Abstract
Multiple myeloma develops primarily inside the bone marrow microenvironment, that confers pro-survival signals and drug resistance. 3D cultures that reproduce multiple myeloma-bone marrow interactions are needed to fully investigate multiple myeloma pathogenesis and response to drugs. To this purpose, we exploited the 3D Rotary Cell Culture System bioreactor technology for myeloma-bone marrow co-cultures in gelatin scaffolds. The model was validated with myeloma cell lines that, as assessed by histochemical and electron-microscopic analyses, engaged contacts with stromal cells and endothelial cells. Consistently, pro-survival signaling and also cell adhesion-mediated drug resistance were significantly higher in 3D than in 2D parallel co-cultures. The contribution of the VLA-4/VCAM1 pathway to resistance to bortezomib was modeled by the use of VCAM1 transfectants. Soluble factor-mediated drug resistance could be also demonstrated in both 2D and 3D co-cultures. The system was then successfully applied to co-cultures of primary myeloma cells-primary myeloma bone marrow stromal cells from patients and endothelial cells, allowing the development of functional myeloma-stroma interactions and MM cell long-term survival. Significantly, genomic analysis performed in a high-risk myeloma patient demonstrated that culture in bioreactor paralleled the expansion of the clone that ultimately dominated in vivo. Finally, the impact of bortezomib on myeloma cells and on specialized functions of the microenvironment could be evaluated. Our findings indicate that 3D dynamic culture of reconstructed human multiple myeloma microenvironments in bioreactor may represent a useful platform for drug testing and for studying tumor-stroma molecular interactions.
Introduction
Tumors develop and progress within co-evolving microenvironments that affect both the fate of the tumor and its drug sensitivity. Response to drugs may be overestimated on 2-dimensional (2D) cultures, and the discrepancy between pre-clinical findings and clinical outcomes can also be attributed to the failure of conventional 2D models.1–4 3D culture models closely reproduce tumor within its microenvironment, recapitulating tumor-stroma interactions and signaling.1–4 Multiple myeloma (MM) is a paradigm of tumor-stroma inter-dependence, as it develops almost exclusively within the bone marrow (BM),5–8 where MM cells establish tight contacts with the stroma, that in turn delivers pro-survival, anti-apoptotic signals and confers drug resistance.9 Accordingly, new drugs, including proteasome inhibitors, have been developed to target both MM cells and their BM microenvironment. However, the disease remains incurable predominantly due to development of drug resistance. Along this line, 3D models of MM cells inside their microenvironment to test the impact of drugs in a relevant human context have been recently described.10–13
We have previously contributed to 3D models for MM exploiting the Rotary Cell Culture System (RCCS™) bioreactor technology. By providing a balance between increased mass transfer and reduced shear stress, this dynamic bioreactor generates optimal conditions for long-term ex vivo maintenance of tissue explants.14–16 Specifically, we have shown that the model preserves, for extended time periods, the morphological and functional features of MM tissue components as well as their sensitivity to drugs.16
The aim of the present study was to recreate a surrogate 3D MM microenvironment able to reproduce the functional interactions of the native MM BM. We developed a robust technology, based on the integrated use of cell-repopulated scaffolds and the RCCS™ bioreactor. We demonstrate that our model simulates crucial MM features, in particular BM-MM dynamic interactions and MM survival/proliferation, thus providing a reliable tool to test the impact of drugs on MM cells inside their microenvironment.
Methods
Cell lines and primary cells
Human MM1.S, U266 and RPMI.8226 MM cell lines, HS-5 BM stromal cell line and murine L-fibroblasts transfected with human VCAM1 (L-VCAM) and their wild-type (wt) counterpart were maintained in DMEM or RPMI 1640 plus 10% fetal bovine serum. BM aspirates from MM patients were collected after written informed consent and ethical approval from the Institutional Review Board; primary MM cells from 7 newly diagnosed patients and one relapsed, and BMSC were obtained (see Online Supplementary Methods). Endothelial cells (HUVEC) were propagated as described.17 Osteoblasts were differentiated from BMSC (see Castrén et al.18 and Online Supplementary Methods).
Scaffold preparation and culture in RCCS™ bioreactor
Scaffolds were generated as in Figure 1A in bioreactor (see Ferrarini et al.16 and Online Supplementary Methods), with MM cell lines (500×103/scaffold) and HS-5 cells or L-VCAM1 or their wt counterpart (200×103/scaffold). Alternatively, primary MM cells (200×103/scaffold) were co-cultured in scaffolds with primary pooled allogeneic BMSC and HUVEC (100×103 each/scaffold), unless otherwise indicated. Scaffolds were then fixed or digested for flow cytometric (FACS) analysis. Supernatants were collected.
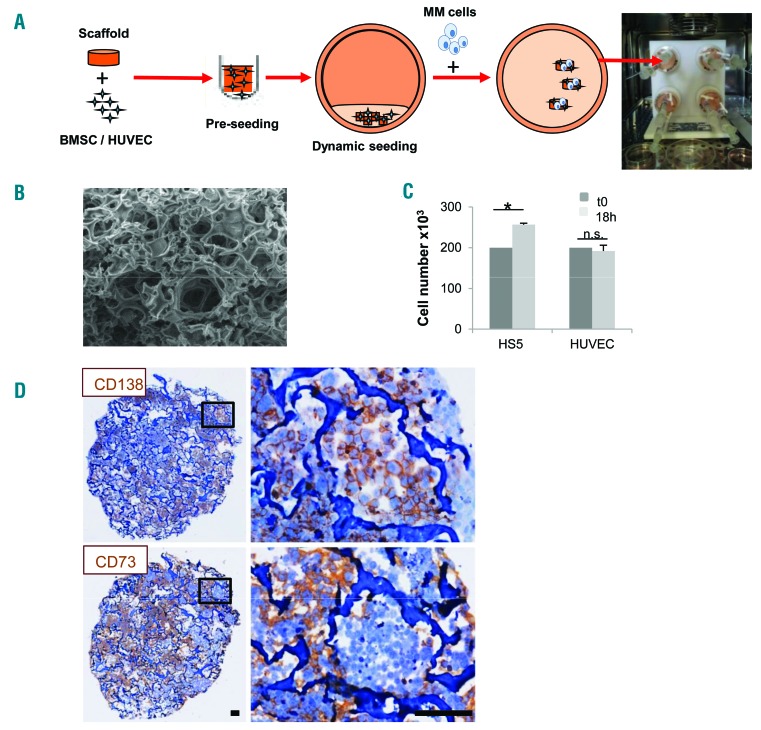
Figure 1.
