This work shows that sphingosine kinase 1 (SphK1) deficiency inhibits human epidermal growth factor 2 (HER2)-positive breast carcinogenesis probably by regulating claudin-2. The results suggest that targeting SphK1 may represent a novel approach for HER2-positive breast cancer chemoprevention and/or treatment.
Abstract
Aberrant sphingolipid metabolism has been reported to promote breast cancer progression. Sphingosine kinase 1 (SphK1) is a key metabolic enzyme for the formation of pro-survival S1P from pro-apoptotic ceramide. The role of SphK1 in breast cancer has been well studied in estrogen receptor (ER)-positive breast cancer; however, its role in human epidermal growth factor 2 (HER2)-positive breast cancer remains unclear. Here, we show that genetic deletion of SphK1 significantly reduced mammary tumor development with reduced tumor incidence and multiplicity in the MMTV-neu transgenic mouse model. Gene expression analysis revealed significant reduction of claudin-2 (CLDN2) expression in tumors from SphK1 deficient mice, suggesting that CLDN2 may mediate SphK1’s function. It is remarkable that SphK1 deficiency in HER2-positive breast cancer model inhibited tumor formation by the different mechanism from ER-positive breast cancer. In vitro experiments demonstrated that overexpression of SphK1 in ER−/PR−/HER2+ human breast cancer cells enhanced cell proliferation, colony formation, migration and invasion. Furthermore, immunostaining of SphK1 and CLDN2 in HER2-positive human breast tumors revealed a correlation in high-grade disease. Taken together, these findings suggest that SphK1 may play a pivotal role in HER2-positive breast carcinogenesis. Targeting SphK1 may represent a novel approach for HER2-positive breast cancer chemoprevention and/or treatment.
Introduction
Breast cancer is the most common malignancy among US women and the second leading cause of cancer mortality in US women (1). Human epidermal growth factor 2 (HER2) amplification is found in approximately 15–30% of primary breast cancer (2), and these patients are noted to harbor more aggressive phenotypes with poor prognosis (3–5). In the past decade, HER2 targeted therapies such as trastuzumab and lapatinib have dramatically improved survival; however, resistance to these agents remains a challenge (6). Thus, there is an urgent need to identify novel targets for the development of chemopreventive and therapeutic agents geared toward HER2 overexpressing breast cancers.
Sphingolipids, specifically ceramide, sphingosine and S1P, are bioactive mediators with an important role in various biological functions. Although these three sphingolipid metabolites are closely interconnected, they possess opposite biological functions; ceramide and sphingosine regulate apoptosis and cell senescence, whereas S1P regulates cell migration, survival, proliferation, angiogenesis and inflammation (7–10). Dysregulation in the ceramide-sphingosine-S1P rheostat leads to disruption in downstream signaling and ultimately of biological functions noted above. Sphingosine kinases (SphKs) are key metabolic enzymes for the formation of pro-survival S1P from pro-apoptotic ceramide. There are two SphK isoforms known: inducible SphK1 and constitutive SphK2 (11,12). Induced SphK1 produces excess S1P, which subsequently promotes tumor growth and survival through increases in cell proliferation, migration and angiogenesis, and protection from apoptosis (7,9,13). Indeed, a number of studies have shown that SphK1 is overexpressed in many human tumors, including breast cancer, where it may contribute to malignant transformation and tumor progression (9,14,15). Therefore, SphK1 plays a crucial role in sphingolipid-mediated functions that can lead to cancer development by regulating the cellular balance between ceramide and S1P.
Previously, our group reported that SphK1 was associated with colon carcinogenesis and with head and neck tumorigenesis (14,16–19). Interestingly, we showed that SphK1 regulated the cyclooxygenase-2 (COX-2)/prostaglandin E2 (PGE2) pathway in colon cancer (14), whereas SphK1 regulated Akt pathway in head and neck cancer (16). In breast cancer, it has been reported that high expression of COX-2 is observed in 37.4% of tumors and high COX-2 levels are associated with a large tumor size, high histological grade, negative hormone receptor status, a high proliferation rate, high p53 expression, axillary node metastasis and the presence of HER2 oncogene amplification (20,21). Additionally, the COX-2 selective inhibitor, celecoxib, and COX-2 deletion have been shown to inhibit HER2-induced breast tumorigenesis in mice (21,22). Thus, we hypothesized that SphK1 also regulates the COX-2/PGE2 pathway in breast cancer.
In breast cancers, high levels of SphK1 mRNA are found in 80% of breast tumors, with an average increase of 2-fold compared with normal tissue from the same patient (15). Additionally, high SphK1 levels correlated with poor prognosis and drug resistance in breast cancer patients (15,23,24). Although recent exciting studies explored the mechanism by which estrogen regulates SphK1 expression, the role of SphK1 in HER2-positive breast cancer remains largely unknown. It is noteworthy that estrogen receptor (ER)-positive and HER2-negative breast tumors with high SphK1 expression are known to be resistant to chemotherapy and to have a poor prognosis (25). On the other hand, ER-positive and HER2-positive tumors with high SphK1 expression were noted to be associated with improve patient survival and reduction in tumor recurrence, when treated with tamoxifen (26). The results suggested that overexpression of SphK1 may have an inhibitory effect on HER2-positive breast cancer. However, in a recent study, Ohotski et al. reported that high expression of SphK1 in ER-negative and HER2-positive tumors was linked with reduced disease-free survival compared with patients with low SphK1 expression (27). Thus, the role and effect of SphK1 expression in HER2 positive tumors is controversial. Mechanistically, the inhibition of SphK1 in HER2-positive breast cancer cell lines has been shown to reduce HER2 and p-ERK1/2 activation (27). Although these studies shed some light on SphK1 in HER2 breast tumors, a gap in knowledge remain as to it is still whether SphK1 have both pro- and anti-tumoral effects in HER2-induced breast tumorigenesis.
Herein, using a MMTV-neu Tg mice model in which breast cancer is driven by overexpression of the wild-type (unactivated) HER2/neu allele, we report that deletion of SphK1 significantly reduced HER2/neu-induced breast tumor development including incidence and multiplicity. We also show that global gene expression analysis of these mice tumors identified claudin 2 (CLDN2) as a potential downstream target of SphK1. Additionally, in vitro studies revealed that forced SphK1 expression enhances cell proliferation, colony formation and migration/invasion. Finally, we confirmed by immunohistochemistry (IHC) that the expression of both SphK1 and CLDN2 proteins was correlated with HER2-positive human breast cancer pathologic features.
