Significance
Microbes are surprisingly social organisms and are providing model systems for the study of the evolution of cooperation and conflict. Despite their many advantages in the laboratory, such as experimental evolution, it is rarely possible to study them in the field. We therefore know little about whether cooperation and conflict are adaptively important in nature. Here we use approaches from population genetics and molecular evolution to test the adaptive relevance of social behavior in a social amoeba. We find signatures of adaptation for both kin selection and social cheating. This provides evidence that these behaviors have been important in the natural evolution of this species and more generally shows a way to study microbial social adaptation in the wild.
Keywords: altruism, cheating, kin selection, Dictyostelium discoideum, social amoeba
Abstract
Many microbes engage in social interactions. Some of these have come to play an important role in the study of cooperation and conflict, largely because, unlike most animals, they can be genetically manipulated and experimentally evolved. However, whereas animal social behavior can be observed and assessed in natural environments, microbes usually cannot, so we know little about microbial social adaptations in nature. This has led to some difficult-to-resolve controversies about social adaptation even for well-studied traits such as bacterial quorum sensing, siderophore production, and biofilms. Here we use molecular signatures of population genetics and molecular evolution to address controversies over the existence of altruism and cheating in social amoebas. First, we find signatures of rapid adaptive molecular evolution that are consistent with social conflict being a significant force in nature. Second, we find population-genetic signatures of purifying selection to support the hypothesis that the cells that form the sterile stalk evolve primarily through altruistic kin selection rather than through selfish direct reproduction. Our results show how molecular signatures can provide insight into social adaptations that cannot be observed in their natural context, and they support the hypotheses that social amoebas in the wild are both altruists and cheaters.
Cooperative behavior, once associated primarily with animals like social insects, is increasingly seen as widespread in nature. Some social microbes are now providing excellent model systems for the study of cooperation and conflict, because they can be genetically manipulated or because their short lifetimes facilitate experimental evolution over many generations. However, these systems have one major disadvantage. Unlike animals, which can be directly observed and assessed in their natural environments, microbes usually need to be taken out of their natural environments for observation. With a few exceptions (1, 2), we therefore know little about microbial social adaptations in nature (3), resulting in multiple controversies over the natural adaptive importance of even some of the best-studied phenomena such as bacterial quorum sensing (4–7), siderophore production (8, 9), and biofilms (10, 11).
The social amoeba Dictyostelium discoideum is a microbial model system for cooperation and conflict (12, 13). In this species, single-celled amoebas join together upon starvation to form multicellular fruiting bodies (14). About 20% of the cells die in the process of forming a stalk, which supports and promotes the dispersal (15) of the other 80%, which differentiate into spores. This appears to be an instance of kin-selected altruism (12, 16, 17). Laboratory studies also show the potential for extensive cheating. Here we use “cheating” as shorthand for any competition within the fruiting body, with the essential point being that when two or more clones aggregate together, they may be in conflict over who gets to produce the reproductive spores (18, 19). However, the relevance of both kin selection and cheating in the natural environment has been questioned (20–23).
It would clearly be useful to develop some alternative methods for understanding microbial social behavior in the wild. Here we deploy theories from population genetics and molecular evolution to search for, and find, molecular signatures that reflect both kin selection and cheating in wild D. discoideum.
Cheating.
In the laboratory, different D. discoideum clones readily join the same fruiting body (18), despite some recognition and segregation (24). Often one clone will show apparent cheating in the sense of getting more than its proportional (fair) share of spores (12, 25). Laboratory evolution under conditions of low kin selection leads to an increase in the frequencies of cheating mutants and a decrease in cooperation, as predicted by theory (17, 26, 27). However, the importance of cheating in the wild is uncertain, partly because relatedness is known to be quite high (16) and partly because of two plausible alternative explanations invoking adaptive trade-offs that would be hard to assess in nature. First, there is a modest number of loner cells that do not join the aggregation (28). A clone that produces fewer loner cells would, other things being equal, contribute more spores in mixtures. It could therefore appear to cheat when selection was really just operating on the trade-off between loner cells and aggregators (20, 29). Second, a clone that makes more, smaller spores could appear to cheat against a clone that makes fewer, larger spores, without necessarily having gained any cheating advantage (21).
If cheating is common in nature and also causes resistance to cheating to evolve, as it does in the laboratory (30–32), this may lead to evolutionary conflict and increased selection pressure. A set of D. discoideum genes whose knockouts cause cheating showed an unusual degree of balancing selection (in which rare alleles are favored), one possible outcome of cycling or stalemate conflict (33). However, this study did not show the more pronounced arms race outcome of rapid adaptation via directional selection. Here we use RNA-seq to screen specifically for genes that change expression in chimeric mixtures of two clones. This is the precise context in which cheating would be adaptive, so it may pinpoint the genes most likely to function specifically in cheating or resistance to cheating. Moreover, these would likely be facultative cheating or resistance genes, the kind that would most likely be favored under high relatedness. (Obligate ones are less likely to be favored because, when alone, they would still pay any cost of cheating without getting any benefits.) We then test for rapid adaptive evolution in these genes relative to other genes in the genome.
Kin Selection or Direct Selection.
