Abstract
Campylobacter jejuni causes more than 2 million cases of gastroenteritis annually in the United States, and is also linked to the autoimmune sequelae Guillan–Barre syndrome (GBS). GBS often results in flaccid paralysis, as the myelin sheaths of nerve cells are degraded by the adaptive immune response. Certain strains of C. jejuni modify their lipooligosaccharide (LOS) with the addition of neuraminic acid, resulting in LOS moieties that are structurally similar to gangliosides present on nerve cells. This can trigger GBS in a susceptible host, as antibodies generated against C. jejuni can cross-react with gangliosides, leading to demyelination of nerves and a loss of signal transduction. The goal of this study was to develop a quantitative PCR (qPCR) method and use whole genome sequencing data to detect the Campylobacter sialyltransferase (cst) genes responsible for the addition of neuraminic acid to LOS. The qPCR method was used to screen a library of 89 C. jejuni field samples collected by the Food and Drug Administration Pacific Northwest Lab (PNL) as well as clinical isolates transferred to PNL. In silico analysis was used to screen 827 C. jejuni genomes in the FDA GenomeTrakr SRA database. The results indicate that a majority of C. jejuni strains could produce LOS with ganglioside mimicry, as 43.8% of PNL isolates and 46.9% of the GenomeTrakr isolates lacked the cst genes. The methods described in this study can be used by public health laboratories to rapidly determine whether a C. jejuni isolate has the potential to induce GBS. Based on these results, a majority of C. jejuni in the PNL collection and submitted to GenomeTrakr have the potential to produce LOS that mimics human gangliosides.
Keywords: Campylobacter jejuni, lipooligosaccharide (LOS), Guillain-Barré syndrome (GBS), ganglioside mimicry, whole genome sequencing, qPCR
Introduction
Campylobacter jejuni is a leading bacterial cause of gastroenteritis in the United States and worldwide (Tauxe, 1992; Kaakoush et al., 2015; CDC, 2017). Infection is usually linked to the consumption of contaminated food or water, especially poultry products (Konkel et al., 2001; Sahin et al., 2015; Skarp et al., 2016). The link between C. jejuni and poultry is due to the fact that both wild and domestic birds can be colonized with high levels of C. jejunia without causing illness to the avian host (Sahin et al., 2015; Johnson et al., 2017). Within commercial chicken flocks, C. jejuni spreads rapidly from bird to bird and remains present in their digestive tract until the time of slaughter (Garcia-Sanchez et al., 2017; Johnson et al., 2017). One gram of cecal contents may contain greater than 108 CFU of C. jejuni, and can cross contaminate C. jejuni-free chicken carcasses during processing (Garcia-Sanchez et al., 2017; Johnson et al., 2017). As a result, raw poultry is often contaminated with C. jejuni at the point of sale. Cross-contamination in the kitchen can then lead to the introduction of C. jejuni into other food products (Sahin et al., 2015; Skarp et al., 2016).
The disease caused by C. jejuni, campylobacteriosis, is characterized by fever, abdominal cramps, vomiting, and diarrhea containing blood and/or fecal leukocytes (Konkel et al., 2001). Most cases are self-limiting and resolve within a few days, but antibiotics such as erythromycin may be administered in severe infections (Konkel et al., 2001). However, approximately one in a 1000 C. jejuni infections leads to autoimmune sequelae such as Guillan–Barre syndrome (GBS) (Tauxe, 1992; Nyati and Nyati, 2013). Campylobacteriosis is responsible for approximately 30% of all GBS cases, and a reduction in C. jejuni incidence can significantly reduce the burden of GBS (Tauxe, 1992; Poropatich et al., 2010; Baker et al., 2012). These autoimmune disorders are caused by the induction of an adaptive immune response that targets cells of the nervous system, and are the leading cause of paralysis in the post-polio era (Amon et al., 2012; Wijdicks and Klein, 2017). In susceptible hosts, antibodies that are generated against the surface of C. jejuni may also bind myelin present on the surface of nerve cells. Destruction of the myelin causes a loss of signal transduction, leading to the flaccid paralysis and loss of motor function that characterizes GBS (Nyati and Nyati, 2013; Wijdicks and Klein, 2017). It is important to note that the development of these autoimmune sequelae is dependent on both a host that is susceptible to self-recognizing antibodies, as well as the bacterial antigens that induce antibody production (Koga et al., 2006; Islam et al., 2012; Huizinga et al., 2015; Lardone et al., 2016; St Charles et al., 2017).
