Abstract
The importance of RNA splicing in numerous cellular processes is well established. However, an underappreciated aspect is the ability of the spliceosome to recognize a set of very small (3–30 nucleotide, 1–10 amino acid) exons named microexons. Despite their small size, microexons and their regulation through alternative splicing have now been shown to play critical roles in protein and system function. Here we review the discovery of microexons over time and the mechanisms by which their splicing is regulated, including recent progress made through deep RNA sequencing. We also discuss the functional role of microexons in biology and disease.
Alternative splicing (AS) is considered to be a major mechanism for expanding the complexity of the proteome of eukaryotic cells, and it is estimated that up to 90–95% of multi-exon genes express numerous transcript isoforms through AS.1, 2 Alternative splicing is a dynamic and well-orchestrated process that defines cellular context, and has been found to play critical roles in diverse functions ranging from maintenance of stem cell pluripotency3 to generation of highly tissue-specific proteins in the brain and muscle.4–6 Misregulation of AS can be devastating, with evidence of aberrant splicing found in cancer and in neurological diseases (reviewed in ref.7).
The splicing machinery, a large RNA-protein complex known as the spliceosome, assembles at the 5′ and 3′ ends of an intron by scanning for the essentially invariant splicing sites, GU and AG, respectively. Splice sites have limited information content and can compete for each other, causing some exons to be skipped over and thus excluded from the resulting transcript. To dictate precise splicing at specific sites, the substrate RNA has to contain additional regulatory sequence or structural features. These include exonic and intronic enhancers and silencers (ESE, ESS, ISE and ISS)—short motifs that are bound by RNA-binding proteins (RBPs) to modulate spliceosome interactions. Exons with stronger splice sites and that are enriched in enhancer elements can be spliced more efficiently than those with opposite features.
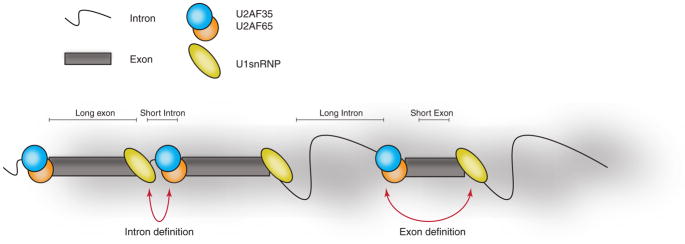
Importantly, splicing efficiency can also be affected by the size of exons and introns. Several models of splicing based on the size of exons and introns have been proposed (reviewed in ref.8). In lower eukaryotes such as yeast, introns are short and coordinated recognition of the 5′ and 3′ splice sites across introns leads to the commitment of the splicing reaction. This model is denoted the “intron-definition model” (Fig 1). In contrast, exons of higher eukaryotes, and mammals in particular, exons are characterized by a much shorter size compared to introns —human exons have a median size of ~120 nucleotides (nt) with 80% less than 200 nt, while introns are on average ten times longer, with some extending to several hundred thousands of nucleotides9 —making intron definition challenging. For this scenario, an “exon-definition model” has been proposed, where the splicing machinery is recruited through coordinated recognition of the sites across exons (Fig 1).10, 11 The exon-definition model suggests a range of optimal exon sizes required to fit all components of the spliceosome and for efficient splicing. Based on splicing assays, it has been reported that exons smaller than 50 nt have a clear disadvantage due to the molecular dynamics of the splicing machinery itself, frequently resulting in exon skipping.12 This model is also consistent with the observation that alternative exons are generally much shorter than constitutive exons.13, 14
Figure 1. Intron- and exon-definition models.
Schematic representation of the two primary models that drive assembly of splicing factors complexes and define the use of the splice sites in different mRNA architectures. The intron definition model is applicable to lower eukaryotes such as yeasts where the length of the intronic regions is rather small. The exon definition model proposes an explanation for more complex splicing regulation in higher eukaryotes due to the complexity of the genome organization and the presence of large intronic areas that are on average ten times longer than coding exons.
This review discusses exons of unusually small size ranging from 3 to 30 nt (up to 51 nt according to some definitions), named microexons. Microexons are particularly interesting models for studying AS from two defining perspectives: at the splicing regulation level—how the small size of the exons is compensated for by additional enhancing signals—and at the functional level—how these small fragments can result in significant impacts on the protein products. We will discuss recent progress in the field, including insights obtained from deep transcriptome sequencing and potential, yet underappreciated, links between microexons and disease.