Materials and methods
Animals
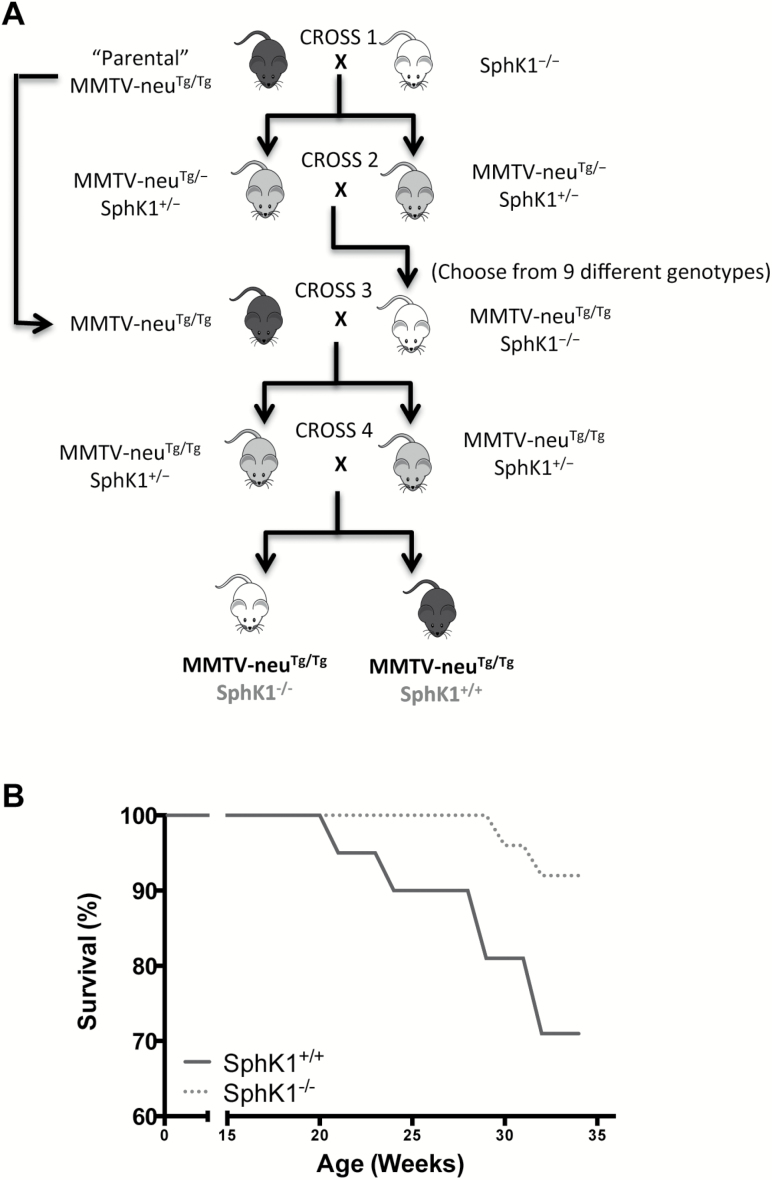
Animals were housed and handled in the animal and veterinary service program of the University of Hawaii (U.H.). All animal experiments were approved by the local institutional animal care and use committee. Animals were maintained under controlled conditions of humidity (50 ± 10%), light (12/12 h light/dark cycle) and temperature (23 ± 2°C) and allowed free access to diet and water. MMTV-neu Tg mice [FVB/N-Tg(MMTVneu)202Mul/J] were purchased from Jackson Laboratory, which overexpresses wild-type (unactivated) form of HER2/neu under MMTV-LTR promoter (28), and bred to produce multiple litters. SphK1 homozygous KO (SphK1−/−) mice of the 129SV-C57BL/6 background were a kind gift from Dr. Richard L. Proia [National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK/NIH)] (29). The background strain of MMTV-neu Tg mice was FVB, whereas the original background stain of SphK1 KO mice was C57BL/6. Because the background of C57BL/6 was well-known modifiers of experimental tumor formation (30), we first backcrossed SphK1 KO mice to FVB/NCrl (Charles River Laboratories) at least 10 times to introduce the targeted SphK1 allele onto an FVB background (29). MMTV-neuTg/Tg (homozygous) and FVB SphK1−/− were intercrossed several generations to produce females of the required test genotypes; SphK1 wild-type (SphK1+/+) and homozygous SphK1 KO (SphK1−/−) with MMTV-neu Tg/Tg (Figure 1A). In brief, MMTV-neuTg/Tg male mice were crossbred to SphK1−/− female mice (cross 1), and the resultant MMTV-neuTg/−, SphK1+/− progeny were bred with each other (cross 2) to generate MMTV-neuTg/Tg, SphK1−/− progeny. These MMTV-neuTg/Tg, SphK1−/− mice were bred with MMTV- neuTg/Tg parental strain (cross 3) and resultant MMTV-neuTg/Tg, SphK1+/−progeny were crossed with each other (cross 4) to generate MMTV-neuTg/Tg mice that are SphK1+/+ or SphK1−/−. All animals in this study were virgin females to eliminate any hormonal concerns. The body weight of SphK1+/+ and SphK1−/− mice were comparable throughout the study (data not shown). Tumor appearance and size were monitored weekly. Animals were killled at 35 weeks of age or regardless of age when the largest diameter of the tumor reached 2 cm.
Figure 1.
Genetic deletion of SphK1 reduced HER2/neu-induced breast tumorigenesis. (A) Breeding schematics of MMTV-neu and SphK1 KO mice. SphK1−/− female mice were crossbred to MMTV-neuTg/Tg transgenic male mice (cross 1), and the resultant SphK1+/−, MMTV-neu Tg/− progeny were bred with each other (cross 2) to generate SphK1−/−, MMTV-neuTg/Tg progeny. SphK1−/−, MMTV-neuTg/Tg mice were bred with MMTV-neuTg/Tg (cross 3) and resultant SphK1+/− with MMTV-neuTg/Tg progeny were crossed with each other (cross 4) to generate SphK1+/+ or SphK1−/− with MMTV-neuTg/Tg mice. (B) Tumor-free survival was compared between SphK1−/− (n = 26) and SphK1+/+ (n = 21) mice using Kaplan-Meier analysis. Mice were considered dead when the largest diameter reached 2 cm. *P < 0.05.
Gene expression microarray analysis
Total RNA from 10 mg of mouse tumor tissues was isolated using RNeasy Plus Mini kit (Qiagen, Valencia, CA) according to the manufacture’s instruction. Total RNA from tumor tissue was reversely transcribed to generate double stranded complementary DNA (cDNA) and was subsequently labeled with biotin [(complementary (cRNA)]. Resulting cRNA was hybridized to MouseWG-6 v2.0 Expression BeadChip Array (Illumina, Inc., San Diego, CA, USA). The array was washed and stained with streptavidin-Cy3 and subsequently scanned to acquire images providing the raw data of gene expression. For analysis, pre-processing and normalization were performed using the Partek Microarray data analysis software (http://www.partek.com/). Partek pre-processes raw intensity files from microarray experiment using GCRMA’s background subtraction and uses quantile normalization as the normalization technique. The samples from different groups were compared using two-way analysis of variance (ANOVA). Significantly changed genes obtained from ANOVA were prioritized by a combination of P-value and fold changes. We used cutoffs for P-value at 0.05 and fold changes at 1.5. Association of gene expression patterns with biological functional and signaling networks was generated through the use of QIAGEN’s Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). The assay was performed by the Genome Analysis Core Mayo Clinic (Rochester, MN, USA). The Biomedical Statistics and Bioinformatics staff at Mayo Clinic performed data analyses.
