To the Editor
The microbiome has emerged as a key aspect of human biology and has been implicated in many disease etiologies. Shotgun metagenomic sequencing is an approach with the highest resolution currently available for studying the taxonomic composition and functional potential of the human microbiome. The increase in publicly available shotgun data theoretically enables hypothesis testing for specific diseases and environmental niches as well as meta-analysis across related studies. However, several factors prevent the research community from taking full advantage of these resources. Barriers include the need for substantial investments of time, computational resources and specialized bioinformatic expertise as well as inconsistencies in annotation and formatting between individual studies.
To overcome these challenges, we developed the curatedMetagenomicData data package (https://waldronlab.github.io/curatedMetagenomicData/ and Supplementary Software) for distribution through the Bioconductor1 ExperimentHub platform (Supplementary Methods). curatedMetagenomicData provides uniformly processed human microbiome data including bacterial, fungal, archaeal, and viral taxonomic abundances, in addition to quantitative metabolic functional profiles and standardized per-participant metadata. Data resources are accessible to users with minimal bioinformatic knowledge; while integration with the R/Bioconductor environment allows flexibility for biologists, clinicians, epidemiologists, or statisticians to perform novel analyses and methodological development, and integration of resources. We produced these resources by (i) downloading raw sequencing data; (ii) processing the data through the MetaPhlAn2 (ref. 2) and HUMAnN2 (ref. 3) pipelines; (iii) manually curating sample and study information (Supplementary Table 1); (iv) representing the above results as documented, integrative Bioconductor objects; and (v) working with the Bioconductor core team to develop the ExperimentHub platform for efficient distribution (Fig. 1). ExperimentHub is a novel platform for scalable distribution of experimental data to the R desktop. It allows distributers and downloaders to access metadata through a Bioconductor-hosted SQL database and to access bulk data through Amazon S3 buckets via a standard R software package. An extension permits command-line access. The current version of curatedMetagenomicData (v1.7.5) includes samples from multiple body sites profiled by the Human Microbiome Project4 and from 25 other metagenomic studies. These total 5,716 samples that span 34 diseases and 28 countries (Supplementary Table 2).
Figure 1.
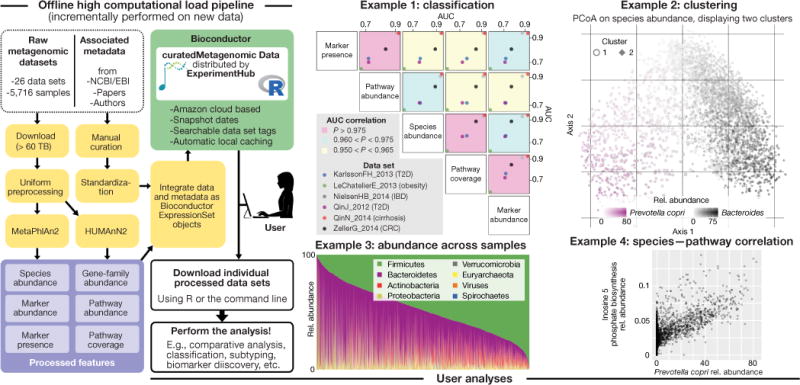
curatedMetagenomicData production pipeline and examples of enabled analyses. The pipeline (left) processes raw metagenomic sequence data to produce taxonomic and functional profiles, integrates these with curated sample data, then documents and packages these for distribution through ExperimentHub as the curatedMetagenomicData package. Example 1: health status was classified for six data sets, five data types each, using a random-forest algorithm and cross-validation to estimate prediction accuracy. Example 2: unsupervised clustering of human gut samples (n = 3,667) using the Bray–Curtis metric and partitioning around medoids. In Cluster 1, Bacteroides relative abundance is shown by point borders and Prevotella copri by centers. Example 3: abundance at the phylum level in human gut samples. Example 4: correlation between Prevotella copri and inosine 5 phosphate biosynthesis, the most correlated species–pathway pair (see Supplementary Fig. 4 for heatmap of most highly correlated species–pathway pairs). These analyses were performed using the script provided in the vignettes/extras package directory.
Using curatedMetagenomicData and the statistical, visualization, and microbial ecology tools available in R/Bioconductor makes many analyses much simpler and more powerful than previous analyses. As a demonstration, we employed a random-forest algorithm on three taxonomic data types (species abundance, genetic marker presence, and abundance) and two functional abundance profiles (pathway abundance and coverage) to develop predictive models of diabetes, inflammatory bowel disease, cirrhosis, colorectal cancer, and obesity. Cross-validation prediction accuracy varied substantially for these different outcomes, but the five data types provided nearly identical prediction accuracy (Fig. 1, example 1). Second, we performed unsupervised clustering of human gut microbiome profiles. In a large combined data set (n = 3,667), we observed that microbial communities are strongly patterned by a continuous gradient between relative abundance of Prevotella copri and Bacteroides spp (Fig. 1, example 2) with little or no support for any discrete clusters (Supplementary Fig. 1). This is consistent with the analysis of Koren et al.5 but stands in contrast with the three-enterotypes hypothesis of Arumugam et al.6. Third, we visualized the continuum of the firmicutes–bacteroidetes gradient in gut microbiomes (Fig. 1, example 3). This continuum has been previously reported4, but not for thousands of microbial species on thousands of individuals as analyzed here. Finally, we ranked all taxa–pathway pairs by magnitude of correlation in samples. The highest correlation pair shown demonstrates a strong relationship between Prevotella copri abundance and inosine 5 phosphate biosynthesis (Fig. 1, example 4), and this suggests functional differences along the gradient shown in example 2. These and other analyses (Supplementary Figs. 1–5) would be very large undertakings using less curated databases such as IMG/M or EBI Metagenomics; but they are straightforward, documented, and reproducible using curatedMetagenomicData.
Our large-scale, curated integration of metagenomic data is well documented and readily usable by broad scientific communities for efficient hypothesis testing and methods development. The automated pipeline allows the resource to be continually expanded by the team and community contributors (Supplementary Methods, package maintenance). By allowing researchers to bring their expertise to the analysis of metagenomic data without the need for extensive bioinformatic experience, curatedMetagenomicData greatly expands the accessibility of public data for study of the human microbiome.
Supplementary Material
Acknowledgments
This work was made possible by the CUNY High Performance Computing Center, College of Staten Island, funded in part by the City and State of New York, CUNY Research Foundation, and National Science Foundation Grants CNS-0958379, CNS-0855217 and ACI 1126113. Support was provided by the European Union H2020 Marie-curie grant (707345) to E.P., the European Research Council (ERC-STG project MetaPG), MIUR “Futuro in Ricerca” RBFR13EWWI_001, the People Programme (Marie Curie Actions) of the European Union Seventh Framework Programme (FP7/2007–2013) under REA grant agreement no. PCIG13-GA-2013-618833, the LEO Pharma Foundation, and by Fondazione CARITRO fellowship Rif.Int.2013.0239 to N.S., the National Institute of Allergy and Infectious Diseases (1R21AI121784-01 to J.B.D. and L.W.) and the US National Cancer Institute (U24CA180996 to M.M. and L.W.).
Footnotes
Data availability statement. The curatedMetagenomicData data package is available online at https://waldronlab.github.io/curatedMetagenomicData/ and through the Bioconductor package installer.
A Life Sciences Reporting Summary is available.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper (doi:10.1038/nmeth.4468).
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Huber W, et al. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truong DT, et al. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 3.Abubucker S, et al. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Human Microbiome Project Consortium. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koren O, et al. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, et al. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


