Abstract
We assessed the potential of Lmna-mRNA repair by spliceosome-mediated RNA trans-splicing as a therapeutic approach for LMNA-related congenital muscular dystrophy. This gene therapy strategy leads to reduction of mutated transcript expression for the benefit of corresponding wild-type (WT) transcripts. We developed 5′-RNA pre-trans-splicing molecules containing the first five exons of Lmna and targeting intron 5 of Lmna pre-mRNA. Among nine pre-trans-splicing molecules, differing in the targeted sequence in intron 5 and tested in C2C12 myoblasts, three induced trans-splicing events on endogenous Lmna mRNA and confirmed at protein level. Further analyses performed in primary myotubes derived from an LMNA-related congenital muscular dystrophy (L-CMD) mouse model led to a partial rescue of the mutant phenotype. Finally, we tested this approach in vivo using adeno-associated virus (AAV) delivery in newborn mice and showed that trans-splicing events occurred in WT mice 50 days after AAV delivery, although at a low rate. Altogether, while these results provide the first evidence for reprogramming LMNA mRNA in vitro, strategies to improve the rate of trans-splicing events still need to be developed for efficient application of this therapeutic approach in vivo.
Keywords: Lmna, lamin A/C, LMNA-related congenital muscular dystrophy, trans-splicing, gene therapy
Introduction
A-type lamins (lamin A and C) are intermediate filament proteins expressed in differentiated cells.1 Together with B-type lamins, they form the nuclear lamina, an organized meshwork found underneath the inner nuclear envelope.2 They are thought to play a key role in nuclear resistance to mechanical stress, thus maintaining cell integrity. A-type lamins are also present in the nucleoplasm,3 where they regulate gene expression through interaction with chromatin, nuclear histones, and transcription factors.4
Lamins A and C (hereafter referred to as lamin A/C) are produced by alternative splicing of the LMNA gene, located on human chromosome 1q21. Since the discovery of the first mutation,5 464 mutations have been reported (http://www.umd.be/LMNA/). These mutations have been related to more than 10 distinct genetic diseases, commonly named laminopathies.4, 6
LMNA-related congenital muscular dystrophy (L-CMD) has been described as an autosomal dominant muscle disorder related to a dominant de novo mutation in LMNA, so far the most severe form of striated muscle laminopathies.7 L-CMD is characterized by an onset before the age of 2 years of major muscle atrophy and weakness mainly affecting axial muscles, leading to complete absence of or limited motor achievements, together with important multiple joint contractures sparing elbows, and severe respiratory insufficiency that requires continuous mechanical ventilation. Identification of cardiac arrhythmias suggested a cardiac involvement in these patients. Our team reported the phenotype of knock-in LmnaΔK32 mice, reproducing an L-CMD mutation.8, 9, 10 Homozygous LmnaΔK32 mice rapidly exhibit generalized growth retardation and develop striated muscles and adipose tissue maturation defects, leading to their premature death in their third week of life.8 Interestingly, while Lmna mRNA levels were unaffected in mutant mice, lamin A/C protein level was strongly reduced, representing only 20% of that observed in the wild-type (WT) mice tissues. Moreover, mutant A-type lamins were absent at the nuclear periphery and found exclusively in the nucleoplasm. Both decreased expression and mislocalization of lamin A/C are thought to contribute to the development of the disease.
In the present study, we investigated the potential interest of spliceosome-mediated RNA trans-splicing (SMaRT) as a therapeutic strategy for L-CMD. The SMaRT approach targets RNA at the pre-mRNA level and converts the endogenous mutated sequences to WT ones.11, 12, 13 This is achieved by expressing pre-trans-spliced molecules (PTMs) containing the WT coding sequence, a strong splicing site and a binding domain to enable the specific binding of the PTMs to the targeted pre-mRNA. This technology has been mainly used to replace 3′ exons, but it can also be used to target the replacement of 5′ or internal exons. Considering the natural alternative splicing of A-type lamins occurring in a distal exon (exon 10 among 12 in total) and the high incidence of LMNA mutations in the first exons in L-CMD patients, we took the option of targeting the intron 5 of Lmna pre-mRNA by a 5′-trans-splicing strategy. We investigate here the feasibility of repairing LMNA transcripts by the SMaRT strategy using AAV delivery both in vitro and in vivo.
Results
PTM Engineering and Selection
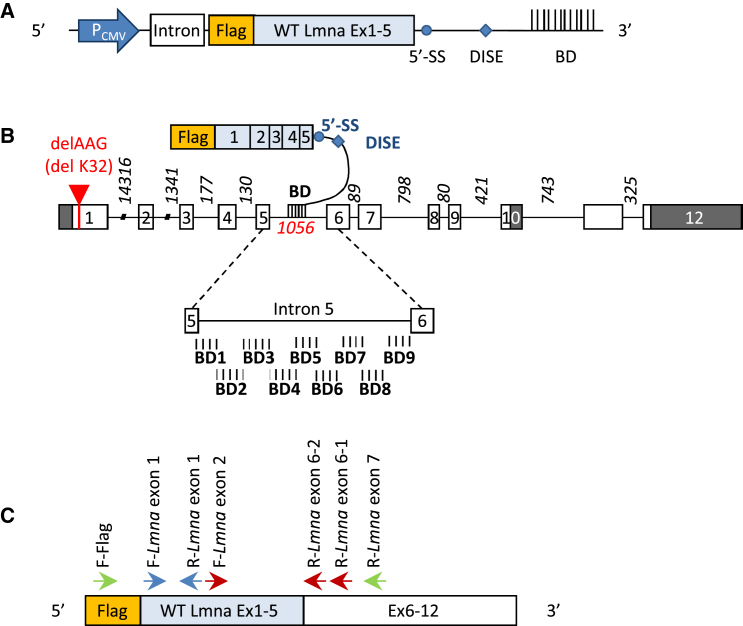
Several plasmids encoding PTMs targeting Lmna intron 5 were designed (Figure 1A). They all harbor the FLAG-WT Lmna exons 1–5 cDNA followed by a canonical 5′ splice donor site sequence and a downstream intronic sequence enhancer element to increase the trans-splicing efficiency14, 15 and various binding domains (BDs). Nine PTMs were designed, differing in their BDs with respect to the position they target in intron 5 (Figure 1B; Table S1) and called PTM-BD. The expected trans-spliced product is composed of exogenous FLAG and exons 1–5 sequences and the endogenous exons 6–12 (Figure 1C). As negative controls that do not bind to intron 5, and thus should not induce 5′ trans-splicing events, we designed a PTM without BD (called PTM-0) and for each PTM-BD a specific negative control harboring an inverted BD sequence and called PTM-BDinv.
Figure 1.
5′ Trans-Splicing Strategy for Lmna Transcripts
(A) Schematic illustration of part of the vector encoding for a pre-trans-splicing molecule (PTM). The PTM is a transcript comprising the FLAG coding sequence followed by the first five Lmna exons (WT Lmna ex 1–5), a 5′ splice site (5′SS) followed by a downstream intronic splicing enhancer (DISE), and a binding domain (BD) of 150 bp complementary to Lmna intron 5. PTM sequences are placed under control of a CMV promoter (PCMV). An intronic sequence was added to stabilize the transcript. (B) Schematic representation of Lmna pre-mRNA and PTMs. The mouse Lmna pre-messenger consists of exons (boxes) and introns (lines). White boxes represent coding sequences, and gray boxes represent non-coding sequence for prelamin A. Intronic size lengths are indicated. The red arrow refers to localization of the ΔK32 mutation in exon 1. Below, an enlarged view of intron 5 shows the localization of the different designed binding domains. (C) Schematic representation of a trans-spliced mRNA molecule and the different primers used to characterize trans-spliced/repaired mRNA (F-FLAG and R-Lmna exon 7), PTMs (F-FLAG and R-Lmna exon 1), and total (endogenous and trans-spliced) Lmna molecules (F-Lmna exon 1 and R-Lmna exon 1). Primers used for nested qPCR of trans-spliced Lmna transcripts are F-FLAG and R-Lmna exon 6-1 for the first PCR, followed by F-Lmna exon 2 and R-Lmna exon 6-2 for the qPCR.
