Abstract
Perturbations in activity and dosage of the UBE3A ubiquitin-ligase have been linked to Angelman Syndrome and autism spectrum disorders. UBE3A was initially identified as the cellular protein hijacked by the human papillomavirus E6 protein to mediate the ubiquitylation of p53, a function critical to the oncogenic potential of these viruses. Although a number of substrates have been identified, the normal cellular functions and pathways affected by UBE3A are largely unknown. Previously we showed that UBE3A associates with HERC2, NEURL4, and MAPK6/ERK3 in a high molecular weight complex of unknown function that we refer to as the HUN complex (HERC2, UBE3A, and NEURL4). In this study, the combination of two complementary proteomic approaches with a rigorous network analysis revealed cellular functions and pathways in which UBE3A and the HUN complex are involved. In addition to finding new UBE3A-associated proteins, such as MCM6, SUGT1, EIF3C, and ASPP2, network analysis revealed that UBE3A associated proteins are connected to several fundamental cellular processes including translation, DNA replication, intracellular trafficking, and centrosome regulation. Our analysis suggests that UBE3A could be involved in the control and/or integration of these cellular processes, in some cases as a component of the HUN complex, and also provides evidence for crosstalk between the HUN complex and CAMKII interaction networks. This study contributes to a deeper understanding of the cellular functions of UBE3A and its potential role in pathways that may be affected in Angelman Syndrome, UBE3A-associated autism spectrum disorders and HPV-associated cancers.
Keywords: Angelman syndrome, autism, human papillomavirus, cervical cancer, proteomics
Graphical Abstract
Introduction
The ubiquitin-ligase UBE3A, also known as E6AP (for E6-associated protein) was originally discovered as the cellular protein that is hijacked by the high risk human papillomavirus (hrHPVs) E6 proteins to target the ubiquitylation and proteolysis of the tumor suppressor p53 [1–4]. UBE3A is a 100-kDa protein encoded by a gene located on chromosome 15q11-q13. Three different isoforms of UBE3A generated by differential splicing differ in their amino-terminal sequences, although it is not yet known whether or how they might differ in function [5]. UBE3A is ubiquitously expressed from both the paternal and maternal alleles but as a consequence of imprinting, only the maternal UBE3A allele is expressed in certain areas of the brain [6, 7].
Defects in UBE3A expression and/or activity are associated with several pathologic disorders. The E6-induced degradation of p53 is a hallmark of hrHPV oncogenicity in HPV-associated cancers. In addition, UBE3A is linked to two different neurological disorders. Increased gene dosage of UBE3A is implicated in some forms of autism spectrum disorders (ASD) [8–10], and the loss of function of UBE3A in the central nervous system is the cause of Angelman syndrome (AS), a neurodevelopmental disorder characterized by severe mental retardation, ataxia, loss of speech, seizures, and other abnormalities [11–14]. Approximately 10% of patients that meet the clinical criteria for AS have no identified UBE3A molecular defect [15] suggesting that in addition to molecular mechanisms directly affecting UBE3A, perturbations in other genes that share a pathway or work through convergent pathways with UBE3A may also cause AS. Also, the pathways downstream of UBE3A affected in AS are still not well understood. For instance, a mouse model for AS showed decreased CAMKII activity due to increased phosphorylation of CAMKII [16]. Restoration of CAMKII activity corrected the behavioral deficits observed in this mouse model [17], yet it is uncertain whether UBE3A and CAMKII might relate to one another.
Despite the fact that UBE3A was discovered over 25 years ago [1], little is known about its biological functions or the cellular processes in which it is involved. Although efforts to identify substrates for its ubiquitin-ligase activity have provided a list of potential candidates, none have yielded much insight into the actual roles of UBE3A in the cell. Some of the reported UBE3A protein interactors and potential substrates include RAD23A [18], UBQLN1 and UBQLN2 [19, 20], the estrogen receptor [21], and PSMD4 [22, 23]. UBE3A also associates with the proteasome [20, 24–28] and with the giant ubiquitin-ligase HERC2 [28–30]. Our recent proteomic analysis of UBE3A binding proteins revealed that UBE3A binds to HERC2 in the context of a high molecular weight complex of unknown function [28] that we will refer to as the HUN (HERC2, UBE3A and NEURL4) complex, based on its major components. Another component, the kinase MAPK6/ERK3, is recruited to the HUN complex through its association with NEURL4 [28]. Impairment of the functions of UBE3A mediated through the HUN complex or defects in other components (e.g. HERC2) of this complex may contribute to the pathological traits observed in AS since a single homozygous mutation in HERC2 segregates in Amish families with a neurodevelopmental disorder with similarities to AS [31, 32] and loss of HERC2 function results in a severe neurodevelopmental phenotype [33].
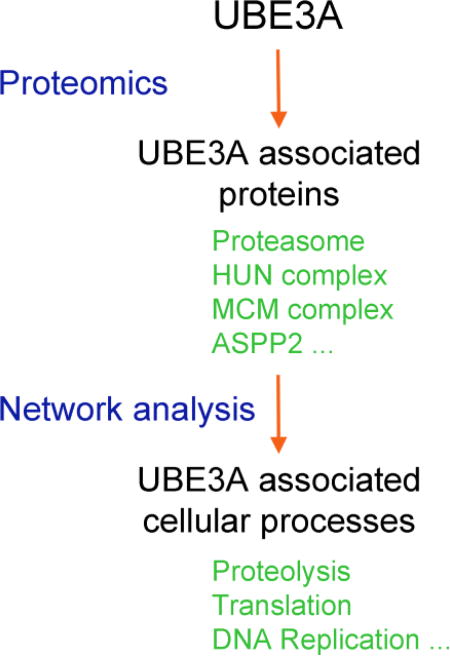
To gain a better understanding of the pathways and processes in which UBE3A and the HUN complex are involved, we employed two complementary proteomic approaches followed by extensive network analysis of the resulting protein interaction data. First, we performed affinity purifications of HA-tagged bait proteins followed by identification of their associated proteins by liquid chromatography-tandem mass spectrometry (AP-MS). For this we used two different cell systems: T-REx 293 cells extending our previous studies on UBE3A and HERC2 [28, 30] and SH-SY5Y human neuroblastoma cells, which have been successfully used in AS and ASD studies [34–36]. Second, AP-MS experiments were complemented with systematic yeast two hybrid (Y2H) [37] and yeast three hybrid [38] screens using UBE3A as bait to specifically identify likely direct interactors of UBE3A and the UBE3A-HPV16 E6 complex respectively.
In this study we identified several new UBE3A-associated or direct interacting proteins, including the minichromosome Maintenance Complex Component 6 (MCM6), Apoptosis-Stimulating Of P53 Protein 2 / Tumor Protein P53 Binding Protein 2 (ASPP2/TP53BP2/P53BP2), the SGT1 Homolog, the MIS12 Kinetochore Complex Assembly Co-chaperone (SUGT1) and the Eukaryotic Translation Initiation Factor 3 Subunit C (EIF3C), as well as two new components of the HUN complex, the Enoyl-CoA Hydratase 1 (ECH1) and the Enoyl-CoA Delta Isomerase 2 (ECI2/PECI). The interactors of UBE3A in SH-SY5Y cells are similar to those previously observed in T-REx 293 cells [28] indicating that these protein associations are conserved across different cell types. Network analysis of the interaction data suggests that UBE3A and its interaction partners might be involved in the control and integration of several fundamental cellular processes, including translation, intracellular trafficking, and cytoskeleton regulation among others. Importantly the interaction data reveals a possible crosstalk between the HUN and CAMKII protein complexes, providing a potential explanation for the mouse phenotypes mentioned above. Our work should contribute to a better understanding of UBE3A and the pathways and cellular processes perturbed in the diseases and conditions with which it has been associated.
Results
Identifying UBE3A interacting proteins in SH-SY5Y cells
Since UBE3A is linked to two different neurological developmental disorders, we identified UBE3A interacting proteins in the human neuroblastoma SH-SY5Y cell line. To avoid toxicity due to the stable expression of a transgene, we generated a SH-SY5Y single cell clone expressing the Tet repressor that can be tightly regulated by doxycycline. We performed coimmunoprecipitations with 64 HA-tagged proteins as baits (Table S2) in SH-SY5Y to build a database for the AP-MS analysis software CompPASS (Comparative Proteomic Analysis Software Suite) to discriminate nonspecific background binding from high-confidence candidate interacting proteins (HCIPs) for each bait [39].
Using the SH-SY5Y database we next employed AP-MS and CompPASS to determine a set of UBE3A HCIPs in SH-SY5Y cells (Table 1). Similar to our previous strategy with the T-REx 293 cell line [28], we used each of the three known isoforms of UBE3A in both the catalytically active and inactive forms as baits, the latest to increase the likelihood of identifying ubiquitin-ligase substrates of UBE3A. As with the T-REx 293 cells [28], there were no major differences among the observed HCIPs for the three UBE3A isoforms (Table 1). In total, 117 HCIPs (including 34 proteasome subunits) were identified for UBE3A combining the HCIPs from both SH-SY5Y AND T-REx 293 cell lines (see Material and Methods) of which 45% were observed in both. Only 10% of the HCIPs were observed exclusively in the SH-SY5Y cell line, from which fewer HCIPs were observed than in the T-REx 293 cell line, probably due to lower protein expression levels. Overall, the set of UBE3A-associated proteins was very similar between the two cell types.
Table 1.
UBE3A interactors in SH-SY5Y cells that were detected as HCIP by CompPASS in more than one immunoprecipitation using UBE3A as bait.
| Symbol | Description | 90% cutoff | 95% cutoff | ||
|---|---|---|---|---|---|
| PDs a | (%)b | PDs a | (%)b | ||
| HERC2* | E3 ubiquitin-protein ligase HERC2 | 6 | 100 | 6 | 100 |
| NEURL4* | Isoform 2 of Neuralized-like protein 4 | 5 | 83 | 4 | 67 |
| ACTBL2 | Beta-actin-like protein 2 | 3 | 50 | ||
| EIF3C* | Eukaryotic translation initiation factor 3 subunit C | 3 | 50 | ||
| HIF1AN* | Hypoxia-inducible factor 1-alpha inhibitor | 3 | 50 | 3 | 50 |
| UCHL5* | Ubiquitin carboxyl-terminal hydrolase isozyme L5 | 3 | 50 | 2 | 33 |
| USP14* | Ubiquitin carboxyl-terminal hydrolase 14 | 3 | 50 | 3 | 50 |
| CEP170* | Isoform 2 of Centrosomal protein of 170 kDa | 2 | 33 | ||
| ECH1* | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | 2 | 33 | 2 | 33 |
| ECI2* | Isoform 2 of Enoyl-CoA delta isomerase 2, mitochondrial | 2 | 33 | ||
| IGLC7 | Ig lambda-7 chain C region | 2 | 33 | ||
| KRT6B | Keratin, type II cytoskeletal 6B | 2 | 33 | ||
| LCN1 | Lipocalin-1 | 2 | 33 | 2 | 33 |
| MTAP* | S-methyl-5'-thioadenosine phosphorylase | 2 | 33 | ||
| PIP | Prolactin-inducible protein | 2 | 33 | ||
| SUGT1* | Isoform 2 of Suppressor of G2 allele of SKP1 homolog | 2 | 33 | ||
| TMCO7* | Transmembrane and coiled-coil domain-containing protein 7 | 2 | 33 | 2 | 33 |
| TUBB2B | Tubulin beta-2B chain | 2 | 33 | ||
| VASP | Vasodilator-stimulated phosphoprotein | 2 | 33 | 2 | 33 |
| Proteasome subunits | |||||
| PSMA2* | Proteasome subunit alpha type-2 | 6 | 100 | ||
| PSMB5* | Proteasome subunit beta type-5 | 6 | 100 | ||
| PSMC4* | 26S protease regulatory subunit 6B | 6 | 100 | 2 | 33 |
| PSMC5* | 26S protease regulatory subunit 8 | 6 | 100 | 2 | 33 |
| PSMD1* | 26S proteasome non-ATPase regulatory subunit 1 | 6 | 100 | 3 | 50 |
| PSMD12* | 26S proteasome non-ATPase regulatory subunit 12 | 6 | 100 | 2 | 33 |
| PSMD13* | 26S proteasome non-ATPase regulatory subunit 13 | 6 | 100 | 2 | 33 |
| PSMD2* | 26S proteasome non-ATPase regulatory subunit 2 | 6 | 100 | 2 | 33 |
| PSMD4* | 26S proteasome non-ATPase regulatory subunit 4 | 6 | 100 | 4 | 67 |
| PSMD7* | 26S proteasome non-ATPase regulatory subunit 7 | 6 | 100 | ||
| PSMA1* | Isoform Long of Proteasome subunit alpha type-1 | 5 | 83 | ||
| PSMC2* | 26S protease regulatory subunit 7 | 5 | 83 | 2 | 33 |
| PSMC3* | 26S protease regulatory subunit 6A | 5 | 83 | 2 | 33 |
| PSMD11* | 26S proteasome non-ATPase regulatory subunit 11 | 5 | 83 | 2 | 33 |
| PSMD3* | 26S proteasome non-ATPase regulatory subunit 3 | 5 | 83 | 2 | 33 |
| PSMD6* | 26S proteasome non-ATPase regulatory subunit 6 | 5 | 83 | 2 | 33 |
| PSMB2* | Proteasome subunit beta type-2 | 4 | 67 | ||
| PSMB4* | Proteasome subunit beta type-4 | 4 | 67 | ||
| PSMB6* | Proteasome subunit beta type-6 | 4 | 67 | ||
| PSMB7* | Proteasome subunit beta type-7 | 4 | 67 | 3 | 50 |
| PSMC1* | 26S protease regulatory subunit 4 | 4 | 67 | 2 | 33 |
| PSMC6* | 26S protease regulatory subunit 10B | 4 | 67 | ||
| PSMB1* | Proteasome subunit beta type-1 | 3 | 50 | ||
| PSMB3* | Proteasome subunit beta type-3 | 3 | 50 | ||
| PSMD8* | 26S proteasome non-ATPase regulatory subunit 8 | 3 | 50 | ||
| PSMA3* | Isoform 2 of Proteasome subunit alpha type-3 | 2 | 33 | ||
| PSMA4* | Proteasome subunit alpha type-4 | 2 | 33 | ||
| PSMA5* | Proteasome subunit alpha type-5 | 2 | 33 | ||
| PSMA6* | Proteasome subunit alpha type-6 | 2 | 33 | ||
| PSMA7* | Proteasome subunit alpha type-7 | 2 | 33 | ||
| PSMB8 | Isoform 2 of Proteasome subunit beta type-8 | 2 | 33 | 2 | 33 |
| PSMD10* | 26S proteasome non-ATPase regulatory subunit 10 | 2 | 33 | 2 | 33 |
| PSMD14* | 26S proteasome non-ATPase regulatory subunit 14 | 2 | 33 | ||
| PSMD5 | 26S proteasome non-ATPase regulatory subunit 5 | 2 | 33 | ||
Number and
percentage of immunoprecipitations in which a UBE3A interactor was detected as HCIP.
Also found as UBE3A interactor in immunoprecipitations from T-Rex 293 cells [28].
