We characterized a panel of 107 structurally distinct pyridylhydroxybutyl DNA phosphate adducts in lung tissues of rats treated chronically with the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). These newly identified adducts could be used as biomarkers in future studies of tobacco carcinogenesis.
Abstract
The tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a powerful lung carcinogen in animal models and is considered a causative factor for lung cancer in people who use tobacco products. NNK undergoes metabolic activation—a critical step in its mechanism of carcinogenesis—to an intermediate which reacts with DNA to form pyridyloxobutyl DNA base and phosphate adducts. Another important metabolic pathway of NNK is its conversion to 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), which similarly forms pyridylhydroxybutyl DNA base adducts that have been characterized previously. In this study, we investigated the potential formation of pyridylhydroxybutyl DNA phosphate adducts. We report the characterization and quantitation of 107 structurally unique pyridylhydroxybutyl DNA phosphate adducts in the lungs of rats treated chronically with a carcinogenic dose of 5 ppm of NNK in their drinking water for up to 70 weeks, by using a novel liquid chromatography-nanoelectrospray ionization-high-resolution tandem mass spectrometry method. Our findings demonstrate that pyridylhydroxybutyl phosphate adducts account for 38–55 and 34–40% of all the measured pyridine-containing DNA adducts in rat lung and liver, respectively, upon treatment with NNK. Some of the pyridylhydroxybutyl DNA phosphate adducts persisted in both tissues for over 70 weeks, suggesting that they could be potential biomarkers of chronic exposure to NNK and NNAL. This study provides comprehensive characterization and relative quantitation of a panel of NNK/NNAL-derived DNA phosphate adducts, thus identifying NNK as the source of the most structurally diverse set of DNA adducts identified to date from any carcinogen.
Introduction
All tobacco products contain 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a tobacco-specific nitrosamine which is a powerful pulmonary carcinogen in rats, mice and hamsters, inducing mainly adenocarcinoma, independent of its route of administration (1–4). For example, in one recent study, F-344 rats were treated with NNK in the drinking water (5 ppm) for up to 90 weeks (5). All treated rats had histologically confirmed lung tumors, most of which were classified as carcinoma; metastases to the pancreas were also observed. In rodents and humans, NNK is extensively metabolized to 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) which is also a potent and selective pulmonary carcinogen with activity similar to that of NNK (4,5). All humans who use tobacco products—either cigarettes or smokeless tobacco—have NNAL in their urine (6–11). NNK is considered as one likely cause of lung cancer—particularly adenocarcinoma of the lung—in cigarette smokers (3,12). Consistent with its organospecificity for the lung, a recent epidemiologic study has also demonstrated a significantly elevated risk of lung cancer in non-smokers who were exclusive users of oral smokeless tobacco products (chewing tobacco and snuff), which have a less complex mixture of carcinogens than cigarette smoke, and in which NNK is one of the most abundant strong carcinogens (13). NNK and the related carcinogen N’-nitrosonornicotine are considered ‘carcinogenic to humans’ by the International Agency for Research on Cancer (1).
A critical step in carcinogenesis by nitrosamines such as NNK and NNAL is formation of DNA adducts, which can cause miscoding and permanent mutations in growth control genes such as K-RAS (14,15). Identification of the relevant DNA adducts can significantly enhance our understanding of mechanisms of cancer induction, potentially leading to biomarkers for the identification of particularly susceptible users of tobacco products and providing insights for cancer prevention.
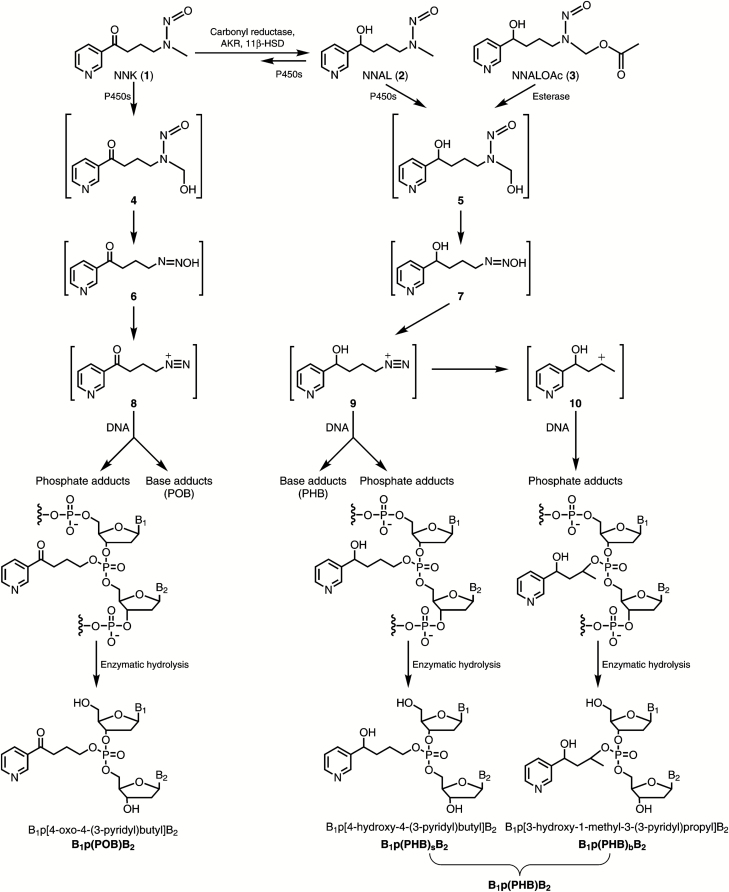
Metabolic activation of NNK to intermediates that react with DNA to form adducts occurs by α-hydroxylation, a cytochrome P450-catalyzed pathway that is typical for nitrosamines (4,16). α-Hydroxylation of both the methylene and methyl carbons is observed; only the latter is considered here. As shown in Figure 1, α-hydroxylation of the NNK methyl group produces the unstable intermediate α-hydroxymethylNNK (4) which spontaneously loses formaldehyde to produce diazohydroxide 6. This intermediate can further decompose to diazonium ion 8, which is highly electrophilic and reacts with DNA to produce pyridyloxobutyl (POB)-base adducts such as 7-[4-(3-pyridyl)-4-oxobutyl]Gua (7-POB-Gua), O6-POB-dGuo and O2-POB-dThd, as well as phosphate adducts such as B1p(POB)B2, which have been previously characterized (and were referred to as B1popB2) (5,17–21). The NNK metabolite NNAL (2) also undergoes α-hydroxylation to similarly produce diazonium ion 9 (4,22). Diazonium ion 9 reacts with DNA bases to form 4-[(3-pyridyl)-4-hydroxybutyl] (PHB)-base adducts which have been previously characterized (23,24), but also has the potential to form phosphate adducts in DNA such as B1p(PHB)sB2, where PHB represents pyridylhydroxybutyl and the subscript s represents ‘straight chain’ (Figure 1). Diazonium ion 9 further rearranges to carbocation 10 (22), which can also form adducts with both DNA bases and phosphate [e,g., B1p(PHB)bB2, where the subscript b represents ‘branched’]. Thus, DNA phosphate adducts of NNAL arising through intermediates 9 and 10, and collectively called B1p(PHB)B2, have not been previously investigated and are the subject of this report.
