ABSTRACT
Sphingobium sp. strain SYK-6 converts four stereoisomers of arylglycerol-β-guaiacyl ether into achiral β-hydroxypropiovanillone (HPV) via three stereospecific reaction steps. Here, we determined the HPV catabolic pathway and characterized the HPV catabolic genes involved in the first two steps of the pathway. In SYK-6 cells, HPV was oxidized to vanilloyl acetic acid (VAA) via vanilloyl acetaldehyde (VAL). The resulting VAA was further converted into vanillate through the activation of VAA by coenzyme A. A syringyl-type HPV analog, β-hydroxypropiosyringone (HPS), was also catabolized via the same pathway. SLG_12830 (hpvZ), which belongs to the glucose-methanol-choline oxidoreductase family, was isolated as the HPV-converting enzyme gene. An hpvZ mutant completely lost the ability to convert HPV and HPS, indicating that hpvZ is essential for the conversion of both the substrates. HpvZ produced in Escherichia coli oxidized both HPV and HPS and other 3-phenyl-1-propanol derivatives. HpvZ localized to both the cytoplasm and membrane of SYK-6 and used ubiquinone derivatives as electron acceptors. Thirteen gene products of the 23 aldehyde dehydrogenase (ALDH) genes in SYK-6 were able to oxidize VAL into VAA. Mutant analyses suggested that multiple ALDH genes, including SLG_20400, contribute to the conversion of VAL. We examined whether the genes encoding feruloyl-CoA synthetase (ferA) and feruloyl-CoA hydratase/lyase (ferB and ferB2) are involved in the conversion of VAA. Only FerA exhibited activity toward VAA; however, disruption of ferA did not affect VAA conversion. These results indicate that another enzyme system is involved in VAA conversion.
IMPORTANCE Cleavage of the β-aryl ether linkage is the most essential process in lignin biodegradation. Although the bacterial β-aryl ether cleavage pathway and catabolic genes have been well documented, there have been no reports regarding the catabolism of HPV or HPS, the products of cleavage of β-aryl ether compounds. HPV and HPS have also been found to be obtained from lignin by chemoselective catalytic oxidation by 2,3-dichloro-5,6-dicyano-1,4-benzoquinone/tert-butyl nitrite/O2, followed by cleavage of the β-aryl ether with zinc. Therefore, value-added chemicals are expected to be produced from these compounds. In this study, we determined the SYK-6 catabolic pathways for HPV and HPS and identified the catabolic genes involved in the first two steps of the pathways. Since SYK-6 catabolizes HPV through 2-pyrone-4,6-dicarboxylate, which is a building block for functional polymers, characterization of HPV catabolism is important not only for understanding the bacterial lignin catabolic system but also for lignin utilization.
KEYWORDS: Sphingobium, lignin, beta-aryl ether, glucose-methanol-choline oxidoreductase, aldehyde dehydrogenase
INTRODUCTION
Lignin, one of the major components of plant cell walls, is a complex phenolic heteropolymer produced from hydroxycinnamyl alcohols by radical coupling (1, 2). Lignin is the second most abundant bioresource on earth after cellulose and is expected to be used as an industrial raw material. However, the current industrial applications of lignin are limited to low-value applications such as the production of solid fuels and concrete additives (3, 4). On the other hand, it has been reported that the building blocks for functional polymers, such as 2-pyrone-4,6-dicarboxylic acid (5–7), cis,cis-muconic acid (8–10), and medium-chain-length polyhydroxyalkanoic acid (11), can be obtained from lignin-derived aromatic compounds such as vanillic acid, vanillin, ferulic acid, and p-coumaric acid through microbial catabolism. Therefore, the production of value-added chemicals from lignin through transformations systems that consist of chemical lignin decomposition and microbial catabolism of lignin-derived aromatics has attracted attention.
β-Aryl ether is the most abundant linkage in lignin, comprising 45 to 50% of all linkages in softwood lignin and 60 to 62% in hardwood lignin (4). Accordingly, degradation of this structure is considered a crucial step in lignin biodegradation. β-Aryl ether-type biaryls have two distinct isomeric forms, erythro and threo, each of which has enantiomeric forms (12). To date, the whole picture of the enzyme system for the cleavage of β-aryl ether in Sphingobium sp. strain SYK-6 has been determined (13). SYK-6 is able to degrade all the stereoisomers of the β-aryl ether-type biaryl, guaiacylglycerol-β-guaiacyl ether (GGE). In SYK-6 cells, four stereoisomers of GGE are converted to two enantiomers of α-(2-methoxyphenoxy)-β-hydroxypropiovanillone (MPHPV) through the oxidation of the GGE α-carbon atom catalyzed by Cα-dehydrogenases (LigD, LigL, and LigN) (Fig. 1) (14). LigD oxidizes (αR,βS)-GGE and (αR,βR)-GGE into (βS)-MPHPV and (βR)-MPHPV, respectively, while LigL/LigN converts (αS,βR)-GGE and (αS,βS)-GGE into (βR)-MPHPV and (βS)-MPHPV, respectively (14). The ether linkage of the resulting MPHPV is cleaved by enantioselective glutathione S-transferases (GSTs)—LigF, LigE, and LigP—to produce α-glutathionyl-β-hydroxypropiovanillone (GS-HPV) and guaiacol via nucleophilic attack of glutathione on the MPHPV β-carbon atom (15, 16). LigF and LigE/LigP attack (βS)-MPHPV and (βR)-MPHPV to produce (βR)-GS-HPV and (βS)-GS-HPV, respectively. Another GST, LigG, catalyzes the cleavage of the thioether linkage in (βR)-GS-HPV by transferring glutathione of (βR)-GS-HPV to another glutathione molecule to produce HPV and glutathione disulfide (15, 17). On the other hand, LigG had little to no activity with (βS)-GS-HPV, suggesting involvement of an alternative GST in the conversion of (βS)-GS-HPV (15, 18). Recently, further detailed biochemical characterization of the β-aryl ether catabolic enzymes of SYK-6 and their orthologs in other bacterial strains, and structural analyses of LigD, LigL, LigE, LigF, and LigG have been performed (17–29). In addition, Erythrobacter sp. strain SG61-1L and Novosphingobium sp. strain MBES04, which are capable of cleaving β-aryl ether, have recently been isolated, and similar enzyme systems have been characterized (23, 24).
FIG 1.
Proposed catabolic pathway of arylglycerol-β-aryl ether in Sphingobium sp. strain SYK-6. The pathway for both guaiacyl (R = H) and syringyl (R = OCH3)-type β-aryl ether compounds is shown. Enzymes: LigD, LigL, and LigN, Cα-dehydrogenases; LigF, LigE, and LigP, β-etherases; LigG, glutathione S-transferase; HpvZ, HPV oxidase; ALDHs, aldehyde dehydrogenases. Abbreviations: GGE, guaiacylglycerol-β-guaiacyl ether; MPHPV, α-(2-methoxyphenoxy)-β-hydroxypropiovanillone; GS-HPV, α-glutathionyl-β-hydroxypropiovanillone; HPV, β-hydroxypropiovanillone; HPS, β-hydroxypropiosyringone; VAL, vanilloyl acetaldehyde; SAL, 3-(4-hydroxy-3,5-dimethoxyphenyl)-3-oxopropanal; VAA, vanilloyl acetic acid; SAA, 3-(4-hydroxy-3,5-dimethoxyphenyl)-3-oxopropanoic acid; CoA, coenzyme A; VAA-CoA, CoA derivative of VAA.
Although many investigations of the cleavage of β-aryl ether have been performed, there are no reports on the characterization of the catabolism of HPV, the product of the cleavage of β-aryl ether. Therefore, in order to understand bacterial β-aryl ether catabolism, it is essential to elucidate the HPV catabolic system. Recently, the production of HPV and β-hydroxypropiosyringone (HPS; an intermediate metabolite of syringyl-type β-aryl ether) from lignin has been attempted through biological and chemical processes for the purpose of obtaining phenolic monomers from lignin (27, 30). Ohta et al. reported that HPV and HPS could be obtained from milled wood lignin from Japanese cedar (Cryptomeria japonica) and Eucalyptus globulus after reactions with MBES04 enzymes (27). Lancefield et al. reported an isolation method for HPV and HPS from Birch lignin via catalytic oxidation of the β-aryl ether linkage in lignin, followed by zinc-mediated cleavage of the ether bonds (30). By combining these decomposition methods with microbial catabolism of HPV and HPS, the development of a production system for value-added chemicals from lignin is expected.
In the present study, we determined the catabolic pathway of HPV and HPS in SYK-6 and characterized the genes involved in the first two steps of the pathway.
RESULTS AND DISCUSSION
Determination of the pathway for the catabolism of HPV and HPS in Sphingobium sp. strain SYK-6.
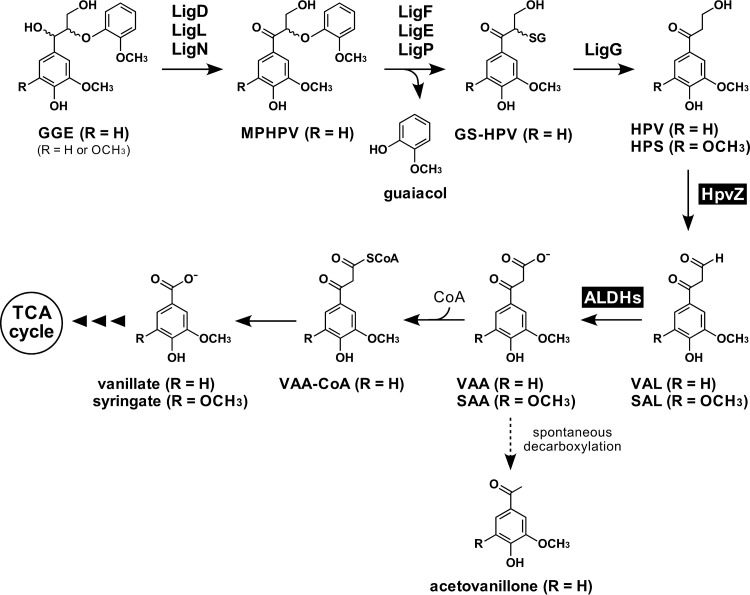
In order to determine the catabolic pathway of HPV in SYK-6, intermediate metabolites generated during the incubation of HPV with resting cells of SYK-6 were identified. Resting cells of SYK-6 grown in Wx minimal medium (31) containing 10 mM sucrose, 10 mM glutamate, 0.13 mM methionine, and 10 mM proline (Wx-SEMP) were incubated with 1 mM HPV for 6 h, and the reaction mixtures were analyzed by high-performance liquid chromatography-mass spectrometry (HPLC-MS). This analysis indicated that HPV was converted into compound I with a retention time of 3.1 min (Fig. 2B). Based on a comparison of the retention time and the m/z value of the deprotonated ion of compound I to those of the authentic sample, this compound was identified as vanillic acid (molecular weight [MW], 168) (Fig. 2C and see Fig. S1A and B in the supplemental material). Next, a cell extract (>10 kDa) of SYK-6 cells grown in Wx-SEMP was incubated with 200 μM HPV for 24 h. HPLC-MS analysis of the reaction mixture showed that HPV was converted into compound II with a retention time of 2.9 min (Fig. 2E). Positive electrospray ionization (ESI)-MS analysis of compound II showed a major fragment at m/z 298 (Fig. 2F). Based on the MW deduced from the major fragment ion and additives in the reaction mixture, compound II was identified as an imine derivative of vanilloyl acetaldehyde (VAL), 2-((3-hydroxy-3-(4-hydroxy-3-methoxyphenyl)allylidene)amino)-2-(hydroxymethyl)propane-1,3-diol or its oxazolidine product (VAL-Tris; MW, 297; see Fig. S1C and D in the supplemental material). It is known that some aldehyde substrates, such as glyceraldehyde 3-phosphate, acetaldehyde, and benzaldehyde, react with Tris to form imine product, which is then trapped by one of the free hydroxyl groups forming an oxazolidine product (32). Furthermore, Fukuzumi et al. attempted to synthesize VAL by Claisen-Wislicenus hydroxymethylene condensation between acetovanillone and ethyl formate in the presence of metallic sodium; however, cis-vanilloyl vinyl alcohol, a tautomer of VAL, was obtained as a major product (33). It was thought that VAL had five possible tautomers, and the form of hydroxyl vinyl ketone structure (vanilloyl vinyl alcohol) was stable (33). These observations suggested that VAL-Tris (imine product) was produced from HPV through oxidation of Cγ-alcohol catalyzed by the SYK-6 cell extract to generate VAL and to result in the isomerization of VAL and condensation between a VAL isomer and Tris in the reaction buffer. VAL-Tris (imine product) was then possibly converted to the oxazolidine product.
FIG 2.
HPLC-MS analysis of HPV metabolites. Resting cells of SYK-6 (OD600 of 5.0) and SYK-6 cell extracts (>10 kDa; 500 μg of protein/ml) were incubated with 1 mM HPV (A and B) and 200 μM HPV (D and E), respectively. The same cell extracts (>10 kDa) were incubated with HPV in the presence of 500 μM NAD+ (G and H) and in the presence of 500 μM NAD+ + 2 mM CoA + 2.5 mM MgSO4 + 2.5 mM ATP (J and K). Portions of the reaction mixtures were collected at the start (A and D) and after 2 h (G and J), 6 h (B), and 24 h (E, H, and K) of incubation and then analyzed by HPLC. The ESI-MS spectra of compounds I (negative mode), II (positive mode), III (negative mode), and IV (positive mode) are shown in panels C, F, I, and L, respectively. The asterisk (*) in panel I indicates an unidentified MS fragment that appeared between retention times of 2.0 and 7.0 min in the HPLC chromatogram (H).