Binary interaction screens
Whereas AP-MS identifies proteins that are components of stable protein complexes, it does not differentiate between direct and indirect interactors. The Y2H system represents a complementary approach to interactome mapping in which mostly direct protein interactions are scored [40]. The combination of these two proteomic approaches thus provides a more comprehensive understanding of UBE3A interacting proteins. We performed a Y2H screen using the three isoforms of UBE3A (both active and inactive forms) as baits to interrogate a library of human open reading frames (ORFs) for proteins that can directly interact with UBE3A. Similar to the results of the AP-MS experiments, the Y2H screen did not reveal major differences among the UBE3A isoforms (Table 2). Included in the 19 proteins identified by the Y2H system were the known UBE3A interactors RAD23A, RAD23B, PSMD4, UBQLN1 and UBQLN2 [18–20, 22, 23]. In addition, we performed a Y3H screen to test how the presence of HPV16 E6, could alter the set of proteins that interact with UBE3A in the Y2H screen. As shown in Table 2, HPV16 E6 does not interfere with the binding of the UBE3A interactors found in the Y2H screen but enables UBE3A to associate with a few additional proteins. These additional proteins include TP53, providing a validation of this approach. There are less proteins that interact with the active UBE3A isoforms in presence of HPV16 E6, but these differences disappeared when we used the inactive isoforms of UBE3A. This is consistent with the fact that E6 works as an allosteric activator of UBE3A [41], therefore E6 may enhance the ubiquitin-ligase activity of UBE3A toward its associated proteins resulting in a more efficient degradation of these proteins by the ubiquitin-proteasome system, making their detection more difficult. This does not happen with the catalytically inactive forms of UBE3A. Additional UBE3A associated proteins in presence of HPV16 E6 were only detectedin the presence of inactive UBE3A isoforms since HPV16 E6 promotes the degradation of these proteins via ubiquitylation by UBE3A. HERC2, an established interactor of UBE3A, was not identified among the UBE3A preys because due to its high molecular weight it was not included in the library used in these experiments. Of note, PSMD4, RAD23B, and MCM6 were identified as UBE3A interactors by both AP-MS, Y2H, and Y3H approaches.
Table 2.
UBE3A interactors detected in the Y2H AND Y3H.
| Bait | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prey | UBE3A isof. I |
UBE3A isof. II |
UBE3A isof. III |
UBE3A isof. I inactive |
UBE3A isof. II inactive |
UBE3A isof. III inactive |
||||||
| − E6 | + E6 | − E6 | + E6 | − E6 | + E6 | − E6 | + E6 | − E6 | + E6 | − E6 | + E6 | |
| CGGBP1 | + | + | + | + | + | + | ||||||
| DAZAP2 | + | + | + | + | + | + | ||||||
| MAGEA12 | + | + | + | + | + | + | + | + | ||||
| MCM6 | + | + | + | + | + | |||||||
| MEOX1 | + | + | + | + | + | + | + | + | ||||
| MEOX2 | + | + | + | + | + | + | + | + | ||||
| MID2 | + | |||||||||||
| PAX5 | + | + | + | + | ||||||||
| PAX6 | + | + | + | + | + | + | ||||||
| PROP1 | + | + | + | + | + | + | + | + | + | |||
| PSMD4 | + | + | + | + | + | + | + | + | + | + | + | + |
| PTCD3 | + | + | + | + | + | + | + | + | + | + | + | + |
| RAD23A | + | + | + | + | + | + | + | + | + | |||
| RAD23B | + | + | + | + | + | + | + | + | + | + | ||
| RPH3A | + | + | + | |||||||||
| TP53 | + | + | + | |||||||||
| TP53BP2 | + | + | + | + | + | + | + | + | + | |||
| TTC23L | + | + | + | + | + | + | ||||||
| UBAC2 | + | |||||||||||
| UBQLN1 | + | + | + | + | + | + | + | + | + | + | ||
| UBQLN2 | + | + | + | + | + | + | + | + | + | + | + | |
| ZNF250 | + | + | + | + | + | + | ||||||
| ZNF446 | + | + | + | + | + | |||||||
+ Indicates that a protein was found as prey using the corresponding bait. + E6 and − E6 signal the presence or absence of the HPV16 E6 protein in the system.
UBE3A Network Analysis
In total, we identified 96 proteins that either directly or indirectly interact with UBE3A in addition to the proteasome. We integrated these data with existing human protein interactome data to gain additional insight into the UBE3A interactors and their potential functional significance. As sources of available human protein interactome data we used four recently published and released proteome-scale human protein interaction datasets that have been generated in systematic screens of the human protein interaction space. BioPlex [42] and QUBIC [43] are two datasets generated from AP-MS screens, CoFrac [44] contains protein complex data obtained from co-fractionation of protein extracts from various cell lines, and HI-union is the total of protein interactions identified at the Center for Cancer Systems Biology (CCSB) using a binary interaction mapping platform that couples Y2H screens with validation in orthogonal mammalian protein interaction assays [45–50]. These four systematically generated datasets cover the human protein interaction space more homogeneously as compared to literature-curated interaction datasets that display strong study biases [45]. Given the low overlaps between the systematic and literature-curated interactome datasets and to add more direct protein-protein interaction (PPI) data of similar high quality as the systematic datasets, we also included a subset of the curated literature interactome, Lit-BM-13, which contains all PPIs from seven public protein interaction databases, that have multiple evidences (publications or methods) of which at least one identifies likely direct PPIs [45]. The union of these five interactome datasets, hereafter referred to as QBCHL, comprised 156,138 PPIs and protein associations involving 15,189 distinct proteins (genes) excluding all homodimeric protein pairs.
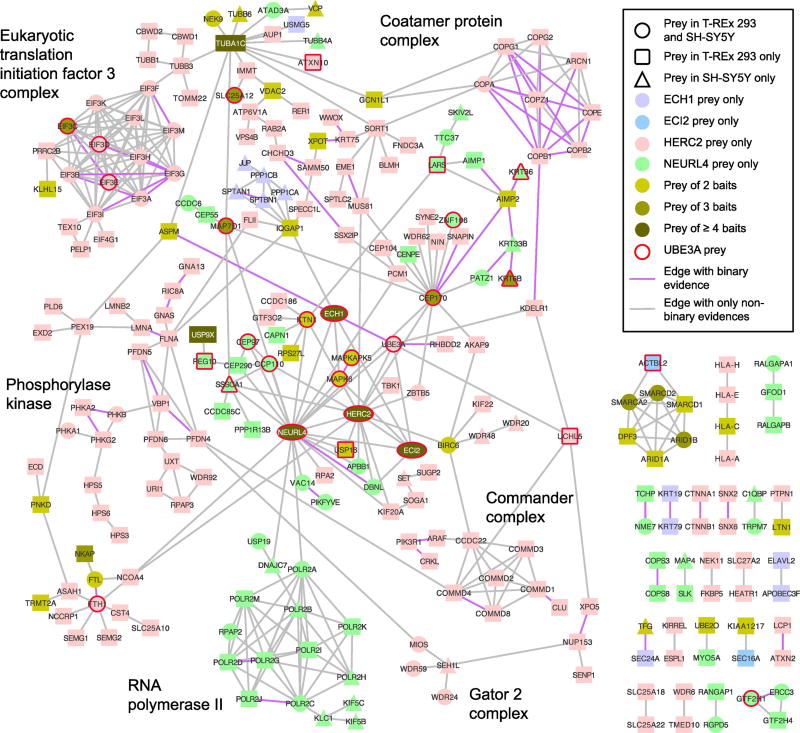
94 of the 96 UBE3A interactors have at least one protein interaction or association in QBCHL. The UBE3A interactors identified from AP-MS are significantly close to each other in the QBCHL interactome (p-value = 0.004, see Material and Methods). However, this signal is lost when combining the AP-MS and Y2H interactors (Table S1 and Figure S1) suggesting that even though both sets of interactors overlap, they seem to be less connected with each other in the QBCHL protein interactome. Starting from the set of 94 UBE3A interactors that had at least one interaction in QBCHL, we next built a network that includes proteins that associate with the UBE3A interactors, if these neighbors were seen to interact with at least two UBE3A interactors. This network was further filtered to retain interactions and the corresponding proteins in the network only if the interactions were supported by at least two independent sources (Figure S2). The network is significantly larger (p-value = 0.011) and contains a significantly higher fraction of UBE3A interactors (p-value = 0.01) than networks built in identical ways but starting from degree-controlled random sampling of proteins from the QBCHL network (Table S1 and Figure S1). We sought to further complete this UBE3A network by including protein interaction and association data around UBE3A from this and other more focused studies [30, 51] (see Methods) if there were at least two lines of evidence supporting the data. To our knowledge, the resulting network (Figure 1) represents the most complete view of the UBE3A interactome module to date confirming previously known interactions and revealing potential new cellular functions of UBE3A.
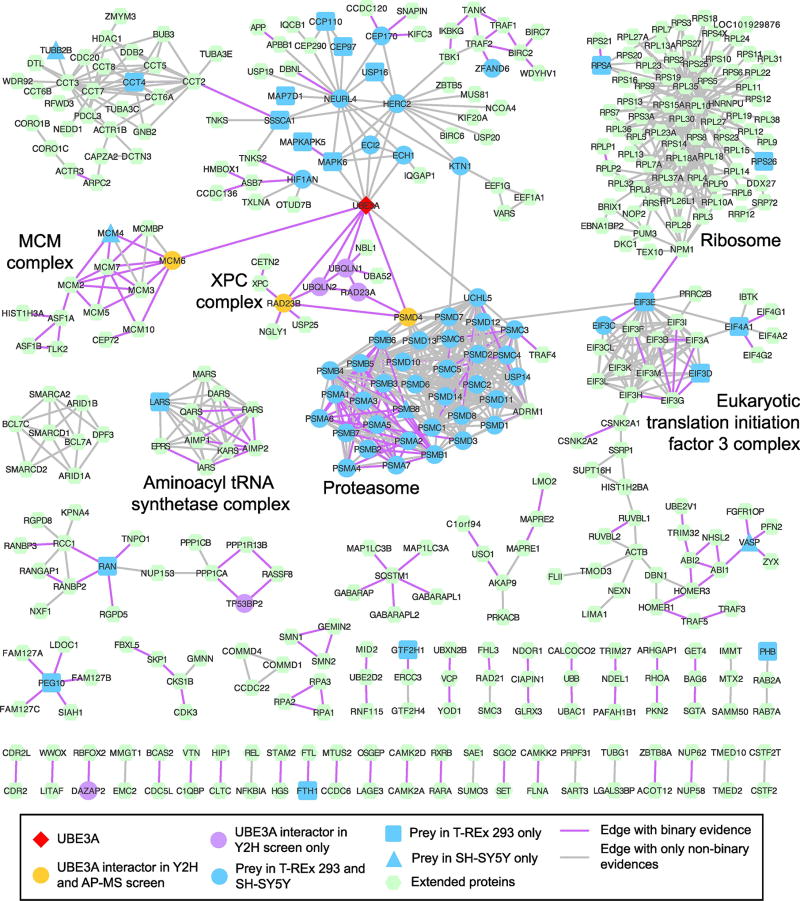
Figure 1. A UBE3A-centered protein interaction network.
This network displays interactors of UBE3A found by AP-MS or Y2H as well as interactors of its associated proteins. All links shown in the network are supported by at least two different sources (see Methods). Highlighted are a few key protein complexes of which all apart from the ribosome are significantly enriched in the systematically built network of UBE3A using only QBCHL as source of interactions (see Figure S2).
The network highlights the strong association of UBE3A with HIF1AN and with the members of the HUN complex, HERC2, NEURL4, MAPK6, ECI2 and ECH1 (see below), of which all but MAPK6 were identified as HCIPs in both T-REx 293 and SH-SY5Y cells. This network also showed a strong and significant enrichment (see Methods) for the Translation Initiation Factor 3 (EIF3) complex and related functions, the Aminoacyl tRNA Synthetase complex, the Nucleotide-Excision Repair complex, Minichromosome Maintenance (MCM) complex and its function in DNA replication, and the Proteasome (Table S2). In addition, there are numerous UBE3A HCIPs that are implicated in centrosome, cytoskeleton or transport-related functions, of which CCP110, CEP97, CEP170, KTN1, and RAN are included in the UBE3A network, however most of them are more closely connected to the other members of the HUN complex, especially NEURL4, which has a demonstrated role in the centrosome [51, 52].
Confirmation of new UBE3A-associated proteins
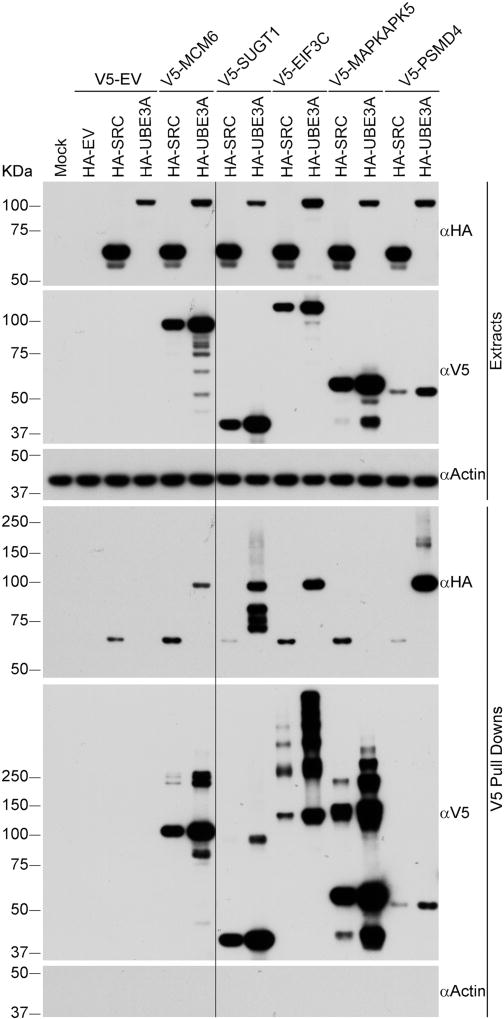
We next confirmed the association of UBE3A with several of the HCIPs identified in our proteomic studies. SUGT1 and EIF3C, that had been previously identified as HERC2 associated proteins [30] were detected as UBE3A HCIPs in several coimmunoprecipitations from both T-REx 293 and SH-SY5Y cell lines. These in addition to MCM6, a component of the mini-chromosome maintenance (MCM) complex, were tested in a reciprocal immunoprecipitation experiment for their ability to bind UBE3A (Figure 2). We observed that V5–tagged EIF3C, SUGT1, and MCM6 coimmunoprecipitated UBE3A in T-REx 293 cells confirming that these proteins associate with UBE3A. In this experiment we also examined whether MAPKAPK5/MK5/PRAK, a known interactor of MAPK6 [53, 54] also coimmunoprecipitates with UBE3A. We had previously observed MAPKAPK5 as a HCIP of UBE3A in T-REx 293 cells [28], however, V5-tagged MAPKAPK5 did not coimmunoprecipitate HA-tagged UBE3A in this experiment. The discrepancy could be due to differences in sensitivity of the assays or could be due to the presence of the V5 tag on MAPKAPK5. Therefore the association of MAPKAPK5 in a complex with UBE3A remains to be determined.
Figure 2. Confirmation of interaction of UBE3A with several HCIPs.
V5-tagged HCIPs were immunoprecipitated from T-REx 293 cells using anti-V5 magnetic beads. Protein extracts and immunoprecipitates were analyzed by SDS-PAGE and Western blot using antibodies against HA-tag, V5-tag and Actin. HA-SRC was used as negative control and V5-PSMD4 as positive control. HA-SRC signal was present in each pulldown, including that done with the empty V5-tag vector and therefore is considered as background for this experiment. The long black line indicates the place where part of the blots was removed because it was irrelevant for the figure. HA-UBE3A: UBE3A isoform 1 C820A, catalytically inactive. EV: empty vector.