Figure 1.
Pyridyloxobutyl and pyridylhydroxybutyl DNA adduct formation by NNK and NNALOAc. B1 and B2 represent the same or different nucleobases.
We developed a novel liquid chromatography (LC)-nanoelectrospray ionization (NSI)-high-resolution tandem mass spectrometry (HRMS/MS) method for analysis of the potentially complex array of DNA phosphate adducts formed in the NNAL pathway of NNK metabolism. DNA phosphate adducts from this pathway were characterized in reactions of NNALOAc (3), a stable precursor to intermediate 5 formed in NNAL metabolism, with DNA, and in lung and liver DNA of rats treated with 5 ppm of NNK in the drinking water for 10, 30, 50 and 70 weeks, conditions which cause lung tumors in all animals as noted above.
Materials and methods
Materials and chemicals
Two diastereomers of dCp[4-oxo-4-(3-pyridyl)butyl]dC [Cp(POB)C-1 and Cp(POB)C-2] (21), and two diastereomers of Tp[4-oxo-4-(3-pyridyl)butyl]T [Tp(POB)T-1 and Tp(POB)T-2] were procured from WuXi AppTec (Hong Kong). The structures of Tp(POB)T-1 and Tp(POB)T-2 were confirmed by one-dimensional and two-dimensional NMR in our laboratory (Supplementary Figure 1, available at Carcinogenesis Online). NNALOAc was synthesized based on our previously published method (22). 5ʹ-Dimethoxytrityl [13C1015N2]2’-thymidine-3ʹ-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite was obtained from Cambridge Isotope Laboratories (Tewksbury, MA). All other nucleoside phosphoramidites, solvents and solid supports required for the solid phase synthesis of dinucleotide [13C1015N2]2ʹ-TpT were acquired from Glen Research Corp. (Sterling, VA). Enzymes and reagents for DNA isolation were purchased from Qiagen Sciences (Germantown, MD). All other chemicals and solvents were obtained from Sigma-Aldrich Chemical Co. (Milwaukee, WI).
Synthesis of chemical standards
Cp(PHB)sC and Tp(PHB)sT
dCp[4-Hydroxy-4-(3-pyridyl)butyl]dC [Cp(PHB)sC] isomers were synthesized by treating Cp(POB)C-1 or Cp(POB)C-2 with sodium borohydride in H2O at room temperature for 60 min. A single peak was detected in each reaction solution by LC-MS in the full scan mode, and the presence of Cp(PHB)sC was confirmed by MS fragmentation data (Supplementary Figure 2, available at Carcinogenesis Online). Two isomers, Cp(PHB)sC-1 [from Cp(POB)C-1] and Cp(PHB)sC-2 [from Cp(POB)C-2], were identified. Similarly, Tp[4-hydroxy-4-(3-pyridyl)butyl]T [Tp(PHB)sT] isomers were synthesized by treating Tp(POB)T-1 or Tp(POB)T-2 with sodium borohydride in H2O at room temperature for 60 min. A single peak was detected in each reaction solution by LC-MS in the full scan mode, and the presence of Tp(PHB)sT was confirmed by MS fragmentation data (Supplementary Figure 3, available at Carcinogenesis Online). Two isomers, Tp(PHB)sT-1 [from Tp(POB)T-1] and Tp(PHB)sT-2 [from Tp(POB)T-2], were identified. The isomers of Cp(PHB)sC and Tp(PHB)sT were further purified on 30 mg Strata-X cartridges (Phenomenex), and their structures were confirmed by one-dimensional and two-dimensional NMR (Supplementary Figures 4 and 5, available at Carcinogenesis Online).
[15N3]Cp(PHB)sC and [13C1015N2]Tp(PHB)sT
A similar synthetic procedure to that used for the preparation of Cp(PHB)sC from Cp(POB)C was used to synthesize [15N3]Cp(PHB)sC from [15N3]Cp(POB)C, which was obtained in our previous study (21). The identities of [15N3]Cp(PHB)sC-1 and [15N3]Cp(PHB)sC-2 were confirmed via LC-MS by comparison with the corresponding unlabeled Cp(PHB)sC standards. To synthesize [13C1015N2]Tp(PHB)sT, [13C1015N2]2ʹ-TpT was first prepared on a DNA synthesizer (ABI 394, Applied Biosystems, Foster City, CA) in accordance with standard solid phase oligodeoxynucleotide synthesis protocols. In this study, 5ʹ-dimethoxytrityl[13C1015N2]2ʹ-thymidine-3ʹ-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite was manually coupled to an Ac-dT-CPG solid support, followed by standard deprotection techniques performed on the DNA synthesizer (Supplementary Figure 6, available at Carcinogenesis Online). [13C1015N2]Tp(POB)T was subsequently synthesized by reacting [13C1015N2]2ʹ-TpT with NNKOAc in 0.01 M NaOH (37°C) for 16 h and purified by reversed phase HPLC using a Waters 510 HPLC system. [13C1015N2]Tp(PHB)sT was then synthesized by treating [13C1015N2]Tp(POB)T with sodium borohydride in H2O at room temperature for 60 min, and purified by 30 mg Strata-X cartridges. The identity of [13C1015N2]Tp(PHB)sT was confirmed via LC-MS by comparison with the corresponding unlabeled Tp(PHB)sT standard.
In vitro and in vivo DNA samples
In the in vitro study, CT-DNA (2 mg) was incubated with NNALOAc (10 mg) in the presence of porcine liver esterase (4 units) in 0.1 M phosphate buffer (1 ml, pH 7.0) at 37°C for 12 h. NNALOAc is a chemically activated form of NNAL, which in the presence of esterase forms the same reactive intermediate 5 as produced in metabolism of NNAL by P450s (Figure 1) (22). The incubation mixture was then washed three times with 4 ml of a CHCl3/isoamyl alcohol mixture (24:1). The treated CT-DNA was precipitated via the addition of cold 2-propanol, washed with 70% EtOH and 100% EtOH sequentially, dried under a stream of N2 and stored at −20°C until analysis. The in vivo DNA samples were from lung and liver tissues of male F-344 rats (n = 5 at each time points) treated chronically with 5 ppm of NNK in drinking water for 10, 30, 50 and 70 weeks, as described previously (5). DNA was isolated using our previously developed protocol (20), and stored at −20°C until analysis.
