Abstract
Due to heterogeneous morphological and immunophenotypic features, approximately 50% of peripheral T-cell lymphomas are unclassifiable and categorized as peripheral T-cell lymphomas, not otherwise specified. These conditions have an aggressive course and poor clinical outcome. Identification of actionable biomarkers is urgently needed to develop better therapeutic strategies. Epigenetic alterations play a crucial role in tumor progression. Histone modifications, particularly methylation and acetylation, are generally involved in chromatin state regulation. Here we screened the core set of genes related to histone methylation (KMT2D, SETD2, KMT2A, KDM6A) and acetylation (EP300, CREBBP) and identified 59 somatic mutations in 45 of 125 (36.0%) patients with peripheral T-cell lymphomas, not otherwise specified. Histone modifier gene mutations were associated with inferior progression-free survival time of the patients, irrespective of chemotherapy regimens, but an increased response to the histone deacetylase inhibitor chidamide. In vitro, chidamide significantly inhibited the growth of EP300-mutated T-lymphoma cells and KMT2D-mutated T-lymphoma cells when combined with the hypomethylating agent decitabine. Mechanistically, decitabine acted synergistically with chidamide to enhance the interaction of KMT2D with transcription factor PU.1, regulated H3K4me-associated signaling pathways, and sensitized T-lymphoma cells to chidamide. In a xenograft KMT2D-mutated T-lymphoma model, dual treatment with chidamide and decitabine significantly retarded tumor growth and induced cell apoptosis through modulation of the KMT2D/H3K4me axis. Our work thus contributes to the understanding of aberrant histone modification in peripheral T-cell lymphomas, not otherwise specified and the stratification of a biological subset that can benefit from epigenetic treatment.
Introduction
Peripheral T-cell lymphomas (PTCL) represent a heterogeneous clinicopathological entity of non-Hodgkin lymphoma with an aggressive disease course and poor clinical outcome. Approximately 50% of PTCL are unclassifiable and categorized as PTCL, not otherwise specified (PTCL-NOS).1 Using gene expression profiling, PTCL-NOS lymphocytes can be distinguished from normal T lymphocytes, with deregulation of genes involved in apoptosis, proliferation, cell adhesion, and transcription regulation.2 Two subgroups of PTCL-NOS have been identified, which are characterized by high expression of either GATA3 or TBX21/T-bet transcription factors and downstream target genes.3 However, actionable biomarkers closely related to the pathogenic mechanism need to be further investigated and may become potential therapeutic targets of PTCL-NOS.4,5
Epigenetic alterations play a crucial role in tumor progression.6 Next-generation sequencing technologies have led to the discovery of epigenetic modifier gene mutations in PTCL, such as the DNA methylation genes TET2, TET1 and DNMT3A,7,8 and chromatin remodeler genes ARID1B and ARID2.9,10 Meanwhile, genes of histone methylation, such as KMT2D, KMT2A, KDM6A, SETD2 and EZH2, and those of histone acetylation, including CREBBP and EP300, have also been found in PTCL and other T-lymphoid malignancies.9,11–13 To further determine their prognostic significance and correlation with clinical treatment, here we assessed the mutational pattern of the main epigenetic modifier genes in patients with PTCL-NOS.
Methods
Patients
A total of 239 patients with previously untreated PTCL-NOS were enrolled in this study. The histological diagnosis was established according to the World Health Organization (WHO) classification.14 The study was approved by the Shanghai Rui Jin Hospital Review Board with informed consent obtained in accordance with the Declaration of Helsinki.
Targeted sequencing
Targeted sequencing was performed on available tumor samples of 125 patients. To determine the mutations of candidate genes, polymerase chain reaction primers were designed by iPLEX Assay Design software (Sequenom, California, USA). Multiplexed libraries of tagged amplicons from patients with PTCL-NOS were generated by the 48×48 Access Array microfluidic platform (Fluidigm, South San Francisco, USA) according to the manufacturer’s protocol. Deep sequencing was performed with established Illumina protocols on the GAIIx and MiSeq platform (Illumina, California, USA). Matched peripheral blood samples were included to exclude germline polymorphisms and the mutations were confirmed by Sanger sequencing.
Cell line and reagents
The Jurkat T-leukemia cell line was obtained from the American Type Culture Collection. Cells were grown in RPMI-1640 medium, supplemented with 10% heat-inactivated fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37°C. Valproic acid (VPA, V3640) was from Sigma (San Francisco, USA). Suberoylanilide hydroxamic acid (SAHA, S1047) and romidepsin (ROMI, S3020) were from Selleck (Houston, USA). Chidamide, the histone deacetylase (HDAC) inhibitor clinically available in China, was kindly provided by Chipscreen (shenzhen, China).
