Abstract
The Arc gene is robustly transcribed in specific neural ensembles in response to experience-driven activity. Upon induction, Arc mRNA is transported to dendrites, where it can be rapidly and locally translated by activation of metabotropic glutamate receptors (mGluR1/5). mGluR-induced dendritic synthesis of Arc is implicated in weakening or elimination of excitatory synapses by triggering endocytosis of postsynaptic AMPARs in both hippocampal CA1 and cerebellar Purkinje neurons. Importantly, CA1 neurons with experience-induced Arc mRNA are susceptible, or primed for mGluR-induced long-term synaptic depression (mGluR-LTD). Here we review mechanisms and function of Arc in mGluR-LTD and synapse elimination and propose roles for these forms of plasticity in Arc-dependent formation of sparse neural representations of learned experience. We also discuss accumulating evidence linking dysregulation of Arc and mGluR-LTD in human cognitive disorders such as intellectual disability, autism and Alzheimer’s disease.
Keywords: Arc/Arg3.1, dynamin, mGluR5, long-term synaptic depression, synapse elimination
Arc/Arg3.1 is one of the most strongly induced immediate early genes in response to experience and neural activity and evidence indicates that Arc-induction marks neuronal ensembles that encode learned behaviors [1–7]. Deletion of Arc in mice leads to deficits in many forms of learning and memory, as well as experience-dependent plasticity of circuits [5, 8–11]. Thus, it is hypothesized that experience-dependent induction of Arc in a neuron, or network of neurons, leads to plasticity circuits that mediate memory of that experience [3–5, 11]. Arc has unique qualities among the immediate early genes that make it well suited to cause plasticity of circuits; its mRNA is rapidly transported to dendrites [12] where it is locally translated in response to glutamate [13–16] and it encodes a protein that affects synapse function directly [17–19]. An accumulating body of work implicates dendritic Arc translation in synaptic weakening, or long-term depression (LTD) [13, 14, 20], and synapse elimination in response to activation of Group 1 metabotropic glutamate receptors (mGluR1/5) [21, 22]. Here we will review the roles of Arc in these related forms of synaptic depression and evidence for their dysfunction in human cognitive disease. We also attempt to integrate roles for mGluR-LTD and/or synapse elimination with recent results revealing roles for Arc in formation of sparse neural representations of learned experience [5].
Arc mRNA is rapidly translated in dendrites in response to Group 1 metabotropic glutamate receptors
The robust transcriptional and translational regulation of the Arc gene and mRNA make it an ideal candidate to couple experience-dependent activation of neuronal circuits to synaptic plasticity within these circuits. For example, in hippocampal CA1 neurons, which encode a memory for place or a spatial environment, Arc is induced rapidly, within 30 s, in response to a novel environment [23]. Once Arc is induced in neurons, its mRNA is promptly transported to dendrites [12, 24, 25] where it can be rapidly translated, within seconds to minutes, in response to activation of glutamatergic synapses [13–16]. A specific agonist for Group 1 metabotropic glutamate receptors (mGluR1/5), DHPG, is sufficient to induce Arc translation [13–15], but glutamate-induced dendritic Arc translation relies on both NMDA receptors and mGluR1/5 [16], which may be more relevant in vivo. Glutamate or DHPG induces remarkably fast (~15s) translation of Arc in dendrites as observed by imaging newly synthesized Arc that was tagged with a bright, rapidly decaying Gaussia-Luciferase (Gluc) [16] or Venus [15]. Interestingly, Arc translation did not occur primarily at synapses, but appeared to be coordinated within a dendritic segment. Although the Arc 3’UTR enhances dendritic trafficking of Arc mRNA [26], the coding region of Arc mRNA is sufficient for glutamate-stimulated local translation [16]. This result, combined with the fact that glutamate stimulates Arc translation in seconds, and occurs in the presence of translation initiation inhibitors, suggested that glutamate stimulates ribosomal movement, or elongation, onto Arc mRNA that is already initiated and ribosomes may be stalled on Arc mRNA in dendrites [16]. In support of this idea, mGluRs potently activate elongation factor 2 kinase and phosphorylation of elongation factor 2 which regulates translational elongation and is necessary for mGluR-induced Arc translation [13]. Furthermore, Arc mRNA interacts with Fragile X Mental Retardation Protein (FMRP), an RNA binding protein implicated in ribosomal stalling and processivity [27, 28] that is necessary for mGluR-induced translation of Arc [15, 29–31].
Dendritic translation of Arc is necessary for an mGluR1/5-induced long-term synaptic depression
A major function of Arc is to weaken synapses by stimulating endocytosis of postsynaptic ionotropic AMPA subtype receptors (AMPARs) and reducing their surface and synaptic expression [16–18]. Arc contributes to multiple forms of activity-induced synaptic weakening; including homeostatic downscaling of synapses [19], mGluR-LTD [13, 14, 20] and synapse elimination [21, 22]. If and how these different forms of Arc-dependent synaptic weakening interact, if they affect the same synapses or utilize the same molecular mechanisms (e.g. endocytosis of AMPARs), but are induced in diverse ways is unclear at present and discussed below. For this review, we focus on forms of Arc-dependent synaptic weakening and elimination that require mGluR1/5.
Brief activation of mGluR1/5 (minutes) leads to a LTD of excitatory and inhibitory synaptic transmission in multiple brain regions, which are expressed through different pre-or postsynaptic loci [32, 33]. The most well characterized forms of mGluR-LTD that is mediated postsynaptically at excitatory synapses, occurs through removal of postsynaptic AMPARs in cerebellar Purkinje (Pkj) neurons and hippocampal CA1 neurons and requires Arc [33–35]. Arc is also necessary for an activity-dependent elimination of inputs onto both Pkj and CA1 neurons, which are mGluR1 or mGluR5 dependent, respectively, suggesting that LTD mechanisms contribute to synapse elimination [36] and these are conserved roles for Arc across distinct brain regions.
In CA1 neurons, brief activation of mGluR1/5 with either the selective agonist, DHPG, or synaptic stimulation (paired-pulse low-frequency (1 Hz) stimulation) induces LTD that requires new protein synthesis from pre-existing mRNA and is mediated by postsynaptic endocytosis and decreases in surface AMPAR subunits GluA1 and GluA2 (reviewed [33, 34]). While de novo protein synthesis is not required to trigger endocytosis, and decreases in surface of AMPARs, it is required to maintain decreases in surface AMPARs and the persistent increases in endocytosis rates that accompany LTD [14, 37]. mGluR-LTD is deficient in area CA1 of acute hippocampal slices from Arc KO mice and postsynaptic inhibition of new Arc translation with antisense oligonucleotides blocks LTD, as well as DHPG-induced decreases in surface AMPARs and persistent increases in AMPAR endocytosis rates [13, 14]. These results suggest that Arc levels are rate limiting for AMPAR endocytosis and mGluR-induced increases in local Arc concentration enhance endocytosis rates which maintain decreased surface AMPARs and synaptic depression [14]. In contrast to this view, recent data demonstrated that DHPG induced a rapid synthesis of Arc, which is then followed by ubiquitination and degradation of Arc by the proteasome, resulting in a long-term decrease in Arc protein levels (> 1hr) [38]. This result suggested that increases in Arc levels trigger LTD, but do not maintain it. Of note, in cultured forebrain or hippocampal neurons, Arc levels are induced and remain elevated for an hour after DHPG suggesting that there is little Arc degradation in cultured neurons after DHPG [13, 31] where persistent increases in AMPAR endocytosis rates are observed to accompany LTD. Remarkably, if proteasomal degradation is blocked with proteasome inhibitor, mGluR-LTD no longer requires new protein synthesis [38]. Although these manipulations are not specific for Arc, it supports the view that new Arc synthesis is not absolutely required for mGluR-LTD and mGluRs trigger posttranslational modifications of Arc, or proteins in complex with Arc to cause LTD. Such mechanisms may include tyrosine dephosphorylation of GluA2, which is implicated in mGluR-LTD [39] and phosphorylation of Arc by ERK [40], a protein kinase required for mGluR-LTD [41].
In cultured cerebellar Purkinje (Pkj) neurons, Arc is necessary mGluR-LTD of granule cell inputs but is specifically required for a transcription-dependent “late-phase” of mGluR-LTD, occurring >1 hr after induction [35]. The late-phase of mGluR-LTD also requires the transcription factor, Serum Response Factor (SRF), and its binding to the Arc promoter [35]. mGluR-LTD inducing stimulation (e.g. DHPG) can induce both transcription and translation of Arc in cultured hippocampal or cortical neurons [13, 42], the former of which likely occurs through regulation of SRF [43]. In contrast to Pkj neurons, mGluR-LTD in area CA1 of acute hippocampal slices is independent of transcription and relies on rapid translation of Arc, within 20–30 minutes, from preexisting mRNA [14, 44]. New Arc transcription may be required for mGluR-LTD in cultured Pkj cells because Arc levels are very low in this culture preparation, whereas, as discussed below, CA1 neurons in acute hippocampal slices express Arc transcripts induced in vivo [20]. An interesting possibility is that mGluR-LTD in acutely prepared cerebellar slices may depend on rapid Arc translation from preexisting mRNA, as observed in CA1. In support of this idea, protein synthesis inhibitors rapidly block mGluR-LTD in Pkj neurons in acute slices [45].
