Abstract
Cyclic adenosine monophosphate (cAMP) response element–binding protein (CREB)–binding protein (CBP) is a histone acetyltransferase that plays a pivotal role in the control of histone modification and the expression of cytokine-encoding genes in inflammatory diseases, including sepsis and lung injury. We found that the E3 ubiquitin ligase subunit FBXL19 targeted CBP for site-specific ubiquitylation and proteasomal degradation. The ubiquitylation-dependent degradation of CBP reduced the extent of lipopolysaccharide (LPS)–dependent histone acetylation and cytokine release in mouse lung epithelial cells and in a mouse model of sepsis. Furthermore, we demonstrated that the deubiquitylating enzyme USP14 (ubiquitin-specific peptidase 14) stabilized CBP by reducing its ubiquitylation. LPS increased the stability of CBP by reducing the association between CBP and FBXL19 and by activating USP14. Inhibition of USP14 reduced CBP protein abundance and attenuated LPS-stimulated histone acetylation and cytokine release. Together, our findings delineate the molecular mechanisms through which CBP stability is regulated by FBXL19 and USP14, which results in the modulation of chromatin remodeling and the expression of cytokine-encoding genes.
INTRODUCTION
Cyclic adenosine monophosphate (cAMP) response element–binding protein (CREB)–binding protein (CBP), a member of the family of histone acetyltransferases (HATs), catalyzes histone acetylation, thereby switching chromatin from the closed state to the open state. This change promotes the binding of RNA polymerase II and basal transcription factors to the open DNA to initiate transcription. The acetylation of histone H4 is correlated with increased expression of genes encoding proinflammatory factors, such as interleukin-8 (IL-8), IL-6, and tumor necrosis factor–α(TNF-α) (1–4). Multiple studies have attempted to investigate the regulation of CBP stability to better understand the molecular basis of its action. Ubiquitylation is a posttranslational modification that alters protein stability. Proteins conjugated to Lys48 (K48)–linked ubiquitin chains are degraded in the proteasome, whereas K63-linked polyubiquitylation triggers protein degradation in the lysosome (5). Siah1 and Siah2 (collectively known as Siah1/2) are E3 ubiquitin ligases that ubiquitylate CBP, leading to its degradation and thereby reducing the extent of CBP-mediated acetylation of p53 (6). However, much less is known about the effect of the Siah proteins on CBP-catalyzed histone acetylation. Sen and Snyder demonstrated that Siah facilitates the acetylation of histone H3 and enhances the expression of CREB-regulated genes (7). These studies suggest that Siah1/2 specifically dampens the CBP that regulates p53 acetylation but not the CBP that modulates histones. In addition, neither Siah1 nor Siah2 is detectable in normal lung tissues (8). To reduce CBP activity in inflammatory diseases, such as lung injury, there must be alternative E3 ubiquitin ligases that target CBP for ubiquitylation and degradation to inhibit its effects on histone acetylation.
The Skp1–Cul1–F-box protein (SCF) ligase complex is one of the largest among the family of E3 ubiquitin ligases. The F-box protein connects the ligase complex and specific substrates through its F-box domain and substrate-binding motif, respectively (9–11). A previously uncharacterized F-box protein, FBXL19, targets the IL-33 receptor ST2L (12) and small guanosine triphosphatases (GTPases) (13–15) for ubiquitylation and proteasomal degradation. We previously showed that FBXL19 reduces inflammatory responses, such as cytokine release, in a mouse model of acute lung injury (12), suggesting that FBXL19 targets an unidentified substrate that plays a pivotal role in regulating the expression of cytokine-encoding genes. Protein ubiquitylation can be reversed by deubiquitylating enzymes (DUBs), which enhance protein stability. Several DUBs [for example, USP3 (ubiquitin-specific peptidase 3), USP22, and Mysm1] remove ubiquitin moieties from ubiquitylated histone H2A proteins (16–18). The HAT P/CAF physically interacts with the DUB Mysm1 (19); however, whether DUBs play a role in the regulation of HAT stability is unclear. We previously showed that USP14 plays a proinflammatory role by enhancing the activity of the transcription factor nuclear factor κB (NF-κB) (20). Xu and Guo showed that inhibition of USP14 by the small-molecule inhibitor IU1 prevents ventilator-induced lung injury in rats (21). Together, these findings suggest that USP14 exhibits proinflammatory properties.
Here, we demonstrated that FBXL19 targets CBP for site-specific ubiquitylation and degradation. FBXL19 functioned as an anti-inflammatory E3 ubiquitin ligase subunit by reducing the abundance of CBP protein, which led to decreased histone acetylation and reduced cytokine release. Together, these data suggest that an E3 ubiquitin ligase of the SCF family ubiquitylates a HAT, thereby leading to its degradation and the reduction of histone acetylation. Furthermore, we revealed that USP14 deubiquitylated CBP and promoted its stability. Inhibition of USP14 decreased the amount of CBP protein, attenuated lipopolysaccharide (LPS)–stimulated histone acetylation and cytokine release, and lessened systemic and lung inflammation in mice.
RESULTS
FBXL19 targets CBP for ubiquitylation and degradation
It is well known that CBP stability is regulated by the ubiquitin-proteasome system; however, the molecular control of CBP stability in the setting of inflammatory responses is unclear, and the ubiquitin acceptor site has not been identified. Here, in experiments with the protein synthesis inhibitor cycloheximide (CHX), we confirmed that CBP was unstable, with a half-life of ~3.4 hours (Fig. 1, A and B). Lysine residues are commonly the targets for ubiquitylation. We found that a lysine-to-arginine mutant of CBP (CBPK2103R) had an extended half-life compared to that of the wild-type (WT) protein (Fig. 1, A and B), suggesting that Lys2103 was the potential ubiquitylation site within CBP. These data are consistent with those from a previous proteomics analysis (22).
Fig. 1. FBXL19 mediates site-specific CBP ubiquitylation and degradation.
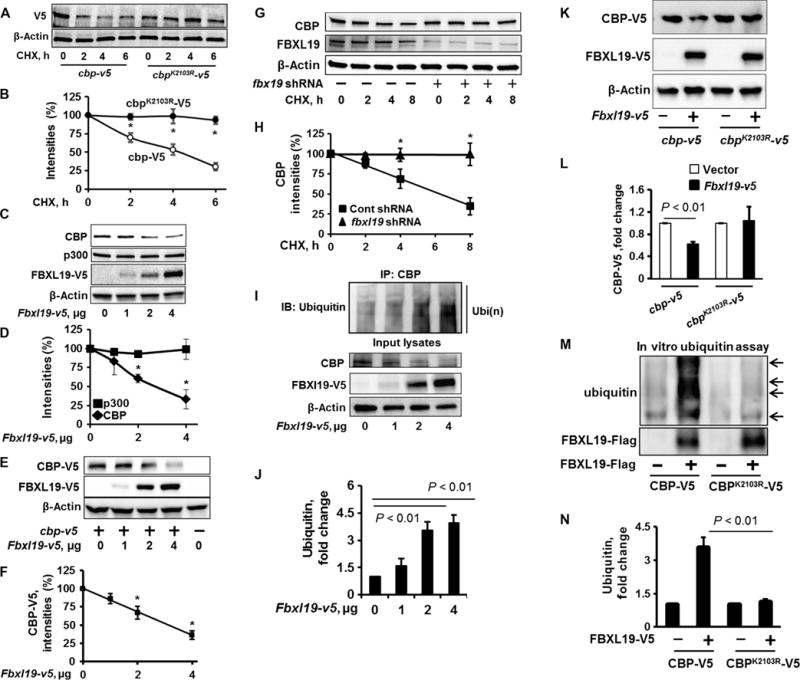
(A and B) MLE12 cells were transfected with the cbp-v5 or cbpK2103R-v5 plasmids. Forty-eight hours later, the cells were treated with CHX (20 μg/ml) for 0, 2, 4, or 6 hours. (A) Cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. (B) Determination of relative protein abundance by densitometric analysis of Western blots with ImageJ software. (B) Data are means ± SEM of three independent experiments. *P < 0.01 by two-way analysis of variance (ANOVA) and post hoc Tukey’s test. (C and D) MLE12 cells were transfected with the indicated amounts of the Fbxl19-v5 plasmid. (C) Forty-eight hours later, cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. (D) The relative amounts of p300 and CBP proteins were determined by densitometric analysis. Data are means ± SEM of three independent experiments. *P < 0.01 by two-way ANOVA and post hoc Tukey’s test. Comparison was made to the amount of CBP in cells that were not transfected with the Fbxl19-v5 plasmid. (E and F) MLE12 cells were transfected with the cbp-v5 and Fbxl19-v5 plasmids as indicated. (E) Forty-eight hours later, cell lysates were analyzed by Western blotting with antibodies against V5 and β-actin. (F) The relative amounts of CBP-V5 