Lentivirus packaging and transduction
Purified plasmids pGV365-KMT2D (WT), pGV365-KMT2D (V5486M), pGV365-EP300 (WT) and pGV365-EP300 (H1377R) were transfected with package vectors into HEK-293T cells using lipofectamine 2000 (Invitrogen, California, USA; 11668019) according to the manufacturer’s protocol. The supernatant fraction of HEK-293T cell cultures was then condensed to a viral concentration of approximately 3×108 transducing units/mL. The lentiviral particles were incubated with Jurkat cells for 72 h. The stably transduced cells were selected by EGFP or mCherry fluorescence protein after transduction.
Statistical analysis
Data were calculated as the mean ± standard deviation from three separate experiments. The Student t-test was applied to compare two normally distributed groups and the Mann-Whitney U test to compare two groups which did not conform to normal distribution. The Bonferroni adjustment was used to perform multiple comparisons. Progression-free survival was calculated from the date when treatment began to the date when the disease progression was recognized or the date of the last follow-up. Overall survival time was measured from the date of diagnosis to the date of death or the last follow-up. Univariate hazard estimates were generated with unadjusted Cox proportional hazards models. Covariates demonstrating statistical significance with P values <0.05 on univariate analysis were included in the multivariate model. All statistical procedures were performed with the SPSS version 20.0 statistical software package or GraphPad Prism 5 software. P<0.05 was considered statistically significant.
Online supplementary methods
DNA preparation, western blot, immunofluorescence, immunohistochemistry, isobolographic analysis, mRNA-seq library preparation and sequencing analysis, ChIP-seq library preparation and sequencing analysis, the TUNEL assay, murine model and micro-positron emission tomography and computed tomography imaging are described in the Online Supplementary Methods.
Results
Histone modifier genes were frequently mutated in peripheral T-cell lymphoma not otherwise specified
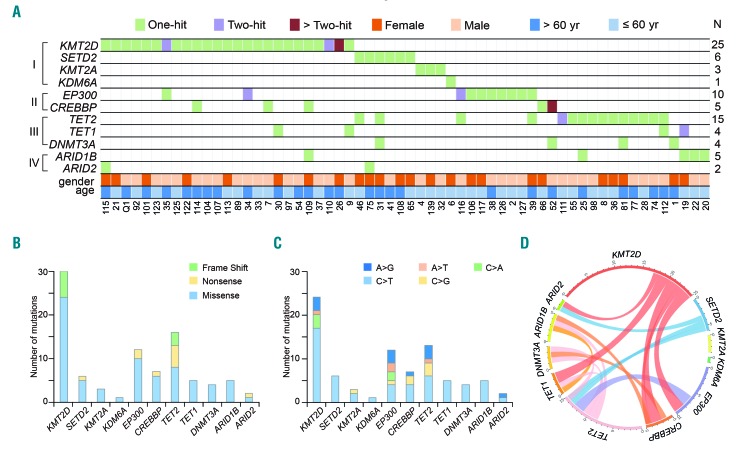
A total of 91 somatic mutations of epigenetic modifier genes were identified in 60 of 125 (48.0%) patients with PTCL-NOS by targeted sequencing (Figure 1A). Most of the somatic mutations were missense mutations (n=72), followed by nonsense (n=10) and frameshift mutations (n=9) (Figure 1B). We observed a preference for C>T/G>A alterations analogous to the somatic single-nucleotide variation spectrum in other cancers (Figure 1C). No correlation was found in terms of age and gender. Mutations of histone methylation genes (category I) most frequently occurred in KMT2D (encoding H3K4 methyltransferase, 25/125 patients, 20.0%), followed by those in SETD2 (encoding H3K36 methyltransferase, 6/125 patients, 4.8%), KMT2A (encoding H3K4 methyltransferase, 3/125 patients, 2.4%) and KDM6A (encoding H3K27 demethylase, 1/125 patients, 0.8%). No EZH2 mutation was detected. Mutations of histone acetylation genes (category II) were found in EP300 (encoding H3K18 acetyltransferase, 10/125 patients, 8.0%) and in CREBBP (encoding H3K18 acetyltransferase, 5/125 patients, 4.0%). DNA methylation genes TET2, TET1 and DNMT3A (category III), as well as chromatin remodeler genes ARID1B and ARID2 (category IV), were also affected in 12.0%, 3.2%, 3.2%, 4% and 1.6% of the patients, respectively (Figure 1A and Online Supplementary Table S1). In accordance with the conceptual classification of the mutated genes, overlap mutations were seldom present among histone methylation, histone acetylation, DNA methylation or chromatin remodeler genes. In particular, histone methylation gene mutations, such as mutations of histone acetylation genes, were mutually exclusive of each other, suggesting that histone modifying genes might be involved in distinct biological processes (Figure 1D).
Figure 1.
Histone modifier gene mutations in peripheral T-cell lymphoma, not otherwise specified. (A) Gene mutations identified by targeted sequencing in 125 patients with peripheral T-cell lymphomas. The number of patients (N) with mutations is listed on the right. The mutations are classified into the categories indicated on the left: I, histone methylation; II, histone acetylation; III, DNA methylation; IV, chromatin remodeler. (B) Number and type of non-silent somatic mutations. (C) Number and percentage of non-silent somatic single nucleotide variants. (D) Circos diagram according to mutation categories.