Generation of a 3D multiple myeloma (MM) microenvironment in bioreactor. (A) Experimental procedure: scaffold is pre-seeded in vitro with bone marrow stromal cells (BMSC)/endothelial cells (HUVEC) and transferred to bioreactor. MM cells are then added and cultured (see Methods section). (B) Scanning electron microscopy analysis of Spongostan (bar=20 mm). (C) Input (t0) (200×103/scaffold) and recovered cell number after 18 hours (h) of 3D culture. Results are mean±Standard Error of Mean of three independent experiments. (D) Immunohistochemistry showing uniform distribution of CD138+ MM cells and CD73+ stroma. Bar=100 mm. *P≤0.05. n.s.: not significant.
Immunohistochemistry
Sections were stained with hematoxylin and eosin (H&E), or with monoclonal antibodies (mAbs): anti-Ki67, anti-CD138 (Ventana Systems); anti-light chains (Immunological Sciences, Italy); anti-CD73 (abcam), anti-cleaved caspase-3 (Cell Signaling Technology).16
Flow-cytometric analysis
Flow cytometric analysis (FACS) (FC500, Beckman Coulter) was performed using the following mAbs: anti-β1integrin/CD29 (BioLegend), followed by ALEXA-488-conjugated goat anti-mouse antibodies (Invitrogen); FITC-conjugated anti-VLA4/CD49d; PE-conjugated anti-VCAM-1/CD106, PC7-conjugated anti-CD38 (both from BD-Pharmingen).
Response to drugs
Bortezomib (Velcade®, Millenium Pharmaceuticals) was used at 10 nM for 24-48 hours (h), melphalan at 1.2 nM for 72 h, dexamethasone at 20 nM for 48 h; IL-6 (R&D Systems) at 10 ng/mL. The anti-VLA-4 mAb natalizumab19 was used at 10 mg/mL. Cells were stained with anti-CD38 mAb and Annexin-V (BD-Pharmingen) for flow cytometric analysis.
Adhesion assay
Multiple myeloma cell lines (200×103) were seeded over HS-5 or L-VCAM1 in 24-well plates. After 3 h, non-adherent cells were removed. Results are expressed as percentage (%) of CD38+ recovered adherent cells over input.
Western blot analysis
Western blot analysis was performed as described in the Online Supplementary Methods with polyclonal anti-pAkt (against S473, R&D Systems), monoclonal anti-pan-Akt (clone C67E7, Cell-Signaling Technology), anti-β1integrin mAb (abcam), anti-β-actin mAb (Santa Cruz Biotechnology), anti-STAT3/p-Stat and survivin (abcam). Proteins were quantified by ImageJ software.20
Scanning electron microscopy analysis
Scaffolds were fixed in 2% glutaraldehyde, post-fixed in 1% OsO4, dehydrated and then sputter coated with gold. Samples were examined by FEI/Philips XL-30 SEM (FEI, the Netherlands).
Determination of soluble factors and metallo-proteasic activities in supernatants
β2-microglobulin concentration was determined by immunonephelometry. Angiopoietin-2 (Ang-2), VEGF, FGF and IL-6 levels were quantified by ELISA (R&D Systems). IL-1β,IL-8/CXCL-8 and TGF-β concentrations were determined by Bio-Plex Multiple-Cytokines Assays (Bio-Rad).21 MMP-2 and MMP-9 activities were assessed through Zymography.16
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH)22 was performed using probes for the detection of trisomy 12, deletions of 11q22.3 (ATM), 13q14.3 (D13S319), 13q34 (LSI13q34), 17p13 (TP53) (Multi-color Probe, Abbott Molecular) and IGH gene rearrangements (DAKO). Microscope observation was performed using a Nikon Eclipse 90i (Nikon Instruments, Japan) and analyzed by Genikon software (Nikon).
Statistical analysis
Statistical analysis was performed using Student t-test or one-way ANOVA. *P≤0.05; **P≤0.01; ***P≤0.001.
Results
Generation of a functional MM 3D microenvironment in RCCS™ bioreactor requires supportive stroma
3D reconstruction of a surrogate MM microenvironment was based upon the proper combination of a scaffold and stromal cells, according to the procedure depicted in Figure 1A. As scaffold we used a commercially available gelatin biomaterial, which was selected according to several parameters (Online Supplementary Figure S1), including efficiency of cell seeding and recovery, and especially its ultrastructure, resembling that of adult bone (Figure 1B). An efficient scaffold seeding with either the HS-5 cell line or HUVEC was achieved in the RCCS™ bioreactor, as indicated by the overall number of viable recovered cells upon 18 h 3D culture (Figure 1C). As a final step, MM cells were added to the bioreactor vessels (Figure 1A) resulting in the successful formation of a homogeneously populated, dense construct where both tumor cells and stroma conserved lineage specific markers (Figure 1D).
