Significance
In this study, we use an experimental imaging–genetics approach for investigating the genetic underpinnings of brain atrophy in Alzheimer’s disease. We successfully combined state-of-the-art imaging–genetics methods and experimental gene expression data to uncover biology in brain atrophy. The experimental paradigm highlighted a significant role of tribbles pseudokinase 3 (TRIB3) in modulating the typical pattern of Alzheimer’s brain pathology. This result corroborates through rigorous data-driven statistical methods evidence emerging from previous studies about the role of TRIB3 in modulating known mechanisms of neurodegeneration, such as neuronal death, cellular homeostasis, and interaction with established genes causing autosomal dominant Alzheimer’s disease: APP and PSEN1. The developed integrated statistical–experimental methodology could serve as a roadmap for investigations in other disorders.
Keywords: imaging–genetics, Alzheimer’s disease, brain atrophy, TRIB3, neuroimaging
Abstract
The joint modeling of brain imaging information and genetic data is a promising research avenue to highlight the functional role of genes in determining the pathophysiological mechanisms of Alzheimer’s disease (AD). However, since genome-wide association (GWA) studies are essentially limited to the exploration of statistical correlations between genetic variants and phenotype, the validation and interpretation of the findings are usually nontrivial and prone to false positives. To address this issue, in this work, we investigate the functional genetic mechanisms underlying brain atrophy in AD by studying the involvement of candidate variants in known genetic regulatory functions. This approach, here termed functional prioritization, aims at testing the sets of gene variants identified by high-dimensional multivariate statistical modeling with respect to known biological processes to introduce a biology-driven validation scheme. When applied to the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort, the functional prioritization allowed for identifying a link between tribbles pseudokinase 3 (TRIB3) and the stereotypical pattern of gray matter loss in AD, which was confirmed in an independent validation sample, and that provides evidence about the relation between this gene and known mechanisms of neurodegeneration.
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder, and its etiology still remains largely concealed. In anticipation of increasing prevalence of AD and other dementias, there is an urgent need for improving the understanding of the disease processes that underlie neurodegeneration. While the knowledge about the genetic and environmental risks underpinning AD is steadily advancing, how these factors interact to lead to the complex pathophysiology that results in dementia is less understood.
Advances in imaging technologies have led to noninvasive or minimally invasive imaging biomarkers that capture various aspects of the disease process, including amyloid deposition (1), tau pathology (2), functional decline (3), and neuronal loss (4). Combining such imaging information with genetic measurements—so-called imaging–genetics—provides the means for investigating the effect of genetic variation on underlying biological mechanisms (5).
Genome-wide association studies (GWAS) query millions of SNPs individually for their association with either case–control status (6) or disease-specific quantitative phenotypes [e.g., in the case of AD, regional brain volumes (7) or brain amyloid burden (8)]. Mass univariate analysis of genetic data is still the predominant method in virtue of its ease of use and well-established theoretical framework, albeit that it suffers from significant limitations, including the requirement for multiple testing, redundancies introduced by linkage disequilibrium (LD), and the lack of analysis of epistatic effects (e.g., SNP–SNP interactions), which have to be explicitly modeled and searched for exhaustively (9). Moreover, more than one quantitative phenotype can be derived from the available imaging data (e.g., dozens or hundreds of regional brain volumes or hundreds of thousands of voxel-level metrics) (10). This potentially large number of genotype–phenotypes features of interest generally complicates the problem of reliably detecting statistical associations and thus, hampers the identification of disease-relevant genetic markers by purely statistical means.
Limitations of classical mass univariate statistical methods have, in recent years, been overcome by using multivariate approaches to data analysis in the context of neuroscience studies (11) and GWAS (12). Likewise, in imaging–genetics, meaningful genotype–phenotype interactions (13) are captured by simultaneously modeling sets of genetic variants that are jointly associated with a given imaging phenotype (14–17). Multivariate GWAS have the potential to shed light on the complex genotype–phenotype relationship and may thus highlight novel links between brain physiology and molecular and biological functions. However, although these methods have proven their ability to identify meaningful SNP combinations associated with brain imaging features, the interpretation and validation of the statistical findings remain very challenging tasks. These problems relate directly to the understanding of the functional role of sets of genetic variants and to the difficulty of replicating the statistical results in unseen cohorts.
We approach this technical bottleneck by leveraging multivariate approaches to explore high-dimensional datasets and to generate hypotheses, which are subsequently tested in downstream experiments. High-quality databases of matched genotype and gene expression measurements, such as the Genotype Tissue Expression project (GTEx; gtexportal.org/home/) (18) and BRAINEAC (www.braineac.org/) (19), facilitate the quantification of effects of SNPs on gene expression in numerous tissues, including various brain tissues. Typically, these databases are used to detail the effect of a genetic variant at the very end of an analysis pipeline and to garner evidence for molecular mechanisms of the genetic locus. However, functional information in “convenience” databases can also be used at an earlier stage in the analysis to prioritize a few candidate hypotheses with a clear functional mechanism [e.g., expression quantitative trait loci (eQTL)] for the validation phase and thus, limit the multiple testing burden.
