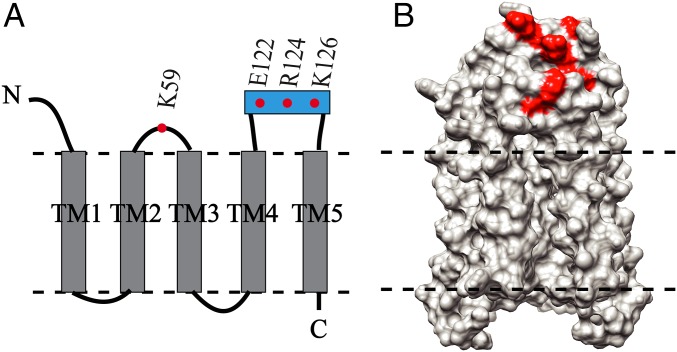
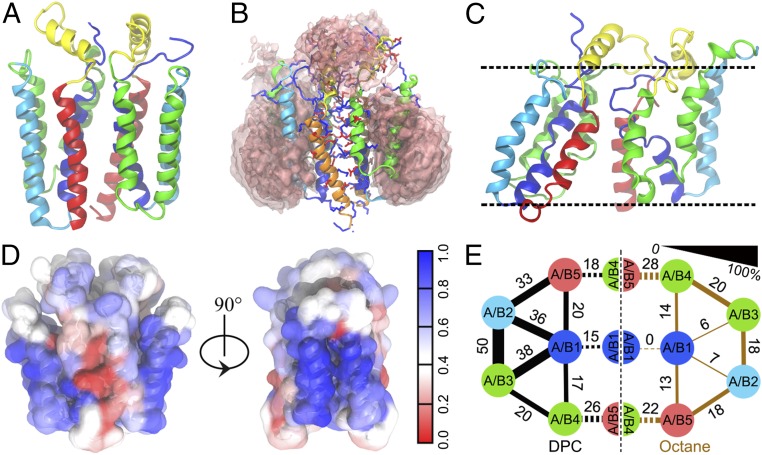
Significance
Mitochondrial respiration is carried out by a chain of protein complexes. Electron transfer through these complexes is coupled to the generation of a proton electrochemical gradient across the mitochondrial inner membrane, which is used, e.g., to synthesize ATP. The components of the respiratory chain are assembled into supercomplexes, presumed to provide functional advantages. The respiratory supercomplex factors (Rcfs), were identified to be required for supercomplex formation in Saccharomyces cerevisiae. To understand the mechanism and dynamics of supercomplex formation, structural information about these Rcfs is needed. Here, we report the solution state NMR structure of Rcf1, which forms a dimer in detergent micelles. The study reveals unique structural features of Rcf1 and provides insights into supercomplex formation.
Keywords: charge zipper, membrane proteins, mitochondria, bicelles, lipids
Abstract
The Saccharomyces cerevisiae respiratory supercomplex factor 1 (Rcf1) protein is located in the mitochondrial inner membrane where it is involved in formation of supercomplexes composed of respiratory complexes III and IV. We report the solution structure of Rcf1, which forms a dimer in dodecylphosphocholine (DPC) micelles, where each monomer consists of a bundle of five transmembrane (TM) helices and a short flexible soluble helix (SH). Three TM helices are unusually charged and provide the dimerization interface consisting of 10 putative salt bridges, defining a “charge zipper” motif. The dimer structure is supported by molecular dynamics (MD) simulations in DPC, although the simulations show a more dynamic dimer interface than the NMR data. Furthermore, CD and NMR data indicate that Rcf1 undergoes a structural change when reconstituted in liposomes, which is supported by MD data, suggesting that the dimer structure is unstable in a planar membrane environment. Collectively, these data indicate a dynamic monomer–dimer equilibrium. Furthermore, the Rcf1 dimer interacts with cytochrome c, suggesting a role as an electron-transfer bridge between complexes III and IV. The Rcf1 structure will help in understanding its functional roles at a molecular level.
Mitochondria are eukaryotic organelles, referred to as the cellular “powerhouses” because of their role in energy conservation. The Saccharomyces cerevisiae (baker’s yeast) respiratory chain consists of three complexes: II (succinate dehydrogenase), III [cytochrome bc1 (Cyt. bc1)], and IV [cytochrome c oxidase (CytcO)]. The membrane-bound complex I (NADH dehydrogenase) found in many higher eukaryotes is replaced by type II NADH dehydrogenases in S. cerevisiae (1, 2). The proton gradient that is maintained by complexes III and IV, is used by complex V (ATP synthase) to generate ATP. These complexes were believed to independently diffuse within the mitochondrial inner membrane (3–5). However, in the last decade it has become increasingly clear that there is a higher organization level of respiratory complexes where individual complexes interact and associate into supercomplexes (6–11). The functional role of these high level interactions in mitochondria is not understood in detail yet. First, the association of respiratory complexes could be important for the assembly and stability of individual complexes. Several mutant studies in Caenorhabditis elegans have reported that complex III and IV are required for the maintenance of complex I (12, 13). Second, supercomplex formation could influence mitochondrial structure. In fact, oligomerization of complex V promotes membrane curvature and formation of the tubular cristae membrane (14, 15). Furthermore, it has been proposed that supercomplex organization could help limit the production of partly reduced O2, referred to as reactive oxygen species (16).