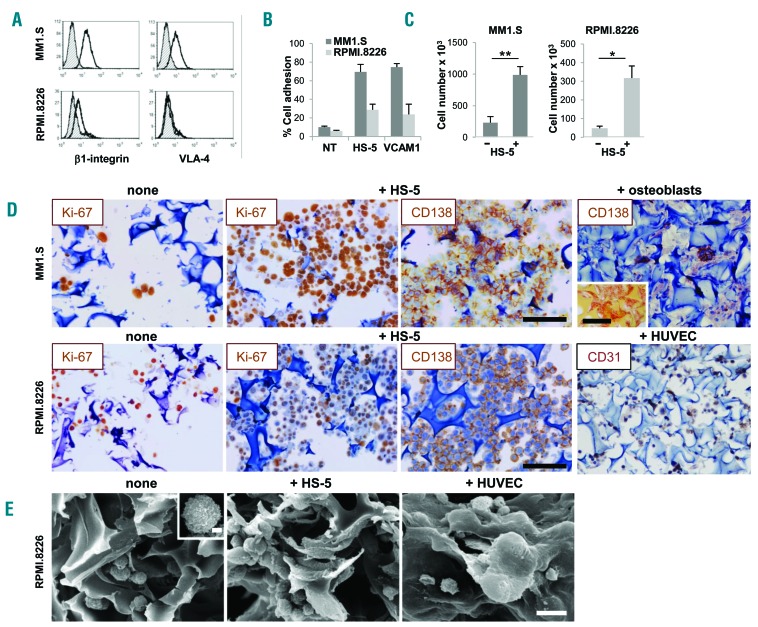
We established our model with MM1.S and RPMI.8266 MM cell lines. These cell lines differ remarkably in the expression of β1-integrin and very late antigen-4 (VLA-4) (Figure 2A) which, through the interaction with its ligand VCAM1, is involved in MM adhesion to BM stroma.23 As a result, adhesion to HS-5 or L-VCAM transfectant parallels these differences (Figure 2B). Stromal cells were also required for MM cell permanence inside the scaffold, as indicated by the significantly higher number of MM cells populating the scaffold in the presence versus the absence (nude scaffold) of stroma; this was particularly evident with MM1.S cells (Figure 2C). Accordingly, immunohistochemistry (IHC) indicated that both MM1.S and RPMI.8266 cells entered, were homogeneously distributed and proliferated inside the scaffolds, prevalently when pre-seeded with the HS-5 stromal cell line (Figure 2D). Other cell types within the MM BM microenvironment, including endothelial cells and osteoblasts, are increasingly recognized as participating in MM pathogenesis and progression.12,24 We then exploited our system to model MM cells-HUVEC and MM cells-osteoblasts co-cultures. The latter were obtained through bone differentiation of BMSC, as reported.18 Upon culture with osteogenic differentiation medium, BMSC underwent morphological changes, increased mineralization and acquired Alizarin staining (Online Supplementary Figure S2A-C). Moreover, parallel cultures performed with differentiated BMSC in 2D and, particularly in 3D conditions, showed alkaline phosphatase release (Online Supplementary Figure S2D). Notably, scaffolds seeded with bone differentiated BMSC and with CD31+HUVEC also supported MM cells homing and permanence (Figure 2D, right panels).
Figure 2.
Stroma is required for multiple myeloma (MM) cell permanence inside scaffolds. β1-integrin and α4 chain expression by flow cytometric (FACS) analysis (A) and in vitro adhesion to HS-5 cells and VCAM1 transfectant (B) of MM1.S and RPMI.8226 cells. Gray histograms represent the isotype controls. (C) Number of MM cells recovered from nude or pre-seeded scaffolds after 24 hours (input number =500×103/scaffold). Data are mean±Standard Error of Mean (SEM) of three independent experiments. (D) Immunohistochemistry (IHC) showing proliferating (Ki67+) CD138+ MM cells over a layer of HS-5 cells or CD31+HUVEC. CD138 staining of MM1.S in the presence of bone-differentiated bone marrow stromal cells is also shown. Insert represents alizarin staining of the osteoblasts-coated scaffold. Bar=100 mm. (E) Scanning electron microscopy analysis shows RPMI.8226 cells without (left panel and insert, bar=2 mm) and with HS-5 cells (middle panel) or endothelial cells (HUVEC) (right panel). Bar=20 mm.
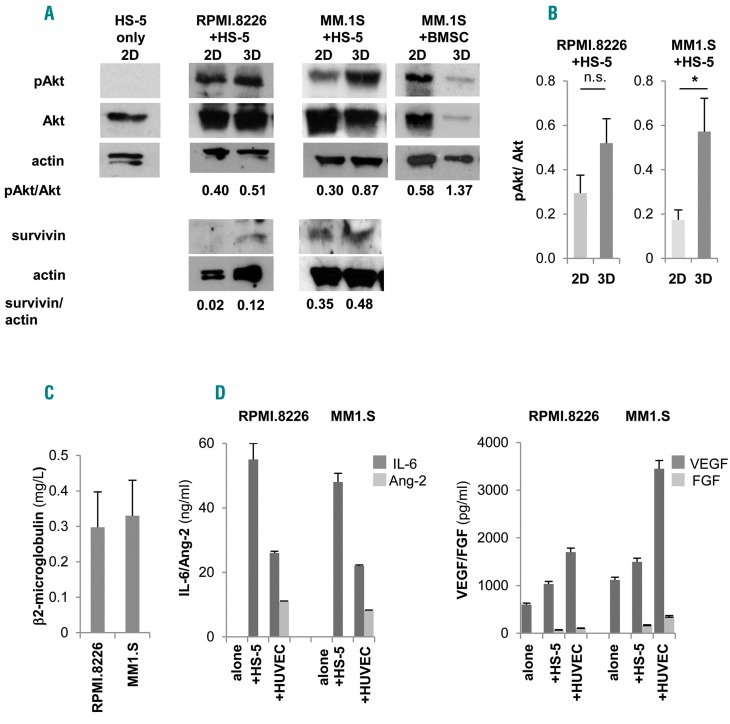
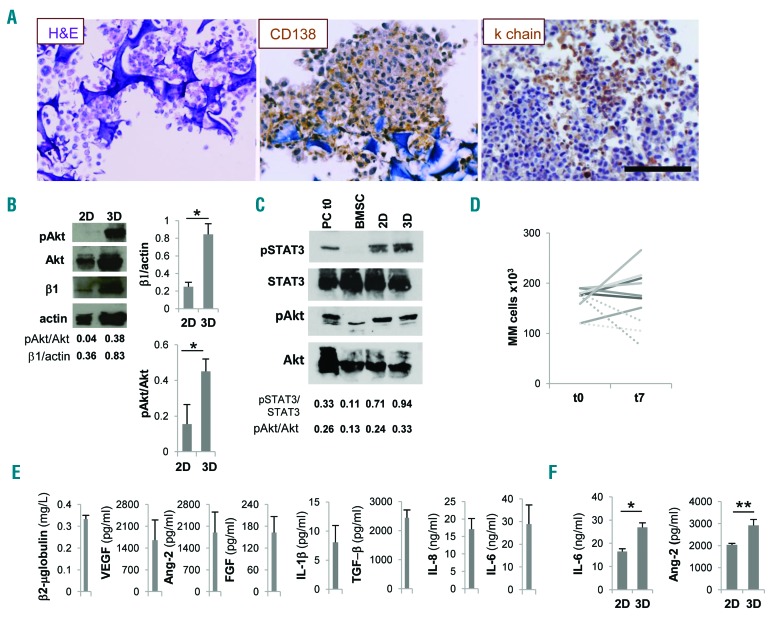
Development of intimate cell-cell contacts between MM cells and microenvironment was visualized at scanning electron microscopy analysis (Figure 2E, middle panel) showing that MM cells acquired a flatter morphology over the stroma, consistent with the induction of adhesion-mediated cytoskeletal rearrangement. Conversely, MM cells exhibited a round shape with few contact points over nude scaffolds (Figure 2E, left panel). Interestingly, the entire 3D cell surface of some MM cells was embedded when HUVEC were used as stroma, suggesting that cell-cell interactions may dynamically evolve upon contacts (Figure 2E, right panel). The recapitulated physical interactions resulted in the activation of pro-survival signals, as indicated by up-regulated Akt phosphorylation in MM cell line-stromal cell co-cultures in 3D compared to 2D co-cultures (Figure 3A and B); instead, pAkt expression was negligible in isolated HS-5 cells. Similarly, the expression of survivin was also increased in 3D co-cultures (Figure 3A).25,26 Specialized functions of both MM cells and stroma were detectable in culture supernatants, including β2-microglobulin (Figure 3C) and growth factor release. The latter varied according to the stromal elements coating the scaffolds: Ang-2 was related to the presence of HUVEC,20 while IL-6, VEGF and FGF were detectable in all co-cultures (Figure 3D).