In this work, we apply this investigative approach to study the genetic functional mechanisms underlying brain atrophy in AD. The framework is composed of two steps.
-
i)
Statistical discovery. Candidate genetic variants are initially identified through data-driven multivariate statistical analysis of the matched imaging and genetics data (Fig. 1). This is achieved by modeling the joint covariation between 1.1 million SNPs and the cortical and subcortical atrophy represented by 327,684 cortical and 27,120 subcortical thickness values of 639 individuals (either healthy older controls or patients with AD) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort.
-
ii)
Functional prioritization. The candidate genetic variants are subsequently screened for functional relevance by querying high-dimensional gene expression databases, such as GTEx.
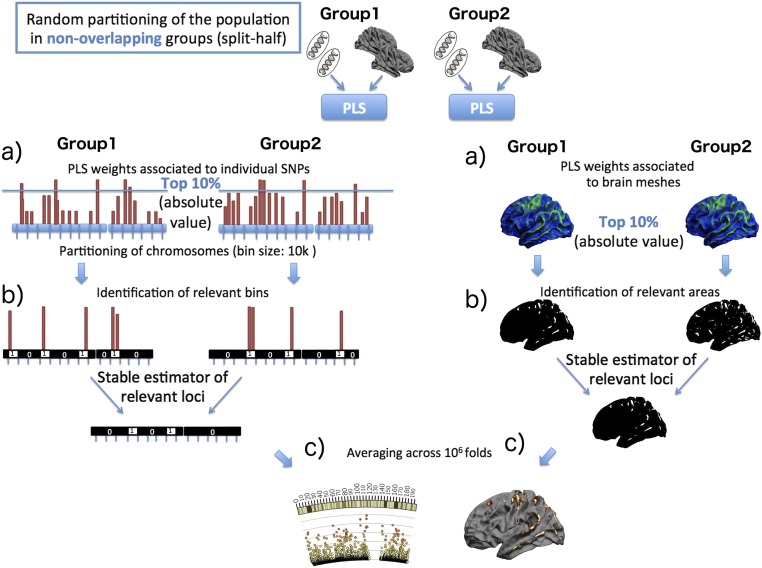
Fig. 1.
Cross-validation scheme for the assessment of the genetic loci of maximal genotype–phenotype correlation identified by the PLS model. The whole procedure is repeated 1 million times, and the resulting array is further analyzed. (A) PLS is applied in a split-half setting. For each of the two nonoverlapping randomly sampled groups, the PLS components of joint phenotype and genotype variation are independently estimated. (B, Left) Each chromosome is partitioned in bins of 10-kb size, which are labeled one if they contain an SNP associated with the largest PLS weights (top 10% of absolute values) or zero otherwise. To obtain stable estimates of the loci of maximal weights, the resulting binary arrays independently estimated in the two groups are merged (binwise and operation). The same procedure is applied on the mesh-based PLS weights associated with the phenotype component. (C) Steps A and B are repeated across 1 million folds, and the results are subsequently averaged to obtain the confidence maps associated with genetic and phenotype components (Figs. 2 and 3).
The resulting small set of genetic loci, which are shown to modify gene expression, is then validated in an independent sample of 553 individuals from the ADNI diagnosed with mild cognitive impairment (MCI), a proportion of whom progressed to AD.
Compared with previous approaches, our work (i) analyzes the whole genome and whole brain in a hypothesis-free fashion (i.e., without preselecting SNPs or brain regions) and (ii) uses a functional prioritization step to select genetic loci for validation in an independent cohort.
Starting from the initial ∼1.1 million SNPs, the multivariate statistical analysis allowed for the identification of a relatively small number of genetic loci that are statistically associated with the typical pattern of AD brain pathology. The subsequent functional prioritization step ultimately identified a significant role of tribbles pseudokinase 3 (TRIB3), a gene showing important connections to known mechanisms of neurodegenerative diseases. Indeed, although a role for TRIB3 in dementia has not been extensively explored, there are several aspects of TRIB3 function that have relevance to mechanisms related to neuronal death, cellular homeostasis, and interaction with established AD genes, such as APP and PSEN1.
This study ultimately offers an illustration of the potential of effectively combining multivariate statistical modeling in imaging–genetics with recent instruments available from computational biology to lead to insights on the pathophysiology of neurodegeneration.
Results
Model Training and Estimated Components.
Figs. 2 and 3 show the relevant areas of the identified joint genetic and phenotype variation, respectively, for the first three partial least squares (PLS) components through stability selection. The components were very robust (100% reproducible) during the stability selection procedure (SI Materials and Methods). The fourth and fifth components did not present any relevant locations (i.e., all bins have P < 0.95) after stability selection for both the genetic modality and the imaging modality.