When a SYK-6 cell extract (>10 kDa) was incubated with 200 μM HPV in the presence of 500 μM NAD+ for 2 h, an accumulation of compound III with a retention time of 4.7 min was observed (Fig. 2G). Negative ESI-MS analysis of compound III showed fragments at m/z 209 and 165 (Fig. 2I). Since these fragments seemed to represent the deprotonated ion and its decarboxylated ion, respectively, compound III was identified as vanilloyl acetic acid (VAA; MW, 210; see Fig. S1E and F in the supplemental material). VAL seems to be transformed to VAA by NAD+-dependent aldehyde dehydrogenase(s) (ALDHs) (Fig. 1). In addition, when the same reaction mixture was incubated for 24 h, compound IV with a retention time of 5.8 min was generated (Fig. 2H). A comparison of the retention time and m/z value of the protonated ion with those of the authentic sample indicated that compound IV was acetovanillone (MW, 166; Fig. 2L and see Fig. S1G and H in the supplemental material). Previously, Niwa and Saburi reported that VAA was spontaneously decarboxylated to acetovanillone (Fig. 1) (34).
Because lignin-derived aromatic acids such as ferulate, p-coumarate, and caffeate are catabolized via coenzyme A (CoA)-dependent pathways, we predicted that VAA is catabolized through its Cγ activation by CoA (35–39). A SYK-6 cell extract (>10 kDa) was therefore incubated with 200 μM HPV in the presence of 500 μM NAD+, 2 mM CoA, 2.5 mM MgSO4, and 2.5 mM ATP. After incubation for 2 h, a decrease in HPV and an accumulation of VAA (compound III) were observed (Fig. 2J). After further incubation for 24 h, vanillic acid (compound I) and acetovanillone (compound IV) were observed (Fig. 2K). Vanillic acid was generated only when CoA, ATP, and MgSO4 were present. This result strongly suggested that VAA was converted to vanillate through the CoA derivative of VAA (Fig. 1).
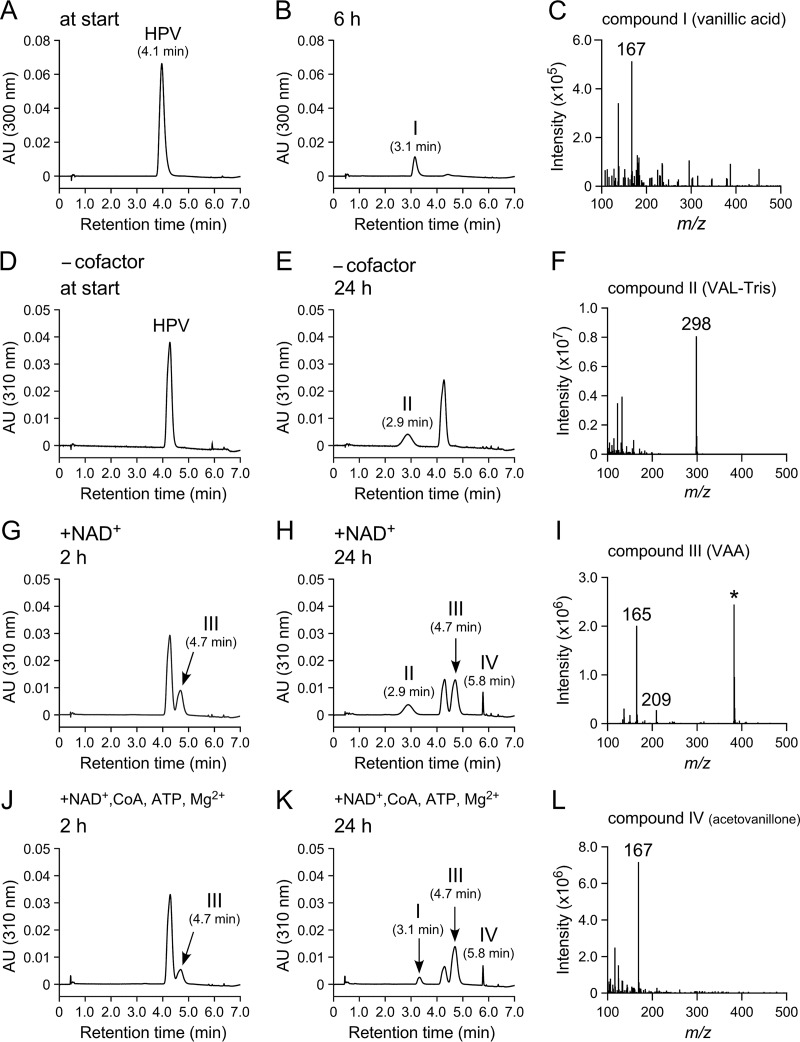
Similarly, we examined the SYK-6 catabolic pathway of HPS. Resting SYK-6 cells grown in Wx-SEMP were incubated with 1 mM HPS for 4 h, and the reaction mixtures were analyzed by HPLC-MS. This analysis indicated that HPS was converted into compound V with a retention time of 2.6 min (Fig. 3B). Negative ESI-MS analysis of compound V showed a major fragment at m/z 197 (Fig. 3C). Based on a comparison of the retention time and m/z value of the deprotonated ion with those of the authentic sample, compound V was identified as syringic acid (MW, 198; see Fig. S1I and J in the supplemental material). In order to clarify the more detailed catabolic pathway of HPS, cell extracts (>10 kDa) prepared from SYK-6 cells grown in Wx-SEMP were incubated with 200 μM HPS for 16 h. An accumulation of compound VI with a retention time of 1.8 min was observed (Fig. 3E). Negative ESI-MS analysis of compound VI showed a major fragment at m/z 326 (Fig. 3F), suggesting the formation of an imine derivative of 3-(4-hydroxy-3,5-dimethoxyphenyl)-3-oxopropanal (SAL), 2-((3-hydroxy-3-(4-hydroxy-3,5-dimethoxyphenyl)allylidene)amino)-2-(hydroxymethyl)propane-1,3-diol, or its oxazolidine product (SAL-Tris; MW, 327).
FIG 3.
HPLC-MS analysis of HPS metabolites. SYK-6 resting cells (OD600 of 5.0) and SYK-6 cell extracts (>10 kDa; 500 μg of protein/ml) were incubated with 1 mM HPS (A and B) and 200 μM HPS (D and E), respectively. The same cell extract (>10 kDa) was incubated with HPS in the presence of 500 μM NAD+ (G and H). Portions of the reaction mixtures were collected at the start (A, D, and G) and after 4 h (B) and 16 h (E and H) of incubation and then analyzed by HPLC. The negative-ion ESI-MS spectra of compounds V, VI, and VII are shown in panels C, F, and I, respectively.
In the presence of 500 μM NAD+, the same cell extract (>10 kDa) converted HPS into SAL-Tris (compound VI) and compound VII with a retention time of 3.3 min (Fig. 3H). Negative ESI-MS analysis of compound VII showed fragments at m/z 239 and 195, which seemed to represent the deprotonated ion and its decarboxylated ion, respectively (Fig. 3I). From these results, compound VII was identified as 3-(4-hydroxy-3,5-dimethoxyphenyl)-3-oxopropanoic acid (designated SAA; MW, 240). These results indicate that HPS was oxidized to SAA via SAL and may be degraded by the same enzyme system involved in HPV catabolism (Fig. 1).
Basic properties of the enzyme involved in the conversion of HPV.
In order to characterize the enzymes involved in the catabolism of HPV in SYK-6, cofactor requirements and induction profiles of the HPV-transforming activities in SYK-6 were examined. The effects of the addition of 1-methoxy-5-methylphenazinium methylsulfate (PMS), flavin adenine dinucleotide (FAD) + PMS, and NAD+ on the enzyme activities in converting 200 μM HPV were investigated. When an extract of SYK-6 cells grown in Wx-SEMP was incubated with HPV in the presence of PMS, the extract showed 1.8-fold-higher activity (4.2 ± 0.7 nmol · min–1 · mg–1) than that in the absence of cofactors (2.4 ± 0.8 nmol · min–1 · mg–1). On the other hand, the addition of FAD or NAD+ had no effect on the activity. These results suggested that an oxidase(s) that requires an electron acceptor is involved in HPV oxidation. The HPV-oxidizing activity of an extract of SYK-6 cells grown with GGE was also measured in the presence of PMS. However, no activation was observed, suggesting the constitutive expression of the gene(s) responsible for the oxidation of HPV.
Isolation of the gene involved in the conversion of HPV.
A cosmid library of SYK-6 constructed in Sphingomonas sanguinis IAM 12578 was screened for clones capable of degrading HPV. Of the 1,000 clones tested, three transconjugants degraded HPV, and cosmids pSA53, pSA88, and pSA684 were isolated. Southern hybridization analysis of the cosmid clones using SalI-digested pSA53 as a probe suggested that a 3.6-kb SalI fragment, a 2.0-kb SalI fragment, and two 1.0-kb SalI fragments were commonly present in the above cosmids. Subcloning and nucleotide sequencing showed that these SalI fragments were present in a 17.9-kb DNA fragment that contained 13 genes corresponding to SLG_12790 through to SLG_12910. In this fragment, SLG_12830 revealed 36 to 39% amino acid sequence identity with the glucose-methanol-choline (GMC) oxidoreductase family enzymes, including (i) 3-(2-(4-hydroxy-3-methoxyphenyl)-3-(hydroxymethyl)-7-methoxy-2,3-dihydrobenzofuran-5-yl)acrylic acid (DCA-C) oxidases (PhcC and PhcD) involved in the catabolism of dehydrodiconiferyl alcohol (DCA) in SYK-6 (40), (ii) AlkJ, which is involved in the oxidation of primary alcohols to aldehydes in Pseudomonas putida GPo1 (41), and (iii) polyethylene glycol dehydrogenase (PegA) of Sphingopyxis terrae (42).
The gene product of SLG_12830 catalyzes oxidation of HPV.
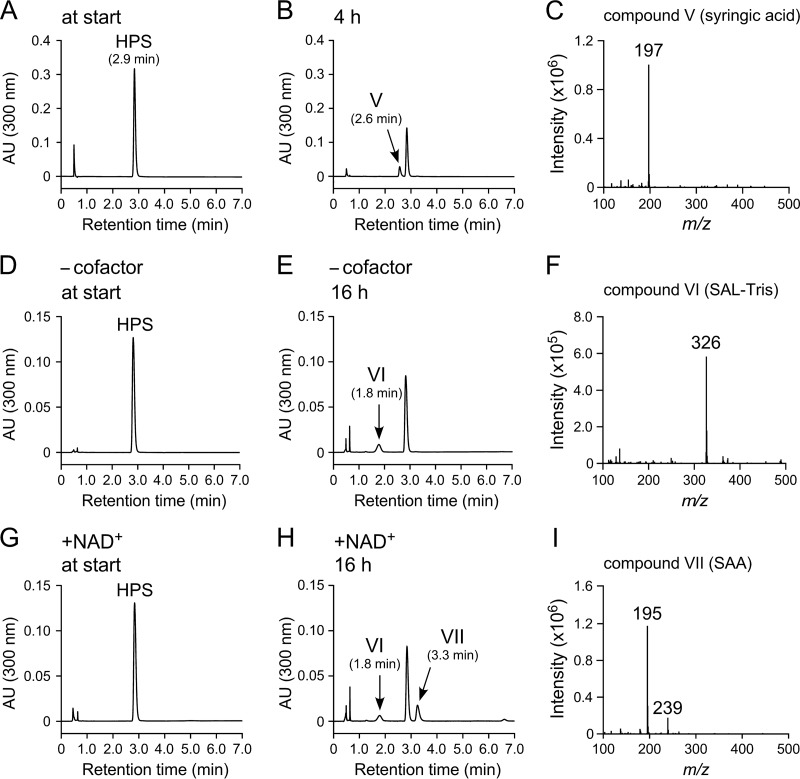
SLG_12830 fused with a His tag at the 5′ terminus was coexpressed in Escherichia coli with the trigger factor chaperone. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of a cell extract prepared from E. coli harboring pCold12830 and pTf16 (hpvZ-expressing E. coli) showed the expression of SLG_12830 (see Fig. S2 in the supplemental material). In order to determine the reaction product, resting cells of the E. coli transformant were incubated with 200 μM HPV in Tris-HCl buffer. HPLC-MS analysis showed that HPV was almost completely converted and that VAL-Tris and compound VIII with a retention time of 5.4 min were produced (Fig. 4A). When the same incubation was performed in water, a significant amount of compound VIII was observed without the generation of VAL-Tris (Fig. 4B). Positive ESI-MS analysis of compound VIII showed a major fragment at m/z 195 (Fig. 4C), suggesting that compound VIII was VAL (MW, 194). These results indicated that the gene product of SLG_12830 has the ability to oxidize HPV into VAL.
FIG 4.
Function and role of hpvZ in SYK-6. (A and B) Conversion of 200 μM HPV by resting cells of E. coli harboring pCold12830 and pTf16 in Tris-HCl buffer (pH 7.5) (A) and water (B). Portions of the reaction mixtures were collected after 6 h of incubation and analyzed by HPLC. The ESI-MS spectra of compound VIII (positive mode) are shown in panel C. (D to F) Conversions of 200 μM HPV (D), GGE (E), and HPS (F) by resting cells of SYK-6 (open symbols) and SME059 (closed symbols). Circles, triangles, and diamonds indicate the concentrations of HPV, GGE, and HPS, respectively. (G) Complementation of SME059 with pJB12830. Cells of SYK-6 harboring pJB864 (circles), SME059 harboring pJB864 (triangles), and SME059 harboring pJB12830 (diamonds) were incubated with 200 μM HPV. Experiments shown in panels D to G were performed in triplicate, and the data represent averages ± the standard deviations.
Role of SLG_12830 in HPV and HPS catabolism.