The importance of kin selection and altruism in the wild has also been questioned. Instead of getting kin-selected benefits by altruistically helping related spores to disperse, stalk cells might instead be making the best of a bad job, doing everything they can to reproduce directly (22, 23, 34). Some evidence consistent with this view comes from the fact that the stalk is made by cells with less glucose (35), that prestalk cells are suppressed and perhaps poisoned by a chlorinated molecule produced by prespore cells (22, 23), and that prestalk cells may actually reproduce on rare occasions (36). An acknowledged weak point of this hypothesis is how an effort to reproduce would lead to producing a complex stalk (23). The question could be settled by evidence on the relative importance of personal and kin effects in the field.
The two hypotheses differ in the proposed role of prestalk cells: are they being selected to reproduce directly or instead indirectly through giving aid to kin? This results in contrasting predictions about the strength of purifying selection in prestalk and prespore cells.
To test the hypothesis that indirect kin selection is irrelevant, and all selection on prespore cells is through direct reproduction (22, 23), we use theory about how selection operates on conditionally expressed genes. Other things being equal, a gene should be selected more weakly and be more variable in proportion to the fraction of individuals that express it (37). For example, genes preferentially expressed in rarer morphs of pea aphids show relaxed purifying selection (38). In D. discoideum, where 80% of the cells in an aggregate become prespore cells, and 20% become prestalk cells, purifying selection against mildly deleterious mutations will be four times less effective in prestalk cells than in prespore cells. Other things being equal, genes expressed primarily in prestalk cells should therefore be four times more polymorphic than genes expressed mainly in prespore cells (37), assuming similar initial distributions of mutant effects on fitness. [Actually, the difference may be more extreme than 4:1 because we have not accounted for the fact that, even in this direct selection hypothesis, prestalk is viewed as a best-of-a-bad-job strategy (22)]. Note that many other genes in the genome may also be conditionally expressed, to unknown degrees. That is why we do not use all genes in this test, but instead compare prestalk genes against prespore genes—these are expressed in the same circumstance (fruiting) but differ in their relative proportions.
The alternative kin-selection hypothesis is that all selection on prestalk cells is indirect selection operating through effects on related spores. Here, theory predicts that the effect of indirect selection, relative to direct selection, is diluted by a factor of the relatedness coefficient (39, 40). A probable empirical example is that honeybee worker genes show lower nonsynonymous variability than queen genes (41). For D. discoideum fruiting bodies, relatedness is high in nature, with two estimates based on molecular markers yielding 0.97 and 0.86 (16). At these levels, prestalk genes under pure kin selection should be only 1.03–1.17 times as variable as prespore genes under direct purifying selection.
Results
Cheating.
Using four pairs of wild clones, we searched for genes changing expression in chimeric mixtures. For each clone pair, the chimeric treatment involved mixing the two clones in equal proportions under starving conditions so they would form fruiting bodies. The controls were identical except that each clone was allowed to form fruiting bodies on its own, starting from the same total number of cells. We harvested RNA at the tight aggregate stage, a key stage for stalk–spore differentiation, after which gene expression patterns switch abruptly (42, 43). Using a generalized linear model (GLM) that accounts for effects of clone pair, library, and sequencing batch, we identified 79 genes that consistently and significantly differed in expression between chimeras and controls at false discovery rate = 0.10 (20 up-regulated in chimeras, 59 down-regulated; Dataset S1). It is interesting to note that the change in expression between chimeras and controls was correlated (Pearson’s r = 0.311, P < 0.001) with expression changes in a previous experiment where chimeras differed only at the tgrB1 and tgrC1 cell adhesion loci that control clonemate recognition (44), suggesting that our response is at least partly influenced by that recognition system.
We tested the hypothesis that these chimera-biased genes would show conflict-generated high rates of adaptive evolution of coding sequence using the program DFE-α (45). It estimates a modified McDonald-Kreitman (46) statistic, α, by measuring the proportion of nonsynonymous sites between species that have been fixed by selection, using within-species polymorphisms to provide an expectation if they were due only to neutral evolution and purifying selection. The program improves on typical McDonald-Kreitman tests by allowing α to be estimated over entire gene sets, yielding greater power, and by using the estimated frequency distribution of polymorphisms to better account for low-frequency deleterious alleles (47).
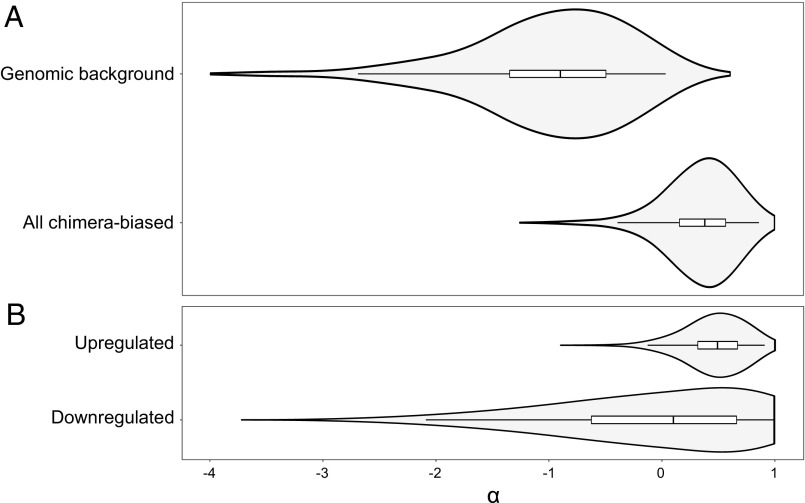
We used 15 D. discoideum genomes from Virginia and Texas to estimate nonsynonymous and synonymous polymorphism within species and the corresponding fixed differences relative to a diverged Costa Rican outgroup clone, S6B, which is probably a different species (48). As predicted by the conflict hypothesis, adaptive evolution is significantly higher in the genes that change expression in chimeras than in genes that do not (Fig. 1, α = 0.149 versus −0.723, P = 0.002 permutation test). This is primarily due to genes up-regulated in chimeras, although they are not significantly different from down-regulated ones (Fig. 1). We found similar results using an alternative measure of adaptive evolution, ωA (49) (Table S1). Balancing selection is another possible outcome of cheating (33), but we found no support for this in three measures of balancing selection. Chimera-biased genes and other genes were not different, using permutation tests, for either fst (P = 0.387) or Tajima’s D (P = 0.514). Fay and Wu’s H (P = 0.0172) did show a difference, but one indicating directional selection, in agreement with our other results. In this case, the signature of selection was entirely due to genes up-regulated in chimeras (P = 0.0096), and not those that were down-regulated in chimeras (P = 0.759) (Table S1).