DNA hydrolysis and sample preparation
The DNA samples were dissolved in 0.6 ml of 10 mM sodium succinate buffer (pH 7.4) containing 5 mM CaCl2 and 10 fmol [15N3]Cp(PHB)sC and [13C1015N2]Tp(PHB)sT as internal standards. The solution was then mixed with deoxyribonuclease I (60 units), phosphodiesterase I (0.03 units) and alkaline phosphatase (40 units), followed by overnight incubation at 37°C. On the next day, 20 μl of hydrolysate was collected for dGuo analysis and DNA quantitation (20). The remaining hydrolysate was filtered through 10 K centrifugal filters (Ultracel YM-10, Millipore), and the filtrates were loaded on 30-mg Strata X cartridges (Phenomenex) activated with 2 ml of MeOH and 2 ml of H2O. The cartridges were washed with 2 ml of H2O and 1 ml of 10% MeOH sequentially and finally eluted with 2 ml of 50% MeOH. The 50% MeOH fraction containing analytes was concentrated to dryness in a centrifugal evaporator. The residue was re-dissolved in 10 μl of deionized H2O prior to analysis by LC-NSI-HRMS/MS.
LC-NSI-HRMS/MS analysis
The analysis was performed on an Orbitrap Fusion Tribrid mass spectro- meter (Thermo Scientific, Waltham, MA) with full scan, selected-ion monitoring (SIM) and product ion scan analysis using Orbitrap detection with internal calibration using the EASY-IC feature of the Fusion instrument. A nanoLC column (75 μm i.d., 360 μm o.d., 17 cm length and 15 μm orifice) packed with Luna C18 bonded separation media (Phenomenex, Torrance, CA) was used. The mobile phase consisted of 5 mM NH4OAc and CH3CN. A 5 μl injection loop was used, and the sample (4 μl) was loaded onto the capillary column with a 900 nl/min flow rate under the initial conditions for 6.5 min at which point the injection valve position was switched to take the injection loop out of the flow path and the flow rate was reduced to 300 nl/min. Separation on the column was performed using a linear gradient with increasing CH3CN from 2 to 18% over 24 min, followed by ramping to 90% CH3CN within 2 min and holding at this composition for an additional 3 min. The gradient was then returned to 2% CH3CN in 1 min, and the system was re-equilibrated at this mobile phase composition for 6 min at 900 nl/min before the next injection. The spray voltage of the mass spectrometer was 2.2 kV. The capillary temperature was 300°C, and the S-Lens RF Level was 60%. A multiplexed SIM analysis was performed whereby each ion mass of interest is sequentially isolated by the qua- drupole and stored in the ion routing multipole, and upon collection of all masses of interest, all ions are transported into the Orbitrap for analysis. The [M + H]+ ions of all possible phosphate adducts listed in Table 1 were analyzed in this fashion using an isolation window of 3 m/z, Automatic Gain Control (AGC) target of 50000, maximum inject time of 100 ms and a resolution of 60000. The product ion scan was performed using higher-energy collisional dissociation (HCD) fragmentation with a normalized collision energy of 25 units, isolation widths of 1 Da for all the precursor ions and product ion analysis performed with a mass range of m/z 100–800 at a resolution of 15000. The accurate mass tolerance used for extraction of precursor and fragment ion signals was 5 ppm.
Table 1.
Numbers of possible isomers of pyridylhydroxybutyl phosphate adducts formed from NNAL (see structures of B1p(PHB)sB2 and B1p(PHB)bB2, Figure 1)
| Base combination | [M + H]+ | [M + 2H]2+ | Number of possible isomers | Number detected | |||
|---|---|---|---|---|---|---|---|
| B1p(PHB)sB2 | B1p(PHB)bB2 | Total | Lung | Liver | |||
| A–A | 714.2508 | 357.6293 | 4 | 8 | 12 | 8 | 4 |
| C–C | 666.2283 | 333.6181 | 4 | 8 | 12 | 7 | 2 |
| G–G | 746.2406 | 373.6242 | 4 | 8 | 12 | 7 | 2 |
| T–T | 696.2276 | 348.6177 | 4 | 8 | 12 | 7 | 4 |
| A–C | 690.2396 | 345.6237 | 8 | 16 | 24 | 12 | 8 |
| A–G | 730.2457 | 365.6268 | 8 | 16 | 24 | 11 | 7 |
| A–T | 705.2392 | 353.1235 | 8 | 16 | 24 | 16 | 7 |
| C–G | 706.2345 | 353.6212 | 8 | 16 | 24 | 18 | 8 |
| C–T | 681.2280 | 341.1179 | 8 | 16 | 24 | 11 | 7 |
| G–T | 721.2341 | 361.1210 | 8 | 16 | 24 | 10 | 5 |
| Total | 64 | 128 | 192 | 107 | 54 | ||
The quantitative analysis of Cp(PHB)sC and Tp(PHB)sT was performed using accurate mass extracted ion chromatograms of the product ion scan data of m/z 328.0945 [C14H19NO6P]+ for Cp(PHB)sC (parent ion m/z 666.2), [15N3]Cp(PHB)sC (parent ion m/z 669.2), Tp(PHB)sT (parent ion m/z 696.2) and [13C1015N2]Tp(PHB)sT (parent ion m/z 708.2) with a mass tolerance of 5 ppm. Quantitation was based on the peak area ratio of Cp(PHB)sC or T(PHB)sT to their corresponding isotope-labeled internal standards, the constructed calibration curves and the amount of internal standards added. Calibration curves were constructed for both Cp(PHB)sC and Tp(PHB)sT before each analysis using a series of standard solutions of Cp(PHB)sC/[15N3]Cp(PHB)sC and Tp(PHB)sT/[13C1015N2]Tp(PHB)sT. The calibration standard solutions contained a constant amount of [15N3]Cp(PHB)sC and [13C1015N2]Tp(PHB)sT (5 fmol on-column) and varying amounts of Cp(PHB)sC and Tp(PHB)sT (0.03, 0.06, 0.15, 0.3, 0.6, 3 and 15 fmol on-column). A semi-quantitative approach was applied to estimate the levels of all other phosphate adducts based on their MS signal intensities compared to Cp(PHB)sC or Tp(PHB)sT under the SIM scan mode. Doubly charged peaks ([M + 2H]2+, Table 1), except for the T–T nucleobase combination, were observed during sample analysis (Supplementary Figure 7, available at Carcinogenesis Online), and their contribution to signal intensity was added when estimating the levels of these adducts. The average signal response of the Cp(PHB)sC and Tp(PHB)sT calibration curves was used to estimate the levels of other phosphate adducts.
All data are presented as mean ± standard deviation (SD). Comparison of adduct levels at different time points were performed using analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons. A P value less than 0.05 was considered significant.