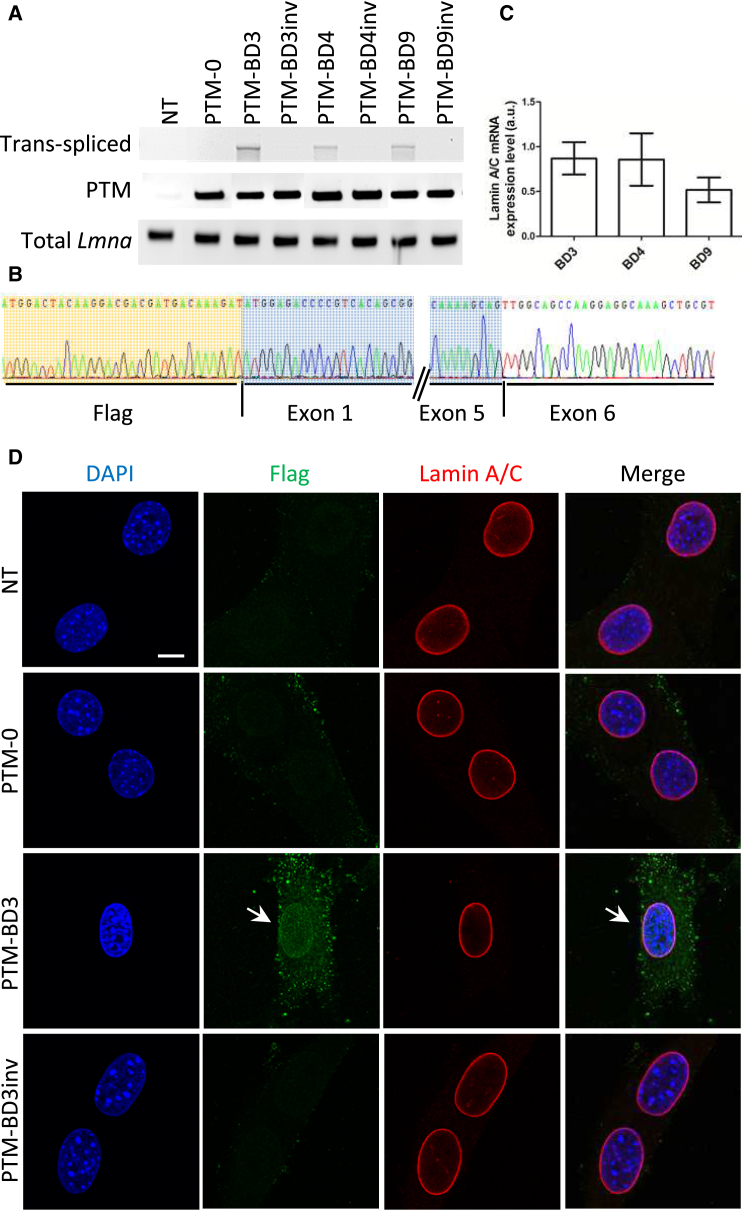
To evaluate the efficacy of Lmna trans-splicing in vitro, we performed transient transfection of the different constructs in C2C12 myoblasts. We did not observe expression of trans-spliced lamin A/C by western blotting using either FLAG or lamin A/C antibodies with any of the PTM, probably because of the low trans-splicing efficiency (Figure S1A). Hence, we chose to assess for the expression of trans-spliced molecules by PCR using primers that specifically amplify the trans-spliced molecules (F-FLAG and R-Lmna exon 7; Figure 1C; Table S2). Using such primers, we identified trans-spliced molecules when using PTM-BD3, -BD4, and -BD9 (Figure 2A), but not when using other PTM-BDs (data not shown) and negative controls (corresponding PTM-BDinv and PTM-0). We excluded the possibility that the absence of trans-splicing events when using PTM-BD1, -BD2, -BD5, -BD6, -BD7, and -BD8 was due to a too low transfection efficiency using primers that recognized PTM and trans-spliced mRNA (F-FLAG and R-Lmna exon 1). Total Lmna mRNA levels were evaluated using primers localized in exon 1 (F-Lmna exon 1 and R-Lmna exon 1) (Figures 1C and 2A). To fully confirm the identity of amplicons from F-FLAG/R-Lmna exon 7 PCR, we cloned and then sequenced the PCR products from C2C12 transfected with PTM-BD3, -BD4, and -BD9. The presence of a FLAG sequence directly followed by Lmna exons including a correct exon 5-to-exon 6 junction attested for a trans-spliced Lmna mRNA (shown for PTM-BD3 in Figure 2B). We then compared the trans-splicing rate between PTM-BD3-, -BD4-, and -BD9-transfected C2C12 by nested qPCR. Although the level of trans-splicing observed after PTM-BD9 transfection is lower than that obtained after PTM-BD3 or -BD4, it is not statistically significant (Figure 2C).
Figure 2.
Evaluation of PTM’s Efficacy in C2C12 Cells
(A) PCR analysis was performed with primers localized on Lmna mRNA (Figure 1C), on RNA extracts from C2C12 myoblasts that were either non-transfected (NT) or transfected for 48 hr with PTM-0, PTM-BD, and PTM-BDinv plasmids. (B) Sequencing of trans-spliced Lmna amplicons from PTM-BD3-transfected cells after subcloning confirmed trans-splicing events with the junction between the exogenous FLAG-Lmna exons 1–5 (highlighted in orange and blue) with endogenous Lmna exon 6 and the following exons. (C) Bar graph showing the level of lamin A/C mRNA expression by nested qPCR. BD3, transfected with pSMD2-PTM-BD3 (n = 4); BD4, transfected with pSMD2-PTM-BD4 (n = 3); BD9, transfected with pSMD2-PTM-BD9 (n = 5). Differences are not statistically significant. Error bars correspond to SEM. (D) Immunofluorescence analysis of NT or transfected C2C12 with PTM-0, PTM-BD3, PTM-BD3inv. Cells were double-stained with anti-FLAG (green) and anti-Lamin A/C (red) antibodies. Nuclei were stained with DAPI (blue). Arrows show a positive FLAG-tagged nucleus. Scale bar: 10 μm.
We investigated whether the trans-spliced Lmna mRNAs were translated into proteins properly localized at the nuclear envelope by immunofluorescence (nine independent experiments performed). Using a FLAG antibody, we successfully detected a FLAG signal in 17% of C2C12 myoblasts (mean from three independent experiments with at least 174 cells counted per experiment). This signal was found at the nuclear rim, colocalized with the A-type lamin signal (Figure 2D for PTM-BD3; Figure S1B for PTM-BD4 and -BD9). No signal was detected by immunofluorescence in non-transfected (NT) conditions or after transfection with control PTM-BD3inv or PTM-0.
In order to better understand the successful trans-splicing using PTM-BD3, -BD4, and -BD9 compared with the other BDs, we first evaluated the GC content that is known to impact on the level of RNA production.16 Interestingly, the GC content of the nine PTM-BDs was not significantly different. We then hypothesize that having less secondary structure in the binding domain might well favor recombination with the intronic sequence. We analyzed RNA folding of each PTM-BD (using RNAfold, The ViennaRNA Package) and found less secondary structures such as loops and hairpins for PTM-BD3, -BD4, and -BD9 compared with other PTM-BDs.
Altogether, results obtained on C2C12 cells using 5′ trans-splicing molecules were very encouraging and we hence decided to test the efficacy of PTM-BD3, -BD4, and -BD9 in myotubes from an L-CMD mouse model.