Alterations of histone modifier genes were primarily located at well-conserved amino acid positions across distinct species (Online Supplementary Figure S1). Typically, KMT2D/KMT2A mutations affected the PHD domain (e.g. residues 134–320, 1378–1556, 5032–5138, and 1433–1624, 1871–1983), HMG domain (residues 2021–2072), undetermined domain (e.g. residues 2487–4658) and SET domain (e.g. residues 5397–5519 and 3825–3969). EP300/CREBBP mutations affected the HAT domain (e.g. residues 1306–1612 and 1342–1649).
Histone modifier gene mutations were associated with disease progression in peripheral T-cell lymphoma not otherwise specified
One hundred and forty patients were treated with CHOP-based chemotherapy in a historical cohort of Shanghai Ruijin Hospital from 1997 to 2011, and referred to as the training cohort. The validation cohort consisted of 99 patients enrolled in two prospective studies (NCT 01746992 and NCT 02533700, randomized trials to compare CHOP-based chemotherapy with sequential chemotherapy with CEOP/IVE/GDP or CTOP/ITE/MTX). Since 2012, 49 and 50 patients have been randomized to CHOP-based or sequential chemotherapy, respectively. No obvious differences in clinical and pathological characteristics or treatment response were observed either between the training and the validation cohort, or between the two arms within the validation cohort (Online Supplementary Table S2). Gene mutation data were available for 73 and 52 patients of the training and validation cohorts with available tissue samples, respectively (Figure 1A).
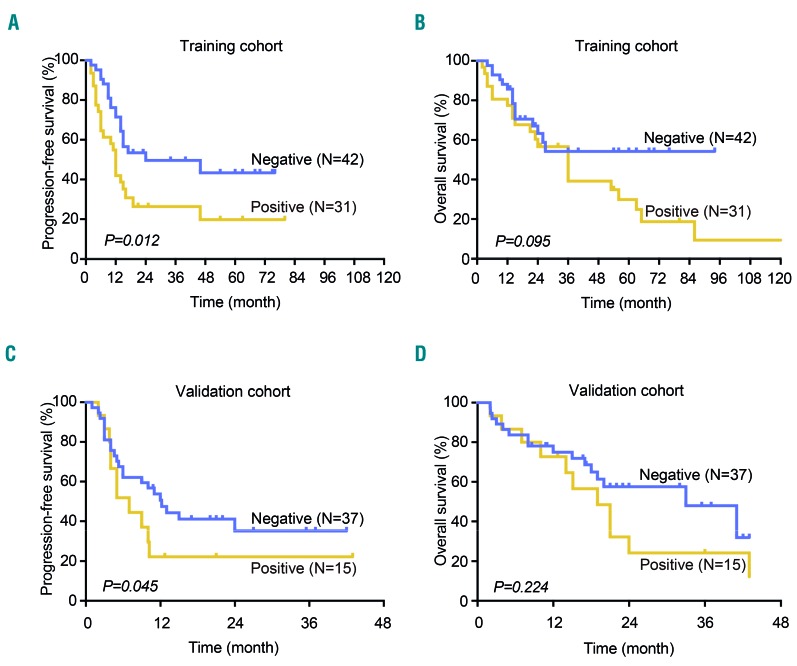
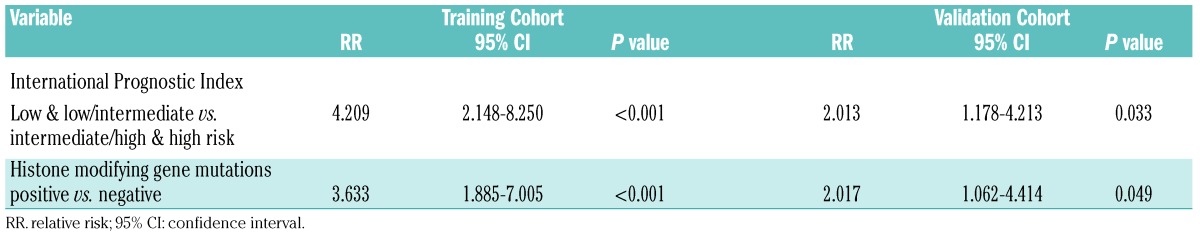
In the training cohort, the median follow-up time was 29.1 months (range, 0.5–162.0 months). The 2-year progression-free and overall survival rates of the patients were 36.7% and 47.1%, respectively. In the univariate analysis, the International Prognostic Index was a significant prognostic factor for both progression-free survival and overall survival (both P<0.001), but histone modifier mutations were only prognostic for progression-free survival and not overall survival (P=0.012 and P=0.095, respectively) (Figure 2A,B). In the multivariate analysis, when the International Prognostic Index was controlled for, the presence of a histone modifier gene mutation was an independent prognostic factor for progression-free survival (P<0.001) (Table 1). The 2-year progression-free and overall survival rates were 26.4% and 56.6% for patients with histone modifier gene mutations and 49.6% and 63.3% for patients without mutations (Figure 2A,B). In the validation cohort, the median follow-up time was 19.5 months (range, 2.1–43.0 months). Histone modifier gene mutations were associated with shorter progression-free survival in multivariate analysis (P=0.049) (Table 1). The 2-year progression-free and overall survival rates were 22.2% and 24.2% for patients with histone modifier gene mutations and 41.1% and 57.5% for patients without mutations (P=0.045 and P=0.224) (Figure 2C,D). Overall, these data expand the prognostic role of histone modification in disease progression in PTCL-NOS.
