)-rich RNAs (U snRNAs). This group of noncoding RNAs catalyze each step of the splicing reaction (for review, (32)) in collaboration with core small nuclear ribonucleoprotein complex subunits (snRNPs), whereas more than 200 non-snRNPs splicing factors (SFs) (33–35) fine-tune complex splicing regulations.
Recently, a growing number of ncRNAs have emerged as modulators of AS of specific genes. In this review, we discuss our current knowledge of long and small ncRNAs identified as splicing regulators and their mechanisms of action.
REGULATION OF ALTERNATIVE SPLICING BY LONG NONCODING RNAs
In the last few years, the identification of large amounts of lncRNAs induced or repressed in particular conditions or diseases suggested their possible implication in a wide range of biological processes. Detailed studies of some of them in different eukaryotic organisms have uncovered several lncRNA-mediated mechanisms fine-tuning almost every step of gene expression, including chromatin remodeling, transcriptional control, co-and posttranscriptional regulation, miRNA processing, and protein stability during different developmental processes (36–40). In particular, a growing number of lncRNAs have been linked to the modulation of AS in both plants and animals (Table 1). The main mechanisms involving lncRNAs in AS modulation can be classified in three ways: (i) lncRNAs interacting with specific SFs; (ii) lncRNAs forming RNA-RNA duplexes with pre-mRNA molecules and (iii) lncRNAs affecting chromatin remodelling, thus fine-tuning the splicing of target genes. Sub-classifications based on similarities and differences among mechanisms are proposed below.
Table 1. Developmental processes and diseases related to splicing-associated lncRNAs from the animal (background in red) and plant (in green) kingdoms.
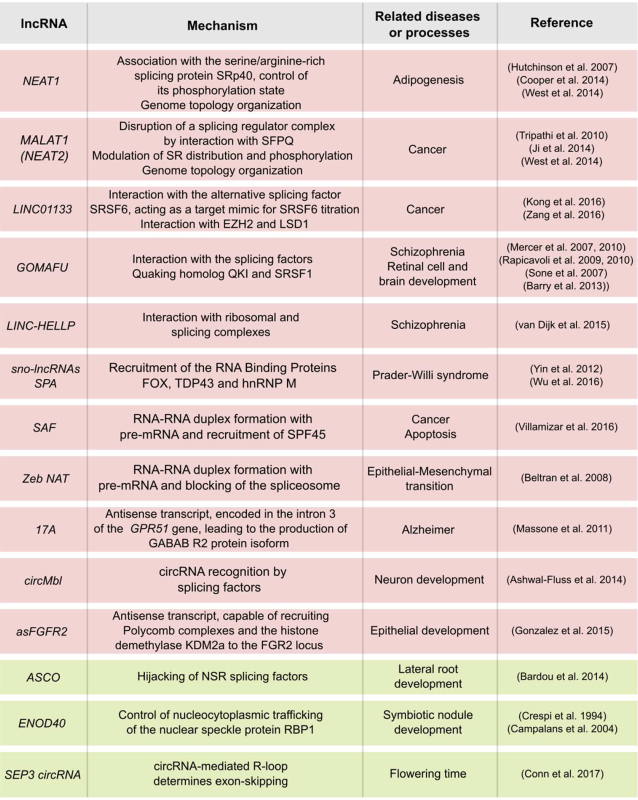
|
LONG NONCODING RNAs AS SPLICING FACTOR INTERACTORS
One of the first presumptions of the role of lncRNAs in splicing was given by a genome-wide screening in human and mouse cells that identified, in both cases, the following nuclear noncoding transcripts: NUCLEAR PARASPECKLE ASSEMBLY TRANSCRIPT 1 (NEAT1) and NUCLEAR PARASPECKLE ASSEMBLY TRANSCRIPT 2 (NEAT2) / METASTASIS ASSOCIATED LUNG ADENOCARCINOMA TRANSCRIPT 1 (MALAT1). RNA FISH analyses revealed an intimate association of NEAT1 and MALAT1 with the SC35 SF-containing nuclear speckles in both human and mouse cells, suggesting their participation in mRNA splicing. These studies also showed that NEAT1 localizes to the speckles periphery, whereas MALAT1 is part of the polyadenylated component of nuclear speckles (41). More recently, further nuclear-localized lncRNAs were linked to splicing regulation both in animals (e.g. GOMAFU, SAF) and plants (e.g. ASCO), whereas other lncRNAs have been found both in the nucleus and the cytoplasm (e.g. LINC01133 and ENOD40). Among them, a subset of lncRNAs seems recognized by SFs, impacting their activity in two ways: (i) modulating their posttranslational modifications (e.g. phosphorylation), or (ii) regulating the interaction with other SFs, and/or with protein-coding (pre)mRNAs.
NEAT1 and MALAT1/NEAT2 regulate the phosphorylation status of splicing factors
Serine/arginine-rich (SR) proteins are a conserved family of proteins largely involved in splicing. These proteins are composed of two domains, the RNA recognition motif (RRM) and the SR domain (42). SR proteins are commonly localized in the nucleus, although several of them are known to shuttle between the nucleus and the cytoplasm. Basically, a continuous phosphorylation/dephosphorylation cycle of SR proteins is required for proper pre-mRNA splicing and the regulation of AS patterns. The hyperphosphorylation of the SR domain influences the binding of SR proteins to target pre-mRNA, impacting the splice site selection. Partially dephosphorylated SR proteins support the first steps of the transesterification reactions (43–45). Also, intranuclear trafficking of SR proteins between nuclear speckles and transcription sites is dependent on their phosphorylation status (46,47). Thus, it is of paramount importance to better understand how lncRNAs may modulate SR phosphorylation, thus affecting AS determination of SR targets.
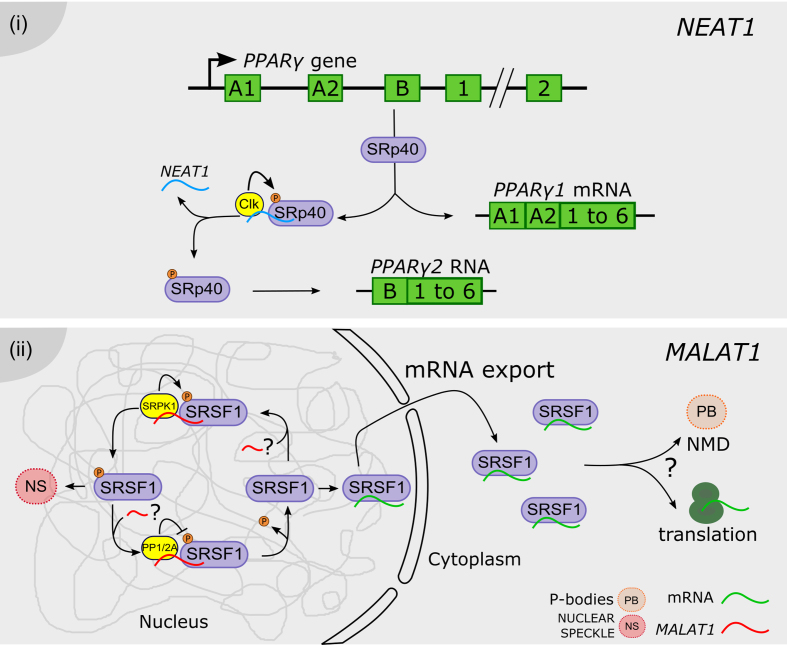
NEAT1 is a highly abundant 4 kb lncRNA found in paraspeckles, nuclear domains that control sequestration of related proteins. NEAT1 accumulation is dynamically regulated during adipocyte differentiation, modulating the AS profile of PPARγ mRNA into both isoforms, PPARγ1 and PPARγ2, which code for the major transcription factor driving adipogenesis. The SR protein SRp40 (SFRS5) is involved in the regulation of PPARγ2 splicing. During differentiation, SRp40 levels are significantly increased when the splicing of PPARγ2 is induced. PPARγ mRNA splicing regulation by SRp40 is modulated by its phosphorylation status determined by the Clk kinase (48). It was demonstrated that SRp40 directly recognizes NEAT1, exhibiting a dynamic association throughout differentiation. NEAT1 depletion causes a decrease of PPARγ1 and PPARγ2, in particular the PPARγ2 isoform. The siRNA-directed NEAT1 depletion impaired the cells ability to phosphorylate SRp40. Furthermore, siRNA treatment for SRp40 resulted in deregulation of PPARγ1 and, primarily, PPARγ2 mRNA levels, whereas the overexpression of SRp40 increased PPARγ2, but not PPARγ1. Therefore, it was proposed that an increased concentration of phosphorylated SRp40 protein is present after being released from NEAT1, promoting the splicing of PPARγ2. This NEAT1-dependent mechanism fine-tunes the relative abundance of mRNA isoforms of key genes during adipogenesis (49) (Figure 1i).
Figure 1.