Several studies have reported that respiratory supercomplex factors (Rcfs), and in particular Rcf1, are required for the formation of respiratory supercomplex (17–19). Rcf1 is an 18.5-kDa (by sequence) integral membrane protein and a member of the hypoxia-inducible gene 1 (HIG1) protein family (20–22). The protein was suggested to be a component that stabilizes the III2IV2 supercomplex, possibly by binding at the interface between Cyt. bc1 and CytcO (18). However, results from another study indicated that Rcf1 can interact independently with Cyt. bc1 and CytcO (19). Furthermore, a more recent study suggested that Cyt. c can bind to Rcf1 to mediate direct electron transfer from Cyt. bc1 to CytcO (23). The Rcf1 protein is not resolved in the currently available, low-resolution (∼20 Å) cryo-EM structure of S. cerevisiae III2IV2 supercomplex (24), and also the homologous protein is not observed in the higher-resolution cryo-EM structures (∼6 Å) determined for mammalian supercomplexes (25, 26). In this study, we determined the solution NMR structure of Rcf1 in detergent micelles. Rcf1 forms a dimer in the presence of dodecylphosphocholine (DPC), and an unusually charged dimer interface was observed. We also studied the interaction between Rcf1 and Cyt. c by NMR titration, which yielded a clear interaction with a defined surface on Rcf1. The Rcf1 dimer structure provides insight into supercomplex formation and dynamics.
Results
Characterization of Rcf1 Purified from Escherichia coli.
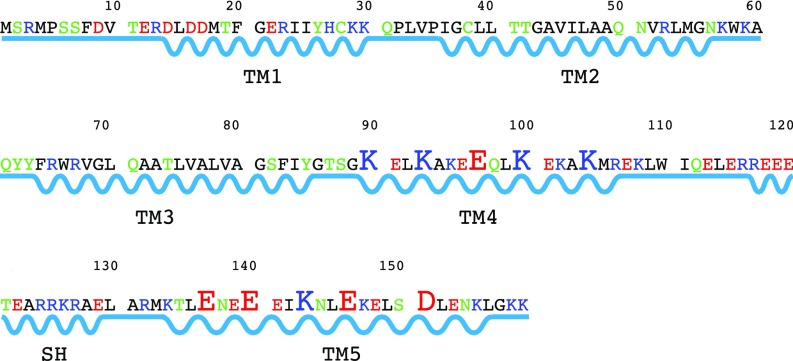
Primary sequence (Fig. 1) analysis shows that Rcf1 lacks a recognizable mitochondrial targeting signal peptide and predicts the presence of two transmembrane (TM) helices in the N-terminal part (Hig1 homologous). The long hydrophilic fungal-specific (only present in Hig1 proteins from yeast and other fungi) region at the C terminus is predicted to be disordered (SI Appendix, Fig. S1 A and B). We cloned the Rcf1 from S. cerevisiae, added a 8× His-tag to the C terminus, and expressed it in E. coli, after which the protein was purified from inclusion bodies and refolded into several different detergents, including DPC. The DPC-refolded Rcf1 eluted from a size exclusion column at an apparent molecular weight of 67 kDa (SI Appendix, Fig. S2A), consistent with a DPC-solubilized dimer [2× Rcf1 monomer (18.5 kDa) + DPC micelle (25 kDa) (27)]. On SDS/PAGE, the Rcf1 monomer runs at an apparent mass of 21 kDa, and there is also a fraction that runs as a dimer (SI Appendix, Fig. S2B). Far-UV circular dichroism (CD) spectroscopy revealed that the refolded Rcf1 in DPC [and n-dodecyl-β-d-maltoside (DDM)] micelles has a predominantly helical profile (SI Appendix, Fig. S3). Rcf1 in DPC micelles was labeled with the amine-reactive Abberior STAR 635 fluorescent probe and reconstituted into giant unilamellar vesicles (GUVs) (SI Appendix, Fig. S4), demonstrating that the refolded Rcf1 protein can be readily exchanged into a more native-like environment.
Fig. 1.
Primary sequence and structural properties of the S. cerevisiae Rcf1. The distribution of residues with different properties in Rcf1. Hydrophobic and polar residues are shown in black and green, respectively, and the positively and negatively charged residues are shown in blue and red. The enlarged charged residues are implicated in the formation of intermolecular salt bridges. Straight and zigzag lines illustrate loop and helical regions.
Rcf1 Dimer Structure in DPC.