The lipooligosaccharide (LOS) found in the outer membrane of C. jejuni is the molecule responsible for inducing GBS autoantibodies (Parker et al., 2005; Koga et al., 2006; Yuki, 2007; Amon et al., 2012). The LOS of C. jejuni can be heavily modified via the addition of carbohydrate moieties to the O-antigen, resulting in a diverse array of LOS antigens amongst C. jejuni strains (Godschalk et al., 2004; Li et al., 2005; Dzieciatkowska et al., 2008; Parker et al., 2008; Semchenko et al., 2012; Stephenson et al., 2013). The addition of sialic (n-acetylneuraminic) acid residues to the O antigen can result in structures that are nearly identical to the GM1, GD1a, GQ1b, and GT1a gangliosides found on the surface of nerve cells (Gilbert et al., 2002; Li et al., 2005; Koga et al., 2006; Yuki, 2007). Sera taken from GBS patients have been shown to recognize one or more of these gangliosides as well as the surface of specific C. jejuni strains (Lardone et al., 2016). While it is not yet possible to determine who is susceptible to these autoimmune disorders, it is possible to predict whether a C. jejuni strain possesses sialylated LOS and has the potential to induce GBS. It is not entirely clear which particular ganglioside mimic is more likely to induce GBS, but it does appear that the addition of sialic acid (sialylation) to LOS is required for ganglioside mimicry (Godschalk et al., 2004; Koga et al., 2006; Yuki, 2007; Parker et al., 2008; Amon et al., 2012; Lardone et al., 2016). Other Campylobacter species, such as Campylobacter coli, may also produce ganglioside mimics although the mechanism and genes required are not as well-characterized (Richards et al., 2013). In this study, the genes necessary for LOS sialylation were chosen as targets for qPCR and whole genome sequencing (WGS)-based assays, so that isolates with the potential to produce ganglioside mimics could be rapidly identified.
Most of the genes responsible for LOS synthesis and modification are clustered in the genome and a typing scheme has been developed to describe the genes present (Figure 1) (Gilbert et al., 2002; Godschalk et al., 2004; Parker et al., 2005, 2008; Richards et al., 2013). There are more than 20 different LOS types described thus far, but LOS types A, B, and C are most commonly associated with GBS (Koga et al., 2006). These three LOS types contain a variant of the cst gene, which encodes a sialyltransferase required for the addition of sialic acid to LOS (Gilbert et al., 2002; Godschalk et al., 2004). LOS types A and B encode the cst-II gene, while type C encodes cst-III (Gilbert et al., 2002). The two cst variants can add sialic acid to a single position or multiple positions within the O-antigen of LOS to create multiple structures with mimicry to GM1, GT1a, or GD1a gangliosides (Koga et al., 2006; Yuki, 2007; Parker et al., 2008; Islam et al., 2012). In addition, interaction between LOS molecules with different sialylation patterns can produce structures that mimic additional gangliosides (Koga et al., 2015), and GBS patients can develop different anti-ganglioside antibodies in response to the same ganglioside mimics (Islam et al., 2012; Lardone et al., 2016). While it is not known which ganglioside mimic(s) are most likely to induce anti-ganglioside antibodies, it is clear that sialylation of LOS is required (Godschalk et al., 2004; Koga et al., 2006; Amon et al., 2012). Thus, the cst genes were chosen for use as markers for C. jejuni strains that can produce LOS with ganglioside mimicry. The waaM gene, which is required for core LOS synthesis, was used as a control as it is found in all strains regardless of LOS type (Gilbert et al., 2002). The presence of cst-II or cst-III indicates that the isolate belongs to LOS types A, B, or C, and has the potential to produce ganglioside mimics (Parker et al., 2005, 2008; Koga et al., 2006).
FIGURE 1.
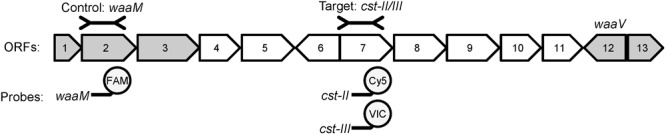
The LOS biosynthetic locus contains variable and conserved genes required for LOS gangliosdide mimicry. An example of the gene organization of LOS type A (cst-II positive) is shown [adapted from Parker et al. (2005)]. ORFs conserved amongst all LOS types are shown in gray, while variable ORFs are shown in white. The control and target regions used in the qPCR are shown above the diagram, and the fluorophores used for each gene are shown below. ORF2 (waaM) encodes an acyltransferase involved in Lipid A biosynthesis and is highly conserved. ORF7 encodes cst-II in LOS types A and B, and cst-III in LOS type C. ORF12 (waaV) is a highly conserved putative glycosyltransferase. The gene content of the variable region between waaM and waaV determines the LOS type.
In this study, a multiplex qPCR method was developed to accurately detect the cst-II or cst-III genes in extracted genomic DNA. The qPCR method was then utilized to determine the prevalence of the cst genes in a collection of C. jejuni strains isolated from food and environmental samples. In silico analyses were also utilized to detect cst in C. jejuni genomes available through GenomeTrakr. The results of this study indicate that the majority of C. jejuni in GenomeTrakr and the FDA Pacific Northwest Laboratory collection have the potential to produce LOS that mimics gangliosides. The screening methods described in this study will enable food safety experts to rapidly identify isolates with sialylated LOS.