Discovery of microexons
The first microexons were described more than three decades ago. In 1985, Beachy et al. described two 5 nt exons in the Drosophila Ubx gene15, and possibly the first use of the term “microexon”. In the same year, Cooper and Ordahl identified a 6 nt constitutive microexon in the chicken Troponin T gene.16 While studying Ncam in rat and mouse brain, researchers were surprised to find multiple alternatively spliced isoforms, including evidence of AS in microexons: Small et al. reported a 30 nt, developmentally regulated microexon in the rat brain in 1988 (ref.17), and Santoni et al. reported a 3 nt microexon in the mouse brain in 1989 (ref.18). In 1992, McAllister et al. reported two 9 nt microexons in the Fasciclin I gene in Drosophila.19
These exons were quite puzzling, and were largely considered to be outliers, studied as peculiar and anecdotic. It is also technically difficult to properly align sequenced transcripts such as expressed sequence tags (ESTs) containing microexons to the genomic DNA. Due to their small size, microexon sequences are easily mis-aligned or ignored as errors, and thus require special consideration when aligning ESTs (and, nowadays, RNA-Seq reads).
Consequently, microexons were not reported as common until 2003, when the first systematic identification of microexons was reported by the Salzberg lab.20 They developed a spliced alignment correction procedure that specifically investigates regions in ESTs/cDNAs with small alignment gaps which might be due to unannotated microexons (Fig 2a). The algorithm considers candidate canonical splice signals flanking these unaligned gaps, and evaluates each possible combination by searching for matches of the unaligned segment in the reference genome. Perfect matches are then labeled as novel microexons. This method significantly outperformed general-purpose cDNA aligners and identified 223 human microexons including 170 exons that were unannotated previously, suggesting that 1.6% of human genes contain microexons20. Wu and Watanabe enhanced this method by incorporating statistical significance scores for the microexons, and included it alongside an improved novel exon detection algorithm as part of GMAP, their EST alignment tool.21 An improved sensitivity was observed relative to other aligners, although specificity is not addressed.
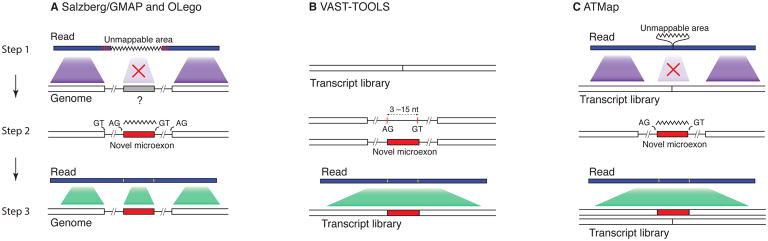
Figure 2. Identification of microexons.
Currently available bioinformatics tools for identifying microexons.
A. Salzberg/GMAP and OLego algorithms: 1) When comparing cDNA sequences (Salzberg/GMAP) or RNA-Seq reads (OLego) to the reference genome, there will be unmappable insertions in the cDNA/RNA-Seq reads corresponding to unannotated microexons. 2) These algorithms search for potential matches to these segments at high resolution and evaluate candidate splice sites in the reference genome to identify novel exons. 3) After identification of the microexon, the reads map correctly.
B. VAST-TOOLS: 1) cDNA libraries are used to build an exon junction database. 2) All possible microexon candidates are enumerated in silico by searching pairs of splice site separated by 3–15nt within known introns. 3) Read mapping for an RNA-Seq library of interest is performed against this exon-microexon-exon junction database to detect microexons.
C. ATMap: 1) Mapping of RNA-Seq reads to a reference cDNA database lacking a microexon results in unmappable insertions in the read. 2) ATMap returns to the reference genome to identify splice sites surrounding a region that matches the read. 3) After identification of the microexon, the reads map correctly.
More recently, high-throughput RNA sequencing (RNA-Seq)22 and the development of splicing-aware aligners (Tophat23, MapSplice24, Olego25, STAR26, and HISAT27) that can identify novel exon junctions have greatly increased our ability to identify alternative exons because of the unprecedented sequencing depth compared to EST data. Some aligners have also made specific optimizations for discovery of microexons.
For example, OLego is designed to identify novel exon junctions and microexons as part of the read mapping process25. It uses a short-seed approach, where reads are divided into small segments, which are mapped independently. The presence of a microexon is frequently indicated by an unaligned gap in the middle of the reads. Similar to the Salzberg algorithm, the unaligned fragment flanked by canonical splice site signals is re-mapped to the genome to search for microexons with high confidence (Fig 2a). Where ambiguity exists, candidate exons were further evaluated by a regression model that considers the strength of splice sites and the size of flanking introns. It was estimated that such an approach can achieve >85% sensitivity and >75% specificity for microexon detection.