Novelty-induced Arc primes CA1 neurons for mGluR-LTD
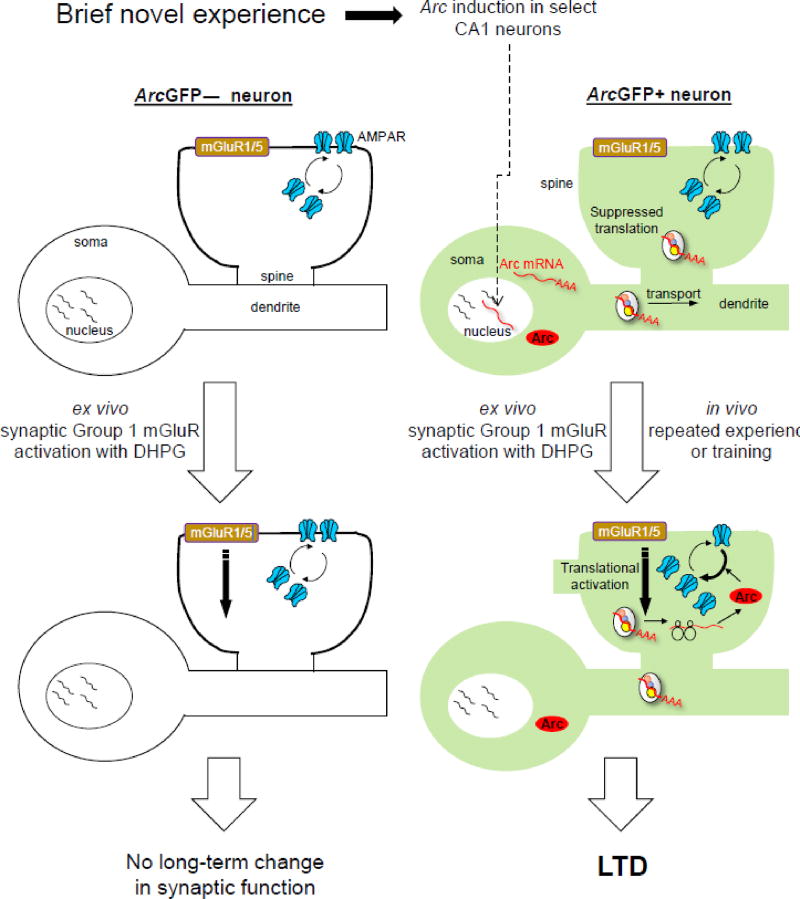
The requirement for Arc in an acute form of synaptic plasticity like mGluR-LTD, suggested that experience-induced Arc induction causes or facilitates mGluR-LTD onto activated neurons and may be a mechanism by which Arc causes plasticity of select circuits activated by salient experience. To determine if Arc induction in vivo results in LTD, we used two different Arc-GFP transcriptional reporter mice (ArcGFP knockin or BAC-ArcGFP), which express destabilized GFP when the Arc gene is induced [9, 46], and allowed visualization and targeting of live neurons with recent Arc induction for measurements of synaptic strength and plasticity [20]. Exposing ArcGFP reporter mice briefly (5 min) to a novel environment, induced GFP expression in a subpopulation (~40%) of CA1 pyramidal neurons, consistent with measurements of endogenous Arc mRNA [20, 47]. Recording from ArcGFP(+) CA1 neurons in acute hippocampal slices of novelty exposed mice did not detect differences in baseline excitatory synaptic function in comparison to neighboring GFP(−) neurons. However, activation of mGluR1/5 onto ArcGFP(+) neurons induced a robust mGluR-LTD, whereas, surprisingly, GFP(−) neurons had little or no mGluR-LTD. Therefore, Arc induction by brief exposure to novelty does not induce LTD, but facilitates or “primes” neurons for subsequent LTD in response to mGluR1/5 activation. Results indicated that mGluR-LTD priming occurs in ArcGFP(+) neurons because they express more dendritic Arc mRNA and display robust mGluR-induced dendritic synthesis of Arc protein in comparison to their GFP(−) neighbors. A model based on this data proposes that Arc mRNA is induced by novelty and transported to dendrites, but may be translationally suppressed (Fig. 1). Subsequent activation of mGluR1/5 derepresses translation of dendritic Arc mRNA, increases dendritic Arc levels and induces mGluR-LTD. In support of this model, Arc and other mRNAs in dendrites are likely translationally suppressed [31, 48–50] and glutamate induces Arc translation in dendrites within seconds [16]. Priming of mGluR-LTD by novelty-induced Arc provides a cellular mechanism for the known facilitation of LTD by exploration of novel objects or environments [20, 51–59]. If novel environment exposure is required to observe robust mGluR-LTD, then why is mGluR-LTD observed in many slice studies without first exposing animals to novelty? Indeed, the standard procedures of mouse handling prior to acute slice preparation, which include removing an unanesthetized mouse from its home cage and transporting to the lab, are sufficient to induce Arc and observe robust mGluR-LTD. Anesthesia of mice in their home cage prevented Arc induction during transportation to the lab and resulted in a small magnitude mGluR-LTD in population field potential recordings in slices [20]. Thus, “standard handling” protocols of mice for slice physiology experiments may constitute a novel, or stressful, experience for the mouse since stress, like novelty, can induce Arc in CA1 [60] and enhance mGluR-LTD [61].
Figure 1. Proposed model of mGluR-LTD priming by novelty-induced Arc in hippocampal CA1 neurons based on data from [20].
A novel experience induces transcription of Arc mRNA in a select population of CA1 neurons (ArcGFP+). After induction, Arc mRNAs are transported to dendrites where evidence suggests it is translationally suppressed [31, 48, 50]. Ex vivo activation of group 1 mGluRs (in slices) activates Arc translation in dendrites, of those neurons with recent Arc mRNA induction (ArcGFP+), but not neighboring ArcGFP(−) neurons [13, 14]. Arc protein increases the endocytosis rate of AMPA receptors, causing long-term synaptic depression only in ArcGFP+ neurons. Repeated experience of the same environment reactivates synapses on the same neurons initially activated when the environment was novel and Arc mRNA was induced (ArcGFP+) [1] which is proposed to suppress synaptic transmission onto these neurons in vivo through a similar mechanism as LTD. Modified from [20].
Arc and activity-dependent, developmental synapse elimination
In addition to Arc’s role in rapid and transient weakening of excitatory synaptic transmission, such as LTD, Arc is also necessary for activity-dependent and developmental pruning or elimination of excitatory synapses onto cerebellar Pkj and hippocampal CA1 neurons [21, 22]. Activity-dependent synapse elimination is a process by which experience and learning remove redundant, or inappropriate connections to form and refine circuit connections for optimal mature function. A well-established model of developmental synapse elimination is the climbing fiber (CF) to Pkj neuron synapse in the cerebellum [62, 63]. At birth, multiple CF axons innervate Pkj neurons, and during the first few postnatal weeks, one CF input becomes progressively strengthened, whereas other CF inputs are weakened and ultimately eliminated by the 3rd postnatal week, as measured by evoked CF EPSCs [64]. Activity of Pkj neurons and/or their inputs, specifically mGluR1, NMDARs or P/Q type voltage-dependent Ca2+ channels (VDCCs) are necessary for CF input elimination and their blockade results in multiply CF innervated Pkj neurons [62, 63, 65, 66]. Conversely, optogenetically increasing firing rates of Pkj neurons in organotypic cultures accelerates CF input elimination by activating of P/Q VDCCs and inducing Arc transcription [21]. Knockdown of P/Q VDCCs or Arc in Pkj neurons prevents activity-accelerated CF input elimination in cultures and developmental CF elimination in vivo [21, 65]. Overexpression of Arc itself is insufficient to eliminate CF inputs in the absence of activity, indicating that other activity-dependent processes function together with Arc to confer CF input elimination [21].
Like its role in Pkj neurons, Arc is required for synapse elimination in CA1 hippocampal neurons [22]. Although synapse elimination in hippocampal CA1 neurons has been less well characterized in comparison to climbing fibers, insight into this process was made when it was discovered that expression of the activity-dependent transcription factor Myocyte-Enhancer Factor 2 (MEF2) induced a rapid (<24 hr) elimination of excitatory synapses onto hippocampal neurons [67, 68]. Four MEF2 proteins (MEF2A-D) belong to the MEF2/MADS box family of transcription factors [69, 70] and MEF2A and MEF2D are expressed in CA1 neurons. In response to neuronal depolarization and Ca2+ influx, MEF2 activates transcription of target genes, including Arc [67, 71]. A consensus binding sequence for MEF2 is present in an upstream enhancer of the Arc gene, that confers robust activity-dependent transcription and thus is termed the synaptic activity response element (SARE) [72]. MEF2A/D bind to the Arc SARE and this binding is necessary for activity-dependent induction of the Arc gene. CREB and SRF also have binding sites in the SARE that are required for activity-induced Arc revealing a combinatorial interaction of 3 transcription factors that are necessary for full induction of Arc [72]. Overexpression of a constitutively active form of MEF2 (MEF2VP16) in organotypic hippocampal slice cultures causes a rapid, within 12–16 hr, and robust (30–50%) elimination of dendritic spines, synaptic markers and depresses evoked EPSC amplitudes as well as the frequency of spontaneous, miniature (m) EPSCs [67, 68] which is consistent with a functional synapse elimination. MEF2VP16 induces transcription of Arc and fails to eliminate functional synapses or spines in CA1 neurons of Arc KO mice [22].
To determine the physiological activity patterns that regulate MEF2 transcriptional induction of Arc in neurons and whether such patterns eliminate synapses through MEF2 and Arc, recent work demonstrated, similar to Pkj neurons [21], that optogenetically driving individual CA1 neurons to fire in bursts at 3 Hz, resulted in a MEF2A/D-dependent induction of Arc and an Arc-dependent functional and structural synapse elimination [73, 74]. Interestingly, the duration of postsynaptic bursting (1 vs. 24 hr) induced distinct forms of synaptic depression that both required Arc. Relatively brief periods of postsynaptic bursting (1 hr) selectively depressed AMPA receptor (R) synaptic transmission, or silenced excitatory synapses, whereas more prolonged (24 hr) firing depressed both AMPAR and NMDAR EPSCs and eliminated spines, indicative of a synapse elimination. Both synapse silencing and elimination required de novo transcription and Arc, but only synapse silencing in response to brief activity bursts required MEF2A/D [74]. This surprising finding indicates that activity and MEF2A/D-induced Arc silence synapses without eliminating them and identifies, perhaps an Arc-dependent intermediate stage of synapse elimination. Furthermore, longer durations of postsynaptic activity likely engage other transcription factors, besides MEF2A/D, to induce Arc and eliminate synapses. Acute, exogenous re-expression of Arc, postsynaptically and cell autonomously, in Arc KO CA1 neurons rescues activity or MEF2VP16-induced synapse silencing or elimination of spines and functional synapses, respectively [22, 74]. However, overexpression of Arc alone is insufficient to silence or eliminate synapses suggesting that other MEF2-induced genes are required [22]. One candidate is Protocadherin 10 (Pcdh10), a MEF2 target gene that functions in synapse elimination to degrade the synaptic scaffold, PSD-95 [75]. Degradation of PSD-95 is necessary for AMPAR endocytosis during LTD [76], thus Pcdh10-dependent degradation of PSD-95 may be necessary for Arc to trigger AMPAR endocytosis and synapse elimination in response to MEF2 activation.