Materials and Methods
Bacterial Strains
All C. jejuni strains used are listed in Supplementary Table 1. Field strains were collected by the Food and Drug Administration Pacific Northwest Laboratory from a variety of food and environmental sources. Clinical isolates were part of an existing archival collection without any patient information. The bacteria were cultured and genomic DNA extracted as described in Liu et al. (2017). Genomic DNA (gDNA) samples used for assay controls (R4B202, R4B208, R7B122) were quantified using the ds DNA BR assay kit and a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Inc.) and diluted to 500, 50, 5, and 0.5 pg/μl. All other genomic DNA samples were tested by qPCR without dilution.
Whole Genome Sequencing
Whole genome sequencing was performed as described in Liu et al. (2016), using genomic DNA isolated from the three assay controls. Briefly, libraries were prepared using the Nextera XT kit (Illumina) and sequenced using a MiSeq instrument (Illumina) with 251 reading cycles. The paired-end reads were imported into CLC Genomics v9.5.3 and de novo assembly was performed using default parameters. The assembled genomic sequences were uploaded to NCBI under BioProject ID PRJNA433413. The waaM, waaV, and cst-III gene sequences from NCTC 11168 (GenBank AL111168.1) and cst-II sequence from RM3196 (GenBank CP012690.1) were used as references to identify homologous genes in the assembled genomes.
Quantitative PCR
Quantitative PCR was performed using EXPRESS qPCR SuperMix Universal (Invitrogen) reagents on an ABI 7500 Fast (Applied Biosystems). The oligonucleotide primers and probes used in the study are listed in Table 1. All primers and the FAM and Cy5 labeled probes were produced by Integrated DNA Technologies (IDT) and the VIC labeled probe was produced by Life Technologies. The primers and probe sequences were based on the PCR strategy described in Parker et al. (2005). New primer and probe sequences for this study were designed using Primer- Basic Local Alignment Search Tool (BLAST) (Ye et al., 2012). Various primer concentrations were tested in simplex (data not shown) to determine optimal reaction conditions. The optimized multiplex oligonucleotide concentrations were: 300 mM cstII-F and cstII-R4, 200 mM cst3-F and cst3-R2, 100 mM waaM-F and waaM-R, 150 mM orf7ab-Cy5-BHQ2, 100 mM cst3-VIC-MGBNFQ, and 50 mM waaM-FAM-BHQ. The final reaction volume contained 15 μl 2X EXPRESS qPCR SuperMix Universal, 0.06 μl ROX dye, 1 μl of DNA template, and water and oligonucleotides to 30 μl total. The thermocycling conditions were: 2 min at 95°C, 1 cycle; 10 s at 95°C, 30 s at 60°C, 40 cycles. Sequence Detection Software v1.4 (Life Technologies) was used to analyze the data with automated baselines and thresholds. The results were plotted in Excel v14.0.7, and PCR efficiency was calculated using the tool at Thermofisher.com.
Table 1.
Oligonucleotides used in this study.
| Name | Target | Sequence (5′–3′) | Modificationsa |
|---|---|---|---|
| waaM-FAM-BHQ | waaM | AAAAAAGGCGGTATAAGACAAATGCTAAG | 5′ FAM, 3′ BHQ-1 |
| orf7ab-Cy5-BHQ2 | cst-II | TATCCCAAATGAGCATCAGGAAAATAATC | 5′ Cy5, 3′ BHQ-2 |
| cst3-VIC-MGBNFQ | cst-III | CAAAACAATTTGATGTATTTAGATGCAATCAG | 5′ VIC, 3′ MGBNFQ |
| waaM-F∗ | waaM | CAAATCTGTTTCCCTCAATACACTCA∗ | None |
| waaM-R∗ | waaM | TTTAATCTTACGCTTTCGTTTTCTAC∗ | None |
| cstII-F∗ | cst-II | ACTACACTTTAAAACATTTAATCCAAAATCA∗ | None |
| cstII-R4 | cst-II | AAGATAAATTTCTTTGTATCCTAGGG | None |
| cst3-F | cst-III | ATGCTTTGGTATGCGGTAATGG | None |
| cst3-R2 | cst-III | TCCACTAGTAATTCTTTGCCTATTG | None |
∗Primer sequence from Parker et al. (2005). aBHQ, Black Hole Quencher; MGBNFQ, Minor groove binding non-fluorescent quencher.
SRA Assembly and BLAST
The SRA files for the GenomeTrakr Campylobacter Umbrella Project (PRJNA258021) were downloaded on 04/25/2017 and imported as paired-end Illumina reads into CLC Genomics v9.5.3. De novo assembly was performed using a workflow consisting of adapter (Nextera adapter sequences) and quality trimming, followed by de novo assembly using default parameters. The BLAST function in CLC Genomics was used to identify contigs that contained waaM, waaV, cst-II, and cst-III using the NCTC11168 and RM3196 reference sequences. Only BLAST hits with an e-value of 0 were used for downstream analysis. The contig lists generated by the BLAST search were imported into Excel v14.0.7 and compared to identify contigs containing multiple genes of interest.