Long noncoding RNAs regulate the phosphorylation status of splicing factors. (i) NEAT1 (blue) modulates SRp40 phosphorylation status by interaction with the Clk kinase. Phosphorylated SRp40 promotes the processing of the PPARy pre-mRNA into the PPARy2 mRNA, whereas the dephosphorylation of SRp40 favors the accumulation of the PPARy1 isoform. ( ii) MALAT1 (red) was proposed to modulate the phosphorylation status of SR proteins in the nucleus, including the MALAT1-interacting SRSF1, likely by interaction with PP1/2A phosphatases or with the SRPK1 kinase. Phosphorylated SRSF1 is accumulated in nuclear speckles (NS), whereas its dephosphorylation promotes the interaction with mRNAs (green), their transport and accumulation in the cytoplasm, likely affecting also protein translation and/or incorporation into P-bodies (PB) hosting the non-sense mediated decay (NMD) machinery. The three question marks indicate that each step was proposed albeit non validated.
The lncRNA MALAT1 (NEAT2) acts as an oncogene transcript and its aberrant expression is involved in the development and progression of many types of cancers (50–52). Also, its accumulation was found to be associated with cancer patient resistance to chemotherapy and radiotherapy (53,54). Studies on human cells indicate that MALAT1 regulates splicing by modulating SR splicing factor distribution and phosphorylation (55). The depletion of MALAT1 enhances the dephosphorylated pool of SR proteins, displaying a more homogeneous nuclear distribution and resulting in the mislocalization of speckle components and changes in AS of pre-mRNAs. It was proposed that the observed changes in AS of endogenous pre-mRNAs could be a consequence of the mislocalization of pre-mRNA processing factors, assuming a critical role exerted by MALAT1 in the shuttling of SR proteins between speckles and the sites of transcription. The control of the levels of phosphorylated SR proteins impacts not only AS but also likely regulates other SR-dependent post-transcriptional regulatory mechanisms, including RNA export, NMD and translation (42,56). Interestingly, it was shown that MALAT1-depleted cells exhibit an increased cytoplasmic pool of poly(A)+ RNA, likely due to the altered cellular levels of dephosphorylated splicing factor SRSF1. It was previously shown that dephosphorylation of SRSF1 is critical for the export of mRNA associated proteins (mRNPs) and is also known to enhance binding of SRSF1 to mRNAs in the cytoplasm (57,58). Furthermore, in hepatocellular carcinoma, MALAT1 oncogenic activity relies on the indirect modulation of AS by transcriptional induction of SRSF1 (51). This induction leads to the over accumulation of active SRSF1 in the cell nucleus and the modulation of SRSF1 splicing targets, including the production of anti-apoptotic AS isoforms of S6K1 (51). However, the precise mechanisms by which MALAT1 depletion alters the ratios of phosphorylated to dephosphorylated SR proteins in the cell remain largely unknown (55). Possibly, MALAT1 modulates the activity of kinases (SRPKs or Clk/STY family), or of phosphatases (PP1 or PP2A), that modify SR proteins (56). Both SRPK1 and PP1 influence AS by regulating SR protein phosphorylation (59,60). Interestingly, the localization of SRPK1 is altered in MALAT1-depleted cells, indicating the potential involvement of MALAT1 in SRPK1 localization and activity. Alternatively, SR protein stability may be modulated by direct interaction with MALAT1, considering the increased cellular levels of SR proteins and the changes observed in AS patterns exhibited by MALAT1-depleted cells (55) (Figure 1ii).
Long noncoding RNAs as splicing factor hijackers
The growth of cancer cells is promoted by the proto-oncogene PTBP2 (POLYPYRIMIDINE TRACT-BINDING PROTEIN 2; 61,62). SFPQ (proline- and glutamine-rich SF; or PSF for PTB-associated SF) is considered a tumor suppressor gene, which regulates the tumor-promoting effects of PTBP2 by direct protein–protein interaction (63–65). More recently, it was demonstrated that MALAT1 is directly recognized by SFPQ, but not by PTBP2, likely disrupting a splicing regulator complex containing this tumor suppressor (66). Considering that SFPQ contains two RNA-binding domains, it was proposed that MALAT1 or other lncRNAs may hijack SFPQ to partially inhibit the interaction between SFPQ and PTBP2, affecting the regulatory role of SFPQ on PTBP2 (67,68). The resulting release of PTBP2 from the SFPQ-PTBP2 complex would allow the promotion of tumor growth and metastasis (66) (Figure 2i).
Figure 2.
Long noncoding RNAs as splicing factor hijackers. ( i) MALAT1 can disrupt the formation of a splicing modulator complex, by directly hijacking the SFPQ factor, thus inhibiting its interaction with the tumor growth factor PTBP2. SFPQ-released PTBP2 promotes the proliferation of cancer cells. (ii) Celf3 and SF1 normally co-localize in the CS nuclear bodies. GOMAFU lncRNA is recognized by both proteins, although it is not accumulated in CS bodies. It was proposed that GOMAFU promotes the re-localization of Celf and SF1 out from CS bodies to the nucleoplasm, modulating their activity in splicing. ( iii) SPA and sno-lncRNAs recruit RNA binding proteins such as FOX, TDP43 and hnRNP M, and may titrate their availability for splicing regulation throughout the nucleus. In PWS patients SPA and sno-lncRNAs loci are deleted or not expressed, thus the related proteins are more uniformly distributed, impacting the AS pattern of their target genes. (iv) ASCO lncRNA is directly recognized by NSRa and b, competing with their binding to NSR-targeted pre-mRNAs. ASCO-mediated modulation of NSRs results in alternative processing of mature mRNAs, exhibiting events of intron retention, exon skipping and alternative 5′ or 3′ ends. (v) ENOD40 lncRNA is directly recognized by the nuclear speckle protein RBP1. ENOD40 participates in the nucleocytoplasmic trafficking of RBP1, inducing its accumulation into cytoplasmic granules, likely modulating RBP1-dependent splicing.
Recently, a SF-interacting lncRNA was also reported in cancer disease. The lncRNA LINC01133 shows a tissue- and organ-specific expression pattern and was linked to different cancers (69,70). This LINC01133 RNA is directly recognized by the AS factor SRSF6. LINC01133 expression inhibits metastasis in a SRSF6-dependent manner. It was proposed that LINC01133 acts as a target mimic to titrate SRSF6 action on other mRNA substrates, leading to the inhibition of epithelial-mesenchymal transition and metastasis in colorectal cancer cells (69). Thus, this lncRNA modulates SRSF6 activity as a decoy element of AS, in order to shape the population of AS isoforms of SRSF6 mRNA targets. Another example of SF-associated lncRNA is GOMAFU, which was involved in schizophrenia-associated AS (71). GOMAFU is expressed in a specific group of neurons in adult mice and has been implicated in retinal cell development (72,73), brain development (74) and post-mitotic neuronal function (75,76). GOMAFU’s downregulation leads to aberrant AS patterns, resembling those observed in typically schizophrenia-associated genes like DISRUPTED IN SCHIZOPHRENIA 1 (DISC1) which exerts a role in the neuronal system, as well as ERYTHROBLASTIC LEUKEMIA VIRAL ONCOGENE HOMOLOG 4 (ERBB4) implicated in mental illness (71). Moreover, GOMAFU was found to be downregulated in post-mortem cortical gray matter from the superior temporal gyrus in schizophrenia, suggesting that aberrant splicing patterns of DISC1 and ERBB4 in schizophrenia could be mediated by GOMAFU. Furthermore, GOMAFU was found to directly interact with the SFs QUAKING homolog QKI and SRSF1 (71), likely modulating AS. QK1 and SRSF1 deregulation could lead to mental disorders such as schizophrenia. GOMAFU is also recognized through a tandem array of UACUAAC motifs by the splicing factor SF1, which participates in the early stages of spliceosome assembly (77).
More recently, Celf3 (CUGBP Elav-like family member 3, also referred to as Tnrc4, Brunol1, CAGH4 or ERDA4; reviewed in (78)) was also found to specifically interact with GOMAFU. Interestingly, Celf3 forms novel nuclear bodies (named CS bodies) in the neuroblastoma cell line Neuro2A, colocalizing with the GOMAFU-interacting protein SF1. However, GOMAFU was not observed in the CS bodies but separately distributed throughout the nucleus. Therefore, it was proposed that GOMAFU indirectly modulates the function of RNA-binding proteins in CS bodies by sequestering them to separate regions of the nucleus (79) (Figure 2ii).
Apart from cancer and neurological disorders, other diseases may be related to AS modulated by lncRNAs. For example, the 205 kb-lncRNA LINC-HELLP, which is implicated in the pregnancy-associated disease HELLP, has also been implicated in splicing regulation. LINC-HELLP purification followed by mass spectrometry revealed that this lncRNA is recognized by components of the splicing and the ribosomal machineries, respectively (80), including the splicing-related factors Y-BOX BINDING PROTEIN 1 (YBX1), and POLY(RC) BINDING PROTEINS 1 and 2 (PCBP1 and PCBP2). Although the molecular mechanisms governing splicing regulation by LINC-HELLP remain largely unknown, it was demonstrated that upon the occurrence of mutations in HELLP patients, the 5′-end up to the middle of the LINC-HELLP transcript loses its ability to interact with its protein partners, whereas binding is gained with mutations at the far 3′-end (80).