Fig. 1.
Genes that change expression in chimeric mixtures show elevated rates of adaptive evolution α. Each violin plot shows a Gaussian kernel-density plot of 1,000 bootstrap replicates of α, the median, the interquartile range, and the 95% range or confidence interval (Table S1) (A) Bootstrap distributions for the 78 chimera-biased genes and for samples of 78 from genomic background genes. The chimera-biased genes show significantly higher α (P < 0.002 permutation test). (B) Bootstrap distributions for the chimera-biased genes separated into the 19 up-regulated genes and 59 down-regulated genes, both of which are significantly different from background genes (up-regulated P = 0.006, down-regulated P = 0.034, permutation tests).
Kin Selection or Direct Selection.
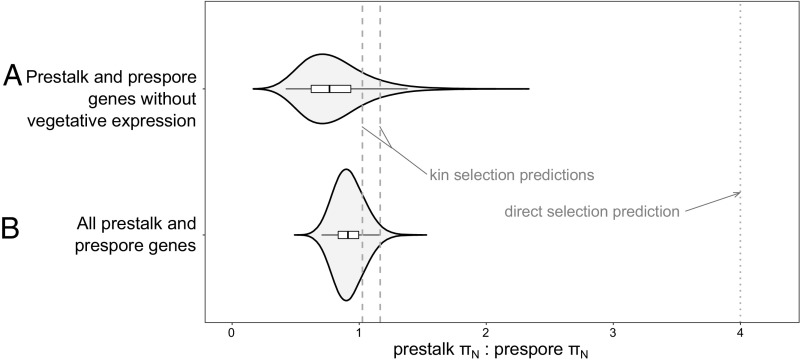
The results above support the hypothesis of cheater-driven molecular evolution, but what about altruism? To test the importance of direct selection versus indirect (kin) selection, we used previously identified (42) genes with significantly greater expression in prestalk cells than prespore cells, or vice versa. To eliminate effects of selection in other contexts, we also tested a second set (n = 145, 113; see Dataset S1), in which we removed genes with any expression during the vegetative (single-cell) stage. We estimated their variability (Table S2) using the same 15 D. discoideum genomes (33). Because the predictions apply only to nonneutral sites (neutral mutations are not subject to purifying selection), we focus on the nonsynonymous diversity, πN. If a large fraction of nonsynonymous mutations were neutral, we would need to also exclude those, but that is not the case. (From the DFE-α program these fractions are low for prestalk and prespore genes combined: f = 0.127 and f = 0.140, with and without vegetative expression, respectively; Table S2.) For the two gene sets, the ratios of nonsynonymous diversity, πN, for prestalk genes to prespore genes (prestalk πN: prespore πN) are 0.914 and 0.771. These are not significantly different from the two predicted values under kin selection (1.03, 1.17) but significantly differ from the value of 4 predicted by the direct selection hypothesis (Fig. 2 and Table S3).
Fig. 2.
Nonsynonymous diversity πΝ supports kin selection, not direct selection, in prestalk cells. Violin plots (Fig. 1) for distributions from 10,000 resamples of the prestalk πN: prespore πN, the ratio of nonsynonymous nucleotide diversity πN for genes expressed significantly more in prestalk to πN for genes expressed significantly more in prespore. (A) All prestalk-biased genes (n = 992) and prespore-biased genes (n = 879). (B) Prestalk-biased genes (n = 145) and prespore-biased genes (n = 113) that are not expressed in the vegetative stage (Table S3).
The conclusion in favor of kin selection is not altered by two potential caveats. First, the expected ratio for two sets of random genes is a ratio of 1, close to our kin selection prediction, but the prestalk and prespore genes are not random sets. Compared with genes in the whole genome (πN = 0.00019), both sets of prespore plus prestalk genes are significantly more variable (including vegetative expression, πN = 0.00021, P = 0.04; without vegetative expression, πN = 0.00027, P = 0.03, permutation tests). Moreover, the data very decisively reject the direct-selection prediction of a ratio of 4 or higher. The maximum values obtained in 10,000 bootstrap samples were only 2.24 and 1.48 for our two prestalk–prespore gene sets (Table S3).