Patients and sample collection
HER2-positive human breast tumors were obtained as part of an institutionally approved study under the direction of CDKC and LWML (CHS#18704). Breast tissue obtained from patients was used to construct a tissue microarray (TMA) in the Pathology Shared Resource laboratory of the University of Hawaii Cancer Center. Each breast cancers case was represented by three tumor cores (0.6 mm in diameter) and, when available, two adjacent normal cores. The TMA included 92 tumor cases and 34 adjacent normal tissues.
Immunohistochemistry (IHC)
At the time on necropsy, mammary tumors were resected and placed in 10% formalin, and then embedded into paraffin. Paraffin blocks were cut (5 uM) and placed on Superfrost Plus slide. IHC was carried out on paraffin-embedded sections using the avidin-biotin complex immunoperoxidase technique as previously described (31,32). Details of the antibodies and conditions are described in Supplementary material, available at Carcinogenesis Online. An experienced pathologist assessed all slides. The proportion of positive cells was scored in four groups and represented the estimated proportion of immunoreactive cells (animal tissues, 0 = 0% of cells; 1 = 1–10%; 2 = 11–50% and 3 = 50–100%).
Similar to the above animal tissue, IHC for SphK1 and CLDN2 was carried out on the HER2-positive human breast cancer TMA using the avidin-biotin complex immunoperoxidase technique as previously described (31,32). Details of the antibodies and conditions are described in Supplementary Material, available at Carcinogenesis Online. An experienced pathologist assessed all specimens. The proportion of immunopositive cells was scored in four groups: 0 = 0% of cells; 1 = 1–10%; 2 = 11–75% and 3 = 75–100%. The intensity was scored and represented the average intensity of immunopositive cells (0 = none; 1 = weak; 2 = intermediate and 3 = strong). The proportion and intensity scores were combined to obtain a total protein staining score, which ranged from 0 to 6. The expression level was determined based on the total staining score as follows: low = 0–2, moderate = 3 or 4 and high = 5 or 6.
RNA extraction and quantitative PCR
RNA extraction was performed using an RNeasy Plus kit (QIAGEN) as described above. cDNA was synthesized from these RNAs using iScript™ Reverse Transcription Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Quantitative PCR (qPCR) was performed using the standard reagents and protocols (Bio-Rad Laboratories). List of primers used for the analysis is provided in Supplementary material, available at Carcinogenesis Online.
Cell culture and transfection
The MDA-MB-453 human breast cancer cell line (ER−, PR−, HER2+) was obtained from ATCC (Manassas, VA, USA) and cultured in DMEM plus 10% FBS and 1% penicillin streptomycin at 37°C and 5% CO2. SKBR3 human breast cancer cell line (ER−, PR−, HER2+) was obtained from ATCC and was cultured in DMEM plus 10% FBS, 1% nonessential amino acids and 1% penicillin streptomycin at 37°C and 5% CO2. The cell lines have been authenticated by Genetic Resources Core Facility at Johns Hopkins (Baltimore, MD) using DNA fingerprinting (Short Tandem Repeat profiling) in 2015.
Human SphK1 expression plasmids were constructed by inserting purified pDONR223-SPHK1 (Addgene, Cambridge, MA, USA (33)) into pcDNA™6.2/V5-DEST Gateway Vectors (Life Technologies, Carlsbad, CA, USA) using LR clonase enzyme (Life Technologies). Next, human SphK1 expression plasmids were constructed in pEGFP-C1 vector as described in the previous study (34). MDA-MB-453 cell (pcDNA6.2) and SKBR3 (pEGFP) cells were transfected with SphK1 or mock control plasmid using jetPRIME (Polyplus Transfection, Illkirch-Graffenstaden, France). Stable transfectant carrying SphK1 in MDA-MB-453 and SKBR3 was selected with Blasticidin (8 µg/ml) and Geneticin (600 µg/ml) (Life Technologies), respectively. Integration of the transfected gene into chromosome was confirmed by RT-PCR, and protein expression of Sphk1 was confirmed by western blot.
Cell proliferation
The breast cancer cells were seeded in 96-well plates at a density of 2.5–5 × 103 cells. After 24, 48, 72 and 96 h of incubation, tetrazolium-based colorimetric assay (MTT assay) was performed according to manufacture’s instruction (Trevigen, Inc., Gaithersburg, MD, USA). Absorbance was read on a microplate reader (BioTek, Winooski, VT, USA) at 562-nm wavelength. The values were normalized to values obtained at 24 h to determine the survival percent. Each assay was performed at least three times in eight replicates.
Anchorage-independent colony formation
Breast cancer cells were suspended in culture medium containing 0.6% agarose and seeded onto a coating composed of culture medium containing 1.2% of agarose in a 96-well plate at a density of 5 × 103 cells. The number of colonies was quantified using CyQuant Cell Proliferation Kit (Life Technologies) according to manufacture’s instruction after solubilization of agarose with 50-units/well agarase. The fluorescence proportional to the number of colonies was read on a microplate reader at excitation/emission wavelengths of 485/528 nm. The fluorescence values were normalized to values obtained in the control group to determine the relative colony formation. Each assay was performed at least three times in four replicates.
Migration and invasion
The breast cancer cells were serum starved overnight. Then, 0.25 × 106 to 0.5 × 106 cells were seeded on polycarbonate membrane (migration) or matrigel-coated (invasion) inserts with 8.0-µm pore size in 24-well plates (Corning, Inc., Corning, NY, USA) and allowed to migrate/invade toward 10% serum. Twenty-four hours later, non-migrated/invaded cells on the upper surface of the filter were removed and the migrated/invaded cells were quantified using CyQuant Cell Proliferation Kit according to manufacture’s instruction. The fluorescence proportional to the number of invaded/migrated cells was read on a microplate reader at excitation/emission wavelengths of 485/528 nm. The fluorescence values were normalized to values obtained in the control group to determine the relative migration/invasion. Each assay was performed at least three times in triplicate.
Statistical analysis
For animal experiments, tumor incidence is presented as mean percentage, whereas tumor multiplicity and tumor volume values are presented as mean ± SEM. Statistical comparisons between groups for tumor incidence, multiplicity and volume were performed using Fisher’s exact test, unpaired Student’s t-test, and one-way ANOVA, followed by Tukey and Dennett’s test, respectively. For cell culture assays, data from control and SphK1 overexpressing cells were analyzed with the unpaired Student’s t test. Differences were considered statistically significant at P < 0.05. The SAS 9.4 (SAS Institute, Inc., Cary, NC, USA) Logistic procedure ran logistic regression (cumulative logit) analyses estimating IHC scores.