In order to examine whether SLG_12830 is indeed involved in the conversion of HPV in SYK-6, an SLG_12830 mutant (SME059) was created (see Fig. S3A and B in the supplemental material). The ability of SME059 to convert HPV was assessed using resting cells. SME059 was no longer able to convert HPV, whereas the wild type completely converted 200 μM HPV within 3 h (Fig. 4D). When GGE was used as a substrate, the conversion rates of the wild-type and SME059 strains were almost identical (Fig. 4E). However, only SME059 accumulated HPV at a concentration approximately equimolar to the added GGE (Fig. 4E). In addition, SME059 also completely lost the ability to convert HPS (Fig. 4F). The HPV conversion defect of SME059 was complemented by the introduction of pJB12830 carrying SLG_12830 (Fig. 4G). These results demonstrated that GGE is catabolized through HPV in SYK-6 and that SLG_12830 is essential for the catabolism of HPV and HPS; thus, we designated this gene hpvZ.
Cellular localization of HpvZ.
In order to determine the cellular localization of HpvZ, HPV-transforming activities of soluble and membrane fractions of SYK-6 cells were compared. The HPV transforming activity in the cytoplasmic and membrane fractions were estimated to be 0.6 ± 0.1 nmol · min−1 (7.5 mg of protein) and 0.5 ± 0.1 nmol · min–1 (0.9 mg of protein), respectively, based on the results that the ratio of the amount of proteins in the soluble and membrane fractions was 75:9.4 (40). These results indicated that HpvZ is localized to cytoplasm and cytoplasmic membrane. Similarly, the GMC oxidoreductase family proteins, PhcC and PhcD from SYK-6, and PegA from S. terrae have been suggested to localize to both the soluble and membrane fractions (40, 43). Another GMC oxidoreductase family protein, AlkJ from P. putida GPo1, and glucose dehydrogenase from Pseudomonas fluorescens, were localized to the membrane (41, 44). Since there are no predicted signal sequences or hydrophobic transmembrane segments in the deduced amino acid sequence of HpvZ, this enzyme appears to be a peripheral cytoplasmic membrane protein like other membrane-associated GMC oxidoreductase family enzymes.
Enzyme properties of HpvZ.
Cell extracts prepared from hpvZ-expressing E. coli were fractionated into the soluble and membrane fractions. SDS-PAGE of both fractions showed the expression of hpvZ (see Fig. S2 in the supplemental material). The specific activity for HPV of the membrane fraction was estimated to be 26 ± 6 nmol · min–1 · mg–1. However, the soluble fraction showed no activity even in the presence of PMS. The membrane fraction was treated with each of 10 different detergents, and the solubilized HpvZ was obtained from the membrane fractions treated with n-dodecylphosphocholine and 5-cyclohexyl-1-pentyl-β-d-maltoside. However, purified HpvZ was not obtained due to the lack of adsorption of the enzyme to a nickel affinity column. Therefore, the enzyme properties of HpvZ were examined using the membrane fraction prepared from extracts of hpvZ-expressing E. coli cells.
The optimum pH and temperature for the activity of HpvZ were determined to be pH 8.5 to 9.0 and at 40 to 45°C, respectively (see Fig. S4 in the supplemental material).
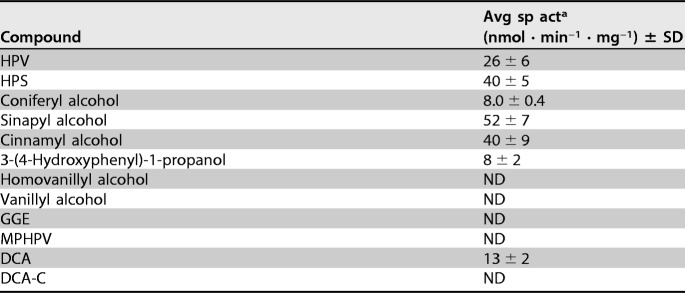
The substrate range of HpvZ was examined using 200 μM HPV, HPS, coniferyl alcohol, sinapyl alcohol, cinnamyl alcohol, 3-(4-hydroxyphenyl)-1-propanol, homovanillyl alcohol, vanillyl alcohol, GGE, MPHPV, DCA, and DCA-C (see Fig. S5 in the supplemental material). HPLC analyses of the reaction mixtures indicated that HpvZ showed no activity for a C6–C2 monomeric alcohol (homovanillyl alcohol), a C6–C1 monomeric alcohol (vanillyl alcohol), and lignin-derived biaryls, including GGE, MPHPV, and DCA-C (Table 1). On the other hand, HpvZ showed activities toward all of the C6–C3 monomeric alcohols (3-phenyl-1-propanol derivatives; Table 1). Generally, GMC oxidoreductase family enzymes act on hydroxyl groups of alcohols, carbohydrates, or sterols (45). For example, an aryl-alcohol oxidase from Pleurotus eryngii is able to oxidize a variety of aromatic alcohols and aldehydes, including coniferyl alcohol and cinnamyl alcohol (46). On the contrary, PhcC and PhcD specifically oxidize the alcohol group at Cγ of the A-ring side chain of DCA-C and DCA, although PhcC has a weak activity for coniferyl alcohol (40). HpvZ was able to oxidize the alcohol group at Cγ of the B-ring side chain of DCA, which is different from the regiospecificity of PhcC and PhcD (data not shown).
TABLE 1.
Substrate range of HpvZ
a The membrane fraction of hpvZ-expressing E. coli (300 μg of protein/ml) was incubated with 200 μM substrate in the presence of 500 μM PMS. The data represent the averages ± standard deviations of three independent experiments. ND, not detected.
Among the enzymes belonging to the GMC oxidoreductase family, an FAD-binding domain (GMC_oxred_N; Pfam PF00732) is conserved in the N-terminal region. This domain includes the typical GxGxxG/A sequence motif, which is indicative of the Rossmann fold involved in binding the ADP moiety of FAD (47). A substrate-binding domain is also conserved in the C-terminal region, although this domain is less conserved. In addition, an active-site histidine, which can assist in substrate oxidation and FAD reoxidation by molecular oxygen, is generally conserved (45, 47). These domains and the residue are also conserved in HpvZ (see Fig. S6 in the supplemental material). To identify the flavin cofactor in HpvZ, a supernatant obtained by heat treating the membrane fraction containing HpvZ was analyzed by HPLC. However, a significant peak was not observed. Furthermore, the specific activities in the presence of FAD (26 ± 8 nmol · min–1 · mg–1) or flavin mononucleotide (27 ± 1 nmol · min–1 · mg–1) were almost equivalent to that in the absence of any flavin cofactors (26 ± 6 nmol · min–1 · mg–1). GMC oxidoreductase family enzymes generally contain either covalently or noncovalently bound FAD (45). Choline oxidase from Arthrobacter globiformis (48) and pyranose dehydrogenase (AmPDH) from Agaricus meleagris (49) have covalently bound FAD. Our results suggested that HpvZ may also covalently bind to FAD. Moreover, although purified inactive HpvZ was obtained from the soluble fraction of the hpvZ-expressing E. coli cells, UV-visible spectra of the enzyme showed no absorption at 454 nm, indicating the absence of flavin cofactor in the enzyme (data not shown). This result suggested that HpvZ produced in the E. coli cytoplasm lacked FAD as a prosthetic group.
In vivo electron acceptor of HpvZ.
AlkJ and PegA are able to use ubiquinone (CoQ10) and its derivatives (CoQ0 and CoQ1) as electron acceptors for the oxidation of their substrates (41, 42). PhcC and PhcD have also been shown to be able to use CoQ0 and CoQ1 as electron acceptors, as well as PMS (40). Furthermore, electron transport from AlkJ to cytochrome c in the presence of CoQ1 has been observed (41). Therefore, the electrons that are removed from the substrate by AlkJ, PegA, and PhcC/PhcD are thought to be transferred to the respiratory chain. Based on these observations, we predicted that HpvZ could use ubiquinone as an electron acceptor in the oxidation of HPV. When using a membrane fraction of SYK-6 cells harboring pJB12830 (hpvZ-expressing SYK-6), HpvZ showed 1.2- and 1.6-fold-higher specific activities in the presence of CoQ0 (27 ± 2 nmol · min–1 · mg–1) and CoQ1 (36 ± 6 nmol · min–1 · mg–1), respectively, than that in the absence of cofactors (22 ± 1 nmol · min–1 · mg–1). The specific activity in the presence of CoQ1 was almost equivalent to that obtained using PMS (40 ± 3 nmol · min–1 · mg–1). These results suggested that HpvZ is able to use ubiquinone as an electron acceptor in vivo. However, no increase in HpvZ activity was observed when the membrane fraction of hpvZ-expressing E. coli was used. The difference in the increase in HpvZ activities between the membrane fractions of hpvZ-expressing SYK-6 and E. coli cells in the presence of ubiquinone derivatives may be caused by a difference in the abundance of ubiquinone in the membrane fractions.
Identification of the ALDH genes responsible for the conversion of VAL.
To obtain more information on the properties of the enzymes involved in the conversion of VAL in SYK-6, cofactor requirements of the enzyme activity were examined. Cell extracts (>10 kDa) of SYK-6 grown in lysogeny broth (LB) were incubated with 200 μM HPV in the presence of FAD, NAD+, and NADP+ for 2 h. In the presence of NAD+ and NADP+, VAA accumulated to levels of 95 ± 9 μM and 86 ± 6 μM, respectively, while the accumulation of VAA was not observed in the presence of FAD. These results suggested that NAD+/NADP+-dependent ALDHs play a major role in VAL oxidation.
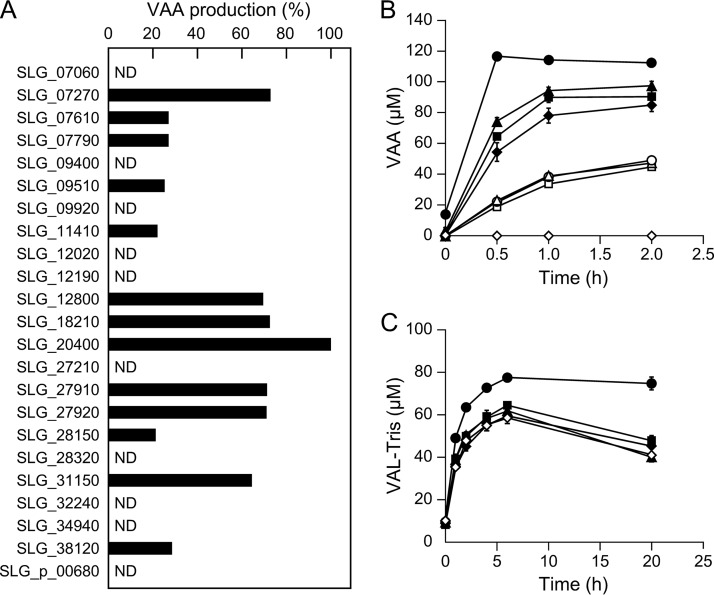
Previously, 23 ALDH genes were predicted to be present in the SYK-6 genome (50, 51). To examine the ability of the putative 23 ALDH gene products to oxidize VAL, these genes were expressed in E. coli using expression plasmids constructed in a previous study (51). SDS-PAGE showed sufficient gene expression, except for SLG_32240 and SLG_34940 (see Fig. S7 in the supplemental material). Since HPV was converted to VAL-Tris during incubation with HpvZ in Tris-HCl buffer, we used HEPES buffer (pH 7.5) and sodium phosphate buffer (pH 7.5) to prepare VAL from HPV using HpvZ. However, HPV was converted to unknown products in HEPES buffer, and the reaction product was not detected in sodium phosphate buffer (data not shown). In addition, the HPV conversion rate was significantly decreased in HEPES buffer (approximately 60%) and sodium phosphate buffer (approximately 50%). Therefore, we examined the ability of these 21 ALDHs to convert VAL by measuring the amount of VAA produced from HPV when HPV was reacted with both HpvZ and each ALDH in Tris-HCl buffer (pH 7.5). Resting cells of E. coli expressing each ALDH gene and hpvZ-expressing E. coli were mixed and incubated with 100 μM HPV for 12 h. When resting cells of E. coli harboring a vector and the hpvZ-expressing E. coli were incubated with HPV, VAA was not generated, whereas VAL-Tris accumulated. On the other hand, when E. coli cells carrying each of the 13 ALDH genes (SLG_07270, SLG_07610, SLG_07790, SLG_09510, SLG_11410, SLG_12800, SLG_18210, SLG_20400, SLG_27910 [bzaA], SLG_27920 [bzaB], SLG_28150, SLG_31150, and SLG_38120) were used instead of the control vector, HPV was converted into VAA (Fig. 5A). Of the 13 ALDH genes, when incubating with E. coli carrying SLG_07270, SLG_12800, SLG_18210, SLG_20400, bzaA, bzaB, and SLG_31150, larger amounts of VAA accumulated (Fig. 5A). We therefore measured the VAA production time course using E. coli carrying these seven ALDH genes and the hpvZ-expressing E. coli. Of these, E. coli carrying SLG_07270, SLG_12800, SLG_20400, and bzaB produced a greater amount of VAA than E. coli carrying one of the three other ALDH genes (Fig. 5B). Specifically, when using E. coli carrying SLG_20400, the amount of VAA produced was the greatest, and no VAL-Tris was detected at any of the sampling points (Fig. 5B).
FIG 5.
