Abstract
WUSCHEL-related homeobox (WOX) genes are key regulators of meristem activity and plant development, the chromatin mechanism of which to reprogram gene expression remains unclear. Histone H3K27me3 is a chromatin mark of developmentally repressed genes. How the repressive mark is removed from specific genes during plant development is largely unknown. Here, we show that WOX11 interacts with the H3K27me3 demethylase JMJ705 to activate gene expression during shoot development in rice. Genetic analysis indicates that WOX11 and JMJ705 cooperatively control shoot growth and commonly regulate the expression of a set of genes involved in meristem identity, chloroplast biogenesis, and energy metabolism in the shoot apex. Loss of WOX11 led to increased H3K27me3 and overexpression of JMJ705 decreased the methylation levels at a subset of common targets. JMJ705 is associated with most of the WOX11-binding sites found in the tested common targets in vivo, regardless of presence or absence of the JMJ705-binding motif. Furthermore, wox11 mutation reduced JMJ705-binding to many targets genome-wide. The results suggest that recruitment of JMJ705 to specific developmental pathway genes is promoted by DNA-binding transcription factors and that WOX11 functions to stimulate shoot growth through epigenetic reprogramming of genes involved in meristem development and energy-generating pathways.
INTRODUCTION
Plant postembryonic development is dependent on meristems that have the capacity to replenish cells for organogenesis and at the same time to maintain meristem cell identity. The stem cell pool of Arabidopsis shoot apical meristem (SAM) is promoted by the homeodomain transcriptional regulator WUSCHEL (WUS) (1). Apart from function in SAM stem cells (1,2), the WUSCHEL-RELATED HOMEOBOX (WOX) family genes play diverse roles in plant development, such as embryogenesis, vascular differentiation, lateral organ patterning and maintenance of the root apical meristem (3–9). The WOX gene family can be phylogenetically identified as three major branches: the WUS clade, the ancient clade consisting of WOX13-related genes found throughout all land plants and in some green algae, and the intermediate WOX9 clade (10).
Although much progress has been made in functional studies of Arabidopsis WOX genes, developmental functions of cereals orthologues remain largely unclear. In addition, there is evidence that developmental functions of WOX family members between rice and Arabidopsis are not always conserved (11). For instance, the rice orthologue of WUS, TILLERS ABSENT1 (TAB1, also known as OsWUS), is shown to be required to initiate axillary meristem development (12), whereas rice WOX4, belonging to the WUS clade, is shown to be involved in the SAM activity (13), and to regulate axillary meristem maintenance after establishment of the meristem by TAB1 (12). Rice WOX3, another member of the WUS clade, is shown to regulate KNOX and YABBY genes required for rice leaf development (14). Rice WOX11, a member of the WOX9 clade, is shown to establish gene expression programs to promote cell division of shoot-borne crown root meristem (15,16), which is originated from the first internodes. Arabidopsis WUS clade members WUS and WOX5 are shown to interact with TOPLESS (TPL)-like co-repressors and a histone deacetylase to repress gene expression programs of cell differentiation in Arabidopsis shoot or root apical meristems, respectively (17,18). The molecular and chromatin basis of rice WOX genes-regulated meristem activities and developmental processes is largely unknown.
Chromatin-based epigenomes are the basis of cell type-specific gene expression patterns and chromatin modification is essential for gene expression reprogramming during development. It is well established that H3K27me3 plays an important role in maintaining gene repression during development in both plants and animals. In plants including rice, the H3K27me3-marked genes have very low expression levels and often exhibit a high degree of tissue specificity (19–21). In fact, a remarkable number of developmental regulators or transcription factors are targeted by H3K27me3 for repression (22,23). Chromatin regulators involved in H3K27me3 deposition and/or recognition have been implicated in regulation of many plant developmental pathways, including seed development, flowering time, vernalization, organ identity, and meristem function (22–24). There is a massive reprogramming of H3K27me3 with many genes shown to gain or lose H3K27me3 during cell differentiation or responding to environmental signals in Arabidopsis and rice (25–27). In particular, it is shown that in rice SAM a large number of genes are marked by H3K27me3 and that dynamic change of H3K27me3 was critical for genome-wide gene expression reprogramming during the transitions to leaf organogenesis and to reproductive inflorescence meristem formation, where thousands of genes gained or lost the mark (27). H3K27me3 is established by SET-domain-containing histone methyltransferases CLF and SWN in Arabidopsis and SDG711 and SDG718 in rice (27–29), and is removed by Jumonji (jmjC) domain-containing histone demethylases RELATIVE OF EARLY FLOWERING 6 (REF6/JMJ12) in Arabidopsis (30) and JMJ705 in rice (26). Recent results have shown that REF6/JMJ12 can be recruited to target loci by binding to the ‘CTCTGYTY, Y = T/C’ motif and is required for recruiting the chromatin remodeler BRM (31,32). However, this motif is found in a very large number of genes genome-wide (31,32). It remains unclear how this kind of enzymes is recruited to remove H3K27me3 from genes in a tissue or organ-specific manner and to regulate a specific developmental program in plants.
In this work, we show that besides its function in crown root development, WOX11 is also required for rice shoot development. Importantly, we show that WOX11 interacts with JMJ705 to cooperatively regulate rice shoot development. Both wox11 and jmj705 mutants show similar shoot growth defects. There were significant overlaps of mis-expressed genes in the shoot apex of wox11 and jmj705 mutants. The common target genes are enriched for function in early light signalling, chloroplast biogenesis, and primary metabolism, in addition to transcription factor genes involved in shoot meristem identity, circadian rhythm, and stress responses. Furthermore, we provide evidence that although JMJ705 can bind to genes containing the ‘CTCTGYTY’ motif, WOX11 is involved in recruitment of JMJ705 to common target genes which can be independent of binding to the ‘CTCTGYTY’ motif. The results reveal a mechanism of JMJ705 recruitment to targets of specific developmental pathways and indicate that WOX11 has a function to promote rice shoot growth, which involves removal of H3K27me3 from a set of genes that function in shoot development in rice.
MATERIALS AND METHODS
Plant materials and growth conditions
Rice (Oryza sativa subsp. japonica) varieties used in this study are Zhonghua11 (ZH11) and Hwayoung (HY). WOX11 mutant (wox11) used in this study is described in (15). JMJ705 overexpression (line oxJ5–5) and jmj705 mutant plants are described in (26).
The Ubi:WOX11:FLAG transgenic plants were produced in the ZH11 background. For that, WOX11 full-length cDNA was amplified using the primer set WOX11-flag-F and WOX11-flag-R (Supplementary Table S1), then inserted into the overexpression vector pU1301-3XFLAG under the control of the maize (Zea mays) ubiquitin gene promoter (33).
wox11 and jmj705 double mutant was generated by genetic cross between wox11 mutant plants with jmj705 mutants and was selected by PCR using previously reported primers (15,26). We also crossed the wox11 mutant with JMJ705 overexpression line oxJ5–5 to generate the segregated wild type (WT), JMJ705 overexpression plants (oxJ5) and wox11/oxJ5 plants for analysis in this study.
For in vitro cultures, seeds were surface-sterilized and germinated in media containing 0.3% phytagel supplemented with 2% (w/v) sucrose at 28°C (in light) and 24°C (in dark) with a 14 h light/10 h dark cycle. For field growth, all indicated plants were grown in Wuhan area in summer.
In situ hybridization and histological observation
All tested materials were fixed, dehydrated, embedded, sliced, and attached to slides as previously reported (15). For preparation for the digoxigenin-labeled RNA probes, a specific coding region of WOX11 was amplified via PCR using primers (WOX11-in situ-F and WOX11-in situ-R, Supplementary Table S1). The PCR products were cloned into pGEM-T Easy vectors, linearized, and used as templates for amplifying digoxigenin-labeled sense and antisense RNA probes. Tissue sections were cleared, dehydrated, dried, hybridized, and washed. The labelled probes were detected and images were photographed with a microscope (Carl Zeiss, Axio Scope.A1).
For histological analysis of SAM, 5-day-old shoot apices were sampled for histological section. Images of SAM were photographed with a microscope (Carl Zeiss, Axio Scope.A1). SAM sizes were determined by the dome area delimited by drawing a straight line between the basal edges of the two opposing youngest leaf primordia according to the method described in http://www.bio-protocol.org/e2055. Ten SAM per genotype were measured with Photoshop.
Protein-Protein interaction assay
For yeast two-hybrid analysis, WOX11 cDNA was cloned into pGADT7 vector and JMJ705 was cloned into pGBKT7 vector. Truncated WOX11 and JMJ705 domains were obtained by PCR using specific primers (Supplementary Table S1) and tested in yeast two-hybrid assays.