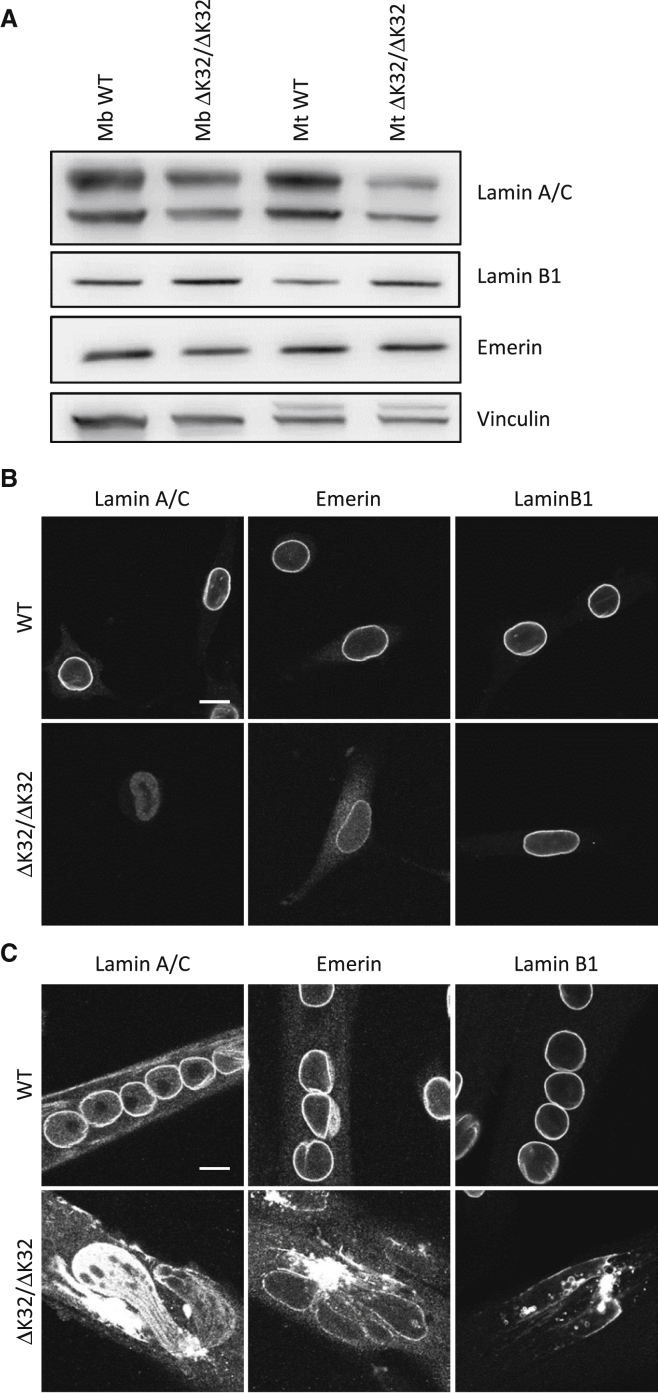
Cellular and Molecular Defects of Homozygous LmnaΔK32 Primary Mouse Myoblasts in Culture
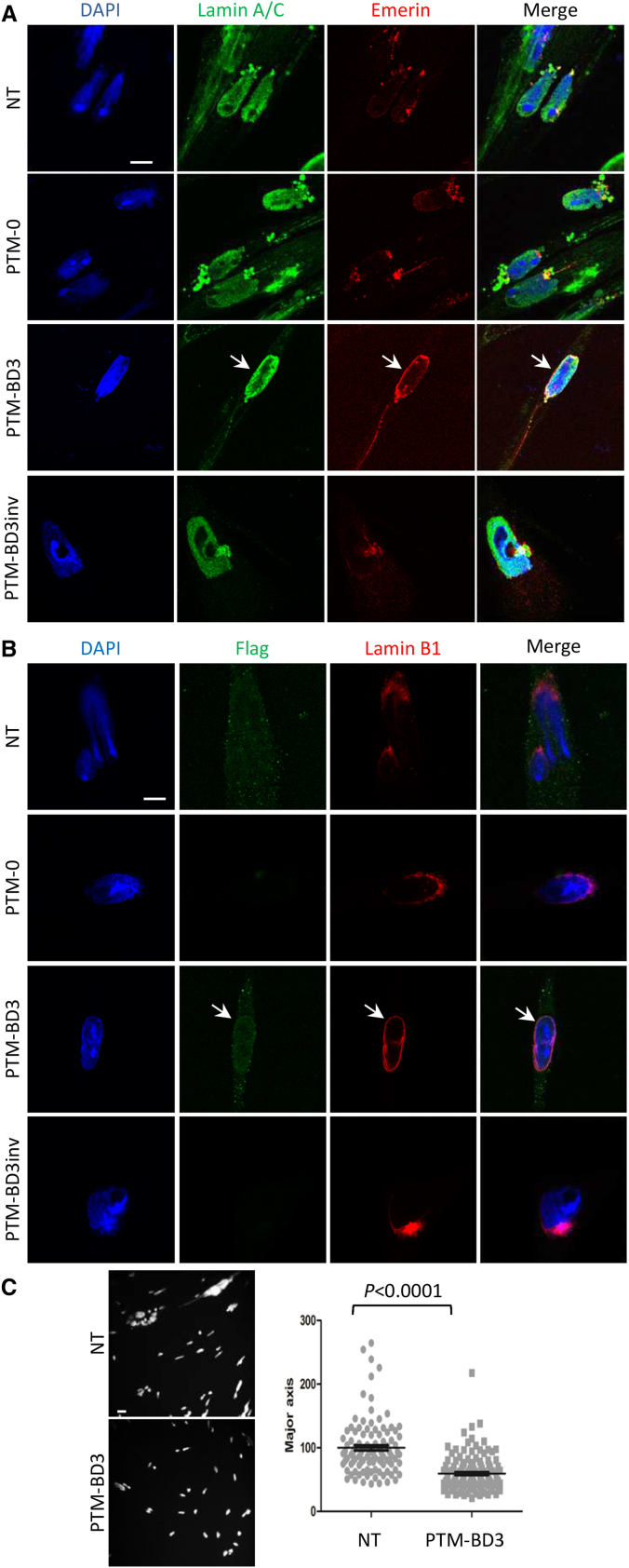
We have reported previously that lamin A/C protein levels are highly reduced in homozygous LmnaΔK32 mouse tissues and that the mutant lamin is unable to assemble under the inner nuclear membrane, and hence is only found throughout the nucleoplasm.8, 17 We checked here on cultured primary myoblasts and myotubes isolated from WT and homozygous LmnaΔK32 mouse tibialis anterior (TA) muscles. Western blot analysis showed a decrease in lamin A/C protein levels in mutant myoblasts compared with WT cells, while other nuclear envelope proteins such as emerin or lamin B1 levels were unchanged (Figure 3A). Despite a moderate differentiation-dependent increase of lamin A/C expression in WT, the decreased lamin A/C expression level was maintained in mutant myotubes with only 40%–60% residual protein expression. We also investigated the localization of WT and mutant lamin A/C proteins by immunostaining. In proliferating myoblasts, ΔK32-lamin A/C was found dispersed throughout the nucleoplasm, whereas WT-lamin A/C was abundantly located at the nuclear envelope and sparsely in the nucleoplasm (Figure 3B). Nuclei of ΔK32-myoblasts also showed an elongated shape, as reported previously in human cells and other animal models with mutations in lamin A/C. In addition, if no defect was found for lamin B1 localization, we observed a decreased emerin staining at the nuclear periphery and an increase in the cytoplasm of mutant compared with WT myoblasts. During myoblast differentiation, mutant myotubes displayed a unique and severely misshapen DNA mass stained with DAPI (data not shown) that could correspond to the aggregation or to the fusion of extremely soft and deformable nuclei. Its nucleoplasm is strongly stained with lamin A/C antibody, while its nuclear envelope only poorly reacts with both emerin and lamin B1 antibodies that mainly mark perinuclear aggregates (Figure 3C). The defects observed in these cells therefore represent good readouts for trans-splicing gene therapy.
Figure 3.
Decreased Expression and Abnormal Localization of Δ32-Lamin A/C in Mouse Mutant Muscle Cells
(A) Representative western blot analysis for mouse primary WT and mutant (ΔK32/ΔK32) myoblasts (Mb) and myotubes (Mt; 24 hr of differentiation) probed for lamin A/C, lamin B1, and emerin proteins relative to vinculin. (B and C) Immunofluorescent confocal micrographs of WT and mutant myoblasts (B) and 24-hr differentiated myotubes (C), stained for lamin A/C, emerin, and lamin B1. Scale bars: 10 μm.