Results and Discussion
A rapid qPCR method was developed to detect the cst-II and cst-III genes based on previous work demonstrating that these genes are required for the addition of sialic acid to Campylobacter LOS (Gilbert et al., 2002; Godschalk et al., 2004; Yuki, 2007; Parker et al., 2008). Primers specific to the cst genes were adapted from the typing strategy described by Parker et al. (2005) (Table 1). Probes within the amplicons were designed using PrimerBLAST, such that they recognize all cst-II, cst-III, and waaM sequences in the National Center for Biotechnology Information (NCBI) database. Primer and probe concentrations were adjusted for maximal detection sensitivity of the cst genes. A limiting concentration of primers was used for the waaM control gene, so that amplification would not interfere with detection of the cst targets. The C. jejuni strains used for testing the specificity of the qPCR assay are listed in Supplementary Table 1. The LOS gene content in each control strain (R4-B2-02, R4-B2-08, and R7-B1-22) was confirmed by WGS. All qPCR reactions yielded the result expected based on the genomic sequences of the C. jejuni strains. The qPCR method was tested using dilutions of gDNA from the three control strains. DNA quantities of 500, 50, 5, and 0.5 pg were used as template to determine the sensitivity and efficiency of the qPCR amplification (Figure 2). The amplification efficiency for all three targets was greater than 95% across the four-log magnitude concentration range. Importantly, detection of the cst-II and cst-III genes was more sensitive than detection of the waaM gene used as a control. These results indicate that the qPCR assay is specific across a wide range of DNA concentrations, and that the waaM control is appropriate.
FIGURE 2.
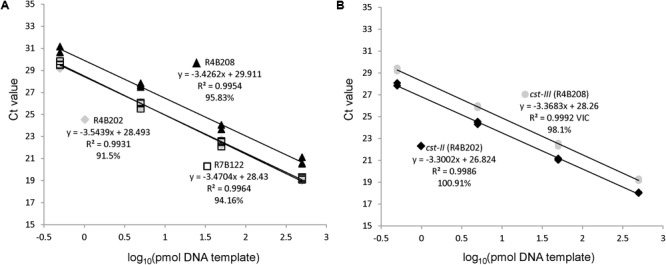
The qPCR assay efficiently amplifies cst-II, cst-III, and waaM. Serial 10-fold dilutions of control strains R4B202 (cst-II), R4B208 (cst-III), and R7B122 (cst-negative) were used as template in the multiplex qPCR assay to determine the PCR efficiency of the waaM (A) and cst-II/cst-III (B) targets.
The qPCR assay was then used to screen 89 C. jejuni strains in an archive at Food and Drug Administration Pacific Northwest Laboratory (FDA-PNL). These strains were isolated from field (i.e., food and environmental sources) and clinical samples, and collected over a 20-year period (Supplementary Table 1). Using the qPCR assay, cst-II was found in 36 samples (40.5%), cst-III was found in 13 samples (14.6%), and waaM was found in all samples tested (Table 2). Interestingly, the 38 clinical samples had more cst-negative isolates (57.9%) than the 51 field samples (33.3%). The qPCR screening method could be employed in future studies to determine the prevalence of cst-II and cst-III isolates in other geographic regions or environmental sources.
Table 2.
Multiplex qPCR screen of 89 C. jejuni isolates.
| Target | # Positive | Percent |
|---|---|---|
| All isolates | ||
| waaM | 89 | 100 |
| cst-II | 36 | 40.5 |
| cst-III | 13 | 14.6 |
| Neg.a | 39 | 43.8 |
| Field isolates | ||
| waaM | 51 | 100 |
| cst-II | 26 | 51.0 |
| cst-III | 7 | 13.7 |
| Neg. | 17 | 33.3 |
| Clinical isolates | ||
| waaM | 38 | 100 |
| cst-II | 10 | 26.3 |
| cst-III | 6 | 15.8 |
| Neg. | 22 | 57.9 |
aNeg., no cst genes detected.
The multiplex qPCR presented in this study has several major differences compared to sequencing or PCR based approaches previously described (Gilbert et al., 2002; Parker et al., 2005, 2008). Our multiplex qPCR method is faster to perform than Sanger sequencing or next generation sequencing, which require extensive time for library preparation, sequencing, and data analysis. The qPCR assay is also multiplexed and uses specific fluorescent probes for detection, so the reaction can be performed and analyzed using a single instrument in less than an hour of runtime. However, the qPCR assay presented here only determines if a strain possesses one of the two sialyltransferase genes; determination of the LOS class type or cst-II polymorphisms would require additional information. Based on the requirement of cst-II or cst-III for sialylation of LOS (Gilbert et al., 2002; Godschalk et al., 2004), we believe that our qPCR assay provides a simplified alternative method for the detection of C. jejuni with the potential to produce sialylated LOS. WGS could then be performed to fully characterize the LOS locus in isolates of interest.