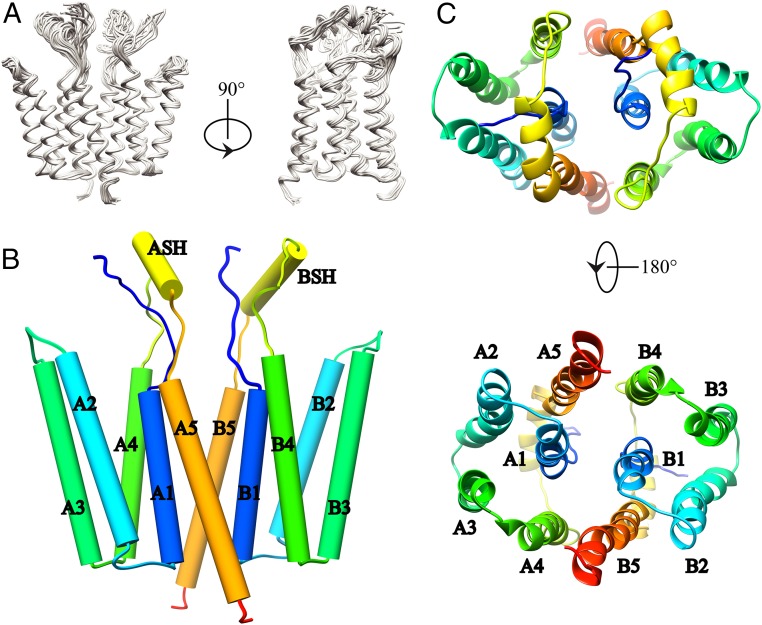
DPC was shown to be a suitable detergent for NMR experiments by 2D 1H-15N transverse relaxation-optimized heteronuclear single-quantum correlation spectroscopy (2D [15N, 1H]-TROSY-HSQC) experiments in which Rcf1 has a well-dispersed fingerprint for a protein of mostly helical content (SI Appendix, Fig. S5) (28, 29). The combination of triple-resonance backbone and side-chain correlation experiments, together with nuclear Overhauser effect (NOE) experiments on differently labeled samples allowed an assignment completeness of 93% for backbone and 70% for side-chain resonances. Specifically, the resonances from methyl groups from 6 of 7 isoleucines, 6 of 7 valines, 16 of 19 leucines, 10 of 13 alanines, 3 of 5 methionines, and 7 of 8 threonines were assigned (SI Appendix, Fig. S6). Chemical shift analysis clearly indicated the presence of six helices in Rcf1 (SI Appendix, Fig. S1C). Dihedral angle restraints were predicted from backbone chemical shifts, and a large number of short-, medium- and long-range distance restraints (SI Appendix, Table S1) were derived from a combination of different NOE experiments (SI Appendix, Fig. S7A). Under these conditions, Rcf1 is not monomeric, as evident from the intermonomer NOE distance restraints obtained from the 3D 13C, 15N-filtered/edited NOE experiments (SI Appendix, Fig. S7B). All together, these restraints enabled the calculation of the Rcf1 dimer structure (see Fig. 1 for sequence and Fig. 2 A and B for the structure).
Fig. 2.
NMR structure of the dimeric Rcf1 in DPC micelles. (A) Backbone ribbon trace of the 15 lowest-energy structures determined by solution state NMR. (B) Cylindrical representation of the Rcf1 dimer structure. The five TM helices of monomer A (A1–A5), monomer B (B1–B5) and the short flexible soluble helices (ASH, BSH) are shown (see labels). (C) Top view (Upper) and bottom view (Lower) of the Rcf1 dimer.
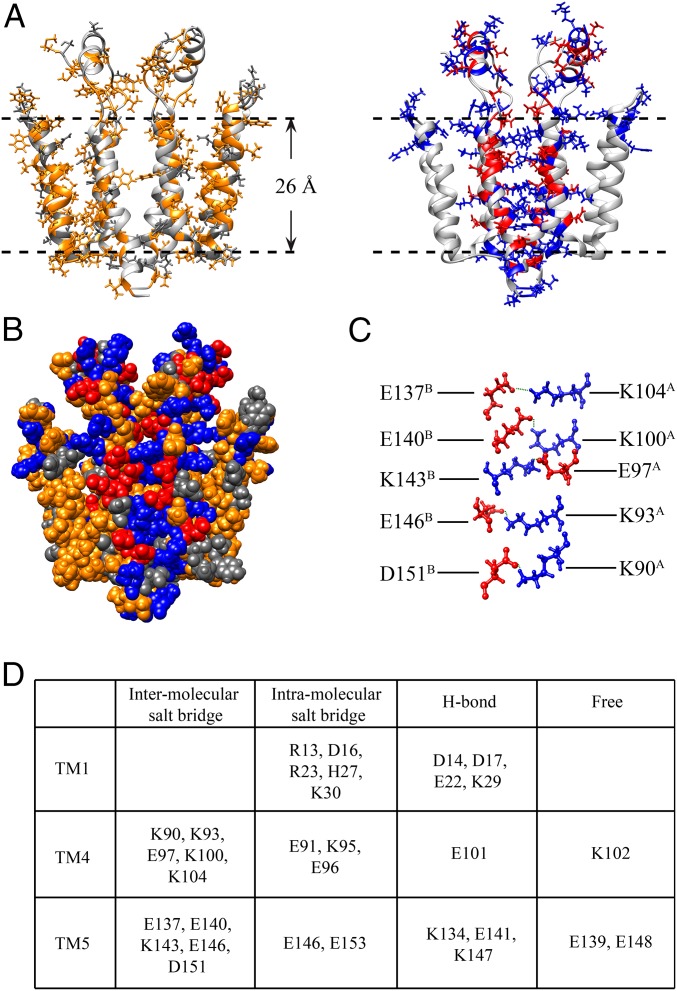
The Rcf1 dimer structure is formed from two identical compact monomers composed of five TM helices (TM1–TM5), which pack together in a clockwise order TM5–TM1–TM4–TM3–TM2 when viewed from “underneath” the C-terminal end (Fig. 2C). TM2 (I36–K57) and TM3 (F64–Y85), located on the lateral side, are hydrophobic and correspond to the two predicted TM helices (SI Appendix, Fig. S1A) that are also present in the human homolog Higd1a protein (30) (SI Appendix, Fig. S8). The Rcf1 monomer has 70 hydrophobic, 31 polar, and 58 charged residues. The distribution in the structure of these residues (Figs. 1 and 3A) is notably different from prediction (SI Appendix, Fig. S1). The Rcf1 dimer structure has two unusual features. First, the stretch of charged residues in the N and C termini fold into three helices, TM1 (D14–K30), TM4 (G89–R106), and TM5 (K134–L156). Second, the charged TM helices mediate the dimer formation. TM1, TM4, and TM5 thus form a central charged dimer interface, while the hydrophobic TM2 and TM3 are located at the lateral side (Fig. 3B). TM4 and TM5 interact with the corresponding charged helical side of the other monomer through the interaction of TM4A–TM5B and TM5A–TM4B (Fig. 2 B and C), with the possibility of 10 salt bridges in total, as shown in Fig. 3. E146 is positioned in such a way that it has a possibility to participate in