Identification of ALDH genes involved in VAL conversion. (A) Resting cells of E. coli carrying each of the 23 SYK-6 ALDH genes (OD600 of 1.0) were incubated with 100 μM HPV in the presence of resting cells of E. coli harboring pCold12830 and pTf16 (OD600 of 5.0). The amounts of VAA produced in the reaction mixtures containing each of the ALDH gene-expressing cells are shown as the relative ratio to that in the reaction mixture containing SLG_20400-expressing cells. ND, VAA was not detected. (B) Time course of the production of VAA during incubation of 100 μM HPV with cells of E. coli harboring pCold12830 and pTf16 (OD600 of 10.0) and E. coli carrying the following ALDH genes (OD600 of 1.0): SLG_20400 (●), SLG_07270 (▲), bzaB (■), SLG_12800 (◆), SLG_18210 (○), bzaA (△), and SLG_31150 (□). E. coli cells harboring pET-21a(+) were used as a negative control (♢). (C) Accumulation of VAL-Tris during incubation of 1 mM HPV with cells of E. coli harboring pCold12830 and pTf16 (OD600 of 10.0) and the following mutants of the ALDH genes (OD600 of 0.5): SME061 (ΔSLG_20400; ●), SME045 (ΔbzaB; ■), SME092 (ΔSLG_07270; ▲), and SME031 (ΔSLG_12800; ◆). SYK-6 cells were used as a control (♢). These experiments were performed in triplicate, and the data represent averages ± the standard deviations.
In order to examine whether SLG_07270, SLG_12800, SLG_20400, and bzaB are involved in the conversion of VAL in SYK-6, SLG_07270 mutant (SME092) and SLG_20400 mutant (SME061) were created (see Fig. S3C to F in the supplemental material). Resting cells of SME092, SME061, and the previously created SLG_12800 mutant (SME031) and bzaB mutant (SME045) were incubated with 1 mM HPV in the presence of hpvZ-expressing E. coli cells. SME061 accumulated a 1.8-fold greater amount of VAL-Tris than the wild type or the other mutants after 20 h of incubation (Fig. 5C). These results suggested that SLG_20400 is involved in VAL oxidation. However, SME061 accumulated only 75 μM VAL-Tris at 20 h of incubation. Therefore, multiple ALDHs, including SLG_20400, appear to be involved in the conversion of VAL. Our previous phylogenetic analysis of the 23 ALDH genes in SYK-6 and other known ALDH genes indicated that 13 SYK-6 ALDH genes, the products of which showed VAL oxidation activities, are phylogenetically diverse (51). SLG_20400 clustered with calB, which encodes coniferyl aldehyde dehydrogenase from Pseudomonas sp. strain HR199, which shared 33% amino acid sequence identity with SLG_20400 (51, 52). The involvement of multiple ALDH genes in SYK-6 was also shown in the oxidation of vanillin, syringaldehyde, and an intermediate metabolite of DCA (DCA-L) (50, 51). Another example of the involvement of multiple ALDH genes in the conversion of vanillin has also been reported for P. putida KT2440 (53). Since ALDHs exhibit broad substrate ranges in general, multiple ALDHs are likely to play roles in the oxidation of aromatic aldehydes to their acids.
Candidate genes for the catabolism of VAA.
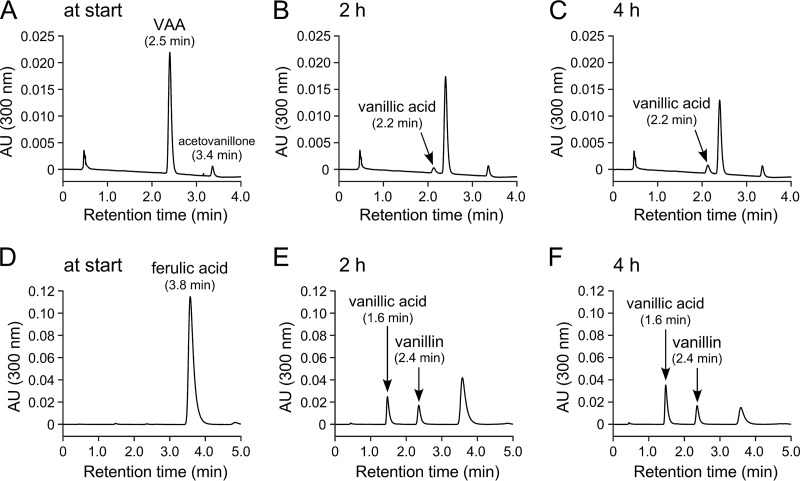
In a previous report, Palamuru et al. detected vanillin as a metabolite when SYK-6 cells were incubated with GGE (24). However, vanillin was not observed during the conversion of HPV (Fig. 2). In order to examine whether vanillin is an actual intermediate in HPV catabolism, resting cells of a desV ligV double mutant (SME077), which has a weak ability to convert vanillin (51), were incubated with 100 μM VAA or 1 mM ferulate. Only the accumulation of vanillate and acetovanillone was observed at any of the sampling points (1, 2, 4, 6, and 24 h) in the mixture for the VAA conversion, whereas the mixture for the ferulate conversion accumulated a significant amount of vanillin in addition to vanillate (Fig. 6; chromatograms at 2 and 4 h of incubation are shown). In addition, when a cell extract (>10 kDa) of SME077 was incubated with 100 μM VAA in the presence of CoA, MgSO4, and ATP, only the accumulation of vanillate and acetovanillone was observed (see Fig. S8 in the supplemental material). These results suggested that VAA was catabolized to vanillate without passing through vanillin as an intermediate.
FIG 6.
Conversion of VAA by a desV ligV double mutant. Resting cells of the desV ligV double mutant (SME077) were incubated with 100 μM VAA (A to C) and 1 mM ferulate (D to F). Portions of the reaction mixtures were collected at the start (A and D) and after 2 h (B and E) and 4 h (C and F) of incubation and then analyzed by HPLC using the two different analytical conditions described in Materials and Methods. The retention times of vanillin separated under the analytical conditions for the reaction mixtures of VAA (A to C) and ferulate (D to F) were 2.9 and 2.4 min, respectively.
We hypothesized that the feruloyl-CoA synthetase gene (ferA) and feruloyl-CoA hydratase/lyase genes (ferB and ferB2) may be involved in VAA catabolism based on the structural similarity between VAA and ferulic acid (36). These genes were adequately expressed in E. coli (see Fig. S9 in the supplemental material). When crude FerA was incubated with 100 μM VAA in the presence of CoA, MgSO4, and ATP for 60 min, compound IX with a retention time of 1.7 min was generated (see Fig. S10B in the supplemental material). Negative ESI-MS analysis of compound IX showed fragments at m/z 959 ([M − H]−) and 479 ([M − 2H]2−), suggesting that compound IX was the CoA derivative of VAA (VAA-CoA; MW, 960) (see Fig. S10E in the supplemental material). However, no other peak except VAA-CoA was observed when crude enzymes of FerA + FerB and FerA + FerB2 were incubated with VAA, respectively (see Fig. S10C and D in the supplemental material). Therefore, FerB and FerB2 appear to be not involved in the conversion of VAA-CoA.
In order to examine whether ferA is indeed involved in the conversion of VAA in SYK-6, resting cells of a previously created ferA mutant (SME009) grown in LB were incubated with 100 μM VAA. SME009 showed a higher conversion rate for VAA than that of the wild type (see Fig. S11 in the supplemental material), suggesting that ferA is not essential for the catabolism of VAA. The reason for the high conversion rate of VAA of SME009 is not clear but disruption of ferA may cancel the substrate competition between FerA and (an) unidentified true VAA-converting enzyme(s).
Recently, the catabolic pathway of p-hydroxycinnamate derivatives, such as dihydroferulate, ferulate, and p-coumarate in Rhodococcus jostii RHA1, were characterized (37). In this pathway, dihydroferulate was catabolized to vanillate via VAA-CoA. VAA-CoA was converted into vanillate and acetyl-CoA by the gene product of couO (ro0512), which encodes 4-hydroxyphenoxy-β-ketoacyl-CoA hydrolase. CouO was predicted to be a zinc-dependent metalloenzyme belonging to amidohydrolase superfamily. Orthologs of couO, showing 53 to 58% amino acid sequence identity, have also been found and characterized in Agrobacterium fabrum C58 (Atu1421) (38) and Corynebacterium glutamicum (phdC) (39). In the SYK-6 genome, we found SLG_12680, which exhibited 50% amino acid sequence identity with CouO. We are currently investigating the function of the gene product of SLG_12680 and exploring the actual VAA-converting enzyme gene.
Genome search for orthologs of hpvZ in other bacteria.
Since HpvZ is essential for the catabolism of HPV and HPS, the presence of this gene determines whether bacteria can utilize the A-ring portion of β-aryl ether compounds. BLAST searches of hpvZ were carried out to determine the distribution of its orthologs among bacteria. The hpvZ orthologs that showed high amino acid sequence identity (62%–92%) were found in Altererythrobacter sp. strain Root672 (ASD76_15935), Altererythrobacter atlanticus (WYH_02786), Erythrobacter sp. strain SG61-1L (SZ64_15220), Sphingomonas hengshuiensis WHSC-8 (TS85_07880), and Sphingobium sp. strain 66-54 (BGP16_16810). All of these bacteria possess orthologs of the genes responsible for the conversion of GGE into HPV. Among these strains, SG61-1L was reported to be able to utilize GGE as the sole source of carbon and energy (24). In contrast, Novosphingobium sp. strain MBES04 accumulated HPV from GGE (23). Consistent with this observation, MBES04 possesses GMC oxidoreductase family enzyme genes, whose products showed less than 37% amino acid sequence identity with HpvZ. Similarly, no hpvZ orthologs were found in Novosphingobium sp. strain PP1Y and Novosphingobium aromaticivorans DSM 12444, which possess orthologs of the genes responsible for converting GGE into HPV. Due to the lack of hpvZ, PP1Y and DSM 12444 also appear to be able to utilize only the portion of the B-ring of β-aryl ether compounds as carbon and energy sources.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
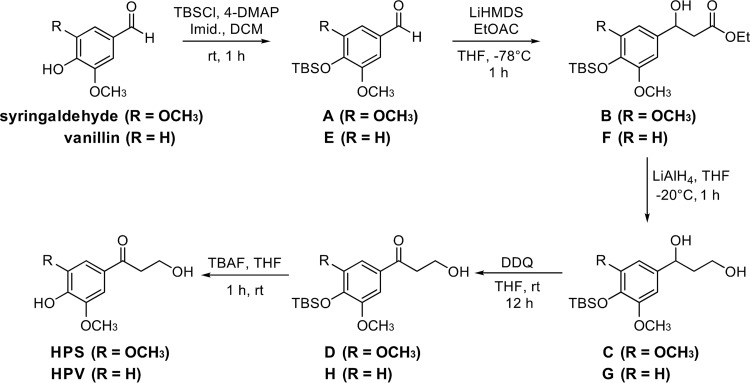
The strains and plasmids used in this study are listed in Table 2. Sphingobium sp. strain SYK-6 and its mutants were grown in LB, Wx-SEMP, and Wx-SEMP containing 5 mM GGE at 30°C. S. sanguinis IAM 12578 was grown in LB at 30°C. When necessary, 50 mg kanamycin/liter, 100 mg streptomycin/liter, or 300 mg carbenicillin/liter were added to the cultures. E. coli strains were grown in LB at 37°C. For cultures of cells carrying antibiotic resistance markers, the media for E. coli transformants were supplemented with 100 mg ampicillin/liter, 25 mg kanamycin/liter, or 12.5 mg chloramphenicol/liter. The synthesis of each HPS and HPV compound is described in detail below (Fig. 7).