An in silico approach was used to screen 827 C. jejuni genomes sequenced as part of the GenomeTrakr project, available from NCBI BioProject PRJNA258021. Fortunately, the genes involved in LOS sialylation are contained within a defined LOS biosynthetic locus flanked by the waaV and waaM genes, which are conserved among C. jejuni strains. The genome of C. jejuni-type strain NCTC 11168 was used to obtain reference sequences for waaM, waaV, and cst-III, while strain 81-176 was used for cst-II. Using the BLAST, sequences were identified within the RefSeq genome database that contained both waaM and waaV, indicating that the complete LOS locus was represented in the available sequence. BLAST searches were then performed to identify genomic sequences containing cst-II or cst-III (Figure 3). In total, 405 complete LOS loci were identified, with 177 (43.7%) containing cst-II, 38 (9.4%) containing cst-III, and 190 (46.9%) that were negative for either cst gene (Tables 3A,B). For the 347 food isolates and 33 environmental isolates, 49.0 and 48.5% were cst-negative, respectively. In contrast, only 16% of the 25 clinical isolates were cst-negative. For the 422 genomic sequences without a complete LOS locus (Table 3C), 123 (29.1%) contained cst-II and 118 (28.0%) contained cst-III. No genome assemblies were found to contain both cst-II and cst-III. It is important to note that more than half sequences in the GenomeTrakr databases did not have enough coverage to complete the LOS locus, which ranges in size from ∼7.8 kb (LOS class F) to 15.2 kb (LOS class E). This is possibly due to the low GC content or homopolymeric tracts in this region which can affect sequencing. Although this database is not a fully accurate representation of C. jejuni strain diversity in nature, it is interesting to note that greater than 50% of strains collected from food production areas and clinical samples have the potential to produce ganglioside mimics.
FIGURE 3.
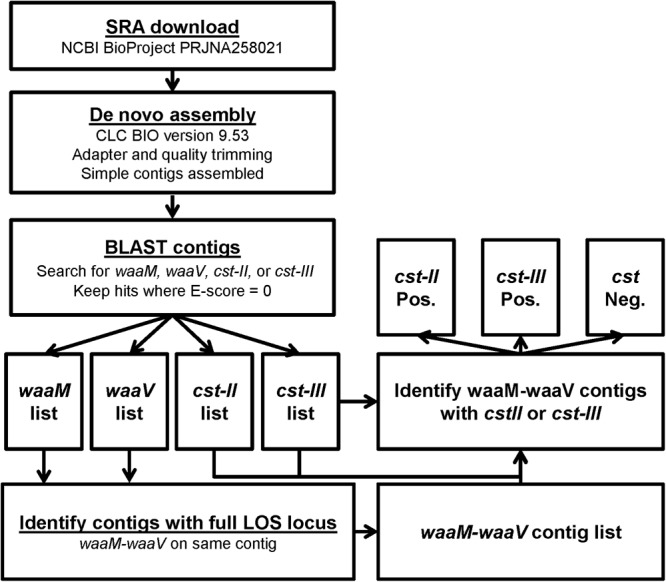
Workflow for the determination of cst-type from GenomeTrakr SRA data. The strategy for identifying cst genes from SRA files starts with de novo assembly, followed by BLAST searches for the four genes of interest. To ensure that the variable genes of the LOS locus were present in an SRA, contigs were identified that contained both flanking genes, waaM and waaV. The waaM-waaV contigs were then determined to be cst-II positive or cst-III positive if the gene was found in the same contig, and cst negative if the genes were not detected.
Table 3.
In silico analysis of 829 GenomeTrakr Sequence Read Archives (SRAs) from NCBI BioProject PRJNA258021.