Like novelty-induced priming of mGluR-LTD, MEF2-induced synapse elimination requires a coordinated transcriptional and dendritic translational control of Arc. Pharmacological block or genetic deletion of mGluR5 prevents MEF2-induced elimination of spines and functional synapses [22]. One role of mGluR5 in MEF2 triggered synapse elimination is to stimulate synthesis of Arc protein in dendrites. mGluR5 antagonists do not interfere with MEF2-induced transcription of Arc or increases in dendritic Arc mRNA, but block MEF2-induced increases in dendritic Arc protein. Culturing hippocampal neurons in microfluidic chambers allowed selective blockade of mGluR5 on dendrites, but not the soma, which was sufficient to block MEF2-induced increases in dendritic Arc protein and synapse elimination [22]. Therefore, this data suggests that that two forms of neural “activity” are required to induce synapse elimination, one in the form of action potentials to trigger Ca2+ influx in the cell soma and transcription of Arc via MEF2, and perhaps other transcription factors. Synaptic stimulation of mGluR5 is then required to translate dendritic Arc mRNA into protein for synapse elimination. As in LTD, mGluR5 may also regulate posttranslational modifications of AMPARs [39] that may function together with Arc to stimulate endocytosis. Roles for mGluR5 in synapse elimination in forebrain are suggested by findings of increased frequency of spontaneous excitatory synaptic events (mEPSCs) and/or dendritic spines in CA1 neurons [22] as well as layer 2/3 and 4 neocortical neurons; with cell autonomous deletion of mGluR5 [77, 78] or decreased spines in Nucleus Accumbens with mGluR5 positive allosteric modulators [79]. Whether Arc also functions in synapse elimination in these brain regions is yet to be determined. Similarly, in Pkj neurons, mGluR1 is necessary for CF input elimination [66], although a role for mGluR1 in regulation of Arc synthesis in Pkj neurons is unknown. The common roles of mGluR1/5 and Arc in mGluR-LTD and synapse elimination in Pkj and CA1 neurons suggest that mGluR-LTD mechanisms, such as AMPAR endocytosis, may be an initial trigger for long-term synapse elimination as at other synapses [36, 80]. In support of this idea, structural synapse elimination is a consequence of repeated episodes of LTD induction by mGluR or NMDAR activation [81–83].
Molecular Mechanisms of Arc regulation of AMPAR endocytosis
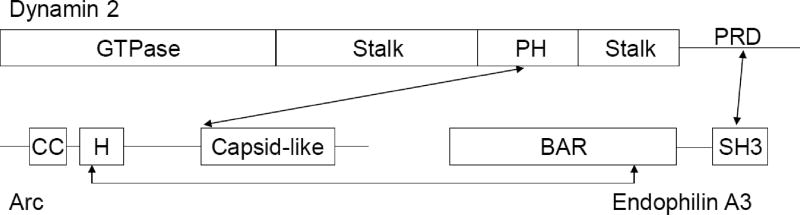
As stated, an important mechanism underlying LTD is the removal of AMPARs from the post-synaptic surface, effected primarily by increasing the rate of endocytosis. A potential mechanism to explain Arc’s role in AMPAR endocytosis was provided by Chowdhury et al. [18], who showed that Arc binds directly to two elements of the endocytic machinery, dynamin and endophilin, and by the more recent finding of DaSilva et al. [84] that Arc also interacts with the clathrin adaptor protein, AP-2 (reviewed in [85]). Dynamins are ~100 kDa GTPases that self-assemble around the necks of invaginating vesicles and promote membrane scission in a GTPase-dependent manner [86, 87]. Mammals express three forms of dynamin: dynamin 2 is ubiquitously expressed; dynamins 1 and 3 are particularly abundant in neurons. All three forms of dynamin share a common domain structure, including an N-terminal GTPase domain, a central phosphoinositide-binding pleckstrin homology (PH) domain flanked by two “stalk” domains, and a C-terminal proline-rich domain (PRD). In the three-dimensional structure, the two stalk domains fold back and interact with each other to form a four-stranded helix involved in dimerization, tetramerization, and higher-order dynamin oligomerization. Dynamin self-assembly, either on biological membranes or, in vitro on the surface of liposomes, stimulates its GTPase activity from a basal rate of approximately 1–10 min−1 for unassembled (tetrameric) dynamin to maximal rates greater than 200 min−1 for polymers. We confirmed, using pure proteins, that Arc interacts with dynamins 2 and 3 [88]. We further showed that Arc increases the rate and extent of dynamin self-assembly, and promotes assembly-dependent GTPase activation of dynamins 2 and 3 to levels of approximately 90 min−1 and 120 min−1, respectively. Interestingly, binding of Arc to dynamin 1, which is largely pre-synaptic and functions in synaptic vesicle recycling [87], was nearly undetectable and Arc had no effect on the enzymatic or assembly properties of this isoform. Dynamins 2 and 3 are found in both pre- and postsynaptic nerve terminals.
By analyzing a series of truncation and deletion mutants, Chowdhury et al. [18] identified a segment comprising amino acids 195–214 of Arc as a critical dynamin-binding determinant. We observed that deletion of this segment weakened, but did not completely abolish, the Arc-dynamin interaction [88], suggesting that additional dynamin-binding sites in Arc remain to be identified. Chowdhury et al. [18] also identified the PH domain as the most likely Arc-interacting determinant in dynamins. Structural studies revealed that dynamins can exist in “closed” and “open” conformations [89–91]. In the closed conformation, the PH domain is folded back onto the stalk, thereby acting as an intra-molecular suppressor of dynamin self-assembly and activation. In the open conformation, the PH domain is extended away from the stalk and is now free to associate with phosphoinositides and to polymerize on the membrane surface. By interacting with the PH domain, Arc may disrupt the autoinhibitory stalk-PH domain interface. It is important to note that the stalk-binding and phosphoinositide-binding residues are on opposite surfaces of the PH domain. Thus, dynamin binding to Arc and to membranes are not mutually exclusive.
Endophilins comprise a family of five (endophilins A1–3 and B1–2) ~40 kDa proteins (endophilins A1–3 and B1–2) that contain N-terminal BAR (Bin/amphiphysin/Rvs) domains and C-terminal SH3 domains connected by variable linker regions. Dimerization of their BAR domains create crescent-shaped structures that induce and/or stabilize membrane curvature [92]. Endophilins interact with dynamin PRDs via their Src homology 3 (SH3) domains, and these interactions promote dynamin oligomerization in vitro and in cells [93]. Arc interacts via residues 89–100 with the BAR domains of endophilins A2 and A3 (Fig. 2), which are abundant in post-synaptic compartments, but it does not interact with endophilin A1, which is predominantly localized to pre-synaptic nerve terminals. Importantly, surface expression of GluR1 was significantly reduced in neurons upon overexpression of full-length Arc but not Arc-Δ91–100 (defective endophilin binding) or Arc-Δ195–214 (defective dynamin binding) [18].
Figure 2. Scheme showing the interaction sites of Arc with Endophilin 3 and Dynamin 2.
Based on secondary structure predictions, Arc is divided into two domains connected by an extended intrinsically disordered region. The N-terminal half contains two stretches predicted to fold into α-helices, the first (~residues 30–60) with coiled-coil (CC) forming potential, the second (~residues 85–130; designated “H” in the figure) containing potential amphipathic segments. The C-terminal half of Arc contains a segment (residues 217–362) that is structurally similar to the retroviral Gag capsid [165]. The domains of dynamin and endophilin are described in the text.
A deeper understanding of the physical properties of Arc in live neurons is likely to provide valuable insights into its role in synaptic plasticity. For example, it will be important to know whether Arc self-assembles in neurons, as it does in vitro [88, 94]. If so, Arc may function as a scaffold to recruit and/or stabilize endocytic proteins, including dynamin and endophilin, on clathrin-coated pits. The mechanism and function of Arc’s interaction with the actin cytoskeleton also remains to be elucidated. In this context, we note that dynamin has an endocytosis-independent role in the regulation of actin polymerization [95] and that endophilin A2 controls a newly discovered form of clathrin-independent but actin- and dynamin-dependent endocytosis termed fast endophilin-mediated endocytosis (FEME) [96, 97]. Perhaps also relevant is the finding that direct binding of endophilins A2 and A3 (but not A1) to oligophrenin 1, a BAR domain-containing Rho GAP (GTPase activating protein), promotes mGluR-induced LTD and the reduction of surface AMPARs [98]. Arc and oligophrenin 1 bind to distinct sites on endophilins, potentially allowing for simultaneous interactions.
Arc-dependent synaptic weakening may contribute to the formation of sparse neural representations of learned experience
As discussed, Arc is rapidly and robustly induced in select neuronal populations during salient experience, such as novelty or context-dependent fear and in response to learning [3, 4]. Using the Arc gene regulatory elements to optogenetically silence Arc-induced hippocampal neuron populations during learning inhibits memory formation [6]. These results suggest that these Arc-induced neuronal ensembles mediate learned behavior. If or how Arc-dependent synaptic weakening (LTD or synapse elimination) contribute to plasticity of these neuronal populations during learning is unclear. Arc-dependent synaptic weakening may mediate formation of a sparse neural representation of an experience that develops during learning. For example, during exploration of a novel environment a subset (~40%) of hippocampal CA1 neurons fire with some spatial specificity and induce Arc [1, 99, 100]. Re-exposure to the same environment re-induces Arc in the same overlapping population of CA1 suggesting that these Arc+ neurons are activated by and encoding the new environment [1] (Fig. 1). As the environment becomes familiar with repeated exposures over days, the average population firing rate of CA1 neurons declines, as well as the number of CA1 neurons that are active [99] and induce Arc [47]. Interestingly, CA1 neurons that are less spatially-tuned and with lower firing rates (<12 Hz) show reduced activity upon repeated exposures, whereas more active neurons (>12 Hz firing rate) develop increased firing rates. Therefore, during familiarity or habituation to a novel environment, an initially diffuse ensemble of activated CA1 neurons is sculpted into a sparse, precise network of highly spatially-tuned CA1 place cells that form the neural representation for that environment [99]. Arc induction may tag and bind together CA1 neurons that were active in response to a specific environment. The CA1 neurons in the network are then primed for plasticity of their synaptic inputs by virtue of the fact that they have Arc mRNA in their dendrites. With repeated exposure to the same environment, neurons that are less spatially tuned and fire at lower rates (<10Hz); rates that promote LTD [101], would be expected to have weakening of active synaptic inputs carrying sensory information about that environment. The Arc-dependent LTD priming may contribute to the reduced excitation of those neurons with subsequent environment exposure and generation of a sparse CA1 representation of that environment. In a test of this model, ArcGFP reporter mice were first exposed to a novel environment to mark novelty-induced CA1 neuron ensembles and then repeatedly exposed to the same environment over next few hours and acute brain slices prepared to determine if LTD occurred with repeated experience or learning of a new environment [20]. ArcGFP(+) CA1 neurons in “repeat environment exposure mice” had depressed excitatory synaptic transmission, in comparison to GFP(−) neighbors and mGluR-LTD was occluded or blocked. This result contrasts with the enhanced mGluR-LTD on ArcGFP+ neurons in mice exposed once to a novel environment [20] and suggest that repeated environment exposure depresses synaptic transmission onto CA1 neurons through a mechanism shared with mGluR-LTD. Such a depression of synaptic transmission with repeated environment exposure may contribution to habituation or familiarity of a novel environment. In support of this idea, mGluR5, Arc and LTD mechanisms such as AMPAR endocytosis are required for behavioral habituation to novel environments [8, 56, 102, 103].