Small nucleolar RNAs (snoRNAs) are a family of conserved nuclear RNAs located in Cajal bodies or nucleoli. They participate in the modification of snRNAs or rRNA, or participate in the processing of rRNA during ribosome subunit maturation (81–83). SnoRNAs are processed from excised and debranched introns by exonucleolytic trimming (84). The snoRNA HBII-52 was implicated in the congenital disease Prader-Willi syndrome (PWS), by modulating the AS of the Serotonin Receptor 2C (85). More recently, a novel class of nuclear-enriched intron-derived lncRNAs was identified, which are processed on both ends by the snoRNA machinery (sno-lncRNAs). Sno-lncRNAs derived from the PWS critical region of chromorome 15 are recognized by the FOX2 SF. It was proposed that these sno-lncRNAs can recruit FOX proteins, regulating their availability for splicing of their targets. In patients suffering PWS, particular sno-lncRNAs are deleted or not expressed. As a result, FOX proteins are more uniformly distributed in the cell nucleus, and modify the AS of specific genes during early embryonic development and adulthood (84). An additional sub-class of snoRNA-derived lncRNAs generated from the PWS locus was recently identified. SPA lncRNAs are snoRNA-5′ capped and 3′ polyadenylated accumulated in the nucleus. They were shown to recruit key RNA binding proteins, such as TDP43, RBFOX2, and hnRNP M, involved in multiple aspects of mRNA metabolism regulation (86). Therefore, SPA lncRNAs also fine-tune the availability of RNA binding proteins, as the previously characterized sno-lncRNAs (Figure 2iii).
In Arabidopsis thaliana, the lncRNA ASCO (ALTERNATIVE SPLICING COMPETITOR) is recognized in vivo by the plant-specific NUCLEAR SPECKLE RNA-BINDING PROTEINS (NSRs), involved in splicing (39). A transcriptomic dataset served to identify RNA processing events in nsra/b double mutants using a bioinformatic approach. This analysis revealed an important number of intron retention events and differential 5′ start or 3′ end in a subset of genes in the nsra/b mutant compared to wild type plants (22). NSRs are positively regulated by the phyto-hormone auxin, to which the nsra/b mutant exhibits a decreased sensitivity, e.g. lower lateral root number than wild type plants in response to auxin treatment. This phenotype was related to the one observed for ASCO overexpressing lines. Interestingly, the splicing of a high number of auxin-related genes was perturbed in nsra/b mutants and some of them behaved accordingly in the ASCO overexpressing lines (39). An in vitro approach showed that ASCO ‘competes’ with other mRNA-target for its binding to the NSR regulators, suggesting that this competition would modulate the affinity of NSRs for their targets. The ASCO-NSR interaction could then regulate AS during auxin response in roots (39) (Figure 2iv). In the model legume Medicago truncatula, NSRs closest homolog, RNA-BINDING PROTEIN 1 (RBP1), is localized in nuclear speckles where many components of the splicing machinery are hosted in plant cells. Remarkably, RBP1 interacts with a highly structured lncRNA, EARLYNODULIN40 (ENOD40), which participates in root symbiotic nodule organogenesis (87). ENOD40 is highly conserved among legumes and was also found in other species such as rice (Oryza sativa; (88)). In contrast to the nuclear localization of Arabidopsis ASCO, ENOD40 was found both in the nucleus and the cytoplasm, and it is able to relocalize RBP1 from nuclear speckles into cytoplasmic granules during nodulation. These observations hint a role of the lncRNA ENOD40 in nucleocytoplasmic trafficking, likely modulating RBP1-dependent splicing (89) (Figure 2v).
ANTISENSE lncRNAs FORMING RNA-RNA DUPLEXES
One particular class of lncRNAs is defined by natural antisense transcripts (NATs). NATs are transcribed from the opposite strand of a protein-coding gene and may or may not overlap with portions of coding sequences (90). A pioneering work in the early 1990s showed that the population of the oncogene N-myc transcriptional isoforms correlates with the presence of a NAT encoded across exon 1 in cell cultures. RNA-RNA duplexes between sense and antisense transcripts were detected in vivo. Furthermore, duplex formation appeared to occur with only a subset of the multiple isoforms of the N-myc mRNA. The precise transcriptional initiation site of the NAT was suggested to play a role in determining this selectivity. It was proposed that duplex formation could modulate pre-mRNA processing by preserving a population of intron 1-retained N-myc mRNA (91). It was recently shown that the N-myc NAT is indeed a protein-coding RNA, the resultant product of which acts as an onco-promoting factor in human cancer (92). Soon after the association of N-myc NAT with AS, it was demonstrated in rats that transcription of overlapping antisense transcripts can modulate the preference of splicing sites, thus impacting the population of alternatively spliced variants of a single locus (93). The erbAα locus encodes two overlapping mRNAs, corresponding to the two α-thyroid hormone receptor isoforms, TR α1 and TR α2, which arise from alternative polyadenylation sites of the locus. The erbAα locus also codes for a third mRNA, Rev-erbAα, which is transcribed in the opposite direction from α1 and α2. Rev-erbAα encodes another protein belonging to the same steroid/thyroid hormone receptor superfamily. Rev-erbAα overlaps with the 3′ end of α2, but not α1 mRNA. Interestingly, in vitro assays demonstrated that the relative abundance of the Rev-erbAα RNA is capable of blocking the accurate splicing of erbAα2, likely by RNA-RNA base pairing. This is consistent with the observation that in tissues where Rev-erbAα mRNA levels are high, the ratio of α2/α1 isoforms is relatively low (93,94). More recently, it was shown that TRα2 is not expressed in marsupials and that the antisense overlap between erbAα and Rev-erbα is unique to eutherian mammals (95). Although Rev-erbAα and N-myc NAT are both protein-coding genes, these paradigmatic cases of co-regulation of overlapping antisense transcripts hinted the existence of RNAs with dual roles (96), e.g. to code for proteins on one hand, and to exert a role as an RNA molecule itself, modulating the splicing of a neighboring gene, on the other.
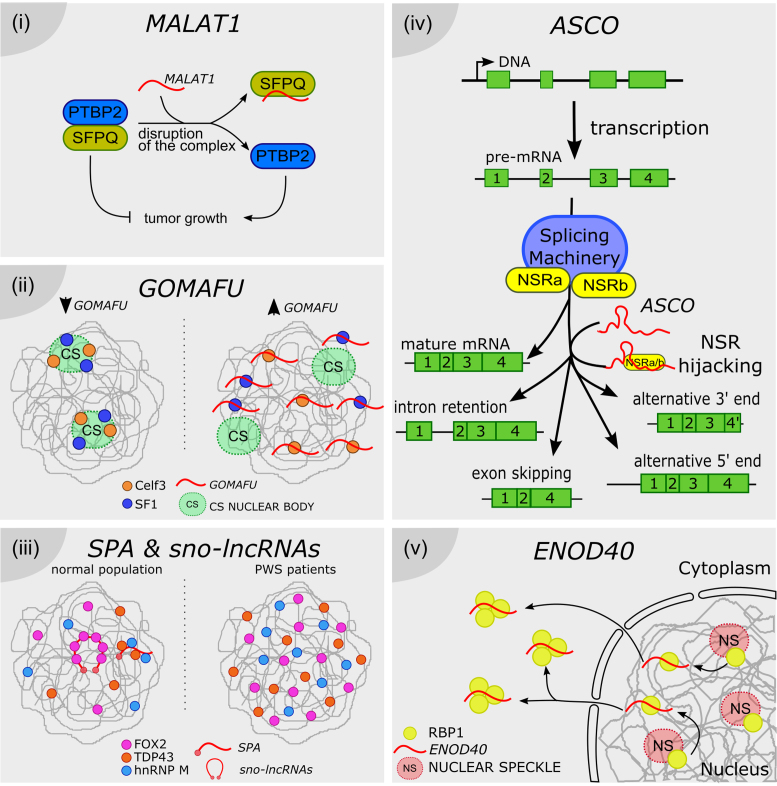
More recently, the NAT named SAF was linked to programmed cell death or apoptosis. One major apoptotic pathway is triggered by the interaction between the cellular Fas receptor (Fas) and its Fas ligand (FasL). Tumor cells are frequently resistant to Fas-mediated apoptosis, because they produce soluble Fas proteins that bind FasL, blocking apoptosis. Soluble Fas (sFas) is encoded by the exon 6-skipped alternatively spliced variant of Fas pre-mRNA (FasΔEx6), which lacks the transmembrane domain. It was shown that the nuclear-enriched NAT SAF is transcribed in reverse orientation and from the opposite strand of intron 1 of the Fas locus. An RNA pull-down assay using in vitro transcribed biotin-labeled SAF served to demonstrate that it can directly interact with the sense transcript Fas. Furthermore, an RNAse A protection assay revealed that the interaction between SAF lncRNA and Fas pre-mRNA occurs predominantly at exon 5–6 and exon 6–7 junctions. Strikingly, RNA pull-down followed by mass spectrometry revealed that SAF is recognized by the human SF 45 (SPF45), likely facilitating AS and exclusion of exon 6. The regulation of FasΔEx6 mRNA accumulation leads to the production of soluble sFas protein, which protects tumor cells against FasL-induced apoptosis (97) (Figure 3i).
Figure 3.