Second, our direct-selection prediction of fourfold greater variation in prestalk genes may be too extreme, given that even our prestalk-enriched genes have some expression in prespore cells. Selection on these prestalk genes may therefore include a minority component of direct selection in prespore cells, which should tend to make selection in the two gene sets somewhat more similar. However, a far more conservative prediction is available concerning the correlation between diversity πN and degree of prestalk versus prespore expression. The kin selection hypothesis predicts there should be little or no correlation because, with relatedness near 1, selection intensity would be roughly equal in the two tissues. In contrast, since the direct selection hypothesis implies stronger purifying selection on prespore genes, prestalk genes should be more variable, and the correlation should be positive. In fact, the correlation is weak and negative (with vegetative expression τ = −0.062, P = 0.00025, without τ = −0.096, P = 0.039; Kendall’s tau correlation between πN and log2 of the prestalk: prespore expression ratio), again strongly rejecting the direct selection hypothesis.
Discussion
Testing adaptation is rarely simple because it requires understanding how the organism interacts with its natural environment. For social behavior, this is particularly difficult because it requires understanding the natural social context. For animals, we can at least observe their behavior in their natural environment. Microbes, however, are more difficult to observe, and they are typically studied in laboratory environments that may not accurately reflect their natural contexts.
For example, the social amoeba Dictyostelium discoideum has usually been studied in a uniclonal social context, with most work on the species being carried out on the clonal descendants of a single natural isolate, NC4. This obscured the possibility of interesting social behaviors, like cheating (18, 19) and kin recognition (24, 50, 51), that only revealed themselves when multiple clones were studied in mixtures. However, the studies are still carried out in the laboratory and might therefore miss important elements of the natural context. This has led to controversies about the adaptiveness of both cheating and altruism in D. discoideum (20–23).
Although one cannot usually observe microbial social adaptations operating in nature, molecular signatures of population genetics and molecular evolution can sometimes provide an alternative approach. Social conflict is expected to lead to rapid evolution of genes involved in the conflict. In D. discoideum the genes most likely to be specialized for cheating conflict are those that change expression in chimeric mixtures. We show that these genes do indeed show more rapid adaptive evolution, supporting the natural importance of cheating. Note that these have not been confirmed as cheating genes. However, that seems the only obvious reason why this particular set of genes should show more adaptive evolution. Similarly, the alternative hypotheses of kin-selected altruism versus direct reproduction are predicted to leave different signatures with respect to the amount of nonsynonymous variation in genes that are particularly expressed in prestalk cells. The results strongly reject the direct selection prediction and support the kin selection prediction. Thus, it appears that both cheating and kin selection are not just laboratory phenomena but are also important in the wild.
The main assumption underlying our analyses is that the observed differences in evolution are due primarily to the relative strengths of selection between the gene sets. Because we are comparing gene categories in the same population, it is reasonable to assume that other forces like drift and migration are equal. It is less certain that mutations must be equal, specifically, the distribution of selective coefficients of mutants. Although there is no specific reason to believe this assumption should fail for our gene sets, it is more questionable, and it is therefore good that kin selection and cheating are supported by other kinds of studies.
With respect to kin selection, there is evidence for all three components of kin selection. Stalk cells pay the large cost of sacrificing their lives to produce a stalk. Other cells have been assumed to benefit from the stalk by gaining more access to dispersers, an assumption supported using a model arthropod disperser in the laboratory (15). Finally, we know that relatedness within fruiting bodies is high in nature (16) in part due to kin recognition (50) but probably also due to passive population structure (25, 52, 53).
However, high relatedness makes our other finding—evidence for cheating in the wild—more surprising because most fruiting bodies are clonal. However, it should be remembered that selective forces that operate rarely, for example, certain pathogens, can still exert important selective forces. There is also some prior evidence with respect to cheating. We know that unequal contribution to spores is common among laboratory clones (18, 19), that mutants in many genes affect this (27), and that cheating mutants spread readily under conditions of low relatedness (17, 27). High relatedness in the field must prevent some cheating, especially from high-cost obligate cheaters that cannot fruit on their own (16). However, this high cost does not apply to facultative cheaters that cheat by changing expression only when a foreign partner is present, so selection might still favor some of these cheaters. Our results suggest that this is indeed the case. Additional supporting data come from several sources. A mutation accumulation experiment showed that random mutations tend to decrease cheating ability, which is the result expected if cheating is a fitness component (54). The presence of kin recognition and segregation seems best explained as a partial solution to the problem of foreign clones that might do harm (55). Finally, clones in chimeras show possible cheating adaptations. Chimeric slugs travel less far, consistent with cells trying to stay out of the front region that will form the stalk (56). Chimeras also produce more spores and higher spore-to-stalk ratios (19). Alternative explanations are possible for most of these phenomena. For example, chimeras could have reduced slug migration due to lower cell–cell adhesion, a side-effect of mismatches at their kin-recognition tgrB1/tgrC1 loci (57). Collectively, however, all these phenomena build a consistent case for the importance of cheating in the wild.
The use of molecular signatures like these might also be employed in other controversies about microbial social evolution (4–11). To be useful, it is necessary to identify a target set of genes hypothesized to be subject to a particular kind of selection and then measure a reliable signature of that kind of selection. This might not always be feasible for some systems and questions, but this approach does add a valuable tool to other approaches such as making the laboratory setting more natural and conducting experiments in the field (3). When it is feasible, the method of using population-genetic or molecular-evolution signatures is superior in one important respect. It yields a more comprehensive record of selection, one that is automatically integrated over the full geographical range sampled and over very long periods of time.
Materials and Methods
Amoeba Samples.