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Sphingobium sp. | ||
| SYK-6 | Wild type; Nalr Smr | 65 |
| SME009 | SYK-6 derivative; ferA::kan; Nalr Smr Kmr | 36 |
| SME031 | SYK-6 derivative; SLG_12800::kan; Nalr Smr Kmr | 66 |
| SME045 | SYK-6 derivative; bzaB::tet; Nalr Smr Tcr | 67 |
| SME059 | SYK-6 derivative; hpvZ::kan; Nalr Smr Kmr | This study |
| SME061 | SYK-6 derivative; ΔSLG_20400; Nalr Smr | This study |
| SME077 | SYK-6 derivative; desV::cat ligV::kan; Nalr Smr Cmr Kmr | 51 |
| SME092 | SYK-6 derivative; SLG_07270::kan; Nalr Smr Kmr | This study |
| Sphingomonas sanguinis IAM 12578 | Nalr | 68 |
| Escherichia coli | ||
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3); T7 RNA polymerase gene under the control of the lacUV5 promoter | 69 |
| HB101 | recA13 supE44 hsd20 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 70 |
| NEB 10-beta | araD 139 Δ(ara-leu)7697 fhuA lacX74 galK (ϕ80 ΔlacZ ΔM15) recA1 endA1 nupG rpsL (Smr) Δ(mrr-hsdRMS-mcrBC) | New England Biolabs |
| Plasmids | ||
| pVK100 | Broad-host-range cosmid vector; Kmr Tcr | 62 |
| pRK2013 | Tra+ Mob+ ColE1 replicon; Kmr | 71 |
| pT7Blue | Cloning vector; Apr | Novagen |
| pBluescript II KS(+) and SK(+) | Cloning vector; Apr | 72 |
| pUC19 | Cloning vector; Apr | 73 |
| pET-16b | Expression vector; T7 promoter; Apr | Novagen |
| pET-21a(+) | Expression vector; T7 promoter; Apr | Novagen |
| pCold I | Expression vector; cspA promoter; Apr | TaKaRa Bio |
| pTf16 | Expression vector for tig; araB promoter; Cmr | TaKaRa Bio |
| pK18mobsacB | oriT sacB; Kmr | 74 |
| pK19mobsacB | oriT sacB; Kmr | 74 |
| pIK03 | KS(+) with a 1.3-kb EcoRV fragment carrying kan of pUC4K; Apr Kmr | 75 |
| pJB864 | RK2 broad-host-range expression vector; Apr Cbr Pm xylS | 76 |
| pAK405 | Plasmid for allelic exchange and markerless gene deletions in sphingomonads; Kmr | 64 |
| pSA53 | pVK100 with partially SalI-digested fragments of SYK-6 carrying hpvZ | This study |
| pSA88 | pVK100 with partially SalI-digested fragments of SYK-6 carrying hpvZ | This study |
| pSA684 | pVK100 with partially SalI-digested fragments of SYK-6 carrying hpvZ | This study |
| pT7B12830 | pT7Blue with a 1.7-kb PCR amplified fragment carrying hpvZ | This study |
| pCold12830 | pCold I with a 1.7-kb NdeI-BamHI fragment carrying hpvZ from pT7B12830 | This study |
| pBH37F | SK(+) with a 3.7-kb HindIII fragment carrying hpvZ from pSA53 | This study |
| pUC12830 | pUC19 with a 2.0-kb SalI fragment of pBH37F | This study |
| pUC12830K | pUC12830 with a 1.3-kb SalI-BamHI fragment of pIK03 carrying kan | This study |
| pKmb12830K | pK19mobsacB with a 3.1-kb SalI fragment of pUC12830K | This study |
| pJB12830 | pJB864 with a 2.3-kb BamHI-SacII (blunted) fragment carrying hpvZ from pBH37F | This study |
| pLVH | pET-21a(+) with a 1.9-kb NdeI-XhoI fragment carrying ligV | 77 |
| pT21-0727 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_07270 | 51 |
| pT21-0761 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_07610 | 51 |
| pT21-0779 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_07790 | 51 |
| pT21-0940 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_09400 | 51 |
| pT21-0951 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_09510 | 51 |
| pT21-0992 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_09920 | 51 |
| pT21-1141 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_11410 | 51 |
| pT21-1202 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_12020 | 51 |
| pT21-1219 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_12190 | 51 |
| pT21-1280 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_12800 | 51 |
| pT21-1821 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_18210 | 51 |
| pT21-2040 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_20400 | 51 |
| pT21-2721 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_27210 | 51 |
| pT21-2791 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying bzaA | 51 |
| pT21-2792 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying bzaB | 51 |
| pT21-2815 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_28150 | 51 |
| pT21-2832 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_28320 | 51 |
| pT21-3115 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_31150 | 51 |
| pT21-3224 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_32240 | 51 |
| pT21-3494 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_34940 | 51 |
| pT21-3812 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_38120 | 51 |
| pT21-p0068 | pET-21a(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_p_00680 | 51 |
| pKS0727 | KS(+) with a 1.5-kb NdeI-BamHI fragment carrying SLG_07270 | 51 |
| pKmb07270 | pK18mobsacB with a 1.5-kb HindIII-XbaI carrying SLG_07270 from pKS0727 | This study |
| pKmb07270K | pKmb07270 with a 1.3-kb EcoRV fragment carrying kan from pIK03 into NruI site of SLG_07270 | This study |
| pAK20400 | pAK405 with a 2.3-kb deletion cassette carrying up- and downstream regions of SLG_20400 | This study |
| pE16FA | pET-16b with a 2.2-kb NdeI-BamHI fragment carrying ferA | 31 |
| pE16FB | pET-16b with a 0.9-kb NdeI-BamHI PCR amplified fragment carrying ferB | This study |
| pE16FB2 | pET-16b with a 1.1-kb NdeI-BamHI PCR amplified fragment carrying ferB2 | This study |
Kmr, Nalr, Smr, Apr, Tcr, Cmr, and Cbr, resistance to kanamycin, nalidixic acid, streptomycin, ampicillin, tetracycline, chloramphenicol, and carbenicillin, respectively.
FIG 7.
Synthetic routes to HPS and HPV.
Synthesis of HPS (S-Hibbert-Westwood-Lancefield ketone). (i) Synthesis of 4-((tert-butyldimethylsilyl)oxy)-3,5-dimethoxybenzaldehyde (compound A).
To synthesize compound A, 4-dimethylaminopyridine (4-DMAP; 13.4 g, 109.8 mmol, 1.0 eq) and imidazole (Imid; 14.9 g, 219.6, 2.0 eq) were added to a stirring solution of syringaldehyde (20.0 g, 109.8 mmol, 1.0 eq) in dichloromethane (DCM; 600 ml, c = 0.18 M). The resulting mixture was allowed to stir for 5 min, and then tert-butyldimethylchlorosilane (TBSCl; 17.3 g, 115.3 mmol, 1.1 eq) was added. The mixture was stirred at room temperature for 1 h. After the reaction had reached completion, it was neutralized with a saturated aqueous solution of NH4Cl (2 × 300 ml). The organic layer was further washed with water (500 ml) and brine (300 ml), dried with MgSO4, filtered, and concentrated in vacuo. Purification by silica gel chromatography using 5 to 10% ethyl acetate in petroleum ether afforded compound A as a white solid (26.6 g, 89.8 mmol, 82%). Analytical data for compound A agreed with those reported previously (54). 1H NMR (500 MHz, CDCl3) δ 9.79 (s, 1H), 7.07 (s, 2H), 3.84 (s, 6 H), 0.98 (s, 9H), 0.13 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 191.1, 152.0, 140.7, 129.4, 106.7, 55.8, 25.8, 18.9, −4.46.
(ii) Synthesis of ethyl 3-(4-((tert-butyldimethylsilyl)oxy)-3,5-dimethoxyphenyl)-3-hydroxypropanoate (compound B).
Ethyl acetate (4.2 g, 47.5 mmol, 1.3 eq) was added to a cooled solution of lithium bis(trimethylsilyl)amide (LiHMDS; 1.0 M in tetrahydrofuran [THF]; 47.5 ml, 47.5 mmol, 1.3 eq) in THF (100 ml) at −78°C. After 15 min, a solution of compound A (10.8 g, 36.6 mmol, 1.0 eq) in THF (20 ml, overall c = 0.30 M) was added at −78°C, and the resulting mixture was left to stir at this temperature for 1 h. After the reaction had reached completion, it was quenched with saturated aqueous solution of NH4Cl (2 × 300 ml) and extracted with ethyl acetate (500 ml). The organic layer was further washed with water (500 ml) and brine (300 ml), dried with MgSO4, filtered, and concentrated in vacuo. Purification by silica gel chromatography using 10 to 20% ethyl acetate in petroleum ether afforded compound B as a light-yellow solid (12.5 g, 32.7 mmol, 89%). 1H NMR (500 MHz, CDCl3) δ 6.51 (s, 2H), 5.00 (dt, J = 9.0, 3.5 Hz, 1H), 4.13 (q, J = 7.0 Hz, 2H), 3.74 (s, 6H), 3.36 (m, 1H), 2.70 (dd, J = 16.0, 9.0 Hz, 1H), 2.63 (dd, J = 16.0, 3.5 Hz, 1H), 1.22 (t, J = 7.0 Hz, 3H), 0.97 (s, 9H), 0.08 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 172.4, 151.5, 135.2, 133.6, 102.5, 70.6, 60.8, 55.7, 43.7, 25.8, 18.7, 14.2, −4.7.
(iii) Synthesis of 1-(4-((tert-butyldimethylsilyl)oxy)-3,5-dimethyoxyphenyl)propane-1,3-diol (compound C).
A suspension of LiAlH4 (1.4 g, 37.8 mmol, 2.2 eq) in THF (100 ml) was cooled to −20°C. After 15 min, a solution of compound B (6.6 g, 17.2 mmol, 1.0 eq) in THF (20 ml, overall c = 0.14 M) was added at −20°C, and the resulting mixture was left to stir at this temperature for 1 h. After the reaction had reached completion, it was poured slowly into a stirring mixture of ethyl acetate (300 ml) and saturated aqueous solution of Na2S2O3 (2 × 300 ml). The organic layer was washed brine (300 ml), dried with MgSO4, filtered, and concentrated in vacuo. Purification by silica gel chromatography using 5 to 10% methanol in DCM afforded compound C as a colorless oil (5.62 g, 16.4 mmol, 96%). 1H NMR (500 MHz, CDCl3) δ 6.49 (s, 2H), 4.79 (dd, J = 8.5, 3.5 Hz, m, 1H), 3.77 (m, 2H), 3.75 (s, 6H), 3.32 (s, 1H), 2.93 (s, 1H), 1.94 (dddd, J = 14.0, 8.5, 7.0, 5.0 Hz, 1H), 1.84 (ddt, J = 14.0, 5.5, 4.0 Hz, 1H), 0.98 (s, 9H), 0.09 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 151.6, 137.2, 133.5, 102.6, 74.6, 61.5, 55.8, 40.6, 25.9, 18.8, −4.6.
(iv) Synthesis of 1-(4-((tert-butyldimethylsilyl)oxy)-3,5-dimethyoxyphenyl)-3-hydroxypropan-1-one (compound D).
2,3-Dichloro-5,6-dicyanobenzoquinone (DDQ) (4.1 g, 18.1 mmol, 1.05 eq) was added to a stirring solution of compound C (5.6 g, 16.4 mmol, 1.0 eq) in THF (160 ml, c = 0.10 M) at 22°C, and the resulting mixture was left to stir at this temperature for 12 h. Afterward, the reaction mixture was diluted with ethyl acetate (300 ml) and washed with saturated aqueous solution of Na2S2O3 (2 × 300 ml). The organic layer was washed with water (300 ml) and brine (300 ml), dried with MgSO4, filtered, and concentrated in vacuo. Purification by silica gel chromatography using 2 to 5% methanol in DCM afforded compound D as a white solid (5.1 g, 14.9 mmol, 91%). 1H NMR (500 MHz, CDCl3) δ 7.16 (s, 2H), 3.97 (app q, J = 5.5 Hz, 2H), 3.80 (s, 6H), 3.15 (t, J = 5.5 Hz, 2H), 3.01 (app t, J = 6.0 Hz, 1H), 0.97 (s, 9H), 0.11 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 199.1, 151.4, 139.9, 129.4, 105.6, 58.3, 55.8, 40.0, 25.7, 18.8, −4.6.
(v) Synthesis of HPS.
Tetrabutylammonium fluoride (TBAF; 1.0 M in THF, 8.8 ml, 8.8 mmol, 3.0 eq) was added to a stirring solution of compound D (1.0 g, 2.9 mmol, 1.0 eq) in THF (15 ml, c = 0.20 M) at 22°C, and the resulting mixture was left to stir at this temperature for 1 h. Afterward, the reaction mixture was diluted with ethyl acetate (300 ml) and washed with saturated aqueous solution of NH4Cl (2 × 300 ml). The organic layer was washed water (300 ml) and brine (300 ml), dried with MgSO4, filtered, and concentrated in vacuo. The crude material was recrystallized in petroleum ether and washed with a minimum amount of ethyl acetate to afford compound HPS as a white solid (0.5 g, 2.3 mmol, 77%). Spectroscopic data agreed with those reported previously (30). High-resolution MS [M − H]+ calculated. For C11H13O5 225.0800; found 225.0766; 1H NMR (500 MHz, DMSO-d6) δ 9.34 (s, 1H), 7.24 (s, 2H), 4.59 (s, 1H), 3.83 (s, 6H), 3.77 (t, J = 6.5 Hz, 2H), 3.10 (t, J = 6.5 Hz, 2H); 13C NMR (125 MHz, DMSO-d6) δ 197.4, 147.6, 140.8, 127.5, 106.0, 57.3, 56.1, 41.0.
Synthesis of HPV (G-Hibbert-Westwood-Lancefield ketone). (i) Synthesis of 4-((tert-butyldimethylsilyl)oxy)-3-methoxybenzaldehyde (compound E).
Compound E was synthesized using the same experimental procedure as that described for the synthesis of compound A. To vanillin (19.2 g, 126.0 mmol, 1.0 eq) in DCM (500 ml, c = 0.25 M) was added 4-DMAP (15.4 g, 126.0 mmol, 1.0 eq), imidazole (17.1 g, 252.0 mmol, 2.0 eq), and TBSCl (1.2 eq). Purification by silica gel chromatography using 5 to 10% ethyl acetate in petroleum ether afforded compound E as a colorless oil (30.0 g, 113.1 mmol, 90%). The spectroscopic data were in agreement with previously reported findings (55). 1H NMR (400 MHz, CDCl3) δ 9.73 (s, 1H), 7.27 (m, 2H), 6.85 (d, J = 8.0 Hz, 1H), 3.74 (s, 3H), 0.90 (s, 9H), 0.09 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 190.5, 151.4, 151.0, 130.9, 125.8, 120.5, 110.0, 55.1, 25.4, 18.3, −4.78.
(ii) Synthesis of ethyl 3-(4-((tert-butyldimethylsilyl)oxy)-3-methoxyphenyl)-3-hydroxypropanoate (compound F).
Compound F was synthesized using the same experimental procedure as that described for the synthesis of compound B. Compound E (10.9 g, 41.0 mmol, 1.0 eq) in THF (140 ml, c = 0.29 M), LiHMDS (1.0 M in THF; 53.2 ml, 53.2 mmol, 1.3 eq), and ethyl acetate (4.7 g, 53.2, 1.3 eq) were combined. Purification by silica gel chromatography using 10 to 30% ethyl acetate in petroleum ether afforded compound F as a light-yellow oil (11.9 g, 33.6 mmol, 81%). 1H NMR (500 MHz, CDCl3) δ 6.84 (d, J = 1.5 Hz, 1H), 6.72 (m, 2H), 4.98 (dt, J = 5.0, 3.5 Hz, 1H), 4.09 (q, J = 7.0 Hz, 2H), 3.73 (s, 3H), 3.50 (m, 1H), 2.68 (dd, J = 16.0, 9.5 Hz, 1H), 2.59 (dd, J = 16.0, 4.0 Hz, 1H), 1.18 (t, J = 7.0 Hz, 3H), 0.95 (s, 9H), 0.09 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 172.3, 150.9, 144.4, 136.3, 120.6, 118.0, 109.5, 70.2, 60.7, 55.3, 43.6, 25.7, 18.4, 14.1, −4.7.