For in vitro pull-down assay, the pGEX4T-1-JMJ705N+C was constructed and expressed in the Escherichia coli BL21 (DE3) strain using primer sets JMJ705N+C-4T-F and JMJ705N+C-4T-R (Supplementary Table S1). His-tagged WOX11 protein was expressed as previously described (15). GST or JMJ705N+C-GST coupled GST beads (GE Healthcare, 17–5132-01) were used to pull down WOX11–6XHis and an anti-His antibody was applied to detect WOX11–6XHis (Abcam, ab19256).
BiFC assays were performed as previously described (34). For generation of the BiFC vectors, the full-length cDNA of JMJ705 was cloned into the BamHI-SalI sites of pVYCE(R) using primer sets JMJ705-BiFC-CF and JMJ705-BiFC-CR (Supplementary Table S1). The full length of WOX11 was cloned into pVYNE(R) (16). Rice mesophyll protoplasts were prepared and the two fusion proteins were transiently co-transfected into rice protoplasts. Fluorescence in the transformed protoplasts was imaged after incubation at 25°C for 12 to 20 h using a confocal laser scanning microscope (Leica, TCS SP2).
Co-Immunoprecipitation assays were performed in tobacco (N. benthamiana) and in rice. The WOX11 full-length cDNA was cloned into pCambia 1301 vector using the primer sets WOX11-1301S-F and WOX11-1301S-R (Supplementary Table S1). The truncated 35S-FLAG-JMJ705-N+C-GFP (see primers in Supplementary Table S1) was co-transfected with 1301S-WOX11 into tobacco leaf cells as described previously (35). After 36 h infection, the leaves were harvested. Rice calli from the wild type (ZH11) and JMJ705 overexpression plants (line oxJ5–5) were also harvested. Those samples were ground into fine powder in liquid nitrogen. Nuclear proteins were extracted as described (16). 10% volume of the total nuclear supernatant was preserved as input and applied to the western blot for detection of WOX11 and JMJ705 expression using anti-WOX11 (16) and anti-FLAG antibodies (Sigma, F3165). Equal volume of anti-WOX11 or anti-FLAG antibody was incubated with 80 μl Dynabeads® Protein A/G for immunoprecipitation (Thermo Fisher Scientific, 10001D/10003D) at 4°C by gentle rotation (>2 h). Then the supernatant was added to the beads and incubated at 4°C with gentle rotation for 8 h. After 5 times wash, the protein was eluted from the beads with 2× sample buffer (125 mM Tris–HCl, pH 6.8, with 4% SDS, 20% (v/v) glycerol and 0.004% bromophenol blue) and was applied to western blot with corresponding antibodies.
RNA extraction for RNA-seq
For RNA-seq samples, shoot apex of ∼2–3 mm without leaf tissues were taken from 5-day-old shoots with a scalpel under a dissecting microscope. About 40 apex per sample were harvested in liquid nitrogen for RNA extraction or RNA-seq. TRIzol-based RNA extraction method was conducted according to the manufacturer's instruction (Invitrogen, 10296-010).
RNA-seq and data analysis
RNA-seq libraries were constructed at the sequencing platform of Huazhong Agriculture University. Briefly, total RNA (10 μg) for each sample was used to purify poly (A) mRNA, and this mRNA was used for the synthesis and amplification of complementary DNA. The libraries were prepared using the TruSeq RNA Sample Preparation Kit from Illumina and sequenced on an Illumina HiSeq 2000.
Sequence reads were filtered using Trimmomatic (version 0.32) (36). The clean data were mapped against the MSU 7.0 rice genome with corresponding annotation by Tophat 2.0 using default parameters. These data were then analyzed for differentially expressed genes using Cufflinks (version 2.2.1) (37). Genes with an expression change fold >2 with P < 0.05 were defined as differentially expressed. For Gene Ontology (GO) analysis of RNA-seq data, we used singular enrichment analysis (http://bioinfo.cau.edu.cn/agriGO/analysis.php) and choose P < 0.001 as the cut-off for significant GO terms.
ChIP
The ChIP experiment was performed as described (38). About 2 g of 5-day-old seedling was cross-linked in 1% formaldehyde under vacuum. Chromatin was extracted and fragmented to 200–500 bp by sonication, and ChIP was performed using the following antibodies: H3K27me3 (ABclonal, a2363), anti-FLAG (Sigma, F3165) anti-IgG (Promoter). The precipitated and input DNA samples were analyzed by quantitative RT-PCR with gene-specific primers listed in Supplementary Table S1. All assays were performed at least three times from three biological replicates.
ChIP-seq and data analysis
About 500 shoot apices were isolated for each sample as described above and were applied to ChIP assay using the anti-FLAG antibody. The precipitated and input DNA were used to construct sequencing libraries following the protocol of Illumina TruSeq ChIP Sample Prep Set A and sequenced by Illumina Hiseq X Ten. For this experiment, 4 ng DNA were used for each library construction.
Sequence reads from all libraries were filtered by Trimmomatic (version 0.32) (36). Clean reads were mapped to the MSU 7.0 rice genome using Bowtie 2 (version 2.1.0) (37) by default. Samtools (version 0.1.17) was used to remove potential PCR duplicates, and MACS software (39) was used to call peaks with the default parameters (bandwidth, 300 bp; model fold, 10, 30; P value, 1.00e–5) (40). After the positions of the peaks on the chromosomes were found, genes (including genebody and the 2-kb upstream and 2-kb downstream regions) overlapping with the peaks were considered to have the protein occupancy. The wig files of the analysis pipeline were used for viewing the data in the Integrative Genomics Viewer software.
Histone extraction
About 60 shoot apices were harvested from 5-day-old shoot in liquid nitrogen for each sample as described above. Histones were extracted using the The EpiQuik™ Total Histone Extraction Kit by following the manufacturer's instruction (Epigentek, OP-0006-100) and then detected by western blot with anti-H3 (Abcam, ab1791) and anti-H3K27me3 (ABclonal, a2363).
Gel shift assay
To conduct the gel shift assay, the fragment containing zinc finger domain of JMJ705 was cloned into the pGEX6P-1 vector using JMJ705-ZF-F and JMJ705-ZF-R separately (Supplementary Table S1). The protein was expressed in the E. coli BL21 (DE3) strain and purified using the Glutathione Sepharose 4 Fast Flow (GE Healthcare, 17-5132-01) according to the manufacturer's instruction. 5′ FAM oligonucleotides were synthesized and labelled by the Shanghai Sangon Company (Supplementary Table S1). Protein and DNA binding reactions were performed according to the Light Shift™ Chemiluminescent EMSA Kit (Thermo Fisher Scientific, 20148) followed by image acquisition with Fujifilm FLA 5100 system as described (41).
RT-PCR
Four micrograms of total RNA was reverse-transcribed in a reaction of 20 μl using DNase I and M-MLV reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer's instructions to obtain cDNA. Real-time PCR was performed in an optical 96-well plate on a PRISM 7500 PCR instrument (Applied Biosystems). The reactions were performed at 95°C for 10 s, 45 cycles of 95°C for 5 s and 60°C for 40 s. Disassociation curve analysis was performed as follows: 95°C for 15 s, 60°C for 20 s and 95°C for 15 s. Data were collected using the ABI PRISM 7500 sequence detection system following the instruction manual. The relative expression levels were analyzed using the 2−ΔΔCT method. The rice Actin gene was used as the internal control. The primers for quantitative RT-PCR are listed in (Supplementary Table S1).
Plant phenotype observation
Seeds with indicated genotype were surface-sterilized and germinated in media. Two days after germination, seeds with similar growth vigorous were transferred to normal growth medium with another 5 days or 7 days growth. The shoot length, primary root length and crown root number were analyzed.