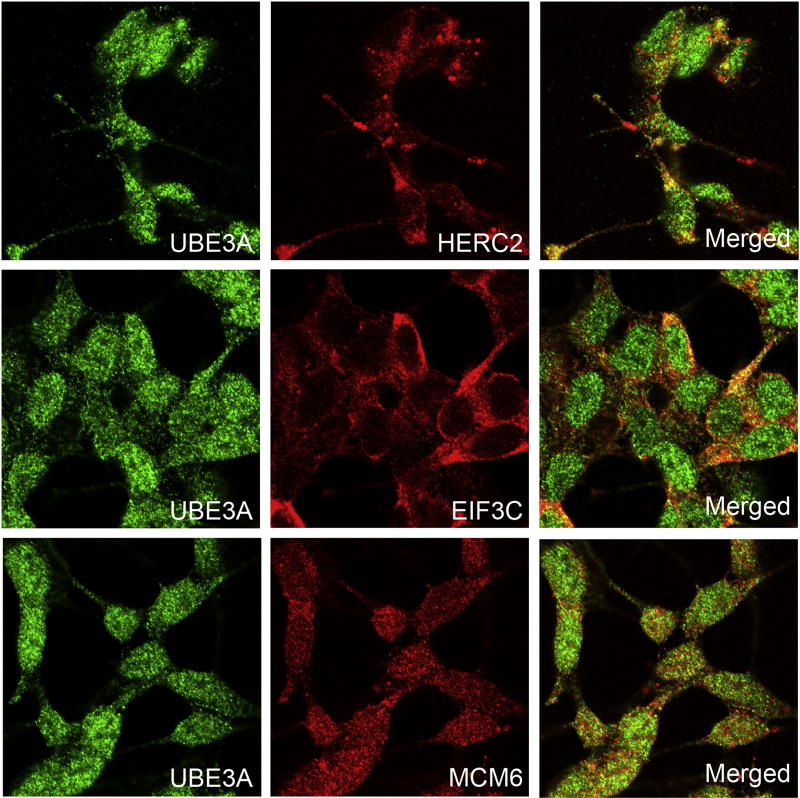
We also performed colocalization experiments in SH-SY5Y cells between UBE3A and HERC2, EIF3C, and MCM6 (Figure 3). As previously reported [55, 56], UBE3A is distributed throughout the cell, with higher levels in the nucleus. UBE3A partially colocalized with HERC2, EIF3C and MCM6 supporting the association of UBE3A with these proteins. However, UBE3A colocalized with HERC2 and EIF3C in the cytoplasm whereas the colocalization with MCM6 was observed predominantly in the nuclei. This pattern was also observed in HaCaT, HeLa, and U2OS cells (Figures S7, S8, and S9) suggesting that it is conserved among different cell types. Also, further support for the association of UBE3A with MCM6 and EIF3C comes from a recent report that identified MCM6 and several components of the EIF3 complex as potential substrates of the ubiquitin-ligase activity of UBE3A [57].
Figure 3. UBE3A partially colocalizes with HERC2, EIF3C, and MCM6 in SH-SY5Y cells.
UBE3A colocalizes with HERC2 and EIF3C in the cytosol and with MCM6 predominantly in the nuclei.
Taken together, our data identified multiple connections of UBE3A to a variety of cellular functions, although it remains to be determined which specific rolesUBE3A may play in these cellular processes.
The HUN complex
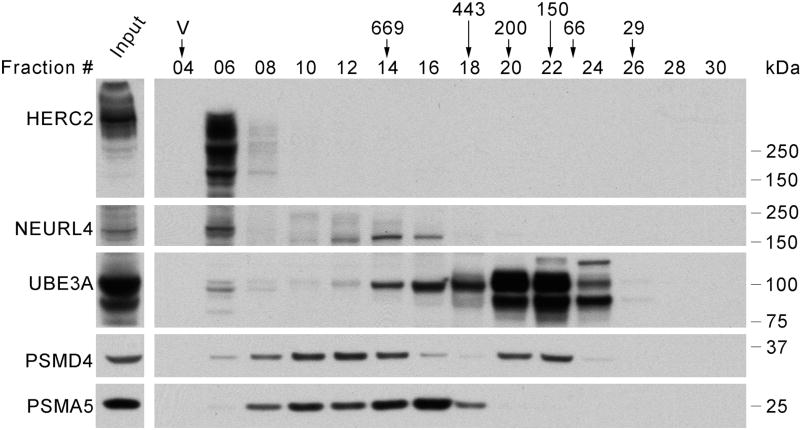
From the accumulated UBE3A interaction data it remains unclear which UBE3A functions might be performed as a component of the HUN complex. We validated that UBE3A interacts with HERC2 and NEURL4 in SH-SY5Y as part of a high molecular weight complex by gel filtration (Figure 4) similarly to what was observed in T-REx 293, C33A, and SiHa cells [28] indicating that the HUN complex is present in SH-SY5Y cells.
Figure 4. UBE3A co-fractionates with HERC2 and NEURL4 in high molecular weight fractions in a size-exclusion experiment with SH-SY5Y cells protein extract.
8 mgs. of protein extract from SH-SY5Y cells were loaded on a Superose 6 column for size-exclusion chromatography. After fractionation, 20 µl of the collected fractions (500 µl) were analyzed by SDS-PAGE and Western blot using antibodies against the UBE3A, HERC2, NEURL4, PSMD4 and PSMA5. 20 µg of protein extract were loaded as input. The position where the peaks of protein standards eluted (669, 443, 200, 150, 66 and 29 kDa) and the void volume (V) are indicated over the fractions number.
Then, to gain further insight into the composition of the HUN complex and other proteins associated to UBE3A, we performed AP-MS experiments using several HA-tagged UBE3A interactors as baits. We immunoprecipitated HA-tagged NEURL4, MAPK6, ECH1, ECI2, and HIF1AN from both SH-SY5Y and T-REx 293 cells, and HERC2 only from SH-SY5Y cells since coimmunoprecipitations from T-REx 293 cells have already been published [30]. Due to its high molecular weight (527 kDa), HERC2 was expressed as six overlapping fragments comprising the complete HERC2 ORF as baits in coimmunoprecipitation experiments to identify HCIPs for this protein, as we previously did for T-REx 293 cells [30]. As shown in Table 3, there was extensive cross association between UBE3A, HERC2, NEURL4, MAPK6, ECH1 and ECI2, suggesting that they are all components of the HUN complex across cell types. Supporting this notion UBE3A colocalizes with HERC2 in HaCaT, HeLa, and U2OS cells (Figures S7, S8, and S9) and their interaction has also been shown in H1299 cells [29]. In contrast, HIF1AN, which associates with UBE3A independently of the HUN complex [28], did not associate with any protein other than UBE3A.
Table 3.
Reciprocal immunoprecipitations of components of the high molecular weight complex as detected by mass spectrometry and CompPASS analysis.
| Prey | ||||||||
|---|---|---|---|---|---|---|---|---|
| UBE3A | HERC2 | NEURL4 | MAPK6 | ECH1 | ECI2 | HIF1AN | ||
| Bait | UBE3A | ++ | ++ | + | ++ | ++ | ++ | |
| HERC2 | ++a | ++b | − | − | − | − | ||
| NEURL4 | − | ++ | ++ | ++ | ++ | − | ||
| MAPK6 | − | ++ | ++ | + | + | − | ||
| ECH1 | − | − | − | − | − | − | ||
| ECI2 | − | + | + | − | ++ | − | ||
| HIF1AN | + | − | − | − | − | − | ||
Detected only in immunoprecipitations from T-Rex 293 cells.
Detected in immunoprecipitations from both T-Rex 293 and SH-SY5Y cells.
Using the fragment 4 of the HERC2 protein.
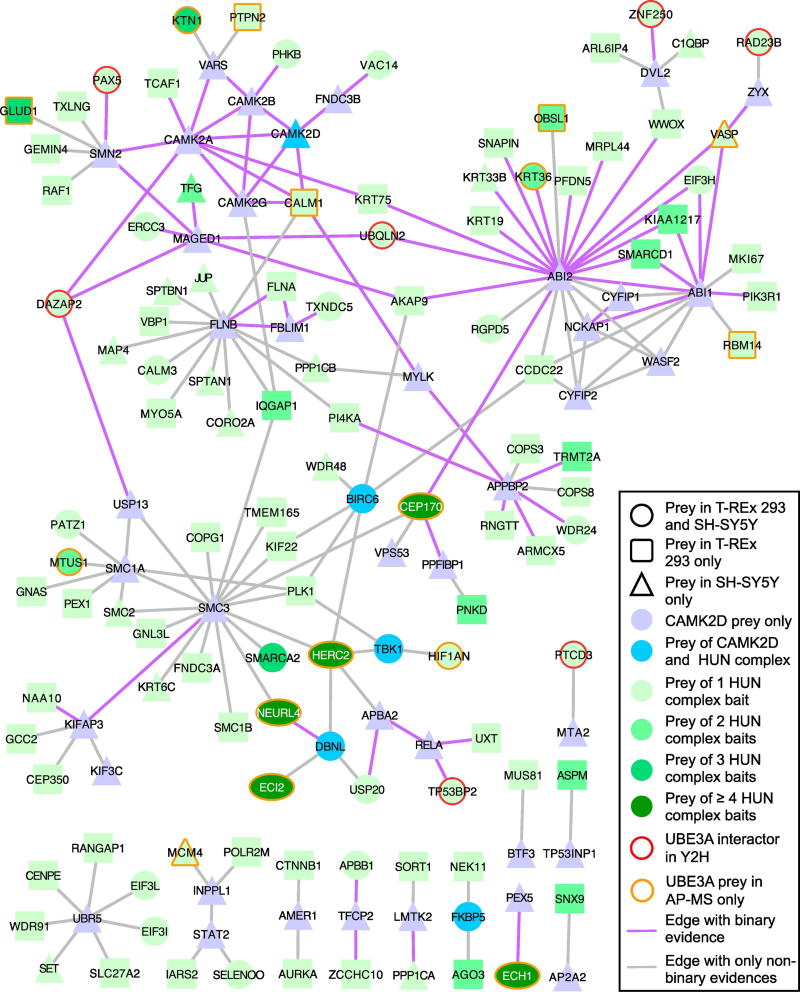
In general, there was good correspondence between the HCIPs detected for HUN component proteins from both T-REx 293 cells and SH-SY5Y cells. Five members of the HUN complex, NEURL4, HERC2, MAPK6, ECH1 and ECI2, coimmunoprecipitated 138, 283, 29, 67, and 34 HCIPs, respectively, comprising 464 distinct HCIPs in total between both cell lines. Out of these 464 HCIPs, 15% appeared as HCIPs in both cell lines while 13% were coimmunoprecipitated by at least two different baits (not counting the baits themselves). Using QBCHL as a source of external interaction data we found that each individual set of HCIPs from HERC2, NEURL4, MAPK6, and ECI2 (p-values < 0.001), apart from ECH1 (p-value=0.173), as well as the union of all HCIPs (p-values < 0.001) was significantly connected within each other in the human protein interactome (Table S1 and Figure S3). To obtain a clearer view of the cellular functions played by the components of the HUN complex and their relationship to UBE3A, we built a network starting from the HCIPs of HERC2, NEURL4, MAPK6, ECI2, and ECH1. We connected any two HCIPs with each other if they shared at least one bait and there was evidence for interaction or association between them from QBCHL, reciprocal IP data from this study, or from a published list of proteins that associate with NEURL4 [51] (Figure 5). This HUN network is dominated by HERC2 HCIPs and to a lesser extent by HCIPs identified from NEURL4 coimmunoprecipitations. The network makes apparent a variety of known protein complexes including the RNA polymerase II complex, EIF3 complex, the Coatomer protein complex, the Commander complex, the Phosphorylase kinase complex and the GATOR 2 complex. Consistent with the presence of these protein complexes the HUN network (for control built using only QBCHL, Figure S4) shows clear enrichment for transcription, translation, vesicle formation and transport as well as metabolism-related functions (Table S3).
Figure 5. The HUN complex interactome.
Preys of HERC2, NEURL4, MAPK6, ECI2, and ECH1 were connected if there was evidence for association or interaction from QBCHL, reciprocal IP from this study or [Al-Hakim et al] and if any pair of preys had at least one bait in common. Key protein complexes are highlighted. Oval shapes are equivalent to circles, they were used with the sole purpose of facilitating the reading of the gene symbols in the corresponding nodes.
HERC2 coimmunoprecipitated about 14 members of the EIF3 complex compared to four members coimmunoprecipitated by UBE3A and one by NEURL4 suggesting that the association of the HUN complex with the EIF3 complex most likely occurs through HERC2. Also, the fact that that HERC2 and EIF3C show a similar colocalization pattern with UBE3A (Figure 3) is in good agreement with the idea of UBE3A associating with the EIF3 complex through HERC2. EIF3C, the member of the EIF3 complex that was coimmunoprecipitated by HERC2, NEURL4 and UBE3A may represent the direct contact point for this association. The EIF3 complex has been shown to serve as a scaffold to connect the kinase mTORC1 with its targets S6K1 and 4E-BP1 [58]. Interestingly, HERC2 coimmunoprecipitated four (MIOS, WDR59, WDR24, SEH1L) of the five known members of the GATOR2 complex, a positive regulator of mTORC1 [59]. Furthermore, NEURL4 co-precipitated the kinase PIKFYVE and its activator VAC14 from both cell lines. PIKFYVE has been shown to be necessary for mTORC1 activation in 3T3-L1 adipocytes [60]. Taken together, our data strongly suggests a link between the HUN complex and mTORC1 signaling.
In addition, the link observed in the UBE3A network between UBE3A/the HUN complex and centrosomal and cytoskeleton-related functions becomes even more apparent in the HUN network, which highlights the association of NEURL4 with the centrosomal proteins CEP97, CCP110, CEP290, CEP170 and the pericentriolar protein PCM1. Furthermore, MAPK6, known for its regulatory role in actin cytoskeleton remodeling and cell migration [61–64], showed a strong functional enrichment for spindle and centriole-related GO terms due to the presence of the kinases AURKA and AURKB as well as the proteins ASPM, CEP170, and MAP7D1 amongst its HCIPs. ASPM, a centrosomal protein previously identified as interactor of UBE3A [65] was also coimmunoprecipitated by HERC2 [30] (Figure 5). Our data clearly reinforces the association of UBE3A with centrosomal functions through its participation in the HUN complex.