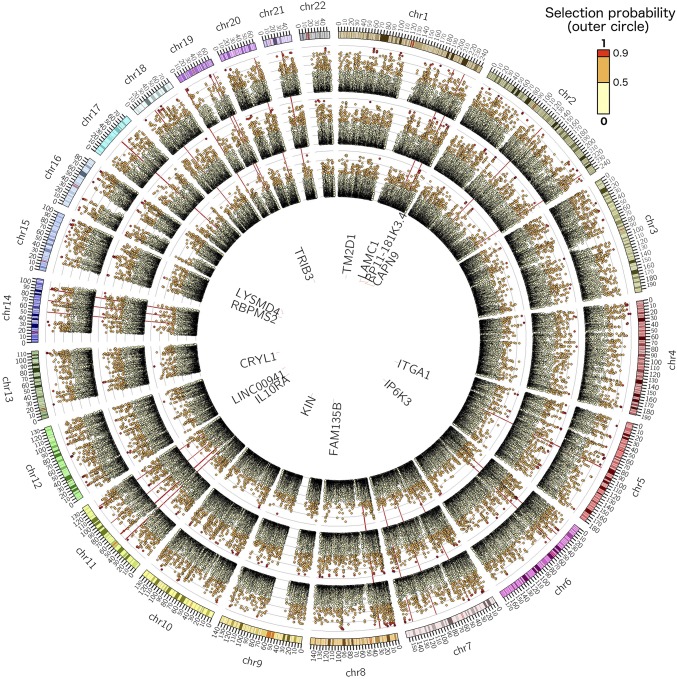
Fig. 2.
PLS genotype component: the circular plots show the probability of a given genetic locus being associated with the phenotype components shown in Fig. 3. Outer to inner circles represents components 1–3, respectively. The plots show the probability of a given genetic bin of size 10 kb being relevant in the PLS model (i.e., to contain an SNP that is ranked in the top 10% of the absolute weights of the genotype component). The genes with eQTL close to the important loci (P > 0.95) are listed in the innermost circle depending on their genomic position. The red radial lines are located in correspondence of known AD genes: ABCA7, APOE, APP, BIN1, CASS4, CD2AP, CD33, CELF1, CLU, CR1, DSG2, EPHA1, FERMT2, HLA-DRB5, INPP5D, MAPT, MEF2C, MS4, NME8, PICALM, PSEN1, PSEN2, PTK2B, SLC24A4, SORL1, and ZCWPW1. High-resolution circular plots for each component are provided in Figs. S2–S4.
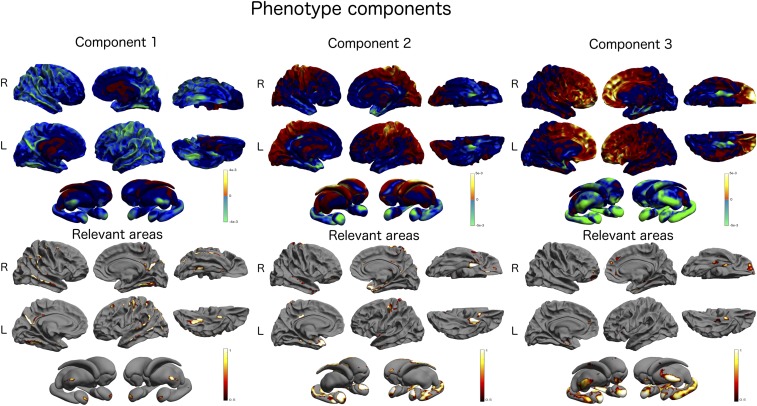
Fig. 3.
PLS phenotype component: Upper shows the topographical distribution of the PLS weights associated with the cortical and subcortical brain areas. The absolute value of the weights is proportional to the importance of the underlying brain areas. The relevance of the brain areas is quantified in Lower. The colors (red to white) indicate the probability of a brain area being associated with the genotype component shown in Fig. 2 and quantify the probability of each cortical mesh point being relevant in the PLS model (i.e., to be ranked among the top 10% of the absolute weights of the phenotype component). L, left; R, right.
Genetic Components.
The circular Manhattan plot [Circos v0.96 (20)] of Fig. 2 shows the selection frequency for the PLS genotype components and describes the importance of the genetic loci associated with cortical thickness variation for components 1–3. The plot shows the probability of a given genetic bin of size 10 kb being relevant in the PLS model (i.e., to contain an SNP that is ranked in the top 10% of the absolute weights of the genotype component). Spatially contiguous loci generally show similar importance values, which are caused by LD of these regions.
In the genetic components 1–3, a total of 118 bins exceeded the selection frequency threshold (61, 50, and 7 for components 1–3, respectively). From these bins, 402 (196, 181, and 25) influential SNPs were extracted and annotated with 98 genes through the Ensembl Variant Effect Predictor (VEP) for GRCh37 (date accessed: October 17, 2016) (21). The extended Apolipoprotein E (APOE) locus comprising APOE and TOMM40 was selected as the highest scoring region in component 1. A total of 3,956 candidate SNP–gene pairs were considered for the GTEx-based eQTL analysis in six tissues. However, a few genes did not show sufficient expression levels in some tissues, and these combinations were excluded from the analysis, resulting in 1,598 unique SNP–gene tissue tests, of which 104 were significant at the Bonferroni-corrected P value threshold (P = 3.1e−5) (Dataset S1) linking to 14 genes (Dataset S2 and Fig. S5): CAPN9, CRYL1, FAM135B, IL-10 receptor subunit alpha (IL10RA), IP6K3, ITGA1, KIN, LAMC1, LINC00941, LYSMD4, RBPMS2, RP11-181K3.4, TM2 domain-containing 1 (TM2D1), and TRIB3. These genes are listed in the innermost circle of Fig. 2 depending on their genomic position.