Figure 2.
Progression-free survival and overall survival curves of patients with peripheral T-cell lymphoma not otherwise specified according to histone modifier gene mutations. (A) Progression-free survival and (B) overall survival curves of the training cohort. (C) Progression-free survival and (D) overall survival curves of the validation cohort.
Table 1.
Multivariate analysis of predictors of progression-free survival in patients with PTCL-NOS controlled by International Prognostic Index.
Histone modifier gene mutations sensitized T-lymphoma cells to the histone deacetylase inhibitor chidamide and/or the hypomethylating agent decitabine
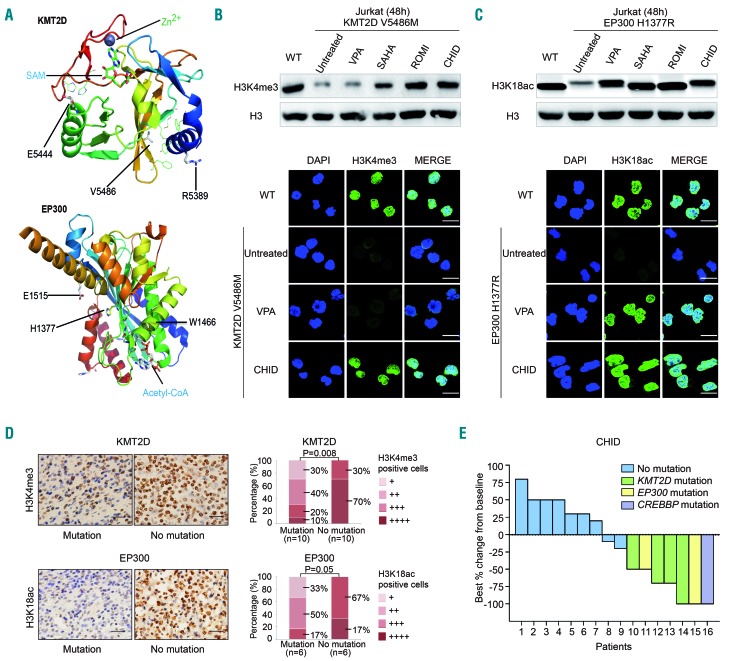
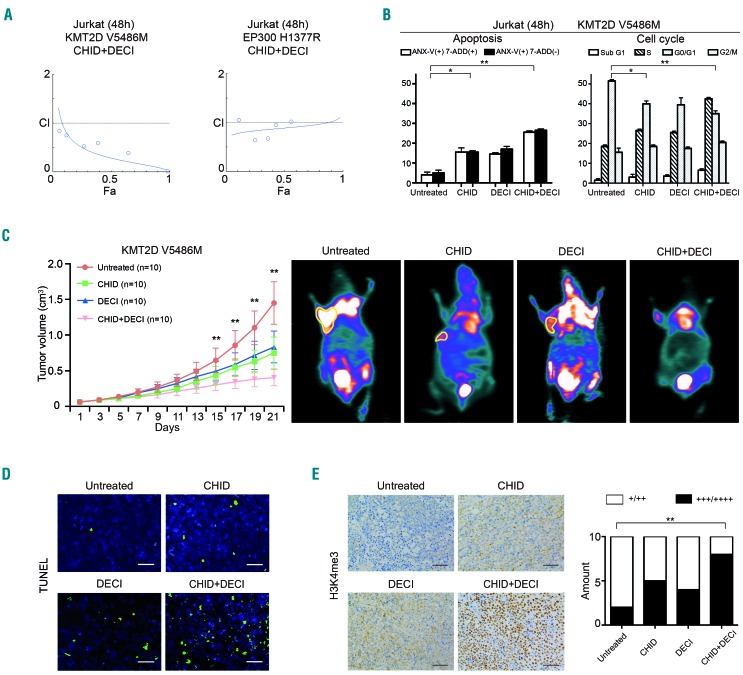
A possible structure-function relationship of the mutants was addressed using the crystal structure of the proteins encoded by KMT2D (PDB4Z4P) and EP300 (PDB4PZR), the two most frequently mutated histone modifier genes. As shown in Figure 3A, KMT2D R5389W, E5444K and V5486M might destabilize the SET domain and reduce histone methylation activity, EP300 E1377R, W1466_ and E1515V might disrupt the acetyl-CoA binding pocket, destabilize the HAT domain and reduce histone acetylation activity. Next, representative missense mutants KMT2D (V5486M) and EP300 (H1377R), as well as WT KMT2D and EP300, were established and transfected into Jurkat cells. Compared with WT protein, while KMT2D mutant reduced the level of H3K4me3, this reduction was restored by the HDAC inhibitors romidepsin and chidamide (Figure 3B and Online Supplementary Figure S2). On the other hand, the level of H3K18ac was reduced in EP300 mutant, but this effect was restored by the HDAC inhibitors valproic acid, suberoylanilide hydroxamic acid, or romidepsin and chidamide (Figure 3C). These results were observed by western blot and by immunofluorescence assay (Figure 3B,C).