Long noncoding RNAs form RNA-RNA duplexes with pre-mRNAs. (i) SAF NAT is encoded in the first intron of the Fas locus. In tumor cells, SAF is transcribed and it specifically interacts with the exon 6 flanking regions of the pre-mRNA, conforming RNA-RNA duplexes. SAF recruits SPF46, promoting the exclusion of exon 6. The resulting mRNA encodes for soluble Fas (sFas), which lacks the transmembrane domain. As a result, the presence of the Fas receptor is reduced in the cell surface whereas sFas sequesters Fas ligand (FasL), rendering cells less sensitive to Fas-mediated apoptosis. (ii) In epithelial cells, the Zeb2 locus is transcribed into a full 5′UTR-including pre-mRNA. The spliceosome mediates the removal of a 3 kb-long intron in the 5′UTR. The resulting mRNA contains a stable secondary structure before the AUG, which is capable of blocking translation in the polysomes. On the other hand, after EMT, the Snail1 transcription factor induces the transcription of Zeb2 NAT in mesenchymal cells. A specific RNA-RNA duplex encompassing the 5′ splice site of the 5′UTR intron prevents the binding of the spliceosome. Thus, the mRNA contains the full isoform of the 5′UTR, including an internal ribosome entry site (IRES) proximal to the Zeb2 AUG, favoring translation. Zeb2 transcription factor extends the repression of the E-cadherin gene initiated by Snail1 during EMT.
Epithelial–mesenchymal transition (EMT) is first triggered by the expression of the Snail1 transcription factor in epithelial cells. Snail1 down-regulates specific epithelial genes, including E-cadherin, and induces the expression of Zeb1 and two transcriptional regulators, which might extend the repression of E-cadherin initiated by Snail1. Normally, the Zeb2 5′-UTR is spliced, conserving a structured region that inhibits scanning by the ribosomes and therefore prevents translation of the Zeb2 protein. However, Snail1 promotes the transcription of a NAT encoded in the opposite strand of the Zeb2 locus, covering the 5′ splice site of the Zeb2 5′-UTR. It was proposed that Zeb2 NAT prevents the recognition of the spliceosome by RNA-RNA duplex conformation, promoting the inclusion of the intron present in the Zeb2 5′-UTR. This intron contains an internal ribosome entry site (IRES) located close to the start of translation. IRES recognition by the ribosomes promotes Zeb2 translation, activating EMT (98) (Figure 3ii).
Although the underlying molecular mechanisms remain largely unknown, another NAT modulating AS also came to light in relation to brain illness. The Pol III-dependent lncRNA 17A is transcribed in an antisense way from the intron 3 of the GPR51 gene (G-PROTEIN-COUPLED RECEPTOR 51, coding GABA B2 receptor or GABAB R2) and is induced by inflammatory molecules. This lncRNA was found to regulate AS of GPR51 (99). Expression of 17A leads to the production of the GABAB R2 protein isoform without transduction activity together with a dramatic down-regulation of the expression of the canonical full-length GABAB R2 variant, thus impairing GABAB signaling. This changing ratio of AS was found to be linked to Alzheimer's disease. Also, 17A expression was increased in patient brains, suggesting an eventual role of this lncRNA in GPR51 splicing regulation to preserve cerebral function (99).
LONG NONCODING RNAs AS CHROMATIN REMODELERS
In the last decade, chromatin structure and histone modifications have emerged as key regulators of AS. The interaction between histone modifications, chromatin-binding proteins and SFs possibly constitutes a complex network of communication between chromatin and RNA (100). Also, it was demonstrated that the chromatin context influences RNA polymerase II (Pol II) elongation rate which in turns affects AS (101–102). This means that epigenetic regulation not only determines which parts of the genome are expressed, but also how they are spliced (100). Several lncRNAs participate in chromatin structure determination and dynamics, which may then impact the splicing output, notably by: (i) direct interaction between lncRNAs and DNA forming heteroduplexes, (ii) recruitment of chromatin modifiers to specific loci, or (iii) shaping the 3D organization of chromatin conformation across the cell nucleus. Finally, we discuss how lncRNA-derived small RNAs control chromatin remodeling and may determine AS patterns through this mechanism.
Splicing regulation by lncRNA-driven DNA–RNA duplexes
Circular RNAs (circRNAs) are covalently-closed circular molecules of single-stranded RNA, resulting from a non-canonical splicing event, the so-called back-splicing. This event consists in the ligation of a downstream splice donor site reversely with an upstream splice acceptor site from the pre-mRNA, generating a circular lncRNA molecule. These circular transcripts are abundant and highly stable, and they may efficiently compete with the linear pre-mRNA for the recognition of related splicing protein complexes (103). For instance, in flies and humans, the SF MUSCLEBLIND can strongly and specifically bind to the circRNA derived from its own locus, called circMbl (104). In human cells, circRNAs are dynamically modulated by the SF QKI during human EMT (105), and it was shown that in human endothelial cells circRNAs occurrence correlates with exon skipping throughout the genome (106). However, the molecular mechanisms involving circRNAs in animals remain largely unknown.
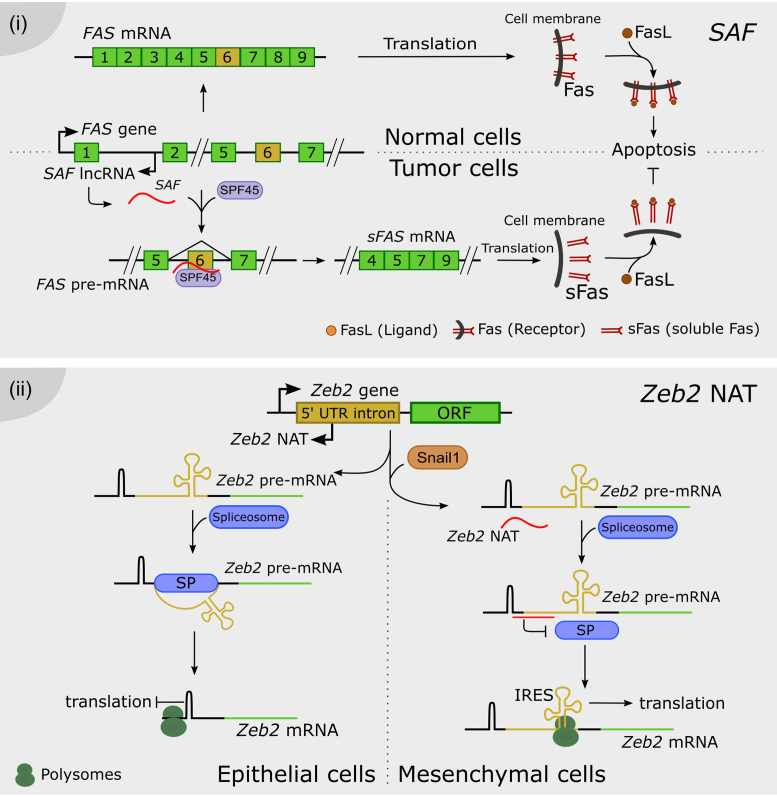
It was recently demonstrated in Arabidopsis that a circRNA can modulate the AS of its own parent gene by directly interacting with the DNA, forming an RNA-DNA hybrid known as an R-loop (107). The overexpression of the circRNA from exon 6 of the SEPALLATA 3 (SEP3) gene enhances the accumulation of the naturally-occurring SEP3.3 isoform, which consists of the exon 6-skipped transcript. SEP3 belongs to the MADS-box family (named after the founder members MCM1-AGAMOUS-DEFICIENS-SRF) of DNA-binding proteins, and it is involved in flower development in Arabidopsis. The modulation of SEP3 splicing gives rise to homeotic phenotypes in the flower. Strikingly, the exon 6 circRNA is capable of generating an R-loop by direct interaction with its own genomic locus, further supporting the idea that chromatin conformation plays a major role in splicing pattern determination (Figure 4i). Genome-wide characterization of R-loops will be needed to assess how widely this mechanism occurs throughout the Arabidopsis genome (108).
Figure 4.
Long noncoding RNAs as chromatin remodelers. (i) The exon 6 of the SEP3 gene is transcribed and back-spliced into a circular RNA. The SEP3 circRNA directly interacts with its parent gene DNA, conforming a DNA–RNA duplex known as an R-loop and promoting the exon 6 skipping, thus the accumulation of the SEP3.3 mRNA isoform. (ii) The antisense transcript of the FGFR2 gene, called asFGFR2 (in red), recruits the PRC2 proteins EZH2 and SUZ12 to its parent locus, triggering the deposition of H3K27me3 and the recruitment of the H3K36 demethylase KDM2a. This complex enhances the deposition of H3K36me3 and impairs the binding of the chromatin-splicing adaptor complex MRG15–PTB to the exon IIIb, which is finally included in the mature mRNA (in green). (iii) NEAT1 and MALAT1 bind to common and distinct actively transcribed loci across the genome. Their binding on the gene body is different between them, e.g. NEAT1 binding peaks at the transcription start site as well as the end of the locus, whereas MALAT1 preferentially binds only at the end of the gene. It was proposed that MALAT1 and NEAT1 promote the formation of splicing-related nuclear speckles and paraspeckles, respectively, around its site of transcription of targeted loci.