The independent validation of those 14 genes in the MCI cohort confirmed TRIB3 (P = 0.0034) (Table 1). Three additional genes were close to nominal significance: TM2D1 (P = 0.053), LAMC1 (P = 0.062), and RP11-181K3.4 (P = 0.053) (Table 1). Of note, the top eQTL SNP for TRIB3 rs4813620 received a P = 0.06175 in stage 1 of a large AD GWAS (6). However, rs62191440, an SNP in strong LD with rs4813620 (D′ = 0.8469; r2 = 0.6559) in the European population (22), received a P value of 0.00601 (Fig. S6) and also constitutes an eQTL for TRIB3 in various tissues in GTEx, including brain tissues cortex and caudate ganglia (Fig. S7). Interestingly, when estimating the PLS components on the subcohort of 279 training individuals positive to amyloid in the cerebrospinal fluid (CSF) (Table 2), we achieved compatible validation results in the independent MCI group. Within this setting, TRIB3 still leads to marginally significant differences (P = 0.0134) between progressing and stable MCI (Table S1).
Table 1.
Statistical comparison of the genes scores in training and testing groups (Kruskal–Wallis nonparametric test)
| Gene | P value training (AD vs. healthy controls) | P value testing (MCI converter vs. MCI stable) |
| TM2D1 | 0.0050 | 0.0528 |
| IL10RA | 0.1069 | 0.6198 |
| TRIB3 | 0.0032 | 0.0034 |
| ZBTB7A | 0.0360 | 0.9135 |
| LYSMD4 | 0.0000 | 0.2057 |
| CRYL1 | 0.6213 | 0.1176 |
| FAM135B | 0.0000 | 0.5588 |
| IP6K3 | 0.0000 | 0.4646 |
| ITGA1 | 0.0993 | 0.7310 |
| KIN | 0.0014 | 0.2061 |
| LAMC1 | 0.0019 | 0.0618 |
| LINC00941 | 0.0000 | 0.6896 |
| RBPMS2 | 0.0000 | 0.2149 |
| RP11-181K3.4 | 0.0017 | 0.0527 |
The score for TRIB3 leads to significant differences in the MCI testing group after Bonferroni correction for multiple comparisons.
Table 2.
Sociodemographic, clinical, and genetic characteristics of the study cohort [mean (SD)]
| Diagnosis at imaging | Discovery | Validation | ||
| Healthy | AD | MCI stable | Progressing MCI | |
| N | 401 | 238 | 341 | 212 |
| Age, y | 74.45 (5.5) | 74.72 (7.9) | 72.91 (7.6) | 73.61 (7.51) |
| Education, y | 16.36 (2.66) | 15.34 (2.9) | 16.05 (2.87) | 15.82 (2.82) |
| Sex, % females | 49 | 45 | 37 | 39 |
| MMSE | 29.1 (1.11) | 23.2 (2) | 27.91 (1.73) | 26.87 (1.74) |
| ADAS11 | 5.98 (2.86) | 19.85 (6.63) | 9.29 (3.9) | 13.31 (4.51) |
| Apoe 4, % zero/one/two alleles | 72/26/2 | 31/48/21 | 54/36/10 | 33/51/16 |
| CSF Aβ1–42, % positives (no. of subjects with baseline measure) | 43 (282) | 93 (166) | 62 (244) | 85 (135) |
Positivity to Aβ1–42 was defined with respect to the nominal cutoff of 192 pg/mL.
Morphometric Components.
Fig. 3 shows the PLS phenotype components 1–3 along with the associated selection frequency describing the loci of brain atrophy associated with genetic variation. The first component is mainly associated with the thinning of the cortical mantle and is localized in temporal and posterior cingulate cortices. The relevant areas at the subcortical level are primarily associated with amygdalae and thalami. The second component is mostly associated with the thinning of the subcortical areas (hippocampi and amygdalae) and with the cortical thinning of the temporal areas at the cortical level. The third component is similar to component 2 and describes a subcortical thickness pattern prevalent in hippocampi, amygdalae, and thalami. At the cortical level, the component is associated with the thinning of frontal cortice and with isolated spots located in the parahippocampal gyrus.
Discussion
In this work, we modeled high-dimensional genome-wide SNP data and brain-wide cortical thickness data via joint multivariate statistical modeling and functional prioritization of genes through bioinformatics annotation and a large eQTL database.