both an intramolecular interaction (with H27) and an intermolecular interaction (with K93) (Fig. 3C). The loops connecting the TM helices are short except for the one connecting TM4 and TM5, where 13 residues, R117 to E129, fold into a short flexible SH. TM1 has eight charged residues and is located in the core of the dimer, making interactions between TM1A and TM1B but is not implicated in any salt bridges. Moreover, five charged residues in both TM4 and TM5 appear not to be involved in salt bridges. These “free” charged residues appear to be stabilized by both intramonomer and intermonomer hydrogen bonds. Furthermore, there are five intramonomer salt bridges: R13–D16, R23–E96, H27–E146, K30–E153, and K95–E91.
Fig. 3.
Distribution of types of amino acids in the Rcf1 dimer structure and analysis of the dimer interface. (A, Left) Stick representation of Rcf1 with hydrophobic (A, I, L, F, V, P, M, W, and G) side chains shown in yellow and polar side chains (Q, N, S, T, Y, and C) in gray. (Right) Positively charged (R, K, and H) and negatively charged (D and E) side chains are shown in blue and red, respectively. Dashed horizontal lines indicate the suggested membrane boundaries. (B) Sphere representation of the hydrophobic, polar, and charged residues in the dimer, color scheme as in A. (C) Sphere representation of the 20 (only 10 are shown, homodimeric C2 symmetry) residues involved in forming the 10 salt bridges at the dimer interface. (D) Charged residues grouped according to whether they are implicated in the formation of intermolecular salt bridges, intramolecular salt bridges, H bonds, or as free charged residues in the Rcf1–DPC structure.
Positioning of Rcf1 in DPC.
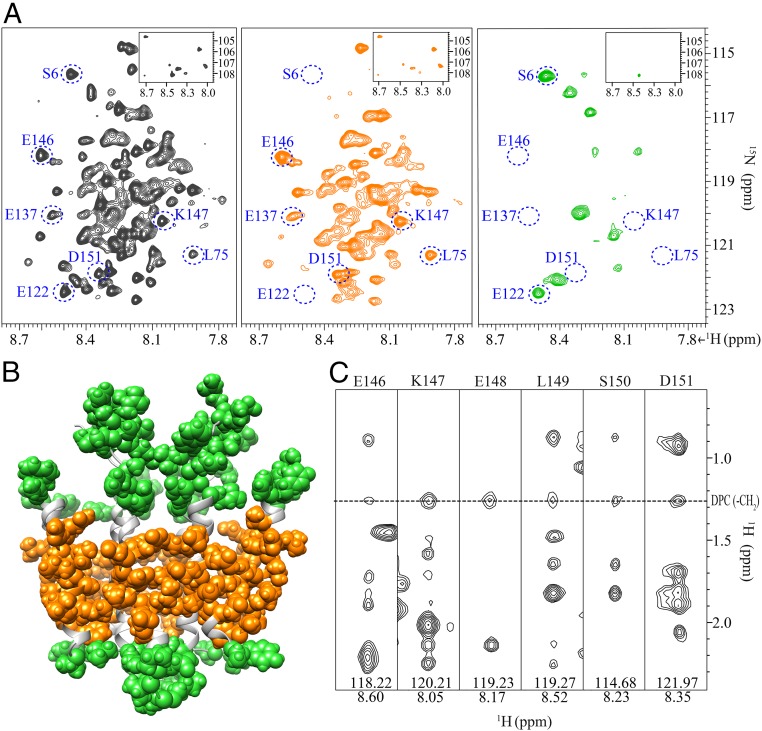
To further verify the position of the charged TM helices with respect to the DPC micelle, we performed titration experiments using two different paramagnetic probes (31); water-soluble gadodiamide (Gd) and detergent-soluble 16-doxyl-stearic acid (16-DSA). The resonances from residues in the loop regions and the short flexible SH were greatly broadened by Gd (Fig. 4A). Conversely, the resonances from residues in the hydrophobic TM2 and TM3, as well as residues in the charged TM1, TM4, and TM5, were broadened beyond detection after addition of 10 mM 16-DSA (Fig. 4A). These results demonstrate that the hydrophobic TM2 and TM3 as well as the charged TM1, TM4, and TM5 are in the proximity to the hydrophobic tails of DPC (Fig. 4B). Signals stemming from most residues in these helices were severely broadened by 16-DSA, indicating that the 16-DSA–labeled chain is flexible. This is presumably due to the protein–detergent complex being dynamic as a whole. Furthermore, NOE cross-peaks between the amide protons in the charged TM4 and TM5, as well as for the amide protons in the hydrophobic TM2 and TM3, and the methylene protons of DPC micelles were also observed, confirming that the surface of the Rcf1 helical bundle is close to the micelle hydrocarbon tails (Fig. 4C). However, NOE cross-peaks between DPC and the amide protons in TM1 were not observed, in agreement with its location in the dimer core. This is also consistent with the observation that the resonances from TM1 were broadened with higher concentration of 16-DSA than resonances in the other helices.
Fig. 4.