An alternative approach, to search for all possible candidate microexons, was taken by Irimia et al. in their VAST-TOOLS software.28 These microexons were defined by two splice sites separated by 3–15 nt within known intronic regions, creating a database of exon-microexon-exon junctions (Fig 2b). RNA-Seq reads were aligned to this junction database to find microexons with experimental evidence.
Another approach, used by Li et al., named ATMap, searches for novel microexons by aligning RNA-Seq reads to a library of transcript models and searching for small insertions (3–51 nt) (Fig 2c).29 These insertions are then compared with the reference genome to define high confidence junctions based on canonical splice site motifs. For microexon quantification, a reference containing the inclusion and exclusion isoforms is generated. In practice, Li et al. found that the results obtained using ATMap and OLego are highly similar.
Leveraging the depth of RNA-Seq data and the computational tools described above, these groups have defined a large number of microexons: 13,145 constitutive and AS microexons 6–51 nt were reported by Li et al.29, 696 AS microexons (in 603 genes) 3–27 nt were reported by Irimia et al.28, and 2,008 AS microexons (in 1,587 genes) 6–30 nt were reported in an analysis of human brain transcriptomes at different developmental stages.30 These three studies reveal that microexons are more prevalent and present in a larger number of genes than previously appreciated. Furthermore, since the focus of these studies is biased towards the brain, investigation of deep RNA-Seq data from other tissues should reveal additional unannotated microexons.
Regulation of microexons
Microexons have at least two apparent disadvantages in efficient splicing compared to exons of regular size: 1) allosteric hindrance based on the exon-definition model, and 2) the difficulty to accommodate necessary splicing enhancers in the exon (ESEs). In this section, we review evidence of compensatory mechanisms that allow microexons to turn the odds of inclusion in their favor.
Li et al. analyzed a number of regulatory features surrounding 7,949 microexons (≤51nt) expressed in the brain and made several interesting observations.29 First, compared to exons of regular size (either alternatively or constitutively spliced), microexons tend to possess shorter flanking introns (median 955 nt vs. 1161 nt). This observation suggests that inefficient exon definition might be compensated for by more efficient intron definition.31 Second, both 3′ and 5′ splice sites of constitutive microexons are significantly stronger compared to alternative exons and other constitutive exons of larger sizes. Third, microexons possess a much higher density of ESEs, despite the fact that the absolute number is limited by their short length. Interestingly, an increased co-occurrence of certain motifs was observed in introns proximally to the microexon splicing sites. High cytosine and uridine elements are the most enriched within 10–20 nucleotides upstream of exon 3′ splice site compared to the constitutive exons. Those can potentially stimulate the binding of multiple RBPs including PTBP, TIA1, HNRNPC, ELAVL1, or U2AF2. Experimental evidence also exists for some sequence motifs that can promote inclusion of microexon in vertebrates. For instance Carlo et al. identified a repeated heptanucleotide intronic enhancer (GGGGCUG) located in the downstream intron that promoted the inclusion of a 6 nt microexon in the chicken cTNT gene.32 This inclusion was stimulated by binding of the SF1 protein, which the authors suggested defines the exon.33
A prominent feature of microexons is the dynamic changes during development or cellular differentiation, which is particularly true in the central nervous system. For example, during in vitro differentiation of mouse embryonic stem cells to glutamatergic excitatory neurons, 30% of exons go from near complete exclusion to near complete inclusion (or vice versa) and 69% showing a change > 50%.28 This includes the 12 nt microexon of the Enah gene (enabled homolog (Drosophila), a phosphoprotein involved in the regulation of actin filament assembly), which reaches 50% inclusion in neurons, but the exon is completely skipped in embryonic stem cells. These dramatic, highly-controlled changes suggest that the microexons perform critical functions in neurodevelopment, although their exact function is not yet clear in most cases.
Many microexons show high neuron-specific splicing compared to the other cell types within the human central nervous system, with more than 90% of regulated microexons having their highest inclusion levels in neurons,28 and such specificity is generally highly conserved in mouse brain.29 It was also reported that microexons represent 1/3 of all the neuron-specific AS events conserved between human and mouse28, suggesting that neural-specific microexon programs are conserved and functional. Strikingly, 55 microexons are deeply conserved in vertebrates that span 450 million years of evolution from zebrafish to human.28 Intronic regions surrounding in-frame microexons are highly conserved, supporting both the importance of the exons and the difficulty in accommodating a sufficient number of regulatory elements within the exon.29 Interestingly, the intronic regions of frame-shifting microexons are less conserved.