To detect genes changing expression in chimeras, we tested four pairs of D. discoideum strains or clones, originally isolated from soil from Mt. Lake Biological Station in Virginia: QS6 with QS160, QS4 with QS174, QS18 with QS154, and QS17 with QS157, a sufficient number to exclude effects that are idiosyncratic to particular clone pairs. For molecular evolution analyses we used the genomes of 16 strains. For polymorphism data, we used 15 D. discoideum strains, eight strains from Virginia and seven strains from Texas, all those that were available after excluding populations with only one strain (33). For divergence estimates, we compared these strains to a tropical outgroup clone S6B from Costa Rica, probably a separate species (48).
Chimera-Biased Genes: RNA Sequencing.
We prepared samples from four strain pairs using the following procedures. We grew amoebas on SM/5 agar plates [2 g glucose, 2 g BactoPeptone (Oxoid), 2 g yeast extract (Oxoid), 0.2 g MgCl2, 1.9 g KH2PO4, 1 g K2HPO4 and 15 g agar per liter] with ∼2 × 105 spores and a food bacterium Klebsiella pneumoniae (250 μL at 1.5 optical density). When amoebas were in log-phase growth, we used a sterile plastic spatula to scrape cells from the plates into KK2 buffer and washed three times to remove most of the food bacteria. For each replicate, we spread 108 cells in 1,000 μL KK2 onto 47-mm-diameter nitrocellulose filters (Millipore) for each of the two unmixed clonal strains and 108 total cells for the 50:50 chimeric mix of strains, resulting in a trio of samples (two clonal, one chimeric). When 90% of the cells were in the tight aggregate stage, we washed cells off of each filter with KK2 buffer into a 5× volume of RNAlater for storage at 4 °C . For each strain pair, we repeated this process three times on different dates. We extracted RNA using a protocol for cytoplasmic RNA purification from animal cells with a Qiagen RNeasy Mini Kit, with modifications based on Kaul and Eichinger (58). From here, we prepared sequencing libraries using the standard Illumina protocol for the poly-A–tailed stranded mRNA library prep kit. We constructed three batches of libraries, each run in one sequencing lane, with each containing a full replicate of the experiment: two clonal and one chimeric sample for all four strain pairs. Sequencing was done on an Illumina Hiseq2500 for 50-bp single-end reads at the Washington University in St. Louis Genome Technology Access Center (GTAC).
Chimera-Biased Genes: Alignment and Differential Expression.
After quality control of raw reads (removal of reads shorter than 12 bp and those with any N nucleotides), reads from each library were mapped onto the D. discoideum reference genome (downloaded Dec 2014 from Ensembl Protist v1.25). Before alignment, we masked the known duplicated region on chromosome 2 of the AX4 reference genome (2: 3016083–3768654) using bedtools v2.19.1 (59). We used GSNAP v2014-12–17 (60) using default alignment parameters, except for only allowing a single alignment path to be followed to avoid chimeric reads (npaths = 1). GSNAP uses an oligomer chaining method combined with dynamic programming to align transcript reads to genomic sequence and is splice junction aware. We derived splice junctions based on the D. discoideum GFF3 gene feature annotations (downloaded September 2015) from dictybase.org (61). We used Picard v1.128 (downloaded from broadinstitute.github.io/picard) to sort alignments and fix read groups. We used R v3.2.1 (62) and Bioconductor package ShortRead v1.26.0 (63) to assess sequence read quality statistics. We excluded one replicate of the strain pair QS6 and QS160 from our analyses because the bamQA report generated by ShortRead indicated it did not meet quality standards. We then used RSeQC v2.5 (64) to look at alignment statistics and read distributions across genomic features. We had aligned 5.7–28.1 million (median 10.3) read tags or reads split by indels per library, and 4.9–27.3 million of these were aligned to annotated coding genes.
We extracted read counts from uniquely mapped reads using HTSeq v0.5.4p5 (65). Only reads with the correct strand orientation and mapping quality above 20 were counted. We imported these counts into R and examined the correlation between replicates within each strain pair across all expressed genes. The correlations across pairwise comparisons of replicates within strains were generally very high (mean r = 0.94), while the excluded sample showed a much lower correlation (r = 0.61), justifying our decision to omit it from further analysis.
We used DESeq2 v1.8.1 (66) to test for evidence of significant differential expression. We tested 9,089 genes, using a GLM model (count ∼ batch + pair + condition), with sequencing lane and library preparation batch as the factor batch, strain pair identity as the factor pair, and the clonal vs. chimeric condition of aggregation as the factor condition. DESeq2 uses a negative binomial distribution to model read counts and correct for sequencing library size using median-of-ratios size factors and uses empirical Bayes shrinkage estimators that correct count variance in individual genes based on other genes with similar expression levels (66).
Prespore and Prestalk Genes.
Our tests of the roles of direct and indirect (kin) selection in the evolution of stalk cells require identification of sets of genes with expression that is relatively specialized in prestalk and prespore cells. We used the candidate prespore and prestalk genes reported by Parikh et al. (42) with slightly reduced sample sizes after removing noncoding elements (see below). This study had separated prestalk and prespore cells and performed RNA-seq to determine which genes were significantly more expressed by each cell type (42). To reduce the influence of selection that occurs during the vegetative stage, we also tested a more restrictive set of 113 prespore and 145 prestalk genes that had no gene expression detected in vegetative cells.
Polymorphism and Divergence.