Results
Characterization and quantitation of pyridylhydroxybutyl phosphate adducts
Table 1 summarizes the possible numbers of DNA phosphate adduct isomers which could be formed from intermediates produced in the metabolism of NNAL. There can be 10 different combinations of the four nucleobases, and there can be different stereoisomers for each combination due to the chiral centers in the structure. Thus, because of the tetrahedral structure of the phosphate group and the chiral carbinol carbon, there can be four possible isomers of B1p(PHB)sB2 with the same nucleobase combination, and eight isomers with different nucleobase combinations, resulting in a total of 64 different possible B1p(PHB)sB2 adducts. For B1p(PHB)bB2 adducts, an additional chiral center is introduced on the methyl-bearing carbon, so the adducts with the same nucleobase combination can potentially form eight isomers, while adducts with a different nucleobase combination can produce 16 isomers, resulting in a total of 128 different possible B1p(PHB)bB2 adducts. Collectively, these straight chain and branched adducts are called B1p(PHB)B2. Therefore, a total of 192 B1p(PHB)B2 adducts could be formed from intermediates produced in the metabolism of NNAL. By analyzing the data obtained from both SIM and product ion scans, as described below, we observed at least seven isomers of each combination of B1p(PHB)B2 phosphate adducts, and a total of 107 and 54 out of 192 possible adducts were detected in rat lung and liver, respectively (Table 1).
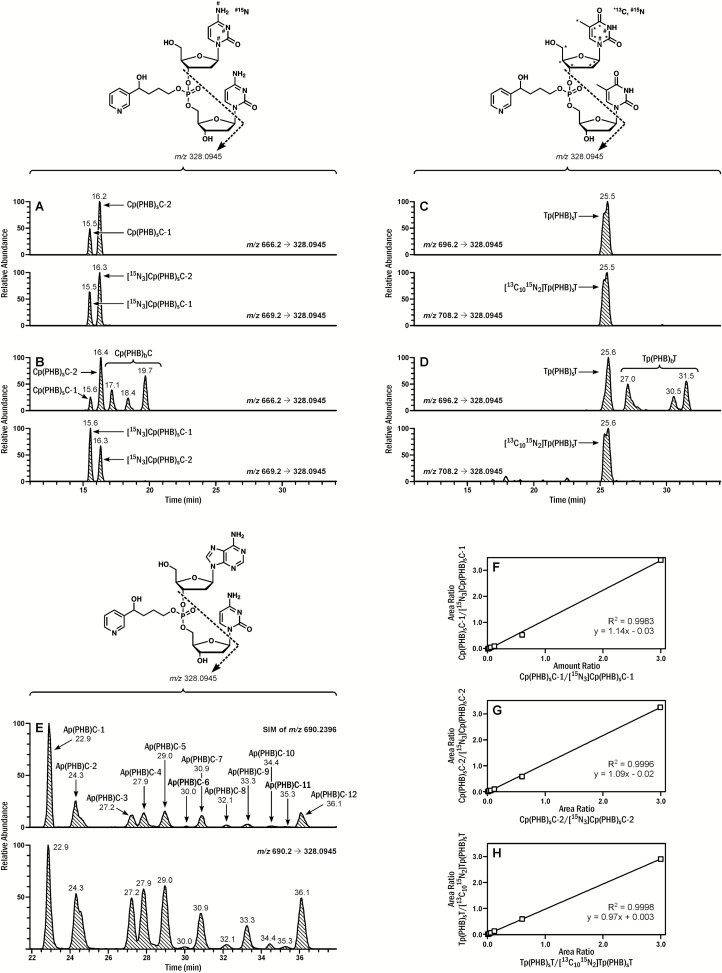
The presence of Cp(PHB)sC and Tp(PHB)sT in rat tissue DNA was confirmed by comparison of their LC-MS retention times and product ion scan spectra to those of the synthesized standards. Figure 2A shows the chromatogram for standard Cp(PHB)sC and Figure 2B the corresponding chromatogram of a DNA sample from rat lung upon treatment with NNK in the drinking water for 30 weeks. The first two peaks in Figure 2B match the standard straight chain Cp(PHB)sC adducts while the remaining peaks arise from the rearranged intermediate 10 and correspond to the branched Cp(PHB)bC adducts, which were also observed in the reactions of DNA with NNALOAc (see below). Similarly, both Tp(PHB)sT and Tp(PHB)bT were observed in the same DNA sample (Figure 2C and D). Detection of 1-(3-pyridyl)-1,3-butanediol (22), which is released from B1p(PHB)bB2 adducts upon acid hydrolysis, was used to further confirm the presence of the branched phosphate adducts. To ensure that 1-(3-pyridyl)-1,3-butanediol was from phosphate adducts but not base adducts, the tissue DNA hydrolysate was fractionated by HPLC to separate out PHB base adducts, and the fraction containing only phosphate adducts was subjected to acid hydrolysis (Supplementary Figure 8, available at Carcinogenesis Online). Indeed, 1-(3-pyridyl)-1,3-butanediol was detected in the phosphate adduct-containing fraction upon acid hydrolysis, which confirmed the presence of B1p(PHB)bB2 adducts in the rat tissue DNA (Supplementary Figure 9, available at Carcinogenesis Online).
Figure 2.
Typical chromatograms obtained upon analysis of (A) Cp(PHB)sC standards, (B) Cp(PHB)C in lung DNA of a rat treated with NNK in drinking water (5 ppm) for 30 weeks, (C) Tp(PHB)sT standards, (D) Tp(PHB)T in lung DNA of a rat treated with NNK in drinking water (5 ppm) for 30 weeks; (E) LC-NSI-HRMS/MS extracted precursor (m/z 690.2396, SIM, selected-ion monitoring) and fragment ion (m/z 328.0945) chromatograms corresponding to Ap(PHB)C obtained upon analysis of lung DNA of a rat treated with NNK in drinking water (5 ppm) for 10 weeks; Linearity of (F) Cp(PHB)sC-1, (G) Cp(PHB)sC-2 and (H) Tp(PHB)sT calibration curves. The amount of Cp(PHB)sC or Tp(PHB)sT in the calibration curve was increased from 0.03 fmol to 0.06, 0.15, 0.3, 0.6, 3 and 15 fmol, with a constant amount of internal standards (5 fmol).
The structures of the other eight nucleobase combinations were characterized based on the accurate masses of the precursor ions and the corresponding fragment ions. With Ap(PHB)C as an example, the exact mass of its precursor ion of m/z 690.2396 was extracted from the SIM scan data upon analysis of lung DNA from a 10 week-treated rat, and 12 peaks were detected (Figure 2E), indicating the presence of at least 12 isomers. The peak shapes and retention times of the major fragment ion, m/z 328.0945, and the other major fragment ions of each of the 12 peaks matched the SIM data from the parent ion (Figure 2E and Supplementary Figure 7, available at Carcinogenesis Online), further confirming the identity and the structure of the Ap(PHB)C adducts. Since the standards of Ap(PHB)C were not available, discrimination between the straight chain and branched isomers was not possible. The nomenclature of the isomers [Ap(PHB)C-1 to Ap(PHB)C-12] was based on the sequence of their retention times in the chromatogram. The extracted ion chromatograms in rat lung DNA and proposed fragmentation patterns for structural characterization of all the other B1p(PHB)B2 phosphate adducts are presented in Supplementary Figure 7, available at Carcinogenesis Online.