Figure 3.
Effect of histone deacetylase inhibitor in KMT2D-mutated and EP300-mutated T-lymphoma. (A) Structure prediction of the missense mutations. The crystal structure of the complex of KMT2D and EP300 is PDB: 4Z4P and PDB: 4PZR, respectively. SAM, S-adenosyl-L-methionine. (B and C) Western blot and immunofluoresence assay of Jurkat cells transfected with wild-type (WT), KMT2D mutants (V5486M) (B) and EP300 mutants (H1377R) (C) upon treatment with different HDAC inhibitors. Jurkat cells were treated for 48 h at IC50. Histone 3 (H3) was used as a loading control. VPA, valproic acid, 3.7 mm; SAHA, suberoylanilide hydroxamic acid, 10 μm; ROMI, romidepsin, 5 nm; CHID, chidamide, 5 μm (48 h). Bar=10 μm. (D) Immunostaining of H3K4me3 and H3K18ac in tumor samples of PTCL-NOS patients with or without KMT2D or EP300 mutations. Bar=20 μm. (E) Response rate in relapsed PTCL-NOS patients treated with CHID according to the mutation status of histone modifier genes.
In tumor samples from PTCL-NOS patients, a significantly lower fraction of nuclear H3K4me3 positivity (+++~++++, 30%) was observed in cases with the KMT2D mutation than in those without mutations. Similarly, a lower fraction of nuclear H3K18ac positivity (+++~++++, 17%) was present in cases with EP300/CREBBP mutations than in those without mutations (Figure 3D). Interestingly, upon treatment with chidamide (administered orally at a dose of 30 mg twice per week), relapsed patients with a histone modifier gene mutation showed a remarkably increased response rate (complete or partial remission), as compared to those without mutations (Figure 3E and Online Supplementary Table S3). Thus, such mutations might alter the protein function on chromatin state regulation, sensitizing PTCL-NOS patients to HDAC inhibitors.
In vitro, Jurkat cells bearing the KMT2D V5486M or EP300 H1377R mutant were treated with different concentrations of chidamide and/or the hypomethylating agent decitabine for 48 h. The combination index (CI) curve yielded most of the data points to the area <1, denoting synergistic interactions in KMT2D V5486M mutated cells. Meanwhile, the inhibitory effect on EP300 H1377R mutated cells was achieved by chidamide alone (Figure 4A). Flow cytometry revealed that chidamide and decitabine synergistically induced KMT2D V5486M mutated cell apoptosis and G0/G1 arrest (Figure 4B). The in vivo anti-tumor activity of dual treatment on T-cell lymphoma was further evaluated in a murine xenograft model in which KMT2D V5486M mutated Jurkat cells subcutaneously injected into nude mice. The tumors formed in mice co-treated with chidamide and decitabine were significantly smaller than those that formed in untreated animals or those treated with the single agents, starting from 15 days of treatment (Figure 4C, left panel), as visualized by 18F-fluorodeoxyglucose small-animal positron emission tomography – computed tomography at 21 days of treatment (Figure 4C, right panel). To search for more evidence of tumor cell apoptosis, a TUNEL assay was performed on mice tumor sections. Compared with the untreated group and the groups treated with single agents, the number of apoptotic tumor cells was increased following combined treatment (Figure 4D). In accordance with in vitro data, upregulation of H3K4me3 was more significant in the combination treatment group than in the single-agent and the untreated group (Figure 4E).
Figure 4.