RESULTS
WOX11 has a function to regulate rice shoot development
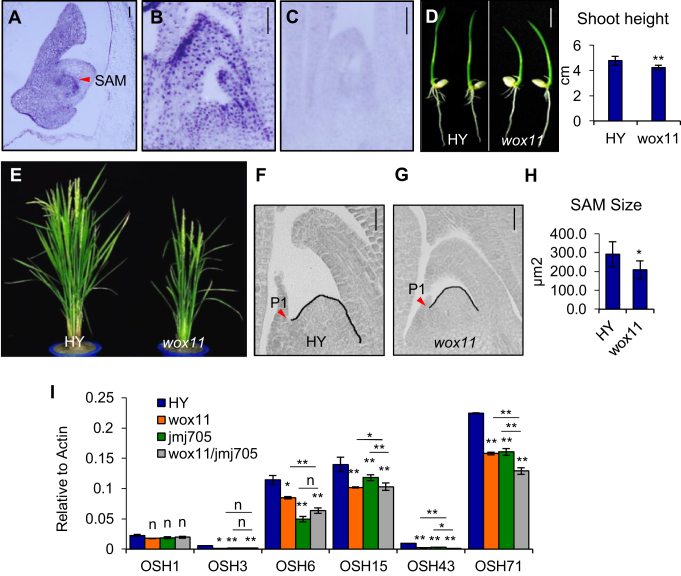
WOX11 has been shown to be required to activate shoot-borne crown root development in rice (15,16). Besides in root meristems, WOX11 transcript was also detected in embryo (particularly in the embryonic SAM), seedling SAM, leaf primordia and young leaves (Figure 1A–C). Compared to wild type (HY), the wox11 mutant displayed reduced shoot growth (Figure 1D). The overexpression of WOX11 controlled by the ubiquitin promoter (Ubi:WOX11) also severely inhibited shoot growth, in addition to the production of a much larger root system (15). This suggested that WOX11 expression level is important for normal shoot development. At the mature stage, wox11 mutant was still semi-dwarf (Figure 1E). In addition, panicle length, spikelet numbers per panicle and secondary branches per panicle were significantly reduced in the mutant (Table 1). These observations indicated that WOX11 had a function to regulate shoot development from juvenile to mature stages.
Figure 1.
WOX11 is involved in plant shoot development. (A–C) In situ hybridization detection of WOX11 transcripts in developing embryo (4 days after fertilization) (A) and 5-day-old shoot apex (B) with an antisense or a sense probe (C). Embryonic SAM in (A) is indicated by a red arrow. Bar = 50 μm. (D) Phenotype of wild type (HY) and wox11 at 7 days after germination (left), shoot height measures are shown on the right. Bar = 1 cm. **P < 0.01, t-test, two-sided (N = 30). (E) Morphology of HY and wox11 at heading stage in the field. (F and G) Shoot apex sections of HY and wox11 at 5 days after germination. Black line traces the SAM contour between leaf primordia P1 and P2. Red arrow indicates P1 leaf primordia. Bar = 20 μm. (H) Statistical analysis of HY and wox11 SAM size determined by measuring the dome area delimited by drawing a straight line between the basal edges of the two opposing youngest leaf primordia. *P < 0.05, t-test, two-sided (N = 10). (I) qRT-PCR analysis of OSH genes expression (relative to Actin transcripts) in shoot apex of HY, wox11, jmj705 and the double mutant (wox11/jmj705). Bar indicates mean ± SD from three replicates. *P< 0.05, **P< 0.01, n, not significant, t-test, two-sided.
Table 1. Phenotype comparisons between wild type (HY), wox11, jmj705, and the double mutant (wox11 /jmj705).
| HY | wox11 | jmj705 | wox11/jmj705 | ||||
|---|---|---|---|---|---|---|---|
| P HY | Pwox11 | Pjmj705 | |||||
| Length of panicle (cm) | 24.45±1.49 | 21.35±0.76 ** | 21.53±0.81 ** | 20.09±1.19 | ** | ** | ** |
| Length of Internode I (cm) | 36.26±1.85 | 35.03±2.37 | 31.95±2.55* | 30.05±1.90 | ** | ** | ** |
| Length of Internode II (cm) | 15.53±1.28 | 15.09±0.96 | 12.50±1.95** | 13.04±1.66 | ** | ** | |
| Length of Internode III (cm) | 6.48±0.62 | 6.22±1.12 | 5.95±1.03 | 5.22±0.75 | ** | * | * |
| Length of Internode IV (cm) | 3.57±0.84 | 3.58±1.09 | 3.74±0.84 | 2.99±1.25 | |||
| Spikelets per panicle | 153±12 | 129±7** | 122±10** | 113±19 | ** | * | |
| Primary branches per panicle | 11±1 | 11±1 | 11±1 | 12±1 | |||
| Secondary branches per panicle | 31±5 | 22±3** | 20±4** | 16±6** | ** | * | * |
Data are presented as mean ± SE (stand error). *P < 0.05, **P < 0.01, t-test, two sided (N = 15).
Analysis of histological sections of 5-day-old shoot apex revealed a smaller SAM of the wox11 mutant than the wild type (Figure 1F–H). Analysis of shoot meristem identity genes revealed that the transcript levels of several Oryza sativahomeobox (OSH) genes (e.g. OSH3, OSH6, OSH15, OSH43, and OSH71) were reduced in wox11 shoot apex (Figure 1I). These observations suggested that in addition to promote crown root development, WOX11 was also required for shoot growth.
WOX11 interacts with histone H3K27me3 demethylase JMJ705
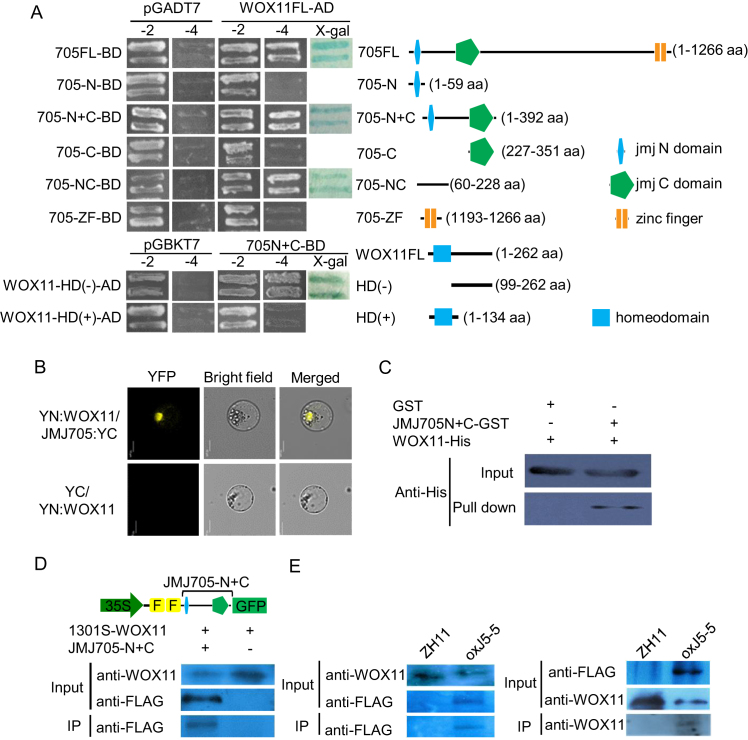
To study molecular mechanisms of WOX11-regulated shoot development, a yeast two-hybrid screening was performed to identify WOX11-interacting proteins. Among the interacting candidates was a previously reported rice H3K27me3 demethylase JMJ705 (26). Deletion analysis revealed that the JMJ705 region located between the jmjN and jmjC domains (amino acids 60–228) interacted with the WOX11 region (amino acids 99–262) downstream the homeodomain (HD) (Figure 2A). Bimolecular fluorescence complementation experiments (BiFC) further confirmed interaction of the two proteins in the nucleus (Figure 2B). Pull-down assays with E. coli-produced JMJ705 N-terminal half (amino acids 1–351) fused with GST and WOX11–6XHis proteins confirmed the interaction (Figure 2C). The interaction was also detected in tobacco (N. benthamiana) cells by co-immunoprecipitation assays, in which 35S:WOX11 was transfected into tobacco leaf cells together with the region comprising the jmjN and jmjC domains of JMJ705 (FLAG-JMJ705-N+C-GFP). The truncated JMJ705 fusion protein was found to be precipitated by the anti-WOX11 antibody (Figure 2D). The interaction was further confirmed in JMJ705-FLAG transgenic calli (dividing and undifferentiated cells) by co-immunoprecipitation with anti-WOX11 and anti-FLAG separately (Figure 2E). Together, those data indicated that WOX11 and JMJ705 could interact in vitro and in vivo.
Figure 2.
WOX11 interacts with the histone H3K27me3 demethylase JMJ705 in vitro and in vivo. (A) Detection of WOX11 interaction with JMJ705 by yeast two-hybrid assay (left). Schematic structures of full length and truncated domains of JMJ705 and WOX11 were indicated on the right. (B) Interaction of WOX11 and JMJ705 in rice protoplasts. Representative cells are shown, as imaged by laser-scanning confocal microscopy. Detection in rice protoplasts of YN:WOX11 and JMJ705:YC interaction shown as yellow signal. The empty vector YC was co-expressed with YN:WOX11 and used as a control. Bar = 10 μm. (C) Pull-down assay of WOX11 and JMJ705. WOX11–6XHis was incubated with GST or JMJ705N+C-GST in GST beads and was pulled down from the JMJ705N+C-GST conjugated GST beads. (D) In vivo co-immunoprecipitation assay of WOX11 and JMJ705 interaction in tobacco. 35S-WOX11 construct was transiently transfected into tobacco leaf cells alone or co-transfected with 35S-FLAG-JMJ705-N+C-GFP. Nuclei isolated from leaves were inspected for expression of WOX11 and JMJ705 protein and then precipitated with the anti-WOX11 antibody. Anti-FLAG was used to detect the JMJ705 protein by western blots. (E) In vivo co-immunoprecipitation assay of WOX11 and JMJ705 interaction in rice. Nuclei isolated from wild type (ZH11) and JMJ705-FLAG (oxJ5–5) calli were precipitated with anti-WOX11 (left) and anti-FLAG antibodies (right) and analysed by immunoblots with anti-FLAG to detect the JMJ705 protein (left) and with the anti-WOX11 antibody to detect the WOX11 protein (right).
