Abstract
Circulating polypeptides and proteins have been implicated in reversing or accelerating aging phenotypes, including growth/differentiation factor 8 (GDF8), GDF11, eotaxin, and oxytocin. These proteoforms, which are defined as the protein products arising from a single gene due to alternative splicing and post-translational modifications, have been challenging to study. Both GDF8 and GDF11 have known antagonists such as follistatin (FST), and WAP, Kazal, immunoglobulin, Kunitz and NTR domain-containing proteins 1 and 2 (WFIKKN1, WFIKKN2). We developed a novel multiplexed selected reaction monitoring (SRM) assay using liquid chromatography-tandem mass spectrometry to measure five proteins related to GDF8 and GDF11 signaling, and in addition, eotaxin and oxytocin. Eighteen peptides consisting of 54 transitions were monitored and validated in pooled human plasma. In twenty-four adults, the mean (SD) concentrations (ng/mL) were as follows: GDF8 propeptide, 11.0 (2.4); GDF8 mature protein, 25.7 (8.0); GDF11 propeptide, 21.3 (10.9); GDF11 mature protein, 16.5 (12.4); FST, 29.8 (7.1); FST cleavage form FST303, 96.4 (69.2); WFIKKN1, 38.3 (8.3); WFIKKN2, 32.2 (10.5); oxytocin, 1.9 (0.9); and eotaxin, 2.3 (0.5). This novel multiplexed SRM assay should facilitate the study of the relationships of these proteoforms with major aging phenotypes.
Keywords: aging, eotaxin, follistatin, growth/differentiation factor 8, growth/differentiation factor 11, oxytocin, WFIKKN1, WFIKKN2
1 Introduction
Studies have attempted to identify circulating signaling proteins that change systematically with aging, correlate with main aging phenotypes independent of chronological age, and respond to stress or trauma by compensation, repair, or adaptation differently in younger and older individuals. However, measuring and defining the role of these proteins in human aging has proven difficult because of at least two reasons. First, the protein product of a single gene takes multiple proteoforms because of alternative splicing or post-translational modifications that affect proteolytic cleavage or terminal degradation [1]. Measuring these alternative proteoforms with conventional immunoassays or aptamers in the blood or other biological material has proven to be difficult [2]. Second, the biological activity of these proteins is often modulated by other proteins that may act as activators, enhancers, or inhibitors.
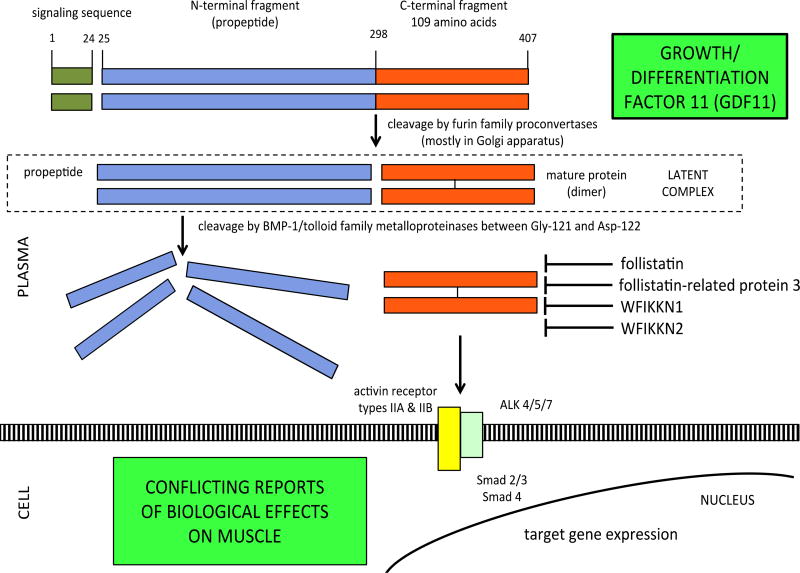
The suggestion in the literature that modulating growth/differentiation factor 11 (GDF11) may reverse or accelerate aging in muscle, heart, and brain is a good example of such complexity. Growth/differentiation factor 11 (GDF11) exists as a single isoform. After cleavage from the signal peptide, intact GDF11 is cleaved by furin family proconvertases into propeptide and mature GDF11 protein. The other product of this cleavage is a disulfide-linked mature protein. The propeptide and mature protein dimers form a non-covalently bound latent complex in the circulation. The latent complex is activated through cleavage of the propeptide by BMP-1/tolloid family astacin metalloproteases [3] (Figure 1).
Figure 1.
Cleavage of intact GDF11 into propeptide and mature protein, followed by dimerization of mature protein. Propeptide and mature protein form latent complex which is activated by metalloproteinase cleavage. Follistatin, FSTR3, WFIKKN1, WFIKKN2, and GDF11 propeptide antagonize mature GDF11.
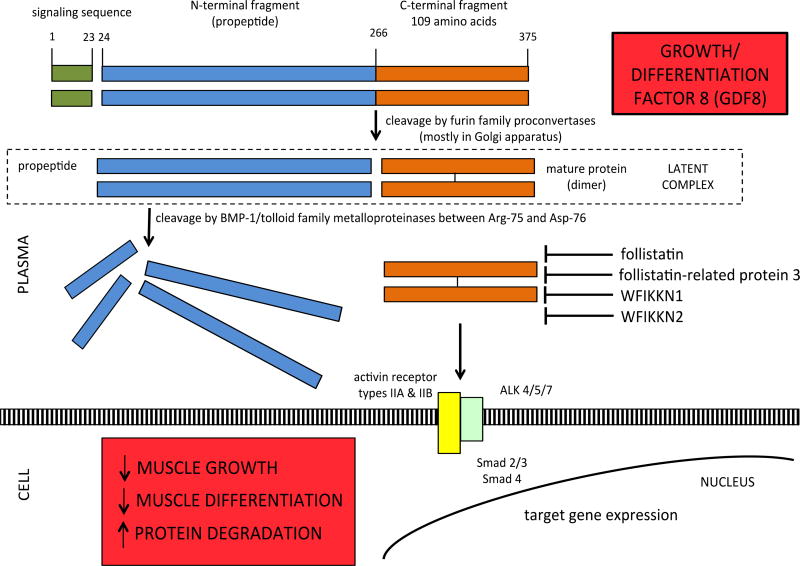
Similar to GDF11, intact growth/differentiation factor 8 (GDF8; also known as myostatin) is cleaved by furin family proconvertases into propeptide and mature GDF8 protein. The other product of this cleavage is a disulfide-linked mature protein, which is the receptor-binding molecule [4]. The propeptide and mature protein dimers form a non-covalently bound latent complex in the circulation [5,6]. The latent complex, which comprises the major circulating form of GDF8, is activated through cleavage of the propeptide by BMP-1/tolloid family astacin metalloproteases [7] (Figure 2).