Based on our unexpected observation of more than eight peaks for each combination of different DNA bases, we examined the formation of B1p(PHB)B2 in CT-DNA treated with NNALOAc, to further confirm their origin. The B1p(PHB)B2 adducts detected in vivo were observed in the in vitro NNALOAc-treated CT-DNA sample. As an example, typical extracted precursor and fragment ion (m/z 328.0945) chromatograms corresponding to Cp(PHB)C and Tp(PHB)T obtained upon analysis of an NNALOAc-treated CT-DNA sample are presented in Supplementary Figure 10, available at Carcinogenesis Online. A total of 97 out of 192 possible B1p(PHB)B2 adducts were detected (Supplementary Table 1, available at Carcinogenesis Online). At least six isomers were observed for each combination of B1p(PHB)B2 adducts. The three most abundant adduct types formed were Cp(PHB)T, Ap(PHB)T and Ap(PHB)C, which accounted for 24, 21 and 18% of the total B1p(PHB)B2 adducts, respectively (Supplementary Figure 10, available at Carcinogenesis Online).
Using HCD fragmentation, the product ion spectra of both Cp(PHB)sC and Tp(PHB)sT generated several major fragment ions (Supplementary Figure 11, available at Carcinogenesis Online). Because of the high signal intensity in the spectra of both Cp(PHB)sC and Tp(PHB)sT, the fragment ion of m/z 328.0945 was selected for quantitative analysis, with the transitions of m/z 666.2→328.0945 for Cp(PHB)sC, m/z 669.2→328.0945 for [15N3]Cp(PHB)sC, m/z 696.2→328.0945 for Tp(PHB)sT, and m/z 708.2→328.0945 for [13C1015N2]Tp(PHB)sT. Typical chromatograms obtained upon analysis of Cp(PHB)sC and Tp(PHB)sT standards are presented in Figure 2. Only two of the possible four isomers were detected for Cp(PHB)sC [Cp(PHB)sC-1 and Cp(PHB)sC-2], likely due to insufficient chromatographic resolution of all isomers under our conditions. Similarly, only two peaks were detected for the four possible isomers of Tp(PHB)sT without baseline separation, and the total area of the two peaks was used for quantitative analysis. By using this method, a limit of detection of 0.03 fmol on-column was obtained for Cp(PHB)sC-1, Cp(PHB)sC-2 and Tp(PHB)sT. The instrument response and the analyte/internal standard ratio were linear in the range of 0.03−15 fmol on-column with typical R2 of 0.9983, 0.9996 and 0.9998 for Cp(PHB)sC-1, Cp(PHB)sC-2, and Tp(PHB)sT (Figure 2F–H), respectively. The calibration curves were then used for quantitation of B1p(PHB)B2 adducts in the rat tissue DNA samples.
B1p(PHB)B2 phosphate adducts in rat tissue DNA
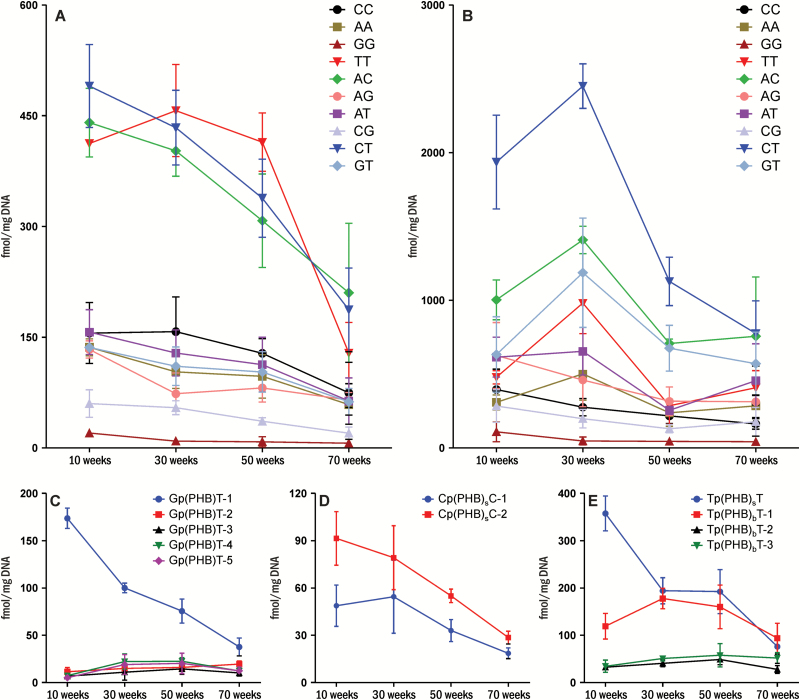
Three major B1p(PHB)B2 phosphate adducts were observed in rat liver DNA—Cp(PHB)T, Ap(PHB)C and Tp(PHB)T—which accounted for 21–23, 19–24 and 15–25% of the total B1p(PHB)B2 phosphate adducts, respectively (Figure 3A). A decrease in adduct levels over the course of the study was observed for all the combinations. For example, the level of Ap(PHB)C decreased from 441 ± 38 fmol/mg DNA at 10 weeks of treatment to 210 ± 77 fmol/mg DNA at 70 weeks of treatment (P < 0.05). In the lung, the three most abundant adducts were Cp(PHB)T, Ap(PHB)C and Gp(PHB)T, which accounted for 20–30, 16–19 and 10–17% of the total B1p(PHB)B2 phosphate adducts, respectively (Figure 3B). However, the formation patterns were different between the two tissues. While the adduct levels of most combinations were maximal at 10 weeks in the liver, the levels of most combinations were the highest at 30 weeks in the lung. For example, the level of Ap(PHB)C in the lung at 30 weeks was 1410 ± 76 fmol/mg DNA, significantly higher than the levels at 10, 50 or 70 weeks (P < 0.05).
Figure 3.
Levels of pyridylhydroxybutyl DNA phosphate adducts in (A) liver DNA, (B) lung DNA, and levels of each isomer of (C) Gp(PHB)T, (D) Cp(PHB)C and (E) Tp(PHB)T in liver DNA of rats (n = 5) treated with 5 ppm of NNK in drinking water for 10, 30, 50 or 70 weeks. Values are presented as means ± SD.