JMJ705 is required for shoot development
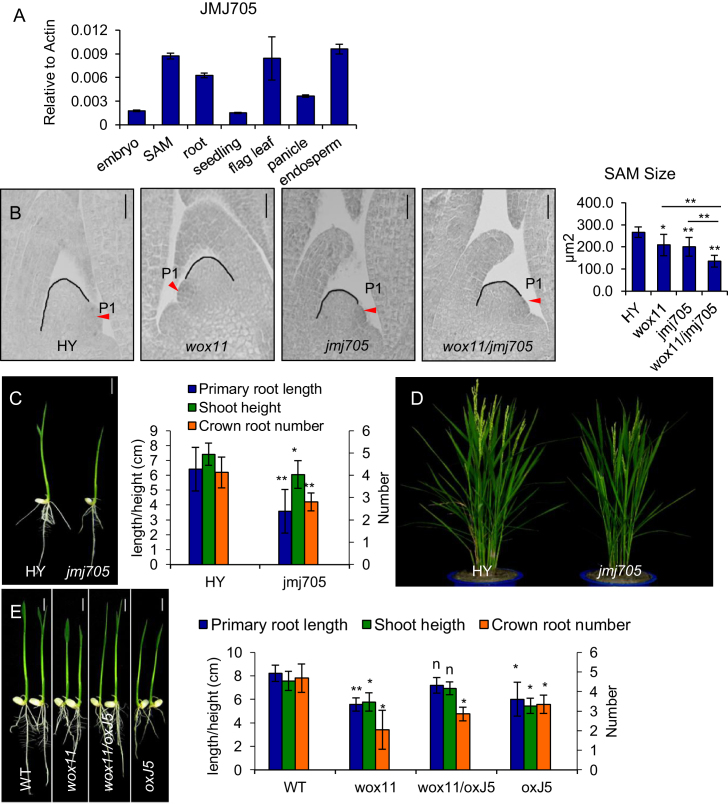
JMJ705 is required for H3K27me3 demethylation and expression of a large number of developmental and stress-responsive genes in rice (26). JMJ705 transcript was generally detected in all examined organ/tissues including SAM by qRT-PCR (Figure 3A) (26). We have previously characterized a T-DNA line in which the insertion is located in the C-terminal end disrupting the Zinc finger (ZnF) domain (26). This mutant displayed similar phenotypes as wox11: a smaller SAM (Figure 3B), a smaller shoot and less developed root system at the seedling stage (Figure 3C), and reduced plant height and panicle size at the mature stage (Figure 3D and Table 1). Expression of OSH genes was also reduced in jmj705 shoot apex (Figure 1I).
Figure 3.
JMJ705 is involved in shoot development. (A) qRT-PCR analysis of JMJ705 transcript levels (relative to Actin transcripts) in SAM, 2-day germinated embryo (embryo), root, 14-day-old seedling (seedling), flag leaf, panicle and endosperm. (B) Shoot apex sections of 5 days HY, wox11, jmj705, and the double mutant (wox11/jmj705) (left). Statistical analysis of SAM sizes is shown on the right. Black line traces the SAM contour between leaf primordia P1 and P2. Red arrow indicates P1 leaf primordia. Bar = 20 μm. *P < 0.05, **P < 0.01, t-test, two sided (N = 10). (C) Phenotype of HY and jmj705 mutant at 7 days after germination. Bar = 1 cm. Statistical analysis is shown on the right. *P < 0.05, **P < 0.01, t-test, two sided (N = 25). (D) Morphology of HY and jmj705 at heading stage in the field. (E) Phenotype of the segregated wild type (WT), the JMJ705-FLAG in wild type background (oxJ5), the wox11 mutant in wild type background (wox11) and JMJ705-FLAG in wox11 background (wox11/oxJ5), from F2 of the crosses between wox11 and oxJ5–5 (left) at 7 days after germination. Bar = 1 cm. Statistical analysis of phenotypes are shown on the right. *P < 0.05, **P < 0.01, n, not significant, t-test, two sided (N = 30).
To study whether JMJ705 and WOX11 were involved in the same regulatory pathway of shoot development, we made wox11/jmj705 double mutants by genetic crosses. The double mutants showed normal germination as the single mutants or wild type, but showed more reduced shoot growth (Supplementary Figure S1A and B). The SAM size was further reduced in the double mutant compared to the single mutants (Figure 3B). The expression of some OSH genes was further reduced (Figure 1I). At heading stage, the double mutants were shorter than the single mutants. The length of each internode was reduced, and sizes of spikelets and secondary branches were smaller than the single mutants (Supplementary Figure S1C–E and Table 1).
To study whether JMJ705 over-expression could compensate for the loss of WOX11 function, we crossed wox11 and oxJ5–5 for phenotypic analysis. As shown in (Figure 3E), shoot growth of wox11/oxJ5 were comparable to that of wild type plants. However, crown root number was further reduced compared to oxJ5–5. The results suggested that elevated JMJ705 levels could complement wox11 shoot but not crown root phenotypes. Thus, JMJ705 and WOX11 appeared to have similar functions in shoot development, but were likely to play different roles in crown root growth.
WOX11 and JMJ705 commonly target many genes in shoot apex
To identify genes that could be commonly regulated by WOX11 and JMJ705, we used RNA-sequencing (RNA-seq) to compare differentially expressed genes in shoot apex (see Methods for sampling) of 5-day-old wox11 and jmj705 mutants relative to wild type (HY). Two biological repeats for each sample were performed (Supplementary Figure S2). In wox11 shoot apex, 209 and 233 genes were respectively down- and up-regulated to >2-fold (P <0.05) compared to wild type (HY). In jmj705 shoot apex, 130 and 457 genes were respectively down and up-regulated to >2-fold (P <0.05) compared to wild type (Figure 4). Analysis of the two sets of data revealed that 73 genes were down-regulated and 139 genes were up-regulated in both wox11 and jmj705 mutants (Figure 4). The overlaps were highly enriched (hypergeometric tests, P < 1.103e–146 and P < 2.230e–234, respectively), suggesting that WOX11 and JMJ705 commonly regulated a subset of genes in the shoot apex. Gene Ontology (GO) analysis revealed that the down-regulated genes in both wox11 and jmj705 were enriched in energy metabolism, photosynthesis, responses to stress and stimuli, chloroplast development and cell parts (P < 0.001, Supplementary Figure S3). Among the mis-expressed genes, many were transcription factors involved in circadian clock, hormone signaling, and stress responses (Supplementary Dataset 1). However, the differential expressions of OSH genes detected by qRT-PCR were not revealed by the RNA-seq analysis, which may be due to relatively lower abundance of the transcripts.
Figure 4.
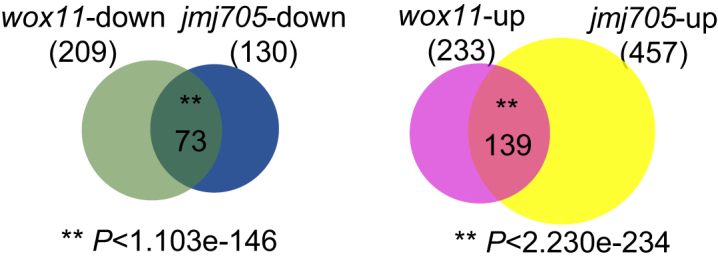
WOX11 and JMJ705 regulate common sets of genes. Venn diagrams showing overlaps between differentially expressed genes in wox11 and jmj705 mutants by RNA-seq. ‘up’ and ‘down’ indicate genes up- and down-regulated in the mutants plants compared to the wild type. Significant enrichments of the overlaps are indicated. **P < 0.01, hypergeometric test.
WOX11 is shown to specifically bind to the WOX-consensus ‘TTAATGG’ motif (15,16). It is recently shown that Arabidopsis REF6/JMJ12, an orthologue of JMJ705, targets genomic loci containing a ‘CTCTGYTY’ motif via its C-terminal ZnF domain (31,32). The ZnF domain is highly conserved between the two proteins (Supplementary Figure S4A). We therefore tested whether JMJ705 could also bind to this sequence by gel shift assays using FAM-labeled DNA probes containing the ‘CTCTGYTY’ motif and E. coli-produced JMJ705 protein. The analysis indicated that JMJ705 could specifically bind to this motif (Supplementary Figure S4B), and the ‘CTCTGYTY’ motif was found in 18 984 genes in rice and was highly enriched at the 5′ region of the genes genome-wide (Supplementary Figure S4C).