Results
SphK1 deficiency reduces HER2/neu-induced breast tumor development in mice
To induce HER2/neu-dependent tumorigenesis in animals, we employed a MMTV-neu transgene-induced breast cancer mouse model. MMTV-neu (FVB background) mice and SphK1 KO (FVB background) mice were intercrossed to generate MMTV-neu females that were either wild-type (SphK1+/+) or homozygous KO (SphK1−/−) for SphK1 gene (Figure 1A). In order to avoid neu transgene copy number variance on tumor development and growth, we analyzed copy numbers of neu gene between SphK1+/+ and SphK1−/− groups. The data showed no significant difference in copy number between SphK1+/+ (2.57 ± 0.08) and SphK1−/− (2.47 ± 0.07) mice (P = 0.8889, Supplementary Figure S1A, available at Carcinogenesis Online), indicating that we can exclude the influence of neu transgene copy number on tumor development in the study. We also observed that SphK1−/− mice expressed no to minimal SphK1 mRNA in the tumors (Supplementary Figure S1B, available at Carcinogenesis Online).
We found that SphK1 deficiency significantly reduced tumor incidence 19% (5/26) in SphK1−/− mice compared with 57% (12/21; P = 0.0136) in SphK1+/+ mice (Table 1). Tumor multiplicity was also reduced in SphK1−/− (0.27 tumors per mouse) compared with SphK1+/+ (0.67 tumors per mouse; P = 0.0469) mice (Table 1). There was no significant difference in the size of the tumors between SphK1+/+ (2841 mm3) and SphK1−/− (1185 mm3) mice (P = 0.5285, Table 1). In the reported novel mouse model, SphK1 deficiency significantly improved survival (P = 0.0461, Figure 1B).
Table 1.
SphK1 genetic deletion inhibits HER2/neu-indued breast tumorigenesis in mice
| SphK1 genotype | Incidence | Multiplicity | Volume (mm3) |
|---|---|---|---|
| SphK1+/+ | 57% (12/21) | 0.67 ± 0.14 | 2841 ± 1245 |
| SphK1−/− | 19% (5/26)* | 0.27 ± 0.13* | 1185 ± 871 |
The tumor number and volume were recorded at necropsy (35 weeks of age). Values in parentheses represent percentage of mice with tumors. Multiplicity and tumor volume values are mean ± SEM. *P < 0.05 vs. SphK1+/+ (wild-type); Student’s t-test.
We then analyzed the protein expression status of ER, PR and HER2 of the mouse tumors using immunohistochemical staining. Consistent with previous reports showing that this mouse strain doesn’t express ER alpha (ERα) and PR (28), tumors from both groups expressed no to very weak ERα and PR and high HER2 (Supplementary Figure S2, available at Carcinogenesis Online). Next, to investigate how SphK1 loss inhibits tumor development, we examined proliferation (PCNA), angiogenesis (CD31), immune response (CD45) and p53 expression in tumors by IHC staining between SphK1+/+ and SphK1−/− mice. The results did not show any difference between SphK1+/+ and SphK1−/− mice (Supplementary Figure S2, available at Carcinogenesis Online).
SphK1 deficiency reduces claudin-2 expression in HER2/neu-induced breast tumor in mice
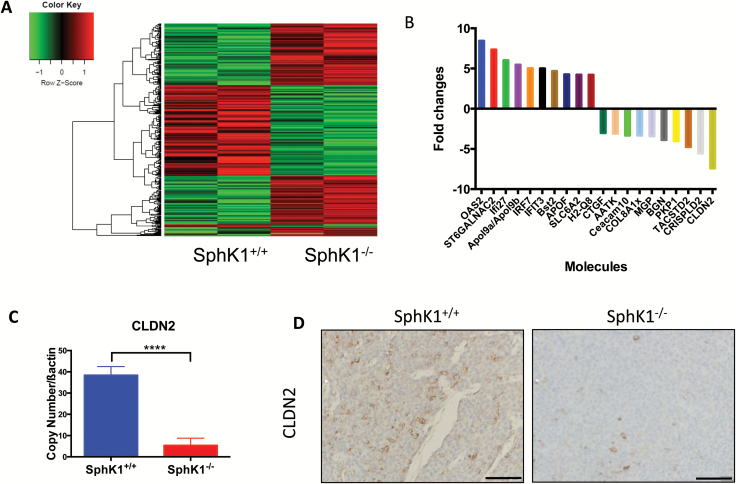
Next, we set out to explore the mechanism by which SphK1 could regulate breast tumorigenesis. Using gene expression microarray analysis, we investigated the effect of SphK1 loss on 45200 transcripts in HER2/neu-induced mouse breast tumors. The gene expression profiles of tumors from MMTV-neu mice with SphK1+/+ (n = 2) and SphK1−/− (n = 2) genotypes were compared and analyzed with a cutoff of P-value 0.05 and fold changes at 1.5. We identified 181 genes up-regulated and 149 genes down-regulated in tumors from SphK1−/− mice compared with tumors from SphK1+/+ mice (Figure 2A, online data supplement and GEO accession number GSE80413). The top 10 ranked genes with the highest degree of differential expression between SphK1+/+ and SphK1−/− tumors are shown in Figure 2B. None of these genes have previously been linked to HER2 expression. Ingenuity Pathway Analysis identified cellular movement (ST6GALNAC2, MGP, Ceacam10, TACSTD2, BGN and IRF7) and cell-to-cell signaling (SLC6A2, BGN, IRF7 and CLDN2) as the highest ranked functional groupings (Supplementary Table S1A, available at Carcinogenesis Online), and associated canonical pathways included the antigen presentation pathway, interferon signaling, hepatic fibrosis/hepatic stellate cell activation, protein ubiquitination pathway and allograft rejection signaling (Supplementary Table S1B, available at Carcinogenesis Online). Additionally, six genes (OAS2, Ifi27, IRF7, IFIT3, Bst2 and CLDN2) demonstrated a dose dependent response in gene expression when we analyzed these expressions in the tumors from SphK1+/− mice by qPCR (data not shown). The data suggest a correlation of these genes with SphK1 expression. Of the genes regulated by SphK1, CLDN2 had the greatest associated change in expression level and was the basis for our subsequent analysis. Specifically, SphK1 deficiency induced downregulation of CLDN2 expression in tumors, and this was confirmed by IHC and qPCR (Figure 2C and D).
Figure 2.
Genetic deletion of SphK1 reduced CLDN2 in HER2/neu-induced mice breast tumors. (A) Whole genome expression profile evaluated by microarray analysis assessed the expression of 39000 gene transcripts. Gene expression microarray heatmap (n = 2 per group) of dysregulated genes. (B) Twenty molecules with the most significant expression changes (10 up-regulation and 20 down-regulation) between SphK1+/+ and SphK1−/− tumors are shown. (C) CLDN2 mRNA expression in tumors from SphK1+/+ and SphK1−/− mice was confirmed by qRT-PCR analysis and analyzed by unpaired t-test. ****P ≤ 0.0001 vs. SphK1+/+. (D) Representative paraffin-tumor sections immunostained for CLDN2 in tumors from SphK1+/+ and SphK1−/− mice (P = 0.1339).