Effect of chidamide and decitabine in KMT2D-mutated and EP300-mutated T-lymphoma. (A) Combination index (CI) curve calculated by Compusyn software in KMT2D-mutated and EP300-mutated Jurkat cells treated with chidamide (CHID, 5 μm) and/or decitabine (DECI, 5 μm) for 48 h. (B) KMT2D-mutated Jurkat cell apoptosis and cell cycle determined by flow cytometry of cells treated with CHID and/or DECI for 48 h. *P<0.05, **P<0.01 compared with the untreated cells. (C) In vivo effect of the CHID and DECI combination in a murine T-lymphoma xenograft model. Tumor volume (left panel) and standardized uptake value (SUV) intensity of micro-positron emission tomograpy-computed tomography (right panel) of xenograft nude mice injected subcutaneously with KMT2D V5486-mutated Jurkat cells treated with CHID (12.5 mg/kg, twice weekly for 3 weeks), DECI (0.5 mg/kg, twice weekly for 3 weeks), either alone or in combination. **P<0.01 compared with the untreated group that received RPMI1640. (D) Apoptotic cells detected by the TUNEL assay (×400). Bar=20 μm. (E) Immunohistochemical assay of H3K4me3 in murine tumor samples treated with CHID and/or DECI. **P<0.01 compared with the untreated group. Bar=50 μm.
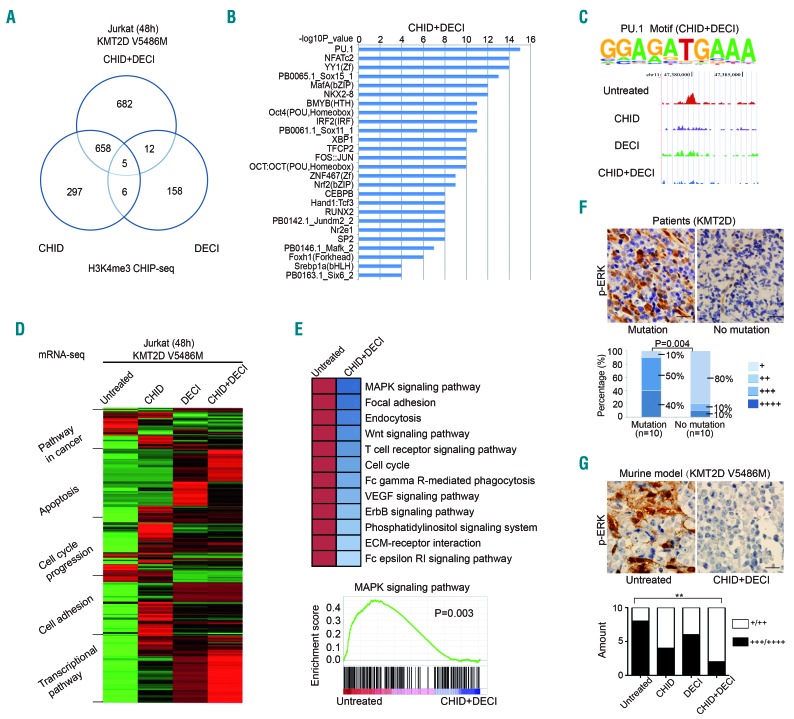
To determine KMT2D-H3K4me3 DNA binding targets, ChIP-seq was performed using H3K4me3 antibody in KMT2D V5486M mutated Jurkat cells treated with chidamide (5 μm) alone or in combination with decitabine (5 μm) for 48 h. Presentation of the data in a Venn diagram identified a significant non-overlapping portion of H3K4me3 binding promoters in the combination group, excluding 663 promoters overlapping with the chidamide group and 17 with the decitabine group (Figure 5A,B). Consistent with previous studies, H3K4me3 peaks were found at gene promoters. The group of promoters, whose H3K4me3 levels were affected by combined chidamide and decitabine treatment, but not by either chidamide or decitabine treatment alone, was enriched with binding site motifs for PU.1, a transcription factor that activates gene expression during myeloid and B-cell lymphoid cell development15,16 (Figure 5C). Furthermore, RNA sequencing analysis indicated that, in comparison with the untreated group and the single-agent groups, combined treatment led to significant modulation of multiple signaling pathways associated with cancer, including those of apoptosis, cell cycle progression, cell adhesion, and transcriptional regulation (Figure 5D). Particularly, PU.1 was included in both the cancer pathway and the transcriptional pathway in the combined treatment group. Pathway enrichment analysis of the overlapping genes of the RNA-Seq and ChIP-Seq in the combination group was then performed. Significant pathways relevant to T-cell biology are shown in Figure 5E. As revealed by gene set enrichment analysis, the MAPK pathway was inactivated in the combination treatment group compared with the untreated group. Accordingly, p-ERK upregulation was observed not only in tumor samples of PTCL-NOS patients with KMT2D mutations, but also in those of xenografted T-lymphoma mice bearing KMT2D V5486M mutants, the latter being inhibited by combined treatment with chidamide and decitabine (Figure 5F,G).
Figure 5.