protein were determined by densitometric analysis. Data are means ± SEM of three independent experiments. *P < 0.01 by one-way ANOVA and post hoc Tukey’s test. Comparison was made to cells that were not transfected with the Fbxl19-v5 plasmid. (G and H) MLE12 cells were transfected with control (Cont) shRNA (−) or fbxl19-specific shRNA (+), and then cells were treated for the indicated times with CHX (20 μg/ml). (G) Cell lysates were analyzed by Western blotting with antibodies against CBP, FBXL19, and β-actin. (H) The relative amounts of CBP protein were determined by densitometric analysis. Data are means ± SEM of three independent experiments. *P < 0.01 by two-way ANOVA and post hoc Tukey’s test. Comparison was made to cells treated with control shRNA. (I and J) MLE12 cells were transfected with the indicated amounts of the Fbxl19-v5 plasmid. Forty-eight hours later, cell lysates were subjected to immunoprecipitation (IP) with an anti-CBP antibody and Western blotting analysis with an anti-ubiquitin antibody. Input lysates were analyzed by Western blotting with antibodies against CBP, V5, and β-actin. (J) The relative amounts of ubiquitylated CBP were determined by densitometric analysis. Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. Comparison was made to cells treated with control shRNA. IB, immunoblotting. Ubi(n), polyubiquitination. (K and L) MLE12 cells were transfected with the Fbxl19-v5, cbp-v5, or cbpK2103R-v5 plasmid, as indicated. (K) Cell lysates were then analyzed by Western blotting with antibodies against V5 and β-actin. (L) The relative amounts of CBP-V5 were determined by densitometric analysis. Data are means ± SEM of three independent experiments. P values were determined by the unpaired Student’s t test. (M and N) Troponin T (TnT)–synthesized CBP-V5 or CBPK2103R-V5 were incubated with a full complement of ubiquitylation reaction components with (+) or without (−) purified Flag-tagged FBXL19. (M) Reaction mixtures were subjected to Western blotting analysis with antibodies against ubiquitin and the Flag tag. The relative amounts of ubiquitylated CBP-V5 and CBPK2103R-V5 were determined by densitometric analysis. Data are means ± SEM of three independent experiments. P values were determined by one-way ANOVA and post hoc Tukey’s test. All Western blots are representative of at least three independent experiments.
FBXL19 is a subunit of an SCF family E3 ubiquitin ligase, which exhibits an anti-inflammatory property (23); however, its substrates related to cytokine gene expression have not been identified. We tested whether FBXL19 regulated the protein stability of HATs. Ectopically expressed FBXL19 reduced the abundances of endogenous and overexpressed CBP proteins (Fig. 1, C to F, and fig. S1), without altering p300 abundance (Fig. 1, C and D); however, knockdown of FBXL19 with Fbxl19-specific short hairpin RNA (shRNA) enhanced the stability of CBP (Fig. 1, G and H). In addition, overexpression of FBXL19 resulted in a reduction in the amount of nuclear CBP, but not that of cytoplasmic CBP (fig. S2, A and B).
To investigate whether FBXL19 transferred ubiquitin chains to CBP, MLE12 cells were transfected with the Fbxl19-v5 plasmid, and then the ubiquitylation state of CBP was determined. We found that overexpression of FBXL19 led to the increased polyubiquitylation of CBP (Fig. 1, I and J). The FBXL19-mediated ubiquitylation of CBP was K48-linked, but not K63-linked (fig. S3). The CBPK2103R mutant protein was resistant to degradation in response to the overexpression of FBXL19 (Fig. 1, K and L) and was not subjected to polyubiquitylation (Fig. 1, M and N). The mutation of specific lysine residues in CBP had no effect on its cellular localization (fig. S2C). These data suggest that FBXL19 induces the site-specific ubiquitylation of CBP and its subsequent degradation.
LPS increases CBP stability through the disassociation of the FBXL19-CBP complex and a reduction in the abundance of FBXL19
CBP-mediated histone acetylation plays a crucial role in LPS-induced inflammatory responses (24–26). We examined the effect of LPS on CBP abundance and found that the treatment of MLE12 cells with LPS increased the amount of CBP protein in a time-dependent manner (Fig. 2, A and B). The effect of LPS was likely a result of an increase in CBP stability, because LPS did not substantially affect the abundance of Cbp mRNA (Fig. 2C). As a positive control, LPS increased the abundance of kc mRNA in MLE12 cells (Fig. 2D). To determine the molecular mechanism by which LPS regulated CBP stability, we examined the effect of LPS on CBP ubiquitylation. LPS reduced both the extent of CBP ubiquitylation (Fig. 2, E and F) and the association between CBP and FBXL19 at 3 hours (Fig. 2, G and H). Furthermore, the LPS-induced increase in CBP abundance was attenuated in cells transfected with plasmid expressing FBXL19 (Fig. 2, I and J). Together, these data suggest that LPS increased CBP abundance through dissociation of CBP from FBXL19 at early times, which led to a reduction in the extent of CBP ubiquitylation. We found that LPS increased the extent of serine phosphorylation of CBP at 3 hours (fig. S4A). We previously showed that the phosphorylation of substrates promotes their binding to E3 ubiquitin ligases (23). Thus, the phosphorylation of CBP may inhibit its interaction with FBXL19.
Fig. 2. FBXL19 dampens the LPS-stimulated increase in CBP protein abundance.
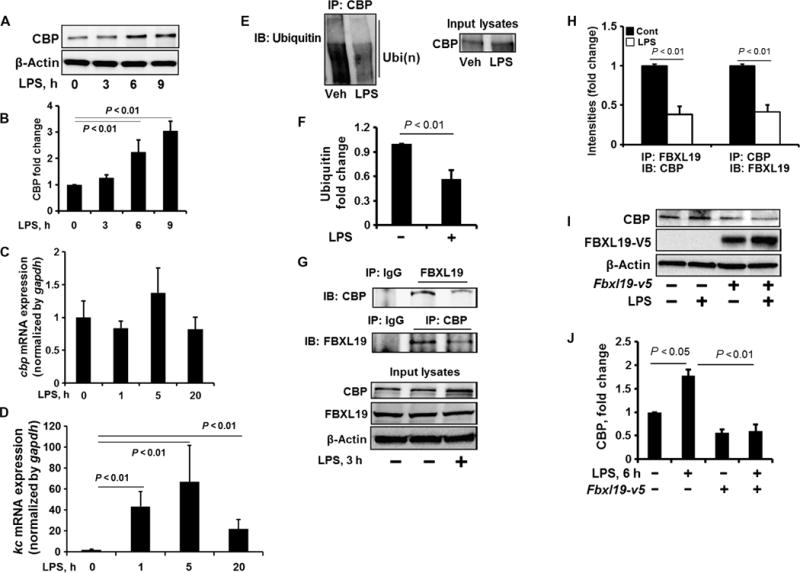
(A and B) MLE12 cells were treated with LPS (10 μg/ml) for 0, 3, 6, and 9 hours. (A) Cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. (B) Determination of relative CBP protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. (C and D) MLE12 cells were treated with LPS (10 μg/ml) for 1, 5, or 20 hours. Total RNAs were extracted and cbp (C) and kc (D) mRNA abundances were measured by reverse transcription polymerase chain reaction (RT-PCR). Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. gapdh, glyceraldehyde-3-phosphate dehydrogenase. (E and F) MLE12 cells were treated with LPS (10 μg/ml, 3 hours). (E) Cell lysates were subjected to immunoprecipitation with an anti-CBP antibody and Western blotting with an anti-ubiquitin antibody. Input lysates were analyzed by Western blotting with an anti-CBP antibody. (F) The relative amounts of ubiquitylated CBP were determined by densitometric analysis. Data are means ± SEM of three independent experiments. P values were calculated by unpaired Student t test. (G and H) MLE12 cells were treated with LPS (10 μg/ml, 3 hours). (G) Cell lysates were subjected to immunoprecipitation with immunoglobulin G (IgG) or with antibody against FBXL19 or CBP, followed by Western blotting with antibodies against CBP and FBXL19. (H) The relative amounts of CBP and FBXL19 were determined by densitometric analysis. Data are means ± SEM of three independent experiments. P values were calculated by unpaired Student t test. (I and J) MLE12 cells were transfected with the Fbxl19-v5 plasmid. Forty-eight hours later, the cells were treated with LPS (10 μg/ml, 6 hours). (I) Cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. (J) Determination of relative protein abundance by densitometric analysis of Western blots with ImageJ software. (J) Data are means ± SEM of three independent experiments. P values were determined by one-way ANOVA and post hoc Tukey’s test. All Western blots are representative of at least three independent experiments.