Locating the detergent-embedded and solvent-exposed regions of the Rcf1 dimer. (A) 2D [15N, 1H]-TROSY-HSQC spectrum of Rcf1 in DPC micelles (black), the same spectrum recorded after addition of 20 mM gadodiamide (orange) and 10 mM 16-DSA (green). The resonances in blue circles, representing residues in loops (S6), soluble helices (E122), charged helices (E137, E146, K147, and D151), and hydrophobic helices (L75) are quenched to below the noise level by gadodiamide or 16-DSA. Inset shows the Gly resonances. (B) Solvent-exposed residues that interact with gadodiamide with a relaxation enhancement ε > 4 mM−1⋅s−1 are shown in green. Detergent-embedded residues that interact with 16-DSA with an enhancement ε > 20 mM−1⋅s−1 are shown in orange. Residues with ε-values below these thresholds are shown in light gray. (C) Six [15N, 1H] strips from the 15N-resolved TROSY [1H, 1H]-NOESY spectrum shows the intermolecular NOEs between methylene groups of DPC and residues E146 to D151 (in TM5) of Rcf1. The DPC–Rcf1 NOE cross-peaks are marked by a dashed line.
Additionally, hydrogen–deuterium (H/D) exchange data revealed that, in contrast to the readily exchanged protons in the soluble and surface regions, the amide protons in the TM helices deeply embedded in the DPC micelles did not exchange with solvent 2H over 24 h (SI Appendix, Fig. S9). Taken together, these results confirm the topology of the Rcf1 dimer structure in detergent micelles.
Influence of Lipids on the Rcf1 Dimer Structure.
To assess whether the unusual structure of Rcf1 in DPC is specific only to the detergent-solubilized form, we measured the far-UV CD spectrum of Rcf1 reconstituted in small unilamellar vesicles (SUVs) and compared with that in DPC micelles (SI Appendix, Fig. S3). The spectra are very similar, but not identical, with both showing predominantly helical profiles, consistent with the NMR-derived structure. The helical content was estimated from the intensity at 222 nm (32) to be ∼85% and 80% for Rcf1 in DPC micelles and in SUVs, respectively. The overall Rcf1 structure may thus depend, to some extent, on the environment. To more specifically examine the effect of lipids on the structure of Rcf1, dimyristoylphosphatidylcholine (DMPC) lipids were gradually added to a sample containing the Rcf1–DPC complex to form small, fast-tumbling bicelles (33). The 2D [15N, 1H]-TROSY-HSQC spectra were recorded to monitor chemical shift changes upon lipid addition (SI Appendix, Fig. S10A). Although the chemical shift changes are small, they are consistent throughout the titration series and the results show a clear correlation between the magnitude of the chemical shift differences and the location of the TM helices (SI Appendix, Fig. S10B), indicating structural rearrangements of these helices upon addition of lipids. Amide 1H and 15N chemical shifts are not normally used to indicate secondary structure, but the dispersion of resonances in the HSQC is an indicator of folded protein. Although chemical shifts move in both directions (SI Appendix, Fig. S10A), overall most 1H amide chemical shift changes indicate less structure, i.e., most resonances shift toward the center of the spectral region. Taken together, these results indicate that the presence of lipids modulates the structure of Rcf1.
Mapping of the Binding Site with Yeast Cytochrome c.