Chip-seq and RNA sequencing data of KMT2D-mutated T-lymphoma cells treated with chidamide and/or decitabine. (A) Venn diagram depicting the overlap between transcription factors bound by H3K4me3 ChIP-seq in the combination group, as compared to the chidamide (CHID)-treated group and the decitabine (DECI)-treated group in KMT2D V5486-mutated Jurkat cells. (B) The top significant transcription factors bound by H3K4me3 in the combination group. (C) ChIP-seq analysis of transcription factors bound by H3K4me3. Enriched H3K4me3-binding motifs for PU.1 analyzed by KMT2D V5486-mutated Jurkat cells treated with CHID and DECI relative to genomic background (upper panel). Genomic snapshots of PU.1 peaks bound by H3K4me3 in different groups (lower panel). (D) Cellular and genetic information processing revealed by RNA-seq on the combination group in KMT2D V5486-mutated Jurkat cells. (E) Pathway analysis of the most differentially expressed genes that overlapped in both RNA-Seq and ChIP-Seq analysis in the combination group (upper panel). Gene-set enrichment analysis of the MAPK pathway (lower panel). (F) Immunohistochemical assay of p-ERK in tumor samples of PTCL-NOS patients with or without KMT2D mutations. (G) Immunostaining of p-ERK in tumor samples of xenografted murine models bearing KMT2D V5486 mutants treated with CHID and/or DECI. **P<0.01 compared with the untreated group. Bar=20 μm.
Discussion
First observed in B-cell lymphoma, recurrent mutations of epigenetic modifier genes have recently been identified in PTCL-NOS.9 In the present study, we performed targeted sequencing of the main epigenetic modifier genes in a large cohort of Chinese PTCL-NOS patients. The results showed that the mutational spectrum of these genes in PTCL-NOS was similar to that in B-cell lymphoma, in which predominantly missense mutations were found.17,18 Importantly, our study provided clinical evidence that histone modifier gene mutations, particularly those involved in histone methylation and acetylation, are significantly associated with tumor chemoresistance and disease progression of PTCL-NOS.
The adverse prognostic effect of histone modifier gene mutations was further proven in a chemotherapy-independent manner, prompting us to explore bio-therapeutic agents that can overcome chemoresistance in PTCL-NOS patients. It is well known that HDAC inhibitors are potent anticancer drugs in hematopoietic malignancies, including lymphoma.19–21 The aim of using HDAC inhibitors is to restore normal histone modification patterns through inhibition of various components of the epigenetic machinery.22,23 In B-cell lymphoma, HDAC inhibitors can rescue deficits in histone acetylation induced by EP300/CREBBP mutations,24 rendering tumor cells more sensitive to suberoylanilide hydroxamic acid.25 This can explain why chidamide also has favorable efficacy on PTCL-NOS patients bearing EP300/CREBBP mutations.
Moreover, KMT2D-mutated PTCL-NOS patients responded to chidamide. Both in vitro and in vivo, the combination of decitabine and chidamide induced apoptosis of Jurkat cells bearing the KMT2D mutant. This is in accordance with previous reports that decitabine and 5-azacytidine produce a marked synergistic effect in combination with suberoylanilide hydroxamic acid and romidepsin in T-lymphoma cell lines by modulating cell cycle arrest and apoptosis.26,27 As a mechanism of action, KMT2D mutations of B-lymphoma cells promote malignant outgrowth by perturbing methylation of H3K4 that affect the JAK-STAT, Toll-like receptor, or B-cell receptor pathway.28,29 Here our study indicated that dual treatment with chidamide and decitabine enhanced the interaction of KMT2D with the transcription factor PU.1, thereby inactivating the H3K4me-associated signaling pathway MAPK, which is constitutively activated in T-cell lymphoma.13,30,31 The transcription factor PU.1 is involved in the development of all hematopoietic lineages32 and regulates lymphoid cell growth and transformation.33 Aberrant PU.1 expression promotes acute myeloid leukemia and is related to the pathogenesis of multiple myeloma via the MAPK pathway.34,35 On the other hand, PU.1 is also shown to interact with chromatin remodeler and DNA methyltransferease to control hematopoiesis and suppress leukemia.36 Our data thus suggested that the combined action of chidamide and decitabine may interfere with the differentiation and/or viability of PTCL-NOS through a PU.1-dependent gene expression program.
In conclusion, histone modifier genes indicate clinical progression of PTCL-NOS and may represent a group of actionable biomarkers of this disease subtype. Characterized as a biological subset of PTCL-NOS, patients with dysregulation of the histone modification machinery may be amenable to therapeutic intervention with HDAC inhibitors, given either alone or in combination with hypomethylating agents.
Supplementary Material
Acknowledgments
The authors would like to thank all the patients involved in this study and their families.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/4/679
Funding
This study was supported, in part, by research funding from the National Natural Science Foundation of China (81325003, 81520108003 and 81670716), Chang Jiang Scholars Program, the Shanghai Commission of Science and Technology (16JC1405800), Shanghai Municipal Education Commission Gaofeng Clinical Medicine Grant Support (20152206 and 20152208), Clinical Research Plan of SHDC (16CR2017A), Multicenter Clinical Research Project by Shanghai Jiao Tong University School of Medicine (DLY201601), Collaborative Innovation Center of Systems Biomedicine and the Samuel Waxman Cancer Research Foundation.