| All GenomeTrakr Isolates | Total | Percent | ||
|---|---|---|---|---|
| (A) | ||||
| SRAs | 827 | 100 | ||
| waaV sequences | 816 | 98.7 | ||
| waaM sequences | 800 | 96.7 | ||
| cstII sequences | 300 | 36.3 | ||
| cstIII sequences | 156 | 18.9 | ||
| Contiguous waam-waaV | 405 | 49.0 | ||
| Non-contiguous waaM-waaV | 422 | 51.0 | ||
|
(B) | ||||
| Total | Food | Enviro. | Clinical | |
| Contiguous waam-waaV | 405 (100%) | 347 (85.7%) | 33 (8.1%) | 25 (6.2%) |
| cstII-positive | 177 (43.7%) | 148 (42.7%) | 16 (48.5%) | 13 (52%) |
| cstIII-positive | 38 (9.4%) | 29 (8.4%) | 1 (3%) | 8 (32%) |
| cst-negative | 190 (46.9%) | 170 (49.0%) | 16 (48.5%) | 4 (16%) |
|
(C) | ||||
| Total | Percent | |||
| Non-contiguous waaM-waaV | 422 | 100 | ||
| cstII-positive | 123 | 29.1 | ||
| cstIII-positive | 118 | 28.0 | ||
| cst-negative | 181 | 42.9 | ||
The strains present in the GenomeTrakr database were collected between 2002 and 2017, with 738 isolates from the United States and 89 isolates from the United Kingdom. All isolates from the United Kingdom were collected from retail meat samples by Fera and sequenced as part of a joint project with the U.S. Food and Drug Administration. In the U.S., food, environmental, and clinical samples were obtained from 12 states (AK, CA, CT, GA, IA, MD, MN, NY, OH, OR, TN, and TX). The dates and sources of isolates uploaded to GenomeTrakr vary widely between contributing labs; for example, all 25 samples from AK were collected from environmental sources between 2009 and2013, whereas all 115 samples from CT were collected from poultry meat between 2002 and2003. Additionally, some genomic sequences may be from identical or highly related isolates collected from the same source. Given these differences in sample collection, it is difficult to assess whether strains that possess cst-II or cst-III are linked to certain sources or geographical locations. However, the GenomeTrakr database is the most representative database of U.S. C. jejuni isolates available, and will continue to grow in size and usefulness as newly isolated sequences are added.
The goal of this study was to examine the prevalence of C. jejuni with the potential for LOS sialylation in the food supply. Interestingly, the qPCR assay and the in silico analysis of WGSs yielded similar numbers of cst-positive results. By qPCR 40.5 and 14.6% of isolates were positive for cst-II and cst-III, respectively, compared to 43.7 and 9.4%, respectively, for the in silico analysis. The percentage of cst-positive isolates is also consistent with previous studies indicating that roughly 50% of isolates contain a cst gene (Hardy et al., 2011; Amon et al., 2012; Ellstrom et al., 2013, 2016; Duarte et al., 2016).
While the qPCR assay differentiates between cst-II and cst-III variants, it does not differentiate between cst-II with Asn51 or Thr51. The cst-III and cst-II Thr51 genes encode for a sialyltransferase with α-2-3 activity, while cst-II Asn51 encodes a bi-functional sialyltransferase with α-2-3 and α-2-8 activity (Koga et al., 2006). As a result, cst-II Asn51 isolates can also produce GT1a-like and GQ1b-like LOS as well as GM1-like and GD1a-like LOS (Hardy et al., 2011). In this study, it was found that of the 300 cst-II sequences in the Campylobacter GenomeTrakr BioProject (PRJNA258021), 230 (76.7%) had Asn51, while 70 (23.3%) had Thr51.
This indicates that the majority of cst-II positive isolates have the ability to produce GT1a/GQ1b-like LOS and could potentially result in a different repertoire of anti-ganglioside antibodies. This could be an important distinction, as the clinical symptoms of GBS are influenced by the ganglioside specificity of the autoantibodies. Specifically, Miller Fisher syndrome, a variant of GBS, is thought to be caused by anti-GQ1b antibodies (Susuki et al., 2001; Yuki, 2007).
Further complicating the link between cst genotype and human disease is the fact that a given isolate of C. jejuni often contains multiple LOS structures (Godschalk et al., 2004; Li et al., 2005; Koga et al., 2006; Dzieciatkowska et al., 2008). Some LOS molecules will be unmodified by cst, while other LOS molecules may have single or multiple sialic acid groups, resulting in a mixture of GD1a, GM1, GT1a, and GQ1b epitopes on the surface of the outermembrane. In addition, it has been proposed that combinations of different LOS epitopes with ganglioside mimicry could stimulate the production of autoantibodies with specificity against additional gangliosides (Li et al., 2005; Koga et al., 2015). Additional research will need to be performed to determine the LOS structures present on C. jejuni with cst-III, cst-II Asn51 or cst-II Thr51, as well as the host immune response to the different ganglioside mimics.
In summary, a qPCR protocol was developed and WGS data available on NCBI analyzed to determine the cst genotype of C. jejuni strains. Using these methods, it was determined that nearly 60% of C. jejuni isolates contain the cst genes required to modify LOS and produce ganglioside mimics. In addition, ∼75% of cst-II isolates have Asn51 and are capable of stimulating additional antibody specificities. Taken together, these results indicate that approximately half of C. jejuni strains have the potential to produce four different ganglioside mimics. Additional studies will need to be performed to determine the role of gene expression and the quantities of each LOS structure present in the outermembrane. In the future, the qPCR assay or WGS could be used to rapidly determine whether a food or outbreak isolate has the potential to produce ganglioside mimics.
Author Contributions
JN-M and KL: bacterial isolates were grown, DNA was extracted, and data analysis was performed. KL and KJ: whole genome sequencing was performed. JN-M: completed the qPCR experiments, as well as SRA assembly and analysis. JN-M, KL, KJ, W-HW, and DR contributed to the experimental design and review of the manuscript.