Increased Proportion of A-Type Lamins at the Nuclear Envelope after Lmna mRNA Trans-Splicing in Mouse Primary LmnaΔK32 Myotubes
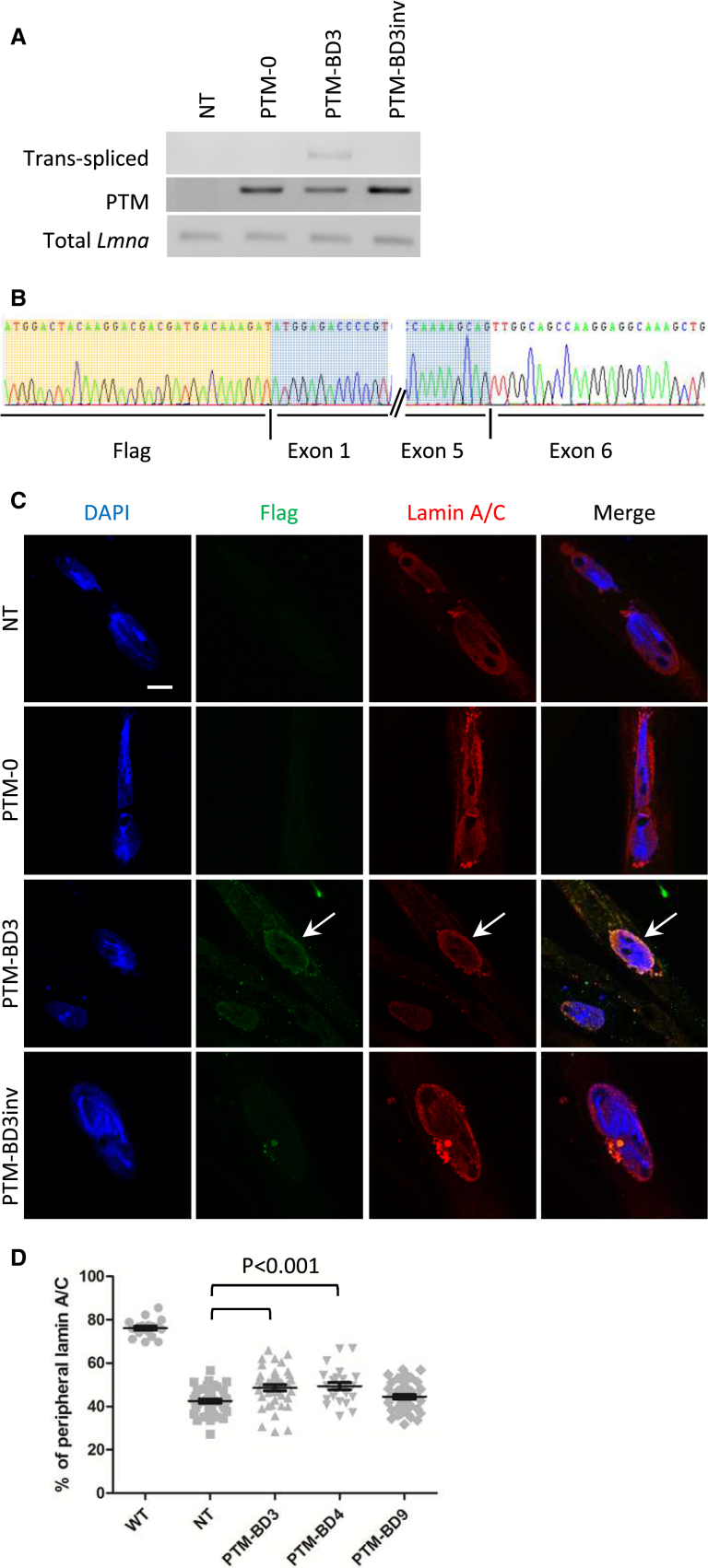
To allow gene transfer in mouse primary myotubes, we produced AAV serotypes 2/9. In our hands, transduction of mouse primary myotubes using a control AAV2/9 encoding for GFP resulted in 90% of GFP+ cells (see Figure S2). In addition to AAV2/9-PTM-BD3, -BD4, and -BD9, we produced control AAVs: one containing the PTM sequence without BD (AAV2/9-PTM-0) and three others with the inverted BD3, BD4, and BD9. Following the transduction of mutant myotubes, we evaluated trans-splicing at the mRNA level. All PTM mRNAs (PTM-0, PTM-BD3, and PTM-BD3inv) were expressed in transduced myotubes, as attested by the presence of a PCR product at the expected molecular weight when using F-FLAG and R-Lmna exon1 primers (Figure 4A). Using primers specific for trans-spliced mRNA, a faint band was observed at the right molecular weight in AAV2/9-PTM-BD3-transduced myotubes corresponding to repaired Lmna transcripts, but not in control transduced myotubes (shown for AAV2/9-PTM-BD3 in Figure 4A). Sequencing of the cloned PCR product showed the presence of exogenous FLAG-Lmna exons 1–5 and endogenous Lmna exon 6 with an exact junction exon 5-exon 6 (Figure 4B).
Figure 4.
Detection of Repaired Lmna mRNA Induced by 5′ Trans-Splicing in Mouse Mutant Primary Myotubes
(A) PCR analysis was performed on RNA extracts of mouse primary mutant myotubes transduced or not (NT) for 7 days with AAV2/9 expressing PTM-0, PTM-BD3, or PTM-BD3inv using primers described in Figure 1C and Table S2. (B) Sequencing of the F-FLAG/R-Lmna exon 7 cloned amplicons from AAV2/9-PTM-BD3-transduced cells confirmed trans-splicing events with the junction between the exogenous FLAG-Lmna exons 1–5 (highlighted in orange and blue) and endogenous Lmna exon 6 and the following exons. (C) Immunofluorescence analysis of mutant myotubes that were non-transduced (NT) or transduced with AAV2/9-PTM-0, -PTM-BD3, and -PTM-BD3inv. Cells were double-stained with anti-FLAG (green) and anti-Lamin A/C (red) antibodies. Nuclei were stained with DAPI (blue). Arrows show a FLAG+ nucleus. Scale bar: 10 μm. (D) Scatterplot representing the percentage of lamin A/C staining found at the nuclear periphery in wild-type (WT; n = 19) and non-transduced (NT; n = 40) or PTM-BD3- (n = 40), PTM-BD4- (n = 24), or PTM-BD9 (n = 41)-transduced mutant myotubes. All groups are statistically different from WT (p < 0.001), while only PTM-BD3 and PTM-BD4 are statistically different from NT (p < 0.01). Error bars correspond to SEM.
We further characterized transduced mutant myotubes by immunostaining against FLAG and observed 49% of positive cells (mean of two independent experiments, with at least 191 nuclei counted per experiment) (Figure 4C). Compared with results obtained in C2C12, we found the FLAG staining to be more broadly distributed within the nucleus, although it is enriched at the nuclear envelope in eight independent experiments. These FLAG+ nuclei also have an increased proportion of lamin A/C at the nuclear envelope compared with non-transduced (NT) or control transduced mutant myotubes (shown for AAV2/9-PTM-BD3 and controls in Figure 4C and for AAV2/9-BD4 and -BD9 in Figure S3A). Measurement of the fluorescence intensity of lamin A/C staining at the nuclear periphery or at the nucleoplasm reveals a significant increase in the proportion of peripheral lamin A/C after PTM-BD3 and PTM-BD4 transduction compared with NT cells (p < 0.01), but not after PTM-BD9 transduction (Figure 4D). Interestingly, even if no signal was obtained by western blot using the FLAG antibody, a faint increase in A-type lamin protein level was observed after transduction of mutant myotubes with AAV2/9-PTM-BD3 and -BD4 (Figure S4), indirectly confirming the occurrence of trans-splicing.
Lmna Trans-Splicing Improves Mutant Myotube Phenotype
While NT or AAV2/9-PTM-0-transduced mutant myotubes displayed abnormal distribution of emerin and lamin B1 proteins, a great proportion of cells transduced with AAV2/9-PTM-BD3 showed a better staining for emerin (Figure 5A; 17% of FLAG+ and FLAG− nuclei; mean of two independent experiments, with at least 83 cells counted per experiment; see a lower magnification and a zoom-in Figure S5A) and lamin B1 (Figure 5B; 20% of FLAG+ and FLAG− nuclei; mean of two independent experiments with at least 90 nuclei counted per experiment; see a lower magnification and zoom-in in Figure S5B) at the nuclear rim that increase in intensity (white arrow) and with a disappearance of protein aggregates. Similar results were obtained with AAV2/9-PTM-BD4 and -BD9 (Figure S3B).
Figure 5.
5′ Trans-Splicing of Lmna Improves the Nuclear Phenotype of Mutant Myotubes
(A and B) Immunofluorescence analysis of mutant myotubes that were non-transduced (NT) or transduced with AAV2/9-PTM-0, -PTM-BD3, and -PTM-BD3inv. Cells were differentiated and transduced for 7 days and then stained with lamin A/C (green)/emerin (red) (A) or FLAG (green)/Lamin B1 (red) (B). Nuclei were stained with DAPI (blue). Arrows point to repaired nuclei. Scale bars: 10 μm. (C) Representative confocal images of DAPI-stained myotubes that were either non-transduced (NT) or transduced with AAV2/9-PTM-BD3, and scatterplot showing the distribution and the mean size of nuclei long axis (NT: n = 107; PTM-BD3: n = 137). Error bars correspond to SEM.