Figure 3.
Pro-survival signals are delivered to multiple myeloma (MM) cell lines. (A) Western blot analyses of pAkt, Akt, survivin and actin in 3D versus 2D co-cultures of HS-5 and RPMI.8266 or MM1.S cells and of primary bone marrow stromal cells (BMSC) and MM1.S cells (one representative experiment out of three) and also on isolated HS-5 cells. (B) Mean±Standard Error of Mean (SEM) of pAkt/totalAkt ratios from three independent experiments. β2-microglobulin (C), IL-6, angiopoietin-2 (Ang-2), VEGF and FGF (D) levels in 3D culture supernatants. *P≤0.05.
Response to anti-myeloma drugs of MM cell lines inside the surrogate microenvironment
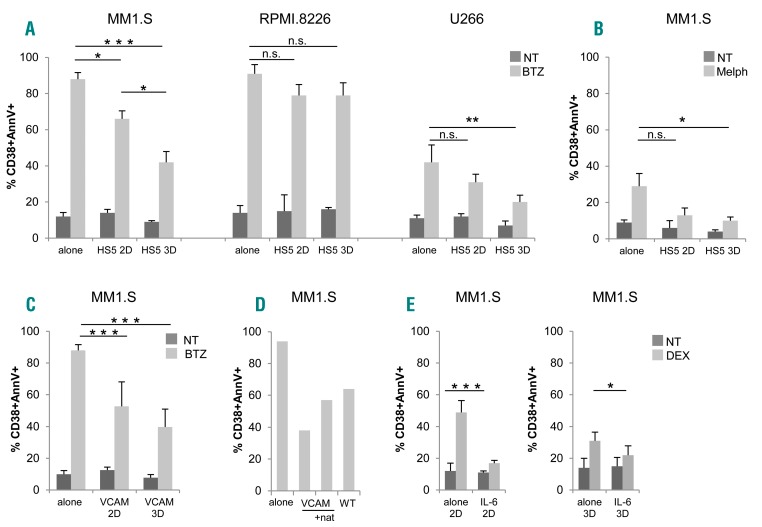
Tumor-stroma interactions affect MM cell behavior, including survival and drug resistance. The latter is induced via two overlapping pathways, i.e. soluble factor-mediated drug resistance (SFM-DR) and cell adhesion-mediated drug resistance (CAM-DR).27,28 To assess the impact of 3D microenvironment on bortezomib sensitivity, we performed parallel experiments in 2D versus 3D using HS-5 cells as stroma. Co-culture of MM1.S with HS-5 cells in 2D conditions conferred higher resistance to bortezomib, compared to MM.1S alone (Figure 4A, left). Significantly, resistance to bortezomib was even more evident when MM1.S were cultured with HS-5 cells in bioreactor, underscoring the role of 3D architecture per se in determining the impact of drugs (Figure 4A, left). The protective effect exerted by HS-5 cells was negligible with the RPMI.8226 cell line (Figure 4A, middle), in agreement with their reduced adhesion to stroma (Figure 2B). Protection conferred by HS-5 cells was not restricted to bortezomib-treated MM1.S since it was also observed with other cell lines (bortezomib-treated U266) (Figure 4A, right) and other drugs (melphalan) (Figure 4B), particularly in 3D conditions.
Figure 4.
Adhesion to stroma and 3D architecture confer drug resistance to multiple myeloma (MM) cells. (A) Bortezomib (BTZ)-induced apoptosis in MM1.S (left), RPMI.8226 (middle) and U266 (right) in 2D versus 3D experiments, evaluated as percentage (%) of CD38+annexinV (AnnV)+ by flow cytometric (FACS) analysis. (B) MM1.S death induced by melphalan (Melph). MM cells were either alone or co-cultured with HS-5 cells. Data are mean±Standard Error of Mean (SEM) of three independent experiments. (C) Comparison between 2D/3D BTZ-induced apoptosis of MM1.S co-cultured with L-VCAM. Data are mean±SEM of three independent experiments. (D) The blocking effect of natalizumab (nat) on BTZ-induced apoptosis on MM1.S co-cultured with L-VCAM is shown. Results are representative of two independent experiments. (E) Dexamethasone-induced apoptosis in MM1.S cells in the presence/absence of IL-6 (10 ng/mL) was evaluated as percentage (%) of CD38+ AnnV+ by flow cytometric analysis in 2D (left) and in 3D (right) conditions. Results are expressed as mean±Standard Deviation of three independent experiments. *P≤0.05; **P≤0.01; ***P≤0.001. WT: wild-type non transfected counterpart; NT: not treated.
HS-5 cells in principle can provide both mechanisms of drug resistance, since they release consistent amounts of IL-6 (Figure 3D) and serve as support to MM cells. Among tumor-stroma cell-cell interactions, we addressed as a paradigm the VLA-4/VCAM1 molecular pathway, which is considered to play a central role.27,28 In experiments performed in 2D conditions, primary VCAM1+ BMSC (Online Supplementary Figure S3A) from MM patients promoted adhesion at least in part via the counter ligand VLA-4 (Online Supplementary Figure S3B), as demonstrated by inhibition experiments with the specific α4 blocking antibody natalizumab,29 and accordingly conferred to MM1.S cells higher resistance to bortezomib compared to the VCAM1 negative HS-5 cell line (Online Supplementary Figure S3C); the release in culture supernatants of β2-microglobulin and of lactate dehydrogenase (LDH), the latter bona fide expression of bortezomib cytotoxicity, paralleled this response (Online Supplementary Figure S3C).