WOX11 is involved in JMJ705 recruitment to common target genes
We validated the down-regulation of 17 genes in shoot apex of the two mutants by qRT-PCR (Supplementary Figure S5). Those validated genes included transcription factors (e.g. ERF3, CCT/B-box, RDD1, LHY, HOX16, bZIP47 and OsLSD1) (42–47), chloroplast related genes (e.g. ATA15, OsChl P, chlorophyll-binding proteins Os03g39610 and Os11g13890, early light-induced gene Os01g14410) (48,49), and RNA recognition motif containing genes.
We then tested WOX11 and JMJ705 binding and H3K27me3 levels of several commonly down-regulated genes (LHY, Os03g19560, Os03g01270, bZIP47, Os11g13890, U2AF and ERF3) in jmj705 and wox11, in addition to OSH3, OSH15 and OSH71. Seven of them (i.e. LHY, Os03g19560, Os03g01270, ERF3, OSH3, OSH15 and OSH71) contained both JMJ705- and WOX11-binding sites, whereas the other three (i.e. bZIP47, Os11g13890 and U2AF) contained only the WOX11-binding site. In addition, LHY, OSH15, and OSH71 contained multiple JMJ705- and/or WOX11-binding sites.
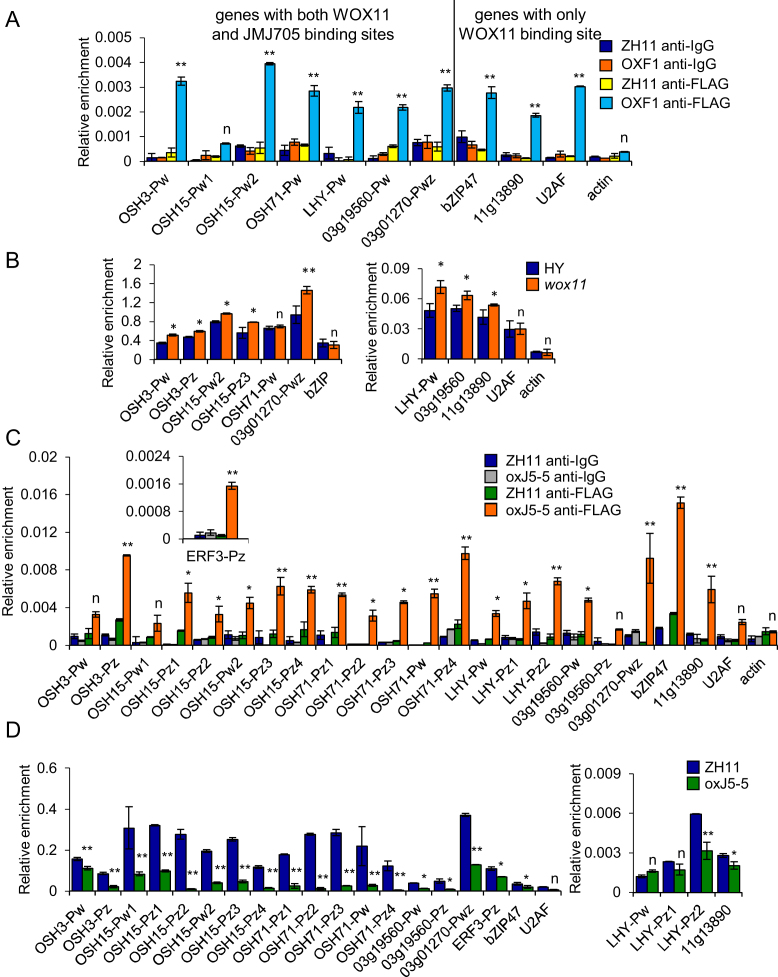
To study WOX11-binding, we produced Ubi:WOX11:FLAG lines, which exhibited similar shoot and root phenotypes as the previously reported Ubi:WOX11 plants (Supplementary Figure S6) (15). ChIP-qPCR analysis of Ubi:WOX11:FLAG shoot with anti-FLAG antibody confirmed WOX11 association to most of the binding sites present in the tested genes in vivo, except the first one in OSH15 (i.e. OSH15-PW1) as well as the negative control Actin (Figure 5A). Due to impossibility to design primers for the ERF3-Pw site, WOX11-binding to this gene was not tested. The wox11 mutation led to moderate but significant increases of H3K27me3 on most of the tested loci (Figure 5B), suggesting that WOX11 might be involved in H3K27me3 removal from the loci. However, western blot analysis using anti-H3K27me3 antibody did not detect any clear difference between wox11 mutant, WOX11 over-expression, and the respective wild type plants (Supplementary Figure S7), suggesting that WOX11 levels did not alter genome-wide H3K27me3.
Figure 5.
Tests of JMJ705 and WOX11 binding and H3K27me3 levels of selected putative common target genes. (A) ChIP assays with anti-FLAG and anti-IgG of 5 days seedling chromatin isolated from wild type (ZH11) and Flag-tagged WOX11 transgenic plants (OXF1). Pw indicates primers contain WOX11 binding site. Actin was used as an internal control. (B) ChIP assays with anti-H3K27me3 of 5 d seedling chromatin isolated from wild type (HY) and wox11 mutant Actin was used as an internal control. (C) ChIP assays with anti-FLAG and anti-IgG of 5 d seedling chromatin isolated from wild type (ZH11) and JMJ705-FLAG plants (oxJ5–5). The first six genes contain both the WOX11 (Pw) and JMJ705-binding (Pz) sites, and the other three genes (bZIP47,Os11g13890, U2AF) contain only WOX11 binding sites. For ERF3 (with Pw and Pz sites), there was no proper primer for Pw and only Pz was detected and presented in the inner box. Actin was used as an internal control. (D) ChIP assays with anti-H3K27me3 of 5 days seedling chromatin isolated from wild type (ZH11) and oxJ5–5 seedlings. Y-axis means assay-site fold enrichment of the signal from immunoprecipitation over the background. Bar indicates mean ± SD from three replicates. Significances of differences are indicated. *P <0.05; **P <0.01, n, not significant, t-test, two-sided.
Interestingly, ChIP-qPCR analysis of oxJ5–5 plants with anti-FLAG revealed that JMJ705 was associated not only with genic regions containing the JMJ705-binding motif, but also with genic regions (i.e. OSH15-Pw2, OSH71-Pw, LHY-Pw, Os03g19560-Pw) or loci (i.e. bZIP47, Os11g13890) that contained only the WOX11-binding site (Figure 5C). The observations suggested that in addition to binding to ‘CTCTGYTY’ motif, JMJ705 might be potentially recruited by WOX11 to loci without this motif. ChIP-qPCR analysis indicated that OxJMJ705 plants display reduced H3K27me3 levels at all JMJ705- and WOX11-binding sites except U2AF, LHY-Pw and LHY-Pz1 (Figure 5D). The reduction appeared more significant for those with higher (P < 0.01) than those with relative lower (P < 0.05) levels of H3K27me3 (Figure 5D).