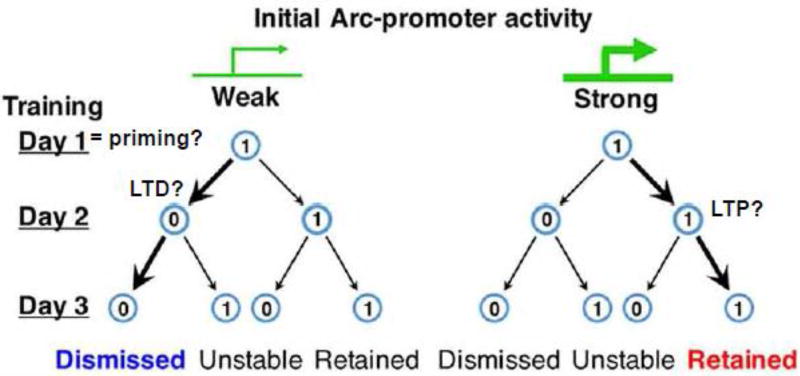
More direct support for a role of Arc in the formation of sparse or consolidated neural representations of learned experience, comes from recent work by Wang and colleagues in the motor cortex [5]. Using Arc-GFP knockin transcriptional reporter mice, they imaged Arc induction in ensembles of higher order motor cortical neurons (M2) in vivo during acquisition of a motor learning task (e.g. walking on a rotating rod; rotarod). The first day of rotarod training increased the number and fluorescence intensity of Arc-GFP(+) M2 neurons in comparison to home caged mice. Repeated, daily rotarod training for the next 2 days, re-induced ArcGFP in a similar, overlapping population of M2 neurons suggesting that motor learning induces Arc in a specific motor cortical neuron ensemble [5]. Like CA1 neuron ensembles during novelty learning, motor learning causes ArcGFP(+) M2 cortical neuron ensembles to consolidate into a sparse network of highly active ArcGFP(+) neurons that mediate the learned skill. Specifically, of the ArcGFP(+) neurons induced on the first rotarod training day, some were either “dismissed”, or not re-activated, on subsequent training days and others were “retained” or consistently activated on repeated training days (Fig. 3). Whether a ArcGFP(+) neuron was dismissed or retained was related to the level of ArcGFP induction on Day 1 of rotarod training. Neurons strongly activated or with elevated levels of ArcGFP on Day 1 were more likely to be retained upon subsequent rotarod training days, whereas neurons with lower levels of ArcGFP on Day 1 were more likely to be dismissed (Fig. 3). Repeating experiments in the homozygous ArcGFP transcriptional reporter knockin mouse (which does not express Arc protein), revealed that Arc itself is necessary for consolidation of the neural ensemble and for motor learning [5]. As observed in CA1 neurons with repeated novelty, low levels of ArcGFP induction in M2 neurons may prime these neurons for Arc-dependent LTD of active inputs the during rotarod training and contribute to their “dismissal” from the ensemble during subsequent training sessions (Fig. 3). In summary, Arc induction occurs in a neuron population with varying levels of activity in response to a learning experience and facilitates formation or consolidation of a sparse, highly active neural ensemble that mediates learning of a new motor task. Evidence that an Arc specific neural ensemble mediates learning comes from experiments that utilized the Arc SARE to inactivate synapses onto Arc-induced neurons in the frontal cortex which erased an acquired rotarod learning skill [7].
Figure 3. Potential role of experience and Arc dependent priming of LTD in refinement of neural ensembles with repeated experience or training.
Cao et al, [5] demonstrated that repeated training on a motor task resulted in a sparse neural ensemble of highly active neurons. Motor cortical neurons in M2 induce Arc on day 1 of training (“1”), as imaged with a GFP transcriptional reporter in vivo which may prime these neurons for synaptic plasticity during repeated training days. Neurons with initially weak levels of Arc promoter activity may be primed for LTD which may cause them to be inactive “0” or “dismissed” on subsequent days of training. Neurons with stronger levels of Arc promotor activity on Day 1 may be more likely to undergo LTP and to “retain” their strong neuronal activity during subsequent days of training. (Figure is modified from [5]).
If or how Arc contributes to the maintenance of the synaptic connectivity onto strongly activated neurons that remain active during repeated environment exposure or repeated motor training is unclear. Recent work demonstrated that Arc is necessary for an mGluR1/5, protein synthesis-dependent LTP in CA1 neurons, which requires extremely high frequencies of presynaptic stimulation (~200 Hz) [104]. These results suggest that the highest frequencies of synaptic activation or firing and strong Arc induction may prime neurons for LTP, or are necessary to promote Arc dependent strengthening and may contribute to retaining highly active neurons in an ArcGFP+ motor cortical neuron ensembles with repeated motor learning.
Roles for Arc-dependent synaptic weakening in neurodevelopmental disorders
Due to the strong links of Arc with learning and memory in healthy animals, perhaps is it not surprising that alterations in Arc and Arc-dependent synaptic plasticity are associated neuropsychiatric diseases, especially those accompanied by cognitive dysfunction. Although mutations in Arc itself are not prevalent in these diseases, dysregulation of Arc mRNA, protein levels or mutations in regulators or interacting proteins of Arc are linked with intellectual disability, autism [13, 29, 31, 104–108], Alzheimer’s disease [109–111], depression [60] and schizophrenia [112–117]. As discussed below, alterations in mGluR-LTD and synapse elimination are associated with these diseases, and some evidence suggests this is mediated by dysregulation of Arc. The earliest example comes from rodent models of Fragile X Syndrome, Fmr1 knockout (KO), the most common inherited form of intellectual disability and leading genetic cause of autism [27, 118] where mGluR-LTD levels in CA1 are enhanced and independent of new protein synthesis [34, 119–123]. Although mGluR-LTD is independent of new protein synthesis in Fmr1 KO mice, it requires Arc, suggesting that mGluR-LTD in Fmr1 KO is mediated by preexisting Arc protein [13]. FXS is caused by loss of function mutations in the FMR1 gene which encodes an RNA binding protein Fragile X Mental Retardation Protein (FMRP) [27, 118]. FMRP binds Arc mRNA and is colocalized with Arc mRNA and translation in dendrites [16, 29, 124]. FMRP suppresses translation of other mRNA targets and likely Arc [27, 31]. Consequently, in the FXS mouse model, Fmr1 KO, Arc protein levels are elevated in dendrites and unresponsive to mGluR stimulation [15, 31]. Results from rescue experiments with FMRP and phosphorylation site mutants of FMRP suggested that a Ser500 phosphorylated FMRP functions to suppress Arc translation in dendrites [31]. In response to mGluR1/5 stimulation, FMRP is dephosphorylated by PP2A [125–127] which derepresses dendritic Arc translation and promotes LTD [31]. Therefore, mGluR-LTD in the Fmr1 KO mouse may be independent of new Arc protein synthesis because Arc levels are elevated to a level to support LTD [31]. This idea is supported by data that inhibition of Arc protein degradation, with proteasome inhibitors, leads to elevated Arc protein levels and mGluR-LTD that is independent of new protein synthesis [38]. Because of the similar effects of proteasome inhibitors and Fmr1 deletion on mGluR-LTD, an interesting possibility is that Arc degradation, in addition to its regulated translation, may be deficient in Fmr1 KO neurons. Remarkably, like mGluR-LTD, mGluR1/5 and Arc-dependent LTP is independent of new protein synthesis in Fmr1 KO CA1 neurons, implying that elevated dendritic Arc levels can support LTP or LTD without the need for de novo synthesis [104]. This result proposes that posttranslational modifications of synaptic proteins caused by LTP or LTD inducing stimulation determine the direction of synaptic strength changes in response to local Arc synthesis. The findings of altered mGluR-LTD in FXS may be relevant clinically because genetic or pharmacological reduction of mGluR5 is able to rescue phenotypes in FXS animal models, including behavioral phenotypes (reviewed in [128–131]). Whether mGluR5 antagonism will be useful as a therapeutic in humans has yet to be demonstrated [132, 133].
A striking commonality among distinct mouse models of autism and intellectual disability is the protein synthesis-independence of mGluR-LTD; first shown in Fmr1 KO mice, but recently demonstrated to occur in mouse models of Rett Syndrome [134, 135], 16p11.2 microdeletion [136] and Syngap haploinsufficiency [137]. Since mGluR-LTD levels are normal in Rett and 16p11.2 mouse models, it is the uncoupling of LTD from synaptically-regulated synthesis of new proteins in dendrites, including Arc, that correlates with pathology. Based on the model of the role of dendritic Arc synthesis in experience-dependent priming of mGluR-LTD (Fig. 1), and its role in consolidation of neural ensembles during learning (Figs. 3), one may predict that in these forms of autism and ID, that there may be an abnormal consolidation of Arc+ neural ensembles during learning which may lead to cognitive deficits.