To validate the suggested role of Rcf1 in binding to S. cerevisiae Cyt. c (23), titration of Rcf1 with Cyt. c was monitored using NMR spectroscopy. Stepwise addition of Cyt. c induced continuous changes for several resonances in the NMR spectrum (SI Appendix, Fig. S11), specifically residues E122, R124, and K126 in the short SH between TM4 and TM5, as well as residue K59 in the connecting loop between TM2 and TM3 (Fig. 5). We note that the presence of high concentrations of Arg and Glu in the NMR buffer presumably shields electrostatic interactions that could be stronger in the native system, and therefore we did not attempt to determine a binding constant for the interaction. Nevertheless, the results demonstrate that Cyt. c interacts with Rcf1 under the experimental conditions used in this study.
Fig. 5.
Mapping of the interaction between Rcf1 and S. cerevisiae Cyt. c. (A) Schematic representation of Rcf1 with the location of the residues involved in the binding with Cyt. c. (B) Surface representation of the Rcf1 dimer structure (light gray, side view) with residues in A highlighted in red.
Molecular Dynamics Simulations.
To examine the stability of the Rcf1 dimer in different environments, we conducted molecular dynamics (MD) simulations of the dimer successively in self-assembled DPC micelles, in a 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) lipid bilayer and in a biphasic octane/water membrane mimetic. The overall dimer structure was well preserved after simulation in DPC where the detergent molecules self-assembled to form micelles around lateral hydrophobic TM2 and TM3, which help to stabilize the dimer structure (Fig. 6 B and D). Simulations of the Rcf1 dimer in a transmembrane orientation in a lipid bilayer preserved the dimer structure, with acyl chains forming extensive contacts with TM2 and TM3, but compromised the structural integrity of the bilayer, with local membrane thinning and lipid headgroups and water molecules reaching across the bilayer to solvate the charged protein interface (SI Appendix, Fig. S12 A and B). These results suggest that the TM orientation may be metastable, with the relaxation of the membrane-embedded protein limited by the slow rearrangement of the bilayer on the sub-µs time scale of the simulations (34). To circumvent this problem, we used a biphasic octane/water slab, a membrane mimetic shown to reproduce the solvation of integral membrane proteins while speeding up structural relaxation (35). In the octane/water slab, the TM orientation was unstable and the overall dimer structure of Rcf1 was not well preserved. The weakest interactions involved central helix TM1, indicative of structural perturbations to the core of the Rcf1 monomer, and much larger structural changes took place (Fig. 6 C and E and SI Appendix, Figs. S12C and S13). Consistent with CD results, helicity was better retained in DPC than in the membrane mimetic (SI Appendix, Fig. S12E). While the Rcf1 dimer retained most of its native nonpolar contacts, especially in the micelles, polar contacts at the highly charged dimer interface underwent significant rearrangements (SI Appendix, Figs. S12F and S13). Within each monomer, the interfaces between TM1, TM2, and TM3 were more stable than interfaces involving TM4 and TM5 in DPC (Fig. 6E). These results suggest that the Rcf1 dimer is more stable in detergent micelles than in membrane (mimetics), where solvation is incompatible with the charged groups on the dimer surface.
Fig. 6.
Structure and solvation of the Rcf1 dimer from MD simulations in micelles and a membrane mimetic. (A) Centroid of the most populated cluster (39.2%) for the Rcf1 dimer in DPC. (B) Spatial distribution function of DPC detergent for the last 20-ns simulations of 10 replicas, with density cutoffs of 0.26 for the core layer and 0.175 for the outer layer. (C) Centroid of the most populated cluster for Rcf1 dimer in an membrane-mimetic octane slab. The mean position of the membrane–water interface is shown with dashed lines. (D) Surface of the Rcf1 dimer structure colored by the fraction of time that each residue made nonpolar contacts with DPC detergent. (E) Schematic representation of the fractions (given as percent) of native contacts between helix interfaces for Rcf1 in DPC (black) and in octane (brown). The interface between monomers is indicated by dashed lines. The fraction is indicated by line thickness.
Discussion
The Rcf1 protein is involved in formation of the S. cerevisiae III2IV2 (or IV1) supercomplex (17–19). Here we showed that in DPC micelles, Rcf1 folds into a five-TM helix arrangement where TM4 and TM5 form a charged interface, interacting to form a dimer (Figs. 2 and 3). Folding of charged, hydrophilic helical segments within detergent micelles is uncommon. One recently noted exception, consistent with our findings, is charged helical peptides that can self-assemble into the membrane via charge pairing, referred to as “charge zipper” (36). We propose that the Rcf1 dimer structure can form such a charge zipper in the membrane, although the population of dimers may be lower in a membrane environment than in DPC micelles. In MD simulations of the Rcf1 dimer in DPC, the overall dimer structure and the interface are largely preserved although individual contacts are more flexible than seen in the NMR structure.
Previous topological analysis of Rcf1 based on the prediction of two TM helices (SI Appendix, Fig. S1A) and proteinase treatment experiments (17–19), suggested that the N and C termini are both exposed to the intermembrane space (IMS). However, our Rcf1 structure has the N and C termini exposed to different sides of the detergent micelle. The previous analysis assumed that the water-soluble C-terminal region included the parts that in our structure folds into TM4, TM5, and the flexible SH. We note that a Nout–Cin orientation of the current structure is also consistent with the previous protease studies, as this would leave the connecting SH exposed to digestion in the IMS. It is also possible that Rcf1 has a Nin–Cout orientation, which would be in agreement with the suggested monomer/dimer equilibrium in the native state discussed below. In this scenario, the charged TM4 and TM5 would “flip out” of the membrane, exposing the (now larger) C terminus to the IMS, accessible to proteases.