We then modeled the VLA-4/VCAM1 pair using the murine fibroblast L-VCAM transfectant (Figure 4C). The capability of VCAM1 expressed by the transfectants (Online Supplementary Figure S3A) to engage specific interactions was demonstrated by inhibition experiments (Figure 4D). Accordingly, the protection conferred by L-VCAM1 was highly significant in both 2D and 3D conditions, particularly in the latter (Figure 4C).
Finally, the involvement of soluble factors in promoting drug resistance in our system was investigated using the well-described model of dexamethasone-treated MM1.S cells, where IL-6 is recognized to exert a protective effect.30 As reported,27 the cytokine specifically triggered STAT3 pathway in MM1.S cells, as indicated by western blot analysis and inhibition experiments with the anti-IL-6R monoclonal antibody tocilizumab (TCZ) (Online Supplementary Figure S4A), and also protected MM1.S cells from dexamethasone-induced death (Figure 4E), both in 2D and in 3D conditions. To further support this finding, a viability assay was performed (Online Supplementary Figure S4B), showing that IL-6 significantly reduced the inhibitory effect of dexamethasone.
3D culture supports primary MM cells survival and functions
Isolated primary MM cells outside their native microenvironment do not survive. We exploited our model using primary MM cells from patients, with the major aim of promoting their survival. We took advantage of MM BM stroma and HUVEC onto which primary isolated CD138-MM cells were seeded and cultured in bioreactor for up to seven days. In the resulting constructs viable MM cells could be identified which retained the expression of lineage-specific markers as well as light chain production (Figure 5A). To address the role of cell-cell interactions, we compared the expression of β1 integrin in 2D and 3D parallel experiments. β1 integrin was up-regulated in scaffolds, and this upregulation was paralleled by downstream Akt phosphorylation (Figure 5B). STAT3 phosphorylation was also increased in 3D compared 2D co-cultures and could be attributed almost exclusively to MM cells (Figure 5C). Accordingly, when primary MM cells from 7 newly diagnosed patients were co-cultured with a pool of allogeneic BMSC plus HUVEC, the number of cells retrieved from the scaffolds at the end of culture matched the input number (Figure 5D), at variance with parallel 2D co-cultures. Moreover, both MM cells and stroma retained their specialized functions, as indicated by β2-microglobulin and soluble factors released in the supernatant from the co-cultures (Figure 5E), recapitulating the native BM microenvironment.31 Of note, cytokine concentrations were significantly higher in 3D than in 2D conditions (Figure 5F).
Figure 5.
Survival and functions of primary multiple myeloma (MM) cells and stroma are promoted in the scaffold. (A) Primary MM cells inside scaffold retain CD138 and light chain (κ chain) expression. (B) Western blot analyses performed on parallel 2D and 3D co-cultures of primary MM cells and MM bone marrow stromal cells (BMSC); pAkt, total Akt, β1 integrin (β1) and actin are depicted in a representative experiment (left) out of three. Right panels represent mean±Standard Error of Mean (SEM) of β1 integrin/actin and pAkt/total Akt ratios in three independent experiments. (C) Representative experiment of western blot analysis of STAT3 and Akt phosphorylation in 2D versus 3D co-cultures of primary MM cells and primary BMSC. Freshly isolated primary MM cells and BMSC are used as controls. (D) Input (t0) and recovered (t7) number of primary CD38+ MM cells from 7 patients after 3D culture for seven days. Dotted lines represent input and recovered number of CD38+ MM cells from 3 patients cultured in parallel 2D experiments. (E) Specialized functions of primary MM cells and stroma are measured in culture supernatants. Data are mean±SEM of six independent experiments. (F) IL-6 release in co-cultures with BMSC (left) and Ang-2 release in co-cultures with HUVEC (right) were determined by ELISA in parallel 2D and 3D experiments. Data are mean±SEM of three independent experiments. *P≤ 0.05; **P≤0.01. H&E: hematoxylin and eosin staining. Bar=100 mm.
Impact of bortezomib on a 3D culture of primary MM cells and stroma
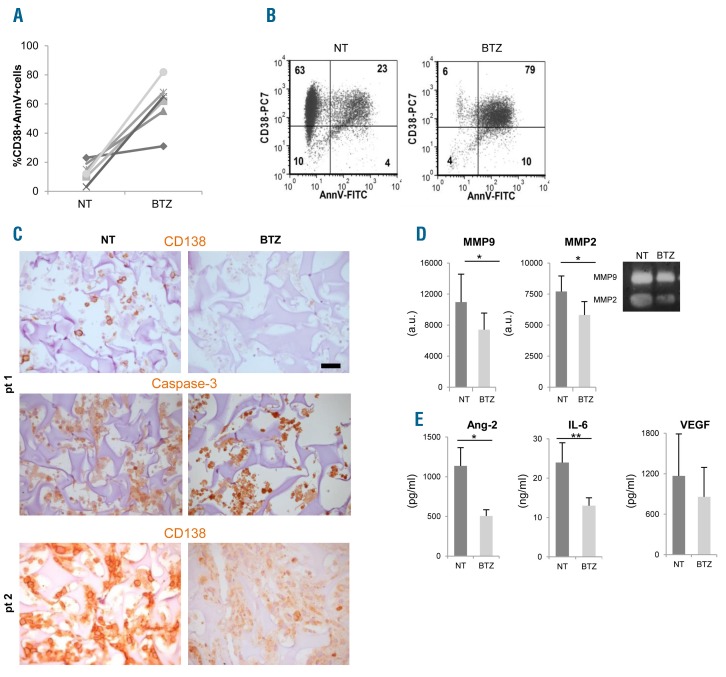
We next investigated whether our reconstructed model was suitable to assess sensitivity to bortezomib of primary MM cells within their microenvironment. To this aim, we performed parallel cultures in bioreactor in the presence/absence of bortezomib, using primary MM cells obtained from 6 patients. Scaffolds were coated with primary MM BMSC and HUVEC to better approximate the native microenvironment.12 After 48 h MM cells were retrieved from scaffolds and submitted to FACS analyses. Bortezomib-induced death, which varied among patients, could be determined as percentage of CD38+AnnV+ MM cells (Figure 6A and B). Bortezomib cytotoxicity could also be evaluated by IHC, showing down-modulated expression of CD138 antigen in caspase-3+ MM cells undergoing apoptosis32 (Figure 6C) and through the assessment of specialized functions. In particular, MMP-2 and MMP-9 activities (Figure 6D), as well as Ang-2 and IL-6 concentrations (Figure 6E), significantly decreased in supernatants from bortezomib-treated scaffolds, underlining the impact of the drug also on stroma.