Long noncoding RNAs as recruiters of chromatin remodelers
An example of cell-specific AS mediated by lncRNA was linked to the antisense transcript called asFGFR2. The lncRNA asFGFR2 is generated from the human FGFR2 locus and it induces epithelial-specific AS of FGFR2 by promoting chromatin modifications in its own FGFR2 locus. It was proposed that asFGFR2 recruits chromatin modifiers specifically to this locus, perhaps via RNA-DNA heteroduplexes. Interestingly, chromatin pulldown of a biotinylated asFGFR2 RNA showed that upon its overexpression, asFGFR2 was targeted to the FGFR2 locus precisely around the differentially spliced intron (109). In epithelial cells, asFGFR2 was found to recruit chromatin modifiers like the Polycomb-group proteins and the H3K36 demethylase KDM2a to the FGFR2 locus. As a result, it generates a chromatin environment that prevents binding of inhibitory splicing regulators and favors exon IIIb inclusion (109). Polycomb-related proteins and KDM2a are differentially recruited along FGFR2 in a cell type–specific manner, correlating with FGFR2 splicing outcome. The hypothesis of a direct role of H3K27me3 and the PRC2 component EZH2 on FGFR2 AS was discarded by expressing EZH2 in cells after knockdown of KDM2a. EZH2 failed to induce exon IIIb inclusion in the absence of KDM2a, indicating that PRC2 promotes exon IIIb inclusion by maintaining low H3K36me2/3 levels via recruitment of KDM2a. This epigenetic landscape impairs binding of the chromatin-splicing adaptor complex MRG15–PTB, which normally inhibits the inclusion of exon IIIb. The antagonistic effect of H3K36me3 and H3K27me3 on FGFR2 splicing points to a lncRNA-mediated cross-talk between these histone modifications (109) (Figure 4ii).
Also, the mentioned splicing-related lncRNA MALAT1 was identified as a key regulator of Polycomb 2 protein (Pc2) methylation status, impacting chromatin conformation (110). It was reported that the methylation/demethylation of Pc2 determines the relocation of growth control genes between Polycomb bodies (PcGs) and interchromatin granules (ICGs). This behavior is ruled by the binding of methylated and unmethylated Pc2 to two lncRNAs, TUG1 and MALAT1, located in PcGs and ICGs, respectively. TUG1- and MALAT1-associated proteins were identified by pull-down using biotinylated RNAs followed by mass spectrometric analysis. This approach revealed that MALAT1 RNA bound not only to pre-mRNA SFs, but also to transcriptional co-activators and histone methyltransferases/demethylases associated with active histone marks; whereas TUG1 RNA also specifically binds to a number of proteins involved in transcriptional repression, including histone methyltransferases/demethylases and chromatin modifiers. It transpired that these lncRNAs mediate the assembly of multiple co-repressors/co-activators, and can alter the histone marks read by Pc2 in vitro. Additionally, binding of MALAT1 to unmethylated Pc2 promotes SUMOylation of the transcription factor E2F1, leading to activation of the growth control gene program. Therefore, MALAT1 also participates in the modulation of the chromatin remodeling environment by selectively interacting with chromatin modifier proteins (110). Although there is no evidence of any direct link between MALAT1- or TUG1-mediated chromatin modulation and splicing, future studies will be needed to determine if the chromatin-related function of these lncRNAs may consequently affect AS of target genes.
Long noncoding RNAs shape the three-dimensional genome organization
The molecular pathway linking the actions of subnuclear structure-specific lncRNAs, such as TUG1 and MALAT1, and non-histone protein methylation to spatial relocation of transcription units in the nucleus, hints the role of lncRNAs in the dynamic 3D configuration of the genome in the cell nucleus. Nuclear spatial organization and chromatin 3D modulation by lncRNAs (37,111–116) as well as AS modulation by chromatin modifications (for review see (100,117)) have long been described in mammalian cells as well as in plants.
The splicing-related lncRNAs NEAT1 and MALAT1 were used as baits to map their binding sites across the human genome (118) by Capture Hybridization Analysis of RNA Targets (CHART; (119)). Strikingly, NEAT1 and MALAT1 localize to hundreds of loci in human cells, primarily on actively transcribed genes. Many of these loci were co-enriched in NEAT1 and MALAT1 CHARTs, although displaying distinct gene body binding patterns, suggesting independent but complementary functions for both lncRNAs. CHART followed by mass spectrometry was also performed to identify NEAT1 and MALAT1 interactors, revealing common nuclear speckle and paraspeckle components. The elucidation of ribonucleoprotein complexes further supports complementary binding and functions exerted by both lncRNAs. The dynamic interactions between nuclear speckles and gene bodies indicate that speckles may serve as a concentrated reservoir of SFs that shuttle to transcribed genes (120,121). Considering that speckles frequently localize within the vicinity of actively transcribed genes undergoing co-transcriptional splicing (122,123), it was proposed that nuclear bodies may be organized around genes regulated by NEAT1 and MALAT1 (118). The previous elucidation of the 3D chromatin organization of human cells indicated that the MALAT1 and NEAT1 genomic loci are located in close proximity in the nucleus (124). According to this model, NEAT1 and MALAT1 could shape the structure of nuclear bodies at highly transcribed loci, as NEAT1 also participates in the organization of paraspeckle formation around its site of transcription (111,112) (Figure 4.iii). Alternatively, NEAT1 and MALAT1 may serve as scaffolds, such as Xist or HOTAIR (119,125,126), bringing proteins that also interact with components of nuclear speckles and paraspeckles, together with RNA and/or DNA binding proteins. This model considers the action of lncRNAs as molecular bridges between specific chromosomal locations and nuclear speckles and paraspeckles (118).
Small RNAs in the interplay between splicing and chromatin compaction
Small ncRNAs (smRNAs) derived from lncRNA precursors act as small molecules of <50 nt. Since the discovery of RNA-mediated gene silencing by small interfering RNAs (siRNAs) (127,128), other classes of small RNAs with multiple functions have been identified, such as microRNAs (miRNAs) (129), small RNA fragments derived from tRNAs (tsRNAs) and small RNA fragments derived from small nucleolar (sno)RNAs (sdRNAs, sno-derived RNAs) (130,131). It has been demonstrated that they regulate gene expression by multiple mechanisms, such as targeting mRNA cleavage, translational or transcriptional repression, decoys of mRNAs or through the generation of other secondary smRNAs (132–134) and protein sequestering or titration (135). During transcriptional gene silencing, siRNAs trigger heterochromatin formation at DNA target sequences. Various plants and yeast studies have reported the relationship between splicing and silencing mediated by smRNA-directed heterochromatin formation. In particular, several mutants in SFs-encoding genes turned out to be also impaired in the silencing of certain genes (136–140). In Schizosaccharomyces pombe, mutation in any of the two SF-encoding genes Cwf10 or Prp39 was found to reduce centromeric siRNAs accumulation and to increase repeated transcripts like dg and dh (136). In the same way, mutation of a single nucleotide in the U4 snRNA gene impairs centromere silencing (137). In both cases, some SFs were found to facilitate siRNA production to modulate heterochromatin formation and induce centromere silencing. Similarly, in Arabidopsis the SF SR45 was found to be involved in de novo methylation by the RNA-directed DNA methylation (RdDM) pathway. In fact, the sr45 mutant decreased siRNAs and DNA methylation in transgenic FWA (FLOWERING WAGENINGEN) in company with the associated late flowering phenotype (138). Another example is the Arabidopsis SMALL NUCLEAR RIBONUCLEOPROTEIN D1 (SmD1) which was proposed to play a role in plants during splicing since a mutant exhibits altered AS of certain genes. This protein was also found to facilitate post-transcriptional gene silencing (PTGS) by protecting transgene aberrant RNAs from degradation by the NMD pathway. As a result, enough template is provided for siRNAs production establishing a link between aberrant RNA and AS (141). Apart from these examples of interaction between the splicing machinery and smRNA-directed heterochromatin formation, new studies also pointed out a link involving smRNAs in the regulation of AS through chromatin remodelling, a process that can be regulated by smRNAs. It was shown in humans, flies and worms that nucleosome density is higher over exons than introns suggesting that nucleosome positioning defines exons at the chromatin level (142–146). The chromatin context influences RNA polymerase II (Pol II) elongation rate which in turns affects AS (100–102). Rapid transcription favors exon skipping whereas slower transcription stimulates the use of weak splice sites of variant exons promoting the intron inclusion process or other alternative sites for splicing reactions (146,147). For instance, the FIBRONECTIN 1 gene (FN1) produces different protein isoforms through AS of exon extra domain I (EDI). In hepatoma and HeLa cells, it was described that exogenous applied siRNAs targeting gene sequences located close to EDI alternative exon lead to a heterochromatic state in the site which affects Pol II elongation efficiency and mediates AS of EDI (148). This regulation was found to be dependent on ARGONAUTE 1 (AGO1), which is a crucial actor in RNA silencing, binding siRNAs to recognize their target RNAs. Another example of AS mediated by siRNAs is the inclusion of exon 18 of the NEURAL CELL ADHESION MOLECULE (NCAM) gene which is regulated by heterochromatin marks after differentiation of mouse N2a neural cells. This process could also be induced by exogenous application of exon-targeted siRNAs in undifferentiated N2a cells (149). These examples suggest that siRNAs could regulate AS through the modulation of heterochromatin in specific sites in order to fine-tune the Pol II elongation rate. Besides the mechanistic implications of using exogenous applied siRNAs to specific alternative exons, a genome-wide approach hinted the potential relevance of this mechanism in physiological conditions (150). Remarkably, purification of AGO1 and AGO2 chromatin associated complexes revealed their interaction with SFs. Furthermore, approximately one-third of smRNAs loaded in these AGO1 and AGO2 complexes align specifically with 3′ ends of introns, the intron–exon junctions (150). These observations suggest that these intron-related smRNAs and the RNAi protein machinery could have a function in AS regulation. Moreover, genome-wide exon arrays on embryonic fibroblasts of Ago2- or Dicer-null mice showed that they have similar altered AS events (150). In this work, the CD44 gene was taken as a model to characterise the underlying mechanism because several of its alternative exons can be highly included by a phorbol-12-myristate 13-acetate (PMA) treatment (150,151). In mammalian cells treated with PMA, smRNAs recruited AGO1 and AGO2 to the transcribed regions of CD44 in a Dicer- and HP1 (HETEROCHROMATIN PROTEIN 1)-dependent manner, which increased local H3K9me3 levels at the region corresponding to the variable exons. Subsequently, the AGO proteins facilitate spliceosome recruitment and modulation of Pol II processivity in order to shape CD44 AS (150), further suggesting an involvement of smRNAs in splicing regulation. Heterochromatin regulation by smRNAs is widely found in different organisms among yeast, plants and mammals. Therefore, it is possible that the interplay between splicing and smRNAs/heterochromatin pathway is a widespread mechanism in eukaryotes to rapidly regulate splicing and AS rates of specific genes throughout growth and differentiation.