The protein interaction networks of the HUN complex and CAMKII are linked
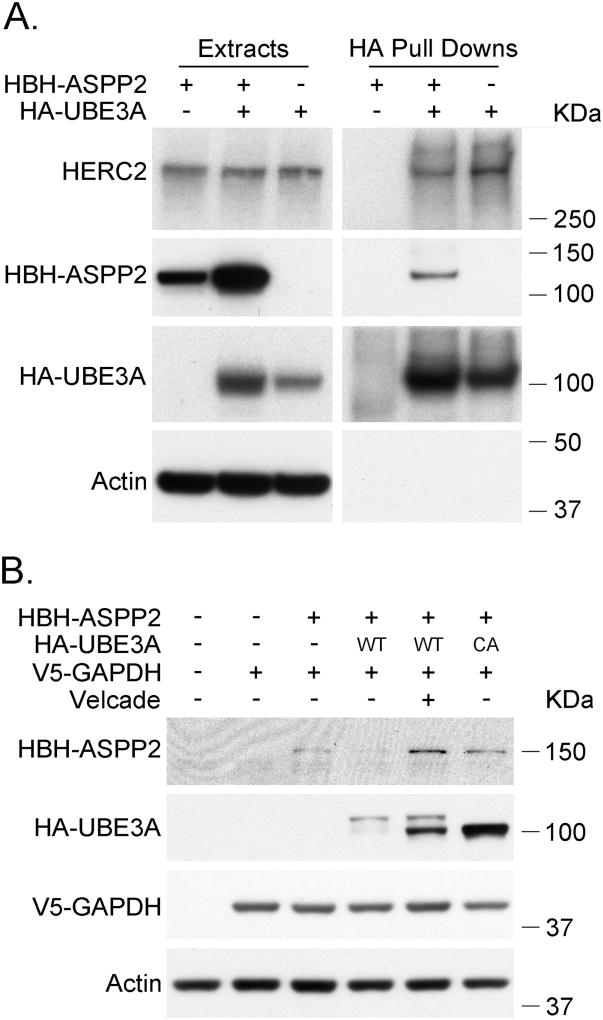
A mouse model for AS with decreased CAMKII activity also has lower protein phosphatase PP1 and PP2A activities, two phosphatases for which CAMKII is a substrate [16]. In this model, it was hypothesized that increased levels of ASPP2, an inhibitor of PP1, could be involved in regulating CAMKII [16]. Interestingly, ASPP2 was identified as a likely direct interactor of UBE3A in our Y2H screen (Figure 2, Table 2). We confirmed the interaction by co-immunoprecipitation (Figure 6A) and examined whether UBE3A affects ASPP2 protein levels in a cotransfection experiment (Figure 6B). We found that coexpression of UBE3A decreased the protein levels of ASPP2. In addition, coexpression of a catalytically inactive UBE3A or proteasomal inhibition increased ASPP2 protein levels. This suggests that ASPP2 could be a target for the ubiquitin-ligase activity of UBE3A that targets ASPP2 for proteasomal degradation, thus providing a potential link between UBE3A and CAMKII.
Figure 6. UBE3A promotes the degradation of ASPP2 by the proteasome.
A.UBE3A coimmunoprecipitates ASPP2. HEK 293T cells were transfected with the indicated vectors. 48 hours after transfection the HA-tagged proteins were immunoprecipitated with anti-HA agarose beads. Protein extracts and immunoprecipitates were analyzed by SDS-PAGE and Western blot using antibodies against HA-tag and Actin. HBH-ASPP2 carrying a biotinylation signal in the HBH tag was detected using streptavidin-HRP. HA-UBE3A: UBE3A isoform 1 C820A, catalytically inactive. B. Coexpression of UBE3A reduces ASPP2 protein levels. HEK 293T cells were transfected with the corresponding vectors. 48 hours post transfection the cells were harvested and the protein extracts were analyzed by SDS-PAGE and Western blot. Proteins were detected using anti HA, V5 and actin antibodies, and streptavidin HRP. Of note, UBE3A runs as a double band while its catalytically inactive form runs as a single band. - indicates that the cells were transfected with the corresponding empty vector. WT: wild type, CA: catalytically inactive form of UBE3A.
We next questioned whether there were functional links between UBE3A and CAMKII that might be dependent on the HUN complex. AP-MS in SH-SY5Y cells using CAMK2D, a subunit of the CAMKII complex, as bait resulted in 61 identified HCIPs. The CAMK2D associated proteins are enriched for members of the cohesin complex, Rac, Ras and neurotrophin signaling, regulation of actin nucleation, organization of the lamellipodium and cell junctions, as well as immune response-related functions. Using degree-controlled randomized QBCHL networks we observe a significant link between the CAMK2D HCIPs and the HUN complex core members UBE3A, HERC2, NEURL4, ECI2, ECH1, and MAPK6 (p-value = 0.002). There was a significant link between the CAMK2D HCIPs and the HUN complex interactors. The same was observed comparing connections between the CAMK2D interactors and the UBE3A interactors or between CAMK2D interactors and the HUN complex interactors with the UBE3A interactors removed (Figure S5). These data therefore suggest connectivity between CAMK2D and the HUN complex beyond that which is mediated by UBE3A.
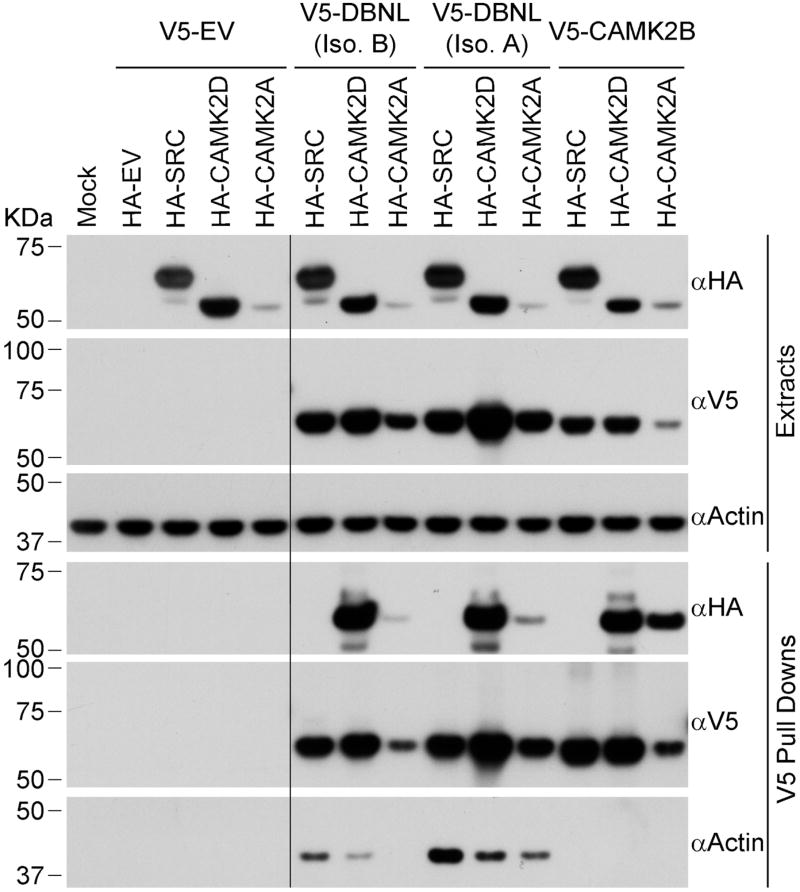
To further examine connections between CAMK2D and the HUN complex, we built a network linking the CAMK2D interactors to each other or to the HUN complex interactors using protein interaction data from QBCHL, reciprocal IP or Y2H data from this study and from Al-Hakim et al. [51] (Figure 7) (network built using only QBCHL in Figure S6). Similar to the list of CAMK2D interactors, the CAMK2D-HUN network shows enrichment for cytoskeleton (spindle, actin nucleation), cell polarity and cell signaling-related functions (EGFR, Ras, FGFR, and neurotrophin) (Table S4 and Methods), all of which have important roles in neurite outgrowth and synapse formation. We found four interactors in common between CAMK2D and the HUN complex (Figure 7). CAMK2D and HERC2 both coimmunoprecipitated as HCIPs the TANK Binding Kinase 1 (TBK1) and FK506 binding protein 5 (FKBP5). In addition, NEURL4 and CAMK2D shared the Baculoviral IAP Repeat Containing 6 (BIRC6) and Drebrin-Like (DBNL) proteins as HCIPs. DBNL functions in receptor-mediated endocytosis, reorganization of the actin cytoskeleton, formation of cell projections such as neurites, neuron morphogenesis, and synapse formation [66–73]. An association between DBNL and NEURL4 has already been described [42] however the association of DBNL with CAMKII has not. Using the V5-tagged DBNL isoforms 1 and 2, we were able to pull down CAMK2D and CAMK2A confirming an association between DBNL and the CAMKII complex (Figure 8). DBNL, thus, represents another possible link between UBE3A/HUN complex and CAMKII. DBNL’s demonstrated role in neuronal functions [67–69, 71–73] suggests that it could mediate a role for UBE3A in AS and ASD.
Figure 7. Interconnectivity of the interactomes of CAMKII and the HUN complex.
The network built between CAMK2D and HUN complex preys using interaction data from QBCHL, reciprocal IP and Y2H data from this study and pulldown data from [Al-Hakim et al.], shows strong connectivity and suggests an involvement of both, the CAMKII and HUN complex, in common biological processes. Interestingly, no distinct protein complexes were identified in this network. Oval shapes are equivalent to circles, they were used with the sole purpose of facilitating the reading of the gene symbols in the corresponding nodes.
Figure 8. Confirmation of interaction of CAMK2D and CAMK2A with several HCIPs.
V5-tagged HCIPs were immunoprecipitated from T-REx 293 cells using anti-V5 magnetic beads. Protein extracts and immunoprecipitates were analyzed by SDS-PAGE and Western blot using antibodies against HA-tag, V5-tag and Actin. HA-SRC was used as negative control and V5-CAMK2B as positive control. The long black line indicates the place where part of the blots was removed because it was irrelevant for the figure. EV: empty vector.
Discussion
A network approach using available systematically generated proteome-scale interaction maps demonstrated the overall coherence of the interaction data obtained in this study, and more specifically identified cellular functions in which UBE3A might have a role. As further discussed below, this systems approach to studying the functions of UBE3A across cell types also provides intriguing insights into its involvement in HPV-associated cancers and the neurodevelopmental disorders AS and ASD.
UBE3A and the Proteasome
Most of the components of the proteasome are UBE3A HCIPs in the AP-MS experiments highlighting the association of UBE3A with the proteasome. The function of UBE3A in the proteasome is still unclear although it has been suggested that it might be involved in controlling the proteolytic activity of the proteasome through its association with PSMD4 [22, 23]. In this study, PSMD4, a member of the regulatory subunit of the proteasome, was also identified as a prey of UBE3A in the Y2H screen indicating that these two proteins directly interact. Interestingly, PSMD4 has been recently reported to mediate the turnover of the proteasome through autophagy when it is ubiquitylated [74, 75] and UBE3A has been shown to ubiquitylate PSMD4 [22, 23]. Therefore it is possible that, under certain conditions, UBE3A may play a role in proteasomal turnover by autophagy through its interaction with PSMD4.
UBE3A and the Centrosome
PSMD4 also plays a role in the centrosome, where it controls proteasome activity and promotes the elaboration of dendrite arbors in the rat brain [76]. Whether UBE3A is involved in those processes has not yet been determined. However UBE3A itself has been reported to associate with ASPM, a centrosomal protein linked to primary microcephaly [65], a condition affecting approximately 80% of the individuals with AS [77]. Our study provides further support for the involvement of UBE3A in centrosomal functions with the identification of SUGT1 as an UBE3A HCIP. SUGT1 is involved in protein ubiquitylation through interaction with SKP1 (a subunit of the SCF complex) in kinetochore assembly, and centrosome organization [78–81]. In addition, the fact that UBE3A associates with centrosomal proteins such as CEP97 and CEP170, which also associate with HERC2 and NEURL4 [51], suggests that UBE3A may contribute to the centrosomal functions ascribed to HERC2 and NEURL4 [51, 52] through its engagement as a component of the HUN complex. It should be noted that the E6 proteins encoded by hrHPVs, which bind UBE3A, have also been implicated in causing centrosome aberrations [82–84] raising the intriguing possibility that HPV E6-associated centrosome aberrations might be mediated through the interaction of E6 with UBE3A and the HUN complex.
UBE3A and the MCM Complex
MCM6 is one of the few proteins that were identified as an UBE3A interactor in both AP-MS and Y2H experiments. MCM6 is part of the MCM complex that is required for initiation and elongation of DNA replication [85]. Although MCM6 was not an HCIP for HERC2, it is noteworthy that HERC2 is a component of the DNA replication fork and regulates its progression as well as DNA origin firing by facilitating the phosphorylation of MCM2, another member of the MCM complex [86–88]. The fact that both HERC2 and UBE3A associate with proteins involved in DNA replication suggests that UBE3A may play a role in some of the functions ascribed to HERC2 in DNA replication and repair. Moreover, recovery of stalled forks is facilitated in part via Trans-Lesion Synthesis (TLS) [89] and template switching by homologous recombination (HR). HERC2 participates in HR repair by coordinating ubiquitin-dependent assembly of DNA repair factors at DNA double-strand breaks [90]. In addition, together with RNF8, HERC2 has been shown to promote TLS at stalled replication forks [91]. Interestingly HPV replication occurs on fragile sites [92], heritable nonrandomly distributed loci on human chromosomes that exhibit an increased frequency of chromosomal breakage under conditions of replication stress. The models proposed to explain how instability at fragile sites arises involve DNA replication and suggest that replication fork stalling along fragile sites during DNA synthesis is a very frequent event (reviewed in [93]). HPV16 E6 has been shown to prevent the recovery of stalled replication forks independently of p53 [94]. HPV16 E6 might achieve this by interfering with the replication functions of HERC2 through the HUN complex and, perhaps also with the functions of UBE3A in the MCM complex. Interestingly, hrHPV E6 proteins have been shown to be necessary for efficient viral replication in keratinocytes [95–97]. In this context the effect of HPV16 E6 on replication forks will result in unreplicated fragile sites moving further through the cell cycle, which might help HPV replication and contribute to E6 oncogenicity by increasing chromosome instability.
UBE3A and Transcription
In addition to its ubiquitin-ligase activity, UBE3A can function in transcription as a coactivator of nuclear hormone receptors [98] or to down regulate the estradiol-induced expression of some genes [99]. One of the down regulated genes is ARC, whose overexpression in AS neurons stimulates the internalization of AMPA receptors resulting in a lower number of these receptors at excitatory synapses [100]. Concerning the transcriptional activity of UBE3A, its interaction with MCM6 might be relevant since the MCM complex interacts with RNA polymerase II and it has been suggested that it could facilitate transcription by remodeling the chromatin template [101]. Of note, NEURL4 consistently coimmunoprecipitated components of the RNA polymerase II as well as the helicase ERCC3/XBP/RAD25, a component of the transcription factor II H, involved in DNA repair and transcription [102, 103]. We do not know whether the UBE3A interaction with MCM6 is physically and/or functionally related to its roles as a component of the HUN complex, or if these are independent events, but all this evidence suggests that both UBE3A and the HUN complex may be involved in DNA replication, repair, and transcription, and that these activities might be of relevance for AS and UBE3A-associated ASD as it has been suggested by others [99, 104].
The HUN Complex
At present we do not know the full composition of the HUN complex or whether there are more than one complex containing HERC2, UBE3A and NEURL4. However, similarly to MAPK6 [28], ECH1 and ECI2 seem to be part of the HUN complex since they are HCIPs for UBE3A, NEURL4 and MAPK6 and, as bait, ECI2 coimmunoprecipitated HERC2, NEURL4 and ECH1. In addition, another study has reported the association of ECI2 with HERC2 and NEURL4 [51]. Both ECH1 and ECI2 are enzymes that participate in the beta-oxidation of fatty acids. We do not yet know whether the HUN complex is involved in the metabolism of fatty acids or whether these enzymes play another role in the context of the HUN complex, but since fatty acid oxidation has been linked to developmental neuropsychiatric disorders, including autism [105–107], a possible role for the HUN complex in that process should be addressed in the future.