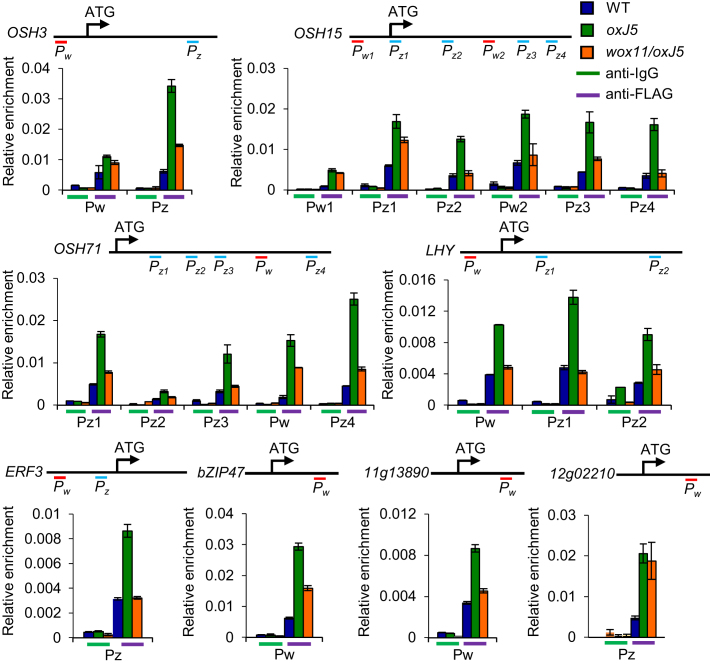
The observations that JMJ705 could be associated to WOX11-binding sites and reduced H3K27me3 from most of WOX11-binding loci and that wox11 mutation increased H3K27me3 levels of most of tested genes, led us to check whether WOX11 was involved in JMJ705 recruitment to common targets for H3K27me3 removal. We therefore tested JMJ705-binding to common target genes in wox11/oxJ5 plants by ChIP-qPCR. In OSH3, OSH15, OSH71, LHY and ERF3 that contained both the WOX11- and JMJ705-binding sites, the wox11 mutation clearly reduced JMJ705 association not only to WOX11-binding sites (i.e. OSH15-PW2, OSH71-PW and LHY-PW), but also to ‘CTCTGYTY’ motifs (i.e. OSH3-PZ, OSH15-PZ1, OSH15-PZ2, OSH15-PZ3, OSH15-PZ4, OSH71-PZ1, OSH71-PZ3, OSH71-PZ4, LHY-PZ1, LHY-PZ2 and ERF3-PZ) (Figure 6 and Supplementary Figure S8). In the two genes (bZIP47 and Os11g13890) that contain only the WOX11-binding site, JMJ705 was enriched on the loci and the wox11 mutation clearly reduced the enrichment (Figure 6 and Supplementary Figure S8). In contrast, JMJ705 enrichment on Os12g02210 locus was not affected, which contains only the JMJ705-binding site (Figure 6 and Supplementary Figure S8). The data suggested that WOX11 was involved in the recruitment of JMJ705 to target genes regardless of presence or absence of the ‘CTCTGYTY’ motif.
Figure 6.
WOX11 modulates JMJ705 binding to targeted genes. ChIP assays with anti-FLAG (anti-IgG as controls) of 5-day-old seedling chromatin isolated from the segregated wild type (WT), the JMJ705-FLAG in wild type background (oxJ5), and JMJ705-FLAG in wox11 background (wox11/oxJ5), from F2 of the crosses between wox11 and oxJ5–5. Seven representative genes were tested. OSH3, OSH15, OSH71, LHY and ERF3 contain both the WOX11 (Pw) and JMJ705-binding (Pz) sites, bZIP47 and Os11g13890 contain only the WOX11-binding site. The Os12g02210 locus that contains only JMJ705-binding site was used as an internal control. Gene structures, Pw and Pz sites were indicated. Three biological replicates of independent chromatin preparations were performed. One of the biological replicate is shown here. The other two biological replicates are in shown in Supplementary Figure S8. Y-axis means assay-site fold enrichment of the signal from immunoprecipitation over the background. Bar = means ± SD from three technical repetitions.
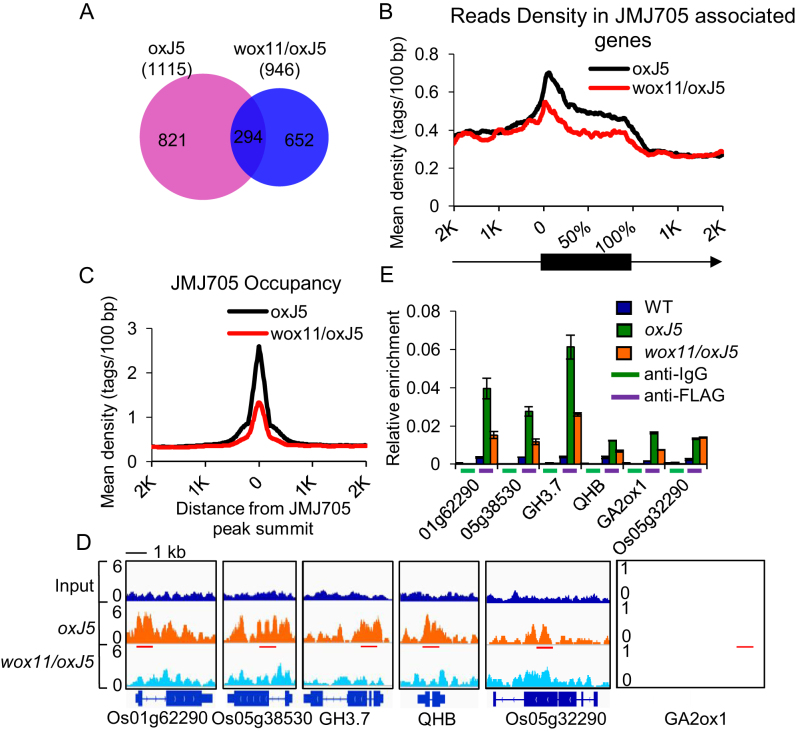
To further study WOX11 function in JMJ705 recruitment, we analyzed genome-wide binding sites of JMJ705–FLAG in shoot apex of oxJ5 and wox11/oxJ5 plants by ChIP-seq using anti-FLAG antibody. The analysis revealed 1551 peaks corresponding to 1115 genes enriched for JMJ705-binding in oxJ5 apex. The relatively smaller number of binding genes identified by anti-FLAG ChIP-seq might be due to the fact that the precipitated peak regions might mainly correspond to ectopic binding sites of JMJ705-FLAG. The JMJ705-FLAG-binding was enriched in gene body regions, showing a distribution pattern similar to that of H3K27me3 in rice (Figure 7B) (50). In wox11/oxJ5 plants, 1309 peaks corresponding to 946 genes were found to be bound by JMJ705-FLAG. Comparison of the two sets of data revealed 294 overlapping genes (Figure 7A). The wox11 mutation reduced the average densities of JMJ705-FLAG-binding in gene bodies (Figure 7B) and in the JMJ705-FLAG-binding peaks (Figure 7C), indicating that WOX11 was required for efficient binding of JMJ705-FLAG. GO analysis indicated that genes that showed loss of JMJ705-FLAG binding in wox11 background were enriched for regulatory function of gene transcription and metabolic processes (Supplementary Figure S9). Among these genes, we found that GH3.7, QHB, GA2ox1, and two heat shock protein family genes (Os01g62290 and Os05g38350) were down-regulated in wox11 and in jmj705 mutants (Supplementary Figure S10A), in addition to genes that are analyzed in Figures 5 and 6. These five genes showed a clear loss of JMJ705-FLAG-binding in wox11 background (Figure 7D), which was validated by ChIP-qPCR, in which Os05g32290 was used as a negative control (Figure 7E, Supplementary Figure S10B). Taking together, the ChIP-seq data suggest that WOX11 is required for efficient targeting of JMJ705 to genomic loci.
Figure 7.
WOX11-dependent recruitment of JMJ705 to genomic loci. (A) Venn diagram displaying significant overlaps between genes occupied by JMJ705 in oxJ5 and wox11/oxJ5. (B) Mean density of JMJ705 occupancy at all JMJ705-associated genes in oxJ5 (1115 genes) as compared to genes with wox11/oxJ5 (946). Numbers of sequenced tags (Y-axis) per 5% of the genic region (black box) or per 100 bp intervals in the 2-kb genomic region flanking the gene (line, X-axis) are shown. Arrow indicates the direction of transcription. (C) Mean density of JMJ705 occupancy at all JMJ705-associated sites (peaks found in oxJ5) in oxJ5 and wox11/oxJ5. The average signal within 2-kb genomic regions flanking the center of the JMJ705 peaks is shown. (D) The Integrative Genomics Viewer (IGV) views of JMJ705 signal on representative genes in oxJ5 and wox11/oxJ5. Gene structures and name are shown underneath. Red lines represent tested regions in ChIP-qPCR. The scale was identical for different tracks, and the Y-axis scales represent normalized signal of shifted merged MACS tag counts for every 10-bp window. Bar = 1 kb. (E) ChIP-qPCR detection of JMJ705 occupancy with anti-IgG (negative control) and anti-FLAG at genes loci listed in (D) in WT, oxJ5 and wox11/oxJ5 apex. Os05g32290 was used as a negative control. Y-axis means assay-site fold enrichment of the signal from immunoprecipitation over the background.
DISCUSSION
WOX11 functions in rice shoot development
The WOX family proteins are essential plant developmental regulators. Members of the WUS clade of the family have been shown to regulate stem cell maintenance and cell fate determination, while members of the WOX9 and WOX13 clades are incapable to sustain stem cells in the SAM (51). WOX11 belongs to the intermediate WOX9 clade. The present work indicates that in addition to promote shoot-borne crown root cell proliferation, WOX11 is also required for shoot development in rice. The expression pattern and the growth phenotypes of the mutant suggest that WOX11 has a general function to stimulate cell division and growth originated from shoot and root meristems.