Figure 2.
Cleavage of intact GDF8 into propeptide and mature protein, followed by dimerization of mature protein. Propeptide and mature protein form latent complex which is activated by metalloproteinase cleavage. Follistatin, FSTR3, WFIKKN1, WFIKKN2, and GDF11 propeptide antagonize mature GDF8.
GDF8 is a negative regulator of skeletal muscle growth and has received attention as a therapeutic target in rejuvenation research since inhibitors of GDF8 can also increase skeletal muscle growth in animal models [4]. GDF11 is closely related to GDF8, as their mature C-terminal domains share ~90% identity [2].
Since other circulating proteins and peptides can modify the biological activity of GDF11 and GDF8, studies aimed at understanding the true relationship of circulating GDF11 and GDF8 with aging phenotypes should include the effect of their known, natural inhibitors. The inhibitors of GDF11 and GDF8 include their respective propeptides [8–10], follistatin [10,11], follistatin-related protein 3 (FSTR3) [9], and WAP, Kazal, immunoglobulin, Kunitz and NTR domain-containing proteins 1 and 2 (WFIKKN1, WFIKKN2) [10,12].
These polypeptides and proteins mentioned above have been difficult to study in the blood using conventional immunoassays or reagents that bind large conformational epitopes, such as aptamers, since some of the peptides or proteins exist in multiple isoforms, undergo post-translational modifications (PTMs) such as cleavage or terminal degradation, or have high portions of homologous sequence [2]. GDF11 and GDF8 circulate as propeptides and mature proteins [9,10]. There are two circulating isoforms of plasma follistatin and one cleaved form [13].
In order to facilitate studies aimed at connecting these circulating proteoforms with aging phenotypes, we developed a novel multiplexed selected reaction monitoring (SRM) assay for the measurement of GDF11 and GDF8 propeptides and mature proteins, WFIKKN1, WFIKKN2, and follistatin. We also included two other proteins in the assay, oxytocin and eotaxin, because they have been identified in animal models as promising candidates with a role in aging. Oxytocin, which circulates as a nonapeptide and as carboxyl-extended forms with biological activity [14], may rejuvenate skeletal muscle [15]. Eotaxin may adversely affect cognition [16].
2 Materials and Methods
2.1. Selection of proteotypic peptides
For SRM assay development, tryptic peptides were selected following the guidelines of Kuzyk and colleagues [17]. Tryptic peptides unique to each protein were identified using PeptideCutter (ExPASy, Swiss Institute of Bioinformatics), NCBI BLAST and UniProt/BLAST searches, with further support for selection of peptides and optimization of transitions through Skyline (Seattle Proteome Center) using the ProteoWizard libraries (Table 1).
Table 1.
Proteotypic peptides for SRM assay
| Protein | Amino acid |
Target peptide sequences | Proteoform quantified |
SRM transitions |
|---|---|---|---|---|
|
| ||||
| GDF8 | 66–74 | LETAPNISK | propeptide | 486.8/558.3(y5);486.8/629.4(y6);486.8/730.4(y7) |
| 210–217 | TVLQNWLK | propeptide | 501.4/560.1(y4);501.4/688.2(y5)501.4/801.3(y6) | |
| 218–228 | QPESNLGIEIK | propeptide | 614.3/260.2(y2);614.3/559.3(y5);614.3/672.4(y6) | |
| 267–280 | DFGLDC[CAM]DEHSTESR | mature | 556.6/579.0(y5);556.6/716.2(y6);556.6/674.7(+3y11+2) | |
|
| ||||
| GDF11 | 90–96 | EAPNISR | propeptide | 393.7/293.7(y5+2);393.7/489.3(y4);393.7/586.3(y5) |
| 232–241 | SGHWQSIDFK | propeptide | 602.8/911.4(b8);602.8/737.4(y6);602.8/923.5(y7) | |
| 253–265 | YPHTHLVQQANPR | mature | 521.0/457.02(+3y4);521.0/585.1(+3y5);521.0/713.1(+3y6) | |
| 299–312 | NLGLDC[CAM]DEHSSESR | mature | 540.4/565.0(y5);540.4/611.1(y10+2);540.4/696.1(y12+2) | |
|
| ||||
| Follistatin | 230–238 | SIGLAYEGK | isoforms 1 & 2 | 469.3/496.0(y4);469.3/567.1(y5);469.3/737.2(y7) |
| 312–331 | HSGSC[CAM]NSISEDTEEEEEDEDQ | FST303 | 809.5/843.3(b8);809.5/7262.0(y2);809.5/764.1(y6) | |
|
| ||||
| WFIKKN1 | 509–521 | DGVAVLDAGSYVR | mature | 661.3/767.1(y7);661.3/880.0(y8);661.3/1050.0(y10) |
|
| ||||
| WFIKKN2 | 32–39 | SLALPPIR | mature | 433.9/482.1(y4);433.9/595.2(y5);433.9/666.2(y6) |
| 298–306 | ADFPLSVVR | mature | 502.3/573.1(y5);502.3/670.2 (y6);502.3/409.1(y7+2) | |
|
| ||||
| Oxytocin | 20–28 | C[CAM]YIQNC[CAM]PLG | mature | 562.8/839.1(b6);562.8/286.1(y3);562.8/446.0(y4) |
| 20–29 | C[CAM]YIQNC[CAM]PLGG | oxytocin-G | 591.3/839.2(b6);591.3/343.1(y4);591.3/503.1(y5) | |
| 20–30 | C[CAM]YIQNC[CAM]PLGGK | oxytocin-GK | 655.4/745.1(y7);655.4/873.2(y8);655.4/986.3(y9) | |
|
| ||||
| Eotaxin | 41–45 | IPLQR | mature | 313.8/257.1(y4+2);313.8/416.1(y3);313.8/513.1(y4) |
| 87–91 | YLDQK | mature | 333.7/275.1(y2);333.7/390.0(y3);333.7/503.1(y4) | |
2.2. Synthesis and purification of peptides
Peptides were obtained from New England Peptide (Gardner, MA). Tryptic fragment peptides were prepared by Fmoc-based solid-phase peptide synthesis using per-15N,13C-labeled (>99% isotopic purity) Arg or Lys as the C-terminal residue attached to the resin. Cysteine side-chain residues were blocked as the carboxyacetamidomethyl thioether. Peptides were cleaved from the resin with ~90% trifluoroacetic acid (TFA) containing appropriate scavengers and isolated by precipitation from ether or by drying of the cleavage cocktail. Peptides were purified by reversed phase chromatography (C18 stationary phase using water-acetonitrile gradients, ion-pairing agent ~0.1% TFA). The purity of the synthetic heavy peptides was ≥ 95% for each by the confirmation of analytical HPLC. MALDI-MS was used to confirm peptide identity. Purified peptide solutions were prepared and the concentration of the solution was determined by amino acid analysis.