Our study ultimately identified a link between TRIB3 and the stereotypical pattern of gray matter loss in AD (cortical thinning in temporal and posterior cingulate regions and subcortical atrophy). TRIB3 is a pseudokinase that acts as a regulator of several signaling pathways. For example, it can interact directly with Akt and inhibit the prosurvival Akt pathway (23). TRIB3 expression is induced during neuronal cell death (24), and recently increased levels of the TRIB3 protein were found in dopaminergic neurons of the substantia nigra pars compacta in patients with Parkinson’s disease (25). TRIB3 expression is stress induced and increases in response to nerve growth factor deprivation, endoplasmatic reticulum stress, and amino acid deprivation (24). Although a role for TRIB3 in dementia has not been extensively explored, there are several aspects of TRIB3 function that have relevance to known mechanisms of neurodegenerative disease. TRIB3 can interact directly with P62 to modulate autophagic flux (26), an important process in maintaining cellular homeostasis that is known to be disrupted in neurodegeneration (27). Knockdown of TRIB3 modulates PSEN1 stability (26), and a yeast two-hybrid screen identified progranulin as a direct interaction partner of TRIB3 (28). Intriguingly, it has recently been shown that TRIB3 induces both apoptosis and autophagy in Aβ-induced neuronal death, and silencing of TRIB3 was strongly neuroprotective (29). These links warrant additional investigation for a functional role of TRIB3 in neuronal death in dementia.
These earlier findings align with our eQTL analysis, where carriers of the minor allele show increased TRIB3 expression (Fig. S5), which potentially lowers the threshold to TRIB3-mediated neuronal cell death. TRIB3 expression was modulated by the identified SNP in various other tissues, including the caudate (Fig. S7), a region affected in Parkinson’s disease and Huntington’s disease. A recent study of Trib3 expression in mice concluded that “Trib3 has a pathophysiological role in diabetes” (30); diabetes itself is a known risk factor for dementia (31), perhaps through shared metabolic processes with AD (32). Interestingly, one of three SNPs (rs1555318) selected in the PLS model and attributed to TRIB3 showed a strong association with type 2 diabetes in stage 1 of a large GWAS (P = 4.4e−4) (Fig. S8) (33). Other GWAS showed links between TRIB3 and information processing speed (P = 1.7e−7) (34) and AD (P = 0.006) (6). An earlier genetic study on AD in Swedish men found an association in TRIB3 as well (P = 0.044) (35), which was replicated in a Canadian cohort (P < 0.001) (36). Lastly, TRIB3 was reported to physically interact with APP (37), and it shares numerous functional annotations for biological processes regarding lipid metabolism with APOE.
The functional prioritization component of the analysis successfully reduced the set of candidate genetic variants for the independent validation; however, this prioritization has a shortcoming: it hypothesizes that identified SNPs alter the expression of a nearby gene. Although this scheme led to the identification of TRIB3 in the cortical thickness phenotype, it did miss a long-established AD risk gene: APOE. SNPs belonging to APOE (rs429358 and rs7412) were selected as the highest-scoring SNPs in component 1. However, none of them were detected as an eQTL, and thus, APOE was excluded from the downstream analysis. Other types of functional prioritizations based on exonic function prediction may have retained APOE and other genes in the pipeline. However, SNP data typically feature only a few nonsynonymous exonic variants, and their high frequency (minor allele frequency > 5%) renders them unlikely to receive significant “damaging” scores in these predictions. Thus, for this scenario, the use of these function predictions would be limited.
The list of genes that we identified contains other interesting candidates. For instance, IL10RA is a receptor for IL-10, a cytokine that controls inflammatory response (38). Carriers of the minor allele show increased IL10RA expression (Fig. S5), and Il10ra expression is increased in affected brain regions with increasing age and presence of AD pathology in transgenic mouse models of AD [MOUSEAC (39)] (Fig. S9). Moreover, a link between down-regulation of IL10RA and TRIB3 in TRIB3-silenced HepG2 cells was reported in ref. 26 along with increased abundance of Presenilin 1, ApoE3, and Clusterin. Finally, blocking IL-10 response was recently suggested as a therapeutic mechanism in AD (40). A gene that showed a statistical trend in the validation sample was TM2D1, which is a beta-amyloid binding protein and may be involved in beta-amyloid–induced apoptosis (41). Furthermore, Myocyte Enhancer Factor 2A (MEF2A), like APOE, was filtered out by the functional prioritization. However, MEF2A is a paralog of MEF2C, which is an established AD gene (6). Noteworthy, bins covering MEF2C only barely missed the selection threshold in component 2 for additional analysis (maximum P = 0.926) (Fig. 2).
This study illustrates the potential of effectively combining multivariate statistical modeling in imaging–genetics with recent instruments available from computational biology to lead to insights on the disease pathophysiology. Thanks to the ever-growing data-driven knowledge based on the vast quantities of information now available to the research community, the paradigm proposed in this study may represent a promising avenue for linking imaging–genetics findings to the current knowledge on functional genetics mechanisms involved in neurodegeneration.
Materials and Methods
This section describes the study data, the statistical setting, and the methodology used in this study. Additional details and discussion about the methodological aspects can be found in SI Materials and Methods.
Study Participants.