(iii) Synthesis of 1-(4-((tert-butyldimethylsilyl)oxy)-3-methyoxyphenyl)propane-1,3-diol (compound G).
Compound G was synthesized using the same experimental procedure as that described for the synthesis of compound C. LiAlH4 (1.6 g, 40.1 mmol, 2.2 eq) in THF (80 ml) and compound F (6.6 g, 18.6 mmol, 1.0 eq) in THF (20 ml, overall c = 0.18 M) were combined. Purification by silica gel chromatography using 5 to 10% methanol in DCM afforded compound G as a light-yellow oil (5.4 g, 17.4 mmol, 94%). 1H NMR (500 MHz, CDCl3) δ 6.89 (d, J = 1.5 Hz, 1H), 6.80 (m, 2H), 4.86 (m, 1H), 3.81 (s, 3H), 3.23 (s, 1H), 2.88 (s, 1H), 2.00 (m, 1H), 1.88 (m, 1H), 1.01 (s, 9H), 0.16 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 151.0, 144.4, 138.1, 120.8, 118.1, 109.6, 74.3, 61.5, 55.6, 40.6, 25.8, 18.6, −4.5.
(iv) Synthesis of 1-(4-((tert-butyldimethylsilyl)oxy)-3-methyoxyphenyl)-3-hydroxypropan-1-one (compound H).
Compound H was synthesized using the same experimental procedure as that described for the synthesis of compound D. DDQ (4.3 g, 19.2 mmol, 1.05 eq) and compound G (5.4 g, 17.4 mmol, 1.0 eq) in THF (170 ml, c = 0.10 M) were combined. Purification by silica gel chromatography using 2 to 5% methanol in DCM afforded compound H as a light-yellow oil (4.1 g, 13.3 mmol, 76%). 1H NMR (400 MHz, CDCl3) δ 7.43 to 7.35 (m, 2H), 6.82 (d, J = 8.0 Hz, 1H), 3.95 (t, J = 5.5 Hz, 2H), 3.79 (s, 3H), 3.12 (t, J = 5.5 Hz, 2H), 0.94 (s, 9H), 0.13 (s, 6H); 13C NMR (125 MHz, CDCl3) δ 199.1, 151.1, 150.4, 130.8, 122.7, 120.3, 110.9, 58.2, 55.4, 40.0, 25.6, 18.5, −4.6.
(v) Synthesis of HPV.
HPV was synthesized using the same experimental procedure as that described for the synthesis of HPS. TBAF (1.0 M in THF; 15.6 ml, 15.6 mmol, 1.2 eq) and compound H (4.13 g, 13.3 mmol, 1.0 eq) in THF (100 ml, c = 0.13 M) were combined. Purification by silica gel chromatography afforded HPV as a white solid (1.0 g, 5.1 mmol, 38%). Spectroscopic analysis was in agreement with the reference (30). High-resolution MS [M + Na]+ calculated. For C10H12O4Na 196.0700; found 219.0625; 1H NMR (500 MHz, CDCl3) δ 7.58 to 7.50 (m, 2H), 6.95 (d, J = 8.0 Hz, 1H), 6.12 (s, 1H), 4.01 (q, J = 5.0 Hz, 2H), 3.95 (s, 3H), 3.18 (t, J = 5.0 Hz, 2H), 2.73 (app t, J 5.9 = Hz, 1H); 13C NMR (125 MHz, CDCl3) δ 199.2, 151.0, 146.9, 129.7, 123.8, 114.1, 109.7, 58.5, 56.2, 39.9.
Preparation of other substrates.
MPHPV, DCA, and DCA-C were prepared as described previously (50, 56). For preparation of VAA, HPV was added into 15 ml of the cell suspensions of hpvZ-expressing E. coli cells (optical density at 600 nm [OD600] of 10.0) and SLG_20400-expressing E. coli cells (OD600 of 2.0) to a final concentration of 200 μM. After incubation with shaking for 12 h at 30°C, the culture was centrifuged, and the supernatant was filtered using an Amicon Ultraspin filter unit (3-kDa cutoff; Millipore). The resulting filtrate was used as a preparation of 200 μM VAA. For the preparation of VAL-Tris, HPV was added to 15 ml of the cell suspension of S. sanguinis IAM 12578 harboring pJB12830 cells (OD600 of 1.0) to a final concentration of 1 mM. After incubation with shaking for 45 h at 30°C, the culture was centrifuged, and the supernatant was collected. The supernatant was extracted with ethyl acetate, and then the extract was finally dissolved in the dimethyl sulfoxide. The compounds obtained were analyzed by HPLC-MS. Other aromatic compounds were purchased from Tokyo Chemical Ind., Co., Ltd.; Sigma-Aldrich Co., Llc.; and Wako Pure Chemical Ind., Ltd.
Preparation of resting cells and cell extracts and fractionation of cell extracts.
Cells of SYK-6 and its mutants grown in Wx-SEMP for 16 h or LB for 24 h were collected by centrifugation (5,000 × g for 5 min) and then washed twice with 50 mM Tris-HCl buffer (pH 7.5; buffer A). The cells were resuspended in the same buffer and used as resting cells. Cells were broken by an ultrasonic disintegrator (57), and the supernatants of cell lysates were obtained as cell extracts after centrifugation (19,000 × g for 15 min). To examine the cofactor requirements, cell extracts were filtered using an Amicon Ultraspin filter unit (10-kDa cutoff; Millipore) and then washed five times buffer with buffer A. The filtrates were used as cell extracts (>10 kDa). For fractionation of cell extracts, they were further centrifuged at 124,000 × g for 30 min at 4°C, and the resulting supernatants were used as the soluble fraction. The pellets were washed twice with buffer A, resuspended in the same buffer, and used as the membrane fraction.
Identification of the metabolites.
SYK-6 resting cells (OD600 of 5.0) were incubated with 1 mM HPV or HPS in buffer A at 30°C with shaking. SME077 resting cells (OD600 of 0.5 or 5.0) were incubated with 100 μM VAA or 1 mM ferulate in buffer A at 30°C with shaking. After incubation, portions of the reaction mixtures were collected, and reactions were stopped by centrifugation. Methanol was added to the resulting supernatants (final concentration, 40%), and the filtered samples were analyzed by HPLC-MS.
SYK-6 cell extracts (>10 kDa; 500 μg protein/ml) were incubated with 200 μM HPV or HPS in the presence or absence of 500 μM NAD+ or of 500 μM NAD+, 2 mM CoA, 2.5 mM MgSO4, and 2.5 mM ATP in buffer A at 30°C. SME077 cell extracts (>10 kDa; 500 μg of protein/ml) were incubated with 100 μM VAA in the presence of 1 mM CoA, 1.25 mM MgSO4, and 1.25 mM ATP in buffer A at 30°C. The reactions were stopped by the addition of methanol or acetonitrile (final concentration, 50%) at various sampling time points. Precipitated proteins were removed by centrifugation at 19,000 × g for 15 min. The resulting supernatants of the reaction mixtures were analyzed by HPLC-MS.
Enzyme assays using cell extracts of SYK-6.
The HPV-oxidizing activities of SYK-6 cell extracts were determined by measuring the decrease in the amount of HPV by HPLC analysis. In order to examine the effect of GGE on enzyme induction, SYK-6 cells grown in LB were inoculated into Wx-SEMP to an OD600 of 0.2 and grown at 30°C. GGE (5 mM) was added when the OD600 of the culture reached 0.5, and the culture was then further incubated for 12 h. SYK-6 cell extracts (500 μg of protein/ml) were incubated with 200 μM HPV in the presence or absence of cofactors (500 μM PMS, 500 μM FAD + PMS, or 500 μM NAD+) in buffer A for 30 min at 30°C. The reaction mixtures were analyzed by HPLC, and HPV was detected at 276 nm. The specific activity was expressed in moles of HPV converted per min per mg of protein.
In order to examine the activity of VAL-oxidizing activity, the production of VAA from HPV was measured when HPV was reacted with SYK-6 cell extracts (>10 kDa). The cell extracts (>10 kDa; 500 μg of protein/ml) were incubated with HPV in the presence of cofactors (500 μM FAD, 500 μM NAD+, or 500 μM NADP+) in buffer A for 2 h at 30°C. The supernatant of the reaction mixtures was analyzed by HPLC, and compounds were detected at 280 nm.
Analytical methods.
HPLC-MS analysis was performed with the Acquity UPLC system (Waters) coupled with an Acquity TQ detector as described previously (58). Reaction products of HPV, coniferyl alcohol, sinapyl alcohol, cinnamyl alcohol, 3-(4-hydroxyphenyl)-1-propanol, homovanillyl alcohol, vanillyl alcohol, GGE, MPHPV, DCA, and DCA-C were analyzed using a TSKgel ODS-140HTP column (2.1 by 100 mm; Tosoh). Reaction products of HPS were analyzed using an Acquity UPLC BEH C18 column (2.1 by 100 mm; Waters). Reaction products of VAA were analyzed using both columns. All analyses were carried out at a flow rate of 0.5 ml/min. The mobile phase was a mixture of solution A (acetonitrile containing 0.1% formic acid) and solution B (water containing 0.1% formic acid) under the following conditions: (i) detection of the reaction products of HPV: 0 to 4.2 min, 5% A; 4.2 to 6.0 min, linear gradient from 5 to 30% A; 6.0 to 6.5 min, decreasing gradient from 30 to 5% A; 6.5 to 7.0 min, 5% A; (ii) detection of VAL generated from HPV: 0 to 4.7 min, 5% A; 4.7 to 4.9 min, linear gradient from 5 to 80%; 4.9 to 7.0 min, 80% A; (iii) detection of VAA-CoA generated from VAA: 0 to 0.8 min, 10% A; 0.8 to 1.0 min, linear gradient from 10 to 25% A; 1.0 to 1.5 min, 25% A; 1.5 to 1.8 min, decreasing gradient from 25 to 10% A; 1.8 to 3.0 min, 10% A; (iv) detection of vanillate generated from VAA: 0 to 3.0 min, linear gradient from 5 to 15% A; 3.0 to 4.0 min, decreasing gradient from 15 to 5%; (v) detection of the reaction product of vanillyl alcohol: 0 to 5.0 min, 5% A; (vi) detection of the reaction products of coniferyl alcohol, sinapyl alcohol, cinnamyl alcohol, homovanillyl alcohol, and HPS: 0 to 7.0 min, 10% A; (vii) detection of the reaction products of 3-(4-hydroxyphenyl)-1-propanol, GGE, and ferulate: 0 to 5.0 min, 15% A; and (viii) detection of the reaction products of MPHPV, DCA, and DCA-C: 0 to 5.0 min, 25% A. HPV, HPS, VAL, VAL-Tris, VAA, VAA-CoA, vanillate, syringate, acetovanillone, coniferyl alcohol, sinapyl alcohol, cinnamyl alcohol, 3-(4-hydroxyphenyl)-1-propanol, homovanillyl alcohol, vanillyl alcohol, GGE, MPHPV, DCA, and DCA-C were detected at 276, 300, 310, 352, 280, 300, 260, 276, 275, 263, 273, 250, 276, 279, 279, 277, 280, 277, and 326 nm, respectively. In ESI-MS analysis, MS spectra were obtained using the positive- and negative-ion modes with the settings described in our previous study (58). Protein concentrations were determined by the Bradford method using the Bio-Rad protein assay kit or by using Lowry's assay with a DC protein assay kit (Bio-Rad Laboratories). The expression of the genes was analyzed by SDS-PAGE. Protein bands in gels were stained with Coomassie brilliant blue.
DNA manipulations and sequence analysis.
The PCR primers used in this study are listed in Table 3. Nucleotide sequences were determined using a CEQ 2000XL genetic analysis system (Beckman Coulter). Sequence analysis was performed with the MacVector program (MacVector, Inc.). Sequence similarity searches, pairwise alignments, and multiple alignments were carried out using the BLASTP program (59), the EMBOSS Needle program through the EMBL-EBI server (60), and the Clustal Omega program (61), respectively.
TABLE 3.
Primer sequences used for construction of plasmids and colony PCR
| Plasmid or strain | Primer | Sequence (5′-3′) |
|---|---|---|
| Plasmids | ||
| pT7B12830 | 12830_pT7B_F | AGACAGGCATATGGTTGATG |
| 12830_pT7B_R | GGGCGGCATGGATCCGC | |
| pAK20400 | dis20400_Top_F | CGGTACCCGGGGATCCGGCTTCGGTGACAATCAT |
| dis20400_Top_R | ATGTCCGTGGTGTTCTGCGT | |
| dis20400_Bot_F | ACGCAGAACACCACGGACATTCCTCCCCGTGATGACCTAT | |
| dis20400_Bot_R | CGACTCTAGAGGATCGTGGCGGCATCAACATATCG | |
| pE16FB | ferB_exp_F | GGAAAATCATATGTCCGAGG |
| ferB_exp_R | ATACTGGCGGATCCAGCC | |
| pE16FB2 | ferB2_exp_F | TGAGGATGCATATGTCGGATG |
| ferB2_exp_R | GCCGGATCCCGGAATGC | |
| Colony PCR (strain) | dis20400_conf_F | CCTTCATCGCCATCATAAAT |
| SME061 | dis20400_Bot_R | CGACTCTAGAGGATCGTGGCGGCATCAACATATCG |
Cloning of hpvZ.
A partially SalI-digested gene library of SYK-6 constructed with pVK100 was introduced into a host strain, S. sanguinis IAM 12578, by triparental mating (62). The ability of 1,000 transconjugants grown in diluted LB to transform 15 μM HPV was analyzed by HPLC. Southern hybridization analysis of the SalI digests of positive clones with pSA53 as a probe was carried out using the digoxigenin system (Roche Diagnostics). The hybridized SalI fragments were cloned in pBluescript II SK(+), and the nucleotide sequences of both ends of the inserts were determined.