2.3. Optimization of the assay
Selection of optimal charge state and collision energy, confirmation of co-elution of endogenous and SIS peptides, and interference detection were conducted as detailed elsewhere [17]. Mass spectrometry optimization was conducted with a continuous injection of individual peptide and internal standard at 100 nM by ramping the following parameters: declustering potential (DP) (0–400 volts), collision energy (CE) (5–130 volts) and collision cell exit potential (CXP) (0–66 volts) from low to high with a step of 1 for all parameters and a fixed setting of 10 volts for entrance potential (EP). Three interference-free SRM transitions constituted the final SRM assay for the respective proteotypic peptides. The SIS peptide spiking concentration was optimized at 100 nM. Details of SRM parameters, linear range of quantification, and lower limit of quantification (LOQ) are shown in Supporting Information Tables 1 & 2.
2.4. Sample preparation and measurement
We measured plasma proteoforms in twenty-four adults who participated the Baltimore Longitudinal Study of Aging (BLSA) or the GESTALT Study. The BLSA and GESTALT protocols were approved by the National Institute of Environmental Health Science Institutional Review Board, and all participants provide written, informed consent. The subjects were 12 males and 12 females, mean (SD) age 55.0 (22.2) years with no history of chronic diseases. The protocol for this study was also approved by the Johns Hopkins School of Medicine Institutional Review Board. Plasma samples were thawed on the day of analysis and centrifuged at 14,000 × g for 15 min at 4°C for delipidation. A volume of 5 µL plasma was aliquoted in 0.1% (w/v) RapiGest buffer containing 100 mM Tris-HCl, pH 8.0, and 100 mM dithiothreitol and incubated at 55°C for 1 h for denaturation and reduction. The samples were then alkylated for 30 min at room temperature in dark with a final concentration of 50 mM iodoacetamide, digested by trypsin/LysC mix (Promega) at an enzyme-to-substrate ratio of 1:50 for 18 h at 37°C. The digestion was terminated with 10% TFA to a final concentration of 1%, followed by centrifugation at 14,000 × g for 15 min to remove RapiGest, and the sample was cleaned up with a 96-well extraction plate vacuum manifold (Waters, Milford, MA, USA) according to manufacturer’s instruction.
SRM assays were run on a 5500 QTrap (Sciex, Framingham, MA, USA) mass spectrometer equipped with an electrospray ionization source, a CBM-20A command module, LC-20ADXR pump, and a CTO-10Ac column oven heater (all Shimadzu, Kyoto, Japan). A sample volume of 10 µL was injected onto the column via a Shimadzu SIL-20AXR autosampler set to 4° C at a flow rate of 0.2 ml/min, with an instrument run time of 18 min/sample including the 5 min column regeneration step. Chromatographic separations were conducted using an XBridge BEH C18 Column, 130Å, 3.5 µm, 2.1 mm × 100 mm (Waters), with a linear gradient starting from 5% phase B increasing to 36% phase B within 10 min with the column oven set at 37° C. Mobile phases consisted of water containing 0.1% formic acid (phase A) and 98% acetonitrile containing 0.1% formic acid (phase B). The mass spectrometry analysis was performed in positive ion mode and a scheduled SRM analysis was performed at unit resolution in both Q1 and Q3 quadrupoles. All sample data were collected using Analyst (version 1.5) software and processed using MultiQuant software (version 3.02 with a scheduled-MRM-Algorithm) (Sciex). Peak review had the parameters: Gaussian smooth width 0 or 1, analyte peak update to an expected retention time, minimum peak width of three points, noise percentage 40.0%, baseline subtraction window 2.00 min, and peak splitting 2 points. In order to monitor quality control (QC), three internal QC plasma standards were prepared at low, middle, and high concentrations respectively.
Pooled plasma samples were prepared from twenty-four adult participants. In order to monitor QC, sixteen internal QC of the pooled plasma standards were used. We generated calibration curves with a 5-point calibration curve within the range of 0.1, 0.20, 0.39, 0.78, 1.56, 3.12, 6.25, and 12.5 nM, depending on the targeted proteoform. For oxytocin-GK, the calibration curve went up to 200 nM because of the higher concentration range of this proteoform. To protect from carry over, we inserted several 20 min washes and blanks between two samples or standards, and monitored QC in each batch of 10 samples. Calibration curves and quality controls were run using the standard addition method. All standards and samples were run with three technical replicates for calculation of mean and CVs. Concentrations below LOQ were excluded from the calibration calculations. Any sample with CV >20% was re-run with the same standard curves and QC arrangement. Although results are reported in ng/mL for each proteoform, the developed SRM method provides a measure of relative abundance, not absolute quantification due to potential matrix effects.
3 Results
A total of 18 peptides were designed and synthesized to monitor plasma propeptides and mature proteins. For each peptide, HPLC and MS parameters of the SRM assay were optimized, and transitions were selected to achieve the greatest sensitivity. HPLC optimization showed a good resolution of peptides in a total time of 18 min, with a linear increasing concentration of acetonitrile from 5% phase B to 36% phase B within 10 min.
The SRM assay included 18 proteotypic peptides and 54 transitions. Optimized MS parameters for the respective peptides were Q1 and Q3 set to dwell time 16 msec and EP 10 V. Peptide-specific tuned CE, DP, CXP voltage, and the retention time for each transition are listed in Supporting Information Table 1.