Figure 6.
Bortezomib affects primary multiple myeloma (MM) cells viability and functions within the scaffolds. (A) Primary MM cells from 6 patients were retrieved from scaffolds after 48 hours of bortezomib (BTZ) treatment; death was calculated as the percentage of CD38+/AnnV+ cells by flow cytometric analysis. (B) A representative experiment is shown. (C) Immunohistochemistry performed on scaffolds populated with primary MM cells from 2 patients reveals the presence of apoptotic MM cells upon bortezomib exposure, as indicated by both down-regulated CD138 expression and intense nuclear caspase-3 immunoreactivity. Bar=50 μm. (D) Metalloprotease (MMPs) activities are measured in supernatants from treated (BTZ) and untreated (NT) samples by zymography and densitometric analyses (left and middle panels). Results are mean±Standard Error of Mean (SEM) of 6 patients. Right panel shows a representative experiment. (E) Angiopoietin-2 (Ang-2), IL-6 and VEGF levels are determined in supernatants of treated and untreated scaffolds. Data are mean±SEM of 6 patients.*P≤ 0.05; **P≤ 0.01. a.u.: arbitrary units.
3D culture supports proliferation of an in vivo expanding MM clone
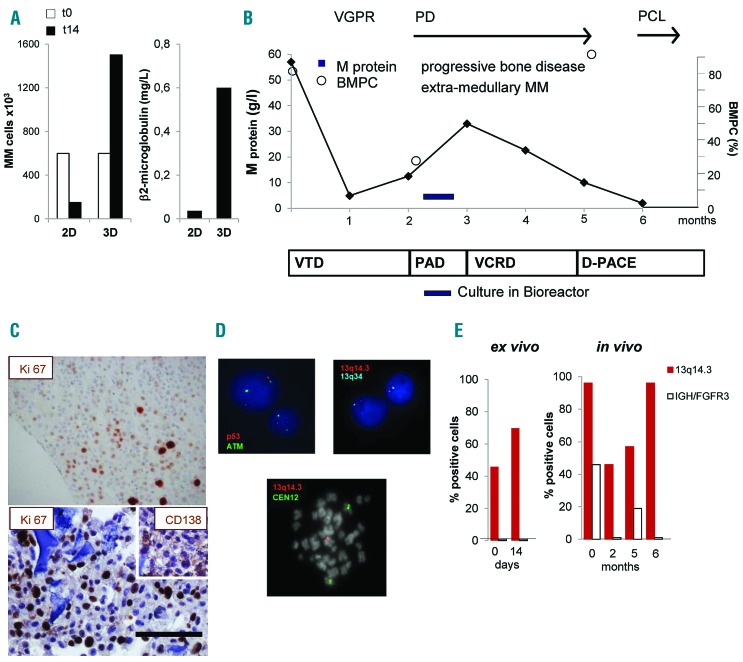
Multiple myeloma cells from 6 patients analyzed survived in 3D culture. In an additional case, primary MM cells not only survived but also significantly proliferated in 3D cultures, but not in parallel 2D conditions (Figure 7A). MM cells were obtained from a patient who initially achieved a very good partial response (VGPR) upon treatment with bortezomib-thalidomide-dexamethasone (VTD) but rapidly progressed to terminal plasma cell leukemia (PCL) despite several lines of therapy (Figure 7B). At progression, a BM biopsy was performed and MM cells were cultured in bioreactor (Figure 7B). MM cell proliferation was confirmed by the high frequency of Ki67+ cells at IHC analysis of the scaffold, which mirrored that observed in the BM biopsy (Figure 7C) and was paralleled by β2-microglobulin levels in supernatants (Figure 7A). Given the unique proliferative behavior of the patient’s MM cells, we compared genomic changes occurring over time in vivo and ex vivo. FISH analysis performed on MM cells retrieved at the end of 3D culture identified cells carrying the 13q14.3 deletion; no deletions of ATM or p53 loci could be detected (Figure 7D, upper panels), nor IGH rearrangements, indicating the absence of the t(4;14) translocation. Cells expressing the 13q14.3 deletion represented 46% of all MM cells at the beginning of culture and became 70% after 14-day 3D culture (Figure 7E), suggesting that this clone preferentially proliferated in bioreactor. Accordingly, few spontaneous metaphases, which are extremely rare in MM samples, could be observed in 3D culture and they all presented the deletion (Figure 7D, lower panel). Notably, at diagnosis, 96% of MM cells carried the 13q14.3 deletion and 46% co-expressed the t(4;14) IGH/FGFR3 translocation, consistent with the reported frequent association between the two cytogenetic lesions.33–35 After an initial tumor burden reduction in response to VTD, MM cells with the 13q14.3 deletion, but not those also carrying the t(4;14) translocation, progressively expanded in vivo, ultimately representing the whole population in PCL cells (Figure 7E).
Figure 7.
Culture in bioreactor mirrors the expansion of an in vivo proliferating multiple myeloma (MM) sub-clone. (A) Primary MM cell number (left panel) and β2 microglobulin release in supernatant (right panel) after 14 days of culture under parallel 2D and 3D co-cultures with HS-5 cells. (B) Schematic representation of patient’s clinical course and treatments. After diagnosis (t0), response to treatment was assessed and defined as Very Good Partial Response (VGPR), Progressive Disease (PD), Plasma Cell Leukemia (PCL). Serum M protein concentration and percentage of bone marrow PC (BMPC) were serially determined. Treatments were: bortezomib-thalidomide-dexamethasone (VTD), bortezomib-doxorubicin-dexamethasone (PAD), bortezomib-cyclophosphamide-lenalidomide-dexamethasone (VCRD), dexamethasone-cisplatin-adriamycin-cyclophosphamide-etoposide (D-PACE). (C) Immunohistochemistry of the proliferation marker Ki-67 inside a scaffold (lower) and in a matched bone biopsy (upper). Insert represents CD138 staining. Bar=100 mm. (D) Interphase fluorescence in situ hybridization analysis performed in purified MM cells retrieved upon culture in bioreactor (upper), showing normal pattern of ATM and p53 (upper left) and 13q14.3/13q34 deletion (upper right); in the lower panel, a spontaneous metaphase of a PC showing 13q14.3/13q34 deletion. (E) Percentage of cells carrying the 13q14.3/13q34 deletion or IGH/FGFR3 translocation ex vivo and in vivo.