SphK1 overexpression induces cancerous cellular functions in HER2-positive breast cancer cell lines
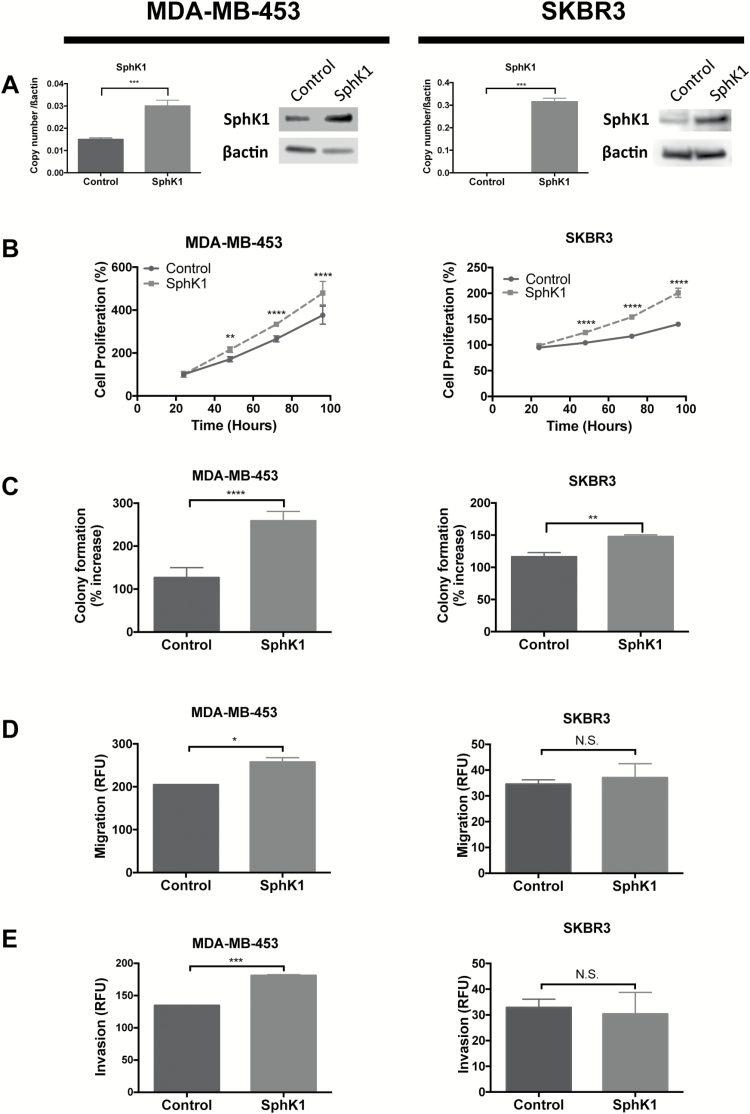
Modulations in the cell’s ability to proliferate, migrate, invade and inhibit apoptosis are key hallmarks of cancers (35). Here, we examined whether SphK1 overexpression induces key cellular functions in ER−/PR−/HER2+ MDA-MB-453 and SKBR3 breast cancer cell lines. First, overexpression of SphK1 mRNA and protein in MDA-MB-453 and SKBR3 cell lines were confirmed by qPCR and western blot, respectively (Figure 3A). Overexpression of SphK1 significantly accelerated cellular proliferation in both MDA-MB-453 and SKBR3 cells, compared with control (P < 0.001 and P < 0.001, respectively; Figure 3B). Furthermore, Sphk1 overexpression significantly enhanced anchorage-independent growth ability in MDA-MB-453 and SKBR3 cells (P < 0.0001 and P < 0.01, respectively; Figure 3C). Overexpression of SphK1 in MDA-MB-453 also increased the ability of cells to migrate and invade (P < 0.05 and 0.001, respectively; Figure 3D and E); however, no significant effects were observed on migration and invasion in SKBR3 cells (P = 0.7932 and P = 0.1755, respectively; Figure 3D and E).
Figure 3.
SphK1 overexpression increased cell growth and aggressiveness MDA-MB-453 and SKBR3 cell lines. MDA-MB-453 and SKBR3 cells transfected with SphK1 and stable cell lines overexpressing SphK1 were generated. (A) qRT-PCR and SphK1 of SphK1 genes in MDA-MB-453 and MDA-MB-231 cells stably transfected with SphK1 or mock control plasmid. (B) Cell proliferation monitored by MTT assay. Cells were seeded at 2.5 × 103 cells or 5 × 103 cells and proliferation was measured at 24, 48, 72 and 96 h. (C) Colony formation measured by soft agar assay. Cells were seeded at 5 × 103 cells and colony formation was measured after 6–10 days. (D) and (E) Migration and invasion assays. Serum starved cells were seeded at 0.25 × 106 to 0.5 × 106 cells in polycarbonate membrane (D) or matrigel coat (E) inserts and allowed to migrate/invade against 10% serum. (A)–(E) Data from one representative experiment are presented as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001 and ***P < 0.001.
SphK1 expression is correlated with CLDN2 expression and associated with clinical stages of HER2-positive human breast cancer
To assess the expression of SphK1 and CLDN2 in a large number of human breast cancer samples, we performed IHC for SphK1 and CLDN2 on a primary breast cancer TMA containing HER2-positive tumors with matched adjacent-normal breast tissues. Information on demographics and clinicopathologic presentation is presented in Table 2.
Table 2.
Characteristics of HER2-positive breast cancer cases
| Tumor (N = 92) | Adjacent normal (N = 34) | |||
|---|---|---|---|---|
| n | Percent of total | n | Percent of total | |
| Age group | ||||
| 25–49 | 33 | 36 | 13 | 38 |
| 50–69 | 49 | 53 | 17 | 50 |
| ≥70 | 9 | 10 | 4 | 12 |
| Unknown | 1 | 1 | 0 | 0 |
| Race/ethnicity | ||||
| Caucasian | 27 | 29 | 10 | 29 |
| Japanese | 36 | 39 | 11 | 32 |
| Native Hawaiian | 28 | 30 | 13 | 38 |
| Unknown | 1 | 1 | 0 | 0 |
| Histology | ||||
| In filtrating ductal carcinoma | 87 | 95 | ||
| Lobular carcinoma | 3 | 3 | ||
| Other | 1 | 1 | ||
| Unknown | 1 | 1 | ||
| Grade | ||||
| 1 | 4 | 4 | ||
| 2 | 20 | 22 | ||
| 3 | 67 | 73 | ||
| Unknown | 1 | 1 | ||
| Stage | ||||
| 1 | 20 | 22 | ||
| 2A | 46 | 50 | ||
| 2B-4 | 25 | 27 | ||
| Unknown | 1 | 1 | ||
| ER | ||||
| Negative | 43 | 47 | ||
| Positive | 48 | 52 | ||
| Unknown | 1 | 1 | ||
| PR | ||||
| Negative | 37 | 40 | ||
| Positive | 54 | 59 | ||
| Unknown | 1 | 1 | ||
| ER and PR | ||||
| ER−PR− | 34 | 37 | ||
| ER−PR+ | 9 | 10 | ||
| ER+PR− | 3 | 3 | ||
| ER+PR+ | 45 | 49 | ||
| Unknown | 1 | 1 | ||
| Vital status | ||||
| Alive | 83 | 90 | ||
| Deceased | 8 | 9 | ||
| Unknown | 1 | 1 | ||
Cancer cases were comprised of adult females over the age of 25 of Caucasian, Japanese or Native Hawaiian ancestry. Tumors were predominantly invasive infiltrating ductal carcinoma (95%), stage 2A (50%) and poorly differentiated/grade 3 (73%). Forty-nine percent of cases were ER+PR+. Staining representing each group is shown in Supplementary Figure S4, available at Carcinogenesis Online.