The dramatic splicing changes in different cellular contexts are regulated by tissue-specific splicing factors such as RBFOX,34, 35,36 NOVA,37, 38 and MBNL.39, 40 These proteins recognize sequence elements frequently located in the intronic regions surrounding alternative exons. There exists a strong correlation between the splicing factor binding position and inclusion or exclusion of the alternative exon, creating an “RNA-map”.41 For example, binding of the RBFOX protein downstream of the AS exon leads to its inclusion (the binding site acts as an ISE) while recognition of the same sequence upstream promotes exon exclusion (the site acts as an ISS).36, 42, 43 It is believed that several AS factors, such as RBFOX, PTBP1, and nSR100 (SRRM4), contribute to the regulation of microexon inclusion in a tissue or cell type-specific manner.28, 29 This was supported by RNA-Seq based transcriptome profiling of splicing factor knockdown and mapping of protein-RNA interactions by CLIP experiments,44 which allow for the investigation of direct regulation of the alternative events by specific RBPs.
Over half of the detected microexons in the study by Irimia et al. were affected in HEK293T kidney cell by overexpression of nSR100.28 The importance of the protein for the direct regulation of these splicing events was also supported by CLIP data, where enrichment of nSR100 binding was observed in the upstream intronic region in close proximity towards the 3′ splice site. Interestingly, Irimia et al. indicated that while other RBPs (RBFOX, MBNL, ESRP1, and PTBP) regulate all exon sizes relatively equally, nSR100 regulates a significantly higher proportion of microexons compared to exons of larger sizes (>27 nt) (ref.28).
Together, these results make clear that proper inclusion of microexons is highly complex, tightly regulated, and not yet fully understood. In addition, tissue or cell type-specific regulation of many microexons suggests that they are likely functional and thus require precise inclusion in specific cellular contexts. The regulatory mechanisms enabling their function, which overcome the steric disadvantages imposed on microexons, are under strong selection pressure to be conserved during vertebrate evolution.
Function of microexons
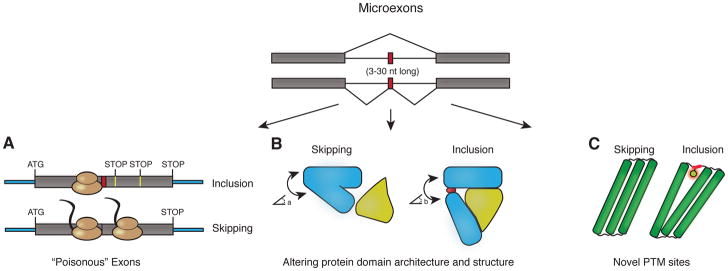
AS in coding regions generally has one of two immediate consequences (Fig 3). If an exon is in-frame (its length is a multiple of three) and contains no stop codon, it leads to alteration of the local amino acid sequence. These exons can change the functional properties of the protein products by modulating protein-protein interactions and enzymatic activity, by including post-transcriptional modification sites, or by influencing protein localization. On the other hand, exons that are not in frame can change the open reading frame during translation, which typically leads to the formation of a premature stop codon (poisonous exons) and subsequent degradation of the transcript via nonsense-mediated decay (NMD, reviewed in ref.45). Interestingly the majority (80–90%) of microexons have a length that is a multiple of three, allowing them to maintain open reading frames.28, 29 The remaining micro-exons will likely cause a frame-shift, and thus trigger NMD as part of the mRNA surveillance pathway or as a mechanism to regulate the steady-state gene expression level. For example, Li et al. identified a conserved, frame-shifting 17 nt microexon in NFKB1 (nuclear factor kappa B subunit 1, encoding a stimulant-activated transcription factor associated with inflammatory disease), which might play a regulatory role.29
Figure 3. Function of microexons.
Inclusion of microexons can: (A) cause a frameshift leading to premature stop codons and degradation of the mRNA via nonsense-mediated decay, (B) change protein structure which may in turn modulate protein-protein interactions, and (C) create sites for post-translational modification of proteins.
Amino acid sequences encoded by neuronal microexons show striking enrichment in protein-domains involved in protein-protein interactions (Fig 3). In fact, microexons are frequently central nodes in the protein interaction networks and are known to be part of the stable protein complexes.28 Several examples indicate that inclusion of microexons leads to the changes in the non-structured and disordered regions of the protein, which remodel protein interaction networks. Often, microexons affect protein function in a tissue-specific fashion. Several examples of microexons affecting protein function are listed below.