We evaluated whether the nuclear elongation was improved after PTM-BD3 transduction by measuring the nuclear long and short axes of all nuclei (FLAG+ and FLAG−). Although the short axis did not vary between the transduced and non-transduced cells, a significant decrease of 55% of the length of the long axis was observed after PTM-BD3 transduction (Figure 5C; mean of two independent experiments, with at least 44 nuclei counted per condition and per experiment). This result showed that abnormally shaped nuclei can regain better shape by a partial lamin A/C rescue following trans-splicing.
In Vivo Evaluation of Trans-Splicing of Lmna mRNA
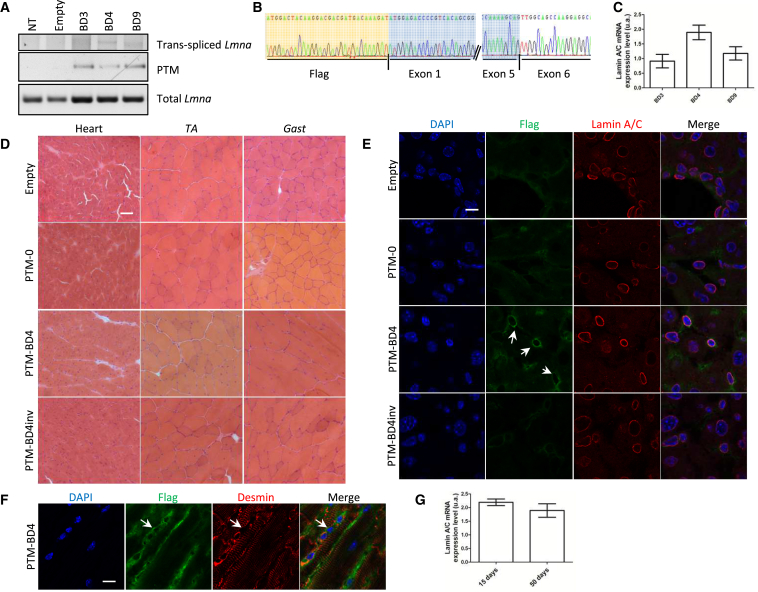
We then performed systemic injections of AAV2/9-empty, AAV2/9-PTM-BD3, -BD4, and -BD9 in newborn WT mice (n = 4 per construct; 1011 virus genomes [vg]/mouse) and sacrificed the mice 50 days post-injection. Despite a good transduction efficiency attested by a strong band after PCR specific for the PTM, we detected an extremely faint band for trans-spliced molecules whatever the virus construct (Figure 6A). As performed previously, to clearly prove the presence of trans-splicing in vivo, we cloned the bands and subsequently sequenced the clones (Figure 6B). Finally, we compared the level of trans-splicing by nested qPCR on heart tissues after AAV2/9-PTM-BD3, -BD4, or -BD9 delivery. Although the level of trans-splicing seems higher after AAV2/9-PTM-BD4 delivery, the differences between the three groups are not statistically significant (Figure 6C).
Figure 6.
Evidence for Reprogramming Lmna mRNA and Protein by 5′ Trans-Splicing In Vivo
(A) PCR analysis was performed on RNA extracts of heart tissue from WT mice 50 days after systemic injection of AAV2/9-empty or expressing PTM-0, PTM-BD4, or PTM-BD4inv using primers described in Figure 1C and Table S2. (B) Sequencing of F-FLAG/R-Lmna exon 7 cloned amplicons from heart from mice 50 days after systemic injection with AAV2/9-PTM-BD4 confirmed trans-splicing events with the junction between the exogenous FLAG-Lmna exons 1–5 (highlighted in orange and blue) and endogenous Lmna exon 6. (C) Bar graph showing the level of lamin A/C mRNA expression by nested qPCR in hearts of WT mice 50 days after systemic injection of AAV2/9-PTM-BD3, -BD4, or -BD9. n = 4 per group, and differences are not statistically significant. Error bars correspond to SEM. (D) H&E staining of striated muscle sections from WT mice 50 days after systemic injection with AAV2/9-empty or expressing PTM-0, PTM-BD4 or PTM-BD4inv. Scale bar: 50 μm. (E) Immunofluorescence analysis of heart sections from WT mice 50 days after systemic injection of AAV2/9-PTM-0, -PTM-BD4, and -PTM-BD4inv. Cells were double-stained with anti-FLAG (green) and anti-Lamin A/C (red) antibodies. Nuclei were stained with DAPI (blue). Scale bar: 10 μm. (F) Immunofluorescence analysis of heart sections from WT mouse 50 days after systemic injection of AAV2/9-PTM-BD4. Cells were double-stained with anti-FLAG (green) and anti-desmin (red) antibodies. Nuclei were stained with DAPI (blue). Arrows point to a positive FLAG-tagged nucleus. Scale bar: 10 μm. (G) Bar graph showing the level of lamin A/C mRNA expression by nested qPCR in hearts of mice 15 (n = 3) or 50 days (n = 4) after systemic injection of AAV2/9-PTM-BD4. Differences are not statistically significant. Error bars correspond to SEM. Gast, gastrocnemius; TA, tibialis anterior.
Observation of transversal sections of different skeletal muscles and heart stained with H&E demonstrated that there was no toxicity consecutive to AAV injections and/or PTM expression (shown for AAV2/9-PTM-BD4 in Figure 6D and for AAV2/9-PTM-BD3 in Figure S6A). Evaluation of trans-splicing at the protein level by immunofluorescence successfully detected positive events in the heart after AAV2/9-PTM-BD4 delivery, but not after transduction with control vectors (Figure 6E). Indeed, a FLAG staining colocalizing with the endogenous lamin A/C was observed at the nuclear envelope in 6% of transduced cells in cardiac sections (mean of two independent experiments with at least 292 nuclei counted per experiment). Unfortunately, a double immunostaining for FLAG and desmin showed that among these FLAG+ nuclei, only a few corresponded to cardiomyocytes (Figure 6F). A low number of positive nuclei was also observed in other tissues, including TA (Figure S6B) or gastrocnemius muscles and liver (data not shown).
Although the trans-splicing efficiency appears to be extremely low in vivo, we decided to test whether our best PTM virus (based on our in vitro and in vivo analyses) was able to alleviate the severe phenotype observed in homozygous LmnaΔK32 mice.8 We hence performed systemic injection of AAV2/9-PTM-BD4 in newborn mice obtained from LmnaΔK32 heterozygous breeders before their genotyping (n = 10; 1011 vg/mouse). Homozygous mice (n = 2) were carefully followed until their natural death. They respectively died at days 14 and 15, hence showing no benefits in terms of survival after AAV2/9-PTM-BD4 injection.8 WT and heterozygous mouse littermates were sacrificed 15 days post-injection, and WT, heterozygous, and homozygous mouse tissues were processed for analysis. PCR on heart, quadriceps, and gastrocnemius muscles using primers specific for PTM mRNA (F-FLAG and R-Lmna exon 1) were all positive, attesting that the different tissues were properly transduced and the virus expressed. PCRs for trans-spliced Lmna mRNA (F-FLAG and R-Lmna exon 7) were all negative (Figure S6C); however, increasing the PCR sensitivity using nested qPCR, we were able to observe trans-splicing events in all 15-day-old transduced animals to a level that is similar to that observed 50 days post-transduction (Figure 6G).
Discussion
Since the time when RNA trans-splicing was discovered,18, 19 this technique has been highlighted in different in vitro and in vivo studies for its safety and specificity to repair genetic defects associated with human diseases.20, 21, 22, 23, 24, 25, 26, 27, 28 In this study, we demonstrate the challenges of RNA repair of Lmna messengers in a context of a particularly severe autosomal dominant genetic disease, as L-CMD. To date, no curative treatment exists for L-CMD, and management of patients is largely supportive. The conversion of the mutant transcript into a normal transcript has many advantages, including a good alternative to classical gene replacement strategies that may lead to toxic effect because of lamin A/C overexpression.29, 30 It also corrects the dominant-negative aspect of the pathology that has been largely described in the past by reducing the expression of the mutant proteins.31 Moreover, expression of trans-spliced molecules is maintained under the endogenous regulation of the targeted mRNA, which avoids undesirable ectopic expression.