Our comparison of the secondary structure in DPC, bicelles and liposomes (SI Appendix, Figs. S3 and S10), indicates that Rcf1 undergoes structural rearrangements in the presence of lipids. The results from CD measurements suggest that there is a small decrease in secondary structure in the presence of lipids, and the NMR data indicate that the changes are located to the TM helices. One such possible structural rearrangement would be a change from dimer to monomer where the charged TM4 and TM5 would flip out of the membrane in the monomeric state. This scenario is also consistent with the MD simulations showing that the Rcf1 dimer is stable in DPC (Fig. 6), but in a membrane (mimetic), it induces significant defects (POPC) (SI Appendix, Fig. S12 A and B) or starts dissociating (octane) (Fig. 6 and SI Appendix, Fig. S12). However, because it has been shown that the native S. cerevisiae Rcf1 can form dimers (37), we suggest that the interconversion is dynamic and reflects a change in dimer/monomer equilibrium.
Evidence for differences in protein dynamics in detergents and in lipid environments, with a pronounced higher degree of dynamic variability in the lipid membrane has recently been presented (38) supporting that the protein dynamics may be constrained in DPC relative to a real membrane, resulting in different monomer/dimer equilibria.
Furthermore, Rcf1 has been suggested to act as a chaperone that stabilizes the newly synthesized Cox3 subunit (17). This function may be exerted in the monomeric state where Rcf1 has the possibility to form the charged surface observed here, but it would in this case be free to interact with Cox3 or other proteins. Such a function is similar to that of the Mistic protein, a chaperone that forms a helical bundle with a charged lipid-facing surface that presumably stabilizes other membrane proteins (39).
The Rcf1 TM2 and TM3 correspond to the two predicted TM helices (SI Appendix, Fig. S1A), which are also present in the human homolog Higd1a protein (30) (SI Appendix, Fig. S8). Higd1a has been reported to interact with CytcO to increase its activity (40), and accordingly, TM2 and TM3 are hypothesized to form the binding surface for S. cerevisiae CytcO. We also note that the charged dimer interface is largely made up from the C terminal of Rcf1 which is not present in Higd1a (not conserved in the Hig1 family) and specific to yeast and other fungi. This observation points to differences in the roles of the Rcf1 protein in yeast compared with higher eukaryotes.
The Rcf1 dimer shows a defined interaction surface for S. cerevisiae Cyt. c (Fig. 5). We previously proposed an Rcf1–Cyt. c interaction based on functional properties of Δrcf1 mitochondria. Rcf1 could either “steer” Cyt. c while it relocates from Cyt. bc1 to CytcO or it could bind another Cyt. c molecule that would mediate direct electron transfer from Cyt. bc1 to CytcO without equilibration with the Cyt. c pool, as previously suggested (23).
In summary, the Rcf1 dimer structure shows an unusually charged putatively membrane-embedded dimer interface, with a charge zipper interaction between the two monomers. The structure will help in understanding its putative dual roles both as a CytcO chaperone and in Cyt. bc1–CytcO supercomplex formation and dynamics in mitochondria.
Materials and Methods
The full length of Rcf1 was cloned into a pET28a vector and expressed in E. coli. Inclusion bodies were refolded into different detergents and screened by 2D [15N, 1H]-TROSY-HSQC experiment. Three-dimensional TROSY-type backbone, side-chain, and NOE experiments on different samples were recorded for the structure calculation. Spectra were analyzed with CcpNmr (41); structure calculations were performed using CNS (version 1.21) (42). Paramagnetic probing and H/D exchanging experiments were carried out to verify Rcf1 topology in micelles. The titration with yeast Cyt. c was monitored by 2D [15N, 1H]-TROSY-HSQC experiments. MD simulations were conducted with the GROMACS simulation package (version 4.5.7) (43). Experimental detail is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank H. Dawitz and M. Ott (Stockholm University) and Chris Ing (University of Toronto) for valuable discussions. This research is supported by the Knut and Alice Wallenberg Foundation and the Swedish Research Council. The computational work was supported by the Natural Sciences and Engineering Council of Canada and by a Restracomp scholarship from The Hospital for Sick Children. Computations were also enabled in part by support provided by Westgrid (www.westgrid.ca), Compute Ontario (www.computeontario.ca), and Compute Canada (www.computecanada.ca). The NMR centers at the University of Gothenburg and the University of Frankfurt are acknowledged for technical help and for acquisition of NMR spectra. The operation and maintenance of the NMR spectrometers used in Frankfurt were supported by the Horizon 2020 Programme Grant iNEXT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates of the NMR structure have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 5NF8) and the NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 34115).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712061115/-/DCSupplemental.