The three paralogs of amyloid-beta precursor protein binding, family B (APBB1, APBB2, and APBB3) contain a 6 nt microexon that maps to the beta-turn loop of phosphotyrosine-binding domain of amyloid-beta precursor that was predicted to influence cytoplasmic tail interaction with amyloid-beta protein. This prediction was confirmed by luminescence-based mammalian interactome mapping and co-precipitation experiments of the Apbb1 and histone-acetyl transferase Kat5/Tip60 as well as with amyloid precursor APP. Inclusion of the microexon (encoding for the amino acids RE) significantly increased interaction between Apbb1 and Kat5 but had little effect on the interaction with APP.28
In vertebrates, the ITSN1 gene (intersectin 1, a neuronal membrane protein involved in membrane and synaptic transport) undergoes AS affecting several domains (SH3, EH, PD, and DH).46 Exon 20 of the ITSN1 gene encodes for five amino acids (VKGEW) within the n-Src loop of the SH3A domain and shows high neuronal specificity. Interestingly inclusion or exclusion of the exon dramatically influences protein interacting preference of the ITSN1 gene. In vitro pull-down experiments with the neuron-specific inclusion isoform showed higher affinity with Dynamin 1, Synaptojanin 1 and CdGAP, while experiments with the ubiquitously-expressed exclusion isoform selectively binds to SOS1 and c-Cbl.47
Finally, a 21 nt microexon in Protrudin (ZFYVE27) is always included in neurons but excluded in oligodendrocytes (astrocytes have a mix of the two isoforms).48 The neuronal isoform promotes neurite outgrowth and axon growth, and establishes neuronal polarity. Microexon 8a in LSD1 gene shows a highly developmentally regulated and neuron specific expression pattern. Interestingly, misregulation of the neuron specific isoform can modulate neurite growth in rat cortical neurons49 by affecting its phosphorylation status on threonine 369b encoded by alternative microexon 8a. This phosphorylation is important for the detachment of the HDAC1/2 and CoREST from LSD1 thus displacing histone deacetylase activity from the co-repressor complex and promoting neuronal maturation.50
These diverse examples represent some of the functions encoded by microexons. With the rapidly increasing list of microexons, these are likely only a tiny fraction of functional microexon events. As biochemical studies on individual alternative exons are performed, we will likely see an increase in microexons with a clear biological role, in turn aiding our understanding of cellular processes.
Microexons in disease
Misregulation of AS can have devastating effects on various cellular functions. Misregulation can occur in one of two ways. First, by perturbation of key splicing regulators, which results in a loss of regulation and fine tuning of various cellular programs including differentiation, maturation, and homeostatic functioning. Second, and more subtly, by perturbation of AS of individual exons through disruption of an RBP binding site or a splice site as a result of a point mutation. An expanding list of diseases are attributed to splicing misregulation, including amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA), frontotemporal dementia (FTD), myotonic dystrophy (DM), and cancer (reviewed in ref.7).
It is inconceivable that microexons would be immune to either of these two types of perturbations. Because they have been shown to play key roles in altering the intrinsically disordered regions of proteins—potentially mediating protein-protein interactions and post-translational modification capacities—it follows that micro-exons should be considered valid candidates in causing genetic diseases.
In fact, several genome-wide studies tried to address the involvement of microexons in the development of neurological disorders. One study focused on autism spectrum disorders (ASD), a highly heritable group of pathologies in children characterized by impaired social response and language development28. Post-mortem samples from the superior temporal gyrus of healthy individuals as well as ASD patients (n=12 for each) were subjected to RNA-Seq and analyzed accordingly, matching for the age and gender. In the studied sample set, 30% of the microexons showed significant splicing changes between the control and ASD groups. A large portion of misregulated microexons appeared to be regulated by the RBP nSR100 (SRRM4) and were located within genes previously linked with ASD, including DTNA, ROBO1, SHANK2, and ANK2. Full loss of nSR100 in mice results in defective neurite outgrowth and cortical layering, and typically (85%) causes death within hours after birth from poor breathing, with the minority that remain alive showing pronounced neurological tremors.51, 52 Interestingly, heterozygous nSR100 knockout mice have features characteristic of autism, including social aversion that is more pronounced in males (a striking observation considering that autism is more prevalent in human males), increased sensitivity to auditory stimuli, and defective dendritic spines.52 These mice also show an increase in excitatory glutamatergic synapses and a decrease in inhibitory GABAergic synapses, possibly to compensate for lowered frequencies of excitatory postsynaptic currents and decreased neuron excitability.