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public–private partnership led by Principal Investigator Michael W. Weiner. Up-to-date information is available at www.adni-info.org. This research mainly involves further processing of previously collected personal data according to the US ethics regulations. Each subject provided signed informed consent before participation. We have explicit authorization for the use of the ADNI dataset, and we have signed the relevant papers guaranteeing that we abide by the ethics standards.
We selected genotype and phenotype data available in the ADNI-1/GO/2 datasets for 1,192 subjects. Summary sociodemographic, clinical, and genetic information is available in Table 2. At the time of study entry, subjects were diagnosed as healthy individuals (n = 401), MCI (n = 553), or AD (n = 238). A total of 212 (38.3%) MCI patients subsequently converted to AD over the course of the study (6 y). All participants were non-Hispanic Caucasian. AD and MCI groups show significant cognitive decline measured by the Mini Mental State Examination (MMSE) and the Alzheimer’s Disease Assessment Scale (ADAS) Cognitive Subscale (COG) compared with the healthy individuals (P < 1e−2, two-sample t test for groupwise comparison). There was also a significant increase in individuals with pathological levels of Aβ1–42 in the CSF (Aβ1–42 < 192 pg/mL) across the clinical groups, with proportions ranging from 43% in healthy individuals to 93% in AD patients (P < 1e−2). Similarly, we observed a higher prevalence of APOE4 carriers in AD and progressing MCI individuals compared with healthy and MCI stable groups. For this analysis, the 639 healthy and AD subjects form the discovery set, while the MCI converters and nonconverters form the independent validation set.
Statistical Discovery.
The joint relationship between the genetic and imaging modalities was investigated through PLS modeling (42–44). Among the several PLS versions proposed in the literature, we focus on the symmetric formulation of PLS computed through the singular value decomposition of the cross-covariance matrix (Fig. S1) (43, 44, 45). Within this setting, the aim of PLS is to estimate the latent components that maximize the global covariance between the two input modalities. Each input feature receives a weight in the latent component that represents its relative importance for describing the global joint multimodal relationship. Analyzing these weights helps identify SNPs that are linked to the patterns of cortical thinning in the brain.
In this study, we applied a robust approach for the stable estimation and interpretation of PLS weights in genome-wide genotyping data aimed at promoting sparsity (i.e., selecting only a few features for simplified interpretation) and regularity (by aggregating SNPs within the same genetic neighborhood). This is achieved through a stability selection procedure, in which the reproducibility and robustness of the PLS parameters are assessed through a split-half cross-validation–based scheme on 1 million repetitions of the models on randomly sampled subgroups (Fig. 1 and SI Materials and Methods). By considering a predefined partition of each chromosome into contiguous loci of size 10 kb, the procedure leads to the estimation of a confidence measure taking values ranging between 0.0 and 1.0, indicating the probability of each genetic loci containing highly reproducible PLS weights and therefore, serving as a measure of importance of the genomic location (Fig. 2). A similar procedure was used to assess the importance of the phenotype component (Fig. 1). However, no regional binning was used (Fig. 3). The procedure was applied to assess the parameter reproducibility of the first five PLS modes; subsequent analyses were performed only on components with relevant genetic and brain regions (i.e., reproducible PLS weights with selection frequency >95%).
Gene Identification.
We analyzed the 10-kb bins (genetic loci) with the selection frequency exceeding 0.95 (i.e., bins selected in 95% or more of the 1 million replications). Within these bins, we then identified the influential SNPs: an SNP was declared influential if it was associated with the weights of greatest magnitude in the PLS components estimated on the full data sample (i.e., SNPs with absolute weights exceeding the 99th quantile of all weights in the component). These weights are the ones contributing to the high selection frequency in the split-half procedure and are representative of the significant variation modeled in the data.
To link SNPs to corresponding genes, we used the computational VEP for GRCh37 with the GENCODE gene annotation. SNPs tagged as “regulatory” were manually investigated and annotated with the nearby genes.
Functional Prioritization.
All SNPs successfully annotated with a gene were subjected to functional prioritization through eQTL analysis based on the GTEx data. The sample size in GTEx for relevant brain tissues in AD was rather small (e.g., n = 81 for hippocampus). Therefore, we added five more tissues with large sample sizes that were more distantly relevant to AD. Nerve tibial (n = 256) was added as a proxy for nervous tissue, whole blood (n = 338) and artery tibial (n = 285) were included to cover blood-based changes and effects on blood vessels (46), and adipose s.c. (n = 298) was selected due to links between AD and obesity, type 2 diabetes, and metabolic disease (47, 48). Finally, transformed fibroblasts (n = 272) were included as a general purpose cell line. P values were corrected for multiple testing using the Bonferroni method.
Model Validation in Independent MCI Subjects.
The genes that were found to be under expression control by the identified SNPs were validated for their capacity to predict clinical conversion in MCI subjects. To this end, for each identified gene, we applied the PLS weights estimated on the discovery set on the validation set, with the genetic component restricted to SNPs ±20 kb from the gene borders. The identified latent projections (i.e., a weighted sum of SNPs) result in one score per subject per gene. For each gene, the association of the projection score with conversion status was assessed by statistically comparing the scores distribution between healthy individuals and AD patients and between MCI converters and nonconverters (Kruskal–Wallis nonparametric test for two-sample comparison, Bonferroni correction for multiple comparisons).