To investigate whether FBXL19 abundance was changed under inflammatory conditions, we measured the relative amounts of FBXL19 in the lungs of mice 24 hours after they were intratracheally challenged with Pseudomonas aeruginosa (strain PA103). We found that abundance of FBXL19 protein in the lungs of infected mice was reduced compared to that in the lungs of uninfected mice (Fig. 3, A and B). To confirm these data, we examined the amount of FBXL19 protein in LPS-treated MLE12 cells. We found that LPS decreased the abundance of FBXL19 in a time-dependent manner (Fig. 3, C and D) and that these effects were attenuated by the proteasomal inhibitor MG132 (N-carbobenzyloxy-L-leucyl-L-leucyl-L-leucinal), but not the lysosomal inhibitor leupeptin (Fig. 3, C and D), which suggests that LPS stimulates the proteasomal degradation of FBXL19. Furthermore, we found that LPS increased the extent of phosphorylation (fig. S4B) and K48-linked polyubiquitylation of FBXL19 (Fig. 3, E and F), without substantially altering the abundance of fbxl19 mRNA (Fig. 3G), suggesting that LPS-induced FBXL19 degradation was mediated through ubiquitylation. It is well known that the phosphorylation of substrate can stimulate protein ubiquitylation (27). Thus, we hypothesized that phosphorylation may be essential for FBXL19 ubiquitylation. We found that overexpression of V5-tagged FBXL19 had no effect on the abundance of myc-tagged FBXL19 (fig. S5), which suggests that the degradation of FBXL19 was not a result of auto-ubiquitylation and degradation by its own activity. Together, these data suggest that LPS increases CBP protein abundance by inhibiting the FBXL19-CBP association at early times and reducing FBXL19 protein abundance at later times, thereby increasing the stability of CBP.
Fig. 3. LPS decreases FBXL19 stability.
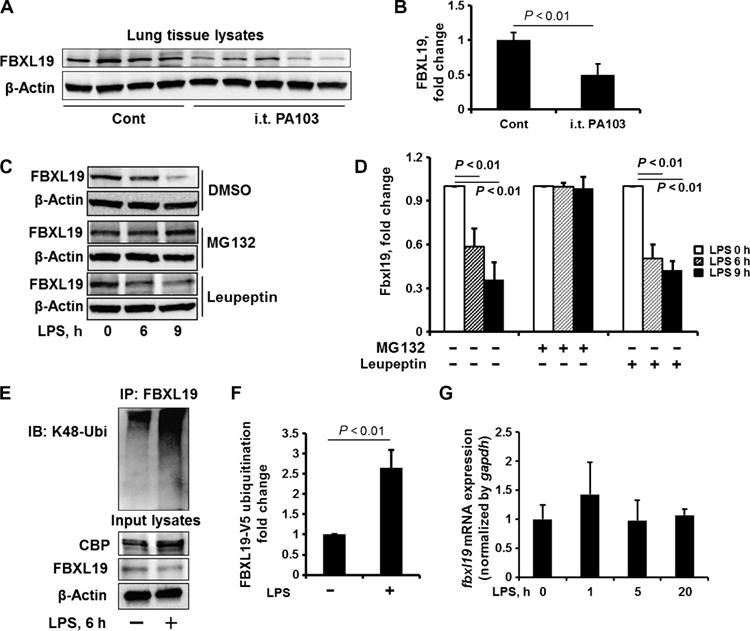
(A and B) C57/BL6 mice were intratracheally administered with P. aeruginosa strain PA103 (104 colony-forming units per mouse). (A) Twenty-four hours later, lung tissues were analyzed by Western blotting with antibodies against FBXL19 and β-actin. i.t., intratracheal. (B) The relative amounts of FBXL19 were determined by densitometric analysis with ImageJ software. Data are means ± SEM of 6 (control) and 11 (PA103) mice. P values were calculated by unpaired Student t test. (C and D) MLE12 cells were treated with DMSO (dimethyl sulfoxide), MG132 (20 μM), or leupeptin (100 μM) for 1 hour before they were treated with LPS (10 μg/ml) for an additional 0, 6, or 9 hours. (C) Cell lysates were analyzed by Western blotting with antibodies against FBXL19 and β-actin. (D) The relative amounts of FBXL19 were determined by densitometric analysis with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (E and F) HBEpCs were treated with LPS (10 μg/ml) for 6 hours. (E) Cell lysates were subjected to immunoprecipitation with an anti-FBXL19 antibody and Western blotting with an anti–K48-ubiquitin antibody. Input lysates were analyzed by Western blotting with antibodies against CBP, FBXL19, and β-actin. (F) The relative amounts of FBXL19 were determined by densitometric analysis with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by unpaired Student t test. (G) MLE12 cells were treated with LPS (10 μg/ml) for 1, 5, or 20 hours. Total RNAs were extracted and fbxl19 mRNA abundance was measured by RT-PCR analysis. Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. All Western blots are representative of at least three independent experiments.
FBXL19 inhibits histone acetylation and cytokine production in lung epithelial cells and a mouse model of sepsis
Histone acetylation is an essential step to control the expression of cytokine-encoding genes in response to LPS (28–30). To investigate whether FBXL19 affected histone acetylation, FBXL19 and CBP were overexpressed in lung epithelial cells. Overexpression of CBP alone slightly increased the extent of acetylation of histone H4 at Lys8 (H4K8ac), whereas the effect was attenuated in cells in which FBXL19 was overexpressed (Fig. 4, A and B). LPS stimulated the acetylation of histone H4K8 in lung epithelial cells (Fig. 4, C and D); however, the effects were attenuated by the expression of FBXL19, whereas H4K8 acetylation was promoted by a reduction in the abundance of FBXL19 (Fig. 4, E and F). Furthermore, we found that FBXL19 reduced the extent of acetylation of histone H4K12 (fig. S6, A and B) and the binding of acetylated H4K8 to the Il8 promoter (fig. S7) in response to LPS. These data suggest that FBXL19 reduced histone acetylation through the destabilization of CBP. We then examined the effects of FBXL19 on cytokine release by lung epithelial cells. The knockdown of FBXL19 with fbxl19-specific shRNA promoted the LPS-stimulated release of the cytokine KC (keratinocyte-derived cytokine) by murine lung epithelial cells (Fig. 4G). Overexpression of FBXL19 attenuated the LPS-induced production of IL-8 by human lung epithelial cells overexpressing WT CBP, but not CBPK2103R (Fig. 4H), suggesting that the FBXL19 exerted its anti-inflammatory properties by reducing CBP stability and the extent of histone acetylation. These data reveal previously uncharacterized biological functions for FBXL19 and suggest that FBXL19 limits LPS-induced histone acetylation and cytokine release.
Fig. 4. The reduction of CBP stability by FBXL19 impairs histone acetylation and cytokine release.