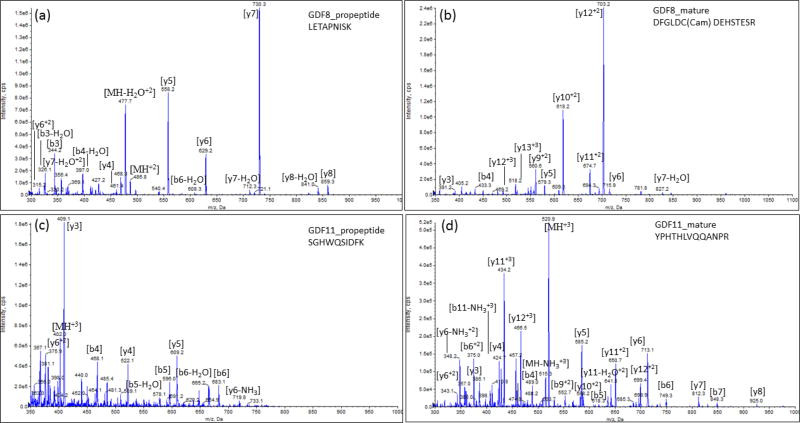
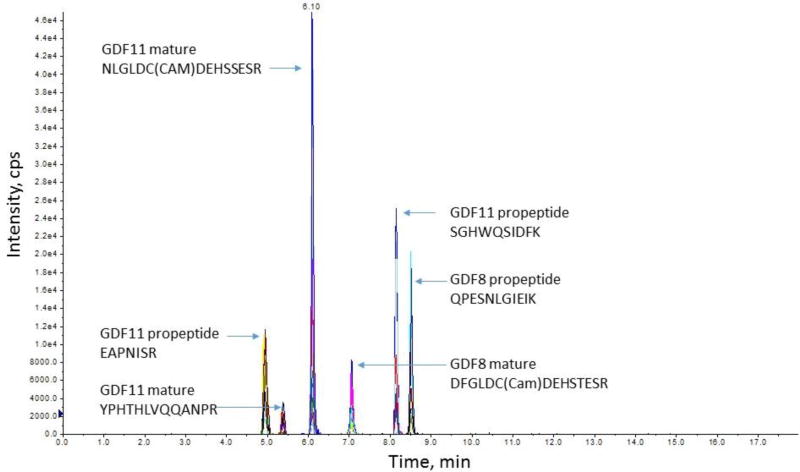
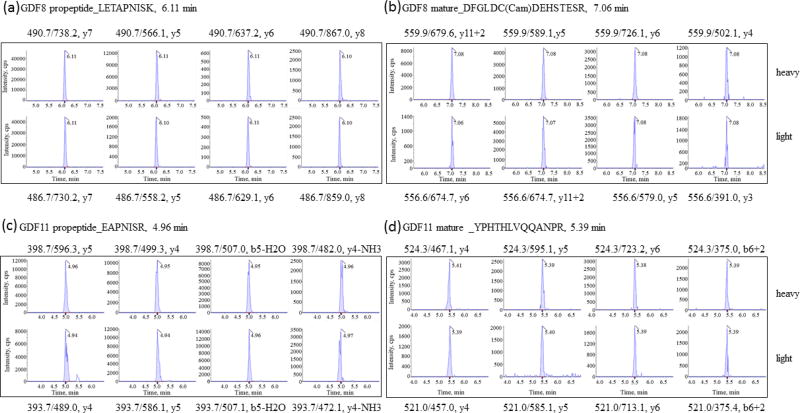
An SRM assay was developed for all peptides using a standard addition method with pooled plasma on the 5500 QTrap. No significant matrix effects were found for these peptides (P >0.05). Figure 3 shows the representative MS/MS spectra from GDF8 propeptide of LETAPNISK, and mature protein DFGLDC(Cam)DEHSTESR, GDF11 propeptide of SGHWQSIDFK, and mature protein YPHTHLVQQANPR. In the plasma matrix, all the targeted peptides were resolved by HPLC separation with good sensitivity, shown in a representative chromatogram of propeptides and mature of GDF8 and GDF11 proteins (Figure 4) and coelutions of four transitions with the light or heavy ion pairs in the SRM reaction (Figure 5).
Figure 3.
MS/MS spectra from proteotypic peptides of (a) GDF8 propeptide, (b) GDF8 mature protein, (c) GDF11 propeptide, and (d) GDF11 mature protein.
Figure 4.
Chromatographic separation of representative propeptides and mature GDF8 and GDF11 proteins.
Figure 5.
Representative SRM reaction in transitions with light and heavy labeled propeptides and mature GDF8 and GDF11 proteins.
Calibration curves of each proteoform were generated before running samples. The lower limit of detection (LOD) and LOQ were determined for each peptide. LOQ was defined as the lowest detected concentration with coefficient of variation (CV) ≤ 20% and a signal-to-noise (S/N) ratio ≥10; the instrument LOD was based on S/N >3. The LOQ are reported in both ng/mL and nM in Supporting Information Table 2. The plasma concentrations of proteoforms in 24 healthy adult men and women are shown in Table 2.
Table 2.