References
- 1.Luttik MA, et al. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 2.Iwata M, et al. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc Natl Acad Sci USA. 2012;109:15247–15252. doi: 10.1073/pnas.1210059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackenbrock CR, Chazotte B, Gupte SS. The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J Bioenerg Biomembr. 1986;18:331–368. doi: 10.1007/BF00743010. [DOI] [PubMed] [Google Scholar]
- 4.Chazotte B, Hackenbrock CR. The multicollisional, obstructed, long-range diffusional nature of mitochondrial electron transport. J Biol Chem. 1988;263:14359–14367. [PubMed] [Google Scholar]
- 5.Chazotte B, Hackenbrock CR. Lateral diffusion as a rate-limiting step in ubiquinone-mediated mitochondrial electron transport. J Biol Chem. 1989;264:4978–4985. [PubMed] [Google Scholar]
- 6.Acin-Perez R, Enriquez JA. The function of the respiratory supercomplexes: The plasticity model. Biochim Biophys Acta. 2014;1837:444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Genova ML, et al. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta. 2008;1777:740–746. doi: 10.1016/j.bbabio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Lenaz G, Genova ML. Structural and functional organization of the mitochondrial respiratory chain: A dynamic super-assembly. Int J Biochem Cell Biol. 2009;41:1750–1772. doi: 10.1016/j.biocel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Guo R, Gu J, Wu M, Yang M. Amazing structure of respirasome: Unveiling the secrets of cell respiration. Protein Cell. 2016;7:854–865. doi: 10.1007/s13238-016-0329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837:427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Enríquez JA. Supramolecular organization of respiratory complexes. Annu Rev Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- 12.Suthammarak W, Morgan PG, Sedensky MM. Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. J Biol Chem. 2010;285:40724–40731. doi: 10.1074/jbc.M110.159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suthammarak W, Yang YY, Morgan PG, Sedensky MM. Complex I function is defective in complex IV-deficient Caenorhabditis elegans. J Biol Chem. 2009;284:6425–6435. doi: 10.1074/jbc.M805733200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velours J, Dautant A, Salin B, Sagot I, Brèthes D. Mitochondrial F1F0-ATP synthase and organellar internal architecture. Int J Biochem Cell Biol. 2009;41:1783–1789. doi: 10.1016/j.biocel.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schägger H. Respiratory chain supercomplexes of mitochondria and bacteria. Biochim Biophys Acta. 2002;1555:154–159. doi: 10.1016/s0005-2728(02)00271-2. [DOI] [PubMed] [Google Scholar]
- 17.Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Chen YC, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedo G, Vargas M, Ferreiro MJ, Chalar C, Agrati D. Characterization of hypoxia induced gene 1: Expression during rat central nervous system maturation and evidence of antisense RNA expression. Int J Dev Biol. 2005;49:431–436. doi: 10.1387/ijdb.041901gb. [DOI] [PubMed] [Google Scholar]
- 21.Gracey AY, Troll JV, Somero GN. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc Natl Acad Sci USA. 2001;98:1993–1998. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem. 2005;280:20580–20588. doi: 10.1074/jbc.M501894200. [DOI] [PubMed] [Google Scholar]
- 23.Rydström Lundin C, Ballmoos C, Ott M, Ädelroth P, Brzezinski P. Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2016;113:4476–4485. doi: 10.1073/pnas.1601196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mileykovskaya E, et al. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 26.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 27.Warschawski DE, et al. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim Biophys Acta. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehm T, Huber R, Holak TA. Application of NMR in structural proteomics: Screening for proteins amenable to structural analysis. Structure. 2002;10:1613–1618. doi: 10.1016/s0969-2126(02)00894-8. [DOI] [PubMed] [Google Scholar]
- 30.Klammt C, et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9:834–839. doi: 10.1038/nmeth.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilty C, Wider G, Fernández C, Wüthrich K. Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. ChemBioChem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- 32.Provencher SW, Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 33.Mäler L. Solution NMR studies of peptide-lipid interactions in model membranes. Mol Membr Biol. 2012;29:155–176. doi: 10.3109/09687688.2012.683456. [DOI] [PubMed] [Google Scholar]
- 34.Neale C, Hsu JC, Yip CM, Pomès R. Indolicidin binding induces thinning of a lipid bilayer. Biophys J. 2014;106:L29–L31. doi: 10.1016/j.bpj.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulleperuma K, et al. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J Gen Physiol. 2013;141:445–465. doi: 10.1085/jgp.201210856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walther TH, et al. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell. 2013;152:316–326. doi: 10.1016/j.cell.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Garlich J, Strecker V, Wittig I, Stuart RA. Mutational analysis of the QRRQ motif in the yeast Hig1 type 2 protein Rcf1 reveals a regulatory role for the cytochrome c oxidase complex. J Biol Chem. 2017;292:5216–5226. doi: 10.1074/jbc.M116.758045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frey L, Lakomek NA, Riek R, Bibow S. Micelles, bicelles, and nanodiscs: Comparing the impact of membrane mimetics on membrane protein backbone dynamics. Angew Chem Int Ed Engl. 2017;56:380–383. doi: 10.1002/anie.201608246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roosild TP, et al. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi T, et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci USA. 2015;112:1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 42.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.