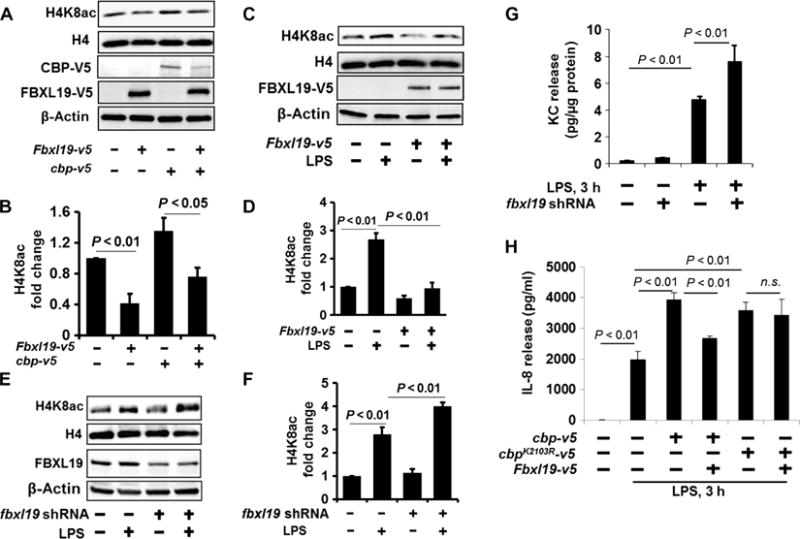
(A and B) MLE12 cells were transfected with cbp-v5 and Fbxl19-v5 plasmids. (A) Forty-eight hours later, isolated histone lysates were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. Cell lysates were analyzed by Western blotting with antibodies against V5 and β-actin. (B) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (C and D) HBEpCs were transfected with the Fbxl19-v5 plasmid, and then cells were treated with LPS (10 μg/ml, 3 hours). (C) Isolated histone lysates were analyzed by Western blotting with antibodies against H4K8ac and H4. (D) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (E and F) MLE12 cells transfected with fbxl19 shRNA (shfbxl19) plasmid. Seventy-two hours later, cells were treated with LPS (10 μg/ml) for 3 hours. (E) Isolated histone lysates were analyzed by Western blotting with antibodies against H4K8ac and H4. (F) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (G) MLE12 cells transfected with or without the fbxl19 shRNA plasmid. Seventy-two hours later, cells were treated with LPS (10 μg/ml) for 3 hours. KC release was measured by enzyme-linked immunosorbent assay (ELISA). Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (H) HBEpCs were transfected with the Fbxl19-v5, cbp-v5, or cbpK2103R-V5 plasmid as indicated. Forty-eight hours later, cells were treated with LPS (10 μg/ml, 3 hours). The amount of IL-8 secreted into the medium was measured by ELISA. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. All Western blots are representative of at least three independent experiments. n.s., not significant.
We previously showed that overexpression of FBXL19 in mouse lungs attenuates intratracheally instilled, LPS-induced cytokine release into the bronchoalveolar lavage (BAL) fluid (BALF) (12). To investigate whether FBXL19 lessened systemic inflammation, we intravenously injected a lentiviral vector expressing Fbxl19 into mice before they were subjected to cecal ligation and puncture (CLP), a well-established murine model of sepsis. CLP increased the acetylation of histone H4K8 in mouse lungs in a time-dependent manner (Fig. 5, A and B), whereas these effects were reduced in the lungs of mice overexpressing FBXL19 (Fig. 5, C and D). CLP-induced KC release in plasma and BALFs and myeloperoxidase activity (a marker of lung inflammation) in the lungs were reduced in mice infected with lentivirus expressing Fbxl19 (Fig. 5, E to G). These data suggest that FBXL19 exhibits an anti-inflammatory effect in murine models of both local and systemic inflammatory diseases.
Fig. 5. FBXL19 mitigates histone acetylation and inflammatory responses in a CLP-induced murine model of sepsis.
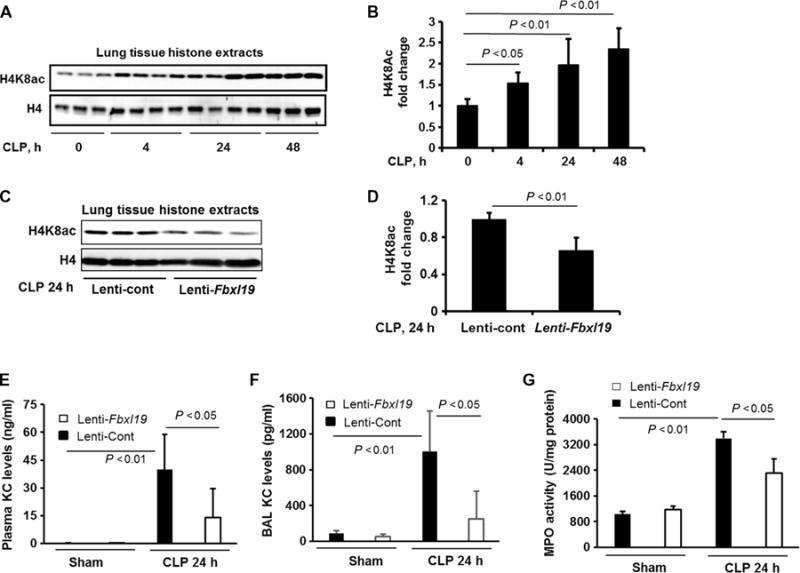
(A and B) C57/BL6 mice were subjected to CLP for 4, 24, or 48 hours. (A) Isolated histone lysates of lung tissues were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. (B) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of six to eight samples per group. P values were calculated by one-way ANOVA and post hoc Tukey’s test. (C and D) C57/BL6 mice were intravenously infected with control lentivirus (Lenti-cont) or lentivirus encoding Fbxl19 (Lenti-Fbxl19) before they were subjected to CLP. (C) Twenty-four hours later, isolated histone lysates of lung tissues were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. (D) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of 11 samples per group. P values were calculated by unpaired Student t test. (E and F) Plasma and BAL KC concentrations were measured by ELISA. Data are means ± SEM of five to eight samples per group. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (G) Myeloperoxidase (MPO) activity in lung tissues was measured by ELISA. Data are means ± SEM of five to eight samples per group. P values were calculated by two-way ANOVA and post hoc Tukey’s test.
USP14 deubiquitylates and stabilizes CBP
Deubiquitylation, the reverse process of ubiquitylation, is enacted by DUBs, which control protein stability, localization, and enzyme activity. We previously showed that USP14 decreases the stability of inhibitor of NF-κB (IκB) protein (20), indicating that USP14 promotes inflammatory responses. Here, we tested whether USP14 regulated CBP stability, because CBP plays a critical role in proinflammatory responses. We found that overexpression of USP14 resulted in the increased abundance of CBP, without altering the abundance of p300 (Fig. 6, A and B). Knockdown of USP14 with usp14-specific shRNA (Fig. 6, C and D) or inhibition of USP14 activity with IU1 (Fig. 6, E and F) resulted in reduced CBP abundance. Furthermore, pretreatment of MLE12 cells with IU1 attenuated the LPS-dependent increase in CBP protein abundance (Fig. 6, G and H). To investigate whether USP14 deubiquitylated CBP, we transfected MLE12 cells to overexpress USP14 or treated them with IU1, and then we examined the ubiquitylation of CBP. We found that overexpression of USP14 reduced CBP ubiquitylation, whereas IU1 had the opposite effect (Fig. 6, I and J). Furthermore, coimmunoprecipitation studies showed an association between USP14 and CBP (Fig. 6K), which was unaffected by LPS (Fig. 6L). These data suggest that USP14 stabilizes CBP through the deubiquitylation of CBP.
Fig. 6. USP14 deubiquitylates and stabilizes CBP.