Plasma concentrations of proteoforms in 24 healthy adults measured by SRM compared with studies in the published literature
| Analyte | This study | Published Literature | |||
|---|---|---|---|---|---|
|
| |||||
| Concentration ng/mL1 |
Concentration ng/mL2 |
Subjects | Method & Sample |
Ref. | |
|
| |||||
| GDF8 mature protein | 25.7 (8.0) | 3.42 (1.61) | n = 58, men | ELISA, plasma | 25 |
| 3.27 (1.43) | n = 66, women | ||||
|
| |||||
| ~4.5, mean | n = 20, healthy men | ELISA, serum | 26 | ||
|
| |||||
| 23.6 (5.2) | n = 30, healthy adults | ELISA, serum | 27 | ||
|
| |||||
| 32 (25–41) | n = 20, healthy adult controls | ELISA, plasma | 28 | ||
| median (IQR) | |||||
|
| |||||
| ~120, mean | n = 27, healthy men | ELISA, serum | 29 | ||
|
| |||||
| ~2.9, mean | n = 55, men with CVD | Immunoaffinity LC-MS/MS serum | 30 | ||
| ~2.7, mean | n = 41, women with CVD | ||||
| 0.64–6.27, range | |||||
|
| |||||
| 3.05 (1.27) | n = 18, juveniles | Immunoaffinity LC-MS/MS serum | 31 | ||
| 8.68 (3.36) | n = 30, adults | ||||
|
| |||||
| 10.5 (9.0–14.4) | n = 40, younger men | Immunoaffinity LC-MS/MS serum | 32 | ||
| 11.8 (7.6–15.7) | n = 40, older men | ||||
| 5.5 (3.2–7.3) | n = 40, younger women | ||||
| 7.3 (5.7–11.7) | n = 40, older women | ||||
| median (IQR) | |||||
|
| |||||
| GDF8 propeptide | 11.0 (2.4) | 15.7 (11.7–19.4) | n = 40, younger men | Immunoaffinity LC-MS/MS serum | 32 |
| 11.8 (7.6–15.7) | n = 40, older men | ||||
| 7.1 (5.2–10.8) | n = 40, younger women | ||||
| 11.4 (7.3–20.1) | n = 40, older women | ||||
| median (IQR) | |||||
|
| |||||
| GDF11 mature protein | 16.5 (12.4) | ~0.5, mean | n = 55, men with CVD | Immunoaffinity LC-MS/MS serum | 30 |
| ~0.5, mean | n = 41, women with CVD | ||||
| 0.22–0.84 range | |||||
|
| |||||
| ~0.4, median | n = 10, young controls | ELISA, serum | 2 | ||
| 0.2–12, range | |||||
| ~1.5 median | n = 9, old controls | ||||
| 0.2–110, range | |||||
|
| |||||
| GDF11 propeptide | 21.3 (10.9) | -- | -- | -- | -- |
|
| |||||
| Follistatin | 29.8 (7.1) | ~1.5, mean | n = 20, healthy men | ELISA, serum | 26 |
|
| |||||
| 0.68, median | n = 10 controls | ELISA, serum | 34 | ||
| 0.32–3.70, range | |||||
|
| |||||
| ~1.8, median | n = 17, young women | ELISA, serum | 35 | ||
| ~2.0, median | n = 98, older women | ||||
|
| |||||
| 33 | PeptideAtlas database | spectral counts | 36 | ||
|
| |||||
| 1328, mean | n = 103, healthy controls | ELISA, plasma | 37 | ||
|
| |||||
| FST303 | 96.4 (69.2) | -- | -- | -- | -- |
|
| |||||
| WFIKKN1 | 38.3 (8.3) | -- | -- | -- | -- |
|
| |||||
| WFIKKN2 | 32.2 (10.5) | 9.4 (7.5–10.6) | n = 40, younger men | Immunoaffinity LC-MS/MS serum | 32 |
| 9.4 (7.3–10.7) | n = 40, older men | ||||
| 7.7 (6.5–10.1) | n = 40, younger women | ||||
| 9.4 (7.2–11.0) | n = 40, older women | ||||
| median (IQR) | |||||
|
| |||||
| ~8.5, mean | n = 20, healthy men | ELISA, serum | 26 | ||
|
| |||||
| ~108, mean | n = 8, healthy young men | ELISA, serum | 29 | ||
|
| |||||
| Oxytocin | 1.9 (0.9) | 0.00735 (0.00143) | n = 28, healthy married couples | ELISA, plasma | 40 |
| mean (SEM) | |||||
|
| |||||
| 0.023 (0.011) | n = 11, women breast CA | ELISA, plasma | 41 | ||
|
| |||||
| 0.198 (0.165) | n = 38, university students | ELISA, not specified | 42 | ||
|
| |||||
| 0.240 (0.224) | n = 7, healthy controls | ELISA, plasma | 43 | ||
|
| |||||
| 0.375 (0.264) | n = 473, men and women | ELISA, plasma | 44 | ||
| 0.051–2.752, range | |||||
|
| |||||
| 35 | PeptideAtlas database | spectral counts | 37 | ||
|
| |||||
| 1.2 | pooled human cord serum | LC-MS/MS, serum | 22 | ||
|
| |||||
| Oxytocin-G | 3.8 (2.4) | 0.002 (0.002) | n = 31, children, 6–12 y | RIA, plasma | 14 |
|
| |||||
| Oxytocin-GK | 216.6 (80.8) | -- | -- | -- | -- |
|
| |||||
| Eotaxin | 2.3 (0.5) | 0.101 (0.073–0.347) | n = 36, adults, 40–50 y | Luminex, serum | 45 |
| 0.135 (0.102–0.172) | n = 75, adults, 61–65 y | ||||
| 0.154 (0.116–0.194) | n = 16, adults, 76–80 y | ||||
| median (IQR) | |||||
|
| |||||
| 0.829 (0.842) | n = 14, controls | ELISA, serum | 46 | ||
Mean (SD).
Values are mean (SD) unless otherwise noted. Means noted with ~, were from papers in which mean results were reported in bar graphs with no numerical results given in text.
Abbreviations: IQR (interquartile range), CVD (cardiovascular disease), CA (cancer), SEM (standard error of the mean), RIA (radioimmunoassay)
4 Discussion
We developed an SRM assay using LC-MS/MS for quantification of GDF8 and GDF11 mature proteins, the known antagonists of GDF8 and GDF11 (the respective GDF8 and GDF11 propeptides, follistatin, WFIKKN1, and WFIKKN2), and two additional candidate rejuvenating factors, oxytocin and eotaxin. In general, SRM offers advantages over immunoassays in that (i) no antibodies are required, and SRM overcomes many of the limitations of Western blotting and ELISA that are related to antibody availability and performance, and issues related to antibody recognition of highly similar homologues and sequence variants, and inability of antibodies to recognize epitopes that are hidden in circulating protein or peptide complexes; (ii) the assay can be systematically configured for a set of proteins of interest in a single multiplexed analysis [18], and (iii) SRM is linear over a 1,000-fold concentration range and proteins present in unfractionated plasma in the low to sub ng/mL (attomole) range can be detected and measured by SRM using recent platforms [19,20].
The present SRM assay, to our knowledge, is the first multiplexed LC-MS/MS assay that is capable of measuring GDF8 and GDF11 mature proteins and five of their known circulating antagonists. The assay requires only 5 µL of plasma to measure 12 proteoforms, which is much less sample volume than would be required for conventional immunoassays. The strengths of our SRM assay included the use of both light and heavy labeled peptide standards for quantification of all the individual proteoforms and the use of three transitions to monitor each peptide in order to give high confidence of detection.