References
- 1.Broccoli A, Zinzani PL. Peripheral T-cell lymphoma, not otherwise specified. Blood. 2017;129(9):1103–1112. [DOI] [PubMed] [Google Scholar]
- 2.Pileri SA, Piccaluga PP. New molecular insights into peripheral T cell lymphomas. J Clin Invest. 2012;122(10):3448–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iqbal J, Wright G, Wang C, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123(19):2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenquist R, Rosenwald A, Du MQ, et al. Clinical impact of recurrently mutated genes on lymphoma diagnostics: state-of-the-art and beyond. Haematologica. 2016;101(9):1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Xu W, Liu H, Li J. Therapeutic options in peripheral T cell lymphoma. J Hematol Oncol. 2016;9:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy DM, Walsh LA, Chan TA. Driver mutations of cancer epigenomes. Protein Cell. 2014;5(4):265–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemonnier F, Couronne L, Parrens M, et al. Recurrent TET2 mutations in peripheral T- cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–1469. [DOI] [PubMed] [Google Scholar]
- 8.Vallois D, Dobay MP, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128(11):1490–1502. [DOI] [PubMed] [Google Scholar]
- 9.Schatz JH, Horwitz SM, Teruya-Feldstein J, et al. Targeted mutational profiling of peripheral T-cell lymphoma not otherwise specified highlights new mechanisms in a heterogeneous pathogenesis. Leukemia. 2015;29(1):237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odejide O, Weigert O, Lane AA, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, He F, Zeng H, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46(3):287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva Almeida AC, Abate F, Khiabanian H, et al. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet. 2015;47(12):1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, Gu ZH, Yan ZX, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47(9):1061–1066. [DOI] [PubMed] [Google Scholar]
- 14.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201(2):221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokalski KM, Li SK, Welch I, Cadieux-Pitre HA, Gruca MR, DeKoter RP. Deletion of genes encoding PU.1 and Spi-B in B cells impairs differentiation and induces pre-B cell acute lymphoblastic leukemia. Blood. 2011;118(10):2801–2808. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–691. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Jia B, Xu W, et al. Chidamide in relapsed or refractory peripheral T cell lymphoma: a multicenter real-world study in China. J Hematol Oncol. 2017;10(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wozniak MB, Villuendas R, Bischoff JR, et al. Vorinostat interferes with the signaling transduction pathway of T-cell receptor and synergizes with phosphoinositide-3 kinase inhibitors in cutaneous T-cell lymphoma. Haematologica. 2010;95(4):613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2(5):405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatopoulos B, Meuleman N, De Bruyn C, Delforge A, Bron D, Lagneaux L. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis, down-regulates the CXCR4 chemokine receptor and impairs migration of chronic lymphocytic leukemia cells. Haematologica. 2010;95(7):1136–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Ortega-Molina A, Geng H, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 2017;7(1):38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen CL, Asmar F, Klausen T, Hasselbalch H, Gronbaek K. Somatic mutations of the CREBBP and EP300 genes affect response to histone deacetylase inhibition in malignant DLBCL clones. Leuk Res Rep. 2012;2(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchi E, Zullo KM, Amengual JE, et al. The combination of hypomethylating agents and histone deacetylase inhibitors produce marked synergy in preclinical models of T-cell lymphoma. Br J Haematol. 2015. 10.1111/bjh.13566 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Kalac M, Scotto L, Marchi E, et al. HDAC inhibitors and decitabine are highly synergistic and associated with unique gene-expression and epigenetic profiles in models of DLBCL. Blood. 2011;118(20):5506–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10): 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones CL, Gearheart CM, Fosmire S, et al. MAPK signaling cascades mediate distinct glucocorticoid resistance mechanisms in pediatric leukemia. Blood. 2015;126(19): 2202–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty AR, Robey RW, Luchenko VL, et al. MAPK pathway activation leads to Bim loss and histone deacetylase inhibitor resistance: rationale to combine romidepsin with an MEK inhibitor. Blood. 2013;121(20): 4115–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang G, Zhang P, Hirai H, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40(1):51–60. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbauer F, Owens BM, Yu L, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38(1):27–37. [DOI] [PubMed] [Google Scholar]
- 34.Will B, Vogler TO, Narayanagari S, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015;21(10):1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iseki Y, Nakahara M, Kubo M, Obata F, Harigae H, Takahashi S. Correlation of PU.1 and signal regulatory protein alpha1 expression in PU.1 transgenic K562 cells. Int J Mol Med. 2012;29(2):319–323. [DOI] [PubMed] [Google Scholar]
- 36.van Riel B, Rosenbauer F. Epigenetic control of hematopoiesis: the PU.1 chromatin connection. Biol Chem. 2014;395(11):1265–1274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.