Different levels of isomers of the same combination of B1p(PHB)B2 were observed for all 10 combinations in both liver and lung (Figure 3C–E and Supplementary Figures 12 and 13, available at Carcinogenesis Online). For example, the level of Gp(PHB)T-1 was significantly higher than the other isomers of Gp(PHB)T in the liver throughout the study (Figure 3C). The formation patterns of each isomer from some combinations were similar. For example, the levels of both Cp(PHB)sC-1 and Cp(PHB)sC-2 in the liver were maximal at 10 weeks and decreased afterwards (Figure 3D). However, the formation patterns of isomers from some other combinations were different. For example, the level of Tp(PHB)sT-1 in the liver was maximal at 10 weeks and decreased afterwards (P < 0.05, Figure 3E), while the levels of Tp(PHB)bT-1 and Tp(PHB)bT-2 reached a peak at 30 and 50 weeks, respectively. The level of Tp(PHB)bT-3 was not significantly changed over the course of the study (P = 0.14, Figure 3E).
Comparison of DNA phosphate and base adducts formed by NNK
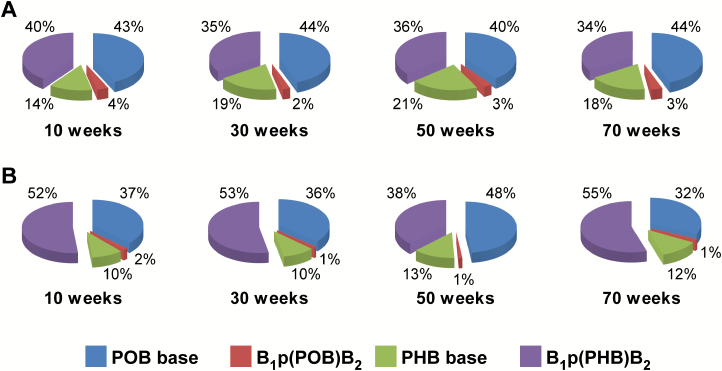
To determine the relative proportion of B1p(PHB)B2 adducts to the total known pyridine-containing DNA adducts formed by NNK, the levels of the other three types of DNA adducts [PHB base adducts, POB base adducts and B1p(POB)B2 adducts], were measured or obtained from our previous studies of rats from the same treatment groups (Table 2) (5,21). The levels of the PHB base adducts—O2-[4-(3-pyridyl)-4-hydroxybut-1-yl]thymidine (O2-PHB-dThd) and 7-[4-(3-pyridyl)-4-hydroxybut-1-yl]guanine (7-PHB-Gua)—were measured in rat liver DNA in this study using our previously developed method (24). The total amount of the two major PHB base adducts ranged from 466 to 1050 fmol/mg DNA in liver DNA over the course of the study. The levels of POB base adducts and B1p(POB)B2 phosphate adducts in the liver and lung were reported previously (5,21). In rat liver, the most abundant type of DNA adducts were POB base adducts, which accounted for 40–44% of the total measured DNA adducts, followed by B1p(PHB)B2 phosphate adducts (34–40%), PHB base adducts (14–21%) and B1p(POB)B2 adducts (2–4%) (Figure 4A; Table 2); while in the lung, the most abundant DNA adducts were B1p(PHB)B2 phosphate adducts, which accounted for 38–55% of the total measured DNA adducts, followed by POB base adducts (32–48%), PHB base adducts (10–13%) and B1p(POB)B2 phosphate adducts (1–2%) (Figure 4B and Table 2). Methyl-DNA base adducts are also formed by NNK due to α-hydroxylation on its methylene carbon (4,5), and the formation and characterization of methyl-DNA phosphate adducts in NNK-treated rats will be reported separately.
Table 2.
Levels of three major POB base adducts, two major PHB base adducts, total B1p(PHB)B2 phosphate adducts and total B1p(POB)B2 phosphate adducts in liver and lung DNA of rats treated with 5 ppm of NNK in drinking water for 10, 30, 50 or 70 weeks
| Tissue | Adduct | Adduct level (fmol/mg DNA) | ||||
|---|---|---|---|---|---|---|
| Liver | POB basea | 10 weeks | 30 weeks | 50 weeks | 70 weeks | |
| O 2-POB-dThd | 1810 ± 840 | 2140 ± 26 | 1530 ± 161 | 910 ± 6 | ||
| 7-POB-Gua | 501 ± 53 | 331 ± 36 | 257 ± 32 | 230 ± 90 | ||
| PHB base | ||||||
| O 2-PHB-dThd | 390 ± 185 | 737 ± 186 | 658 ± 107 | 272 ± 86 | ||
| 7-PHB-Gua | 335 ± 40 | 313 ± 79 | 295 ± 42 | 194 ± 32 | ||
| Total B1p(PHB)B2 phosphate | 2140 ± 131 | 1930 ± 81 | 1630 ± 166 | 878 ± 210 | ||
| Total B1p(POB)B2 phosphatea | 190 ± 49 | 134 ± 64 | 120 ± 36 | 89 ± 4 | ||
| Lung | POB baseb | O 6-POB-dG | 34 ± 21 | 9 ± 9 | 9 ± 2 | 4 ± 2 |
| O 2-POB-dThd | 3590 ± 414 | 4810 ± 193 | 4410 ± 320 | 1980 ± 467 | ||
| 7-POB-Gua | 970 ± 148 | 751 ± 29 | 688 ± 65 | 315 ± 75 | ||
| PHB baseb | ||||||
| O 2-PHB-dThd | 968 ± 28 | 1370 ± 94 | 1260 ± 66 | 807 ± 142 | ||
| 7-PHB-Gua | 235 ± 40 | 164 ± 44 | 129 ± 34 | 62 ± 15 | ||
| Total B1p(PHB)B2 phosphate | 6390 ± 457 | 8160 ± 654 | 4000 ± 291 | 3950 ± 223 | ||
| Total B1p(POB)B2 phosphatea | 475 ± 95 | 417 ± 43 | 346 ± 41 | 218 ± 15 | ||
Values are presented as means ± SD (n = 3 or 5).
The data of total B1p(POB)B2 phosphate adduct levels in both tissues, and POB base adduct levels in rat liver are from Ma et al. (21).
The data of POB and PHB base adduct levels in rat lung are from Balbo et al. (5).
Figure 4.
Relative levels of DNA phosphate and base adducts in (A) liver and (B) lung DNA of rats treated with 5 ppm of NNK in drinking water for 10, 30, 50 or 70 weeks.
In addition to O2-PHB-dThd and 7-PHB-Gua, O6-[4-(3-pyridyl)-4-hydroxybut-1-yl]deoxyguanosine (O6-PHB-dGuo) was also detected and quantified in NNALOAc-treated CT-DNA. The relative levels of total B1p(PHB)B2 adducts and total PHB base adducts were comparable to those in the DNA samples from treated rats, accounting for 48 and 52% of the total measured DNA adducts, respectively (Supplementary Figure 10, available at Carcinogenesis Online).