In the present SRM assay, we did not use antibodies for immunoaffinity purification prior to SRM for four reasons. First, the proteins and polypeptides targeted in this assay are found in the human circulation in the ng/mL range or higher, which is a sufficient concentration for detection by SRM without immunoaffinity purification. Second, previous immunoassays to measure GDF8, GDF11, and their antagonists did not disrupt non-covalent binding complexes such as the latent GDF8 propeptide-GDF8 mature protein complex [9,10], WFIKKN2-GDF8 mature protein complex [10], or FSTR3-GDF8 mature protein complex [9] prior to measurement. Pre-treatment of human plasma with acid to disrupt non-covalent binding complexes increases GDF8 mature protein concentrations that are measured using ELISA [21]. Third, recombinant proteins do not have the same epitopes as native proteins. When recombinant proteins are used as immunogens to generate antibodies used for immunoaffinity purification, the antibodies may not recognize native proteins with high efficiency because of differences in folding and structure between recombinant and native protein. It is difficult to generate disulfide bonds in recombinant proteins that are expressed by E. coli due to the reduced environment of the bacterial cytoplasm. E. coli-derived recombinant GDF8 and GDF11 may not contain the four disulfide bonds that are required for proper protein folding. In addition, recombinant proteins are unlikely to have the same post-translational modifications as native proteins, such as glycosylation or phosphorylation, which affect structural conformation of the protein. Notably, GDF8, GDF11, FST, WFIKKN1, and WFIKKN2 all contain one or more glycosylation sites. GDF8 and FST contain one or more phosphorylation sites.
Finally, immunoaffinity purification of GDF8, GDF11, and their antagonists is likely to be further compromised by non-covalent binding complexes as noted above, since epitopes recognized by the antibodies can be blocked or altered by binding with antagonists. In addition, oxytocin is largely bound to albumin [22], and eotaxin forms heterodimers or homodimers [23,24] which could interfere with the recognition of specific epitopes by antibodies. The denaturation, reduction, alkylation, and digestion of plasma proteins disrupts protein complexes, albumin binding, and dimers, and allows the quantification of specific proteotypic peptides of the proteins and polypeptides targeted in the assay.
The plasma concentrations of proteoforms in twenty-four adults as measured by our SRM assay are shown in comparison with other values from the published literature in Table 2. The plasma concentrations for the proteins and polypeptides reported in the present paper are generally higher than other reports in the literature that used antibody-based approaches, including ELISA and immunoaffinity-SRM. Circulating GDF8 concentrations by ELISA-based approaches have yielded highly discrepant results for healthy adults, with means or medians ranging from about 3 to 120 ng/mL [25–32]. The mean protein concentrations of both GDF8 mature protein and propeptide reported in the present study somewhat similar to published GDF8 concentrations using ELISA [25–29] and immunoaffinity SRM [30–32].
In the present study, plasma GDF11 mature protein concentrations were higher than reported in an assay that used immunoaffinity SRM [30] or ELISA [2]. In contrast to previous reports in the literature, our results suggest that both GDF11 mature protein and propeptide circulate in concentrations that are comparable to GDF8 mature protein and propeptide. To our knowledge, our study is the first to report GDF11 propeptide concentrations in humans. Plasma concentrations for FST in the present study are higher than most previous reports by immunoassay [26,34,35], similar to a report based upon spectral counting [36], but less than 1328 ng/ml reported by ELISA [37]. The present study is the first, to our knowledge, to report circulating FST303 concentrations in humans. Importantly, our results suggest that the follistatin cleavage form FST303 is the dominant proteoform of FST in the circulation, as FST303 was found at more than three-fold higher concentrations than FST. Distinguishing FST from FST303 may be biologically important since their binding affinities are different [38]. The present SRM assay has advantages over immunoassays as it can distinguish and quantify both FST and FST303.
To our knowledge, the present study is the first to quantify plasma WFIKKN1 concentrations in humans. The concentrations of WFIKKN2 in the present study are higher than reported elsewhere as measured by immunoaffinity SRM [32] or one ELISA [26], but less than another report by ELISA [29]. A limitation of the present SRM assay is that it does not include FSTR3, a known antagonist of GDF8 and GDF11. A proteotypic peptide for follistatin-related protein 3, QATC[CAM]FLGR, was originally included in assay development but did not pass quality control. Studies are in progress to test alternative proteotypic peptides such LQV[CAM]GSDGATYR and GHPDLSVMYR in order to include FSTR3 in future updates of our assay.
Plasma oxytocin has been particularly challenging to measure. Plasma oxytocin measurements using ELISA or radioimmunoassay (RIA) are problematic and unreliable [39]. Some commercial kits (Assay Designs, Alpco) require C18 column extraction of oxytocin from a large amount of plasma (3 mL) prior to ELISA. Without extraction (as used by some investigators), plasma oxytocin levels are 100-fold higher than extracted plasma, with no correlation between measurements done with or without extraction [39]. RIA lacks sensitivity; many subjects are below the limit of detection [39]. Circulating levels for oxytocin in the published literature range from low pg/mL [40,41] to 0.05 to 2.75 ng/mL [42–44] as measured by ELISA, or even as high as 35 ng/mL reported using relative quantitation and spectral counting [37]. Only one study, to our knowledge, attempted to measure oxytocin-X (the C-terminal extended forms of oxytocin) and found 0.002 ng/mL using radioimmunoassay [14]. Immunoassays may be inappropriate for measuring oxytocin, since most circulating oxytocin is bound to plasma proteins such as albumin [22]. Oxytocin levels described in the present study are similar to another study using LC-MS/MS [22]. In the latter assay, the precursor mass used for oxytocin was 1007.475, which is incorrect. If cysteine alkylation is ignored, the correct precursor is 1010.4434 (mono). However, the alkylation of the oxytocin nonapeptide CYIQNCPLG definitely must be considered as carboxyamidomethylcysteine (CAM) from Cys modification by iodoacetamide results in C[CAM]YIQNC[CAM]PLG, in which case the correct precursor is MH+2 at m/z 562.8 (native, mono).
The present SRM assay allows the quantification of mature oxytocin as well as two C-terminal extended forms of oxytocin. Using our method, we show that the C-terminal extended forms of oxytocin, such as oxytocin-G and oxytocin-GK, account for a substantial amount of circulating oxytocin. A limitation of our SRM assay is that the third C-terminal extended form of oxytocin, oxytocin-GKR, cannot be measured using our approach, since the C-terminal arginine (-R) is removed during trypsin digestion. The plasma eotaxin concentrations in the present study are higher than values reported using a Luminex assay [45] or ELISA [46]. Plasma eotaxin concentrations may be underestimated by ELISA, since E. coli-derived eotaxin was used as the immunogen to generate antibodies to eotaxin [46], which is problematic for eotaxin, since it contains two disulfide bonds. E. coli-derived eotaxin which lack disulfide bonds may not fold like native eotaxin, thus giving rise to differences in epitopes between recombinant and native eotaxin.