The neuron-specific microexon 27 of the L1CAM gene (L1 cell adhesion molecule, an axonal glycoprotein involved in neuron migration and differentiation) translates to four amino acids (RSLE), which together with its upstream sequence form a critical YRSLE motif that is necessary for axonal growth cone sorting and membrane incorporation during axon growth of DRG neurons.53 The same sequence motif is necessary for the interaction with AP-2 protein and clathrin mediated endocytosis.54 Surprisingly, microexon 2 of the same gene contains a protein motif (YEGHHV) that modulates L1 interaction with protein ligands, in turn modulating nervous system development.55 It was reported that exon 2 truncated neurons have lower neurite promoting activity.56 Understanding the role and function of the microexon splicing in the context of the L1 protein is particularly important due to the number of diseases that are associated with the mutations in L1CAM gene. These are usually combined under the term L1 syndrome and include hydrocephalus and CRASH syndrome (Corpus callosum hypoplasia, Retardation, Adducted thumbs, Spasticity and Hydrocephalus), which in certain cases are directly linked to deletions in the regions containing the above discussed microexons.57
Interleukin-18 (IL-18) is a pro-inflammatory cytokine that is expressed in the form of a precursor which is processed to its active form by caspase-1 and caspase-4. Ovarian carcinoma cell lines as well as epithelial cells from fresh ovarian tumors contained a large portion of IL-18 precursor that was resistant to caspase mediated activation compared to healthy ovarian epithelial cells. This was found to be associated with the expression of a truncated isoform that lacks microexon 3, which encodes four amino acids, despite the fact that the caspase binding domain, located in the downstream exon 4, is intact. Such alteration of microexon inclusion potentially leads to establishment of the immune privilege during neoplastic transformation.58
Finally, the GABAa receptor subunit gamma has two distinct splicing isoforms: the long (γ2L) and short (γ2S) isoforms, which differ by a 24 base pair coding microexon (LLRMFSFK). This microexon introduces a protein kinase C phosphorylation site.59, 60 The phosphorylation state is dependent on inclusion of the microexon and was shown to influence GABA activated current61. In schizophrenia patients, a statistically significant imbalance between the long and short isoform was observed: in affected individuals brain levels of the γ2S isoform were drastically reduced by 51.7% compared to control while expression of the longer isoform was reduced non-significantly by 16.9%62, thus affecting the current mediated by one of the major receptors.
We are currently in the early stages of estimating the extent to which individual microexons contribute to the development of disease and in distinguishing between microexon misregulation as a primary cause or a secondary effect of disease progression. In the case of LSD1, for example, animals with a deletion of the neuron-specific microexon experience milder symptoms of pilocarpine-induced status epilepticus (PISE), 63 display a reduced anxiety phenotype64, and survive at a higher rate compared to control animals.63 Along the same line, global alteration of microexon expression as a result of reduced levels of nSR100/SRRM4 also points toward a causal role of microexons in contributing to ASD-like phenotypes, as evidenced by the prevalence towards male animals reminiscent of the gender bias of autism incidence in humans.52 These examples show that the functional significance of this under-investigated class of exons could be substantial.
Conclusions
Recent technological innovations have allowed us to identify thousands of microexons, highlighting an underappreciated type of functional genomic elements. In this review, we have discussed both individual and global roles for sets of microexons that reveal their potential for modulating cellular biology in normal physiological settings and genetic diseases.
Despite their small size, microexon inclusion levels are tightly regulated, and many microexons are constitutively expressed. This is achieved by a combination of stronger canonical splice signals and the regulatory motifs recognized by splicing factors. In addition, microexons are in general flanked by shorter and well conserved introns that can potentially influence and promote their inclusion level. Importantly, the presence of splicing enhancers suggests that microexons are intentionally included by the cell, even if they serve as yet unknown functions.
Several diseases directly attributed to microexon perturbations have been described in the literature. It is apparent that, similar to the already established model of AS in disease, microexons encoding for even a few amino acids can have critical effects on global physiological processes such as tumorigenesis and development. Perturbation of the factors that are involved in those mechanisms can have dramatic effects on pathology establishment and disease progression. As microexons are identified and characterized more comprehensively, it is likely that more direct causes of disease will be uncovered.