Discussion
The results of this study were unexpected with respect to both the extent and diversity of DNA phosphate adducts—B1p(PHB)B2 —formed in the NNAL pathway of NNK metabolism. Thus, our previous study of B1p(POB)B2 adducts formed in NNK metabolism demonstrated that they comprised a relatively small percentage of total adduct formation (Figure 4). In contrast, the present results show that B1p(PHB)B2 adducts represent a sizable slice of the NNK-DNA adduct pie in rat liver and lung (Figure 4). Notably, our LC-NSI-HRMS/MS method revealed a surprising variety of NNAL-derived DNA phosphate adducts, explainable by the recognition that rearrangement of diazonium ion 9 to carbocation 10 is a substantial in vivo pathway of NNAL-DNA adduct formation. Thus, we demonstrate an unprecedented diversity of structurally unique DNA adducts formed from a single carcinogen—NNK. Taken together with our previous study in which we characterized 30 B1p(POB)B2 adducts, we have now identified 137 novel and structurally distinct DNA-phosphate adducts in tissues of rats treated with NNK, far greater than reported to date from any other carcinogen.
Most DNA adduct studies have focused on the interaction of chemicals with DNA nucleobases rather than the inter-nucleotide phosphate groups. Studies of DNA phosphate adducts have demonstrated however that they have longer half-lives than their corresponding base adducts in animals treated with alkylating agents (25–28). For example, after administration of a single dose of ethylnitrosourea to mice, the half-lives of ethyl phosphate adducts in the major tissues were 10−15 weeks, compared to the half-lives of 30−220 h of the corresponding O6-ethylguanine base adduct (28). While the levels of most DNA base adducts decreased over time upon chronic treatment of rats with NNK (5), our previous study demonstrated that the levels of some NNK-derived B1p(POB)B2 adducts were persistently high in the same rats over the 70 weeks of treatment (21). Consistent with those observations, some of the B1p(PHB)B2 phosphate adducts in the current study demonstrated persistence in rats over the 70 weeks of treatment. These findings indicate that these DNA phosphate adducts could potentially be used as biomarkers for chronic exposure to tobacco-specific carcinogens in humans. Exploring this approach would most likely involve focusing on a particular isomer such as Tp(PHB)bT-3 in lung. The first step would be further characterization of the selected isomer. This could be accomplished by synthesis of the appropriate diastereoisomeric precursors related to 1-(3-pyridyl)-1,3-butanediol, then reaction of these with the selected dinucleotide phosphate followed by isolation and characterization. With specific standards in hand, analytical methods could be developed for quantitation in cellular DNA from human lung or oral tissue. Based on the high sensitivity and selectivity of LC-NSI-HRMS/MS analysis, this research direction appears feasible.
As noted above, B1p(PHB)B2 phosphate adducts are formed to a greater extent than B1p(POB)B2 phosphate adducts in rats treated with NNK. In rat liver, the levels of B1p(PHB)B2 phosphate adducts (878–2140 fmol/mg DNA) were significantly higher than B1p(POB)B2 phosphate adducts (89–190 fmol/mg DNA). Similarly, in the lung, the levels of B1p(PHB)B2 phosphate adducts were 3950–8160 fmol/mg DNA, compared to 218–475 fmol/mg DNA of B1p(POB)B2 phosphate adducts. These differences were possibly due to the pharmacokinetic properties of NNK. When NNK is administered to rats, it is rapidly converted to NNAL. Although NNAL and NNK are in equilibrium, the equilibrium is strongly shifted toward NNAL in rats (4,29). Moreover, the half-life of NNAL in rats is 298 min, while that of NNK is only 25 min (30). Therefore, NNAL predominates over NNK in the rat circulation system (30,31). Since B1p(PHB)B2 phosphate adducts were formed from NNAL, while B1p(POB)B2 adducts were from NNK, this is one explanation for the higher levels of B1p(PHB)B2 than B1p(POB)B2 adducts. Indeed, the relative amount of total pyridine-containing DNA adducts from NNAL [B1p(PHB)B2 plus PHB-base, 51–67%] was higher than that from NNK [B1p(POB)B2 plus POB-base, 33–49%] in the lung. Similarly, slightly higher levels of NNAL-derived adducts (52–57%) than NNK-derived adducts (43–48%) were observed in the liver. Interestingly, POB base adducts were more abundant than B1p(POB)B2 phosphate adducts, while PHB base adducts were less abundant than B1p(PHB)B2 phosphate adducts. This suggests that NNK-derived base and phosphate adducts are either formed or repaired in a different manner than NNAL-derived base and phosphate adducts. However, the repair mechanism of the NNK/NNAL-derived DNA phosphate adducts is still unclear, and further studies are warranted.
The biological significance of the newly characterized NNK/NNAL-derived B1p(PHB)B2 phosphate adducts is unclear. Studies on other modifications of phosphate groups in DNA suggest some potential effects, including inhibiting DNA metabolic enzymes (25,32,33), affecting DNA helicases (34–36) and interfering with the binding of DNA to other macromolecules (37). Similar effects could be caused by the NNK-derived phosphate adducts, potentially leading to detrimental biological consequences; therefore, further studies of their effects on DNA repair processes and other biological effects are needed.
In summary, we applied our highly sensitive and selective LC-NSI-HRMS/MS methodology to characterize and quantify 54 and 107 previously unknown pyridylhydroxybutyl DNA phosphate adducts formed in liver and lung, respectively, of F-344 rats as the result of exposure to the tobacco-specific carcinogen NNK. These adducts are formed via NNK metabolism to NNAL and subsequent bioactivation of NNAL. Levels of some of these newly identified adducts in lung and liver of NNK-treated rats were persistent over the 70 weeks of treatment. These novel adducts could potentially serve as biomarkers of NNK exposure and carcinogenesis in humans.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This work was supported by National Cancer Institute (CA-81301). Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by Cancer Center Support Grant from National Cancer Institute (CA-77598). Salary support for P.W.V. was provided by National Cancer Institute (CA-211256).
Acknowledgements
We thank Xun Ming for his help with the mass spectrometry analysis, and Dr. Silvia Balbo for providing rat lung DNA samples. We thank Christopher Seiler in Department of Medicinal Chemistry, University of Minnesota for useful discussions on the synthesis of [13C1015N2]2’-TpT. We also thank Robert Carlson for editorial assistance.
Conflict of Interest Statement: None declared.