Like too much Arc may give rise to enhanced mGluR-LTD in FXS, reduced mGluR-LTD, too little Arc or dysregulation of Arc is associated with other genetic causes of autism and ID, such as Tuberous Sclerosis Complex Syndrome (TSC). TSC results from loss of function mutations in either TSC1 or TSC2 which form a complex and regulates signaling to mTORC1 [138]. Tsc2+/− mice have reduced mGluR-LTD, reduced levels and protein synthesis rates of Arc in hippocampal area CA1 [139]. Similarly, cell autonomous deletion of Tsc1 or a dominant negative mutant of TSC2 result in reduced mGluR-LTD [140, 141] Remarkably, enhancing mGluR5 activity, using a positive-allosteric modulator, rescues behavioral phenotypes in in Tsc2+/− mice [139]. These results, together with the successful rescue of FXS phenotypes with mGluR5 antagonism, suggest that optimal levels of mGluR5 activity, and perhaps mGluR-LTD, are critical for normal cognitive function. Whether mGluR5 therapeutic strategies restore normal Arc levels in TSC or FXS, has not been reported. The cellular mechanisms by which Tsc1/2 regulate Arc levels is unclear. Tsc2+/− neurons have overall reduced protein synthesis rates [139], which may lead to reduced Arc protein. Conversely, cultured Tsc1 KO neurons are hyperexcitable which leads to enhanced steady state levels of Arc mRNA and protein, but reduced activity-induced Arc transcription [142, 143]. In vivo, one would expect this to lead to blunted experience-induced Arc in relevant neurons which may contribute to the reduced mGluR-LTD observed with Tsc1 deletion in hippocampal slices ex vivo.
Arc is also dysregulated in a mouse model of Angelman Syndrome, a neurodevelopmental disorder with symptoms of autism and intellectual disability caused by lack of expression of the maternal copy of UBE3A, a E3 ubiquitin ligase [144]. In AS mice, Ube3A KO, synaptic Arc levels are elevated which results in decreased synaptic AMPAR expression and reduced synaptic transmission in cultured neurons [105]. Ube3a regulates ubiquitination of Arc and promotes its degradation [105]. Although Ube3A may not directly ubiquitinate Arc, it also inhibits transcription of Arc [107, 145]. In either mechanism, Ube3A inhibits Arc levels. Although elevated levels of Arc are found in both FXS and AS, mGluR-LTD is deficient in AS mice [146], in contrast to FXS where mGluR-LTD levels are elevated. The deficient mGluR-LTD in AS mice may be because synaptic transmission is depressed and mGluR-LTD mechanisms are saturated or “occluded”. Alternatively, or in addition, a deficit in mGluR5 signaling and association with scaffolding proteins is observed in AS mice which may prevent normal mGluR5 signaling to LTD mechanisms [146, 147]. The findings in FXS, TSC and AS indicate that mGluR-LTD phenotypes are not always predicted from steady state Arc levels, but abnormal levels of Arc and mGluR-LTD are associated with cognitive disorders.
Arc, mGluR5 and LTD in age-related cognitive ability and Alzheimer’s disease
In addition to neurodevelopmental disorders, dysregulation of Arc and Arc-dependent mGluR-LTD is implicated in age related-cognitive function and Alzheimer’s disease (AD) [148]. mGluR-LTD is enhanced in areas CA1 and CA3 in aged (2-year-old) rats that have maintained cognitive performance in comparison to their counterparts experiencing age-related memory impairment [149, 150]. Although the correlation of Arc expression with successful cognitive aging in individual animals is unknown, experience-dependent induction of Arc in CA1 and other hippocampal areas declines with age [151, 152]. While maintaining the ability to express normal levels of mGluR-LTD promotes cognition in aging, excessive or unregulated LTD is associated with dementia and Alzheimer’s disease (AD). Accumulating evidence indicates that AD is a disease of synapse loss or failure which may be caused in part by unregulated or excess LTD or synapse elimination mechanisms [153–159]. The pathogenic, soluble amyloid β (Aβ) peptides inhibit LTP, as well as promote LTD-like decreases in postsynaptic AMPA and NMDARs and eventually lead to synapse loss [157–159]. Aβ -induced synaptic depression requires activation of mGluR5, and NMDARs in some cases, and prevents or occludes subsequent mGluR-LTD suggesting that Aβ is activating an mGluR-LTD mechanism to cause synaptic pathology [153, 157–159]. Furthermore, Aβ peptides bind to mGluR5 and stimulate signaling [160] and mGluR5 antagonists reverse AD-related brain pathology and cognitive impairment in mouse models [161–163]. Surprisingly, whether Arc is necessary for amyloid β -induced synaptic depression or loss has not been reported. There are also alterations in basal and activity-induced Arc expression in AD animal models and humans, likely reflecting altered circuit function in these models (reviewed in [148]). The combination of excess synaptic depression, due to Aβ accumulation, together with abnormal experience-regulated induction of Arc+ neural ensembles likely contribute to the devastating cognitive deficits associated with AD.
Although the role of Arc in AD-associated synaptic weakening is unclear in AD, Arc may contribute directly to AD pathogenesis by facilitating the activity-dependent generation of Amyloid β (Aβ). Sequential cleavage of APP by β-secretase (BACE1) and γ-secretase yields Aβ peptides that assemble and accumulate in AD. Arc binds directly to presenilin 1 (PS1), the catalytic subunit of the γ-secretase complex, and disruption of the Arc-PS1 interaction prevents activity-dependent increase in Aβ [109]. Arc had previously been shown to play a similar role in generating the transcriptional regulator, NICD (Notch intracellular domain), by γ-secretase cleavage of Notch [164]. Interestingly, Arc does not affect the rate of internalization of APP, BACE-1, or γ-secretase from the plasma membrane. Thus, Arc’s role in APP processing may not involve its interactions with either dynamin or AP-2, which function primarily in endocytic budding from the plasma membrane. Instead, Arc and PS1 co-localize in dendritic puncta that contain endocytic markers, including EEA1 (early endosomes) and Rab11 (recycling endosomes) [109]. APP itself colocalizes with Arc and endophilin 3 in early (Rab5-positive), late (Rab7-positive), and especially recycling (Rab11-positive) endosomes. Importantly, the extent of colocalization of APP and γ-secretase in the same endocytic structures was significantly reduced in neurons from Arc knockout mice. These results suggest a model wherein Arc assists in the sorting of γ-secretase to APP-containing endosomes [109]. The finding that Arc induction can generate Aβ, together with studies showing that Aβ contributes to Arc induction raises the possibility of an Arc-mediated positive feedback mechanism in amyloidogenesis [109, 148].
Concluding Remarks
Arc is a fascinating gene and has captured the interest of neuroscientists for decades due to its links to learning and memory. The precise experience-dependent induction of Arc in select neural ensembles combined with the transport of Arc mRNA to dendrites provides mechanisms to control plasticity of specific synaptic inputs onto these neural ensembles during learning. Recent work has surprisingly revealed roles of Arc in synaptic weakening mechanisms such as LTD and synapse elimination suggesting important roles for these forms of plasticity in learning and memory. The accumulating evidence of alterations in mGluR-LTD and Arc with diseases of cognition further support their key roles. Challenges for the future include linking what we know about Arc-dependent synaptic plasticity mechanisms to the plasticity of experience-induced, Arc+, neural ensembles and how this mediates learning, as well as understanding the cellular and molecular mechanisms by which Arc is necessary for so many diverse forms of synaptic plasticity. Such basic understanding of these processes will be necessary to know how mGluR-LTD and Arc dysregulation contribute to cognitive disease.
Highlights.
mGluR1/5-induced dendritic synthesis of Arc causes LTD and synapse elimination
Arc stimulates endocytosis of postsynaptic AMPA receptors and dynamin assembly
Experience-induced Arc primes CA1 neurons for mGluR-induced LTD
Dysregulation of mGluR-LTD and Arc synthesis are linked with neuropsychiatric disease
Acknowledgments
This work was supported by grants from the National Institutes of Health (HD052731; KMH; MH110223; JPA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature neuroscience. 1999;2(12):1120–4. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- 2.Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, Chowdhury S, Mikhael D, Worley PF, Guzowski JF, Barnes CA. Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. The Journal of comparative neurology. 2006;498(3):317–29. doi: 10.1002/cne.21003. [DOI] [PubMed] [Google Scholar]
- 3.Miyashita T, Kubik S, Lewandowski G, Guzowski JF. Networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiology of learning and memory. 2008;89(3):269–84. doi: 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minatohara K, Akiyoshi M, Okuno H. Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Frontiers in molecular neuroscience. 2015;8:78. doi: 10.3389/fnmol.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao VY, Ye Y, Mastwal S, Ren M, Coon M, Liu Q, Costa RM, Wang KH. Motor Learning Consolidates Arc-Expressing Neuronal Ensembles in Secondary Motor Cortex. Neuron. 2015;86(6):1385–92. doi: 10.1016/j.neuron.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, Hen R. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83(1):189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, Loshbaugh AL, Kuhlman B, Hahn KM, Kasai H. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525(7569):333–8. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52(3):437–44. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126(2):389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 10.McCurry CL, Shepherd JD, Tropea D, Wang KH, Bear MF, Sur M. Loss of Arc renders the visual cortex impervious to the effects of sensory experience or deprivation. Nature neuroscience. 2010;13(4):450–7. doi: 10.1038/nn.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nature neuroscience. 2011;14(3):279–84. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21(4):741–51. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 13.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59(1):70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59(1):84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatavarty V, Ifrim MF, Levin M, Korza G, Barbarese E, Yu J, Carson JH. Single-molecule imaging of translational output from individual RNA granules in neurons. Molecular biology of the cell. 2012;23(5):918–29. doi: 10.1091/mbc.E11-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Na Y, Park S, Lee C, Kim DK, Park JM, Sockanathan S, Huganir RL, Worley PF. Real-Time Imaging Reveals Properties of Glutamate-Induced Arc/Arg 3.1 Translation in Neuronal Dendrites. Neuron. 2016;91(3):561–73. doi: 10.1016/j.neuron.