CONCLUSIONS
Generally, regulation of AS by lncRNAs seems to be a common emerging event in many species although the underlying mechanisms differ among them. In this review, we have classified and sub-classified splicing-related lncRNAs characterized from different species and kingdoms, according to the commonalities and singularities of their interaction with SFs, their direct association with pre-mRNAs, their roles in nuclear localization processes or their impact on chromatin conformation. The involvement of lncRNAs in AS regulation seems to be critical under specific conditions, as impairing the production of lncRNAs could lead to diseases in mammals or developmental disorders in plants.
LncRNA-derived small RNAs have also been described here, as their action in the modulation of the chromatin context in the gene body in order to modify Pol II elongation rate, may privilege inclusion or exclusion of specific exon/introns. The identification of certain proteins involved in both, splicing and siRNA production, supports the implication of smRNAs in the regulation of AS.
Some lncRNAs and their mode of action are conserved in closely related species, as it is the case of MALAT1. Nevertheless, the sequence of the majority of lncRNAs are not conserved across species and common roles of different lncRNAs in AS regulation suggests that structural patterns may be conserved for proper recognition by SFs. Further studies on the structure of lncRNAs interacting with SFs across species should shed light on the evolution of these mechanisms. Novel methods developed in the last few years will certainly continue to cope with this challenge. For instance, Cross-Linking and ImmunoPrecipitation (CLIP) assays are used to identify specific RNA sequences bound by protein partners (152,153). Also, RNA Hybrid and Individual-nucleotide resolution UV Cross-Linking and ImmunoPrecipitation (hiCLIP) allows us to identify RNA duplexes, particular RNA intramolecular structures or intermolecular interactions at nucleotide resolution level, bound by specific RNA-associated proteins (154). It is worth considering the differential accumulation and sub-cellular localization of lncRNAs in response to external or intrinsic stimuli. This suggests that, for a given organism, different lncRNAs with similar structures may be recognized by common proteins and exert the same role in alternative developmental contexts.
The link between ncRNAs and diseases clearly highlights the interest in further studies about the so-called dark matter of the genome. A deeper knowledge about lncRNAs activity will help us to detect and hopefully treat a wider range of diseases. For instance, the antisense transcript of the EPIDERMAL GROWTH FACTOR RECEPTOR (EGFR) coding locus, the NAT EGFR-AS1, was recently shown to modulate the EGFR isoforms abundance. Targeting EGFR is a validated approach in the treatment of squamous-cell cancers (SCCs). Notably, a silent punctual mutation impacts the accumulation of EGFR-AS1, what was proposed as a predictive biomarker for SCCs. However, the molecular mechanisms governing EGFR splicing regulation by AGFR-AS1 remain unknown (155).
In plant biology, the identification of differentially expressed lncRNAs in crops, including variation in different ecotypes, may help us decipher how different varieties can adapt to changing environments. A more comprehensive knowledge about the plasticity of plant genomes and the impact of lncRNA in the modulation of the protein-coding genome will certainly allow us to develop novel strategies for sustainable agriculture.
As the noncoding transcriptome, mainly composed of introns and ncRNAs, is the major component of the eukaryotic genome, there may be a large reservoir of mechanisms through which ncRNAs can interact with the splicing machinery to modulate and boost the proteome plasticity, contributing to the expansion of diversity in life. Mechanistic insight into how long and small noncoding RNAs are recruited and fine-tune AS may open large perspectives as novel tools in gene therapy for several diseases including cancer, as well as in biotechnological applications in agriculture and human health.
FUNDING
Funding for open access charge: grant from ANPCyT, Argentina, PICT 2016-0289.
Conflict of interest statement. None declared.
REFERENCES
- 1. Marquez Y., Brown J.W.S., Simpson C., Barta A., Kalyna M.. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012; 22:1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gerstein M.B., Rozowsky J., Yan K.-K., Wang D., Cheng C., Brown J.B., Davis C.A., Hillier L., Sisu C., Li J.J. et al. Comparative analysis of the transcriptome across distant species. Nature. 2014; 512:445–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 4. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Syed N.H., Kalyna M., Marquez Y., Barta A., Brown J.W.S.. Alternative splicing in plants–coming of age. Trends Plant Sci. 2012; 17:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Djebali S., Davis C.A., Merkel A., Gingeras T.R.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boon K.-L., Grainger R.J., Ehsani P., Barrass J.D., Auchynnikava T., Inglehearn C.F., Beggs J.D.. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat. Struct. Mol. Biol. 2007; 14:1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanackovic G., Ransijn A., Thibault P., Elela S.A., Klinck R., Berson E.L., Chabot B., Rivolta C.. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 2011; 20:2116–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011; 478:64–69. [DOI] [PubMed] [Google Scholar]
- 10. Scotti M.M., Swanson M.S.. RNA mis-splicing in disease. Nat. Rev. Genet. 2015; 17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan K., Liu P., Wu C.-A., Yang G.-D., Xu R., Guo Q.-H., Huang J.-G., Zheng C.-C.. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell. 2012; 48:521–531. [DOI] [PubMed] [Google Scholar]
- 12. Reddy A.S.N., Marquez Y., Kalyna M., Barta A.. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013; 25:3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding F., Cui P., Wang Z., Zhang S., Ali S., Xiong L.. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 2014; 15:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhan X., Qian B., Cao F., Wu W., Yang L., Guan Q., Gu X., Wang P., Okusolubo T.A., Dunn S.L. et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015; 6:8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Palusa S.G., Reddy A.S.N.. Differential recruitment of splice variants from SR Pre-mRNAs to polysomes during development and in response to stresses. Plant Cell Physiol. 2015; 56:421–427. [DOI] [PubMed] [Google Scholar]
- 16. Rauch H.B., Patrick T.L., Klusman K.M., Battistuzzi F.U., Mei W., Brendel V.P., Lal S.K.. Discovery and Expression Analysis of Alternative Splicing Events Conserved among Plant SR Proteins. Mol. Biol. Evol. 2013; 31:605–613. [DOI] [PubMed] [Google Scholar]
- 17. Palusa S.G., Ali G.S., Reddy A.S.N.. Alternative splicing of pre-mRNAs of Arabidopsis serine / arginine-rich proteins: regulation by hormones and stresses. Plant J. 2007; 49:1091–1107. [DOI] [PubMed] [Google Scholar]
- 18. Seo P.J., Park M.-J., Park C.-M.. Alternative splicing of transcription factors in plant responses to low temperature stress: mechanisms and functions. Planta. 2013; 237:1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Severing E.I., Van Dijk A.D.J., Morabito G., Busscher-lange J.. Predicting the Impact of Alternative Splicing on Plant MADS Domain Protein Function. PLoS One. 2012; 7:e30524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lareau L.F., Brenner S.E.. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 2015; 32:1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reddy A.S.N., Rogers M.F., Richardson D.N., Hamilton M., Ben-Hur A.. Deciphering the Plant Splicing Code: Experimental and Computational Approaches for Predicting Alternative Splicing and Splicing Regulatory Elements. Front. Plant Sci. 2012; 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tran V.D.T., Souiai O., Romero-Barrios N., Crespi M., Gautheret D.. Detection of generic differential RNA processing events from RNA-seq data. RNA Biol. 2016; 13:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matlin A.J., Clark F., Smith C.W.J.. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005; 6:386–398. [DOI] [PubMed] [Google Scholar]
- 24. Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006; 126:37–47. [DOI] [PubMed] [Google Scholar]
- 25. Wang G.-S., Cooper T.A.. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007; 8:749–761. [DOI] [PubMed] [Google Scholar]
- 26. Faial T. RNA splicing in common disease. Nat Genet. 2015; 47:105. [Google Scholar]
- 27. Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W., Mockler T.C.. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010; 20:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanabe N., Yoshimura K., Kimura A., Yabuta Y.. Differential Expression of Alternatively Spliced mRNAs of Arabidopsis SR Protein Homologs, atSR30 and atSR45a, in Response to Environmental Stress. Plant Cell Physiol. 2007; 48:1036–1049. [DOI] [PubMed] [Google Scholar]
- 29. Wang K.C., Chang H.Y.. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2016; 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guttman M., Rinn J.L.. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moran V.A., Perera R.J., Khalil A.M.. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. 2012; 40:6391–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rappsilber J., Ryder U., Lamond A.I., Mann M.. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002; 12:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herold N., Will C.L., Wolf E., Kastner B., Urlaub H., Lu R.. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell Biol. 2009; 29:281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang B., Brendel V.. The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004; 5:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., Zhang X.-O., Chen T., Xiang J.-F., Yin Q.-F., Xing Y.-H., Zhu S., Yang L., Chen L.-L.. Circular intronic long noncoding RNAs. Mol. Cell. 2013; 51:792–806. [DOI] [PubMed] [Google Scholar]
- 37. Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., Crespi M.. Noncoding transcription by alternative rna polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell. 2014; 55:383–396. [DOI] [PubMed] [Google Scholar]