HERC2 associated proteins have already been discussed [30], however it is noteworthy to highlight its association with mTORC1, a key regulator of metabolism in eukaryotes [108–110] whose dysregulation is associated with neurodevelopmental and neuropsychiatric disorders such as ASD (reviewed in [111]). Although HERC2 itself does not associate with mTORC1, it associates with the mTORC1 regulatory complex GATOR 2 [59] and with the EIF3 complex, which is necessary for mTORC1 signaling through the translational regulators S6K1 and 4E-BP1 [58]. In addition, NEURL4 and UBE3A associate with components of the EIF3, probably through HERC2, suggesting that the HUN complex may play a role in mTORC1 signaling and regulation of translation. Interestingly, the hippocampus of a mouse model for AS has been shown to have increased mTORC1 and decreased mTORC2 activities. Restoring the activities of mTORC1 and 2 to normal levels resulted in improved long-term potentiation (LTP), restored actin polymerization (see below) and reduced ARC levels [112] suggesting that any effect of UBE3A on this pathway may be relevant for neural function and AS. On the other side, E6 proteins of mucosal HPVs activate mTORC1 and enhance cap-mediated transcription and these effects correlate, at least in part, with the ability of the E6 proteins to bind UBE3A [113, 114], raising the possibility that E6 proteins influence the mTORC1 pathway and translation through its recruitment into the HUN complex by binding to UBE3A.
Mouse models for AS show characteristics of the human disease including abnormal dendritic spine morphology and density [55, 115], impaired LTP and learning deficits [17, 116]. The induction of long-term potentiation or depression is associated with the enlargement or shrinkage of the spine, respectively. The F-actin cytoskeleton is the main driving force of dendritic spine remodeling and sustains synaptic plasticity [117, 118]. It has been recently proposed that AS is associated with a defect in activity-driven spine cytoskeletal reorganization what results in loss of synaptic plasticity necessary for long-term memory [119]. Interestingly, MAPK6 regulates the actin cytoskeleton and promotes cell migration of several cell types [62–64]. Furthermore, the deubiquitinating enzyme USP20 has been shown to regulate the actin cytoskeleton organization and cell migration by deubiquitylation of MAPK6, altering its protein levels [61]. The same study also identified USP16 as a deubiquitinating enzyme for MAPK6 but with moderate effects on its protein levels. Intriguingly, both deubiquitinating enzymes associate with HERC2 [120–122] but while MAPK6 associates with fragment 1 of HERC2 through its association with NEURL4, USP20 associates with fragment 4 of HERC2 [30] indicating that HERC2 might work as scaffold to bring these proteins together. This suggests that the HUN complex may also play a role in the reorganization of the actin cytoskeleton and therefore in spine morphology and synaptic plasticity in neurons, and that disruption of this function could contribute to AS and UBE3A-associated ASD.
UBE3A and CAMKII
In this study we confirm the association of HIF1AN, a known interactor of UBE3A in T-REx 293 cells, with UBE3A in SH-SY5Y cells. HIF1AN is a negative regulator of HIF1A [123, 124] and NOTCH signaling [125, 126]. The NOTCH pathway controls multiple cell differentiation processes including neuron differentiation [127–132], neurite development [133–136], and epithelial cell differentiation [137–142]. In addition, Notch signaling is thought to play a role in cervical cancer [143, 144] and is altered by hrHPV E6 proteins [145, 146]. Altogether this suggests that control of NOTCH signaling by UBE3A through HIF1AN could be relevant for AS, ASD, and HPV-associated cancers. Interestingly HIF1AN also regulates the binding of ASPP2 to specific interactors by hydroxylation [147]. Increased levels of ASPP2, an inhibitor of PP1, has been postulated to be a potential cause of decreased CAMKII activity in an AS mouse model [16]. The decrease in CAMKII activity seems to be very relevant for AS since restoring its activity corrected the behavioral deficits observed in the AS mouse [17]. Here we have shown that UBE3A directly interacts with and regulates ASPP2 protein levels through degradation by the proteasome, supporting the notion that ASPP2 might be involved in AS. In this case, the lack of UBE3A would result in higher levels of ASPP2 which, in turn, would inhibit PP1 resulting in the increased phosphorylation and lower activity of CAMKII observed in the AS mouse model.
The relationship between CAMKII and UBE3A is not limited to ASPP2. The recruitment of the proteasome to the NMDA receptors by CAMKII is necessary for LTP and synaptic plasticity [148–150]. UBE3A is present in neuronal proteasomes, including synaptic proteasomes, and disengages from those proteasomes after NMDA treatment [27], however, the significance of this event for LTP and plasticity is not yet understood. In addition, there is a significant overlap between the CAMKII and UBE3A/HUN complex protein networks including some shared HCIPs such as TBK1 and DBNL (Figure 7). TBK1 plays a role in autophagy [151, 152] and in the innate immune response as an inducer of type-1 interferons [153, 154], and genetic alterations of TBK1 have been associated to amyotrophic lateral sclerosis and frontotemporal dementia [155, 156]. The association of the HUN complex and CAMKII with DBNL might have functional implications for AS and ASD since DBNL is known to be involved in neuronal functions [67–69, 71–73]. Additionally, many known components of the postsynaptic density and other cargos destined to the dendritic spine are transported down the dendrites along microtubules by various kinesin family members [157–161] and CAMKII has been shown to be a regulator of microtubule based motors of the kinesins super family [162, 163]. Interestingly one of the most prominent HCIPs of NEURL4 is KTN1, which is a receptor for kinesin involved in kinesin-driven vesicle motility [164–166]. All this suggests an intricate net of associations between CAMKII and UBE3A-associated pathways that may be relevant for AS and UBE3-Aassociated ASD. On the other side, CAMKII has been shown to have increased activity or expression in several cancers and to be important for proliferation, differentiation and survival of several cancer cells [167–170]. Therefore, some of the interactors/pathways shared by the CAMKII- and UBE3A-associated networks might contribute to HPV-associated cervical cancer via dysregulation by the hrHPV E6 proteins through their interaction with UBE3A.
Summary
Using an approach that involves proteomics and network analysis we have identified new UBE3A interacting and associated proteins that we were able to place within a larger network comprising various cellular processes, some of which are known to be altered in HPV-associated cervical cancer, AS and ASD. We also found a possible connection between UBE3A and CAMKII through ASPP2 and possibly HIF1AN as there is a significant crosstalk between their associated proteomes, suggesting that UBE3A and CAMKII must, at least partially, converge in a common network. We provide strong evidence for UBE3A to be connected to several cellular processes suggesting that UBE3A might be involved in their coordination to ensure adequate cellular responses. Such a role for UBE3A would be advantageous for HPVs, since binding to UBE3A would grant to E6 proteins of hrHPVs, which have a small genome and limited encoding capability, access to several key cellular processes in addition to the ability to degrade TP53. The fact that UBE3A is connected to several cellular processes also has important implications for AS and UBE3A-associated ASD since changes in UBE3A protein levels and activities could affect a variety of cellular pathways contributing to the aspects of the phenotypes associated with those neurodevelopmental disorders. To design effective therapies for HPV-associated cancers, AS, and ASD, it is necessary to identify these cellular processes and understand how they are related to each other. We believe that the results presented in this study should contribute further insight into the role of UBE3A in these disorders and build an important foundation for future work towards an even more mechanistic understanding of the cellular processes altered in HPV-associated cancer, AS, and UBE3A-associated ASD.
Materials and Methods
Cloning and plasmids
The generation of the UBE3A and HERC2 expression vectors was described before [28, 30]. Entry clones in pDONR223 from the CCSB Human ORFeome collection (http://horfdb.dfci.harvard.edu/index.php?page=home) were recombined into the pHAGE expression vectors using the Gateway LR II clonase Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. To obtain a pHAGE vector containing an N-terminal V5 tag, the expression vector pHAGE-P CMVt N-HA GAW [28] was digested with PmeI and XhoI (New England Biolabs) to remove the HA tag and then it was ligated with the following annealed oligonucleotides 5’-AAACGCTAGCCACCATGGCGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGC-3’ and 5’-TCGAGCCGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCCGCCATGGTGGCTAGCGTTT-3’ to generate pHAGE-P CMVt N-V5 GAW. PHAGE-P CMVt N-HBH GAW was generated by replacing the HA tag in PHAGE-P CMVt N-HA GAW with an HBH tag, which contains a biotinylation signal between two RGS-6×His tags [171, 172], using the PmeI and XhoI restriction sites.
Cell culture
All cell lines were maintained in Dulbecco’s modified Eagle’s medium −10% fetal bovine serum at 37°C and 5% CO2. Culture conditions for T-REx 293 cells (Invitrogen) have been described previously [28, 30]. The human neuroblastoma SH-SY5Y cells were a kind gift from Bruce Yankner. The SH-SY5Y is a neuroblast clone isolated from the human neuroblastoma cell line SK-N-SH that was established in 1970 from a metastatic tumor. The SH-SY5Y cell line is widely used in studies of neurologic disorders [173]. In order to have tetracycline-regulated expression the SH-SY5Y cells were transfected with the plasmid pcDNA6/TR (Invitrogen) linearized with FspI (New England Biolabs) and cultured in the presence of 5 µg/ml Blasticidin for several passages to ensure integration of the plasmid into the genomic DNA. A single cell clone was selected based on its ability to repress expression of pHAGE-P CMVt N-HA EGFP. After infection with lentiviruses carrying pHAGE-P CMVt NHA GAW-derived vectors, transduced SH-SY5Y cells were selected with 5 µg/ml Blasticidin and 2 µg/ml puromycin. To induce the expression of the HA-tagged proteins, the cells were kept for 24 h in medium without antibiotics and then induced with 1 µg/ml doxycycline for 24 h. HEK 293T cells were transfected using TransIT®-293 Transfection Reagent (Mirus) using a transfection reagent to DNA ratio of 3:1.
Immunoprecipitations
Immunopurification from T-REx 293 cells for AP-MS experiments have been described previously [28, 30]. The same protocol was followed for SH-SY5Y cells with few modifications. Briefly, cells from four 500 cm2 culture plates (approximately 80 to 90% confluent) expressing HA-tagged proteins were washed once with phosphate-buffered saline (PBS) at room temperature and then lysed on the plates with 5 ml ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150mMNaCl, 0.5% NP-40, 12.5mMNaF, 1mMNa3VO4, 12.5mM β-glycerophosphate, cOmplete™ protease inhibitor cocktail [Roche]) per plate. Cleared lysates were filtered through a 20-µm SFCA-PF syringe filter (Corning) and immunoprecipitated with 50 µl (settled) anti-HA–agarose beads (Sigma) for 2 h at 4°C. The beads were then washed twice with 10 ml lysis buffer and twice with 10 ml PBS, transferred with 1 ml PBS into a microcentrifuge tube, and then eluted with three washes of 0.5 N NH4OH, 0.5 mM EDTA. For reciprocal immunoprecipitation experiments HEK293T cells were transfected on 6 well plates with 1 µg of each vector DNA and harvested for analysis 48 h later. Only for the UBE3A and ASPP2 coimmunoprecipitation experiment 2 µg of each vector were used. Cells were lysed on ice in 1 ml of the same lysis buffer used for the AP-MS experiments and the V5-tagged proteins were coimmunoprecipitated with anti-V5 magnetic beads (M167-41, MBL) (30 µl of 50% slurry per sample). After 6 washes with 1 ml of ice cold lysis buffer the proteins were eluted by incubating the beads for 60 minutes at 65°C in 30 µl of gel loading buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10 mM TCEP, 5 mM EDTA, 10% Glycerol, Serva Blue G). For HA immunoprecipitations the same protocol was used with anti-HA agarose beads (A2095, Sigma).
MS and CompPASS analysis
The MS run and analysis have already been described in detail [30]. Then the AP-MS data for each bait were analyzed by CompPASS, which relies on an unbiased comparative approach for identifying HCIPs as previously described [39, 174]. For the analysis of coimmunoprecipitations from T-REx 293 cells we used a table with the data of 171 HA coimmunoprecipitations from the same cells produced by the Harper lab [174]. To do the CompPASS analysis of AP-MS data from SH-SY5Y cells we used a table with the data of 64 HA coimmunoprecipitations from those cells generated in this study. The D and WD score threshold was calculated in a way that 90% of the data falls below it for coimmunoprecipitations from SH-SY5Y cells and 95% of the data is under it in the case of T-REx 293 cells. Using a 90% cutoff might produce more false negatives but seem to work better with the data obtained from SH-SY5Y cells based on the UBE3A coimmunoprecipitations (Table 1) since it includes more proteasomal subunits as well as several proteins that were also detected as UBE3A HCIPs in the coimmunoprecipitation experiments done with T-REx 293 cells [28]. The analyses of the AP-MS data from SH-SY5Y cells using a 95% cutoff are also provided in the supplementary material. The normalized D (ND) and WD (NWD) scores results from dividing each score by the calculated threshold. We considered HCIPs those proteins who have a ND or NWD equal to or higher than 1 (normalized score equal to or higher than the calculated threshold). In most cases considering the ND score in addition to the NWD score adds no more than a couple of proteins to the list of HCIPs. A complete set of data including the number of peptides detected for each protein is presented in the Supplementary AP-MS Tables at Mendeley datasets (https://data.mendeley.com/datasets/5w3b4y4cwr/draft?a=98bf0268-3fa9-4854-8b76-b80b8fa2f97f, doi:10.17632/5w3b4y4cwr.1).
Identification of binary protein-protein interactions by Y2H screens
Y2H screens were performed as described previously [37], with some modifications.
Library preparation
ORF clones in pDONR223 containing sequences encoding the three catalytically active and inactive forms of UBE3A [28] were transferred to the Y2H destination vector pQZ212 (DB vector) by Gateway recombinational cloning (Invitrogen) as described in Dreze et al. [37]. pQZ212 is a modified version of pVV212, a high-copy 2-micron (2µ) vector, engineered to contain the LEU2 and CAN1 selection markers.
Competent yeast cells of the Y8930c strain (MATα leu2-3,112 trp1-901 his3Δ200 ura3-52 gal4Δgal80ΔGAL2::ADE2 GAL1::HIS3@LYS2 GAL7::lacZ@MET2 cyh2R) were transformed with the cloned DB-ORF constructs [37]. Yeast cells were plated on selective synthetic complete (SC) solid medium lacking leucine (SC-Leu) to select for transformants.
Y2H screening
Y2H screens were then conducted as described in Rolland, et al. [45] and Sahni et al. [47]. Briefly, saturated cultures of Y8930c containing the vector for each DB-ORF fusion were mated against Y8800 containing AD-ORF mini-pools from human ORFeome 9.1. Each mini-pool contained 96 different AD-ORF yeast strains. After mating in rich liquid media (YEPD), cells were transferred to SC medium lacking leucine and tryptophan (SC-Leu-Trp) to select for diploid cells. Finally, cells were plated on selective solid medium also lacking histidine (SC-Leu-Trp-His + 1mM 3-amino-1,2,4-triazole [3AT] agar plates), allowing for selection of yeast cells which have activated the GAL1::HIS3 reporter gene.