Disclaimer
The views presented in this work do not necessarily reflect those ofthe U.S. Food and Drug Administration nor do the authors specifically endorse the listed instrumentation or products.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Microbiology Branch at Pacific Northwest Laboratory for technical assistance. We also thank the FDA GenomeTrakr team and all participating laboratories for their efforts.
Footnotes
Funding. This study was funded by the U.S. Food and Drug Administration as all authors are FDA employees.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00408/full#supplementary-material
References
- Amon P., Klein D., Springer B., Jelovcan S., Sofka D., Hilbert F. (2012). Analysis of Campylobacter jejuni isolates of various sources for loci associated with Guillain-Barre Syndrome. Eur. J. Microbiol. Immunol. 2 20–23. 10.1556/EuJMI.2.2012.1.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. G., Kvalsvig A., Zhang J., Lake R., Sears A., Wilson N. (2012). Declining Guillain-Barre Syndrome after campylobacteriosis control, New Zealand, 1988–2010. Emerg. Infect. Dis. 18 226–233. 10.3201/eid1802.111126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2017). Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet 2015 Surveillance Report (Final Data). Atlanta, GA: CDC. [Google Scholar]
- Duarte A., Seliwiorstow T., Miller W. G., De Zutter L., Uyttendaele M., Dierick K., et al. (2016). Discriminative power of Campylobacter phenotypic and genotypic typing methods. J. Microbiol. Methods 125 33–39. 10.1016/j.mimet.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Dzieciatkowska M., Liu X., Heikema A. P., Houliston R. S., van Belkum A., Schweda E. K., et al. (2008). Rapid method for sensitive screening of oligosaccharide epitopes in the lipooligosaccharide from Campylobacter jejuni strains isolated from Guillain-Barre Syndrome and Miller Fisher syndrome patients. J. Clin. Microbiol. 46 3429–3436. 10.1128/JCM.00681-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrom P., Feodoroff B., Hanninen M. L., Rautelin H. (2013). Characterization of clinical Campylobacter jejuni isolates with special emphasis on lipooligosaccharide locus class, putative virulence factors and host response. Int. J. Med. Microbiol. 303 134–139. 10.1016/j.ijmm.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Ellstrom P., Hansson I., Nilsson A., Rautelin H., Olsson Engvall E. (2016). Lipooligosaccharide locus classes and putative virulence genes among chicken and human Campylobacter jejuni isolates. BMC Microbiol. 16:116. 10.1186/s12866-016-0740-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez L., Melero B., Jaime I., Hanninen M. L., Rossi M., Rovira J. (2017). Campylobacter jejuni survival in a poultry processing plant environment. Food Microbiol. 65 185–192. 10.1016/j.fm.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Gilbert M., Karwaski M. F., Bernatchez S., Young N. M., Taboada E., Michniewicz J., et al. (2002). The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277 327–337. 10.1074/jbc.M108452200 [DOI] [PubMed] [Google Scholar]
- Godschalk P. C. R., Heikema A. P., Gilbert M., Komagamine T., Ang C. W., Glerum J., et al. (2004). The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barré Syndrome. J. Clin. Invest. 114 1659–1665. 10.1172/jci200415707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. G., Lackey L. G., Cannon J., Price L. B., Silbergeld E. K. (2011). Prevalence of potentially neuropathic Campylobacter jejuni strains on commercial broiler chicken products. Int. J. Food Microbiol. 145 395–399. 10.1016/j.ijfoodmicro.2010.12.027 [DOI] [PubMed] [Google Scholar]
- Huizinga R., van den Berg B., van Rijs W., Tio-Gillen A. P., Fokkink W. J., Bakker-Jonges L. E., et al. (2015). Innate immunity to Campylobacter jejuni in Guillain-Barre Syndrome. Ann. Neurol. 78 343–354. 10.1002/ana.24442 [DOI] [PubMed] [Google Scholar]
- Islam Z., Gilbert M., Mohammad Q. D., Klaij K., Li J., van Rijs W., et al. (2012). Guillain-Barre Syndrome-related Campylobacter jejuni in Bangladesh: ganglioside mimicry and cross-reactive antibodies. PLoS One 7:e43976. 10.1371/journal.pone.0043976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Shank J. M., Johnson J. G. (2017). Current and potential treatments for reducing Campylobacter colonization in animal hosts and disease in humans. Front. Microbiol. 8:487. 10.3389/fmicb.2017.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush N. O., Castano-Rodriguez N., Mitchell H. M., Man S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28 687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M., Gilbert M., Li J., Yuki N. (2015). Complex of GM1- and GD1a-like lipo-oligosaccharide mimics GM1b, inducing anti-GM1b antibodies. PLoS One 10:e0124004. 10.1371/journal.pone.0124004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M., Gilbert M., Takahashi M., Li J., Koike S., Hirata K., et al. (2006). Comprehensive analysis of bacterial risk factors for the development of Guillain-Barre Syndrome after Campylobacter jejuni enteritis. J. Infect. Dis. 193 547–555. [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Monteville M. R., Rivera-Amill V., Joens L. A. (2001). The pathogenesis of Campylobacter jejuni mediated enteritis. Curr. Issues Intest. Microbiol. 2 55–71. [PubMed] [Google Scholar]