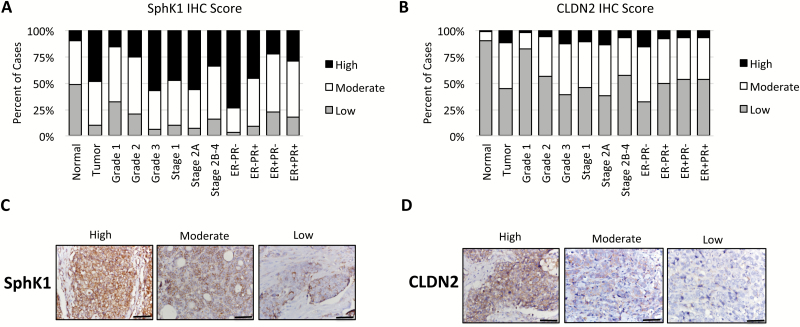
The SAS 9.4 program (SAS Institute, Inc., Cary, NC, USA) was used to perform logistic regression (cumulative logit) analyses for IHC scores. These IHC models estimated that 48% of the tumors expressed SphK1 at high levels, whereas only 10% of adjacent-normal tissues highly expressed SphK1 (P < 0.0001; Figure 4A). Similarly, an estimated 55% of tumors expressed moderate-high levels of CLDN2, whereas only 9% of adjacent-normal tissues expressed CLDN2 at moderate-high (P < 0.0001; Figure 4B).
Figure 4.
SphK1 and CLDN2 levels are increased in HER2-positive breast cancers in humans. Tissue microarray containing breast cancer samples from 92 HER2-positive breast cancer patients and matched adjacent normal breast tissues (n = 34) are stained for SphK1 and CLDN2. Staining were scored based on the intensity (0–3) and proportion (0–3), and the total scores were used for the analysis. [(A) and (B)] SphK1 and CLDN2 expressions were compared with pathological features including grade, stage and hormone receptor status. [(C) and (D)] Representative image of high, moderate and low SphK1 and CLDN2 expressing tumors.
Furthermore, we estimated significantly enhanced SphK1 expression in high-grade tumors compared with low-grade tumors (grade 1, 16%; grade 2, 25% and grade 3, 57%; P = 0.0004; Figure 4A). The expression of SphK1 correlated with ER-negativity, where high SphK1 expressions were estimated in 74% of ER−/PR− and 46% of ER−/PR+ tumors compared with 23% and 29% in ER+/PR− and ER+/PR+ tumors, respectively (P < 0.0001). These results are consistent with a previous report showing high SphK1 mRNA expression among ER-negative tumors (23). Expression of SphK1 was slightly higher in lower stage tumors (Stages 1 and 2A) compared with stage 2B-4, suggesting that SphK1 may play a key role in the early stages of malignant transformation. Similar to SphK1, a higher proportion of moderate-high CLDN2 expression was in high-grade tumors (grade 1, 18%; grade 2, 43% and grade 3, 60%; P < 0.01; Figure 4B). Additionally, although 68% of ER−/PR− tumors showed moderate-high CLDN2 expression, only 50, 47 and 46% of ER−/PR+, ER+/PR− and ER+/PR+ tumors expressed CLDN2 at the same level, respectively (P < 0.0001). Importantly, SphK1 expression was strongly correlated with CLDN2 expression in HER2 positive primary breast tumor tissues (r = 0.71, P < 0.0001; Supplementary Table S2, available at Carcinogenesis Online).
Discussion
To the best of our knowledge, this is the first report to show that genetic deletion of SphK1 protects against HER2/neu dependent mammary tumorigenesis. Using MMTV-neu transgenic mice, we demonstrated that loss of SphK1 caused a decrease in tumor incidence and tumor multiplicity. Furthermore, using gene expression microarray analysis of mice tumors, we identified CLDN2 as a potential downstream target of SphK1. Using ER-/HER2+ human breast cancer cell lines, we demonstrated that SphK1 overexpression enhances cell proliferation, colony formation and migration/invasion. Lastly, we confirmed that the expression of both SphK1 and CLDN2 was correlated with HER2-positive human breast cancer pathologic features.
The major finding in this study is the role of SphK1 in HER2-positive human breast cancer. We first showed that SphK1 deficiency reduced tumor incidence and tumor multiplicity, suggesting that SphK1 is involved in HER2 transgene-induced breast cancer. The tumor sizes between mice with and without SphK1 were not significantly different; however, this is not surprising when considering the fact that the tumors were allowed to grow until they reached any diameter of 20 mm, at which they were killed regardless of age. Our data provide an important complement to the work of Ohotski et al. who showed that inhibition of SphK1 reduced ERK1/2 activation (27). Additionally, to overcome issues of consistency with early activated neu models (36,37), a Tg mouse model expressing the wild-type (unactivated) form of HER2/neu under MMTV-LTR promoter control has been developed by Guy et al. (28). These mice develop focal breast tumors with a latency period of 29 weeks and first detectable tumors are seen at 16 weeks. It is this model that we employed in this study to recapitulate the role of SphK1 in human HER2-positive breast cancer tumorigenesis.
We identified CLDN2 as a potential downstream target of SphK1 in HER2/neu dependent mammary tumorigenesis. Our laboratory previously showed that SphK1 inhibits colon tumorigenesis in mice via the COX-2/PGE2 pathway (14). However, our analysis in this study showed no significant difference in the COX-2 expression levels in mammary tumors from mice with or without SphK1 (Supplementary Figure S3, available at Carcinogenesis Online), suggesting that SphK1 utilizes a different mechanism to regulate tumorigenesis in breast cancer (14,16,18). Therefore, we sought to find the mechanism(s) by which loss of SphK1 inhibits breast tumorigenesis in HER2/neu-induced breast tumorigenesis. Global gene expression analysis revealed that CLDN2 levels were significantly reduced in tumors from animals lacking SphK1. The reduction of CLDN2 expression was dependent on SphK1 expression levels; tumors from SphK1+/− mice expressed CLDN2 at a level between SphK1+/+ and SphK1−/− mice (data not shown). In support of this finding, CLDN2 was overexpressed in HER2-positive human primary breast tumors, especially in high-grade tumors, compared with adjacent normal tissue. Interestingly, we found that SphK1 expression is strongly correlated with CLDN2, which has not been reported in any disease.