De novo DB-ORF auto-activators, which activate GAL1::HIS3 reporter gene expression without an AD-ORF construct and which can arise during the Y2H screens, were detected using the same method as described in Dreze et al. [37] and Rolland et al. [45]. Diploid yeast cells were spotted in parallel on SC-Leu-Trp-His + 1 mM 3AT and SC-Leu-His + 1mM 3AT + 1 mg/L cycloheximide (CHX) agar plates. Yeast cells containing an AD-ORF plasmid are sensitive to CHX due to the presence of the counter selection marker cyh2S in the plasmid. Only autoactivator DB-ORF cells, which have lost the AD-ORF plasmid by plasmid shuffling, will exhibit a growth phenotype on SC-Leu-His + 1 mM 3AT + 1 mg/L CHX agar plates. Yeast strains containing the DB-ORF auto-activators are therefore excluded from the screen data.
To identify the interacting protein pairs, specifically the interacting AD-ORF from the 96 different strains of the mini-pool, we followed the same protocol as in Rolland et al. [45]. Up to five yeast colonies were picked per spot and subsequently lysed. The lysates were then used as a template for PCR amplification. PCR products were subjected to Sanger sequencing and human ORFs were identified by BLASTing the resulting sequence against the human ORFeome v9.1. This generated a list of first-pass pairs (FiPPs) from primary yeast two-hybrid screens.
Pairwise testing to verify primary screen results
FiPPs from primary Y2H screens were then pairwise tested as described in Dreze et al. [37]. Yeast strains expressing each interacting pair of DB-ORF and AD-ORF were freshly inoculated in SC-Leu and SC-Trp, respectively. Saturated yeast cultures are then mated in quadruplicate in YEPD, followed by a diploid enrichment in SC-Leu-Trp 24 hours later. Diploid yeast cells were spotted in quadruplicate on SC-Leu-Trp-His + 1 mM 3AT and once on SC-Leu-His + 1 mM 3AT + 1 mg/L CHX agar plates. Protein-protein interactions (PPIs) were scored as positive when yeast colonies developed in at least three out of four spots on SC-Leu-Trp-His + 1mM 3AT but did not grow on SC-Leu-His + 1 mM 3AT + 1 mg/L CHX. Colonies were picked and then lysed. The lysates were used as template for duplex PCR amplification using Platinum Taq Polymerase High Fidelity (Life Technologies). For duplex PCR amplification, an indexed AD and DB primers set was used, in which each primer contains a unique barcode. This allowed for the pooling of up to 96 different DB-ORF and 96 different AD-ORF PCR products per well. The barcodes and ORF sequences were then determined using Illumina sequencing by seqWell, Inc. (Beverly, MA) and identified by BLASTing the resulting sequence against the human ORFeome 9.1 [175].
Y3H screen
For the Y3H screen, the destination vector pAG416GPD (Addgene Plasmid #14148) expressing HA-HPV16 E6 and the selection marker URA3 was selected. Y8930c strain was co-transformed with pQZ212, expressing one of the three catalytically active and inactive isoforms of UBE3A, and pAG416GPD (Addgene Plasmid #14148). To select for transformants expressing HA-HPV16 E6 and UBE3A, yeast cells were platted on SC solid medium lacking leucine and uracil (SC –Leu-Ura).
To perform the Y3H screen, saturated cultures of Y8930c expressing UBE3A and HPV16 HA-E6 were mated against Y8800 containing the AD-ORF minipools. To test for functional expression of HA-HPV16E6 the obtained cultures were mated with a Y8800 clone expressing TP53. In contrast to a culture only expressing inactive UBE3A which did not show any growth, the clones containing wt UBE3A and HA-16E6 showed detectable growth of colonies which was strongly increased when using the strains expressing inactive UBE3A and HA-HPV16E6, verifying that our Y3H system can be used to detect UBE3A/HPV16E6 complexes. All the following screening steps were identical to the Y2H procedure, except that the SC medium lacked uracil to maintain selection of yeast expressing HA-HPV16 E6. The Y3H screen was performed in parallel with a Y2H screen of Y8930c expressing only UBE3A mated against Y8800 containing the AD-ORF minipools.
Western Blots and Size exclusion chromatography
Western blots and size exclusion chromatography have already been described in detail [28, 30]. Blots were incubated with the following primary antibodies: HA conjugated to horseradish peroxidase (HRP) (clone 3F10, Roche), V5 conjugated to HRP (R961-25, Invitrogen), beta actin conjugated to HRP (ab49900, Abcam), UBE3A (clone E6AP-330; Sigma), HERC2 (BD Transduction Laboratories), NEURL4 (ProteinExpress), PSMD4 (S5a-18; Enzo), PSMA5 (PA1-1962; Thermo). HRP-conjugated anti-mouse or anti-rabbit antibodies (GE Healthcare) were used as secondary antibodies when needed. Biotinylated proteins were detected with Streptavidin-HRP (#21130, Pierce).
Immunofluorescence
HaCat, HeLa, SH-SY5Y, and U2OS cells were plated onto glass cover slips and fixed with 3% formaldehyde in PBS for 10 minutes at room temperature. Cells were then washed 3 times with PBS and stored at 4°C until stained. For staining, cells were permeabilized and blocked for 15 minutes with 0.2% Triton X-100 and 5% normal donkey serum in PBS (blocking buffer). Cover slips were then incubated for 1 hour at room temperature with blocking buffer containing the rabbit polyclonal antibody to UBE3A (E6AP H-182) (sc-25509; Santa Cruz Biotechnology), together with the mouse monoclonal antibodies to EIF3C (eIF3 p110 B-6) (sc-74507; Santa Cruz Biotechnology) (1:50), MCM6 (H-8) (sc393618; Santa Cruz Biotechnology) (1:50), or Herc2 (612366; BD Biosciences) (1:50) as indicated. The primary antibodies were washed 5 times with 0.2% Triton X-100, 1% normal donkey serum in PBS (washing buffer) and incubated for 1 hour at room temperature with donkey anti-mouse IgG conjugated to alexa 594 (A-21203; ThermoFisher Scientific) and donkey anti-rabbit IgG conjugated to alexa 488 (A-21206; ThermoFisher Scientific) diluted in blocking buffer. Cover slips were then washed 5 times with washing buffer, 1 time with water, and mounted onto glass slides using Prolong Gold Antifade Mountant (ThermoFisher Scientific). All samples were analyzed by confocal microscopy using an Olympus FV3000 confocal laser scanning microscope. The specificity of the antibodies against UBE3A, HERC2, and MCM6 was tested and established in cells treated with the corresponding siRNAs. The antibody against EIF3C has been previously used in immunofluorescence experiments [176].
UBE3A mediated degradation of ASPP2
HEK 293T cells were transfected with 0.5 µg of pHAGE-P CMVt N-HA UBE3A isoform 1, pHAGE-P CMVt N-HA UBE3A isoform 1 C820A, pHAGE-P CMVt N-HBH ASPP2, pHAGE-P CMVt N-V5 GAPDH, or the corresponding empty vectors. Proteasomal inhibition was achieved by 4 hours treatment with 500 nM Velcade (Active Biochem, A1014). 48 hours after transfection the cells were lysed on the plates with 50 mM Tris-HCL pH 6.8, 2% SDS. The protein extracts were analyzed by SDS-PAGE and Western blot.
Software
All code was written in Python 2.7 and is being made publicly available at github.com/CCSB-DFCI/UBE3A_HUN_paper_pub (doi:10.5281/zenodo.846870) along with most input and output files and all plots generated for this project. The code makes use of the additional python libraries igraph for network manipulations [177], numpy for numeric operations [178], pyjsonrpc for easy access to the FuncAssociate JSON-RPC based web service (github.com/gerold-penz/python-jsonrpc), and matplotlib for plotting [179]. Cytoscape v3.4.0 was used for network visualization [180]. Jupyter python notebooks were used to provide interactive python code for plotting [181]. The Funcassociate web service with default settings was used to perform all Gene Ontology [182] functional enrichment analyses described in this paper [183].
Source of gene annotation data
Entrez gene IDs along with gene symbols, synonyms and descriptions were obtained from the Homo sapiens gene info file downloaded from ftp://ftp.ncbi.nih.gov/gene/DATA/GENE_INFO/Mammalia/ on November 13th 2016.
Assembly of final list of HCIPs for every bait
UBE3A: The HEK293T pulldown data was obtained from [28]. Gene symbols were mapped to Entrez gene IDs and combined with pulldown data obtained in this study from the SY5Y cell line for a 90 and 95% HCIP confidence cutoff as determined from CompPASS. A final HCIP cutoff was determined for every prey by combining the pulldown data from the different isoforms and variants of UBE3A that were used as bait in this study. At least half of all prey occurrences had to be found at a 95% cutoff in order to assign the 95% confidence to that prey. All HCIPs that were only observed in one of the coimmunoprecipitations per cell line were removed unless that HCIP was also observed at least once in the other cell line or if it was a gene of the MCM complex. Based on this filtering, three sets of HCIPs were built for UBE3A: i) AP-MS based HCIPs, ii) AP-MS based HCIPs with all but the PSMD4 proteasome subunits removed based on finding the substring "PSM" in the official gene symbol of the prey, iii) combining ii) with all UBE3A interactors identified in the Y2H screen in this study.
HERC2
The HEK293T pulldown data for the HERC2 fragments was obtained from [30] and combined with the pulldown data obtained for the HERC2 fragments from the SY5Y cell line. No other filtering than the removal of proteasome subunits was performed.
All other baits
The HEK293T and SY5Y pulldown data obtained in this study for the baits ECH1, ECI2, HIF1AN, MAPK6, NEURL4, and CAMK2D (this one only from SH-SY5Y) were combined (per bait) without any additional filtering apart from removing any proteasome subunits that appeared as prey.
The final list of HCIPs for each bait is provided in GitHub (github.com/CCSB-DFCI/UBE3A_HUN_paper_pubdoi:10.5281/zenodo.846870).
External protein interaction data
The QBCHL network (named after the initials of the different protein interaction data sets used) was built from the following five different sources: HI-union was built on March 6th 2017 from downloading all published and unpublished, test space verified and test space validated datasets from http://interactome.baderlab.org/ [45–50]. BioPlex was obtained from downloading the dataset with time stamp 06/12/2015 from http://bioplex.hms.harvard.edu/downloadInteractions.php [42]. CoFrac was obtained from downloading Supplementary Table 2 from Wan et al [44]. QUBIC was obtained from downloading Supplementary Table 2 from Hein et al [43]. Lit-BM-13 was obtained from downloading Supplementary Table 1 from Rolland et al [45]. The HI-union and QUBIC PPIs and protein pairs were mapped from UniProt accession numbers (ACs) to Entrez gene IDs using the mapping file HUMAN_9606_idmapping_selected.tab downloaded from UniProt on March 3rd, 2017 [184]. The CoFrac protein pairs were mapped from Ensembl gene IDs to Entrez gene IDs using a mapping file obtained from the BioMart tool at http://www.ensembl.org/ (release 88) [185].
Additional sources of PPI data were obtained as follows: DuB_AP was built from downloading the PPI data in MI-TAB from IntAct [186] for the PubMed identifiers 19615732 [39] and 20562859 [174] on April 24th 2017. PPIs were mapped from Uniprot ACs to Entrez gene IDs as described above. AlHakim_IPs was built from curating protein associations published in Al-Hakim et al [51]. Howley_map was built from building pairs between the baits UBE3A, HERC2, ECI2, ECH1, NEURL4, MAPK6, CAMK2D, and HIF1AN and their respective final sets of HCIPs. These additional sources of PPI data as well as the reciprocal IPs obtained during this and previous studies [28, 30] and the UBE3A interactors identified in the Y2H screen described in this study are available on GitHub (github.com/CCSB-DFCI/UBE3A_HUN_paper_pubdoi:10.5281/zenodo.846870).
All PPI datasets were made non-redundant (any directionality removed) and any homodimer pairs were removed prior to be merged and used for analysis.
Network generation
UBE3A
The UBE3A networks were built using either as source for PPI data only QBCHL or QBCHL, DuB_AP, Howley_map, Y2H_UBE3A, reciprocal_IP, and AlHakim_IP. The final set of UBE3A HCIPs (without proteasome subunits and with the interactors from Y2H) were connected between each other and two their neighbors if the neighbors interacted with at least two UBE3A HCIPs using the indicated sources of PPI data. Next, all PPIs in this network with only one source of evidence were removed as well as all proteins that ended up being unconnected from any other protein in the network.
HUN complex network
The final set of HCIPs from HERC2, NEURL4, MAPK6, ECH1, and ECI2 were connected with each other using either QBCHL or QBCHL, reciprocal IP, AlHakim and Y2H_UBE3A, if both HCIPs connected to each other had at least one of these five baits in common.
CAMK2D network
The CAMK2D networks were built using the same two sets of source networks as described above for the HUN complex network. The CAMK2D HCIPs were connected with each other and to any neighbors as defined by the PPI data in the set of source networks used, if these neighbors were HCIPs from at least one of the baits HERC2, NEURL4, ECI2, ECH1, MAPK6, or UBE3A. Interactions between these neighbors were not added to the network.
The list of interactions and interaction partners, the Cytoscape session file as well as the network images are all available on GitHub (github.com/CCSB-DFCI/UBE3A_HUN_paper_pubdoi:10.5281/zenodo.846870).
Generation of randomized QBCHL networks
The python package igraph and function Degree_Sequence() was used to obtain degree-controlled randomized QBCHL networks [187].
Randomization of sets of HCIPs
For some statistical analyses where network randomization was not appropriate, starting sets of HCIPs to build networks were randomized instead. We observed that the sets of HCIPs obtained for each of the baits displayed a slightly elevated degree distribution (a higher fraction of proteins with more interaction partners) compared to the degree distribution of the QBCHL network. Therefore, simple random drawing of proteins from the QBCHL network to build random sets of HCIPs was inappropriate. Degree-controlled random protein sampling was achieved by applying a sliding window around a given degree d for which a random protein needed to be sampled. The size of the window was determined by the degree plus/minus the square of the log base 2 of d (log(d,2)^2). The sufficient number of proteins to sample from for a certain degree was put to 1,000 in line with the number or total randomizations performed. If there were < 1,000 proteins of degree d available from the QBCHL network to randomly sample from, proteins from d+1 and d-1 degrees were added and so forth until the limits of the window were reached or the minimum number of 1,000 proteins to sample from was obtained. This procedure allowed to perform degree-controlled random protein sampling even for high degree nodes where only a few proteins in the QBCHL network are available for a given degree. Overlaps of random sets of HCIPs with the original set of HCIPs was possible but remained small (< 3).
Empirical testing for significance of enriched GO terms in expanded networks
Functional enrichment tests using the Fisher's exact test and correction for multiple testing are not sufficient to assess significance of functional enrichments observed for the UBE3A and CAMK2D networks (built only with QBCHL as source) because these networks were built by extension to neighbors of UBE3A or CAMK2D HCIPs. Network expansion will lead to preferential addition of hubs to the network, which might represent a class of proteins of certain biological function, therefore leading to potential functional bias, which is not captured by the null hypothesis of the Fisher's exact test.
Gene Ontology (GO) terms that were found to be enriched in any of these two networks as defined by an adjusted p-value cutoff of 0.05 determined by FuncAssociate [188] were further tested for significance using the following empirical approach. For each network (CAMK2D and UBE3A) degree-controlled random sets of HCIPs were built 1,000 times. For each random set of HCIPs, a network was built using the exact same protocol as used for the original set of HCIPs (as described above) using QBCHL as only source of PPI data. Each randomly built network was then assessed for functional enrichments using FuncAssociate. Any GO term that was significantly enriched in the original network was assessed by how often that term was observed to be at least as significantly enriched in the randomly built networks (using the log odds ratio as a measure for strength of enrichment). Only enriched GO terms from the original UBE3A or CAMK2D network that were observed at most 5% of the time (p-value cutoff of 0.05) to be as much or more enriched in the randomly built corresponding networks were retained.