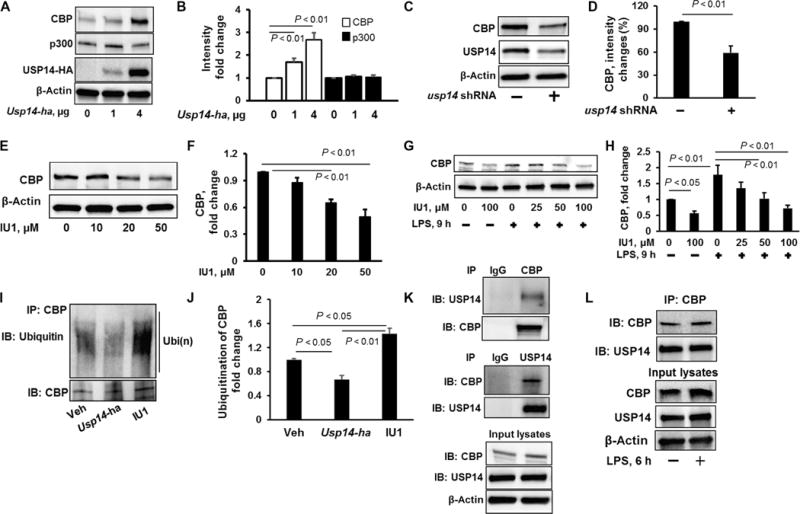
(A and B) MLE12 cells were transfected with the indicated amounts of Usp14-ha plasmid. (A) Forty-eight hours later, cell lysates were analyzed by Western blotting with antibodies against indicated proteins. (B) Determination of relative CBP and p300 protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (C and D) MLE12 cells were transfected with Cont shRNA or usp14 shRNA. (C) Seventy-two hours later, cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. (D) Determination of relative CBP protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by unpaired Student’s t test. (E and F) MLE12 cells were treated with IU1 (0, 10, 20, or 50 μM). (E) Twenty-four hours later, cell lysates were analyzed by Western blotting with antibodies against CBP and β-actin. (F) Determination of relative CBP protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. (G and H) MLE12 cells were treated with increasing concentrations of IU1 for 16 hours before they were treated with LPS for a further 9 hours. (G) Cell lysates were analyzed by Western blotting with antibodies against CBP and β-actin. (H) Determination of relative CBP protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. (I and J) MLE12 cells were transfected with Usp14-ha plasmid for 48 hours or were pretreated with IU1 (50 μM) for 4 hours. (I) Cells were then subjected to immunoprecipitation with an anti-CBP antibody and Western blotting analysis with antibodies against ubiquitin and CBP. Veh, vehicle. (J) The relative amounts of ubiquitylated CBP were determined by densitometric analysis with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by one-way ANOVA and post hoc Tukey’s test. (K) Top: MLE12 cell lysates were subjected to immunoprecipitation with control IgG or an anti-CBP antibody, followed by Western blotting with antibodies against USP14 and CBP. Middle: MLE12 cells were subjected to immunoprecipitation with control IgG or an anti-USP14 antibody, followed by Western blotting with antibodies against CBP and USP14. Bottom: Input lysates were analyzed by Western blotting with antibodies against CBP, USP14, and β-actin. (L) HBEpCs were treated with LPS (10 μg/ml, 6 hours), and cell lysates were subjected to immunoprecipitation with an anti-CBP antibody, followed by Western blotting with antibodies against CBP and USP14. Input lysates were analyzed by Western blotting with antibodies against CBP, USP14, and β-actin. All Western blots are representative of at least three independent experiments.
Inhibition of USP14 attenuates LPS-induced histone acetylation and cytokine release and lessens systemic and lung inflammation
We next found that LPS increased the deubiquitylating activity of USP14 and confirmed the inhibitory effect of IU1 on USP14 activity (Fig. 7A). Because USP14 stabilized CBP, we investigated whether USP14 promoted CBP-mediated histone acetylation. The LPS-induced acetylation of histone H4K8 was enhanced in MLE12 cells overexpressing USP14 (Fig. 7, B and C), whereas knockdown of USP14 with usp14-specific shRNA attenuated the acetylation of histone H4K8 (Fig. 7, D and E). Furthermore, transfection of MLE12 cells with usp14-specific shRNA (Fig. 8A) or treatment with IU1 (Fig. 8B and Fig. S8, A and B) attenuated the LPS-induced release of KC and IL-6 by MLE12 cells and RAW264.7 cells (a mouse macrophage cell line). The anti-inflammatory effect of IU1 was also confirmed in experiments with LPS-treated peripheral blood mononuclear cells (fig. S8C).
Fig. 7. Inhibition of USP14 impairs histone acetylation.
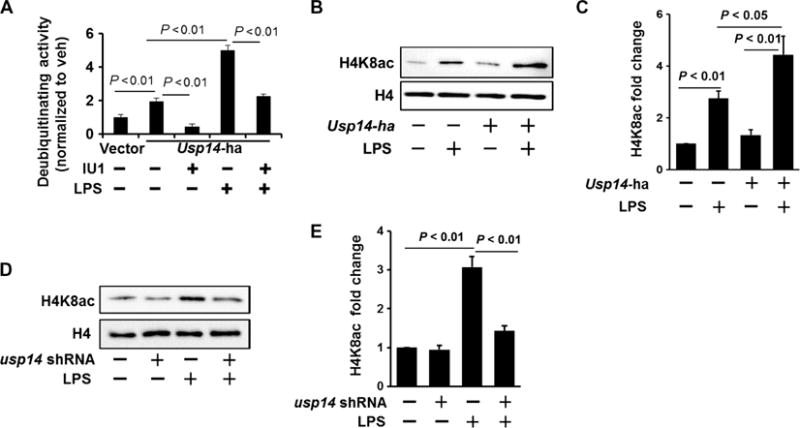
(A) MLE12 cells were transfected with Usp14-ha plasmid. Forty-eight hours later, cells were treated with LPS (10 μg/ml, 1 hour). USP14-HA (hemagglutinin) was immunoprecipitated with an anti-HA antibody, and then the immunoprecipitated complex was incubated with or without IU1. A deubiquitylation assay was performed according to the manufacturer’s instructions. Deubiquitylating activity was normalized by the control. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (B and C) MLE12 cells were transfected with Usp14-ha plasmid. (B) Forty-eight hours later, cells were treated with LPS (10 μg/ml, 3 hours). Isolated histone lysates were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. (C) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (D and E) MLE12 cells were transfected with usp14 shRNA plasmid. (D) Seventy-two hours later, cells were treated with LPS (10 μg/ml, 3 hours). Isolated histone lysates were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. (E) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. All Western blots are representative of at least three independent experiments.
Fig. 8. Inhibition of USP14 reduces cytokine release in vitro and in vivo.
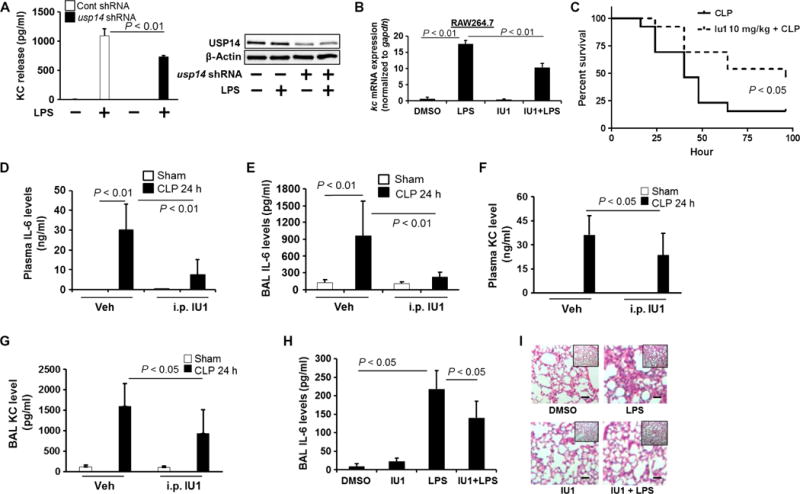
(A) MLE12 cells were transfected with Cont shRNA or usp14 shRNA before they were treated with LPS (10 μg/ml) for 6 hours. KC release was measured by ELISA. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. Cell lysates were analyzed by Western blotting with antibodies against USP14 and β-actin. (B) Raw264.7 cells were treated with IU1 (50 μM) for 16 hours before they were treated with LPS (200 ng/ml) for 3 hours. The relative abundance of kc mRNA was analyzed by real-time PCR. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (C) C57/BL6 mice were subjected to CLP, which was followed by intraperitoneal injection of IU1 (10 mg/kg body weight). Survival rates were determined for up to 100 hours (n = 13 mice per group), and P values were calculated by Mantel-Cox log-rank test. (D to G) C57/BL6 mice were subjected to CLP, which was followed by intraperitoneal (i.p.) injection of IU1 (10 mg/kg body weight). Twenty-four hours later, the concentrations of plasma IL-6 (D), BAL IL-6 (E), plasma KC (F), and BAL KC (G) were analyzed by ELISA. Data are means ± SEM of five to seven samples per group. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (H) C57/BL6 mice were subjected to intratracheal injection of LPS (5 mg/kg body weight), followed by intratracheal injection of IU1 (10 mg/kg body weight). Twenty-four hours later, the concentration of IL-6 in the BAL was measured by ELISA. Data are means ± SEM of six to eight samples per group. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (I) Lung tissues from the experiments depicted in (H) were stained with hematoxylin and eosin. Scale bars, 20 μm. Images are representative of lung tissues from six to eight mice.
To determine the anti-inflammatory effects of IU1 in murine models of inflammatory diseases, such as sepsis and lung injury, IU1 was intraperitoneally injected into mice after CLP. IU1 increased the survival rate of these mice (Fig. 8C) and reduced the concentrations of IL-6 and KC in both plasma and BALF (Fig. 8, D to G). Intratracheally administered IU1 reduced the LPS-induced increase in IL-6 abundance in the BAL (Fig. 8H) and inhibited the influx of neutrophils into the alveolar spaces (Fig. 8I). These data indicate that the inhibition of USP14 by IU1 reduces inflammatory responses by reducing CBP stability and histone acetylation.