An interesting extension to the study of microexons is that they may potentially be the answer to some unexplained intronic variants. There are currently many identified mutations in non-coding, deep-intronic regions that are associated with human disease. Their function has been unclear in the absence of any coding potential, but given the existence of very small microexons (3nt or 1 amino acid), it is possible that these mutations are in fact located within or near an undefined, coding microexon. Even these small perturbations can change protein-protein interactions as a result of remodeling of flexible inter-domain linkers, protein localization, and post-transcriptional modifications.
Further work will be needed to systematically identify candidate causal splicing events. As with AS in general, the combination of extensive throughput techniques, genome editing, and deep RNA sequencing will provide a powerful toolkit for precision medicine and enabling the understanding of individual AS events and their impact on disease. We hope that future studies will take advantage of the progress made in identifying the presence and importance of microexons and fully consider them as candidate causal exons.
Acknowledgments
We apologize to authors whose relevant work we could not cite due to space limit. We thank members of the Zhang laboratory for helpful discussion related to this review. Our work was supported by grants from the National Institutes of Health (NIH) (R00GM95713, R01NS089676 and R21NS098172) and the Simons Foundation Autism Research Initiative (307711).
References
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K, Dai X, Wu J. Alternative splicing: an important mechanism in stem cell biology. World J Stem Cells. 2015;7:1–10. doi: 10.4252/wjsc.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 9.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 10.Robberson BL, Cote GJ, Berget SM. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 12.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorek R, Shemesh R, Cohen Y, Basechess O, Ast G, Shamir R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Krainer AR, Zhang MQ. Evolutionary impact of limited splicing fidelity in mammalian genes. Trends Genet. 2007;23:484–488. doi: 10.1016/j.tig.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- 16.Cooper TA, Ordahl CP. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985;260:11140–11148. [PubMed] [Google Scholar]
- 17.Small SJ, Haines SL, Akeson RA. Polypeptide variation in an N-CAM extracellular immunoglobulin-like fold is developmentally regulated through alternative splicing. Neuron. 1988;1:1007–1017. doi: 10.1016/0896-6273(88)90158-4. [DOI] [PubMed] [Google Scholar]
- 18.Santoni MJ, Barthels D, Vopper G, Boned A, Goridis C, Wille W. Differential exon usage involving an unusual splicing mechanism generates at least eight types of NCAM cDNA in mouse brain. EMBO J. 1989;8:385–392. doi: 10.1002/j.1460-2075.1989.tb03389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McAllister L, Rehm EJ, Goodman GS, Zinn K. Alternative splicing of micro-exons creates multiple forms of the insect cell adhesion molecule fasciclin I. J Neurosci. 1992;12:895–905. doi: 10.1523/JNEUROSCI.12-03-00895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volfovsky N, Haas BJ, Salzberg SL. Computational discovery of internal micro-exons. Genome Res. 2003;13:1216–1221. doi: 10.1101/gr.677503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallieres M, Tapial J, Raj B, O’Hanlon D, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YI, Sanchez-Pulido L, Haerty W, Ponting CP. RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome Res. 2015;25:1–13. doi: 10.1101/gr.181990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci U S A. 2015;112:3445–3450. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sterner DA, Carlo T, Berget SM. Architectural limits on split genes. Proc Natl Acad Sci U S A. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlo T, Sterner DA, Berget SM. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA. 1996;2:342–353. [PMC free article] [PubMed] [Google Scholar]
- 33.Carlo T, Sierra R, Berget SM. A 5′ splice site-proximal enhancer binds SF1 and activates exon bridging of a microexon. Mol Cell Biol. 2000;20:3988–3995. doi: 10.1128/mcb.20.11.3988-3995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 42.Sun S, Zhang Z, Fregoso O, Krainer AR. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA. 2012;18:274–283. doi: 10.1261/rna.030486.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 46.Tsyba L, Skrypkina I, Rynditch A, Nikolaienko O, Ferenets G, Fortna A, Gardiner K. Alternative splicing of mammalian Intersectin 1: domain associations and tissue specificities. Genomics. 2004;84:106–113. doi: 10.1016/j.ygeno.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Tsyba L, Gryaznova T, Dergai O, Dergai M, Skrypkina I, Kropyvko S, Boldyryev O, Nikolaienko O, Novokhatska O, Rynditch A. Alternative splicing affecting the SH3A domain controls the binding properties of intersectin 1 in neurons. Biochem Biophys Res Commun. 2008;372:929–934. doi: 10.1016/j.bbrc.2008.05.156. [DOI] [PubMed] [Google Scholar]