- 38. Ariel F., Romero-Barrios N., Jegu T., Benhamed M., Crespi M.. Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 2015; 20:362–371. [DOI] [PubMed] [Google Scholar]
- 39. Bardou F., Ariel F., Simpson C.G., Romero-Barrios N., Laporte P., Balzergue S., Brown J.W.S., Crespi M.. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell. 2014; 30:166–176. [DOI] [PubMed] [Google Scholar]
- 40. Song P., Ye L.-F., Zhang C., Peng T., Zhou X.-H.. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016; 592:8–14. [DOI] [PubMed] [Google Scholar]
- 41. Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A.. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007; 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Long J.C., Caceres J.F.. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009; 417:15–27. [DOI] [PubMed] [Google Scholar]
- 43. Cao W., Jamison S.F., Garcia-Blanco M.A.. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997; 3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao S.H., Manley J.L.. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997; 11:334–344. [DOI] [PubMed] [Google Scholar]
- 45. Xiao S.H., Manley J.L.. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998; 17:6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cáceres J.F., Misteli T., Screaton G.R., Spector D.L., Krainer A.R.. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997; 138:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Misteli T., Cáceres J.F., Clement J.Q., Krainer A.R., Wilkinson M.F., Spector D.L.. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998; 143:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang K., Patel N.A., Watson J.E., Apostolatos H., Kleiman E., Hanson O., Hagiwara M., Cooper D.R.. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCβJII messenger ribonucleic acid. Endocrinology. 2009; 150:2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cooper D.R., Carter G., Li P., Patel R., Watson J.E., Patel N.A.. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARγ2 splicing during Adipogenesis in 3T3-L1 cells. Genes (Basel). 2014; 5:1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X., Sehgal L., Jain N., Khashab T., Mathur R., Samaniego F.. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016; 14:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malakar P., Shilo A., Mogilavsky A., Stein I., Pikarsky E., Nevo Y., Benyamini H., Elgavish S., Zong X., Prasanth K. V et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 up-regulation and mTOR activation. Cancer Res. 2016; 77:1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li R.-Q., Ren Y., Liu W., Pan W., Xu F.-J., Yang M.. MicroRNA-mediated silence of onco-lncRNA MALAT1 in different ESCC cells via ligand-functionalized hydroxyl-rich nanovectors. Nanoscale. 2017; 9:2521–2530. [DOI] [PubMed] [Google Scholar]
- 53. Chen W., Xu X., Li J., Kong K., Li H., Chen C.. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017; 8:22783–22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Z., Zhou Y., Tu B., Bu Y., Liu A., Xie C.. Long noncoding RNA MALAT1 affects the efficacy of radiotherapy for esophageal squamous cell carcinoma by regulating Cks1 expression. J. Oral Pathol. Med. 2016; 46:583–590. [DOI] [PubMed] [Google Scholar]
- 55. Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010; 39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 2008; 283:1223–1227. [DOI] [PubMed] [Google Scholar]
- 57. Huang Y., Yario T.A., Steitz J.A.. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sanford J.R., Ellis J.D., Cazalla D., Cáceres J.F.. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15042–15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi Y., Manley J.L.. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol. Cell. 2007; 28:79–90. [DOI] [PubMed] [Google Scholar]
- 60. Zhong X.Y., Ding J.H., Adams J.A., Ghosh G., Fu X.D.. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009; 23:482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patton J.G., Mayer S.A., Tempst P., Nadal-Ginard B.. Characterization and molecular cloning of polypymiridine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991; 5:1237–1251. [DOI] [PubMed] [Google Scholar]
- 62. He X., Pool M., Darcy K.M., Lim S.B., Auersperg N., Coon J.S., Beck W.T.. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007; 26:4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Patton J.G., Porto E.B., Galceran J., Paul T., Nadal-ginard B.. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993; 7:393–406. [DOI] [PubMed] [Google Scholar]
- 64. Gozani O., Patton J.G., Reed R.. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994; 13:3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meissner M., Dechat T., Gerner C., Grimm R., Foisner R., Sauermann G.. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J. Cell. Biochem. 2000; 76:559–566. [PubMed] [Google Scholar]
- 66. Ji Q., Zhang L., Liu X., Zhou L., Wang W., Han Z., Sui H., Tang Y., Wang Y., Liu N. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ / PTBP2 complex. Br. J. Cancer. 2014; 111:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang G., Cui Y., Zhang G., Garen A., Song X.. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:16794–16798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li L., Feng T., Lian Y., Zhang G., Garen A., Song X.. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12956–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kong J., Sun W., Li C., Wan L., Wang S., Wu Y., Xu E., Zhang H., Lai M.. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016; 380:476–484. [DOI] [PubMed] [Google Scholar]
- 70. Zang C., Nie F., Wang Q., Sun M., Li W., He J., Zhang M., Lu K.. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget. 2016; 7:11696–11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Barry G., Briggs J., Vanichkina D., Poth E., Beveridge N., Ratnu V., Nayler S., Nones K., Hu J., Bredy T. et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry. 2013; 19:486–494. [DOI] [PubMed] [Google Scholar]
- 72. Rapicavoli N. a, Poth E.M., Blackshaw S.. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010; 10:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rapicavoli N.A., Blackshaw S.. New meaning in the message: Noncoding RNAs and their role in retinal development. Dev. Dyn. 2009; 238:2103–2114. [DOI] [PubMed] [Google Scholar]
- 74. Mercer T.R., Qureshi I.A., Gokhan S., Dinger M.E., Li G., Mattick J.S., Mehler M.F.. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010; 11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S.. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 2007; 105:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sone M., Hayashi T., Tarui H., Agata K., Takeichi M., Nakagawa S.. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007; 120:2498–2506. [DOI] [PubMed] [Google Scholar]
- 77. Tsuiji H., Yoshimoto R., Hasegawa Y., Furuno M., Yoshida M., Nakagawa S.. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011; 16:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ladd A.N. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol. Cell Neurosci. 2013; 29:456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ishizuka A., Hasegawa Y., Ishida K., Yanaka K., Nakagawa S.. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cells. 2014; 19:704–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van Dijk M., Visser A., Buabeng K.M.L., Poutsma A., van der Schors R.C., Oudejans C.B.M.. Mutations within the LINC-HELLP non-coding RNA differentially bind ribosomal and RNA splicing complexes and negatively affect trophoblast differentiation. Hum. Mol. Genet. 2015; 24:5475–5485. [DOI] [PubMed] [Google Scholar]
- 81. Boisvert F.M., Van Koningsbruggen S., Navascués J., Lamond A.I.. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007; 8:574–585. [DOI] [PubMed] [Google Scholar]
- 82. Kiss T., Agris P., Bachellerie J., Michot B., Nicoloso M., Balakin A., Ni J., Fournier M., Balakin A., Smith L. et al. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001; 20:3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Matera A.G., Terns R.M., Terns M.P.. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007; 8:209–220. [DOI] [PubMed] [Google Scholar]
- 84. Yin Q.F., Yang L., Zhang Y., Xiang J.F., Wu Y.W., Carmichael G.G., Chen L.L.. Long Noncoding RNAs with snoRNA Ends. Mol. Cell. 2012; 48:219–230. [DOI] [PubMed] [Google Scholar]
- 85. Kishore S., Stamm S.. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006; 311:230–232. [DOI] [PubMed] [Google Scholar]
- 86. Wu H., Yin Q.F., Luo Z., Yao R.W., Zheng C.C., Zhang J., Xiang J.F., Yang L., Chen L.L.. Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol. Cell. 2016; 64:534–548. [DOI] [PubMed] [Google Scholar]
- 87. Crespi M.D., Jurkevitch E., Poiret M., d’Aubenton-Carafa Y., Petrovics G., Kondorosi E., Kondorosi A.. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994; 13:5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gultyaev A.P., Roussis A.. Identification of conserved secondary structures and expansion segments in enod40 RNAs reveals new enod40 homologues in plants. Nucleic Acids Res. 2007; 35:3144–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Campalans A., Kondorosi A., Crespi M.. Enod40, a short open reading frame – containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004; 16:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Khorkova O., Myers A.J., Hsiao J., Wahlestedt C.. Natural antisense transcripts. Hum. Mol. Genet. 2014; 23:R54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Krystal G.W., Armstrong B.C., Battey J.F.. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol. Cell. Biol. 1990; 10:4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Suenaga Y., Islam S.M.R., Alagu J., Kaneko Y., Kato M., Tanaka Y., Kawana H., Hossain S., Matsumoto D., Yamamoto M. et al. NCYM, a cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3β resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet. 2014; 10:e1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Munroe S.H., Lazar M.A.. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol. Chem. 1991; 266:22083–22086. [PubMed] [Google Scholar]