Abbreviations
- HCD
higher-energy collisional dissociation
- LC
liquid chromatography
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNAL
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- NSI
nanoelectrospray ionization
References
- 1. International Agency for Research on Cancer. (2007)Smokeless tobacco and tobacco-specific nitrosamines. IARC monographs on the evaluation of carcinogenic risks to humans. Vol. 89 International Agency for Research on Cancer: Lyon, France. [Google Scholar]
- 2. Appleton S., et al. (2013)TSNA levels in machine-generated mainstream cigarette smoke: 35 years of data. Regul. Toxicol. Pharmacol., 66, 197–207. [DOI] [PubMed] [Google Scholar]
- 3. Hecht S.S. (2014)It is time to regulate carcinogenic tobacco-specific nitrosamines in cigarette tobacco. Cancer Prev. Res. (Phila)., 7, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hecht S.S. (1998)Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol., 11, 559–603. [DOI] [PubMed] [Google Scholar]
- 5. Balbo S., et al. (2014)Carcinogenicity and DNA adduct formation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in F-344 rats. Carcinogenesis, 35, 2798–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hecht S.S., et al. (2016)Exposure and metabolic activation biomarkers of carcinogenic tobacco-specific nitrosamines. Acc. Chem. Res., 49, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Appleton S., et al. (2014)TSNA exposure from cigarette smoking: 18 years of urinary NNAL excretion data. Regul. Toxicol. Pharmacol., 68, 269–274. [DOI] [PubMed] [Google Scholar]
- 8. Hecht S.S., et al. (2002)Quantitation of metabolites of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone after cessation of smokeless tobacco use. Cancer Res., 62, 129–134. [PubMed] [Google Scholar]
- 9. Hecht S.S., et al. (2007)Similar exposure to a tobacco-specific carcinogen in smokeless tobacco users and cigarette smokers. Cancer Epidemiol. Biomarkers Prev., 16, 1567–1572. [DOI] [PubMed] [Google Scholar]
- 10. Kresty L.A., et al. (1996)Metabolites of a tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), in the urine of smokeless tobacco users: relationship between urinary biomarkers and oral leukoplakia. Cancer Epidemiol. Biomarkers Prev., 5, 521–525. [PubMed] [Google Scholar]
- 11. Stepanov I., et al. (2005)Tobacco-specific nitrosamines and their pyridine-N-glucuronides in the urine of smokers and smokeless tobacco users. Cancer Epidemiol. Biomarkers Prev., 14, 885–891. [DOI] [PubMed] [Google Scholar]
- 12. Burns D.M., et al. (2011)Do changes in cigarette design influence the rise in adenocarcinoma of the lung?Cancer Causes Control, 22, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andreotti G., et al. (2017)Tobacco use and cancer risk in the Agricultural Health Study. Cancer Epidemiol. Biomarkers Prev., 26, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ronai Z.A., et al. (1993)G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis, 14, 2419–2422. [DOI] [PubMed] [Google Scholar]
- 15. Jasti V.P., et al. (2011)Tobacco-specific nitrosamine-derived O2-alkylthymidines are potent mutagenic lesions in SOS-induced Escherichia coli. Chem. Res. Toxicol., 24, 1833–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jalas J.R., et al. (2005)Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem. Res. Toxicol., 18, 95–110. [DOI] [PubMed] [Google Scholar]
- 17. Wang M., et al. (2003)Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco specific carcinogens. Chem. Res. Toxicol., 16, 616–626. [DOI] [PubMed] [Google Scholar]
- 18. Hecht S.S., et al. (2004)Identification of O2-substituted pyrimidine adducts formed in reactions of 4-(acetoxymethylnitrosamino)- 1-(3-pyridyl)-1-butanone and 4-(acetoxymethylnitros- amino)-1-(3-pyridyl)-1-butanol with DNA. Chem. Res. Toxicol., 17, 588–597. [DOI] [PubMed] [Google Scholar]
- 19. Sturla S.J., et al. (2005)Mass spectrometric analysis of relative levels of pyridyloxobutylation adducts formed in the reaction of DNA with a chemically activated form of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem. Res. Toxicol., 18, 1048–1055. [DOI] [PubMed] [Google Scholar]
- 20. Lao Y., et al. (2006)Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol., 19, 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma B., et al. (2015)Comprehensive high-resolution mass spectrometric analysis of DNA phosphate adducts formed by the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem. Res. Toxicol., 28, 2151–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spratt T.E., et al. (1990)Solvolysis of model compounds for alpha-hydroxylation of N’-nitrosonornicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone: evidence for a cyclic oxonium ion intermediate in the alkylation of nucleophiles. Chem. Res. Toxicol., 3, 350–356. [DOI] [PubMed] [Google Scholar]
- 23. Upadhyaya P., et al. (2008)Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol., 21, 1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang S., et al. (2009)Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol., 22, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones G.D., et al. (2010)Phosphotriester adducts (PTEs): DNA’s overlooked lesion. Mutagenesis, 25, 3–16. [DOI] [PubMed] [Google Scholar]
- 26. Singer B. (1985)In vivo formation and persistence of modified nucleosides resulting from alkylating agents. Environ. Health Perspect., 62, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Den Engelse L., et al. (1987)O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis, 8, 751–757. [DOI] [PubMed] [Google Scholar]
- 28. Shooter K.V., et al. (1977)The stability of methyl and ethyl phosphotriesters in DNA in vivo. Chem. Biol. Interact., 19, 353–361. [DOI] [PubMed] [Google Scholar]
- 29. Upadhyaya P., et al. (2000)Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis, 21, 1233–1238. [PubMed] [Google Scholar]
- 30. Adams J.D., et al. (1985)On the pharmacokinetics of tobacco-specific N-nitrosamines in Fischer rats. Carcinogenesis, 6, 509–511. [DOI] [PubMed] [Google Scholar]
- 31. Adams J.D., et al. (1985)Pharmacokinetics of N’-nitrosonornicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in laboratory animals. Cancer Lett., 28, 195–201. [DOI] [PubMed] [Google Scholar]
- 32. Yashiki T., et al. (1992)Sequence specific block of in vitro DNA synthesis with isopropyl phosphotriesters in template oligodeoxyribonucleotides. Nucleic Acids Symp. Ser., 27, 197–198. [PubMed] [Google Scholar]
- 33. Miller P.S., et al. (1982)Synthesis and template properties of an ethyl phosphotriester modified decadeoxyribonucleotide. Biochemistry, 21, 5468–5474. [DOI] [PubMed] [Google Scholar]
- 34. Eoff R.L., et al. (2005)Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry, 44, 666–674. [DOI] [PubMed] [Google Scholar]
- 35. Suhasini A.N., et al. (2012)DNA repair and replication fork helicases are differentially affected by alkyl phosphotriester lesion. J. Biol. Chem., 287, 19188–19198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khan I., et al. (2015)Close encounters for the first time: helicase interactions with DNA damage. DNA Repair (Amst)., 33, 43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Makishi S., et al. (2014)Modulation of binding properties of amphiphilic DNA containing multiple dodecyl phosphotriester linkages to lipid bilayer membrane. Bioorg. Med. Chem. Lett., 24, 3578–3581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.