- 48.Ohnishi T, Shirane M, Hashimoto Y, Saita S, Nakayama KI. Identification and characterization of a neuron-specific isoform of protrudin. Genes Cells. 2014;19:97–111. doi: 10.1111/gtc.12109. [DOI] [PubMed] [Google Scholar]
- 49.Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, et al. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- 51.Quesnel-Vallieres M, Dargaei Z, Irimia M, Gonatopoulos-Pournatzis T, Ip JY, Wu M, Sterne-Weiler T, Nakagawa S, Woodin MA, Blencowe BJ, et al. Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol Cell. 2016;64:1023–1034. doi: 10.1016/j.molcel.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 52.Quesnel-Vallieres M, Irimia M, Cordes SP, Blencowe BJ. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 2015;29:746–759. doi: 10.1101/gad.256115.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamiguchi H, Lemmon V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J Neurosci. 1998;18:3749–3756. doi: 10.1523/JNEUROSCI.18-10-03749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamiguchi H, Long KE, Pendergast M, Schaefer AW, Rapoport I, Kirchhausen T, Lemmon V. The neural cell adhesion molecule L1 interacts with the AP-2 adaptor and is endocytosed via the clathrin-mediated pathway. J Neurosci. 1998;18:5311–5321. doi: 10.1523/JNEUROSCI.18-14-05311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Angelis E, Brummendorf T, Cheng L, Lemmon V, Kenwrick S. Alternative use of a mini exon of the L1 gene affects L1 binding to neural ligands. J Biol Chem. 2001;276:32738–32742. doi: 10.1074/jbc.M105156200. [DOI] [PubMed] [Google Scholar]
- 56.Jacob J, Haspel J, Kane-Goldsmith N, Grumet M. L1 mediated homophilic binding and neurite outgrowth are modulated by alternative splicing of exon 2. J Neurobiol. 2002;51:177–189. doi: 10.1002/neu.10052. [DOI] [PubMed] [Google Scholar]
- 57.Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S. X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet. 1994;7:402–407. doi: 10.1038/ng0794-402. [DOI] [PubMed] [Google Scholar]
- 58.Gaggero A, De Ambrosis A, Mezzanzanica D, Piazza T, Rubartelli A, Figini M, Canevari S, Ferrini S. A novel isoform of pro-interleukin-18 expressed in ovarian tumors is resistant to caspase-1 and -4 processing. Oncogene. 2004;23:7552–7560. doi: 10.1038/sj.onc.1208036. [DOI] [PubMed] [Google Scholar]
- 59.Whiting P, McKernan RM, Iversen LL. Another mechanism for creating diversity in gamma-aminobutyrate type A receptors: RNA splicing directs expression of two forms of gamma 2 phosphorylation site. Proc Natl Acad Sci U S A. 1990;87:9966–9970. doi: 10.1073/pnas.87.24.9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moss SJ, Doherty CA, Huganir RL. Identification of the cAMP-dependent protein kinase and protein kinase C phosphorylation sites within the major intracellular domains of the beta 1, gamma 2S, and gamma 2L subunits of the gamma-aminobutyric acid type A receptor. J Biol Chem. 1992;267:14470–14476. [PubMed] [Google Scholar]
- 61.Krishek BJ, Xie X, Blackstone C, Huganir RL, Moss SJ, Smart TG. Regulation of GABAA receptor function by protein kinase C phosphorylation. Neuron. 1994;12:1081–1095. doi: 10.1016/0896-6273(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 62.Huntsman MM, Tran BV, Potkin SG, Bunney WE, Jr, Jones EG. Altered ratios of alternatively spliced long and short gamma2 subunit mRNAs of the gamma-amino butyrate type A receptor in prefrontal cortex of schizophrenics. Proc Natl Acad Sci U S A. 1998;95:15066–15071. doi: 10.1073/pnas.95.25.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rusconi F, Paganini L, Braida D, Ponzoni L, Toffolo E, Maroli A, Landsberger N, Bedogni F, Turco E, Pattini L, et al. LSD1 neurospecific alternative splicing controls neuronal excitability in mouse models of epilepsy. Cereb Cortex. 2015;25:2729–2740. doi: 10.1093/cercor/bhu070. [DOI] [PubMed] [Google Scholar]
- 64.Rusconi F, Grillo B, Ponzoni L, Bassani S, Toffolo E, Paganini L, Mallei A, Braida D, Passafaro M, Popoli M, et al. LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A. 2016;113:3651–3656. doi: 10.1073/pnas.1511974113. [DOI] [PMC free article] [PubMed] [Google Scholar]