Discussion
The availability of suitable models that recapitulate the complex tumor-host interplay is central to understanding cancer biology and developing appropriate treatments. This is especially true in MM,36 where MM-BM interactions are crucial to disease progression and responsiveness to drugs.7,8 We here show that our ex vivo 3D co-culture model in bioreactor meets the requirements of recapitulated MM-BM dialogue, permanence and survival of primary MM cells for an extended time period, thereby also incorporating the temporal dimension. The model relies on the integrated use of a gelatin scaffold seeded with tumor cells and stroma and the RCCS™ bioreactor technology. Scaffolds are a key component for the reconstitution of MM microenvironment as they provide cells with mechanical support;37–39 we selected a gelatin biomaterial that allows morphological investigations and also mimics 3D ultrastructure of MM BM. Stroma is required to achieve ex vivo cell seeding efficiency and to re-create tumor-stroma contacts and signaling, both with MM cell lines and primary MM cells. Overall, the construct reproduces the tumor-stroma 3D ‘dynamic reciprocity’40 which is lost in conventional 2D co-culture. Both MM cells and stroma retain the expression of lineage specific markers as well as their specialized functions, including the release of β2-microglobulin, cytokines and growth factors, thus recapitulating the profile of the native BM.29,31
The model was established with MM cell lines and successfully applied to primary MM cells. Notably, primary MM cells from patients survived for up to seven days when cultured inside scaffolds. We used as stroma primary allogeneic BMSC from MM patients which possess the repertoire of adhesion molecules and have been previously validated for the reconstruction of an MM BM niche.12 Given the high flexibility of our model, the contribution of additional cellular elements of MM microenvironment can be addressed. As an example, the engagement of intimate EC-MM contacts inside the scaffold, documented by morphological and functional analyses, indicates that the system is suitable to elicit and study dynamic EC-MM targetable interactions. Moreover, the feasibility of co-culturing MM cells and bone-differentiated BMSC underscores the potential to fulfill the unmet need for a model to study the relationship between MM progression and bone disease.41 Finally, development of repopulated scaffolds could be further exploited to address the contribution of circulating MM cells in humanized in vivo scaffold-mouse models.42
Multiple myeloma-BM functional interactions and down-stream signaling are promoted inside our surrogate microenvironment. Indeed, higher levels of pAkt, pSTAT and survivin are more appreciable in 3D than in 2D culture with both MM cell lines and primary MM cells, mimicking the activation of pro-survival signaling pathways in MM cells from patients.25,43,44 Akt pathway is crucial for MM survival and drug resistance, and has been proposed as a promising target for future molecular-based therapies.43 Tumor-stroma interactions are considered a major determinant in drug resistance in MM via the release of soluble factors and cell-to-cell adhesion. Our data on the protective effect of stroma against bortezomib-induced apoptosis are consistent with the induction of CAM-DR; in this regard, experiments conducted with the L-VCAM transfectant indicate that the system can be exploited to model and elucidate specific molecular interactions. The additional contribution by 3D culture further supports the importance of tissue architecture per se in drug resistance. The achievement of a construct where MM cells survive via the establishment of proper 3D interactions with a compliant microenvironment is a prerequisite to test the impact of drugs in a relevant human context. Accordingly, the impact of bortezomib on primary MM cells and their microenvironment could be assessed by means of FACS and IHC analyses, and also through determination of specialized functions in supernatants. In particular, the decrease of Ang-2 can monitor the cytotoxic effect of the drug on EC,45 while variations in IL-6 and MMP-2/-9 activities may result from disruption of MM-stroma interplay.7,9
The dissection of clonal dynamics during disease progression and in response to therapy is increasingly emerging as a central issue of MM investigation.34,35 In a patient with high-risk MM, sequential BM sampling allowed the evolutionary path to be defined along the clinical course. Two sub-clones co-existing at diagnosis initially responded to first-line therapy; subsequently, only one evolved over time. Significantly, the bioreactor culture could anticipate the expansion of the same clone. These data suggest that, in selected cases, the model can be exploited to monitor the dynamics of clones inside the whole MM cell population, and possibly to identify new potential targets.
Altogether, our findings indicate that 3D dynamic culture of reconstructed human MM microenvironments in RCCS™ bioreactor may represent an important platform for drug testing and the study of tumor-stroma molecular interactions.
Supplementary Material
Acknowledgments
We would like to thank Dr. Andrea Motta and Dr. Tania Carenzo for technical assistance, Dr. Cristina Tresoldi, San Raffaele Scientific Institute, for providing MM samples, and Prof. V. Gattei, Aviano, Italy, for providing L-VCAM.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/4/707
Funding
This work was supported by AIRC-Special Program Molecular Clinical Oncology AIRC 5×1000 project n. 9965 (to FCC) and from its extension (to PG).