It has been shown that several members of the WUS clade (WUS, WOX2 and WOX4) interact with the histone deacetylase (HDAC)-containing TOPLESS (TPL) complex and the loss of the WUS-TPL interaction impairs WUS function (17). In particular, WOX5, a key regulator of root meristem, recruits histone deacetylase HDA19 to repress cell differentiation program in root stem cells (18). These data indicate that recruiting repressive chromatin modifiers is a mechanism for WOX protein-regulated developmental gene repression. The present data showing recruitment of JMJ705 by WOX11 to remove H3K27me3 reveal a novel chromatin mechanism of WOX proteins-mediated gene activation. It is recently shown that WOX11 also interacts, through the homeodomain, with a histone acetylation complex to activate gene expression required for crown root cell proliferation (52). However, the present data suggest that JMJ705 is unlikely to be involved in the pathway of WOX11-controlled crown root development (Figure 3E). Thus, WOX11 may interact through different domains with distinct epigenetic regulators to establish permissive chromatin of gene expression required for cell proliferation during shoot and root growth.
Being expressed also in leaf primordia and young leaves, WOX11 function in shoot development is likely to be beyond its function in meristems. Our data suggests that activation of light-controlled energy metabolic production by WOX11 is an important functional aspect of the protein to stimulate SAM and shoot development, which is consistent with the role of light and metabolic signaling in SAM development (53–56). The observation that key transcription factors involved in circadian clock (LHY, CCT/B-box, RDD1) (Supplementary Figure S5 and Supplementary Dataset 1), are among the mis-expressed genes in wox11 suggests that WOX11 may act as a higher hierarchical regulator of shoot development programs that involves circadian regulation of gene expression and cellular and metabolic activities.
JMJ705-mediated removal of H3K27me3 is essential for WOX11-controlled shoot developmental programs
Chromatin reprogramming that leads to massive changes of gene expression is a prerequisite for cell differentiation during meristem development. The repressive mark H3K27me3 is shown to play a critical role in maintaining gene repression until a specific transition of plant development (51–53). Previous work suggests that H3K27me3 level is higher in rice vegetative SAM compared to young leaves and inflorescence meristem (27), suggesting that removal of the mark from many genes must occur during meristem transition and leaf/shoot development. The present work uncovers a mechanism of targeted removal of H3K27me3 from specific sets of genes involved in shoot development in rice. The data showing that WOX11/JMJ705-mediated H3K27me3 removal targets mostly transcription factors and energy metabolic genes corroborate with the recent results showing that these genes are particularly enriched for H3K27me3 in rice SAM and are reprogrammed during SAM to leaf/inflorescence transitions (27). The observation that JMJ705 over-expression could complement wox11 seedling defects supports the hypothesis that removal of H3K27me3 from common targets is essential for WOX11-controlled shoot development in rice. The significant overlaps between down- or up-regulated genes in wox11 and jmj705 mutants suggest that appropriate expression levels of the WOX11 and JMJ705 target genes are essential for proper SAM activity and shoot development.
The observation that a large numbers of genes were commonly up-regulated in the wox11 and jmj705 mutants suggests that WOX11 and JMJ705 may cooperate for gene repression in germinating seedlings. Considering that the JMJ705-mediated removal of H3K27me3 is supposed to mediate gene activation, the activation of genes in jmj705 mutant may be of an indirect effect, as the expression of many transcription factors was controlled by WOX11-JMJ705. Nevertheless it is not excluded that WOX11-JMJ705 may also mediate gene repression. Indeed, we have previously shown that WOX11 represses RR genes in crown root to stimulate cytokinin signaling (15,16).
Mechanism of H3K27me3 demethylases locus-specific recruitment
The Jumonji C (jmjC) group demethylases remove preferentially di- and tri-methylated histone lysines through ferrous ion (Fe (II)) and alpha-ketoglutaric acid-dependent oxidative reactions (57). JmjC proteins are classified into several phylogenetic subgroups, each of which demethylates specific lysine residues (58). Plant JmjC proteins are conserved with yeast and animal homologs, but there exist subgroups of plant-specific JmjC proteins (33). The mammalian JMJD3/UTX subgroup proteins exhibit H3K27me3 demethylase activities and have pleiotropic functions in gene expression required for different types of cell differentiation and oncogenesis (59,60), while a precise mechanism of recruiting JMJD3/UTX proteins to specific genes remains unclear in mammalian cells. The JMJD3/UTX group is not found in plants. Instead members from the plant JMJD2 subgroup, such as Arabidopsis REF6/JMJ12 and rice JMJ705, display H3K27me3 demethylase activity (26,30). REF6/JMJ12 has been shown to bind to ‘CTCTGYTY’ sequence via the ZnF domain of the protein (31,32). The observation that ‘CTCTGYTY’ is found in a very large number of genes in the rice genome suggests that additional factors are required to selectively target JMJ705 to specific genes in specific cell/tissue types during development. The present results indicate that, although this DNA-binding activity is conserved in rice JMJ705, interaction with a DNA-binding transcription factor is a key mechanism for JMJ705-mediated locus-specific removal of H3K27me3. The present work indicates that interaction with WOX11 not only facilitated JMJ705 binding to genes without the ‘CTCTGYTY’ motif, but also helped the protein to bind to the ‘CTCTGYTY’ motif. Likely, interaction with a DNA-binding transcription factor may allow precise and efficient targeting of JMJ705 to a specific set of genes, which may represent a general mechanism for JMJ705-mediated H3K27me3 removal and gene activation of specific developmental pathways.
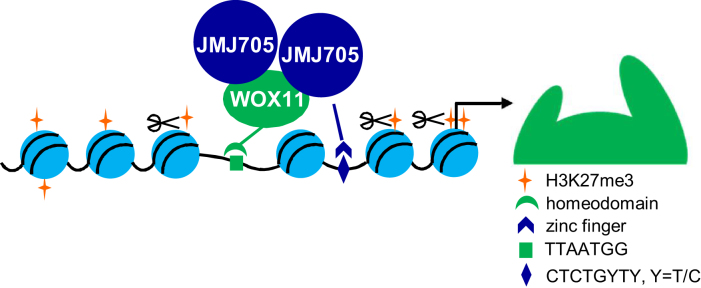
In conclusion, the present results show that WOX11 has a function to promote shoot growth in rice, which involves removal of H3K27me3 by recruiting the demethylase JMJ705 from genes involved in shoot developmental and energy generating pathways (Figure 8).
Figure 8.
A working model of WOX11 and JMJ705 regulated shoot development in rice.