Claudins are tight junction proteins that play key roles in maintaining cell-to-cell adhesion and communication (38). Accumulating evidence suggests changes in the expression patterns of CLDNs during developments of various cancers (39–42). For example, CLDN2 expression is increased in colon and lung tumors compared with normal tissues from the same patient (43,44). In breast cancers, early studies showed that lower expression of CLDN2 in breast carcinoma is associated with poor prognosis (39,45). However, recent studies reported that CLDN2 is highly expressed in breast cancer that has metastasize to the liver. Furthermore, CLDN2 has been associated with shorter metastasis-free interval (40,42). Thus, the role of CLDN2 in breast cancer, particularly in HER2, remains unclear.
HER2/neu is a potent oncoprotein that forms homodimers or heterodimers with other EGF receptors and helps sustain ERK pathway activation, leading to increased cell proliferation, migration and resistance to apoptosis (46,47). Previously, an in vitro study has shown that ERK activation by S1P is accompanied by enhanced tyrosine phosphorylation of HER2 (48). Furthermore, inhibition of HER2 (ERBB2 inhibitor II and siRNA) blocked S1P-induced ERK activation, results which suggest that SphK1/S1P acts upstream of HER2–ERK pathways (48). On the other hand, ERK has also been shown to interact with CLDN2 in colon and lung cancers (43,44). CLDN2 is an integral membrane protein, which has been implicated in various cancers including colon, lung, gastric and liver (43,44,49–51). In the Caco-2 colon cancer cell line, overexpression of CLDN2 increased cell proliferation, anchorage-independent growth and tumor growth (43). Mechanistically, EGF enhanced CLDN2 expression via EGFR/MAPK/ERK pathway in Caco-2 colon and A549 lung cancer cell lines (43,44). In both colon and lung cancer patients, CLDN2 expression is enhanced in tumors compared with normal tissue. In the current study, we showed that CLDN2 expression is higher in HER2-positive breast tumors compared with matched normal tissue.
In order to confirm the results from animal models, we used human cell lines in which SphK1 expression was manipulated. Overexpression of SphK1 significantly enhanced cellular functions associated with tumorigenesis, including cell proliferation, colony formation and migration/invasion. Results from the expression patterns of SphK1 expression on the human breast cancer TMA demonstrate that SphK1 is overexpressed in a large cohort of HER2-positive primary breast tumors and is associated with ER negativity and high grade. Together, the results suggest that SphK1 has a critical function in HER2-positive breast tumorigenesis and may serve as a potential target for therapeutics in HER2-positive breast cancer patients.
Conclusion
In summary, we show that SphK1 deficiency inhibits HER2/neu-induced breast carcinogenesis and may regulate CLDN2 to promote tumor development, probably by regulating HER2 function. We also show that SphK1 and CLDN2 are concordantly highly expressed in HER2-positive human breast cancers. Collectively, these results suggest that SphK1 plays a pro-tumoral role in HER2-positive breast carcinogenesis, and that targeting SphK1 and/or CLDN2 may present a novel approach for HER2-positive breast cancer chemoprevention and/or treatment.
Ethical approval
All mouse experiments were approved by the institutional animal care and use committee at UH (IACUC# 12–1113). The manuscript does not contain any individual person’s data in any form. The datasets supporting the conclusions of this article are included within the article and its additional file, and available in GEO accession number GSE80413.
Supplementary material
Supplementary data is available at Carcinogenesis online.
Funding
This work was supported by U.S. National Institutes of Health (NIH) grants [R01CA124687 and P01CA97132 to TK], NIH/NCI [5P30CA071789-6071 to CJR and P30 CA071789—12S6 to LWML].
Supplementary Material
Acknowledgements
We thank the Lipidomics Core Facility, the Hollings Cancer Center, Medical University of South Carolina and M.D. Anderson Science Park Histology and Tissue Processing core facility, and the University of Hawaii Cancer Center Pathology Shared Resource and Animal Carcinogenesis Shared Resource for their support.
Conflict of Interest Statement: No potential conflicts of interest were disclosed by all authors.
Abbreviations
- CLDN2
claudin-2
- COX-2
cyclooxygenase-2
- DMEM
Dulbecco’s Modified Eagle’s medium
- FBS
fetal bovine serum
- IPA
ingenuity pathway analysis
- KO
knockout
- NIDDK
National Institute of Diabetes and Digestive and Kidney Diseases
- PCR
polymerase chain reaction
- PGE2
prostaglandin E2
- S1P
sphingosine 1-phosphate
- SphK1
sphingosine kinase 1
- SphKs
sphingosine kinases
- TMA
tissue microarray
- UH
University of Hawaii
References
- 1. Siegel R.L., et al. (2016)Cancer statistics, 2016. CA. Cancer J. Clin., 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Mitri Z., et al. (2012)The HER2 receptor in breast cancer: pathophysiology, clinical use, and new advances in therapy. Chemother. Res. Pract., 2012, 743193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borg A., et al. (1990)HER-2/neu amplification predicts poor survival in node-positive breast cancer. Cancer Res., 50, 4332–4337. [PubMed] [Google Scholar]
- 4. Ursini-Siegel J., et al. (2007)Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat. Rev. Cancer., 7, 389–397. [DOI] [PubMed] [Google Scholar]
- 5. Gabos Z., et al. (2006)Prognostic significance of human epidermal growth factor receptor positivity for the development of brain metastasis after newly diagnosed breast cancer. J. Clin. Oncol., 24, 5658–5663. [DOI] [PubMed] [Google Scholar]
- 6. Rimawi M.F., et al. (2015)Targeting HER2 for the treatment of breast cancer. Annu. Rev. Med., 66, 111–128. [DOI] [PubMed] [Google Scholar]
- 7. Hannun Y.A., et al. (2008)Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol., 9, 139–150. [DOI] [PubMed] [Google Scholar]
- 8. Alvarez S.E., et al. (2007)Autocrine and paracrine roles of sphingosine-1-phosphate. Trends Endocrinol. Metab., 18, 300–307. [DOI] [PubMed] [Google Scholar]
- 9. Furuya H., et al. (2011)Sphingolipids in cancer. Cancer Metastasis Rev., 30, 567–576. [DOI] [PubMed] [Google Scholar]
- 10. Le Stunff H., et al. (2004)Role of sphingosine-1-phosphate phosphatase 1 in epidermal growth factor-induced chemotaxis. J. Biol. Chem., 279, 34290–34297. [DOI] [PubMed] [Google Scholar]
- 11. Spiegel S., et al. (2003)Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol., 4, 397–407. [DOI] [PubMed] [Google Scholar]
- 12. Hait N.C., et al. (2006)Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim. Biophys. Acta., 1758, 2016–2026. [DOI] [PubMed] [Google Scholar]