Animal studies suggest that GDF8 and GDF11 and their antagonists, and two other candidate rejuvenating factors, oxytocin and eotaxin, may be relevant to human aging. The relationship of these factors with aging phenotypes in humans has not been well characterized and remains a major gap in knowledge. The concentrations of GDF8, GDF11, and their antagonists as measured by this SRM assay are generally higher than reported by antibody-based assays that did not take measures to disrupt circulating protein and polypeptide complexes. A recent protocol for disrupting protein complex binding increases GDF8 concentrations measured using ELISA by approximately four-fold, but the ELISA is still prone to interference by follistatin and WFIKKN2 [21]. Concomitant measurement of antagonists to GDF8 is underscored by the observation that that latent GDF8 complex still has biological activity that is controlled by WFIKKN1 and WFIKKN2 binding [47]. The SRM assay present in this paper should facilitate future studies of twelve proteoforms that represent seven proteins of great interest to human aging. The use of this SRM assay, combined with precision robotic sample preparation [48], could be applied to cohorts of human aging to determine whether these circulating proteoforms and their antagonists are relevant to major adverse aging outcomes, such as change in muscle strength and mass, physical performance, mobility disability, cognition, renal function, and all-cause mortality. This SRM assay should facilitate the testing of the hypothesis that plasma concentrations of these candidate proteoforms change with aging, and, beyond chronological age, predict the development of specific aging phenotypes in humans.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants R56 AG052973, R01 EY024596, R01 AG027012, and the Intramural Research Program of the National Institute on Aging.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Smith LM, Kelleher NL, Consortium for Top Down Proteomics Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egerman MA, Cadena SM, Gilbert JA, Meyer A, et al. GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab. 2015;22:164–174. doi: 10.1016/j.cmet.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho DM, Yeo CY, Whitman M. The role and regulation of GDF11 in Smad2 activation during tailbud formation in the Xenopus embryo. Mech. Dev. 2010;127:485–495. doi: 10.1016/j.mod.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McPherron AC. Metabolic functions of myostatin and GDF11. Immunol. Endocr. Metab. Agents Med. Chem. 2010;10:217–231. doi: 10.2174/187152210793663810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thies RS, Chen T, Davies MV, Tomkinson KN, et al. GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors. 2001;18:251–259. doi: 10.3109/08977190109029114. [DOI] [PubMed] [Google Scholar]
- 7.Wolfman NM, McPherron AC, Pappano WN, Davies MV, et al. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc. Natl. Acad. Sci. USA. 2003;100:15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha M, Jang YC, Oh J, Khong D, et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science. 2014;344:649–652. doi: 10.1126/science.1251152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill JJ, Davies MV, Pearson AA, Wang JH, et al. The myostatin propeptide and the follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J. Biol. Chem. 2002;277:40735–40741. doi: 10.1074/jbc.M206379200. [DOI] [PubMed] [Google Scholar]
- 10.Hill JJ, Qui Y, Hewick RM, Wolfman NM. Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol. Endocrinol. 2003;17:1144–1154. doi: 10.1210/me.2002-0366. [DOI] [PubMed] [Google Scholar]
- 11.Schneyer AL, Sidis Y, Gulati A, Sun JL, et al. Differential antagonism of activin, myostatin and growth and differentiation factor 11 by wild-type and mutant follistatin. Endocrinology. 2008;149:4589–4595. doi: 10.1210/en.2008-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondás K, Szláma G, Trexler M, Patthy L. Both WFIKKN1 and WFIKKN2 have high affinity for growth and differentiation factors 8 and 11. J. Biol. Chem. 2008;283:23677–23684. doi: 10.1074/jbc.M803025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimasaki S, Koga M, Esch F, Cooksey K, et al. Primary structure of the human follistatin precursor and its genomic organization. Proc. Natl. Acad. Sci. USA. 1988;85:4218–4222. doi: 10.1073/pnas.85.12.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green L, Fein D, Modahl C, Feinstein C, et al. Oxytocin and autistic disorder: alterations in peptide forms. Biol. Psychiatry. 2001;50:609–613. doi: 10.1016/s0006-3223(01)01139-8. [DOI] [PubMed] [Google Scholar]
- 15.Elabd C, Cousin W, Upadhyayula P, Chen RY, et al. Oxytocin is an age-specific circulating hormone that is necessary for muscle maintenance and regeneration. Nat. Commun. 2014;5:4082. doi: 10.1038/ncomms5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villeda SA, Luo J, Mosher KI, Zou B, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzyk MA, Parker CE, Domanski D, Borchers CH. Development of MRM-based assays for the absolute quantitation of plasma proteins. Methods Mol. Biol. 2013;1023:53–82. doi: 10.1007/978-1-4614-7209-4_4. [DOI] [PubMed] [Google Scholar]
- 18.Liebler DC, Zimmerman LJ. Targeted quantitation of proteins by mass spectrometry. Biochemistry. 2013;52:3797–3806. doi: 10.1021/bi400110b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domanski D, Percy AJ, Yang J, Chambers AG, et al. MRM-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics. 2012;12:1222–1243. doi: 10.1002/pmic.201100568. [DOI] [PubMed] [Google Scholar]
- 20.Ceglarek U, Dittrich J, Becker S, Baumann F, et al. Quantification of seven apolipoproteins in human plasma by proteotypic peptides using fast LC-MS/MS. Proteomics Clin. Appl. 2013;7:794–801. doi: 10.1002/prca.201300034. [DOI] [PubMed] [Google Scholar]
- 21.Quantikine ELISA. GDF8/Myostatin Immunoassay. R & D Systems; Minneapolis, MN: 2016. [Accessed February 7, 2017]. p. 14. [ https://resources.rndsystems.com/pdfs/datasheets/dgdf80.pdf] [Google Scholar]
- 22.Brandtzaeg OK, Johnsen E, Roberg-Larsen H, Seip KF, et al. Proteomics tools reveal startlingly high amounts of oxytocin in plasma and serum. Sci. Rep. 2016;6:31693. doi: 10.1038/srep31693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crump MP, Rajarathnam K, Kim KS, Clark-Lewis I, et al. Solution structure of eotaxin, a chemokine that selectively recruits eosinophils in allergic inflammation. J. Biol. Chem. 1998;273:22471–22479. doi: 10.1074/jbc.273.35.22471. [DOI] [PubMed] [Google Scholar]