Expression of SYK-6 genes in E. coli and SYK-6.
A DNA fragment carrying hpvZ was amplified by PCR using SYK-6 total DNA as a template. The amplified fragment was ligated into pT7Blue, and the NdeI-BamHI fragment of the resulting plasmid was then inserted in pCold I to generate pCold12830. DNA fragments carrying ferB and ferB2 were amplified by PCR. The amplified fragments were ligated into pET-16b to obtain pE16FB and pE16FB2. Nucleotide sequences of their inserts were confirmed by nucleotide sequencing. Expression plasmids for SYK-6 ALDH genes and ferA were prepared in previous studies (31, 51). The expression plasmids were introduced into E. coli BL21(DE3), and the transformed cells were grown in LB. In the case of hpvZ expression, pTf16 encoding the trigger factor chaperone was introduced into E. coli BL21(DE3) in addition to pCold12830, and the resulting transformant was grown in the presence of 0.5 mg/ml l-arabinose. The expressions of hpvZ and other genes were induced for 24 h at 16°C and for 4 h at 30°C, respectively, by adding 0.1 to 1 mM isopropyl-β-d-thiogalactopyranoside when the OD600 of the cultures reached 0.5. Cells were then harvested by centrifugation and washed with buffer A. pJB12830 was created by inserting the 2.3-kb BamHI-SacII (blunted) fragment carrying hpvZ from pBH37F into pJB864. pJB12830 was introduced into SYK-6, and the transformed cells were grown in LB containing 1 mM m-toluate for 24 h. Resting cells and cell extracts were prepared as described above.
Construction of mutants.
To construct pKmb12830K, the 1.3-kb SalI-BamHI fragment carrying the kanamycin resistance gene (kan) of pIK03 was inserted into the XhoI-BglII sites in hpvZ of pUC12830. The 3.1-kb SalI fragment of the resulting plasmid was ligated into the same site of pK19mobsacB to obtain pKmb12830K. To construct pKmb07270K, the 1.5-kb HindIII-XbaI fragment of pKS0727 was ligated into the same sites of pK18mobsacB, yielding pKmb07270. pKmb07270K was constructed by inserting kan into the NruI site in SLG_07270 of pKmb07270. pKmb12830K and pKmb07270K were independently introduced into SYK-6 cells by electroporation, and candidate mutants were isolated as described previously (63). Disruption of each gene was examined by Southern hybridization analysis (see Fig. S3 in the supplemental material). To construct pAK20400, upstream and downstream regions (ca. 1.0 kb each) of SLG_20400 were amplified by PCR. The resulting fragments were cloned into pAK405 by In-Fusion cloning (In-Fusion HD cloning kit; TaKaRa Bio). pAK20400 was introduced into SYK-6 cells by triparental mating (62), and the mutant strain was selected as described previously (64). Disruption of the gene was confirmed by colony PCR. For the complementation of hpvZ in SME059, pJB12830 was introduced into cells by electroporation.
Resting cell assays.
Resting cells of E. coli harboring pCold12830 and pTf16 (OD600 of 10.0), SYK-6 (OD600 of 0.5, 1.0, or 5.0), SYK-6 harboring pJB864 (OD600 of 1.0), SME059 (OD600 of 0.5, 1.0, or 5.0), SME059 harboring pJB864 (OD600 of 1.0), SME059 harboring pJB12830 (OD600 of 1.0), and SME009 (OD600 of 1.0) prepared from LB-grown cultures were incubated with substrates (200 μM HPV, 200 μM HPS, 200 μM GGE, or 100 μM VAA) at 30°C with shaking. Portions of the cultures were collected, and the amounts of substrates were measured by HPLC.
Enzyme properties of HpvZ.
To determine the cellular localization of HpvZ, the HPV-oxidizing activities of the soluble and membrane fractions prepared from cell extracts of SYK-6 were measured. The cell extracts (500 μg of protein/ml), soluble fraction (500 μg of protein/ml), and membrane fraction (500 μg of protein/ml) were incubated with 200 μM HPV and 500 μM PMS in buffer A for 10 or 30 min at 30°C. The amounts of HPV were measured by HPLC.
The enzyme reaction was typically carried out by incubating the membrane fraction (300 μg of protein/ml) of E. coli cells harboring pCold12830 and pTf16 with 200 μM HPV and 500 μM PMS in buffer A for 10 min at 30°C. After incubation, the amount of substrate was measured by HPLC. The optimum pH was determined at pH ranges from 7.0 to 10.0 using 50 mM GTA buffer (50 mM 3,3-dimethylglutaric acid, 50 mM Tris, and 50 mM 2-amino-2-methyl-1,3-propanediol; pH 7.0 to 9.0) and 50 mM CHES (N-cyclohexyl-2-aminoethanesulfonic acid) buffer (pH 8.6 to 10) at 30°C. The optimum temperature was determined at temperature ranges from 25 to 50°C using buffer A. To determine the substrate range, 200 μM HPV, HPS, coniferyl alcohol, sinapyl alcohol, cinnamyl alcohol, 3-(4-hydroxyphenyl)-1-propanol, homovanillyl alcohol, vanillyl alcohol, GGE, MPHPV, DCA, and DCA-C were used for the reaction, and the decrease in the amount of substrate was measured by HPLC. To examine the effect of flavin cofactors on HpvZ activity, the activities of HpvZ in the presence of 500 μM FAD or flavin mononucleotide were determined. To examine the effect of ubiquinone derivatives on HpvZ activity, the activities of HpvZ in the presence of CoQ0 and CoQ1 were determined. The enzyme reactions were carried out by incubating membrane fractions of E. coli cells harboring pCold12830 and pTf16 (300 μg of protein/ml) or SYK-6 cells harboring pJB12830 (300 μg of protein/ml) with 200 μM HPV and 500 μM CoQ0 or CoQ1 in buffer A for 10 and 5 min, respectively, at 30°C. The decrease in the substrate was determined by HPLC analysis.
Identification of the ALDH genes involved in VAL conversion.
The abilities of the 23 ALDHs of SYK-6 to convert VAL were examined by measuring the amount of VAA produced from HPV when HPV was reacted with both HpvZ and each ALDH. Resting cells of E. coli expressing each ALDH gene (OD600 of 1.0) and E. coli harboring pCold12830 and pTf16 (OD600 of 5.0 or 10.0) were incubated with 100 μM HPV in buffer A at 30°C with shaking. Portions of the cultures were collected at various sampling time points, and the supernatants of the reaction mixtures were analyzed by HPLC.
The mixtures of resting cells of SYK-6, SME031, SME045, SME061, or SME092 (OD600 of 0.5) and resting cells of E. coli harboring pCold12830 and pTf16 (OD600 of 10.0) were incubated with 1 mM HPV in buffer A at 30°C with shaking. Portions of the reaction mixtures were collected at various sampling time points. The supernatants prepared were analyzed by HPLC.
Conversion of VAA by enzymes for ferulate catabolism.
Crude enzymes of FerA, FerB, and FerB2 were prepared from the E. coli transformants described above. FerA, FerA + FerB, and FerA + FerB2 (100 μg protein/ml of each) were incubated with 100 μM VAA, 1 mM CoA, 1.25 mM MgSO4, and 1.25 mM ATP in buffer A for 60 min at 30°C. The supernatants prepared were analyzed by HPLC.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daisuke Sato for assistance with the construction of mutants. We also thank Daniel Miles-Barrett and Amol Thakkar for their contributions.
This study was supported in part by a grant from the JSPS KAKENHI (grant 26850046) and the Japan Science and Technology Agency (Advanced Low Carbon Technology Research and Development Program).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02670-17.
REFERENCES
- 1.Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 2.Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, Boerjan W. 2004. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3:29–60. doi: 10.1023/B:PHYT.0000047809.65444.a4. [DOI] [Google Scholar]
- 3.Doherty WOS, Mousavioun P, Fellows CM. 2011. Value-adding to cellulosic ethanol: lignin polymers. Ind Crop Prod 33:259–276. doi: 10.1016/j.indcrop.2010.10.022. [DOI] [Google Scholar]
- 4.Zakzeski J, Bruijnincx PCA, Jongerius AL, Weckhuysen BM. 2010. The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev 110:3552–3599. doi: 10.1021/cr900354u. [DOI] [PubMed] [Google Scholar]
- 5.Otsuka Y, Nakamura M, Shigehara K, Sugimura K, Masai E, Ohara S, Katayama Y. 2006. Efficient production of 2-pyrone 4,6-dicarboxylic acid as a novel polymer-based material from protocatechuate by microbial function. Appl Microbiol Biotechnol 71:608–614. doi: 10.1007/s00253-005-0203-7. [DOI] [PubMed] [Google Scholar]
- 6.Michinobu T, Bito M, Yamada Y, Tanimura M, Katayama Y, Masai E, Nakamura M, Otsuka Y, Ohara S, Shigehara K. 2009. Fusible, elastic, and biodegradable polyesters of 2-pyrone-4,6-dicarboxylic acid (PDC). Polym J 41:1111–1116. doi: 10.1295/polymj.PJ2009045R. [DOI] [Google Scholar]
- 7.Michinobu T, Hiraki K, Inazawa Y, Katayama Y, Masai E, Nakamura M, Ohara S, Shigehara K. 2011. Click synthesis and adhesive properties of novel biomass-based polymers from lignin-derived stable metabolic intermediate. Polym J 43:648–653. doi: 10.1038/pj.2011.40. [DOI] [Google Scholar]
- 8.Sonoki T, Morooka M, Sakamoto K, Otsuka Y, Nakamura M, Jellison J, Goodell B. 2014. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J Biotechnol 192 (Pt A):71–77. doi: 10.1016/j.jbiotec.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 9.Johnson CW, Salvachúa D, Khanna P, Smith H, Peterson DJ, Beckham GT. 2016. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab Eng Commun 3:111–119. doi: 10.1016/j.meteno.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonoki T, Takahashi K, Sugita H, Hatamura M, Azuma Y, Sato T, Suzuki S, Kamimura N, Masai E. 2018. Glucose-free cis,cis-muconic acid production via new metabolic designs corresponding to the heterogeneity of lignin. ACS Sustainable Chem Eng 6:1256–1264. doi: 10.1021/acssuschemeng.7b03597. [DOI] [Google Scholar]
- 11.Linger JG, Vardon DR, Guarnieri MT, Karp EM, Hunsinger GB, Franden MA, Johnson CW, Chupka G, Strathmann TJ, Pienkos PT, Beckham GT. 2014. Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci U S A 111:12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyama T, Magara K, Matsumoto Y, Meshitsuka G, Ishizu A, Lundquist K. 2000. Proof of the presence of racemic forms of arylglycerol-β-aryl ether structure in lignin: studies on the stereo structure of lignin by ozonation. J Wood Sci 46:414–415. doi: 10.1007/BF00776407. [DOI] [Google Scholar]
- 13.Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, Higuchi Y, Masai E. 2017. Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep 9:679–705. doi: 10.1111/1758-2229.12597. [DOI] [PubMed] [Google Scholar]
- 14.Sato Y, Moriuchi H, Hishiyama S, Otsuka Y, Oshima K, Kasai D, Nakamura M, Ohara S, Katayama Y, Fukuda M, Masai E. 2009. Identification of three alcohol dehydrogenase genes involved in the stereospecific catabolism of arylglycerol-β-aryl ether by Sphingobium sp. strain SYK-6. Appl Environ Microbiol 75:5195–5201. doi: 10.1128/AEM.00880-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masai E, Ichimura A, Sato Y, Miyauchi K, Katayama Y, Fukuda M. 2003. Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J Bacteriol 185:1768–1775. doi: 10.1128/JB.185.6.1768-1775.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanamura K, Abe T, Kamimura N, Kasai D, Hishiyama S, Otsuka Y, Nakamura M, Kajita S, Katayama Y, Fukuda M, Masai E. 2011. Characterization of the third glutathione S-transferase gene involved in enantioselective cleavage of the β-aryl ether by Sphingobium sp. strain SYK-6. Biosci Biotechnol Biochem 75:2404–2407. doi: 10.1271/bbb.110525. [DOI] [PubMed] [Google Scholar]
- 17.Meux E, Prosper P, Masai E, Mulliert G, Dumarçay S, Morel M, Didierjean C, Gelhaye E, Favier F. 2012. Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class. FEBS Lett 586:3944–3950. doi: 10.1016/j.febslet.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 18.Pereira JH, Heins RA, Gall DL, McAndrew RP, Deng K, Holland KC, Donohue TJ, Noguera DR, Simmons BA, Sale KL, Ralph J, Adams PD. 2016. Structural and biochemical characterization of the early and late enzymes in the lignin β-aryl ether cleavage pathway from Sphingobium sp. SYK-6. J Biol Chem 291:10228–10238. doi: 10.1074/jbc.M115.700427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiter J, Strittmatter H, Wiemann LO, Schieder D, Sieber V. 2013. Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem 15:1373–1381. doi: 10.1039/c3gc40295a. [DOI] [Google Scholar]
- 20.Gall DL, Kim H, Lu F, Donohue TJ, Noguera DR, Ralph J. 2014. Stereochemical features of glutathione-dependent enzymes in the Sphingobium sp. strain SYK-6 β-aryl etherase pathway. J Biol Chem 289:8656–8667. doi: 10.1074/jbc.M113.536250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gall DL, Ralph J, Donohue TJ, Noguera DR. 2014. A group of sequence-related sphingomonad enzymes catalyzes cleavage of β-aryl ether linkages in lignin β-guaiacyl and β-syringyl ether dimers. Environ Sci Technol 48:12454–12463. doi: 10.1021/es503886d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picart P, Müller C, Mottweiler J, Wiermans L, Bolm C, de María PD, Schallmey A. 2014. From gene towards selective biomass valorization: bacterial β-etherases with catalytic activity on lignin-like polymers. ChemSusChem 7:3164–3171. doi: 10.1002/cssc.201402465. [DOI] [PubMed] [Google Scholar]