Test for significances of LCC sizes
The LCC of a given network was determined as the largest cluster returned from the python package igraph clusters() function. 1,000 random networks around each set of HCIPs as negative control were built using either degree-controlled randomized QBCHL networks wherever applicable (individual sets of HCIPs and HUN network) or starting from randomized sets of HCIPs (UBE3A and CAMK2D network).
Test for significance of closeness between CAMK2D HCIPs and HUN complex HCIPs
Significances of closeness of CAMK2D HCIPs to the union of HCIPs from HERC2, NEURL4, ECI2, ECH1, MAPK6, and UBE3A were empirically determined using 1,000 degree-controlled randomized QBCHL networks.
Supplementary Material
Highlights.
UBE3A is associated with Angelman syndrome, autism, and HPV-associated cancers.
Several new UBE3A associated proteins were identified (e.g. MCM6 and ASPP2).
UBE3A associated proteins are connected to several fundamental cellular processes.
There is evidence for crosstalk between UBE3A and CAMKII interaction networks.
Network analyses provide new insights into the cellular functions of UBE3A.
Acknowledgments
The authors thank J.W. Harper and his laboratory in Cell Biology for assistance with the AP-MS experiments and analyses, T. Rolland for input in network analyses performed in this study, N. Jailkhani for help with the Y2H screens, and T. Hao for help with the processing of sequencing data. AD was supported by a fellowship from FRS-FNRS-Télévie. SK was supported by the German Research Foundation (DFG) through a scholarship grant (KU 3196/2-1). This work was supported by grants from the Angelman Syndrome Foundation and the Simons Foundation (to PMH) as well as National Institutes of Health grants U41HG001715 (to DEH and MV), P50HG004233 (to MV), and R35CA197262 (to PMH).
MV serves as a member of the SAB of seqWell, Inc., Beverly, MA
Abbreviations
- AP-MS
affinity purifications followed by identification of proteins by liquid chromatography-tandem mass spectrometry
- AS
Angelman syndrome
- ASD
autism spectrum disorders
- CompPASS
Comparative Proteomic Analysis Software Suite
- HCIPs
high confidence interacting proteins
- HPV
human papillomavirus
- HPV16
human papillomavirus type 16
- hrHPVs
high risk human papillomavirus
- HUN complex
protein complex containing HERC2, UBE3A, and NEURL4
- Y2H
yeast two hybrid
- Y3H
yeast three hybrid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–35. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–84. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huibregtse JM, Scheffner M, Howley PM. E6-AP directs the HPV E6-dependent inactivation of p53 and is representative of a family of structurally and functionally related proteins. Cold Spring Harb Symp Quant Biol. 1994;59:237–45. doi: 10.1101/sqb.1994.059.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto Y, Huibregtse JM, Howley PM. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41:263–6. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- 6.Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–3. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 7.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–5. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 8.Guffanti G, Strik Lievers L, Bonati MT, Marchi M, Geronazzo L, Nardocci N, et al. Role of UBE3A and ATP10A genes in autism susceptibility region 15q11-q13 in an Italian population: a positive replication for UBE3A. Psychiatry Res. 2011;185:33–8. doi: 10.1016/j.psychres.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 9.Nurmi EL, Bradford Y, Chen Y, Hall J, Arnone B, Gardiner MB, et al. Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics. 2001;77:105–13. doi: 10.1006/geno.2001.6617. [DOI] [PubMed] [Google Scholar]
- 10.Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3:103ra97. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelman H. ‘Puppet’ Children A Report on Three Cases. Developmental Medicine & Child Neurology. 1965;7:681–8. doi: 10.1111/j.1469-8749.2008.03035.x. [DOI] [PubMed] [Google Scholar]
- 12.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 2011;34:293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genet Med. 2010;12:385–95. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- 14.Buiting K, Williams C, Horsthemke B. Angelman syndrome - insights into a rare neurogenetic disorder. Nat Rev Neurol. 2016;12:584–93. doi: 10.1038/nrneurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 15.Sadikovic B, Fernandes P, Zhang VW, Ward PA, Miloslavskaya I, Rhead W, et al. Mutation Update for UBE3A variants in Angelman syndrome. Hum Mutat. 2014;35:1407–17. doi: 10.1002/humu.22687. [DOI] [PubMed] [Google Scholar]
- 16.Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–44. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila Freire R, et al. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–2. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–92. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- 19.Kleijnen MF, Alarcon RM, Howley PM. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell. 2003;14:3868–75. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–19. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Li Z, Howley PM, Sacks DB. E6AP and calmodulin reciprocally regulate estrogen receptor stability. J Biol Chem. 2006;281:1978–85. doi: 10.1074/jbc.M508545200. [DOI] [PubMed] [Google Scholar]
- 22.Tomaic V, Banks L. Angelman syndrome-associated ubiquitin ligase UBE3A/E6AP mutants interfere with the proteolytic activity of the proteasome. Cell Death Dis. 2015;6:e1625. doi: 10.1038/cddis.2014.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SY, Ramirez J, Franco M, Lectez B, Gonzalez M, Barrio R, et al. Ube3a, the E3 ubiquitin ligase causing Angelman syndrome and linked to autism, regulates protein homeostasis through the proteasomal shuttle Rpn10. Cell Mol Life Sci. 2014;71:2747–58. doi: 10.1007/s00018-013-1526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–65. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 25.Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–49. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanlon TC, Gottlieb B, Durcan TM, Fon EA, Beitel LK, Trifiro MA. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp Cell Res. 2009;315:176–89. doi: 10.1016/j.yexcr.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the Brain 26S Proteasome and its Interacting Proteins. Front Mol Neurosci. 2010;3 doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, et al. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol. 2012;32:3095–106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhnle S, Kogel U, Glockzin S, Marquardt A, Ciechanover A, Matentzoglu K, et al. Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J Biol Chem. 2011;286:19410–6. doi: 10.1074/jbc.M110.205211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galligan JT, Martinez-Noel G, Arndt V, Hayes S, Chittenden TW, Harper JW, et al. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J Proteome Res. 2015;14:953–66. doi: 10.1021/pr501005v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puffenberger EG, Jinks RN, Wang H, Xin B, Fiorentini C, Sherman EA, et al. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum Mutat. 2012;33:1639–46. doi: 10.1002/humu.22237. [DOI] [PubMed] [Google Scholar]
- 32.Harlalka GV, Baple EL, Cross H, Kuhnle S, Cubillos-Rojas M, Matentzoglu K, et al. Mutation of HERC2 causes developmental delay with Angelman-like features. J Med Genet. 2013;50:65–73. doi: 10.1136/jmedgenet-2012-101367. [DOI] [PubMed] [Google Scholar]
- 33.Morice-Picard F, Benard G, Rezvani HR, Lasseaux E, Simon D, Moutton S, et al. Complete loss of function of the ubiquitin ligase HERC2 causes a severe neurodevelopmental phenotype. Eur J Hum Genet. 2016;25:52–8. doi: 10.1038/ejhg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thatcher KN, Peddada S, Yasui DH, Lasalle JM. Homologous pairing of 15q11-13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum Mol Genet. 2005;14:785–97. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- 35.Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasui DH, Scoles HA, Horike S, Meguro-Horike M, Dunaway KW, Schroeder DI, et al. 15q11.2-13.3 chromatin analysis reveals epigenetic regulation of CHRNA7 with deficiencies in Rett and autism brain. Hum Mol Genet. 2011;20:4311–23. doi: 10.1093/hmg/ddr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, et al. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Lautar S. A yeast three-hybrid method to clone ternary protein complex components. Anal Biochem. 1996;242:68–72. doi: 10.1006/abio.1996.0429. [DOI] [PubMed] [Google Scholar]
- 39.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luck K, Sheynkman GM, Zhang I, Vidal M. Proteome-Scale Human Interactomics. Trends Biochem Sci. 2017 doi: 10.1016/j.tibs.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortensen F, Schneider D, Barbic T, Sladewska-Marquardt A, Kuhnle S, Marx A, et al. Role of ubiquitin and the HPV E6 oncoprotein in E6AP-mediated ubiquitination. Proc Natl Acad Sci U S A. 2015;112:9872–7. doi: 10.1073/pnas.1505923112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–9. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–23. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 44.Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–44. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–26. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell. 2016;164:805–17. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–60. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 49.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, et al. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478–80. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Hakim AK, Bashkurov M, Gingras AC, Durocher D, Pelletier L. Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Kim S, Kobayashi T, Liang FX, Korzeniewski N, Duensing S, et al. Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres. EMBO Rep. 2012 doi: 10.1038/embor.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schumacher S, Laass K, Kant S, Shi Y, Visel A, Gruber AD, et al. Scaffolding by ERK3 regulates MK5 in development. EMBO J. 2004;23:4770–9. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seternes OM, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, et al. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J. 2004;23:4780–91. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–8. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- 56.Gustin RM, Bichell TJ, Bubser M, Daily J, Filonova I, Mrelashvili D, et al. Tissue-specific variation of Ube3a protein expression in rodents and in a mouse model of Angelman syndrome. Neurobiol Dis. 2010;39:283–91. doi: 10.1016/j.nbd.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Liu X, Zhou L, Duong D, Bhuripanyo K, Zhao B, et al. Identifying the ubiquitination targets of E6AP by orthogonal ubiquitin transfer. Nat Commun. 2017;8:2232. doi: 10.1038/s41467-017-01974-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–80. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 59.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–6. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–62. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathien S, Deleris P, Soulez M, Voisin L, Meloche S. Deubiquitinating Enzyme USP20 Regulates Extracellular Signal-Regulated Kinase 3 Stability and Biological Activity. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00432-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Al-Mahdi R, Babteen N, Thillai K, Holt M, Johansen B, Wetting HL, et al. A novel role for atypical MAPK kinase ERK3 in regulating breast cancer cell morphology and migration. Cell Adh Migr. 2015;9:483–94. doi: 10.1080/19336918.2015.1112485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W, Bian K, Vallabhaneni S, Zhang B, Wu RC, O'Malley BW, et al. ERK3 promotes endothelial cell functions by upregulating SRC-3/SP1-mediated VEGFR2 expression. J Cell Physiol. 2014;229:1529–37. doi: 10.1002/jcp.24596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, et al. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Invest. 2012;122:1869–80. doi: 10.1172/JCI61492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singhmar P, Kumar A. Angelman syndrome protein UBE3A interacts with primary microcephaly protein ASPM, localizes to centrosomes and regulates chromosome segregation. PLoS One. 2011;6:e20397. doi: 10.1371/journal.pone.0020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boateng LR, Cortesio CL, Huttenlocher A. Src-mediated phosphorylation of mammalian Abp1 (DBNL) regulates podosome rosette formation in transformed fibroblasts. J Cell Sci. 2012;125:1329–41. doi: 10.1242/jcs.096529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fenster SD, Kessels MM, Qualmann B, Chung WJ, Nash J, Gundelfinger ED, et al. Interactions between Piccolo and the actin/dynamin-binding protein Abp1 link vesicle endocytosis to presynaptic active zones. J Biol Chem. 2003;278:20268–77. doi: 10.1074/jbc.M210792200. [DOI] [PubMed] [Google Scholar]
- 68.Haag N, Schwintzer L, Ahuja R, Koch N, Grimm J, Heuer H, et al. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J Neurosci. 2012;32:17842–56. doi: 10.1523/JNEUROSCI.0843-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haeckel A, Ahuja R, Gundelfinger ED, Qualmann B, Kessels MM. The actin-binding protein Abp1 controls dendritic spine morphology and is important for spine head and synapse formation. J Neurosci. 2008;28:10031–44. doi: 10.1523/JNEUROSCI.0336-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kessels MM, Engqvist-Goldstein AE, Drubin DG, Qualmann B. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J Cell Biol. 2001;153:351–66. doi: 10.1083/jcb.153.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koch N, Kobler O, Thomas U, Qualmann B, Kessels MM. Terminal axonal arborization and synaptic bouton formation critically rely on abp1 and the arp2/3 complex. PLoS One. 2014;9:e97692. doi: 10.1371/journal.pone.0097692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qualmann B, Boeckers TM, Jeromin M, Gundelfinger ED, Kessels MM. Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J Neurosci. 2004;24:2481–95. doi: 10.1523/JNEUROSCI.5479-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell. 2015;58:1053–66. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A. 2016;113:E7490–E9. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Puram SV, Kim AH, Park HY, Anckar J, Bonni A. The ubiquitin receptor S5a/Rpn10 links centrosomal proteasomes with dendrite development in the mammalian brain. Cell Rep. 2013;4:19–30. doi: 10.1016/j.celrep.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan WH, Bacino CA, Skinner SA, Anselm I, Barbieri-Welge R, Bauer-Carlin A, et al. Angelman syndrome: Mutations influence features in early childhood. Am J Med Genet A. 2011;155A:81–90. doi: 10.1002/ajmg.a.33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bansal PK, Abdulle R, Kitagawa K. Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol Cell Biol. 2004;24:8069–79. doi: 10.1128/MCB.24.18.8069-8079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 80.Martins T, Maia AF, Steffensen S, Sunkel CE. Sgt1, a co-chaperone of Hsp90 stabilizes Polo and is required for centrosome organization. EMBO J. 2009;28:234–47. doi: 10.1038/emboj.2008.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steensgaard P, Garre M, Muradore I, Transidico P, Nigg EA, Kitagawa K, et al. Sgt1 is required for human kinetochore assembly. EMBO Rep. 2004;5:626–31. doi: 10.1038/sj.embor.7400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci U S A. 2000;97:10002–7. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002;62:7075–82. [PubMed] [Google Scholar]
- 84.Schaeffer AJ, Nguyen M, Liem A, Lee D, Montagna C, Lambert PF, et al. E6 and E7 oncoproteins induce distinct patterns of chromosomal aneuploidy in skin tumors from transgenic mice. Cancer Res. 2004;64:538–46. doi: 10.1158/0008-5472.can-03-0124. [DOI] [PubMed] [Google Scholar]
- 85.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–93. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 86.Izawa N, Wu W, Sato K, Nishikawa H, Kato A, Boku N, et al. HERC2 Interacts with Claspin and regulates DNA origin firing and replication fork progression. Cancer Res. 2011;71:5621–5. doi: 10.1158/0008-5472.CAN-11-0385. [DOI] [PubMed] [Google Scholar]
- 87.Deegan TD, Diffley JF. MCM: one ring to rule them all. Curr Opin Struct Biol. 2016;37:145–51. doi: 10.1016/j.sbi.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 88.Hyrien O. How MCM loading and spreading specify eukaryotic DNA replication initiation sites [version 1; referees: 4 approved] 2016 doi: 10.12688/f1000research.9008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–53. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 90.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 2010;12:80–6. doi: 10.1038/ncb2008. sup pp 1-12. [DOI] [PubMed] [Google Scholar]