The vast majority of L-CMD cases harbor mutations in exons 1 (36.5%) and 4 (32.5%) (G. Bonne, personal communication) (http://www.umd.be/LMNA/). While the majority of published reports has been concentrated on the use of 3′ trans-splicing approach,20, 22, 28, 32 we took the option of the 5′ trans-splicing. This strategy has the advantage to leave the natural alternative splicing of the Lmna mRNA (taking place in exon 10) unaffected. Moreover, the Lmna gene has several introns that are too small to be targeted for trans-splicing (Figure 1A). For this proof of principle, we chose to target the Lmna intron 5, which is more than 1,000 base length and allows to target 51% of the described LMNA mutations (G. Bonne, personal communication) (http://www.umd.be/LMNA/).
In this study, PTM-BD screening in mouse myoblasts evaluated at the mRNA and protein levels has displayed positive trans-splicing events for three out of the nine designed PTM-BDs. There are no rules to design BD sequences.12, 33 All designed BDs in our study specifically target intron 5 of Lmna pre-mRNA and are 150-bp-long sequences, which have been shown to limit off-targets.34 The toxicity of the peptides translated from PTM, described in 3′ trans-splicing strategies,25, 35 was avoided in our 5′ strategy by the deletion of the poly-adenylation sequence. Moreover, we have shown that having binding domains less prone to form secondary structures might increase the rate of trans-splicing events. Finally, different studies have suggested that binding domains targeting 5′ regions of introns are more efficient than the ones targeting 3′ regions.26, 35 Interestingly, two of our three PTM-BDs giving positive results are indeed targeting the 5′ region of Lmna intron 5.
In a pathological context, PTM-BD3 and -BD4 are able to rescue part of the nuclear phenotypes of homozygous LmnaΔK32 myotubes. Indeed, the localization of lamin A/C was increased at the nuclear periphery in FLAG+ cells. We suppose that the FLAG signal is less well localized to the nuclear envelope of mutant myotubes compared with experiments performed in C2C12 myoblasts, because corrected/trans-spliced lamin A/C proteins are interacting and polymerizing with mutant lamin A/C. Furthermore, both emerin and lamin B1 mislocalization and nuclear deformation were partially restored in those FLAG+ cells. However, the absence of detection of trans-spliced lamin A/C proteins by western blot, as well as the faint FLAG staining on immunofluorescence, underlines the low trans-splicing rate already described by others.15, 28 Using primary mouse myoblasts that restrict the transduction time to only few days may also explain this latter point. Interestingly, PTM-BD9 that is not able to increase significantly the proportion of lamin A/C found at the nuclear periphery is also the PTM that leads to the lower level of lamin A/C mRNA expression after nested qPCR.
Considering the ubiquitous expression of lamin A/C and the potential interest of modulating the ratio of mutant versus WT proteins in striated muscles of L-CMD patients, we chose to use AAV2/9 vectors that show a good tropism for heart and skeletal muscles.36 Evaluation of trans-splicing events in tissues from systemic injected pups showed an extremely modest increase in lamin A/C mRNA expression, observed only after a nested PCR, as well as the presence of a few FLAG+ cells dispersed throughout the various analyzed tissues (i.e., heart, skeletal muscles, liver). This level is way too low to lead to any obvious benefits for the homozygous LmnaΔK32 mice like an increase in the survival. Increasing the time between AAV delivery and analysis does not result in the accumulation of trans-spliced protein, probably because of the fast turnover of lamin A/C. In addition, if systemic administration of AAV2/9 particles is known to efficiently transduce the heart,37 we found the FLAG staining to be more in the nuclei of vessel cells than cardiomyocytes. The mechanism underlying the impaired trans-splicing in cardiomyocytes has yet to be elucidated; a possibility exists that changing the promoter from CMV to a cell-specific promoter might solve this issue. Several other options may be tested to increase Lmna trans-splicing efficiency: (1) the use of self-complementary AAV, which is already double stranded and hence needs less time to be expressed and may also lead to higher expression in tissues;38 (2) a combo treatment of PTMs with antisense oligonucleotides to reduce cis-splicing and hence favor the trans-splicing of Lmna gene;39 and (3) the use of codon-optimized PTMs to increase translation of the trans-spliced mRNA.28
Overall, this proof of principle pinpoints onto the crucial importance of reducing the expression of the highly toxic mutated lamin A/C and increasing the expression of WT lamin A/C in an extremely short time. In addition, the safety and the advantages of using PTM-BD molecules that may target more than 70% of L-CMD patients have to be taken in account. Despite the low efficiency of repaired mutated lamin A/C by trans-splicing that we and other teams have observed, this technique remains a promising and hopeful RNA therapy for dominant genetic diseases.
Materials and Methods
Plasmid Construction and Adeno-Associated Virus Production
The coding sequence of the PTM was generated by PCR of mouse WT cDNA with a forward primer containing a SacII restriction site, the Kozak sequence, followed by the FLAG sequence and the first 19 nucleotides of Lmna exon 1. The reverse primer contained a SacII restriction site, as well as: (1) downstream intronic splicing enhancer element (DISE) from the rat fibroblast growth factor receptor 2 (Fgfr2) gene, followed by (2) the 5′ canonical splice donor site sequence and (3) the last 26 nucleotides of Lmna exon 5. The different binding domains were obtained by PCR of WT mouse genomic DNA using a forward primer containing an EcoRI restriction site and 21 nucleotides of Lmna intron 5 (Table S1). The reverse primer contained an EcoRI restriction site and 21 nucleotides complementary to Lmna intron 5. PCR products were sequentially cloned into pSMD240 under a CMV promoter and deleted of its SV40 polyA signal. Control plasmid for evaluation of primary myotube transduction efficiency was performed using the same vector backbone with GFP as a coding sequence and its SV40 polyA signal. All constructs were sequenced and quantified using a spectrophotometer. Adeno-associated virus (AAV2/9) vectors were generated using a three-plasmid transfection protocol as described previously.15, 41 Virus titers measured by qPCR ranged from 7.1 × 1012 to 3.1 × 1013 vg/mL.
Cell Culture, Plasmid Transfection, and AAV Transduction
The mouse C2C12 myoblast and NIH 3T3 fibroblast cell lines were cultured in DMEM (Thermo Fisher Scientific, Courtaboeuf, France) supplemented with 1% penicillin/streptomycin (Thermo Fisher Scientific, Courtaboeuf, France) and 10% fetal bovine serum (Thermo Fisher Scientific, Courtaboeuf, France). Cells were grown to 70% of confluence in four-well plates, and 1–3 μg of DNA was transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Courtaboeuf, France) according to manufacturer’s instructions for 48 hr. A pSMD2-CMV plasmid encoding FLAG-human Prelamin A sequence was used as a transfection control. Primary mouse myoblasts from WT and LmnaΔK32/ΔK32 mutant mice were cultured in DMEM supplemented with 20% fetal bovine serum, 10% donor horse serum (VWR International, Fontenay sous Bois, France), and 1% Chicken Embryo Extract (Sera Laboratories International, West Sussex, UK), as described previously.42 Differentiation medium consisted of DMEM supplemented with 2% donor horse serum and 0.5% Chicken Embryo Extract. All media contained 1% penicillin/streptomycin. Tissue culture plastic dishes were coated with 10% Matrigel (Corning, Avon, France). Twenty-four hours after plating, cells were transduced with AAV2/9 vectors at 2.6 × 105 to 6.0 × 105 MOI for 7 days in differentiation medium. All cells were grown in a humidified incubator at 37°C in 5% CO2.
Mouse Lines
LmnaΔK32 (corresponding to Lmnatm2.1Gbon according to MGI nomenclature) mice were described previously.8, 9, 10 All mouse procedures were done according to protocols conformed to the French laws, and regulations concerning the use of animals for research were approved by an external ethical committee (approval no. 00972.03 and 00971.02; delivered by the French Ministry of Higher Education and Scientific Research).
Systemic injections of the different AAV2/9 (1011 vg/mouse) were performed into the temporal vein of 2-day-old mice. WT pups were sacrificed 15 or 50 days post-injection. Tissue sampling and processing were done post-mortem.