We tested whether our candidate chimera-biased genes show high rates of adaptive evolution consistent with an arms race scenario driven by social conflict. We cleaned and clipped raw Illumina reads from the 16 D. discoideum strains using ngsShoRT v2.2 (https://research.bioinformatics.udel.edu/genomics/ngsShoRT/). We generated mpileup files that merged strains within each geographic location using samtools v0.1.19 (67, 68), with adjusted mapping quality (-C 50) and a minimum basecall quality of 30 (-Q 30). We used Varscan v2.3.9 (69) to call variants from these merged mpileup files. We specified a minimum coverage of 20 reads per SNP and filtered for strand bias at a P value of 0.01. We then resplit the VCF file by strain and reconstructed the sequences of over 12,000 genes using GATK FastaAlternateReferenceMaker. We used custom scripts to convert these genomic FASTA files into coding sequences by removing introns and reverse complementing as necessary. Because our downstream tests assume that genes are coding, we removed noncoding RNAs, pseudogenes, and transposable elements as annotated in the D. discoideum genome (61, 70). This eliminated one of our chimerism genes from further analyses.
We used PolyMORPHOrama (71) to estimate average pairwise nucleotide diversity (π) using a Jukes-Cantor correction (72) and counted the numbers of polymorphisms per site class, both nonsynonymous (Pn) and synonymous (Ps). PolyMORPHOrama also generated the allele frequency spectra that we used in estimates of Tajima’s D (73), Fay and Wu’s H (74), and other downstream analyses of molecular evolution (see below). Next, we created a consensus FASTA of the 15 wild clones for each gene for comparison with S6B as outgroup, using ancestral sequence reconstruction method implemented by codeml (runmode = 0, CodonFreq = 2) in PAML v4.8 (75) and a custom Perl script. From this, we used codeml (runmode = −2, CodonFreq = 2) to generate our pairwise estimates of Dn and Ds. We used vcftools v0.1.12a (76) to estimate Weir-Cockerham’s fst (77) directly from VCF files. We imported these data into R and identified the genes associated with each variant using ChIPpeakAnno v3.2.2 Anno (78).
Molecular Evolution Analyses.
We assessed the relative strength of purifying selection on prestalk and prespore genes by taking the ratio of their nonsynonymous π’s: prestalk πN: prespore πN. The mean π’s are calculated for the numerator and denominator before dividing to reduce variance and eliminate zero denominators. This ratio was tested against predicted values of 4 for direct selection on prestalk genes, versus 1.025 and 1.165 for indirect selection on prestalk genes (the reciprocals of two relatedness estimates).
To test for adaptive selection on genes up-regulated in chimeras, we used tests of selection based on the McDonald-Kreitman test (46), originally instituted as a 2 × 2 Fisher’s Exact test to compare nonsynonymous (Pn) to synonymous (Ps) polymorphism to nonsynonymous (Dn) to synonymous (Ds) divergence for a single gene. Related metrics have been developed to summarize the effects of numerous selective events over multiple genes. These include: α, the proportion of nonsynonymous substitutions driven to fixation by positive selection (45, 79, 80); ωa, the rate of adaptive fixation relative to neutral fixation (80); and ƒ, the proportion of nonsynonymous mutations that are effectively neutral. We generated these four parameters for our gene sets with the maximum likelihood method of Eyre-Walker and Keightley (45), implemented in the command-lined version of DFE-α v2.15 (www.homepages.ed.ac.uk/pkeightl/). We used a custom Perl wrapper to sum the allele frequency spectra generated by PolyMORPHOrama, incorporate divergence information, and perform either permutations or bootstrapping.
Statistics.
Confidence intervals for all molecular evolution parameters are obtained by bootstrapping. From the i genes contributing to a statistic X, we repeatedly drew samples (either 1,000 or 10,000; see below) of i genes with replacement, recomputed X from each sample, and defined the 95% confidence interval as between the upper and lower 2.5% of the distribution.
Statistical tests for molecular evolution parameters were either bootstrap tests (tests against a predicted value Y) or permutation tests for differences between two samples. For a test of a difference in a statistic between two samples, X1–X2, based on i and j genes, we randomly drew, without replacement, samples (either 1,000 or 10,000; see below) of i and j genes from the total of i + j genes and recalculated the difference X1–X2 for each. For comparisons against the genomic background, we randomly drew, without replacement, samples of i genes from the total of i + j genes. P values were calculated as the proportion of times the permuted difference was more extreme than zero in the direction predicted. For two-tailed bootstrap tests of an estimate X1 against a predicted value Y, we repeatedly drew with replacement samples of i genes from the i original genes and recalculated X1. From this distribution, the percentage in the shorter tail cut off by Y, doubled, is the two-tailed P value.
For π, Tajima’s D, Fay and Wu’s H, and fst, we drew 10,000 resamples. Because α and other site frequency spectrum metrics required extensive computation (rerunning the DFE-α program) for each replicate, we drew 1,000 resamples.
Supplementary Material
Acknowledgments
We thank Tracy Douglas for the sequence of clone S6B as well as Tyler Larsen and two referees for comments on the manuscript. This material is based upon work supported by the National Science Foundation under Grants DEB1146375 and IOS-1656756.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All data, including genome alignments for the 16 published Dictyostelium discoideum genomes, along with the code that generated the statistics, are deposited in Dryad Digital Repository (doi: 10.5061/dryad.43cp320).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720324115/-/DCSupplemental.