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52(3):461–74. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52(3):445–59. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52(3):475–84. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakkamsetti V, Tsai NP, Gross C, Molinaro G, Collins KA, Nicoletti F, Wang KH, Osten P, Bassell GJ, Gibson JR, Huber KM. Experience-induced Arc/Arg3.1 primes CA1 pyramidal neurons for metabotropic glutamate receptor-dependent long-term synaptic depression. Neuron. 2013;80(1):72–9. doi: 10.1016/j.neuron.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikuni T, Uesaka N, Okuno H, Hirai H, Deisseroth K, Bito H, Kano M. Arc/Arg3.1 is a postsynaptic mediator of activity-dependent synapse elimination in the developing cerebellum. Neuron. 2013;78(6):1024–35. doi: 10.1016/j.neuron.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkerson JR, Tsai NP, Maksimova MA, Wu H, Cabalo NP, Loerwald KW, Dictenberg JB, Gibson JR, Huber KM. A role for dendritic mGluR5-mediated local translation of Arc/Arg3.1 in MEF2-dependent synapse elimination. Cell reports. 2014;7(5):1589–600. doi: 10.1016/j.celrep.2014.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pevzner A, Miyashita T, Schiffman AJ, Guzowski JF. Temporal dynamics of Arc gene induction in hippocampus: relationship to context memory formation. Neurobiology of learning and memory. 2012;97(3):313–20. doi: 10.1016/j.nlm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30(1):227–40. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 25.Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. The Journal of comparative neurology. 2007;500(3):433–47. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi H, Yamamoto S, Maruo T, Murakami F. Identification of a cis-acting element required for dendritic targeting of activity-regulated cytoskeleton-associated protein mRNA. The European journal of neuroscience. 2005;22(12):2977–84. doi: 10.1111/j.1460-9568.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- 27.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nature neuroscience. 2013;16(11):1530–6. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112(3):317–27. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 30.Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, Brosius J, Denman RB, Khandjian EW, Kindler S, Tiedge H. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci U S A. 2008;105(2):734–9. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(17):5924–36. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gladding CM, Fitzjohn SM, Molnar E. Metabotropic glutamate receptor-mediated long-term depression: molecular mechanisms. Pharmacological reviews. 2009;61(4):395–412. doi: 10.1124/pr.109.001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luscher C, Huber KM. Group 1 mGluR-dependent synaptic long-term depression: mechanisms and implications for circuitry and disease. Neuron. 2010;65(4):445–59. doi: 10.1016/j.neuron.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol. 2009 doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith-Hicks C, Xiao B, Deng R, Ji Y, Zhao X, Shepherd JD, Posern G, Kuhl D, Huganir RL, Ginty DD, Worley PF, Linden DJ. SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nature neuroscience. 2010;13(9):1082–9. doi: 10.1038/nn.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piochon C, Kano M, Hansel C. LTD-like molecular pathways in developmental synaptic pruning. Nature neuroscience. 2016;19(10):1299–310. doi: 10.1038/nn.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder EM, Philpot BD, Huber KM, Dong X, Fallon JR, Bear MF. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nature neuroscience. 2001;4(11):1079–85. doi: 10.1038/nn746. [DOI] [PubMed] [Google Scholar]
- 38.Klein ME, Castillo PE, Jordan BA. Coordination between Translation and Degradation Regulates Inducibility of mGluR-LTD. Cell reports. 2015 doi: 10.1016/j.celrep.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gladding CM, Collett VJ, Jia Z, Bashir ZI, Collingridge GL, Molnar E. Tyrosine dephosphorylation regulates AMPAR internalisation in mGluR-LTD. Molecular and cellular neurosciences. 2009;40(2):267–79. doi: 10.1016/j.mcn.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Nikolaienko O, Eriksen MS, Patil S, Bito H, Bramham CR. Stimulus-evoked ERK-dependent phosphorylation of activity-regulated cytoskeleton-associated protein (Arc) regulates its neuronal subcellular localization. Neuroscience. 2017;360:68–80. doi: 10.1016/j.neuroscience.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 41.Gallagher SM, Daly CA, Bear MF, Huber KM. Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24(20):4859–64. doi: 10.1523/JNEUROSCI.5407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66(1):57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindecke A, Korte M, Zagrebelsky M, Horejschi V, Elvers M, Widera D, Prullage M, Pfeiffer J, Kaltschmidt B, Kaltschmidt C. Long-term depression activates transcription of immediate early transcription factor genes: involvement of serum response factor/Elk-1. The European journal of neuroscience. 2006;24(2):555–63. doi: 10.1111/j.1460-9568.2006.04909.x. [DOI] [PubMed] [Google Scholar]
- 44.Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science (New York, N.Y.) 2000;288(5469):1254–7. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 45.Karachot L, Shirai Y, Vigot R, Yamamori T, Ito M. Induction of long-term depression in cerebellar Purkinje cells requires a rapidly turned over protein. J Neurophysiol. 2001;86(1):280–9. doi: 10.1152/jn.2001.86.1.280. [DOI] [PubMed] [Google Scholar]
- 46.Grinevich V, Kolleker A, Eliava M, Takada N, Takuma H, Fukazawa Y, Shigemoto R, Kuhl D, Waters J, Seeburg PH, Osten P. Fluorescent Arc/Arg3.1 indicator mice: a versatile tool to study brain activity changes in vitro and in vivo. Journal of neuroscience methods. 2009;184(1):25–36. doi: 10.1016/j.jneumeth.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci U S A. 2006;103(4):1077–82. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130(1):179–91. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 49.Wu B, Eliscovich C, Yoon YJ, Singer RH. Translation dynamics of single mRNAs in live cells and neurons. Science (New York, N.Y.) 2016;352(6292):1430–5. doi: 10.1126/science.aaf1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, Doyle M, Bauer KE, Thomas S, Planyavsky M, Arn E, Bakosova A, Jungwirth K, Hormann A, Palfi Z, Sandholzer J, Schwarz M, Macchi P, Colinge J, Superti-Furga G, Kiebler MA. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell reports. 2013;5(6):1749–62. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 51.Xu L, Anwyl R, Rowan MJ. Spatial exploration induces a persistent reversal of long-term potentiation in rat hippocampus. Nature. 1998;394(6696):891–4. doi: 10.1038/29783. [DOI] [PubMed] [Google Scholar]
- 52.Goh JJ, Manahan-Vaughan D. Spatial object recognition enables endogenous LTD that curtails LTP in the mouse hippocampus. Cerebral cortex (New York, N.Y. : 1991) 2013;23(5):1118–25. doi: 10.1093/cercor/bhs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kemp A, Manahan-Vaughan D. Hippocampal long-term depression and long-term potentiation encode different aspects of novelty acquisition. Proc Natl Acad Sci U S A. 2004;101(21):8192–7. doi: 10.1073/pnas.0402650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manahan-Vaughan D, Braunewell KH. Novelty acquisition is associated with induction of hippocampal long-term depression. Proc Natl Acad Sci U S A. 1999;96(15):8739–44. doi: 10.1073/pnas.96.15.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braunewell KH, Manahan-Vaughan D. Long-term depression: a cellular basis for learning? Reviews in the neurosciences. 2001;12(2):121–40. doi: 10.1515/revneuro.2001.12.2.121. [DOI] [PubMed] [Google Scholar]
- 56.Popkirov SG, Manahan-Vaughan D. Involvement of the metabotropic glutamate receptor mGluR5 in NMDA receptor-dependent, learning-facilitated long-term depression in CA1 synapses. Cerebral cortex (New York, N.Y. : 1991) 2011;21(3):501–9. doi: 10.1093/cercor/bhq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goh JJ, Manahan-Vaughan D. Endogenous hippocampal LTD that is enabled by spatial object recognition requires activation of NMDA receptors and the metabotropic glutamate receptor, mGlu5. Hippocampus. 2013;23(2):129–38. doi: 10.1002/hipo.22072. [DOI] [PubMed] [Google Scholar]
- 58.Mukherjee S, Manahan-Vaughan D. Role of metabotropic glutamate receptors in persistent forms of hippocampal plasticity and learning. Neuropharmacology. 2013;66:65–81. doi: 10.1016/j.neuropharm.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Dietz B, Manahan-Vaughan D. Hippocampal long-term depression is facilitated by the acquisition and updating of memory of spatial auditory content and requires mGlu5 activation. Neuropharmacology. 2017;115:30–41. doi: 10.1016/j.neuropharm.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Pehrson AL, Waller JA, Dale E, Sanchez C, Gulinello M. A critical evaluation of the activity-regulated cytoskeleton-associated protein (Arc/Arg3.1)'s putative role in regulating dendritic plasticity, cognitive processes, and mood in animal models of depression. Frontiers in neuroscience. 2015;9:279. doi: 10.3389/fnins.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaouloff F, Hemar A, Manzoni O. Acute stress facilitates hippocampal CA1 metabotropic glutamate receptor-dependent long-term depression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(27):7130–5. doi: 10.1523/JNEUROSCI.1150-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashimoto K, Kano M. Synapse elimination in the developing cerebellum. Cellular and molecular life sciences : CMLS. 2013;70(24):4667–80. doi: 10.1007/s00018-013-1405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe M, Kano M. Climbing fiber synapse elimination in cerebellar Purkinje cells. The European journal of neuroscience. 2011;34(10):1697–710. doi: 10.1111/j.1460-9568.2011.07894.x. [DOI] [PubMed] [Google Scholar]
- 64.Hashimoto K, Kano M. Functional differentiation of multiple climbing fiber inputs during synapse elimination in the developing cerebellum. Neuron. 2003;38(5):785–96. doi: 10.1016/s0896-6273(03)00298-8. [DOI] [PubMed] [Google Scholar]