- 13. Siow D.L., et al. (2011)Sphingosine kinase localization in the control of sphingolipid metabolism. Adv. Enzyme Regul., 51, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawamori T., et al. (2009)Role for sphingosine kinase 1 in colon carcinogenesis. FASEB J., 23, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. French K.J., et al. (2003)Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res., 63, 5962–5969. [PubMed] [Google Scholar]
- 16. Shirai K., et al. (2011)A role of sphingosine kinase 1 in head and neck carcinogenesis. Cancer Prev. Res. (Phila), 4, 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Furuya H., et al. (2013)Effect of sphingosine kinase 1 inhibition on blood pressure. FASEB J., 27, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawamori T., et al. (2006)Sphingosine kinase 1 is up-regulated in colon carcinogenesis. FASEB J., 20, 386–388. [DOI] [PubMed] [Google Scholar]
- 19. Paulette M., et al. (2013)The impact of sphingosine kinase-1 in head and neck cancer. Biomolecules., 3, 481–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ristimäki A., et al. (2002)Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res., 62, 632–635. [PubMed] [Google Scholar]
- 21. Howe L.R., et al. (2002)Celecoxib, a selective cyclooxygenase 2 inhibitor, protects against human epidermal growth factor receptor 2 (HER-2)/neu-induced breast cancer. Cancer Res., 62, 5405–5407. [PubMed] [Google Scholar]
- 22. Harris R.E., et al. (2000)Chemoprevention of breast cancer in rats by celecoxib, a cyclooxygenase 2 inhibitor. Cancer Res., 60, 2101–2103. [PubMed] [Google Scholar]
- 23. Ruckhäberle E., et al. (2008)Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res. Treat., 112, 41–52. [DOI] [PubMed] [Google Scholar]
- 24. Nava V.E., et al. (2002)Sphingosine kinase type 1 promotes estrogen-dependent tumorigenesis of breast cancer MCF-7 cells. Exp. Cell Res., 281, 115–127. [DOI] [PubMed] [Google Scholar]
- 25. Shimizu Y., et al. (2013)Involvement of sphingosine kinases/Sphingosine-1-phosphate (S1P)/S1P receptors in breast cancer subtypes. J Oncobiomarkers., 1, 6. [Google Scholar]
- 26. Long JS, Edwards J, Watson C, Tovey S, Mair KM et al. (2010)Sphingosine kinase 1 induces tolerance to human epidermal growth factor receptor 2 and prevents formation of a migratory phenotype in response to sphingosine 1-phosphate in estrogen receptor-positive breast cancer cells. Mol Cell Biol., 30, 3827–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohotski J., et al. (2012)Expression of sphingosine 1-phosphate receptor 4 and sphingosine kinase 1 is associated with outcome in oestrogen receptor-negative breast cancer. Br. J. Cancer., 106, 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guy C.T., et al. (1992)Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl. Acad. Sci. U.S.A., 89, 10578–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allende M.L., et al. (2004)Mice deficient in sphingosine kinase 1 are rendered lymphopenic by FTY720. J. Biol. Chem., 279, 52487–52492. [DOI] [PubMed] [Google Scholar]
- 30. Rowse G.J., et al. (1998)Genetic modulation of neu proto-oncogene-induced mammary tumorigenesis. Cancer Res., 58, 2675–2679. [PubMed] [Google Scholar]
- 31. Gomes-Giacoia E., et al. (2013)Targeting plasminogen activator inhibitor-1 inhibits angiogenesis and tumor growth in a human cancer xenograft model. Mol. Cancer Ther., 12, 2697–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyake M., et al. (2015)Angiogenin promotes tumoral growth and angiogenesis by regulating matrix metallopeptidase-2 expression via the ERK1/2 pathway. Oncogene., 34, 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johannessen C.M., et al. (2010)COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature., 468, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson K.R., et al. (2002)PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA). J. Biol. Chem., 277, 35257–35262. [DOI] [PubMed] [Google Scholar]
- 35. Hanahan D., et al. (2011)Hallmarks of cancer: the next generation. Cell., 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 36. Muller W.J., et al. (1988)Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell., 54, 105–115. [DOI] [PubMed] [Google Scholar]
- 37. Bouchard L., et al. (1989)Stochastic appearance of mammary tumors in transgenic mice carrying the MMTV/c-neu oncogene. Cell., 57, 931–936. [DOI] [PubMed] [Google Scholar]
- 38. Ouban A., et al. (2010)Claudins in human cancer: a review. Histol. Histopathol., 25, 83–90. [DOI] [PubMed] [Google Scholar]
- 39. Kim T.H., et al. (2008)Down-regulation of claudin-2 in breast c arcinomas is associated with advanced disease. Histopathology., 53, 48–55. [DOI] [PubMed] [Google Scholar]
- 40. Kimbung S., et al. (2014)Claudin-2 is an independent negative prognostic factor in breast cancer and specifically predicts early liver recurrences. Mol. Oncol., 8, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soini Y. (2004)Claudins 2, 3, 4, and 5 in Paget’s disease and breast carcinoma. Hum. Pathol., 35, 1531–1536. [DOI] [PubMed] [Google Scholar]
- 42. Tabariès S., et al. (2012)Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol. Cell. Biol., 32, 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dhawan P., et al. (2011)Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene, 30, 3234–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikari A., et al. (2012)Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim. Biophys. Acta, 1823, 1110–1118. [DOI] [PubMed] [Google Scholar]
- 45. Soini Y. (2005)Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology, 46, 551–560. [DOI] [PubMed] [Google Scholar]
- 46. Graus-Porta D., et al. (1997)ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J., 16, 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yarden Y., et al. (2001)Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol., 2, 127–137. [DOI] [PubMed] [Google Scholar]
- 48. Long J.S., et al. (2010)Sphingosine 1-phosphate receptor 4 uses HER2 (ERBB2) to regulate extracellular signal regulated kinase-1/2 in MDA-MB-453 breast cancer cells. J. Biol. Chem., 285, 35957–35966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung H., et al. (2011)The expression of claudin-1, claudin-2, claudin-3, and claudin-4 in gastric cancer tissue. J. Surg. Res., 167, e185–e191. [DOI] [PubMed] [Google Scholar]
- 50. Patonai A., et al. (2011)Claudins and tricellulin in fibrolamellar hepatocellular carcinoma. Virchows Arch., 458, 679–688. [DOI] [PubMed] [Google Scholar]
- 51. Kinugasa T., et al. (2000)Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterology, 118, 1001–1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.