References
- 1.Bozdag GO, Greig D. The genetics of a putative social trait in natural populations of yeast. Mol Ecol. 2014;23:5061–5071. doi: 10.1111/mec.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Köhler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci USA. 2009;106:6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarnita CE. The ecology and evolution of social behavior in microbes. J Exp Biol. 2017;220:18–24. doi: 10.1242/jeb.145631. [DOI] [PubMed] [Google Scholar]
- 4.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 5.Keller L, Surette MG. Communication in bacteria: An ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 6.Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: When is a signal not a signal? Philos Trans R Soc Lond B Biol Sci. 2007;362:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20:586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Zhang XX, Rainey PB. Exploring the sociobiology of pyoverdin-producing Pseudomonas. Evolution. 2013;67:3161–3174. doi: 10.1111/evo.12183. [DOI] [PubMed] [Google Scholar]
- 9.Andersen SB, Marvig RL, Molin S, Krogh Johansen H, Griffin AS. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci USA. 2015;112:10756–10761. doi: 10.1073/pnas.1508324112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb JS, Givskov M, Kjelleberg S. Bacterial biofilms: Prokaryotic adventures in multicellularity. Curr Opin Microbiol. 2003;6:578–585. doi: 10.1016/j.mib.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104:876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strassmann JE, Queller DC. Evolution of cooperation and control of cheating in a social microbe. Proc Natl Acad Sci USA. 2011;108:10855–10862. doi: 10.1073/pnas.1102451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaulsky G, Kessin RH. The cold war of the social amoebae. Curr Biol. 2007;17:R684–R692. doi: 10.1016/j.cub.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge Univ Press; Cambridge, UK: 2001. p. 308. [Google Scholar]
- 15.Smith J, Queller DC, Strassmann JE. Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC Evol Biol. 2014;14:105. doi: 10.1186/1471-2148-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert OM, Foster KR, Mehdiabadi NJ, Strassmann JE, Queller DC. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc Natl Acad Sci USA. 2007;104:8913–8917. doi: 10.1073/pnas.0702723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzdzal-Fick JJ, Fox SA, Strassmann JE, Queller DC. High relatedness is necessary and sufficient to maintain multicellularity in Dictyostelium. Science. 2011;334:1548–1551. doi: 10.1126/science.1213272. [DOI] [PubMed] [Google Scholar]
- 18.Strassmann JE, Zhu Y, Queller DC. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408:965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 19.Buttery NJ, Rozen DE, Wolf JB, Thompson CRL. Quantification of social behavior in D. discoideum reveals complex fixed and facultative strategies. Curr Biol. 2009;19:1373–1377. doi: 10.1016/j.cub.2009.06.058. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-García R, Tarnita CE. Lack of ecological and life history context can create the illusion of social interactions in Dictyostelium discoideum. PLoS Comput Biol. 2016;12:e1005246. doi: 10.1371/journal.pcbi.1005246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf JB, et al. Fitness trade-offs result in the illusion of social success. Curr Biol. 2015;25:1086–1090. doi: 10.1016/j.cub.2015.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atzmony D, Zahavi A, Nanjundiah V. Altruistic behaviour in Dictyostelium discoideum explained on the basis of individual selection. Curr Sci. 1997;72:142–145. [Google Scholar]
- 23.Zahavi A, Zahavi A. The Handicap Principle: A Missing Piece of Darwin’s Puzzle. Oxford Univ Press; Oxford: 1997. [Google Scholar]
- 24.Ostrowski EA, Katoh M, Shaulsky G, Queller DC, Strassmann JE. Kin discrimination increases with genetic distance in a social amoeba. PLoS Biol. 2008;6:e287. doi: 10.1371/journal.pbio.0060287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert OM, Strassmann JE, Queller DC. High relatedness in a social amoeba: The role of kin-discriminatory segregation. Proc Biol Sci. 2012;279:2619–2624. doi: 10.1098/rspb.2011.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ennis HL, Dao DN, Pukatzki SU, Kessin RH. Dictyostelium amoebae lacking an F-box protein form spores rather than stalk in chimeras with wild type. Proc Natl Acad Sci USA. 2000;97:3292–3297. doi: 10.1073/pnas.050005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santorelli LA, et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature. 2008;451:1107–1110. doi: 10.1038/nature06558. [DOI] [PubMed] [Google Scholar]
- 28.Dubravcic D, van Baalen M, Nizak C. An evolutionarily significant unicellular strategy in response to starvation in Dictyostelium social amoebae. F1000 Res. 2014;3:133. doi: 10.12688/f1000research.4218.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarnita CE, Washburne A, Martinez-Garcia R, Sgro AE, Levin SA. Fitness tradeoffs between spores and nonaggregating cells can explain the coexistence of diverse genotypes in cellular slime molds. Proc Natl Acad Sci USA. 2015;112:2776–2781. doi: 10.1073/pnas.1424242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khare A, et al. Cheater-resistance is not futile. Nature. 2009;461:980–982. doi: 10.1038/nature08472. [DOI] [PubMed] [Google Scholar]
- 31.Levin SR, Brock DA, Queller DC, Strassmann JE. Concurrent coevolution of intra-organismal cheaters and resisters. J Evol Biol. 2015;28:756–765. doi: 10.1111/jeb.12618. [DOI] [PubMed] [Google Scholar]