DISCUSSION
E3 ubiquitin ligase complexes of the SCF family regulate diverse cellular functions, including inflammatory responses (11–13, 31, 32). Among the subunits of the SCF E3 ligases, the F-box protein links substrates to the E3 ligase complex. We previously showed that FBXL19 regulates the stability of the IL-33R (12) and of small GTPases, such as RhoA (14), Rac1 (13), and Rac3 (15). Here, we demonstrated that CBP is a previously uncharacterized substrate of FBXL19. We found that FBXL19 mediated the site-specific ubiquitylation of CBP, thus reducing its stability and inhibiting histone acetylation and cytokine release. Furthermore, our study revealed that LPS increased the stability of CBP by interfering with the FBXL19-CBP interaction and reducing the abundance of FBXL19. Ubiquitylation is reversed by DUBs. We found that the DUB USP14 targeted CBP for deubiquitylation, resulting in stabilization of CBP. Understanding the regulation of CBP stability in the setting of inflammatory responses is essential to identifying previously uncharacterized molecular targets that could mediate the expression of genes encoding proinflammatory cytokines. We also reported that the USP14 inhibitor IU1 reduced the stability of CBP, thereby mitigating against inflammatory pathogen-induced histone modification and cytokine release.
The acetylation of core histones plays a major role in the pathogenesis of various diseases, including inflammatory diseases (1, 2, 4, 33). CBP, one of the major members of the HAT family, facilitates chromatin remodeling, recruitment of the transcriptional machinery to the promoter region, and initiation of the expression of cytokine-encoding genes (33–35). Studies showed that the E3 ubiquitin ligases Siah1 and Siah2 mediate the ubiquitylation and degradation of CBP, thereby limiting p53 biological function (6). Polekhina et al. demonstrated that Siah1 potentiates the TNF-α–induced activation of NF-κB (36), suggesting that Siah proteins play a proinflammatory role. These data are consistent with the finding of Sen and Snyder that showed that Siah increases histone H3 acetylation and the expression of CREB-regulated genes (7). Together, these studies suggest that Siah specifically targets p53-associated CBP for its degradation, without regulating the CBP-mediated acetylation of histones and subsequent cytokine release. Here, we identified FBXL19 as the E3 ubiquitin ligase subunit responsible for the ubiquitylation and degradation of CBP. The FBXL19-mediated ubiquitylation of CBP was K48-linked, but not K63-linked.
It is common for multiple E3 ubiquitin ligases to target the same substrate under different biological conditions. Our study revealed that FBXL19 is an anti-inflammatory E3 ubiquitin ligase subunit, because it reduced the extents of histone acetylation and cytokine release in vitro and in vivo. We identified Lys2013 within CBP as a ubiquitin acceptor site for FBXL19, whereas the ubiquitin acceptor site from Siah1/2 has not been identified. It is possible that Siah1 and Siah2 target a distinct lysine residue in CBP, which may lead to a biological role distinct from that caused by modification by FBXL19. For example, the stability and activity of p53 are regulated by multiple E3 ubiquitin ligases, including Mdm2, makorin-1, RING finger protein MSL2, and E4F transcription factor 1. The ubiquitylation of p53 at different lysines by these enzymes stimulates distinct biological processes, such as degradation, nuclear export, and translocation to chromatin (37). Our data are consistent with the following pathway: LPS reduces the extent of the association between FBXL19 and CBP and reduces the abundance of FBXL19 protein, which results in increased CBP stability, histone acetylation, and cytokine release. Our study suggests that FBXL19 is a potential target for dampening CBP-modulated inflammatory diseases. To our knowledge, there is no single-nucleotide polymorphism (SNP) that affects CBP Lys2103. Future studies will focus on identifying previously uncharacterized SNPs associated with CBP stability. We showed that that lentiviral expression of FBXL19 reduced lung inflammation in a mouse model; however, the effect of FBXL19 on lung repair and remodeling has not yet been revealed. The generation of lung-specific FBXL19-transgenic mice will be useful to determine the pathogenic role of FBXL19 in lung inflammatory diseases in future studies.
USP14, one of the ubiquitin-specific peptidases, disassembles and recycles the ubiquitin chain from the distal tip of its substrates (38, 39). We previously showed that USP14 decreases the stability of IκB by interacting with the NF-κB subunit RelA (20). This finding is consistent with another study demonstrating that the inhibition of USP14 increases IκB stability in rat lungs and prevents ventilator-induced lung injury (21). Other studies also suggest that USP14 stabilizes its substrates through its deubiquitylating activity (40, 41). Here, we showed that USP14 targets and stabilizes a previously uncharacterized substrate, CBP. Furthermore, we revealed a previously uncharacterized biological function of USP14: Inhibition of USP14 reduced the stability of CBP, thereby lessening the severity of pathogen-induced lung inflammation. Previously, we showed that LPS stimulates the serine phosphorylation of USP14 (20). Here, we provided further evidence showing that LPS increases USP14 activity. Thus, it will be important to determine the role of serine phosphorylation in the regulation of USP14 activity in future studies. USP14 is a major regulator of the proteasome. Inhibition of USP14 by IU1 may cause unexpected cellular responses. This raises concerns regarding the translational potential of IU1 in treating lung inflammatory diseases.
Our study revealed the molecular mechanism of regulation of CBP stability by FBXL19 and USP14. We showed that LPS reduced the interaction between CBP and its degrader (FBXL19) as well as the abundance of FBXL19 and activated USP14, thus increasing CBP stability, enhancing histone acetylation, and promoting the expression of genes encoding proinflammatory cytokines. This study provides a previously uncharacterized therapeutic strategy to treat inflammatory disorders by inhibiting the activity of USP14 with IU1.
MATERIALS AND METHODS
Cells and reagents
Murine lung epithelial (MLE12) cells were obtained from the American Type Culture Collection and cultured with HITES (hydrocortisone, insulin, transferrin, estradiol, and selenium) medium containing 10% fetal bovine serum (FBS) and antibiotics at 37°C in an incubator containing 5% CO2. RAW264.7 cells were cultured in minimum essential medium containing 10% FBS and antibiotics. Human bronchial epithelial cells (HBEpCs, Lonza) were cultured in bronchial epithelial basal medium (BEBM) with growth factors (Lonza). The anti-V5 antibody (catalog #461157), anti-ubiquitin antibody (catalog #131600), the plasmid pcDNA3.1/His-V5-topo, and Escherichia coli Top10 competent cells were obtained from Life Technologies. Antibodies against H4K8ac (catalog #A4028) and H4K12ac (catalog #A4029) were from EpiGentek. The anti-FBXL19 antibody (catalog #AP18447b) was purchased from Abgent. CHX, leupeptin, LPS, the anti–β-actin antibody (catalog #A2228), the anti-HA tag antibody (catalog #H6908), Usp14-specific shRNA, and control shRNA were obtained from Sigma. MG132 and antibodies against H4 (catalog #05-858), p300 (catalog #05-257), and phosphoserine (catalog #05-1000) were from Millipore. The Fbxl19-specific shRNA was obtained from Open Biosystems. Immunobilized protein A/G beads, control IgG, anti-CBP (catalog #sc-369), and anti-USP14 (catalog #sc-393872) antibodies were from Santa Cruz Biotechnology. All materials used in the experiments were of the highest grades commercially available.
Transfection of cells with plasmids and shRNAs
Complementary DNAs (cDNAs) encoding WT and mutant mouse CBP were inserted into pCDNA3.1/V5-His-Topo vector (Invitrogen). Primers used to amplify cbp cDNA were as follows: forward, CACCATGGCCGAGAACTTGCTG; reverse, CAAACCCTCCACAAACTTTTCT. Briefly, cbp-encoding cDNA was synthesized by PCR using the mouse cDNA as a template. The PCR product was purified and ligated into pCDNA3.1/V5-His-Topo vector according to the manufacturer’s instructions. Site-directed mutagenesis was performed to generate cbp mutants according to the manufacturer’s instructions (Agilent Technologies). MLE12 cells were transfected with plasmids or shRNAs with the Amaxa Nucleofector system (Lonza). Lipofectamine 2000 (Life Technologies) was used to transfect HBEpCs with plasmids according to the manufacturer’s instructions.