DATA AVAILABILITY
Sequence data from this article can be found in Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: WOX11, Os11g47580; JMJ705, Os01g67970; OSH1, Os03g51690; OSH3, Os03g51710; OSH6, Os01g19694; OSH10, Os03g47016; OSH15, Os07g03770; OSH43, Os03g56110; OSH71, Os05g03884; LOG, Os01g40630; ERF3, Os12g41060; RDD1, Os01g15900; LHY, Os08g06110; HOX16, Os02g49700; bZIP47, Os06g15480; LSD1, Os08g06280; ATA15, Os05g05600; OsChl P, Os02g51080; U2AF, Os09g31482; GH3.7, Os06g30440; QHB, Os01g63510; GA2ox1, Os05g11810. The high throughput data are available under the GEO accession: GSE104306.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Qinghua Zhang for the mutant RNA-seq and ChIP-seq library construction and sequencing and helps from several members of the lab.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2016YFD0100802]; National Natural Science Foundation of China [31571256; 31730049]; Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2016RC003]; Postgraduate Science and Technology Innovation Project [2013YB04]. Funding for open access charge: National Key Research and Development Program of China [2016YFD0100802].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schoof H., Lenhard M., Haecker A., Mayer K.F., Jurgens G., Laux T.. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000; 100:635–644. [DOI] [PubMed] [Google Scholar]
- 2. Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U.. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005; 438:1172–1175. [DOI] [PubMed] [Google Scholar]
- 3. Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T.. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004; 131:657–668. [DOI] [PubMed] [Google Scholar]
- 4. Wu X.L., Dabi T., Weigel D.. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 2005; 15:436–440. [DOI] [PubMed] [Google Scholar]
- 5. Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T.. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007; 446:811–814. [DOI] [PubMed] [Google Scholar]
- 6. Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T.. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell. 2008; 14:867–876. [DOI] [PubMed] [Google Scholar]
- 7. Hirakawa Y., Kondo Y., Fukuda H.. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010; 22:2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ji J.B., Strable J., Shimizu R., Koenig D., Sinha N., Scanlon M.J.. WOX4 promotes procambial development. Plant Physiol. 2010; 152:1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suer S., Agusti J., Sanchez P., Schwarz M., Greb T.. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell. 2011; 23:3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van der Graaff E., Laux T., Rensing S.A.. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009; 10:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Somssich M., Je B., Simon R., Jackson D.. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016; 143:3238–3248. [DOI] [PubMed] [Google Scholar]
- 12. Tanaka W., Ohmori Y., Ushijima T., Matsusaka H., Matsushita T., Kumamaru T., Kawano S., Hirano H.Y.. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell. 2015; 27:1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohmori Y., Tanaka W., Kojima M., Sakakibara H., Hirano H.Y.. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell. 2013; 25:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dai M.Q., Hu Y.F., Zhao Y., Liu H.F., Zhou D.X.. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007; 144:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y., Hu Y.F., Dai M.Q., Huang L.M., Zhou D.X.. The WUSCHEL-Related Homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 2009; 21:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Y., Cheng S.F., Song Y.L., Huang Y.L., Zhou S.L., Liu X.Y., Zhou D.X.. The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell. 2015; 27:2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Causier B., Ashworth M., Guo W.J., Davies B.. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012; 158:423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pi L.M., Aichinger E., van der Graaff E., Llavata-Peris C.I., Weijers D., Hennig L., Groot E., Laux T.. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell. 2015; 33:576–588. [DOI] [PubMed] [Google Scholar]
- 19. Zhang X.Y., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E.. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007; 5:1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X.S., Zhou D.X.. Rice epigenomics and epigenetics: challenges and opportunities. Curr. Opin. Plant. Biol. 2013; 16:164–169. [DOI] [PubMed] [Google Scholar]
- 21. Makarevitch I., Eichten S., Waters A., Vaughn M., Springer N.M.. Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell. 2013; 25:780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kohler C., Villar C.B.R.. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008; 18:236–243. [DOI] [PubMed] [Google Scholar]
- 23. Zheng B.L., Chen X.M.. Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant. Biol. 2011; 14:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanchez M.D., Aceves-Garcia P., Petrone E., Steckenborn S., Vega-Leon R., Alvarez-Buylla E.R., Garay-Arroyo A., Garcia-Ponce B.. The impact of Polycomb group (PcG) and Trithorax group (TrxG) epigenetic factors in plant plasticity. New Phytol. 2015; 208:684–694. [DOI] [PubMed] [Google Scholar]
- 25. Charron J.B.F., He H., Elling A.A., Deng X.W.. Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell. 2009; 21:3732–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li T.T., Chen X.S., Zhong X.C., Zhao Y., Liu X.Y., Zhou S.L., Cheng S.F., Zhou D.X.. Jumonji C domain protein JMJ705-mediated removal of histone H3 Lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell. 2013; 25:4725–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu X.Y., Zhou S.L., Wang W.T., Ye Y.R., Zhao Y., Xu Q.T., Zhou C., Tan F., Cheng S.F., Zhou D.X.. Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell. 2015; 27:1428–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pien S., Grossniklaus U.. Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta. 2007; 1769:375–382. [DOI] [PubMed] [Google Scholar]
- 29. Liu X.Y., Zhou C., Zhao Y., Zhou S.L., Wang W.T., Zhou D.X.. The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 2014; 5:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu F.L., Cui X., Zhang S.B., Jenuwein T., Cao X.F.. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011; 43:715–719. [DOI] [PubMed] [Google Scholar]
- 31. Cui X., Lu F.L., Qiu Q., Zhou B., Gu L.F., Zhang S.B., Kang Y.Y., Cui X.K., Ma X., Yao Q.Q. et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 2016; 48:694–699. [DOI] [PubMed] [Google Scholar]
- 32. Li C.L., Gu L.F., Gao L., Chen C., Wei C.Q., Qiu Q., Chien C.W., Wang S.K., Jiang L.H., Ai L.F. et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016; 48:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Q.W., Zhou D.X.. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J.. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008; 56:505–516. [DOI] [PubMed] [Google Scholar]
- 35. Chen Q.F., Chen X.S., Wang Q., Zhang F.B., Lou Z.Y., Zhang Q.F., Zhou D.X.. Structural basis of a Histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet. 2013; 9:e1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang L.M., Sun Q.W., Qin F.J., Li C., Zhao Y., Zhou D.X.. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007; 144:1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X.F., Elling A.A., Li X.Y., Li N., Peng Z.Y., He G.M., Sun H., Qi Y.J., Liu X.S., Deng X.W.. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009; 21:1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zong W., Tang N., Yang J., Peng L., Ma S.Q., Xu Y., Li G.L., Xiong L.Z.. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 2016; 171:2810–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang L.J., Pei Z.Y., Tian Y.C., He C.Z.. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol. Plant Microbe Interact. 2005; 18:375–384. [DOI] [PubMed] [Google Scholar]
- 43. Iwamoto M., Higo K., Takano M.. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 2009; 32:592–603. [DOI] [PubMed] [Google Scholar]
- 44. Ogiso E., Takahashi Y., Sasaki T., Yano M., Izawa T.. The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 2010; 152:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rashid M., He G.Y., Yang G.X., Hussain J., Yan X.. AP2/ERF transcription factor in rice: genome-wide canvas and syntenic relationships between monocots and eudicots. Evol. Bioinform. Online. 2012; 8:321–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chi W.C., Chen Y.A., Hsiung Y.C., Fu S.F., Chou C.H., Trinh N.N., Chen Y.C., Huang H.J.. Autotoxicity mechanism of Oryza sativa: transcriptome response in rice roots exposed to ferulic acid. BMC Genomics. 2013; 14:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harrop T.W.R., Din I.U., Gregis V., Osnato M., Jouannic S., Adam H., Kater M.M.. Gene expression profiling of reproductive meristem types in early rice inflorescences by laser microdissection. Plant J. 2016; 86:75–88. [DOI] [PubMed] [Google Scholar]
- 48. Chen G.H., Liu C.P., Chen S.C.G., Wang L.C.. Role of ARABIDOPSIS A-FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2. J. Exp. Bot. 2012; 63:275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang P.Y., Li C.M., Wang Y., Huang R., Sun C.H., Xu Z.J., Zhu J.Q., Gao X.L., Deng X.J., Wang P.R.. Identification of a geranylgeranyl reductase gene for chlorophyll synthesis in rice. Springerplus. 2014; 3:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu Y.F., Liu D.N., Zhong X.C., Zhang C.J., Zhang Q.F., Zhou D.X.. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dolzblasz A., Nardmann J., Clerici E., Causier B., van der Graaff E., Chen J.H., Davies B., Werr W., Laux T.. Stem cell regulation by Arabidopsis WOX genes. Mol Plant. 2016; 9:1028–1039. [DOI] [PubMed] [Google Scholar]
- 52. Zhou S.L., Jiang W., Long F., Cheng S.F., Yang W.J., Zhao Y., Zhou D.X.. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell. 2017; 29:1088–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fleming A. Metabolic aspects of organogenesis in the shoot apical meristem. J. Exp. Bot. 2006; 57:1863–1870. [DOI] [PubMed] [Google Scholar]
- 54. Stasolla C. Glutathione redox regulation of in vitro embryogenesis. Plant Physiol. Biochem. 2010; 48:319–327. [DOI] [PubMed] [Google Scholar]
- 55. Schuster C., Gaillochet C., Medzihradszky A., Busch W., Daum G., Krebs M., Kehle A., Lohmann J.U.. A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell. 2014; 28:438–449. [DOI] [PubMed] [Google Scholar]
- 56. Schippers J.H., Foyer C.H., van Dongen J.T.. Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol. 2016; 29:121–128. [DOI] [PubMed] [Google Scholar]
- 57. Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y.. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006; 439:811–816. [DOI] [PubMed] [Google Scholar]
- 58. Mosammaparast N., Shi Y.. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010; 79:155–179. [DOI] [PubMed] [Google Scholar]
- 59. Bosselut R. Pleiotropic functions of H3K27Me3 demethylases in immune cell differentiation. Trends Immunol. 2016; 37:102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiao Z.G., Shen J., Zhang L., Li L.F., Li M.X., Hu W., Li Z.J., Cho C.H.. The roles of histone demethylase UTX and JMJD3 (KDM6B) in cancers: current progress and future perspectives. Curr. Med. Chem. 2016; 23:3687–3696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: WOX11, Os11g47580; JMJ705, Os01g67970; OSH1, Os03g51690; OSH3, Os03g51710; OSH6, Os01g19694; OSH10, Os03g47016; OSH15, Os07g03770; OSH43, Os03g56110; OSH71, Os05g03884; LOG, Os01g40630; ERF3, Os12g41060; RDD1, Os01g15900; LHY, Os08g06110; HOX16, Os02g49700; bZIP47, Os06g15480; LSD1, Os08g06280; ATA15, Os05g05600; OsChl P, Os02g51080; U2AF, Os09g31482; GH3.7, Os06g30440; QHB, Os01g63510; GA2ox1, Os05g11810. The high throughput data are available under the GEO accession: GSE104306.