References
- 1.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. [DOI] [PubMed] [Google Scholar]
- 2.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18(1):27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8(10):839–845. [DOI] [PubMed] [Google Scholar]
- 4.Yamada M, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601–610. [DOI] [PubMed] [Google Scholar]
- 5.Hallek M, Bergsagel PL, Anderson KC. Multiple myeloma: increasing evidence for a multistep transformation process. Blood. 1998;91(1):3–21. [PMC free article] [PubMed] [Google Scholar]
- 6.Hussein MA, Juturi JV, Lieberman I. Multiple myeloma: present and future. Curr Opin Oncol. 2002;14(1):31–35. [DOI] [PubMed] [Google Scholar]
- 7.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. [DOI] [PubMed] [Google Scholar]
- 8.Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood. 2009;114(16):3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2009;23(1):10–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirshner J, Thulien KJ, Martin LD, et al. A unique three-dimensional model for evaluating the impact of therapy on Multiple Myeloma. Blood. 2008;112(7):2935–2945. [DOI] [PubMed] [Google Scholar]
- 11.Calimeri T, Battista E, Conforti F, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia. 2011;25(4):707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reagan MR, Mishima Y, Glavey SV, et al. Investigating osteogenic differentiation in multiple myeloma using a novel 3D bone marrow niche model. Blood. 2014;124(22): 3250–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakubikova J, Cholujova D, Hideshima T, et al. A novel 3D mesenchymal stem cell model of the multiple myeloma bone marrow niche: biologic and clinical applications. Oncotarget. 2016;7(47):77326–77341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazzoleni G, Di Lorenzo D, Steimberg N. Modelling tissues in 3D: the next future of pharmaco-toxicology and food research? Genes Nutr. 2009;4(1):13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosmi F, Steimberg N, Dreossi D, Mazzoleni G. Structural analysis of rat bone explants kept in vitro in simulated microgravity conditions. J Mech Behav Biomed Mater. 2009;2(2):164–172. [DOI] [PubMed] [Google Scholar]
- 16.Ferrarini M, Steimberg N, Ponzoni M, et al. Ex-vivo dynamic 3-D culture of human tissues in the RCCS™ bioreactor allows the study of Multiple Myeloma biology and response to therapy. PLoS One. 2013;8(8):e71613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belloni D, Scabini S, Foglieni C, et al. The vasostatin-I fragment of chromogranin A inhibits VEGF-induced endothelial cell proliferation and migration. FASEB J. 2007;21(12):3052–3062. [DOI] [PubMed] [Google Scholar]
- 18.Castrén E, Sillat T, Oja S, et al. Osteogenic differentiation of mesenchymal stromal cells in two-dimensional and three-dimensional cultures without animal serum. Stem Cell Res Ther. 2015;6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Schürpf T, Springer TA. How natalizumab binds and antagonizes α4 integrins. J Biol Chem. 2013;288(45):2314–32325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belloni D, Marcatti M, Ponzoni M, et al. Angiopoietin-2 in bone marrow milieu promotes multiple myeloma-associated angiogenesis. Exp Cell Res. 2015;330(1):1–12. [DOI] [PubMed] [Google Scholar]
- 21.Dagna L, Corti A, Langheim S, et al. Tumor necrosis factor α as a master regulator of inflammation in Erdheim-Chester disease: rationale for the treatment of patients with infliximab. J Clin Oncol. 2012;30(28):286–290. [DOI] [PubMed] [Google Scholar]
- 22.Gole L, Lin A, Chua C, Chang WJ. Modified cIg-FISH protocol for multiple myeloma in routine cytogenetic laboratory practice. Cancer Genet. 2014;207(1–2):31–34. [DOI] [PubMed] [Google Scholar]
- 23.Noborio-Hatano K, Kikuchi J, Takatoku M, et al. Bortezomib overcomes cell-adhesion-mediated drug resistance through down-regulation of VLA-4 expression in multiple myeloma. Oncogene. 2009;28(2):231–242. [DOI] [PubMed] [Google Scholar]
- 24.Mondello P, Cuzzocrea S, Navarra M, Mian M. Bone marrow micro-environment is a crucial player for myelomagenesis and disease progression. Oncotarget. 2017;8(12):20394–20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu J, Shi Y, Krajewski S, et al. The AKT kinase is activated in multiple myeloma tumor cells. Blood. 2001;98(9):2853–2855. [DOI] [PubMed] [Google Scholar]
- 26.Younes H, Leleu X, Hatjiharissi E, et al. Targeting the phosphatidylinositol 3-kinase pathway in multiple myeloma. Clin Cancer Res. 2007;13(13):3771–3775. [DOI] [PubMed] [Google Scholar]
- 27.Furukawa Y, Kikuchi J. Epigenetic mechanisms of cell adhesion-mediated drug resistance in multiple myeloma. Int J Hematol. 2016;104(3):281–292. [DOI] [PubMed] [Google Scholar]
- 28.Nefedova Y, Landowski TH, Dalton WS. Bone marrow stromal-derived soluble factors and direct cell contact contribute to de novo drug resistance of myeloma cells by distinct mechanisms. Leukemia. 2003;17(6):1175–1182. [DOI] [PubMed] [Google Scholar]
- 29.Podar K, Zimmerhackl A, Fulciniti M, et al. The selective adhesion molecule inhibitor Natalizumab decreases multiple myeloma cell growth in the bone marrow microenvironment: therapeutic implications. Br J Haematol. 2011;155(4):438–448. [DOI] [PubMed] [Google Scholar]
- 30.Greenstein S, Krett NL, Kurosawa Y, et al. Characterization of the MM.1 human multiple myeloma (MM) cell lines: a model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells. Exp Hematol. 2003;31(4):271–282. [DOI] [PubMed] [Google Scholar]
- 31.Manier S, Sacco A, Leleu X, Ghobrial IM, Roccaro AM. Bone marrow microenvironment in multiple myeloma progression. J Biomed Biotechnol. 2012;2012:157496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nerini-Molteni S, Ferrarini M, Cozza S, Caligaris-Cappio F, Sitia R. Redox homeostasis modulates the sensitivity of myeloma cells to bortezomib. Br J Haematol. 2008;141(4):494–503. [DOI] [PubMed] [Google Scholar]
- 33.Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14(2):100–113. [DOI] [PubMed] [Google Scholar]
- 34.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrarini M, Mazzoleni G, Steimberg N, et al. Innovative Models to Assess Multiple Myeloma Biology and the Impact of Drugs. In: Multiple Myeloma - A Quick Reflection on the Fast Progress. Hajek R. (Ed.), InTech, 2013. 10.5772/54312. [DOI] [Google Scholar]
- 37.Zhang M, Boughton P, Rose B, Lee CS, Hong AM. The use of porous scaffold as a tumor model. Int J Biomater. 2013;2013:396056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De la Puente P, Azab AK. 3D tissue-engineered bone marrow: what does this mean for the treatment of multiple myeloma? Future Oncol. 2016;12(13):1545–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischbach C, Chen R, Matsumoto T, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4(10):855–860. [DOI] [PubMed] [Google Scholar]
- 40.Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116(12):2377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reagan MR, Liaw L, Rosen CJ, Ghobrial IM. Dynamic interplay between bone and multiple myeloma: emerging roles of the osteoblast. Bone. 2015;75:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu D, Wang Z, Zhao JJ, et al. The Cyclophilin A-CD147 complex promotes the proliferation and homing of multiple myeloma cells. Nat Med. 2015;21(6):572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mimura N, Hideshima T, Shimomura T, et al. Selective and potent Akt inhibition triggers anti-myeloma activities and enhances fatal endoplasmic reticulum stress induced by proteasome inhibition. Cancer Res. 2014;74(16):4458–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsubaki M, Takeda T, Ogawa N, et al. Overexpression of survivin via activation of ERK1/2, Akt, and NF-κB plays a central role in vincristine resistance in multiple myeloma cells. Leuk Res. 2015;39(4):445–452. [DOI] [PubMed] [Google Scholar]
- 45.Belloni D, Veschini L, Foglieni C, et al. Bortezomib induces autophagic death in proliferating human endothelial cells. Exp Cell Res. 2010;316(6):1010–1018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.