Immunoprecipitations and Western blotting analysis
Cells grown on 35-, 60-, or 100-mm dishes were washed with cold phosphate-buffered saline (PBS), collected in cell lysis buffer, and sonicated. An equal amount of cell lysates (20 μg) was subjected to SDS–polyacrylamide gel electrophoresis, electrotransferred to membranes, and analyzed by Western blotting according to standard protocols. For immunoprecipitations, equal amounts of cell lysates (1 mg) were incubated with the appropriate antibody overnight at 4°C, which was followed by the addition of 40 μl of protein A/G agarose and incubation for an additional 2 hours at 4°C. The immunoprecipitated complex was washed three times with 0.1% Triton X-100 in ice-cold PBS and then analyzed by Western blotting.
Immunostaining
MLE12 cells were cultured in glass-bottom dishes and fixed with 3.7% paraformaldehyde for 20 min, which was followed by permeabilization in 0.1% Triton X-100 for 1 min. Cells were exposed to primary antibody, which was followed by incubation with a fluorescently labeled secondary antibody. Immunofluorescent cell imaging was performed with a Nikon A1 confocal microscope according to standard protocols.
In vitro translation of cDNAs for Cbp and WT and mutant Fbxl19
In vitro transcription and translation were performed with a TnT in vitro quick-coupled transcription-translation system (Promega) according to the manufacturer’s instructions. This mammalian-based system expresses soluble, functional proteins that are posttranslationally modified. Translated CBP protein and WT and mutant FBXL19 proteins were confirmed by Western blotting analysis.
In vitro ubiquitin conjugation assay
The ubiquitylation of CBP was performed in a reaction mixture containing synthesized WT or mutant CBP protein in 50 mM tris (pH 7.6), 5 mM MgCl2, 0.6 mM dithiothreitol (DTT), 2 mM adenosine triphosphate, E1 enzyme (1.5 ng/μl, Boston Biochem), Ubc5 (10 ng/μl), Ubc7 (10 ng/μl), ubiquitin (1 μg/μl, Boston Biochem), 1 μM ubiquitin aldehyde, and His-purified recombinant Cullin 1, Skp1, Rbx1, and synthesized FBXL19. The reaction mixture was then subjected to Western blotting analysis with anti-ubiquitin antibody.
Assay of DUB activity
MLE12 cells transfected with plasmid encoding USP14-HA or the appropriate controls were treated with LPS and then subjected to immunoprecipitation of USP14-HA with an anti-HA antibody. The DUB activity in the immunoprecipitates was analyzed with the DUB-Detector deubiquitylation assay kit (Active Motif). Briefly, the immunoprecipitated complex was incubated for 20 min in the presence or absence of 100 nM IU1. After the incubation, fluorescent ubiquitin substrate (100 nM) was added, and the fluorescence intensity of the sample was measured after 30 min with an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
Histone isolation
The EpiQuik total histone extraction kit (EpiGentek) was used to isolate core histones from the cells or tissue samples indicated in the figure legends. Cell pellets or lung tissue were lysed or disaggregated in prelysis buffer containing 400 nM trichostatin A (Cayman Chemical), which was followed by centrifugation of the samples at 10,000g for 1 min at 4°C. The supernatant was discarded and the pellet was resuspended in lysis buffer (3× volume) and incubated on ice for 30 min. After centrifugation of the samples at 12,000g for 5 min at 4°C, the supernatant was collected and neutralized by a balance buffer containing DTT (EpiGentek). Aliquots were taken for the determination of protein concentration and were analyzed by Western blotting to detect acetylated H4K8, H4K12, and H4.
Chromatin immunoprecipitation assays
Cells were cross-linked for 5 min with 0.3% formaldehyde, and the reaction was terminated by the addition of glycine. The EZ Nucleosomal DNA Prep Kit (Zymo Research) was used to isolate nuclei and prepare nucleosomal DNA. Immunoprecipitation was performed with an anti-H4K8ac antibody or with IgG as a control. Chromatin DNA was extracted with the Zymo-Spin ChIP Kit. The Il8 proximal promoter region was detected by real-time PCR with primers specific for a region within the Il8 promoter. The sequences of the PCR primers used for the analysis of the human Il8 proximal promoter were as follows: 5′-GTGTGATGACTCAGGTTTGCCC-3′ and 5′-AGTGCTCCGGTGGCTTTTTATATC-3′.
Mice
Human Fbxl19 cDNA was inserted into the pLVX-IRES-tdTomato vector (Clontech). Lentivirus expressing Fbxl19 and control virus were generated and concentrated with a lentivirus packaging system (Clontech). C57/BL6 mice were housed in a specific pathogen–free barrier facility maintained by the University of Pittsburgh Animal Resources Center. C57/BL6 mice were intravenously administered Lenti-control or Lenti-Fbxl19 (109 plaque-forming units per mouse) for 7 days before they were subjected to CLP to induce sepsis. Briefly, the cecum with the adjoining intestine was ligated at 0.5 cm from its end. The ligated cecum was then punctured with an 18-gauge needle, which enabled entrapped fecal material to leak into the normally sterile peritoneal cavity. The cecum was then repositioned in the peritoneal cavity, and the abdomen was closed. Sham-operated animals received laparotomy only. After the times indicated in the figure legends, plasma, BALF, and lung tissues were collected. Core histones in the lung tissue were isolated and analyzed by Western blotting to detect H4 and H4K8ac.
Cytokine measurement
Cell culture medium, BALF, or mouse plasma was centrifuged at 500g for 10 min to remove cell debris. The amounts of KC, IL-6, and IL-8 in the samples were then measured with specific ELISA kits according to the manufacturer’s instructions (Invitrogen).
Statistical analysis
Statistical analysis was performed by one-way or two-way ANOVA with Tukey’s test for the post hoc test or by unpaired Student’s t test to compare continuous measurements or with the Mantel-Cox log-rank test to compare survival curves. P < 0.05 was considered to be statistically significant.
Supplementary Material
Fig. S1. FBXL19 induces CBP degradation.
Fig. S2. FBXL19 mediates CBP degradation in the nucleus.
Fig. S3. FBXL19 induces the K48-linked polyubiquitylation of CBP.
Fig. S4. LPS stimulates the phosphorylation of CBP and FBXL19.
Fig. S5. FBXL19 is not auto-ubiquitylated.
Fig. S6. The reduction in CBP stability attenuates the LPS-induced acetylation of histone H4 on Lys12.
Fig. S7. FBXL19 attenuates histone H4K8ac–mediated Il8 promoter activity.
Fig. S8. IU1 attenuates LPS-stimulated cytokine release.
Acknowledgments
We thank J. Franz for English editing of the manuscript. We thank J. G. Yabes for help with the statistical analysis.