- 65.Hashimoto K, Tsujita M, Miyazaki T, Kitamura K, Yamazaki M, Shin HS, Watanabe M, Sakimura K, Kano M. Postsynaptic P/Q-type Ca2+ channel in Purkinje cell mediates synaptic competition and elimination in developing cerebellum. Proc Natl Acad Sci U S A. 2011;108(24):9987–92. doi: 10.1073/pnas.1101488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kano M, Watanabe T. Type-1 metabotropic glutamate receptor signaling in cerebellar Purkinje cells in health and disease. F1000Research. 2017;6:416. doi: 10.12688/f1000research.10485.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science (New York, N.Y.) 2006;311(5763):1008–12. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 68.Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66(2):191–7. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development (Cambridge, England) 2007;134(23):4131–40. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 70.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends in biochemical sciences. 2002;27(1):40–7. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 71.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–38. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci U S A. 2009;106(1):316–21. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68(3):512–28. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang CW, Wilkerson J, Hale C, Gibson JR, Huber KM. Distinct stages of synapse elimination are induced by burst firing of CA1 neurons and differentially require MEF2A/D. Elife. 2017;6 doi: 10.7554/eLife.26278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai NP, Wilkerson JR, Guo W, Maksimova MA, Demartino GN, Cowan CW, Huber KM. Multiple Autism-Linked Genes Mediate Synapse Elimination via Proteasomal Degradation of a Synaptic Scaffold PSD-95. Cell. 2012;151(7):1581–94. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40(3):595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen CC, Lu HC, Brumberg JC. mGluR5 knockout mice display increased dendritic spine densities. Neuroscience letters. 2012;524(1):65–8. doi: 10.1016/j.neulet.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ballester-Rosado CJ, Sun H, Huang JY, Lu HC. mGluR5 Exerts Cell-Autonomous Influences on the Functional and Anatomical Development of Layer IV Cortical Neurons in the Mouse Primary Somatosensory Cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36(34):8802–14. doi: 10.1523/JNEUROSCI.1224-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gross KS, Brandner DD, Martinez LA, Olive MF, Meisel RL, Mermelstein PG. Opposite Effects of mGluR1a and mGluR5 Activation on Nucleus Accumbens Medium Spiny Neuron Dendritic Spine Density. PloS one. 2016;11(9):e0162755. doi: 10.1371/journal.pone.0162755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balice-Gordon RJ, Lichtman JW. Long-term synapse loss induced by focal blockade of postsynaptic receptors. Nature. 1994;372(6506):519–24. doi: 10.1038/372519a0. [DOI] [PubMed] [Google Scholar]
- 81.Shinoda Y, Kamikubo Y, Egashira Y, Tominaga-Yoshino K, Ogura A. Repetition of mGluR-dependent LTD causes slowly developing persistent reduction in synaptic strength accompanied by synapse elimination. Brain Res. 2005;1042(1):99–107. doi: 10.1016/j.brainres.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 82.Shinoda Y, Tanaka T, Tominaga-Yoshino K, Ogura A. Persistent synapse loss induced by repetitive LTD in developing rat hippocampal neurons. PloS one. 2010;5(4):e10390. doi: 10.1371/journal.pone.0010390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bastrikova N, Gardner GA, Reece JM, Jeromin A, Dudek SM. Synapse elimination accompanies functional plasticity in hippocampal neurons. Proc Natl Acad Sci U S A. 2008;105(8):3123–7. doi: 10.1073/pnas.0800027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DaSilva LL, Wall MJ, L PdA, Wauters SC, Januario YC, Muller J, Correa SA. Activity-Regulated Cytoskeleton-Associated Protein Controls AMPAR Endocytosis through a Direct Interaction with Clathrin-Adaptor Protein 2. eNeuro. 2016;3(3) doi: 10.1523/ENEURO.0144-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wall MJ, Correa SAL. The mechanistic link between Arc/Arg3.1 expression and AMPA receptor endocytosis. Seminars in cell & developmental biology. 2017 doi: 10.1016/j.semcdb.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 86.Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, Ford M, Frolov VA, Frost A, Hinshaw JE, Kirchhausen T, Kozlov MM, Lenz M, Low HH, McMahon H, Merrifield C, Pollard TD, Robinson PJ, Roux A, Schmid S. Membrane fission by dynamin: what we know and what we need to know. The EMBO journal. 2016;35(21):2270–2284. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nature reviews. Molecular cell biology. 2012;13(2):75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Byers CE, Barylko B, Ross JA, Southworth DR, James NG, Taylor CAt, Wang L, Collins KA, Estrada A, Waung M, Tassin TC, Huber KM, Jameson DM, Albanesi JP. Enhancement of dynamin polymerization and GTPase activity by Arc/Arg3.1. Biochimica et biophysica acta. 2015;1850(6):1310–8. doi: 10.1016/j.bbagen.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477(7366):556–60. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 90.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477(7366):561–6. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Srinivasan S, Dharmarajan V, Reed DK, Griffin PR, Schmid SL. Identification and function of conformational dynamics in the multidomain GTPase dynamin. The EMBO journal. 2016;35(4):443–57. doi: 10.15252/embj.201593477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kjaerulff O, Brodin L, Jung A. The structure and function of endophilin proteins. Cell biochemistry and biophysics. 2011;60(3):137–54. doi: 10.1007/s12013-010-9137-5. [DOI] [PubMed] [Google Scholar]
- 93.Ross JA, Chen Y, Muller J, Barylko B, Wang L, Banks HB, Albanesi JP, Jameson DM. Dimeric endophilin A2 stimulates assembly and GTPase activity of dynamin 2. Biophysical journal. 2011;100(3):729–37. doi: 10.1016/j.bpj.2010.12.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Myrum C, Baumann A, Bustad HJ, Flydal MI, Mariaule V, Alvira S, Cuellar J, Haavik J, Soule J, Valpuesta JM, Marquez JA, Martinez A, Bramham CR. Arc is a flexible modular protein capable of reversible self-oligomerization. The Biochemical journal. 2015;468(1):145–58. doi: 10.1042/BJ20141446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sever S, Chang J, Gu C. Dynamin rings: not just for fission. Traffic (Copenhagen, Denmark) 2013;14(12):1194–9. doi: 10.1111/tra.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boucrot E, Ferreira AP, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517(7535):460–5. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- 97.Renard HF, Simunovic M, Lemiere J, Boucrot E, Garcia-Castillo MD, Arumugam S, Chambon V, Lamaze C, Wunder C, Kenworthy AK, Schmidt AA, McMahon HT, Sykes C, Bassereau P, Johannes L. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature. 2015;517(7535):493–6. doi: 10.1038/nature14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nadif Kasri N, Nakano-Kobayashi A, Van Aelst L. Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron. 2011;72(2):300–15. doi: 10.1016/j.neuron.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karlsson MP, Frank LM. Network dynamics underlying the formation of sparse, informative representations in the hippocampus. The Journal of neuroscience. 2008;28(52):14271. doi: 10.1523/JNEUROSCI.4261-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilson MA, McNaughton BL. Dynamics of the hippocampal ensemble code for space. Science (New York, N.Y.) 1993;261(5124):1055–8. doi: 10.1126/science.8351520. [DOI] [PubMed] [Google Scholar]
- 101.Sjostrom PJ, Turrigiano GG, Nelson SB. Rate, timing, and cooperativity jointly determine cortical synaptic plasticity. Neuron. 2001;32(6):1149–64. doi: 10.1016/s0896-6273(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 102.Dong Z, Gong B, Li H, Bai Y, Wu X, Huang Y, He W, Li T, Wang YT. Mechanisms of hippocampal long-term depression are required for memory enhancement by novelty exploration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(35):11980–90. doi: 10.1523/JNEUROSCI.0984-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qi Y, Hu NW, Rowan MJ. Switching off LTP: mGlu and NMDA receptor-dependent novelty exploration-induced depotentiation in the rat hippocampus. Cerebral cortex (New York, N.Y. : 1991) 2013;23(4):932–9. doi: 10.1093/cercor/bhs086. [DOI] [PubMed] [Google Scholar]
- 104.Wang H, Ardiles AO, Yang S, Tran T, Posada-Duque R, Valdivia G, Baek M, Chuang YA, Palacios AG, Gallagher M, Worley P, Kirkwood A. Metabotropic Glutamate Receptors Induce a Form of LTP Controlled by Translation and Arc Signaling in the Hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36(5):1723–9. doi: 10.1523/JNEUROSCI.0878-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–16. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Husain N, Yuan Q, Yen YC, Pletnikova O, Sally DQ, Worley P, Bichler Z, Shawn Je H. TRIAD3/RNF216 mutations associated with Gordon Holmes syndrome lead to synaptic and cognitive impairments via Arc misregulation. Aging cell. 2017;16(2):281–292. doi: 10.1111/acel.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuhnle S, Mothes B, Matentzoglu K, Scheffner M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc Natl Acad Sci U S A. 2013;110(22):8888–93. doi: 10.1073/pnas.1302792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mandel-Brehm C, Salogiannis J, Dhamne SC, Rotenberg A, Greenberg ME. Seizure-like activity in a juvenile Angelman syndrome mouse model is attenuated by reducing Arc expression. Proc Natl Acad Sci U S A. 2015;112(16):5129–34. doi: 10.1073/pnas.1504809112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu J, Petralia RS, Kurushima H, Patel H, Jung MY, Volk L, Chowdhury S, Shepherd JD, Dehoff M, Li Y, Kuhl D, Huganir RL, Price DL, Scannevin R, Troncoso JC, Wong PC, Worley PF. Arc/Arg3.1 regulates an endosomal pathway essential for activity-dependent beta-amyloid generation. Cell. 2011;147(3):615–28. doi: 10.1016/j.cell.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rudinskiy N, Hawkes JM, Betensky RA, Eguchi M, Yamaguchi S, Spires-Jones TL, Hyman BT. Orchestrated experience-driven Arc responses are disrupted in a mouse model of Alzheimer's disease. Nature neuroscience. 2012;15(10):1422–9. doi: 10.1038/nn.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Landgren S, von Otter M, Palmer MS, Zetterstrom C, Nilsson S, Skoog I, Gustafson DR, Minthon L, Wallin A, Andreasen N, Bogdanovic N, Marcusson J, Blennow K, Zetterberg H, Kettunen P. A novel ARC gene polymorphism is associated with reduced risk of Alzheimer's disease. Journal of neural transmission (Vienna, Austria : 1996) 2012;119(7):833–42. doi: 10.1007/s00702-012-0823-x. [DOI] [PubMed] [Google Scholar]