Supplementary Material
Acknowledgments
M.L., J.M.S., D.C.A., and S.O. received support from European Union’s Horizon 2020 Research and Innovation Programme Grant 666992 (EuroPOND) for this work. A.A. holds an MRC eMedLab Medical Bioinformatics Career Development Fellowship. This work was supported by Medical Research Council Grant MR/L016311/1. The contribution to this work by B.G. and P.M.T. was funded by NIH “Big Data to Knowledge” Grant U54 EB020403 (principal investigator: P.M.T.). S.W. and C.A. are supported by the National Institute for Health Research (NIHR) Queen Square Biomedical Research Unit in Dementia and Alzheimer’s Research UK and the NIHR University College London (UCL) Hospitals Biomedical Research Centre. J.M.S. acknowledges the support of the NIHR UCL Hospitals Biomedical Research Centre; the Wolfson Foundation; Engineering and Physical Sciences Research Council (EPSRC) Grant EP/J020990/1; Medical Research Council (MRC) Grant MR/L023784/1; Alzheimer Research UK (ARUK) Grants ARUK-Network 2012-6-ICE, ARUK-PG2017-1946, and ARUK-PG2017-1946; Brain Research Trust Grant UCC14191; and European Union’s Horizon 2020 Research and Innovation Programme Grant 666992. EPSRC Grants EP/J020990/01 and EP/M020533/1 support the work of D.C.A. and S.O. on this topic. S.O. receives funding from EPSRC Grants EP/H046410/1 and EP/K005278, MRC Grant MR/J01107X/1, EU-FP7 Project VPH-DARE@IT Grant FP7-ICT-2011-9-601055, the NIHR Biomedical Research Unit (Dementia) at the UCL, and NIHR University College London Hospitals Biomedical Research Centre (NIHR BRC UCLH/UCL High Impact Initiative-BW.mn.BRC10269). Data collection and sharing for this project were funded by the ADNI (NIH Grant U01 AG024904) and the Department of Defense (DOD) ADNI (DOD Award W81XWH-12-2-0012). The ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering, and it is also funded through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the NIH (https://fnih.org/). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory for NeuroImaging at the University of Southern California.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All the study data are available through the Alzheimer’s Disease Neuroimaging Initiative (ADNI, adni.loni.usc.edu/). The experimental material, including PLS components and probability measures, has been deposited in Figshare (https://doi.org/10.6084/m9.figshare.5914375.v1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706100115/-/DCSupplemental.
References
- 1.Rabinovici GD, Jagust WJ. Amyloid imaging in aging and dementia: Testing the amyloid hypothesis in vivo. Behav Neurol. 2009;21:117–128. doi: 10.3233/BEN-2009-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villemagne VL, Okamura N. In vivo tau imaging: Obstacles and progress. Alzheimers Dement. 2014;10(3 Suppl):S254–S264. doi: 10.1016/j.jalz.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Mosconi L, et al. Pre-clinical detection of Alzheimer’s disease using FDG-PET, with or without amyloid imaging. J Alzheimers Dis. 2010;20:843–854. doi: 10.3233/JAD-2010-091504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frisoni GB, Fox NC, Jack CR, Jr, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigos KL, Hariri AR, Weinberger DR. Neuroimaging Genetics: Principles and Practices. Oxford Univ Press; Oxford: 2016. [Google Scholar]
- 6.Lambert JC, et al. European Alzheimer’s Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potkin SG, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS ONE. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanan VK, et al. Alzheimer’s Disease Neuroimaging Initiative APOE and BCHE as modulators of cerebral amyloid deposition: A florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19:351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kam-Thong T, et al. GLIDE: GPU-based linear regression for detection of epistasis. Hum Hered. 2012;73:220–236. doi: 10.1159/000341885. [DOI] [PubMed] [Google Scholar]
- 10.Stein JL, et al. Alzheimer’s Disease Neuroimaging Initiative Voxelwise genome-wide association study (vGWAS) Neuroimage. 2010;53:1160–1174. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrouff J, et al. PRoNTo: Pattern recognition for neuroimaging toolbox. Neuroinformatics. 2013;11:319–337. doi: 10.1007/s12021-013-9178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szymczak S, et al. Machine learning in genome-wide association studies. Genet Epidemiol. 2009;33:S51–S57. doi: 10.1002/gepi.20473. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Calhoun VD. A review of multivariate analyses in imaging genetics. Front Neuroinform. 2014;8:29. doi: 10.3389/fninf.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Floch E, et al. Significant correlation between a set of genetic polymorphisms and a functional brain network revealed by feature selection and sparse partial least squares. Neuroimage. 2012;63:11–24. doi: 10.1016/j.neuroimage.2012.06.061. [DOI] [PubMed] [Google Scholar]