Funding: This work was supported by the NIH (R01GM115389 to J.Z. and R01HL112791 and R01HL131665 to Y.Z.), the American Heart Association (16GRNT30660001 to Y.Z.), and American Lung Association Biomedical Research Grant RG350146 (to J.Z.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/10/483/eaak9660/DC1
Author contributions: J.Z. and Y.Z. jointly designed and performed experiments, analyzed the data, and wrote the manuscript; J.W., R.K.B., S.D., A.K., A.M.J., and L.Z. performed experiments and analyzed the data; and J.Z. and Y.Z. oversaw and directed the study.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 2.Modak R, Das Mitra S, Krishnamoorthy P, Bhat A, Banerjee A, Gowsica BR, Bhuvana M, Dhanikachalam V, Natesan K, Shome R, Shome BR, Kundu TK. Histone H3K14 and H4K8 hyperacetylation is associated with Escherichia coli-induced mastitis in mice. Epigenetics. 2012;7:492–501. doi: 10.4161/epi.19742. [DOI] [PubMed] [Google Scholar]
- 3.Schmeck B, Beermann W, van Laak V, Zahlten J, Opitz B, Witzenrath M, Hocke AC, Chakraborty T, Kracht M, Rosseau S, Suttorp N, Hippenstiel S. Intracellular bacteria differentially regulated endothelial cytokine release by MAPK-dependent histone modification. J Immunol. 2005;175:2843–2850. doi: 10.4049/jimmunol.175.5.2843. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Xia X, Weiss RE, Refetoff S, Yen PM. Distinct and histone-specific modifications mediate positive versus negative transcriptional regulation of TSHα promoter. PLOS ONE. 2010;5:e9853. doi: 10.1371/journal.pone.0009853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague MJ, Urbé S. Ubiquitin: Same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Grishina I, Debus K, García-Limones C, Schneider C, Shresta A, Garcia C, Calzado MA, Schmitz ML. SIAH-mediated ubiquitination and degradation of acetyl-transferases regulate the p53 response and protein acetylation. Biochim Biophys Acta. 2012;1823:2287–2296. doi: 10.1016/j.bbamcr.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Sen N, Snyder SH. Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proc Natl Acad Sci USA. 2011;108:20178–20183. doi: 10.1073/pnas.1117820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruzzoni-Giovanelli H, Fernandez P, Veiga L, Podgorniak MP, Powell DJ, Candeias MM, Mourah S, Calvo F, Marín M. Distinct expression patterns of the E3 ligase SIAH-1 and its partner Kid/KIF22 in normal tissues and in the breast tumoral processes. J Exp Clin Cancer Res. 2010;29:10. doi: 10.1186/1756-9966-29-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73. doi: 10.1016/j.tibs.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallampalli RK, Coon TA, Glasser JR, Wang C, Dunn SR, Weathington NM, Zhao J, Zou C, Zhao Y, Chen BB. Targeting F box protein Fbxo3 to control cytokine-driven inflammation. J Immunol. 2013;191:5247–5255. doi: 10.4049/jimmunol.1300456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, Zou C, Mallampalli RK, Zhao Y. F-box protein FBXL19–mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–658. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Mialki RK, Wei J, Coon TA, Zou C, Chen BB, Mallampalli RK, Zhao Y. SCF E3 ligase F-box protein complex SCFFBXL19 regulates cell migration by mediating Rac1 ubiquitination and degradation. FASEB J. 2013;27:2611–2619. doi: 10.1096/fj.12-223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei J, Mialki RK, Dong S, Khoo A, Mallampalli RK, Zhao Y, Zhao J. A new mechanism of RhoA ubiquitination and degradation: Roles of SCFFBXL19 E3 ligase and Erk2. Biochim Biophys Acta. 2013;1833:2757–2764. doi: 10.1016/j.bbamcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong S, Zhao J, Wei J, Bowser RK, Khoo A, Liu Z, Luketich JD, Pennathur A, Ma H, Zhao Y. F-box protein complex FBXL19 regulates TGFβ1-induced E-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol Cancer. 2014;13:76. doi: 10.1186/1476-4598-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma N, Zhu Q, Wani G, He J, Wang QE, Wani AA. USP3 counteracts RNF168 via deubiquitinating H2A and γH2AX at lysine 13 and 15. Cell Cycle. 2014;13:106–114. doi: 10.4161/cc.26814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X-Y, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijnik A, Clare S, Hale C, Raisen C, McIntyre RE, Yusa K, Everitt AR, Mottram L, Podrini C, Lucas M, Estabel J, Goulding D, Sanger Institute Microarray Facility; Sanger Mouse Genetics Projects. Adams N, Ramirez-Solis R, White JK, Adams DJ, Hancock REW, Dougan G. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119:1370–1379. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 19.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mialki RK, Zhao J, Wei J, Mallampalli DF, Zhao Y. Overexpression of USP14 protease reduces I-κB protein levels and increases cytokine release in lung epithelial cells. J Biol Chem. 2013;288:15437–15441. doi: 10.1074/jbc.C112.446682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu YH, Guo NL. USP14 inhibitor IU1 prevents ventilator-induced lung injury in rats. Cell Mol Biol. 2014;60:50–54. [PubMed] [Google Scholar]
- 22.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Wei J, Dong S, Bowser RK, Zhang L, Jacko AM, Zhao Y. Destabilization of lysophosphatidic acid receptor 1 reduces cytokine release and protects against lung injury. EBioMedicine. 2016;10:195–203. doi: 10.1016/j.ebiom.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao H, Hwang J-w, Moscat J, Diaz-Meco MT, Leitges M, Kishore N, Li X, Rahman I. Protein kinase Cζ mediates cigarette smoke/aldehyde- and lipopolysaccharide-induced lung inflammation and histone modifications. J Biol Chem. 2010;285:5405–5416. doi: 10.1074/jbc.M109.041418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller D, Holt L, Kim SC, Schwabe RF, Sartor RB, Jobin C. Transforming growth factor-β1 inhibits non-pathogenic Gram negative bacteria-induced NF-κB recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. J Biol Chem. 2003;278:23851–23860. doi: 10.1074/jbc.M300075200. [DOI] [PubMed] [Google Scholar]
- 26.Garrett S, Fitzgerald MC, Sullivan KE. LPS and poly I:C induce chromatin modifications at a novel upstream region of the IL-23 p19 promoter. Inflammation. 2008;31:235–246. doi: 10.1007/s10753-008-9070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbosa S, Carreira S, Bailey D, Abaitua F, O’Hare P. Phosphorylation and SCF-mediated degradation regulate CREB-H transcription of metabolic targets. Mol Biol Cell. 2015;26:2939–2954. doi: 10.1091/mbc.E15-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bomsztyk K, Flanagin S, Mar D, Mikula M, Johnson A, Zager R, Denisenko O. Synchronous recruitment of epigenetic modifiers to endotoxin synergistically activated Tnf-α gene in acute kidney injury. PLOS ONE. 2013;8:e70322. doi: 10.1371/journal.pone.0070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naito M, Bomsztyk K, Zager RA. Endotoxin mediates recruitment of RNA polymerase II to target genes in acute renal failure. J Am Soc Nephrol. 2008;19:1321–1330. doi: 10.1681/ASN.2007121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aung HT, Schroder K, Himes SR, Brion K, van Zuylen W, Trieu A, Suzuki H, Hayashizaki Y, Hume DA, Sweet MJ, Ravasi T. LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB J. 2006;20:1315–1327. doi: 10.1096/fj.05-5360com. [DOI] [PubMed] [Google Scholar]
- 31.Pilar AVC, Reid-Yu SA, Cooper CA, Mulder DT, Coombes BK. GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLOS Pathog. 2012;8:e1002773. doi: 10.1371/journal.ppat.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka K, Kawakami T, Tateishi K, Yashiroda H, Chiba T. Control of IκBα proteolysis by the ubiquitin-proteasome pathway. Biochimie. 2001;83:351–356. doi: 10.1016/s0300-9084(01)01237-8. [DOI] [PubMed] [Google Scholar]
- 33.Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. The nuclear factor-κB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 34.Nettles KW, Gil G, Nowak J, Métivier R, Sharma VB, Greene GL. CBP is a dosage-dependent regulator of nuclear factor-κB suppression by the estrogen receptor. Mol Endocrinol. 2008;22:263–272. doi: 10.1210/me.2007-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spooren A, Kooijman R, Lintermans B, Van Craenenbroeck K, Vermeulen L, Haegeman G, Gerlo S. Cooperation of NFκB and CREB to induce synergistic IL-6 expression in astrocytes. Cell Signal. 2010;22:871–881. doi: 10.1016/j.cellsig.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DDL. Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-α signaling. Nat Struct Biol. 2002;9:68–75. doi: 10.1038/nsb743. [DOI] [PubMed] [Google Scholar]
- 37.Jain AK, Barton MC. Making sense of ubiquitin ligases that regulate p53. Cancer Biol Ther. 2010;10:665–672. doi: 10.4161/cbt.10.7.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HM, Yu Y, Cheng Y. Structure characterization of the 26S proteasome. Biochim Biophys Acta. 2011;1809:67–79. doi: 10.1016/j.bbagrm.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.R110.003871. R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung H, Kim B-G, Han WH, Lee JH, Cho J-Y, Park WS, Maurice MM, Han J-K, Lee MJ, Finley D, Jho E-H. Deubiquitination of Dishevelled by Usp14 is required for Wnt signaling. Oncogenesis. 2013;2:e64. doi: 10.1038/oncsis.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. FBXL19 induces CBP degradation.
Fig. S2. FBXL19 mediates CBP degradation in the nucleus.
Fig. S3. FBXL19 induces the K48-linked polyubiquitylation of CBP.
Fig. S4. LPS stimulates the phosphorylation of CBP and FBXL19.
Fig. S5. FBXL19 is not auto-ubiquitylated.
Fig. S6. The reduction in CBP stability attenuates the LPS-induced acetylation of histone H4 on Lys12.
Fig. S7. FBXL19 attenuates histone H4K8ac–mediated Il8 promoter activity.
Fig. S8. IU1 attenuates LPS-stimulated cytokine release.