- Lardone R. D., Yuki N., Irazoqui F. J., Nores G. A. (2016). Individual restriction of fine specificity variability in Anti-GM1 IgG antibodies associated with Guillain-Barre Syndrome. Sci. Rep. 6:19901. 10.1038/srep19901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Koga M., Brochu D., Yuki N., Chan K., Gilbert M. (2005). Electrophoresis-assisted open-tubular liquid chromatography/mass spectrometry for the analysis of lipooligosaccharide expressed by Campylobacter jejuni. Electrophoresis 26 3360–3368. 10.1002/elps.200500145 [DOI] [PubMed] [Google Scholar]
- Liu K. C., Jinneman K. C., Neal-McKinney J., Wu W. H., Rice D. H. (2016). Genome sequencing and annotation of a Campylobacter coli strain isolated from milk with multidrug resistance. Genom. Data 8 123–125. 10.1016/j.gdata.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. C., Jinneman K. C., Neal-McKinney J., Wu W. H., Rice D. H. (2017). Simultaneous identification of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari with smartcycler-based multiplex quantitative polymerase chain reaction. Foodborne Pathog. Dis. 14 371–378. 10.1089/fpd.2016.2245 [DOI] [PubMed] [Google Scholar]
- Nyati K. K., Nyati R. (2013). Role of Campylobacter jejuni infection in the pathogenesis of Guillain-Barre Syndrome: an update. Biomed Res. Int. 2013:852195. 10.1155/2013/852195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. T., Gilbert M., Yuki N., Endtz H. P., Mandrell R. E. (2008). Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J. Bacteriol. 190 5681–5689. 10.1128/JB.00254-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. T., Horn S. T., Gilbert M., Miller W. G., Woodward D. L., Mandrell R. E. (2005). Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43 2771–2781. 10.1128/JCM.43.6.2771-2781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poropatich K., Fischer Walker C., Black R. (2010). Quantifying the association between Campylobacter infection and Guillain-Barré Syndrome: a systematic review. J. Health Popul. Nutr. 28 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards V. P., Lefebure T., Pavinski Bitar P. D., Stanhope M. J. (2013). Comparative characterization of the virulence gene clusters (lipooligosaccharide [LOS] and capsular polysaccharide []) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect. Genet. Evol. 14 200–213. 10.1016/j.meegid.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin O., Kassem I. I., Shen Z., Lin J., Rajashekara G., Zhang Q. (2015). Campylobacter in poultry: ecology and potential interventions. Avian Dis. 59 185–200. 10.1637/11072-032315-Review [DOI] [PubMed] [Google Scholar]
- Semchenko E. A., Day C. J., Moutin M., Wilson J. C., Tiralongo J., Korolik V. (2012). Structural heterogeneity of terminal glycans in Campylobacter jejuni lipooligosaccharides. PLoS One 7:e40920. 10.1371/journal.pone.0040920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarp C. P., Hanninen M. L., Rautelin H. I. (2016). Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 22 103–109. 10.1016/j.cmi.2015.11.019 [DOI] [PubMed] [Google Scholar]
- St Charles J. L., Bell J. A., Gadsden B. J., Malik A., Cooke H., Van de Grift L. K., et al. (2017). Guillain Barre Syndrome is induced in Non-Obese Diabetic (n.d.) mice following Campylobacter jejuni infection and is exacerbated by antibiotics. J. Autoimmun. 77 11–38. 10.1016/j.jaut.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Stephenson H. N., John C. M., Naz N., Gundogdu O., Dorrell N., Wren B. W., et al. (2013). Campylobacter jejuni lipooligosaccharide sialylation, phosphorylation, and amide/ester linkage modifications fine-tune human Toll-like receptor 4 activation. J. Biol. Chem. 288 19661–19672. 10.1074/jbc.M113.468298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susuki K., Yuki N., Hirata K. (2001). Fine specificity of anti-GQ1b IgG and clinical features. J. Neurol. Sci. 185 5–9. [DOI] [PubMed] [Google Scholar]
- Tauxe R. V. (1992). “Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations,” in Campylobacter jejuni: Current Status and Future Trends eds Nachamkin I., Tompkins M. J., Blaser L. S. (Washington, DC: American Society for Microbiology; ) 9–19. [Google Scholar]
- Wijdicks E. F., Klein C. J. (2017). Guillain-Barre Syndrome. Mayo Clin. Proc. 92 467–479. 10.1016/j.mayocp.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki N. (2007). Campylobacter sialyltransferase gene polymorphism directs clinical features of Guillain–Barre Syndrome. J. Neurochem. 103 150–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