- 91.Mohiuddin, Kobayashi S, Keka IS, Guilbaud G, Sale J, Narita T, et al. The role of HERC2 and RNF8 ubiquitin E3 ligases in the promotion of translesion DNA synthesis in the chicken DT40 cell line. DNA Repair (Amst) 2016;40:67–76. doi: 10.1016/j.dnarep.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jang MK, Shen K, McBride AA. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014;10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Franchitto A. Genome instability at common fragile sites: searching for the cause of their instability. Biomed Res Int. 2013;2013:730714. doi: 10.1155/2013/730714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen B, Simpson DA, Zhou Y, Mitra A, Mitchell DL, Cordeiro-Stone M, et al. Human papilloma virus type16 E6 deregulates CHK1 and sensitizes human fibroblasts to environmental carcinogens independently of its effect on p53. Cell Cycle. 2009;8:1775–87. doi: 10.4161/cc.8.11.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park RB, Androphy EJ. Genetic analysis of high-risk e6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J Virol. 2002;76:11359–64. doi: 10.1128/JVI.76.22.11359-11364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci U S A. 1999;96:8449–54. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009;23:181–94. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–9. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuhnle S, Mothes B, Matentzoglu K, Scheffner M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc Natl Acad Sci U S A. 2013;110:8888–93. doi: 10.1073/pnas.1302792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–16. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yankulov K, Todorov I, Romanowski P, Licatalosi D, Cilli K, McCracken S, et al. MCM proteins are associated with RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:6154–63. doi: 10.1128/mcb.19.9.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thompson LH, Carrano AV, Sato K, Salazar EP, White BF, Stewart SA, et al. Identification of nucleotide-excision-repair genes on human chromosomes 2 and 13 by functional complementation in hamster-human hybrids. Somat Cell Mol Genet. 1987;13:539–51. doi: 10.1007/BF01534495. [DOI] [PubMed] [Google Scholar]
- 103.Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, et al. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–72. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 104.Catoe HW, Nawaz Z. E6-AP facilitates efficient transcription at estrogen responsive promoters through recruitment of chromatin modifiers. Steroids. 2011;76:897–902. doi: 10.1016/j.steroids.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 105.Richardson AJ, Ross MA. Fatty acid metabolism in neurodevelopmental disorder: a new perspective on associations between attention-deficit/hyperactivity disorder, dyslexia, dyspraxia and the autistic spectrum. Prostaglandins Leukot Essent Fatty Acids. 2000;63:1–9. doi: 10.1054/plef.2000.0184. [DOI] [PubMed] [Google Scholar]
- 106.Clark-Taylor T, Clark-Taylor BE. Is autism a disorder of fatty acid metabolism? Possible dysfunction of mitochondrial beta-oxidation by long chain acyl-CoA dehydrogenase. Med Hypotheses. 2004;62:970–5. doi: 10.1016/j.mehy.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 107.Xie Z, Jones A, Deeney JT, Hur SK, Bankaitis VA. Inborn Errors of Long-Chain Fatty Acid beta-Oxidation Link Neural Stem Cell Self-Renewal to Autism. Cell Rep. 2016;14:991–9. doi: 10.1016/j.celrep.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–64. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–9. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16:1537–43. doi: 10.1038/nn.3546. [DOI] [PubMed] [Google Scholar]
- 112.Sun J, Liu Y, Tran J, O'Neal P, Baudry M, Bi X. mTORC1-S6K1 inhibition or mTORC2 activation improves hippocampal synaptic plasticity and learning in Angelman syndrome mice. Cell Mol Life Sci. 2016;73:4303–14. doi: 10.1007/s00018-016-2269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Spangle JM, Ghosh-Choudhury N, Munger K. Activation of cap-dependent translation by mucosal human papillomavirus E6 proteins is dependent on the integrity of the LXXLL binding motif. J Virol. 2012;86:7466–72. doi: 10.1128/JVI.00487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spangle JM, Munger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol. 2010;84:9398–407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–83. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 117.Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr Opin Neurobiol. 2012;22:383–8. doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chazeau A, Giannone G. Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell Mol Life Sci. 2016;73:3053–73. doi: 10.1007/s00018-016-2214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baudry M, Kramar E, Xu X, Zadran H, Moreno S, Lynch G, et al. Ampakines promote spine actin polymerization, long-term potentiation, and learning in a mouse model of Angelman syndrome. Neurobiol Dis. 2012;47:210–5. doi: 10.1016/j.nbd.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Z, Yang H, Wang H. The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage. J Biol Chem. 2014;289:32883–94. doi: 10.1074/jbc.M114.599605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yuan J, Luo K, Deng M, Li Y, Yin P, Gao B, et al. HERC2-USP20 axis regulates DNA damage checkpoint through Claspin. Nucleic Acids Res. 2014;42:13110–21. doi: 10.1093/nar/gku1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhu M, Zhao H, Liao J, Xu X. HERC2/USP20 coordinates CHK1 activation by modulating CLASPIN stability. Nucleic Acids Res. 2014;42:13074–81. doi: 10.1093/nar/gku978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–71. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–86. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–38. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 126.Zheng X, Linke S, Dias JM, Zheng X, Gradin K, Wallis TP, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci U S A. 2008;105:3368–73. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ben-Shushan E, Feldman E, Reubinoff BE. Notch signaling regulates motor neuron differentiation of human embryonic stem cells. Stem Cells. 2015;33:403–15. doi: 10.1002/stem.1873. [DOI] [PubMed] [Google Scholar]
- 128.Faux CH, Turnley AM, Epa R, Cappai R, Bartlett PF. Interactions between fibroblast growth factors and Notch regulate neuronal differentiation. J Neurosci. 2001;21:5587–96. doi: 10.1523/JNEUROSCI.21-15-05587.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lassiter RN, Ball MK, Adams JS, Wright BT, Stark MR. Sensory neuron differentiation is regulated by notch signaling in the trigeminal placode. Dev Biol. 2010;344:836–48. doi: 10.1016/j.ydbio.2010.05.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vernon AE, Movassagh M, Horan I, Wise H, Ohnuma S, Philpott A. Notch targets the Cdk inhibitor Xic1 to regulate differentiation but not the cell cycle in neurons. EMBO Rep. 2006;7:643–8. doi: 10.1038/sj.embor.7400691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang L, Chopp M, Zhang RL, Zhang L, Letourneau Y, Feng YF, et al. The Notch pathway mediates expansion of a progenitor pool and neuronal differentiation in adult neural progenitor cells after stroke. Neuroscience. 2009;158:1356–63. doi: 10.1016/j.neuroscience.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bonini SA, Ferrari-Toninelli G, Uberti D, Montinaro M, Buizza L, Lanni C, et al. Nuclear factor kappaB-dependent neurite remodeling is mediated by Notch pathway. J Neurosci. 2011;31:11697–705. doi: 10.1523/JNEUROSCI.1113-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ferrari-Toninelli G, Bonini SA, Uberti D, Napolitano F, Stante M, Santoro F, et al. Notch activation induces neurite remodeling and functional modifications in SH-SY5Y neuronal cells. Dev Neurobiol. 2009;69:378–91. doi: 10.1002/dneu.20710. [DOI] [PubMed] [Google Scholar]
- 135.Levy OA, Lah JJ, Levey AI. Notch signaling inhibits PC12 cell neurite outgrowth via RBP-J-dependent and -independent mechanisms. Dev Neurosci. 2002;24:79–88. doi: 10.1159/000064948. [DOI] [PubMed] [Google Scholar]
- 136.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–6. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 137.Lefort K, Dotto GP. Notch signaling in the integrated control of keratinocyte growth/differentiation and tumor suppression. Semin Cancer Biol. 2004;14:374–86. doi: 10.1016/j.semcancer.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 138.Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–42. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–36. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kawano S, Morotomi T, Toyono T, Nakamura N, Uchida T, Ohishi M, et al. Establishment of dental epithelial cell line (HAT-7) and the cell differentiation dependent on Notch signaling pathway. Connect Tissue Res. 2002;43:409–12. doi: 10.1080/03008200290000637. [DOI] [PubMed] [Google Scholar]
- 141.Wang XD, Leow CC, Zha J, Tang Z, Modrusan Z, Radtke F, et al. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol. 2006;290:66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 142.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of postmitotic intestinal epithelial cells. Genes Dev. 2005;19:1686–91. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Maliekal TT, Bajaj J, Giri V, Subramanyam D, Krishna S. The role of Notch signaling in human cervical cancer: implications for solid tumors. Oncogene. 2008;27:5110–4. doi: 10.1038/onc.2008.224. [DOI] [PubMed] [Google Scholar]
- 144.Rong C, Feng Y, Ye Z. Notch is a critical regulator in cervical cancer by regulating Numb splicing. Oncol Lett. 2017;13:2465–70. doi: 10.3892/ol.2017.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chakrabarti O, Veeraraghavalu K, Tergaonkar V, Liu Y, Androphy EJ, Stanley MA, et al. Human papillomavirus type 16 E6 amino acid 83 variants enhance E6-mediated MAPK signaling and differentially regulate tumorigenesis by notch signaling and oncogenic Ras. J Virol. 2004;78:5934–45. doi: 10.1128/JVI.78.11.5934-5945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weijzen S, Zlobin A, Braid M, Miele L, Kast WM. HPV16 E6 and E7 oncoproteins regulate Notch-1 expression and cooperate to induce transformation. J Cell Physiol. 2003;194:356–62. doi: 10.1002/jcp.10217. [DOI] [PubMed] [Google Scholar]
- 147.Janke K, Brockmeier U, Kuhlmann K, Eisenacher M, Nolde J, Meyer HE, et al. Factor inhibiting HIF-1 (FIH-1) modulates protein interactions of apoptosis-stimulating p53 binding protein 2 (ASPP2) J Cell Sci. 2013;126:2629–40. doi: 10.1242/jcs.117564. [DOI] [PubMed] [Google Scholar]
- 148.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–8. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 149.Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–78. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 150.Ferreira JS, Schmidt J, Rio P, Aguas R, Rooyakkers A, Li KW, et al. GluN2B-Containing NMDA Receptors Regulate AMPA Receptor Traffic through Anchoring of the Synaptic Proteasome. J Neurosci. 2015;35:8462–79. doi: 10.1523/JNEUROSCI.3567-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–34. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–6. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 154.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A. 2004;101:233–8. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–6. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 156.Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, Perkersen R, et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 2015;130:77–92. doi: 10.1007/s00401-015-1436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bai X, Karasmanis EP, Spiliotis ET. Septin 9 interacts with kinesin KIF17 and interferes with the mechanism of NMDA receptor cargo binding and transport. Mol Biol Cell. 2016;27:897–906. doi: 10.1091/mbc.E15-07-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ghiretti AE, Thies E, Tokito MK, Lin T, Ostap EM, Kneussel M, et al. Activity-Dependent Regulation of Distinct Transport and Cytoskeletal Remodeling Functions of the Dendritic Kinesin KIF21B. Neuron. 2016;92:857–72. doi: 10.1016/j.neuron.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McVicker DP, Awe AM, Richters KE, Wilson RL, Cowdrey DA, Hu X, et al. Transport of a kinesincargo pair along microtubules into dendritic spines undergoing synaptic plasticity. Nat Commun. 2016;7:12741. doi: 10.1038/ncomms12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Niwa S, Lipton DM, Morikawa M, Zhao C, Hirokawa N, Lu H, et al. Autoinhibition of a Neuronal Kinesin UNC-104/KIF1A Regulates the Size and Density of Synapses. Cell Rep. 2016;16:2129–41. doi: 10.1016/j.celrep.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Willemsen MH, Ba W, Wissink-Lindhout WM, de Brouwer AP, Haas SA, Bienek M, et al. Involvement of the kinesin family members KIF4A and KIF5C in intellectual disability and synaptic function. J Med Genet. 2014;51:487–94. doi: 10.1136/jmedgenet-2013-102182. [DOI] [PubMed] [Google Scholar]
- 162.Guillaud L, Wong R, Hirokawa N. Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat Cell Biol. 2008;10:19–29. doi: 10.1038/ncb1665. [DOI] [PubMed] [Google Scholar]
- 163.Hoerndli FJ, Wang R, Mellem JE, Kallarackal A, Brockie PJ, Thacker C, et al. Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs. Neuron. 2015;86:457–74. doi: 10.1016/j.neuron.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kumar J, Yu H, Sheetz MP. Kinectin, an essential anchor for kinesin-driven vesicle motility. Science. 1995;267:1834–7. doi: 10.1126/science.7892610. [DOI] [PubMed] [Google Scholar]
- 165.Toyoshima I, Yu H, Steuer ER, Sheetz MP. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–31. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu H, Nicchitta CV, Kumar J, Becker M, Toyoshima I, Sheetz MP. Characterization of kinectin, a kinesin-binding protein: primary sequence and N-terminal topogenic signal analysis. Mol Biol Cell. 1995;6:171–83. doi: 10.1091/mbc.6.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chai S, Xu X, Wang Y, Zhou Y, Zhang C, Yang Y, et al. Ca2+/calmodulin-dependent protein kinase IIgamma enhances stem-like traits and tumorigenicity of lung cancer cells. Oncotarget. 2015;6:16069–83. doi: 10.18632/oncotarget.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Britschgi A, Bill A, Brinkhaus H, Rothwell C, Clay I, Duss S, et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci U S A. 2013;110:E1026–34. doi: 10.1073/pnas.1217072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang T, Guo S, Liu Z, Wu L, Li M, Yang J, et al. CAMK2N1 inhibits prostate cancer progression through androgen receptor-dependent signaling. Oncotarget. 2014;5:10293–306. doi: 10.18632/oncotarget.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang C, Li N, Liu X, Zheng Y, Cao X. A novel endogenous human CaMKII inhibitory protein suppresses tumor growth by inducing cell cycle arrest via p27 stabilization. J Biol Chem. 2008;283:11565–74. doi: 10.1074/jbc.M800436200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 171.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, et al. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5:737–48. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 172.Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, Hatfield GW, et al. HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast. 2006;23:623–32. doi: 10.1002/yea.1380. [DOI] [PubMed] [Google Scholar]
- 173.Xicoy H, Wieringa B, Martens GJ. The SH-SY5Y cell line in Parkinson's disease research: a systematic review. Mol Neurodegener. 2017;12:10. doi: 10.1186/s13024-017-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yachie N, Petsalaki E, Mellor JC, Weile J, Jacob Y, Verby M, et al. Pooled-matrix protein interaction screens using Barcode Fusion Genetics. Mol Syst Biol. 2016;12:863. doi: 10.15252/msb.20156660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gal J, Kuang L, Barnett KR, Zhu BZ, Shissler SC, Korotkov KV, et al. ALS mutant SOD1 interacts with G3BP1 and affects stress granule dynamics. Acta Neuropathol. 2016;132:563–76. doi: 10.1007/s00401-016-1601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Csárdi GaN T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;1695 [Google Scholar]
- 178.Walt Svd, Colbert SC, Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science & Engineering. 2011;13:22–30. [Google Scholar]
- 179.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–5. [Google Scholar]
- 180.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kluyver T, Ragan-Kelley B, Perez F, Granger B, Bussonnier M, Frederic J, et al. Jupyter Notebooks-a publishing format for reproducible computational workflows. Amsterdam: Ios Press; 2016. [Google Scholar]
- 182.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–4. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 184.Bateman A, Martin MJ, O'Donovan C, Magrane M, Alpi E, Antunes R, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D69. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–63. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Viger F, Latapy M. Efficient and Simple Generation of Random Simple Connected Graphs with Prescribed Degree Sequence. In: Wang L, editor. Computing and Combinatorics: 11th Annual International Conference, COCOON 2005 Kunming, China, August 16–19, 2005 Proceedings. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005. pp. 440–9. [Google Scholar]
- 188.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–4. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.