- 23.Ohta Y, Nishi S, Hasegawa R, Hatada Y. 2015. Combination of six enzymes of a marine Novosphingobium converts the stereoisomers of β-O-4 lignin model dimers into the respective monomers. Sci Rep 5:15105. doi: 10.1038/srep15105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palamuru S, Dellas N, Pearce SL, Warden AC, Oakeshott JG, Pandey G. 2015. Phylogenetic and kinetic characterization of a suite of dehydrogenases from a newly isolated bacterium, strain SG61-1L, that catalyze the turnover of guaiacylglycerol-β-guaiacyl ether stereoisomers. Appl Environ Microbiol 81:8164–8176. doi: 10.1128/AEM.01573-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picart P, Sevenich M, de María PD, Schallmey A. 2015. Exploring glutathione lyases as biocatalysts: paving the way for enzymatic lignin depolymerization and future stereoselective applications. Green Chem 17:4931–4940. doi: 10.1039/C5GC01078K. [DOI] [Google Scholar]
- 26.Helmich KE, Pereira JH, Gall DL, Heins RA, McAndrew RP, Bingman C, Deng K, Holland KC, Noguera DR, Simmons BA, Sale KL, Ralph J, Donohue TJ, Adams PD, Phillips GN Jr. 2016. Structural basis of stereospecificity in the bacterial enzymatic cleavage of β-aryl ether bonds in lignin. J Biol Chem 291:5234–5246. doi: 10.1074/jbc.M115.694307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta Y, Hasegawa R, Kurosawa K, Maeda AH, Koizumi T, Nishimura H, Okada H, Qu C, Saito K, Watanabe T, Hatada Y. 2017. Enzymatic specific production and chemical functionalization of phenylpropanone platform monomers from lignin. ChemSusChem 10:425–433. doi: 10.1002/cssc.201601235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosini E, Allegretti C, Melis R, Cerioli L, Conti G, Pollegioni L, D'Arrigo P. 2016. Cascade enzymatic cleavage of the β-O-4 linkage in a lignin model compound. Catal Sci Technol 6:2195–2205. doi: 10.1039/C5CY01591J. [DOI] [Google Scholar]
- 29.Higuchi Y, Takahashi K, Kamimura N, Masai E. 2018. Bacterial enzymes for the cleavage of lignin β-aryl ether bonds: properties and applications. In Beckham GT. (ed), Lignin valorization: emerging approaches, in press. Royal Society of Chemistry, London, United Kingdom. [Google Scholar]
- 30.Lancefield CS, Ojo OS, Tran F, Westwood NJ. 2015. Isolation of functionalized phenolic monomers through selective oxidation and C-O bond cleavage of the β-O-4 linkages in lignin. Angew Chem Int Ed Engl 54:258–262. doi: 10.1002/anie.201409408. [DOI] [PubMed] [Google Scholar]
- 31.Kasai D, Kamimura N, Tani K, Umeda S, Abe T, Fukuda M, Masai E. 2012. Characterization of FerC, a MarR-type transcriptional regulator, involved in transcriptional regulation of the ferulate catabolic operon in Sphingobium sp. strain SYK-6. FEMS Microbiol Lett 332:68–75. doi: 10.1111/j.1574-6968.2012.02576.x. [DOI] [PubMed] [Google Scholar]
- 32.Bubb WA, Berthon HA, Kuchel PW. 1995. Tris buffer reactivity with low-molecular-weight aldehydes: NMR characterization of the reactions of glyceraldehyde-3-phosphate. Bioorg Chem 23:119–130. doi: 10.1006/bioo.1995.1010. [DOI] [Google Scholar]
- 33.Fukuzumi T, Oyake S, Matsumoto H. 1974. Synthesis of vanilloyl acetaldehyde. Mokuzai Gakkaishi 20:138–142. [Google Scholar]
- 34.Niwa M, Saburi Y. 2002. Vanilloyl acetic acid as an unstable intermediate from β-hydroxypropiovanillone to acetovanillone. Holzforschung 56:360–362. doi: 10.1515/HF.2002.057. [DOI] [Google Scholar]
- 35.Overhage J, Priefert H, Steinbüchel A. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol 65:4837–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masai E, Harada K, Peng X, Kitayama H, Katayama Y, Fukuda M. 2002. Cloning and characterization of the ferulic acid catabolic genes of Sphingomonas paucimobilis SYK-6. Appl Environ Microbiol 68:4416–4424. doi: 10.1128/AEM.68.9.4416-4424.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otani H, Lee YE, Casabon I, Eltis LD. 2014. Characterization of p-hydroxycinnamate catabolism in a soil actinobacterium. J Bacteriol 196:4293–4303. doi: 10.1128/JB.02247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campillo T, Renoud S, Kerzaon I, Vial L, Baude J, Gaillard V, Bellvert F, Chamignon C, Comte G, Nesme X, Lavire C, Hommais F. 2014. Analysis of hydroxycinnamic acid degradation in Agrobacterium fabrum reveals a coenzyme A-dependent, beta-oxidative deacetylation pathway. Appl Environ Microbiol 80:3341–3349. doi: 10.1128/AEM.00475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallscheuer N, Vogt M, Kappelmann J, Krumbach K, Noack S, Bott M, Marienhagen J. 2016. Identification of the phd gene cluster responsible for phenylpropanoid utilization in Corynebacterium glutamicum. Appl Microbiol Biotechnol 100:1871–1881. doi: 10.1007/s00253-015-7165-1. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi K, Hirose Y, Kamimura N, Hishiyama S, Hara H, Araki T, Kasai D, Kajita S, Katayama Y, Fukuda M, Masai E. 2015. Membrane-associated glucose-methanol-choline oxidoreductase family enzymes PhcC and PhcD are essential for enantioselective catabolism of dehydrodiconiferyl alcohol. Appl Environ Microbiol 81:8022–8036. doi: 10.1128/AEM.02391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirmair L, Skerra A. 2014. Biochemical analysis of recombinant AlkJ from Pseudomonas putida reveals a membrane-associated, flavin adenine dinucleotide-dependent dehydrogenase suitable for the biosynthetic production of aliphatic aldehydes. Appl Environ Microbiol 80:2468–2477. doi: 10.1128/AEM.04297-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohta T, Kawabata T, Nishikawa K, Tani A, Kimbara K, Kawai F. 2006. Analysis of amino acid residues involved in catalysis of polyethylene glycol dehydrogenase from Sphingopyxis terrae, using three-dimensional molecular modeling-based kinetic characterization of mutants. Appl Environ Microbiol 72:4388–4396. doi: 10.1128/AEM.02174-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawai F, Kimura T, Fukaya M, Tani Y, Ogata K, Ueno T, Fukami H. 1978. Bacterial oxidation of polyethylene glycol. Appl Environ Microbiol 35:679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsushita K, Ameyama M. 1982. d-Glucose dehydrogenase from Pseudomonas fluorescens, membrane-bound. Methods Enzymol 89:149–154. doi: 10.1016/S0076-6879(82)89026-5. [DOI] [PubMed] [Google Scholar]
- 45.Romero E, Gadda G. 2014. Alcohol oxidation by flavoenzymes. Biomol Concepts 5:299–318. doi: 10.1515/bmc-2014-0016. [DOI] [PubMed] [Google Scholar]
- 46.Guillén F, Martínez AT, Martínez MJ. 1992. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem 209:603–611. doi: 10.1111/j.1432-1033.1992.tb17326.x. [DOI] [PubMed] [Google Scholar]
- 47.Dijkman WP, de Gonzalo G, Mattevi A, Fraaije MW. 2013. Flavoprotein oxidases: classification and applications. Appl Microbiol Biotechnol 97:5177–5188. doi: 10.1007/s00253-013-4925-7. [DOI] [PubMed] [Google Scholar]
- 48.Quaye O, Lountos GT, Fan F, Orville AM, Gadda G. 2008. Role of Glu312 in binding and positioning of the substrate for the hydride transfer reaction in choline oxidase. Biochemistry 47:243–256. doi: 10.1021/bi7017943. [DOI] [PubMed] [Google Scholar]
- 49.Tan TC, Spadiut O, Wongnate T, Sucharitakul J, Krondorfer I, Sygmund C, Haltrich D, Chaiyen P, Peterbauer CK, Divne C. 2013. The 1.6 Å crystal structure of pyranose dehydrogenase from Agaricus meleagris rationalizes substrate specificity and reveals a flavin intermediate. PLoS One 8:e53567. doi: 10.1371/journal.pone.0053567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi K, Kamimura N, Hishiyama S, Hara H, Kasai D, Katayama Y, Fukuda M, Kajita S, Masai E. 2014. Characterization of the catabolic pathway for a phenylcoumaran-type lignin-derived biaryl in Sphingobium sp. strain SYK-6. Biodegradation 25:735–745. doi: 10.1007/s10532-014-9695-0. [DOI] [PubMed] [Google Scholar]
- 51.Kamimura N, Goto T, Takahashi K, Kasai D, Otsuka Y, Nakamura M, Katayama Y, Fukuda M, Masai E. 2017. A bacterial aromatic aldehyde dehydrogenase critical for the efficient catabolism of syringaldehyde. Sci Rep 7:44422. doi: 10.1038/srep44422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Achterholt S, Priefert H, Steinbüchel A. 1998. Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. strain HR199 and molecular characterization of the gene. J Bacteriol 180:4387–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon O, Klaiber I, Huber A, Pfannstiel J. 2014. Comprehensive proteome analysis of the response of Pseudomonas putida KT2440 to the flavor compound vanillin. J Proteomics 109:212–227. doi: 10.1016/j.jprot.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Cushman M, Nagarathnam D, Gopal D, He HM, Lin CM, Hamel E. 1992. Synthesis and evaluation of analogues of (Z)-1-(4-methoxyphenyl)-2-(3,4,5-trimethoxyphenyl)ethene as potential cytotoxic and antimitotic agents. J Med Chem 35:2293–2306. doi: 10.1021/jm00090a021. [DOI] [PubMed] [Google Scholar]
- 55.Shirai T, Kumihashi K, Sakasai M, Kusuoku H, Shibuya Y, Ohuchi A. 2017. Identification of a novel TRPM8 agonist from nutmeg: a promising cooling compound. ACS Med Chem Lett 8:715–719. doi: 10.1021/acsmedchemlett.7b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hishiyama S, Otsuka Y, Nakamura M, Ohara S, Kajita S, Masai E, Katayama Y. 2012. Convenient synthesis of chiral lignin model compounds via optical resolution: four stereoisomers of guaiacylglycerol-β-guaiacyl ether and both enantiomers of 3-hydroxy-1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxyphenoxy)-propan-1-one (erone). Tetrahedron Lett 53:842–845. doi: 10.1016/j.tetlet.2011.12.016. [DOI] [Google Scholar]
- 57.Fukuhara Y, Kamimura N, Nakajima M, Hishiyama S, Hara H, Kasai D, Tsuji Y, Narita-Yamada S, Nakamura S, Katano Y, Fujita N, Katayama Y, Fukuda M, Kajita S, Masai E. 2013. Discovery of pinoresinol reductase genes in sphingomonads. Enzyme Microb Technol 52:38–43. doi: 10.1016/j.enzmictec.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Fukuhara Y, Inakazu K, Kodama N, Kamimura N, Kasai D, Katayama Y, Fukuda M, Masai E. 2010. Characterization of the isophthalate degradation genes of Comamonas sp. strain E6. Appl Environ Microbiol 76:519–527. doi: 10.1128/AEM.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A 77:7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukuda M. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J Bacteriol 181:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaczmarczyk A, Vorholt JA, Francez-Charlot A. 2012. Markerless gene deletion system for sphingomonads. Appl Environ Microbiol 78:3774–3777. doi: 10.1128/AEM.07347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katayama Y, Nishikawa S, Nakamura M, Yano K, Yamasaki M, Morohoshi N, Haraguchi T. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77–79. [Google Scholar]
- 66.Peng X, Masai E, Kasai D, Miyauchi K, Katayama Y, Fukuda M. 2005. A second 5-carboxyvanillate decarboxylase gene, ligW2, is important for lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl Environ Microbiol 71:5014–5021. doi: 10.1128/AEM.71.9.5014-5021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto Y, Kasai D, Kamimura N, Masai E. 2012. Isolation and characterization of bzaA and bzaB of Sphingobium sp. strain SYK-6, which encode aromatic aldehydes dehydrogenases with different substrate preferences. Trans GIGAKU 1:01009/01001–01006. [Google Scholar]
- 68.Takeuchi M, Kawai F, Shimada Y, Yokota A. 1993. Taxonomic study of polyethylene glycol-utilizing bacteria: emended description of the genus Sphingomonas and new descriptions of Sphingomonas macrogoltabidus sp. nov., Sphingomonas sanguis sp. nov., and Sphingomonas terrae sp. nov. Syst Appl Microbiol 16:227–238. doi: 10.1016/S0723-2020(11)80473-X. [DOI] [Google Scholar]
- 69.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 70.Bolivar F, Backman K. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol 68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 71.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Short JM, Fernandez JM, Sorge JA, Huse WD. 1988. λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res 16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 74.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 75.Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, Katayama Y, Fukuda M. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol 186:2757–2765. doi: 10.1128/JB.186.9.2757-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 77.Masai E, Yamamoto Y, Inoue T, Takamura K, Hara H, Kasai D, Katayama Y, Fukuda M. 2007. Characterization of ligV essential for catabolism of vanillin by Sphingomonas paucimobilis SYK-6. Biosci Biotechnol Biochem 71:2487–2492. doi: 10.1271/bbb.70267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.