Quantification of Trans-Splicing Rate at RNA Level
Total RNA was isolated using the RNeasy fibrous tissue Mini Kit (QIAGEN, Courtaboeuf, France) according to manufacturer’s instructions and reverse transcribed using Superscript III kit (Thermo Fisher Scientific, Courtaboeuf, France) with 1–2 μg of total RNA. Endpoint PCR amplification was performed using PCR Super Mix (Thermo Fisher Scientific, Courtaboeuf, France) with different primer pairs (Table S2). PCR products were visualized on 1% or 2% agarose gels. The band corresponding to the trans-spliced molecule (F-FLAG/R-Lmna exon 7; 1,398 bp) was purified using Nucleospin Gel and PCR clean-up columns (Machery Nagel, Hoerdt, France), cloned in pGEM-T easy vector (Promega, Charbonnières, France), and sequenced.
Nested qPCR was performed as follows: 10 PCR cycles using 2× PCR Reddy Mix (Thermo Fisher Scientific, Courtaboeuf, France) and primers specific for trans-spliced molecules (F-FLAG and R-Lmna exon 6-1; see Table S2). Samples of the first PCR were cleaned using PCR clean-up columns (Macherey-Nagel, Hoerdt, France) to remove the first primer set. Two microliters of a 1/20 dilution was used to perform a qPCR using F-Lmna exon 2 and R-Lmna exon 6-2 primers with SYBR Green I Master Mix on a Roche LightCycler 480 II (both from Roche, Meylan, France).
Western Blot
Cell pellets were homogenized in protein extraction buffer (50 mM Tris-HCl [pH 7.5]; 2% SDS; 250 mM sucrose; 75 mM urea; 1 mM dithiothreitol) containing protease/phosphatase inhibitors [25 μg/mL aprotinin, 10 μg/mL leupeptin; 1 mM 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride, and 2 mM Na3VO4]. Protein extracts were quantified with Pierce BCA Protein Assay Kit (Pierce Biotechnology, USA), separated on a SDS-PAGE, and transferred onto nitrocellulose membrane. After blocking, nitrocellulose blots were incubated with primary antibodies directed against lamin A/C (H-110, sc-20681 or E-1, sc-376248; Santa Cruz Biotechnology, Santa Cruz, CA, USA), lamin B1 (C-20, sc-6216 [Santa Cruz Biotechnology, Santa Cruz, CA, USA] and ab16048 [Abcam, Cambridge, UK]), vinculin (V9131; Sigma-Aldrich, Saint Quentin Fallavier, France), and emerin (NCL-clone 4G5; Novocastra, Newcastle, UK) and then with secondary anti-rabbit, anti-goat, or anti-mouse antibodies coupled to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA, USA). Immunoblots were visualized by Immobilon Western Chemiluminescent HRP Substrate (Millipore, Molsheim, France) on a G Box system using GeneSnap software (Ozyme, Saint Quentin, France).
Histology and Immunofluorescent Analysis
Seven-micrometer sections were made using a cryostat Leica CM3050S. Frozen sections were stained with H&E staining using standard method and visualized under light microscopy.
For immunostaining, cells and tissue sections were fixed for 10 min in 4% paraformaldehyde, permeabilized in 0.5% Triton X-100 in PBS for 5 min, and then blocked for 1 hr in 10% BSA IgG-free. To avoid unspecific staining, we then incubated the sections with Fab fragment solution (0.2 mg/mL in PBS) for 1 hr at room temperature. Primary antibodies against lamin A/C (N-18, sc-6215 or E-1, sc-376248) and lamin B1 (C-20, sc-6216) were purchased from Santa Cruz Biotechnology, anti-FLAG (F1804) from Sigma-Aldrich (Saint Quentin Fallavier, France), and anti-emerin (NCL-clone 4G5) from Novocastra (Newcastle, UK), while anti-desmin (Ab15200) was purchased from Abcam (Cambridge, UK) and anti-GFP (A10262) from Invitrogen (Life Technologies, Saint-Aubin, France). Secondary antibodies against mouse, rabbit, or donkey were coupled with Alexa Fluor 488 or Alexa Fluor 568 (Life Technologies, Saint-Aubin, France). Preparations were mounted with Vectashield mounting medium containing DAPI (Vector Labs, Burlingame, CA, USA), and confocal images were collected using upright confocal laser-scanning microscope (FV-1000 or FV-1200; Olympus) equipped with a UPlanS-Apo 60×/1.35 NA oil immersion objective lens.
Fluorescence intensity of lamin A/C staining at the nuclear periphery versus the nucleoplasm was evaluated by measuring the corrected total fluorescence intensity (CTCF) of total lamin A/C and the CTCF of nucleoplasmic lamin A/C using ImageJ software. CTCF is calculated using the following formula: Integrated density − (area of selected cell × mean fluorescence of background readings). Nuclear dimensions were measured from fluorescence profiles using ImageJ software. Two perpendicular line segments were drawn manually on a nuclear cross section with the longest diameter representing the major axis of the nucleus.
Statistics
Double-blind experiments and result analyses were performed during this study. Measurement of the percentage of peripheral lamin A/C and nuclear dimensions are expressed as means ± SEM. An unpaired two-tailed t test was performed for the nuclear dimension and a one-way ANOVA followed by Tukey’s multiple comparison test using GraphPad Prism software. Equal variances were controlled using Bartlett’s test. p < 0.05 was considered as statistically significant.
Author Contributions
Conceptualization: M. Bitoun, G.B., and A.T.B.; Methodology: F.A., M. Bitoun, S.L., G.B., and A.T.B.; Investigation: F.A., L.A., A.B., M. Beuvin, I.N., A.J., E.Z., B.P., M. Bitoun, and A.T.B.; AAV Production: S.B.-Z; Writing – Original Draft: F.A.; Writing – Review & Editing: F.A., A.B., I.N., M. Bitoun, S.L., G.B., and A.T.B.; Visualization: F.A., A.B., and A.T.B.; Supervision: G.B. and A.T.B.; Project Administration: G.B. and A.T.B.; Funding Acquisition: G.B. and A.T.B.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We are grateful to the Penn Vector Core, Gene Therapy Program, University of Pennsylvania, Philadelphia, for providing the pAAV-9 plasmid (p5E18-VD29) and the vectorology platform of the Centre of Research in Myology-UMRS974 (Paris, France) for AAV production, the Pitié-Salpêtrière Imaging Platform (PICPS) for confocal imaging acquisition facilities, the Animal facility of Pitié-Salpêtrière campus (Centre d’Expérimentation Fonctionnelle, Sorbonne Université-Médecine), and to Joelle Marie for her helpful advice and comments. We would like to thank Delphine Trochet for fruitful discussions and comments. This work was supported by INSERM, the Association Institut de Myologie (AIM), Sorbonne Université-Médecine, the Centre National de la Recherche Scientifique (CNRS), CURE-CMD Foundation, and the Andrés Marcio Foundation.
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.012.