- 94. Hastings M.L., Milcarek C., Martincic K., Peterson M.L., Munroe S.H.. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997; 25:4296–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rindfleisch B.C., Brown M.S., VandeBerg J.L., Munroe S.H.. Structure and expression of two nuclear receptor genes in marsupials: insights into the evolution of the antisense overlap between the α-thyroid hormone receptor and Rev-erbα. BMC Mol. Biol. 2010; 11:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bardou F., Merchan F., Ariel F., Crespi M.. Dual RNAs in plants. Biochimie. 2011; 93:1950–1954. [DOI] [PubMed] [Google Scholar]
- 97. Villamizar O., Chambers C.B., Riberdy J.M., Persons D.A., Wilber A.. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget. 2015; 7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Beltran M., Puig I., Peña C., García J.M., Álvarez A.B., Peña R., Bonilla F., De Herreros A.G.. A natural antisense transcript regulates Zeb2 / Sip1 gene expression during Snail1-induced epithelial – mesenchymal transition. Genes Dev. 2008; 22:756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Massone S., Vassallo I., Fiorino G., Castelnuovo M., Barbieri F., Borghi R., Tabaton M., Robello M., Gatta E., Russo C. et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011; 41:308–317. [DOI] [PubMed] [Google Scholar]
- 100. Luco R.F., Allo M., Schor I.E., Kornblihtt A.R., Misteli T.. Epigenetics in alternative pre-mRNA splicing. Cell. 2011; 144:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Batsché E., Yaniv M., Muchardt C.. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:22–29. [DOI] [PubMed] [Google Scholar]
- 102. Schor I.E., Rascovan N., Pelisch F., Alló M., Kornblihtt A.R.. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016; 17:205–211. [DOI] [PubMed] [Google Scholar]
- 104. Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014; 56:55–66. [DOI] [PubMed] [Google Scholar]
- 105. Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J.. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015; 160:1125–1134. [DOI] [PubMed] [Google Scholar]
- 106. Kelly S., Greenman C., Cook P.R., Papantonis A.. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015; 427:2414–2417. [DOI] [PubMed] [Google Scholar]
- 107. Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C. et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017; 3:17053. [DOI] [PubMed] [Google Scholar]
- 108. Ariel F., Crespi M.. Alternative splicing: the lord of the rings. Nat. Plants. 2017; 3:17065. [DOI] [PubMed] [Google Scholar]
- 109. Gonzalez I., Munita R., Agirre E., Dittmer T. a, Gysling K., Misteli T., Luco R.F.. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015; 22:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D., Dorrestein P.C., Rosenfeld M.G.. NcRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011; 147:773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B.. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of Paraspeckles. Mol. Cell. 2009; 33:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mao Y.S., Sunwoo H., Zhang B., Spector D.L.. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011; 13:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Engreitz J.M., Sirokman K., McDonel P., Shishkin A.A., Surka C., Russell P., Grossman S.R., Chow A.Y., Guttman M., Lander E.S.. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014; 159:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ietswaart R., Wu Z., Dean C.. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012; 28:445–453. [DOI] [PubMed] [Google Scholar]
- 115. Rodriguez-Granados N.Y., Ramirez-Prado J.S., Veluchamy A., Latrasse D., Raynaud C., Crespi M., Ariel F., Benhamed M.. Put your 3D glasses on: plant chromatin is on show. J. Exp. Bot. 2016; 67:3205–3221. [DOI] [PubMed] [Google Scholar]
- 116. Kim D.H., Sung S.. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell. 2017; 40:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhou H.-L., Luo G., Wise J.A., Lou H.. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014; 42:701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E.. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014; 55:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Simon M.D., Pinter S.F., Fang R., Sarma K., Rutenberg-Schoenberg M., Bowman S.K., Kesner B.A., Maier V.K., Kingston R.E., Lee J.T.. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013; 504:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Misteli T., Cáceres J., Spector D.. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997; 387:523–527. [DOI] [PubMed] [Google Scholar]
- 121. Zeng C., Kim E., Warren S.L., Berget S.M.. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997; 16:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Huang S., Spector D.L.. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 1996; 133:719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Smith K.P., Moen P.T., Wydner K.L., Coleman J.R., Lawrence J.B.. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 1999; 144:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B.. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013; 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013; 341:1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y.. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010; 329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. van der Krol A.R., Mur L.A., Beld M., Mol J.N., Stuitje A.R.. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990; 2:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Napoli C., Lemieux C., Jorgensen R.. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990; 2:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 130. Taft R.J., Glazov E.A., Lassmann T., Hayashizaki Y., Carninci P., Mattick J.S.. Small RNAs derived from snoRNAs. RNA. 2009; 15:1233–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Thompson D.M., Lu C., Green P.J., Parker R.. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008; 14:2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A.. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010; 16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Catalanotto C., Cogoni C., Zardo G.. MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 2016; 17:e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhai J., Bischof S., Wang H., Feng S., Lee T.F., Teng C., Chen X., Park S.Y., Liu L., Gallego-Bartolome J. et al. A one precursor one siRNA model for pol IV-dependent siRNA biogenesis. Cell. 2015; 163:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Duss O., Michel E., Yulikov M., Schubert M., Jeschke G., Allain F.H.-T.. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature. 2014; 509:588–592. [DOI] [PubMed] [Google Scholar]
- 136. Bayne E.H., Portoso M., Kagansky A., Kos-Braun I.C., Urano T., Ekwall K., Alves F., Rappsilber J., Allshire R.C.. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008; 322:602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chinen M., Morita M., Fukumura K., Tani T.. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J. Biol. Chem. 2010; 285:5630–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ausin I., Greenberg M.V.C., Li C.F., Jacobsen S.E.. The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics. 2012; 7:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Dou K., Huang C.F., Ma Z.Y., Zhang C.J., Zhou J.X., Huang H.W., Cai T., Tang K., Zhu J.K., He X.J.. The PRP6-like splicing factor STA1 is involved in RNA-directed DNA methylation by facilitating the production of Pol V-dependent scaffold RNAs. Nucleic Acids Res. 2013; 41:8489–8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Huang C.F., Miki D., Tang K., Zhou H.R., Zheng Z., Chen W., Ma Z.Y., Yang L., Zhang H., Liu R. et al. A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013; 9:e1003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Elvira-Matelot E., Bardou F., Ariel F., Jauvion V., Bouteiller N., Le Masson I., Cao J., Crespi M.D., Vaucheret H.. The nuclear ribonucleoprotein SmD1 interplays with splicing, RNA quality control and post-transcriptional gene silencing in Arabidopsis. Plant Cell. 2016; 28:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Spies N., Nielsen C.B., Padgett R.A., Burge C.B.. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009; 36:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Schwartz S., Meshorer E., Ast G.. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009; 16:990–995. [DOI] [PubMed] [Google Scholar]
- 144. Andersson R., Enroth S., Rada-iglesias A., Wadelius C., Komorowski J.. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009; 19:1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcárcel J., Guigó R.. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009; 16:996–1001. [DOI] [PubMed] [Google Scholar]
- 146. de la Mata M., Lafaille C., Kornblihtt A.R.. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA. 2010; 16:904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kornblihtt A.R. Chromatin, transcript elongation and alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:5–7. [DOI] [PubMed] [Google Scholar]
- 148. Alló M., Buggiano V., Fededa J.P., Petrillo E., Schor I., De Mata M., Agirre E., Plass M., Eyras E., Elela S.A. et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009; 16:717–725. [DOI] [PubMed] [Google Scholar]
- 149. Schor I.E., Fiszbein A., Petrillo E., Kornblihtt A.R.. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J. 2013; 32:2264–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.-C. et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012; 19:998–1004. [DOI] [PubMed] [Google Scholar]
- 151. König H., Ponta H., Herrlich P.. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998; 17:2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B.. CLIP identifies nova-regulated RNA networks in the brain. Science. 2003; 302:1212–1215. [DOI] [PubMed] [Google Scholar]
- 153. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sugimoto Y., Chakrabarti A.M., Luscombe N.M., Ule J.. Using hiCLIP to identify RNA duplexes that interact with a specific RNA-binding protein. Nat. Protoc. 2017; 12:611–637. [DOI] [PubMed] [Google Scholar]
- 155. Tan D.S.W., Chong F.T., Leong H.S., Toh S.Y., Lau D.P., Kwang X.L., Zhang X., Sundaram G.M., Tan G.S., Chang M.M. et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 2017; 23:1167–1175. [DOI] [PubMed] [Google Scholar]