- 112.Chuang YA, Hu TM, Chen CH, Hsu SH, Tsai HY, Cheng MC. Rare mutations and hypermethylation of the ARC gene associated with schizophrenia. Schizophrenia research. 2016;176(2–3):106–13. doi: 10.1016/j.schres.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 113.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O'Donovan MC. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ. Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biological psychiatry. 2015;77(1):52–8. doi: 10.1016/j.biopsych.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 115.Huentelman MJ, Muppana L, Corneveaux JJ, Dinu V, Pruzin JJ, Reiman R, Borish CN, De Both M, Ahmed A, Todorov A, Cloninger CR, Zhang R, Ma J, Gallitano AL. Association of SNPs in EGR3 and ARC with Schizophrenia Supports a Biological Pathway for Schizophrenia Risk. PloS one. 2015;10(10):e0135076. doi: 10.1371/journal.pone.0135076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O'Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular psychiatry. 2012;17(2):142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annual review of pathology. 2012;7:219–45. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 119.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99(11):7746–50. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51(4):441–54. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 121.Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95(5):3291–5. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- 122.Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, Darnell JC, Darnell RB. A mouse model of the human Fragile X syndrome I304N mutation. PLoS genetics. 2009;5(12):e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Till SM, Asiminas A, Jackson AD, Katsanevaki D, Barnes SA, Osterweil EK, Bear MF, Chattarji S, Wood ER, Wyllie DJ, Kind PC. Conserved hippocampal cellular pathophysiology but distinct behavioural deficits in a new rat model of FXS. Human molecular genetics. 2015;24(21):5977–84. doi: 10.1093/hmg/ddv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14(6):926–39. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(8):2582–7. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(52):14349–57. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. The Journal of biological chemistry. 2008;283(27):18478–82. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annual review of medicine. 2011;62:411–29. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.D'Antoni S, Spatuzza M, Bonaccorso CM, Musumeci SA, Ciranna L, Nicoletti F, Huber KM, Catania MV. Dysregulation of group-I metabotropic glutamate (mGlu) receptor mediated signalling in disorders associated with Intellectual Disability and Autism. Neuroscience and biobehavioral reviews 46 Pt. 2014;2:228–41. doi: 10.1016/j.neubiorev.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pop AS, Gomez-Mancilla B, Neri G, Willemsen R, Gasparini F. Fragile X syndrome: a preclinical review on metabotropic glutamate receptor 5 (mGluR5) antagonists and drug development. Psychopharmacology. 2014;231(6):1217–26. doi: 10.1007/s00213-013-3330-3. [DOI] [PubMed] [Google Scholar]
- 131.Scharf SH, Jaeschke G, Wettstein JG, Lindemann L. Metabotropic glutamate receptor 5 as drug target for Fragile X syndrome. Current opinion in pharmacology. 2015;20:124–34. doi: 10.1016/j.coph.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 132.Berry-Kravis E, Des Portes V, Hagerman R, Jacquemont S, Charles P, Visootsak J, Brinkman M, Rerat K, Koumaras B, Zhu L, Barth GM, Jaecklin T, Apostol G, von Raison F. Mavoglurant in fragile X syndrome: Results of two randomized, double-blind, placebo-controlled trials. Science translational medicine. 2016;8(321):321ra5. doi: 10.1126/scitranslmed.aab4109. [DOI] [PubMed] [Google Scholar]
- 133.Davenport MH, Schaefer TL, Friedmann KJ, Fitzpatrick SE, Erickson CA. Pharmacotherapy for Fragile X Syndrome: Progress to Date. Drugs. 2016;76(4):431–45. doi: 10.1007/s40265-016-0542-y. [DOI] [PubMed] [Google Scholar]
- 134.Gogliotti RG, Senter RK, Rook JM, Ghoshal A, Zamorano R, Malosh C, Stauffer SR, Bridges TM, Bartolome JM, Daniels JS, Jones CK, Lindsley CW, Conn PJ, Niswender CM. mGlu5 positive allosteric modulation normalizes synaptic plasticity defects and motor phenotypes in a mouse model of Rett syndrome. Human molecular genetics. 2016;25(10):1990–2004. doi: 10.1093/hmg/ddw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tao J, Wu H, Coronado AA, de Laittre E, Osterweil EK, Zhang Y, Bear MF. Negative Allosteric Modulation of mGluR5 Partially Corrects Pathophysiology in a Mouse Model of Rett Syndrome. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36(47):11946–11958. doi: 10.1523/JNEUROSCI.0672-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tian D, Stoppel LJ, Heynen AJ, Lindemann L, Jaeschke G, Mills AA, Bear MF. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nature neuroscience. 2015;18(2):182–4. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barnes SA, Wijetunge LS, Jackson AD, Katsanevaki D, Osterweil EK, Komiyama NH, Grant SG, Bear MF, Nagerl UV, Kind PC, Wyllie DJ. Convergence of Hippocampal Pathophysiology in Syngap+/− and Fmr1-/y Mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35(45):15073–81. doi: 10.1523/JNEUROSCI.1087-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ebrahimi-Fakhari D, Sahin M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Current opinion in neurology. 2015;28(2):91–102. doi: 10.1097/WCO.0000000000000186. [DOI] [PubMed] [Google Scholar]
- 139.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480(7375):63–8. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(24):8862–9. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chevere-Torres I, Kaphzan H, Bhattacharya A, Kang A, Maki JM, Gambello MJ, Arbiser JL, Santini E, Klann E. Metabotropic glutamate receptor-dependent long-term depression is impaired due to elevated ERK signaling in the DeltaRG mouse model of tuberous sclerosis complex. Neurobiology of disease. 2012;45(3):1101–10. doi: 10.1016/j.nbd.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bateup HS, Denefrio CL, Johnson CA, Saulnier JL, Sabatini BL. Temporal dynamics of a homeostatic pathway controlling neural network activity. Frontiers in molecular neuroscience. 2013;6:28. doi: 10.3389/fnmol.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78(3):510–22. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buiting K, Williams C, Horsthemke B. Angelman syndrome - insights into a rare neurogenetic disorder. Nature reviews. Neurology. 2016;12(10):584–93. doi: 10.1038/nrneurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 145.Mabb AM, Je HS, Wall MJ, Robinson CG, Larsen RS, Qiang Y, Correa SA, Ehlers MD. Triad3A regulates synaptic strength by ubiquitination of Arc. Neuron. 2014;82(6):1299–316. doi: 10.1016/j.neuron.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pignatelli M, Piccinin S, Molinaro G, Di Menna L, Riozzi B, Cannella M, Motolese M, Vetere G, Catania MV, Battaglia G, Nicoletti F, Nistico R, Bruno V. Changes in mGlu5 receptor-dependent synaptic plasticity and coupling to homer proteins in the hippocampus of Ube3A hemizygous mice modeling angelman syndrome. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34(13):4558–66. doi: 10.1523/JNEUROSCI.1846-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28(2):543–7. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kerrigan TL, Randall AD. A new player in the "synaptopathy" of Alzheimer's disease - arc/arg 3.1. Frontiers in neurology. 2013;4:9. doi: 10.3389/fneur.2013.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee HK, Min SS, Gallagher M, Kirkwood A. NMDA receptor-independent long-term depression correlates with successful aging in rats. Nature neuroscience. 2005;8(12):1657–9. doi: 10.1038/nn1586. [DOI] [PubMed] [Google Scholar]
- 150.Yang S, Megill A, Ardiles AO, Ransom S, Tran T, Koh MT, Lee HK, Gallagher M, Kirkwood A. Integrity of mGluR-LTD in the associative/commissural inputs to CA3 correlates with successful aging in rats. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(31):12670–8. doi: 10.1523/JNEUROSCI.1086-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiology of aging. 2011;32(12):2198–210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hartzell AL, Burke SN, Hoang LT, Lister JP, Rodriguez CN, Barnes CA. Transcription of the immediate-early gene Arc in CA1 of the hippocampus reveals activity differences along the proximodistal axis that are attenuated by advanced age. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(8):3424–33. doi: 10.1523/JNEUROSCI.4727-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hsieh H, Boehm J, Sato C, Iwatsubo T, Tomita T, Sisodia S, Malinow R. AMPAR removal underlies Abeta-induced synaptic depression and dendritic spine loss. Neuron. 2006;52(5):831–43. doi: 10.1016/j.neuron.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ondrejcak T, Klyubin I, Hu NW, Barry AE, Cullen WK, Rowan MJ. Alzheimer's disease amyloid beta-protein and synaptic function. Neuromolecular medicine. 2010;12(1):13–26. doi: 10.1007/s12017-009-8091-0. [DOI] [PubMed] [Google Scholar]
- 155.Selkoe DJ. Alzheimer's disease is a synaptic failure. Science (New York, N.Y.) 2002;298(5594):789–91. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 156.Stephan AH, Barres BA, Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. Annual review of neuroscience. 2012;35:369–89. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- 157.Rammes G, Hasenjager A, Sroka-Saidi K, Deussing JM, Parsons CG. Therapeutic significance of NR2B–containing NMDA receptors and mGluR5 metabotropic glutamate receptors in mediating the synaptotoxic effects of beta-amyloid oligomers on long-term potentiation (LTP) in murine hippocampal slices. Neuropharmacology. 2011;60(6):982–90. doi: 10.1016/j.neuropharm.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 158.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nature medicine. 2008;14(8):837–42. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang Q, Walsh DM, Rowan MJ, Selkoe DJ, Anwyl R. Block of long-term potentiation by naturally secreted and synthetic amyloid beta-peptide in hippocampal slices is mediated via activation of the kinases c-Jun N-terminal kinase, cyclin-dependent kinase 5, and p38 mitogen-activated protein kinase as well as metabotropic glutamate receptor type 5. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24(13):3370–8. doi: 10.1523/JNEUROSCI.1633-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Um JW, Kaufman AC, Kostylev M, Heiss JK, Stagi M, Takahashi H, Kerrisk ME, Vortmeyer A, Wisniewski T, Koleske AJ, Gunther EC, Nygaard HB, Strittmatter SM. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron. 2013;79(5):887–902. doi: 10.1016/j.neuron.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kumar A, Dhull DK, Mishra PS. Therapeutic potential of mGluR5 targeting in Alzheimer's disease. Frontiers in neuroscience. 2015;9:215. doi: 10.3389/fnins.2015.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hamilton A, Vasefi M, Vander Tuin C, McQuaid RJ, Anisman H, Ferguson SS. Chronic Pharmacological mGluR5 Inhibition Prevents Cognitive Impairment and Reduces Pathogenesis in an Alzheimer Disease Mouse Model. Cell reports. 2016;15(9):1859–65. doi: 10.1016/j.celrep.2016.04.077. [DOI] [PubMed] [Google Scholar]
- 163.Haas LT, Salazar SV, Smith LM, Zhao HR, Cox TO, Herber CS, Degnan AP, Balakrishnan A, Macor JE, Albright CF, Strittmatter SM. Silent Allosteric Modulation of mGluR5 Maintains Glutamate Signaling while Rescuing Alzheimer's Mouse Phenotypes. Cell reports. 2017;20(1):76–88. doi: 10.1016/j.celrep.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alberi L, Liu S, Wang Y, Badie R, Smith-Hicks C, Wu J, Pierfelice TJ, Abazyan B, Mattson MP, Kuhl D, Pletnikov M, Worley PF, Gaiano N. Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron. 2011;69(3):437–44. doi: 10.1016/j.neuron.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. 2015;86(2):490–500. doi: 10.1016/j.neuron.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]