- 15.Vounou M, Nichols TE, Montana G. Alzheimer’s Disease Neuroimaging Initiative Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach. Neuroimage. 2010;53:1147–1159. doi: 10.1016/j.neuroimage.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silver M, Janousova E, Hua X, Thompson PM, Montana G. Alzheimer’s Disease Neuroimaging Initiative Identification of gene pathways implicated in Alzheimer’s disease using longitudinal imaging phenotypes with sparse regression. Neuroimage. 2012;63:1681–1694. doi: 10.1016/j.neuroimage.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009;30:241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carithers LJ, et al. GTEx Consortium A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trabzuni D, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aken BL, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machiela MJ, Chanock SJ. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: A tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 24.Zareen N, Biswas SC, Greene LA. A feed-forward loop involving Trib3, Akt and FoxO mediates death of NGF-deprived neurons. Cell Death Differ. 2013;20:1719–1730. doi: 10.1038/cdd.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aimé P, et al. Trib3 Is elevated in Parkinson’s disease and mediates death in Parkinson’s disease models. J Neurosci. 2015;35:10731–10749. doi: 10.1523/JNEUROSCI.0614-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hua F, et al. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;13:7951. doi: 10.1038/ncomms8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, et al. E3 ubiquitin ligase SIAH1 mediates ubiquitination and degradation of TRB3. Cell Signal. 2008;20:942–948. doi: 10.1016/j.cellsig.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Saleem S, Biswas SC. Tribbles Pseudokinase 3 induces both apoptosis and autophagy in amyloid-β induced neuronal death. J Biol Chem. 2017;292:2571–2585. doi: 10.1074/jbc.M116.744730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, et al. Skeletal muscle TRIB3 mediates glucose toxicity in diabetes and high- fat diet-induced insulin resistance. Diabetes. 2016;65:2380–2391. doi: 10.2337/db16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sims-Robinson C, Kim B, Rosko A, Feldman EL. How does diabetes accelerate Alzheimer disease pathology? Nat Rev Neurol. 2010;6:551–559. doi: 10.1038/nrneurol.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribe EM, Lovestone S. Insulin signalling in Alzheimer’s disease and diabetes: From epidemiology to molecular links. J Intern Med. 2016;280:430–442. doi: 10.1111/joim.12534. [DOI] [PubMed] [Google Scholar]
- 33.Morris AP, et al. Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luciano M, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol. 2011;86:193–202. doi: 10.1016/j.biopsycho.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giedraitis V, et al. Genetic analysis of Alzheimer’s disease in the uppsala longitudinal study of adult men. Dement Geriatr Cogn Disord. 2009;27:59–68. doi: 10.1159/000191203. [DOI] [PubMed] [Google Scholar]
- 36.Li H, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 37.Oláh J, et al. Interactions of pathological hallmark proteins: Tubulin polymerization promoting protein/p25, β-amyloid, and α-synuclein. J Biol Chem. 2011;286:34088–34100. doi: 10.1074/jbc.M111.243907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li MO, Flavell RA. Contextual regulation of inflammation: A duet by transforming growth factor-β and interleukin-10. Immunity. 2008;28:468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Matarin M, et al. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell Rep. 2015;10:633–644. doi: 10.1016/j.celrep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 40.Guillot-Sestier MV, et al. Il10 deficiency rebalances innate immunity to mitigate Alzheimer-like pathology. Neuron. 2015;85:534–548. doi: 10.1016/j.neuron.2014.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kajkowski EM, et al. β -Amyloid peptide-induced apoptosis regulated by a novel protein containing a g protein activation module. J Biol Chem. 2001;276:18748–18756. doi: 10.1074/jbc.M011161200. [DOI] [PubMed] [Google Scholar]
- 42.Wold H. Multivariate Analysis. Academic; New York: 1966. Estimation of principal components and related models by iterative least squares; pp. 391–420. [Google Scholar]
- 43.McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. Neuroimage. 1996;3:143–157. doi: 10.1006/nimg.1996.0016. [DOI] [PubMed] [Google Scholar]
- 44.Worsley KJ. An overview and some new developments in the statistical analysis of PET and fMRI data. Hum Brain Mapp. 1997;5:254–258. doi: 10.1002/(SICI)1097-0193(1997)5:4<254::AID-HBM9>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Friston KJ, Frith CD, Liddle PF, Frackowiak RSJ. Functional connectivity: The principal-component analysis of large (PET) data sets. J Cereb Blood Flow Metab. 1993;13:5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- 46.Kimbrough IF, Robel S, Roberson ED, Sontheimer H. Vascular amyloidosis impairs the gliovascular unit in a mouse model of Alzheimer’s disease. Brain. 2015;138:3716–3733. doi: 10.1093/brain/awv327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luchsinger JA, Gustafson DR. Adiposity, type 2 diabetes, and Alzheimer’s disease. J Alzheimers Dis. 2009;4:693–704. doi: 10.3233/JAD-2009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira ST, Clarke JR, Bomfim TR, De Felice FG. Inflammation, defective insulin signaling, and neuronal dysfunction in Alzheimer’s disease. Alzheimers Dement. 2014;10(1 Suppl):S76–S83. doi: 10.1016/j.jalz.2013.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.