- 24.Crown SE, Yu Y, Sweeney MD, Leary JA, et al. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J. Biol. Chem. 2006;281:25438–25446. doi: 10.1074/jbc.M601518200. [DOI] [PubMed] [Google Scholar]
- 25.Yano S, Nagai A, Isomura M, Yamasaki M, et al. Relationship between blood myostatin levels and kidney function: Shimane CoHRE Study. PLoS. ONE. 2015;10:e0141035. doi: 10.1371/journal.pone.0141035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratkevicius A, Joyson A, Selmer I, Dhanani T, et al. Serum concentrations of myostatin and myostatin-interacting proteins do not differ between young and sarcopenic elderly men. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:620–626. doi: 10.1093/gerona/glr025. [DOI] [PubMed] [Google Scholar]
- 27.Furihata T, Kinugawa S, Fukushima A, Takada S, et al. Serum myostatin levels are independently associated with skeletal muscle wasting in patients with heart failure. Int. J. Cardiol. 2016;220:483–487. doi: 10.1016/j.ijcard.2016.06.231. [DOI] [PubMed] [Google Scholar]
- 28.Wintgens KF, Dschietzig T, Stoeva S, Paulsson M, et al. Plasma myostatin measured by a competitive ELISA using a highly specific antiserum. Clin. Chim. Acta. 2012;413:1288–1294. doi: 10.1016/j.cca.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 29.Saremi A, Gharakhanloo R, Sharghi S, Gharaati MR, et al. Effects of oral creatine and resistance training on serum myostatin and GASP-1. Mol. Cell Endocrinol. 2010;317:25–30. doi: 10.1016/j.mce.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Schafer MJ, Atkinson EJ, Vanderboom PM, Kotajarvi B, et al. Quantification of GDF11 and myostatin in human aging and cardiovascular disease. Cell Metab. 2016;23:1207–1215. doi: 10.1016/j.cmet.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palandra J, Quazi A, Fitz L, Rong H, et al. Quantitative measurements of GDF-8 using immunoaffinity LC-MS/MS. Proteomics Clin. Appl. 2016;10:597–604. doi: 10.1002/prca.201500112. [DOI] [PubMed] [Google Scholar]
- 32.Bergen HR, Farr JN, Vanderboom PM, Atkinson EJ, et al. Myostatin as a mediator of sarcopenia versus homeostatic regulator of muscle mass: insights using a new mass spectrometry-based assay. Skelet. Muscle. 2015;5:21–36. doi: 10.1186/s13395-015-0047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris HN, Ashman K, Vaswani K, Kvaskoff D, et al. Method development for the detection of human myostatin by high-resolution and targeted mass spectrometry. J. Proteome Res. 2014;13:3802–3809. doi: 10.1021/pr5004642. [DOI] [PubMed] [Google Scholar]
- 34.Hughes RD, Evans LW. Activin A and follistatin in acute liver failure. Eur. J. Gastroenterol. Hepatol. 2003;15:127–131. doi: 10.1097/00042737-200302000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann M, Halper B, Oesen S, Franzke B, et al. Serum concentrations of insulin-like growth factor-1, members of the TGF-beta superfamily and follistatin do not reflect different stages of dynapenia and sarcopenia in elderly women. Exp. Gerontol. 2015;64:35–45. doi: 10.1016/j.exger.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Farrah T, Deutsch EW, Omenn GS, Campbell DS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006353. M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen J, Rinnov A, Krogh-Madsen R, Fischer CP, et al. Plasma follistatin is elevated in patients with type 2 diabetes: relationship to hyperglycemia, hyperinsulinemia, and systemic low-grade inflammation. Diabetes Metab. Res. Rev. 2013;29:463–472. doi: 10.1002/dmrr.2415. [DOI] [PubMed] [Google Scholar]
- 38.Sugino K, Kurosawa N, Nakamura T, Takio K, et al. Molecular heterogeneity of follistatin, an activin-binding protein. Higher affinity of the carboxyl-terminal truncated forms for heparan sulfate proteoglycans on the ovarian granulosa cell. J. Biol. Chem. 1993;268:15579–15587. [PubMed] [Google Scholar]
- 39.Szeto A, McCabe PM, Nation DA, Tabak BA, et al. Evaluation of enzyme immunoassay and radioimmunoassay methods for the measurement of plasma oxytocin. Psychosom. Med. 2011;73:393–400. doi: 10.1097/PSY.0b013e31821df0c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holt-Lunstad J, Birmingham WA, Light KC. Influence of a "warm touch" support enhancement intervention among married couples on ambulatory blood pressure, oxytocin, alpha amylase, and cortisol. Psychosom. Med. 2008;70:976–985. doi: 10.1097/PSY.0b013e318187aef7. [DOI] [PubMed] [Google Scholar]
- 41.Billhult A, Lindholm C, Gunnarsson R, Stener-Victorin E. The effect of massage on immune function and stress in women with breast cancer--a randomized controlled trial. Auton. Neurosci. 2009;150:111–115. doi: 10.1016/j.autneu.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Zak PJ, Kurzban R, Matzner WT. Oxytocin is associated with human trustworthiness. Horm. Behav. 2005;48:522–527. doi: 10.1016/j.yhbeh.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Goldman M, Marlow-O'Connor M, Torres I, Carter CS. Diminished plasma oxytocin in schizophrenic patients with neuroendocrine dysfunction and emotional deficits. Schizophr. Res. 2008;98:247–255. doi: 10.1016/j.schres.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisman O, Zagoory-Sharon O, Schneiderman I, Gordon I, et al. Plasma oxytocin distributions in a large cohort of women and men and their gender-specific associations with anxiety. Psychoneuroendocrinology. 2013;38:694–701. doi: 10.1016/j.psyneuen.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Shurin GV, Yurkovetsky ZR, Chatta GS, Tourkova IL, et al. Dynamic alteration of soluble serum biomarkers in healthy aging. Cytokine. 2007;39:123–129. doi: 10.1016/j.cyto.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Panizzutti B, Gubert C, Schuh AL, Ferrari P, et al. Increased serum levels of eotaxin/CCL11 in late-stage patients with bipolar disorder: An accelerated aging biomarker? J. Affect. Disord. 2015;182:64–69. doi: 10.1016/j.jad.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Szláma G, Trexler M, Patthy L. Latent myostatin has significant activity and this activity is controlled more efficiently by WFIKKN1 than by WFIKKN2. FEBS J. 2013;280:3822–3839. doi: 10.1111/febs.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu M, Zhang P, Geng-Spyropoulos M, Moaddel R, et al. A robotic protocol for high-throughput processing of samples for selected reaction monitoring assays. Proteomics. 2016 Nov 15; doi: 10.1002/pmic.201600339. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.