Supplemental Information
References
- 1.Broers J.L., Ramaekers F.C., Bonne G., Yaou R.B., Hutchison C.J. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 2.Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 3.Hozák P., Sasseville A.M., Raymond Y., Cook P.R. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell Sci. 1995;108:635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- 4.Gruenbaum Y., Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 5.Bonne G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 6.Bertrand A.T., Chikhaoui K., Yaou R.B., Bonne G. Clinical and genetic heterogeneity in laminopathies. Biochem. Soc. Trans. 2011;39:1687–1692. doi: 10.1042/BST20110670. [DOI] [PubMed] [Google Scholar]
- 7.Quijano-Roy S., Mbieleu B., Bönnemann C.G., Jeannet P.Y., Colomer J., Clarke N.F., Cuisset J.M., Roper H., De Meirleir L., D’Amico A. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann. Neurol. 2008;64:177–186. doi: 10.1002/ana.21417. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand A.T., Renou L., Papadopoulos A., Beuvin M., Lacène E., Massart C., Ottolenghi C., Decostre V., Maron S., Schlossarek S. DelK32-lamin A/C has abnormal location and induces incomplete tissue maturation and severe metabolic defects leading to premature death. Hum. Mol. Genet. 2012;21:1037–1048. doi: 10.1093/hmg/ddr534. [DOI] [PubMed] [Google Scholar]
- 9.Cattin M.E., Bertrand A.T., Schlossarek S., Le Bihan M.C., Skov Jensen S., Neuber C., Crocini C., Maron S., Lainé J., Mougenot N. Heterozygous LmnadelK32 mice develop dilated cardiomyopathy through a combined pathomechanism of haploinsufficiency and peptide toxicity. Hum. Mol. Genet. 2013;22:3152–3164. doi: 10.1093/hmg/ddt172. [DOI] [PubMed] [Google Scholar]
- 10.Cattin M.E., Ferry A., Vignaud A., Mougenot N., Jacquet A., Wahbi K., Bertrand A.T., Bonne G. Mutation in lamin A/C sensitizes the myocardium to exercise-induced mechanical stress but has no effect on skeletal muscles in mouse. Neuromuscul. Disord. 2016;26:490–499. doi: 10.1016/j.nmd.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell L.G., McGarrity G.J. Gene therapy progress and prospects: reprograming gene expression by trans-splicing. Gene Ther. 2005;12:1477–1485. doi: 10.1038/sj.gt.3302596. [DOI] [PubMed] [Google Scholar]
- 12.Wally V., Murauer E.M., Bauer J.W. Spliceosome-mediated trans-splicing: the therapeutic cut and paste. J. Invest. Dermatol. 2012;132:1959–1966. doi: 10.1038/jid.2012.101. [DOI] [PubMed] [Google Scholar]
- 13.Wood M., Yin H., McClorey G. Modulating the expression of disease genes with RNA-based therapy. PLoS Genet. 2007;3:e109. doi: 10.1371/journal.pgen.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorain S., Peccate C., Le Hir M., Garcia L. Exon exchange approach to repair Duchenne dystrophin transcripts. PLoS ONE. 2010;5:e10894. doi: 10.1371/journal.pone.0010894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mearini G., Stimpel D., Krämer E., Geertz B., Braren I., Gedicke-Hornung C., Précigout G., Müller O.J., Katus H.A., Eschenhagen T. Repair of Mybpc3 mRNA by 5′-trans-splicing in a mouse model of hypertrophic cardiomyopathy. Mol. Ther. Nucleic Acids. 2013;2:e102. doi: 10.1038/mtna.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilat U., Dechat T., Bertrand A.T., Woisetschläger N., Gotic I., Spilka R., Biadasiewicz K., Bonne G., Foisner R. The muscle dystrophy-causing ΔK32 lamin A/C mutant does not impair the functions of the nucleoplasmic lamin-A/C-LAP2α complex in mice. J. Cell Sci. 2013;126:1753–1762. doi: 10.1242/jcs.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985;43:667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- 19.Konarska M.M., Padgett R.A., Sharp P.A. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. doi: 10.1016/s0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- 20.Chao H., Mansfield S.G., Bartel R.C., Hiriyanna S., Mitchell L.G., Garcia-Blanco M.A., Walsh C.E. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- 21.Tahara M., Pergolizzi R.G., Kobayashi H., Krause A., Luettich K., Lesser M.L., Crystal R.G. Trans-splicing repair of CD40 ligand deficiency results in naturally regulated correction of a mouse model of hyper-IgM X-linked immunodeficiency. Nat. Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- 22.Wally V., Klausegger A., Koller U., Lochmüller H., Krause S., Wiche G., Mitchell L.G., Hintner H., Bauer J.W. 5′ Trans-splicing repair of the PLEC1 gene. J. Invest. Dermatol. 2008;128:568–574. doi: 10.1038/sj.jid.5701152. [DOI] [PubMed] [Google Scholar]
- 23.Mansfield S.G., Kole J., Puttaraju M., Yang C.C., Garcia-Blanco M.A., Cohn J.A., Mitchell L.G. Repair of CFTR mRNA by spliceosome-mediated RNA trans-splicing. Gene Ther. 2000;7:1885–1895. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- 24.Puttaraju M., Jamison S.F., Mansfield S.G., Garcia-Blanco M.A., Mitchell L.G. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 25.Puttaraju M., DiPasquale J., Baker C.C., Mitchell L.G., Garcia-Blanco M.A. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Mol. Ther. 2001;4:105–114. doi: 10.1006/mthe.2001.0426. [DOI] [PubMed] [Google Scholar]
- 26.Berger A., Lorain S., Joséphine C., Desrosiers M., Peccate C., Voit T., Garcia L., Sahel J.A., Bemelmans A.P. Repair of rhodopsin mRNA by spliceosome-mediated RNA trans-splicing: a new approach for autosomal dominant retinitis pigmentosa. Mol. Ther. 2015;23:918–930. doi: 10.1038/mt.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He X., Liu F., Yan J., Zhang Y., Yan J., Shang H., Dou Q., Zhao Q., Song Y. Trans-splicing repair of mutant p53 suppresses the growth of hepatocellular carcinoma cells in vitro and in vivo. Sci. Rep. 2015;5:8705. doi: 10.1038/srep08705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorain S., Peccate C., Le Hir M., Griffith G., Philippi S., Précigout G., Mamchaoui K., Jollet A., Voit T., Garcia L. Dystrophin rescue by trans-splicing: a strategy for DMD genotypes not eligible for exon skipping approaches. Nucleic Acids Res. 2013;41:8391–8402. doi: 10.1093/nar/gkt621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bechert K., Lagos-Quintana M., Harborth J., Weber K., Osborn M. Effects of expressing lamin A mutant protein causing Emery-Dreifuss muscular dystrophy and familial partial lipodystrophy in HeLa cells. Exp. Cell Res. 2003;286:75–86. doi: 10.1016/s0014-4827(03)00104-6. [DOI] [PubMed] [Google Scholar]
- 30.Favreau C., Dubosclard E., Ostlund C., Vigouroux C., Capeau J., Wehnert M., Higuet D., Worman H.J., Courvalin J.C., Buendia B. Expression of lamin A mutated in the carboxyl-terminal tail generates an aberrant nuclear phenotype similar to that observed in cells from patients with Dunnigan-type partial lipodystrophy and Emery-Dreifuss muscular dystrophy. Exp. Cell Res. 2003;282:14–23. doi: 10.1006/excr.2002.5669. [DOI] [PubMed] [Google Scholar]
- 31.Azibani F., Muchir A., Vignier N., Bonne G., Bertrand A.T. Striated muscle laminopathies. Semin. Cell Dev. Biol. 2014;29:107–115. doi: 10.1016/j.semcdb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Coady T.H., Lorson C.L. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J. Neurosci. 2010;30:126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koller U., Hainzl S., Kocher T., Hüttner C., Klausegger A., Gruber C., Mayr E., Wally V., Bauer J.W., Murauer E.M. Trans-splicing improvement by the combined application of antisense strategies. Int. J. Mol. Sci. 2015;16:1179–1191. doi: 10.3390/ijms16011179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger A., Maire S., Gaillard M.C., Sahel J.A., Hantraye P., Bemelmans A.P. mRNA trans-splicing in gene therapy for genetic diseases. Wiley Interdiscip. Rev. RNA. 2016;7:487–498. doi: 10.1002/wrna.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monjaret F., Bourg N., Suel L., Roudaut C., Le Roy F., Richard I., Charton K. Cis-splicing and translation of the pre-trans-splicing molecule combine with efficiency in spliceosome-mediated RNA trans-splicing. Mol. Ther. 2014;22:1176–1187. doi: 10.1038/mt.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 37.Inagaki K., Fuess S., Storm T.A., Gibson G.A., Mctiernan C.F., Kay M.A., Nakai H. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarty D.M. Self-complementary AAV vectors; advances and applications. Mol. Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 39.Coady T.H., Baughan T.D., Shababi M., Passini M.A., Lorson C.L. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS ONE. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snyder R.O., Spratt S.K., Lagarde C., Bohl D., Kaspar B., Sloan B., Cohen L.K., Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum. Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 41.Lorain S., Gross D.A., Goyenvalle A., Danos O., Davoust J., Garcia L. Transient immunomodulation allows repeated injections of AAV1 and correction of muscular dystrophy in multiple muscles. Mol. Ther. 2008;16:541–547. doi: 10.1038/sj.mt.6300377. [DOI] [PubMed] [Google Scholar]
- 42.Boudreau É., Labib S., Bertrand A.T., Decostre V., Bolongo P.M., Sylvius N., Bonne G., Tesson F. Lamin A/C mutants disturb sumo1 localization and sumoylation in vitro and in vivo. PLoS ONE. 2012;7:e45918. doi: 10.1371/journal.pone.0045918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.