- 32.Hollis B. Rapid antagonistic coevolution between strains of the social amoeba Dictyostelium discoideum. Proc Biol Sci. 2012;279:3565–3571. doi: 10.1098/rspb.2012.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostrowski EA, et al. Genomic signatures of cooperation and conflict in the social amoeba. Curr Biol. 2015;25:1661–1665. doi: 10.1016/j.cub.2015.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahavi A, Harris KD, Nanjundiah V. An individual-level selection model for the apparent altruism exhibited by cellular slime moulds. J Biosci. 2018;43:49–58. [PubMed] [Google Scholar]
- 35.Thompson CRL, Kay RR. Cell-fate choice in Dictyostelium: Intrinsic biases modulate sensitivity to DIF signaling. Dev Biol. 2000;227:56–64. doi: 10.1006/dbio.2000.9877. [DOI] [PubMed] [Google Scholar]
- 36.Kuzdzal-Fick J, Foster K, Queller D, Strassmann J. Exploiting new terrain: An advantage to sociality in the slime mold Dictyostelium discoideum. Behav Ecol. 2007;18:433–437. [Google Scholar]
- 37.Van Dyken JD, Wade MJ. The genetic signature of conditional expression. Genetics. 2010;184:557–570. doi: 10.1534/genetics.109.110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purandare SR, Bickel RD, Jaquiery J, Rispe C, Brisson JA. Accelerated evolution of morph-biased genes in pea aphids. Mol Biol Evol. 2014;31:2073–2083. doi: 10.1093/molbev/msu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linksvayer TA, Wade MJ. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution. 2009;63:1685–1696. doi: 10.1111/j.1558-5646.2009.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall DW, Goodisman MA. The effects of kin selection on rates of molecular evolution in social insects. Evolution. 2012;66:2080–2093. doi: 10.1111/j.1558-5646.2012.01602.x. [DOI] [PubMed] [Google Scholar]
- 41.Warner MR, Mikheyev AS, Linksvayer TA. Genomic signature of kin selection in an ant with obligately sterile workers. Mol Biol Evol. 2017;34:1780–1787. doi: 10.1093/molbev/msx123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parikh A, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosengarten RD, et al. Leaps and lulls in the developmental transcriptome of Dictyostelium discoideum. BMC Genomics. 2015;16:294. doi: 10.1186/s12864-015-1491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirose S, Benabentos R, Ho H-I, Kuspa A, Shaulsky G. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science. 2011;333:467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 46.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 47.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douglas TE, Kronforst MR, Queller DC, Strassmann JE. Genetic diversity in the social amoeba Dictyostelium discoideum: Population differentiation and cryptic species. Mol Phylogenet Evol. 2011;60:455–462. doi: 10.1016/j.ympev.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Gossmann TI, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27:1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benabentos R, et al. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr Biol. 2009;19:567–572. doi: 10.1016/j.cub.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho H-I, Hirose S, Kuspa A, Shaulsky G. Kin recognition protects cooperators against cheaters. Curr Biol. 2013;23:1590–1595. doi: 10.1016/j.cub.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.smith j, Strassmann JE, Queller DC. Fine-scale spatial ecology drives kin selection relatedness among cooperating amoebae. Evolution. 2016;70:848–859. doi: 10.1111/evo.12895. [DOI] [PubMed] [Google Scholar]
- 53.Buttery NJ, et al. Structured growth and genetic drift raise relatedness in the social amoeba Dictyostelium discoideum. Biol Lett. 2012;8:794–797. doi: 10.1098/rsbl.2012.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall DW, Fox S, Kuzdzal-Fick JJ, Strassmann JE, Queller DC. The rate and effects of spontaneous mutation on fitness traits in the social amoeba, Dictyostelium discoideum. G3 (Bethesda) 2013;3:1115–1127. doi: 10.1534/g3.113.005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert OM. Histocompatibility as adaptive response to discriminatory within-organism conflict: A historical model. Am Nat. 2015;185:228–242. doi: 10.1086/679442. [DOI] [PubMed] [Google Scholar]
- 56.Foster KR, Fortunato A, Strassmann JE, Queller DC. The costs and benefits of being a chimera. Proc Biol Sci. 2002;269:2357–2362. doi: 10.1098/rspb.2002.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gruenheit N, et al. A polychromatic ‘greenbeard’ locus determines patterns of cooperation in a social amoeba. Nat Commun. 2017;8:14171. doi: 10.1038/ncomms14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaul M, Eichinger L. Analysis of gene expression using cDNA microarrays. In: Eichinger L, Rivero F, editors. Methods in Molecular Biology: Dictyostelium Discoideum Protocols. Humana Press; Totowa, NJ: 2006. pp. 75–93. [DOI] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu TD, Watanabe CK. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 61.Fey P, et al. dictyBase–A Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2014. [Google Scholar]
- 63.Morgan M, et al. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, Wang S, Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 65.Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chisholm RL, et al. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haddrill PR, Bachtrog D, Andolfatto P. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol. 2008;25:1825–1834. doi: 10.1093/molbev/msn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nei M. Molecular Evolutionary Genetics. Columbia Univ Press; New York: 1987. [Google Scholar]
- 73.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fay JC, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 76.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 78.Zhu LJ, et al. ChIPpeakAnno: A bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fay JC, Wyckoff GJ, Wu C-I. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gossmann TI, Keightley PD, Eyre-Walker A. The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol Evol. 2012;4:658–667. doi: 10.1093/gbe/evs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.